Introduction
Genomics has data in its DNA. The term “genomics” took root in 1987 when Frank Ruddle and Victor McKusick borrowed Tom Roderick’s neologism to launch a new journal (92). Genomics has since become a field, or at least an approach to biology and biomedical research. It generally describes a style of science and application that features measuring many genes at once, rather than one gene at a time; intensive use of instruments for mapping and sequencing nucleic acids; generation and utilization of large data-sets, including DNA sequences, their underlying mapping markers, and functional analyses of genes and proteins; and computation. The extension from more traditional genetics entailed greater scale and implied a need to share data, because no one laboratory could fully make sense of the deluge of data being generated while mapping and sequencing the genomes of humans and other organisms.
The Human Genome Project (HGP) was conceived in 1985, when Robert Sinsheimer, Renato Dulbecco and Charles DeLisi independently realized that it would be useful to have a reference sequence of the human genome as a tool for research and application (35; 103). DeLisi had the authority to fund a research initiative within the U.S. Department of Energy (DOE). As vigorous debate proceeded through 1988, the goals were broadened to include, in addition to humans, mapping and sequencing the genomes of model organisms: Escherichia coli (bacterium), Arabidopsis thaliana (plant), yeast (initially Saccharomyces cerevisiae, and then others), nematode, although not formally included in the HGP at first (Caenorhabditis elegans), fruit fly (Drosophila melanogaster), and finally Mus musculus (mouse) as the mammalian model. The imagined project also included research to advance—by increased speed, lower cost, and improved accuracy—the instruments and methods of DNA sequencing and mapping, as well as computational methods and algorithms. Finally, the project incorporated research on ethical, legal and social implications (ELSI). The ELSI program was novel and distinctive, setting a precedent for initiatives that had foreseeable impacts well beyond the technical community (155). It appears today that the greatest challenges of data sharing are indeed in law, ethics, and policy. The technical challenges are daunting; the social and legal complexities are even more so.
The HGP left many legacies. The most obvious is a reference sequence of the human genome that continues to be refined; but the Project also drove the creation of new instruments. A major avenue has been the Advanced Sequencing Technology Program, directed by Jeff Schloss of the National Human Genome Research Institute (NHGRI), which helped conceive and develop most of the technologies that gave rise to the hyper-Moore’s curve acceleration of DNA sequencing speed and plummeting costs from 2004 to 2014 (44; 68). The Program supported new mathematical approaches to analyzing data in bioinformatics, and the NHGRI became known as an institution that could manage large-scale, technology- and data-intensive research programs requiring more coordination than the distribution of peer-reviewed grants could provide. One of the signature features of NHGRI has always been an open science ethos associated with the pre-publication sharing of data (33; 60).
The HGP reached its goal of a human reference sequence earlier than predicted, in the period from 2000 to 2003. The initial milestone toward completion was achieved on June 26, 2000, when a draft human sequence was first announced by President Bill Clinton and Prime Minister Tony Blair at the White House and 10 Downing Street in London (97). Articles revealing the HGP’s “public” assembly of a genome and that produced by the private company, Celera Genomics, were then published one day apart in Science and Nature in February 2001 (83; 152). This initiated a cascade of publications on sequences for chromosomes that culminated in April 2003, marking the 50th anniversary of the canonical April 25th, 1953 publication of the double helical structure of DNA by James Watson and Francis Crick (30; 77; 98; 156).
The 2001 publications represented two scientific strategies and two modes of managing data. The publicly funded HGP sequence—authored by a group of laboratories from the U.S., the U.K., France, Japan, Germany, and China under the banner “International Human Genome Sequencing Consortium” (IHGSC)—represented the work of a global coalition. It entailed extensive and systematic data sharing, characterized perhaps most distinctly by the daily release of publicly funded DNA sequences into the public domain. Data from the HGP came primarily from high-throughput sequencing laboratories in the six partner countries, with leadership on assembly centered at the University of California, Santa Cruz and databases at the U.S. National Center for Biotechnology Information (NCBI) and the European Bioinformatics Institute (EBI). This public “hierarchical” sequence depended on genetic and physical maps, which situated DNA markers based on their chromosomal locations, and sequencing coordinated by chromosomal region, assigned amongst the global partners (23; 148; 153). Science, in contrast, published a sequence assembled by “shotgun” methods, characterized by a vertical organization that integrated data generation and computation within a single laboratory and also incorporated data from public sources (151; 154).
Access to the data was open and free from the HGP, and had restrictions at Celera. Celera’s data were available to subscribers for a fee, free for noncommercial use in packets up to 1 megabase (Mb), or by access agreement with the company (158). Divergent access policies were thus present even during the gestational phase of the human reference sequence, and controversy over access was intense (2; 65; 78; 95; 106). It is not an exaggeration to say that debates about sharing have been woven into the fabric of genomics itself, an inextricable part of the new field from its very beginning. In 2003, a National Research Council (NRC) committee was appointed to make recommendations about responsibilities accompanying publication, partly in reaction to the HGP-Celera race. Chaired by Nobel Laureate Tom Cech, this committee laid out the Uniform Principle for Sharing Integral Data and Materials Expeditiously (UPSIDE), and various other committees and reports on sharing have followed since (25; 38; 97–99). The strongest embodiment of the HGP’s open science ethos, however, was not sharing sequences at publication, but rather long before, even as the data were being generated.
Emergence of the Bermuda Principles for Prepublication Data Sharing
By early 1996, the HGP had been proceeding for six years. A workable human genetic linkage map was available, and physical maps of cloned DNA (and the bacterial and yeast cell lines that housed them) were available for most regions of the chromosomes (76; 85; 94; 107; 128). Reference sequences for several yeast chromosomes were published, and a complete genomic sequence was in the offing, with a network of European laboratories leading the way (5; 57). Progress toward a complete 19,000-gene, 97-Mb sequence of C. elegans bred confidence that it would soon be completed, as indeed it was in 1998 (12).
The initial goal of the HGP, however, a reference sequence of the human genome, was more distant, although it was visible over the horizon. The Wellcome Trust—the British biomedical research charity that by 2003 funded one-third of the public human genome project—had already upped its funding substantially, focusing on human sequencing and basing its efforts at the new Sanger Centre (founded in 1993) near the University of Cambridge (41; 51). The U.S. National Center for Human Genome Research (NCHGR, which would become NHGRI in 1997) announced in 1995 a competition for high-throughput sequencing parts of the human genome, and reviewed center-grant proposals later that year (85). The DOE, moreover, was constructing a Joint Genome Institute (JGI) to house its sequencing operations, which would open in 1997 (81). By early 1996, the HGP, with contributions from five national partners (China would join in 1999), was transitioning from mapping to large-scale human genomic sequencing (120). The stage was set for a hard push for the human reference sequence.
It was quickly becoming apparent that detailed coordination was required. Even after earlier mapping efforts, sequencing—of both the human and other large model genomes—would generate more data than any other discrete effort in the history of biology (103). This was predicted in the HGP’s founding reports, and was becoming a tangible reality by 1996. More specifically, the pragmatic tasks at hand included: (a) deciding which chromosomal regions to assign to each team, lest several centers sequence the same juicy bits and waste time and effort; (b) specifying targets for data quality, as the sequencing goals were also set; and (c) verifying sequencing outputs from individual laboratories, to measure progress and to help justify the Project’s large public investment.
As the NIH large-scale sequencing grants were about to commence, Michael Morgan at the Wellcome Trust and Francis Collins, then NCHGR director, decided to hold an organizational meeting amongst those funded (or expected to be funded) to plan the launch phase of large-scale sequencing (57). They looked for a relatively neutral venue, one that would not be perceived as U.S.-dominated (138). They settled on Bermuda, which was accessible from all the HGP centers and appropriately located in the mid-Atlantic between Europe and North America. The meeting took at the “Pink Palace,” the Princess Hotel in Bermuda, February 26th through 28th, 1996.
The weather was dreary, but the conference was not (90). The new NIH sequencing centers had been announced, but had yet to receive their funding. The attendees—including the Hinxton group funded by Wellcome, the leaders of the DOE’s efforts, working mostly on mapping and technology development, the leaders of the NIH centers, and mappers, technology developers, and administrators from Japan, Germany and France—were eager to get started, yet anxious about the long journey ahead. They were, frankly, not sure a human reference genome was yet attainable (19; 27; 138).
In addition to the practical needs to allocate work and measure progress toward a sequence, two intimately related issues loomed as the 1996 meeting was planned: patenting and data-sharing. The patent issue surfaced when a scientist at the National Institute for Neurological Disorders and Stroke, J. Craig Venter, filed patent applications on short segments of DNA that allowed for unique identification of sequences coding for genes in the human brain (1; 22; 123). “Expressed sequence tags” (ESTs) could be used to fish genes out of the genome by identifying unique sequences that are translated into protein. Genentech patent lawyer Max Hensley advised Reid Adler, the lawyer at the NIH’s Office of Technology Transfer, that the NIH should file for patents on Venter’s ESTs, to ensure they could be licensed for further development (35). Such patents could “protect” the DNA fragments, and might be important to preserve private investments in characterizing the corresponding full-length genes and related proteins. These incentives might be important for developing drugs and biologics as treatments, as well as genetic tests for neurological diseases. Controversy erupted in July of 1991, when at a Senate meeting Venter made public that the NIH had filed its first application and was planning others, making claims on more ESTs and the method for obtaining them. The EST method patent was later converted to a Statutory Registration of Invention, effectively preventing anyone from patenting it.
Patenting DNA molecules and methods had become common at universities and biotechnology and pharmaceutical companies, a practice that in the 1980s and 1990s often conflicted with more traditionally minded academic biologists not used to patenting their work (17; 36; 130). One of the foremost patent scholars, Rebecca Eisenberg of the University of Michigan Law School, observed:
The patent system rests on the premise that scientific progress will best be promoted by conferring exclusive rights in new discoveries, while the research scientific community has traditionally proceeded on the opposite assumption that science will advance most rapidly if the community enjoys free access to prior discoveries. (46)
According to some, the NIH patent applications—which claimed both ESTs (gene fragments) and corresponding full-length genes (in the form of complementary DNAs, or cDNAs)—could block downstream research and development requiring the use of many genes in tandem. The NIH Director, Bernadine Healy, supported the patents, though the then-NCHGR Director James Watson, of DNA structure fame, vigorously disagreed with them (69; 155). This became one of several bones of contention between Watson and Healy that culminated in his resignation as head of NCHGR in spring 1992, leaving the door open for Healy to recruit Collins, who assumed the leadership of the NIH’s genome efforts in 1993 (139).
Reflecting contemporary commercialization trends, the debate over gene patents echoed between the public and the private sectors. As the NIH’s applications were pending, Venter left the NIH to direct The Institute for Genomic Research (TIGR), a private nonprofit research institute (35; 131). Some of the patent rights from TIGR would be assigned to a for-profit corporation, Human Genome Sciences (HGS), which itself began sequencing genes and fragments and filing for its own patents while also drawing on TIGR’s output. Meanwhile, another small firm, Incyte, had also become interested in sequencing ESTs and full-length cDNAs. HGS, Incyte, and several other companies were building business models around discovering and sequencing genes and fragments, and patenting parts of the genome likely to contain sequences of keen biological interest and commercial value (104).
Concerns about patent impediments to research, created by thickets of broad EST and cDNA patents on genes of unknown function, haunted debates about genomics and patent policy (47). In 1993, the pharmaceutical giant Merck initiated a partnership with the head of the HGP center at Washington University in St. Louis, the C. elegans expert Robert Waterston (42; 159). The goal of the Merck Gene Index Project was to sequence human ESTs and release them with a minimal delay, usually of only 48 hours, into the public domain, with the logic that “making the EST data freely available should stimulate patentable inventions stemming from subsequent elucidation of the entire sequence, function and utility of each gene” (159). One reason for this policy was a spirit of open science; another was to thwart patents on short DNA fragments by companies like HGS and Incyte; a third was that the Gene Index was funded by a nonprofit unit of Merck, so Merck had to demonstrate it did not have privileged access. Meanwhile at NIH, Harold Varmus soon became NIH Director, appointed by Bill Clinton to replace Healy. Varmus sought expert advice on the NIH’s EST patent applications, which had been initially rejected by the USPTO in 1992 (5; 87). Eisenberg and another legal scholar, California Berkeley’s Robert Merges, drafted a detailed memo for Varmus noting that the NIH’s EST patent strategy made little sense, given that ESTs were primarily research tools (49). Varmus abandoned the applications. This was yet another turn along the tortuous path whereby genomics, and the rules about how and when genomic data should be shared and commercialized, were developing in tandem (71; 72; 79; 80).
The brouhaha over ESTs, coupled to the evolving controversies and worldwide negative press over the patenting of full-length genes like BRCA, colored the Bermuda meeting in 1996 (11; 58). The Bermuda attendees, who hailed from five nations, the European Molecular Biology Laboratory (EMBL) in Heidelberg, and the Brussels-based European Commission (EC), had to contend both with the pragmatic, scientific problems of getting the human genome sequenced on time and accurately, and the more principled problem of how the HGP’s soon-to-be produced deluge of sequence data should be shared, utilized, and commercialized. For the majority of attendees to the 1996 meeting, it was not surprising that developing a clear-cut data-sharing policy was a chief agenda item from the start (57).
The specific historical roots of the daily, online release of HGP-funded DNA sequences—under the policy that came to be known as the “Bermuda Principles”—are complex and contingent. So, too, was the process by which this radical policy, which ran against the norm in most biomedical research of releasing data at publication, was ratified within the HGP and justified as the Project proceeded. In a forthcoming historical article, the authors enumerate and assess these details at length (89). Several points about precedents and the measured agreement that HGP participants reached are relevant here.
The Bermuda Principles filled a policy lacuna, replacing a set of guidelines that had previously applied only to the NIH and DOE. The HGP’s founding reports, produced in 1988 by the NRC and the congressional Office of Technology Assessment (OTA), were notoriously vague about data sharing, noting only that data and materials must be shared rapidly for coordination and quality control, and admitting that this might create conflicts with commercialization (21; 101; 103). The 1990 joint plan for the NIH and DOE echoed this message, but similarly failed to provide a timeline for sharing amongst collaborators (23; 46; 62; 114; 133; 135; 148; 155). By the early-1990s, mapping and sequencing technology development were proceeding impressively around the globe, not just in the U.S. but also in Britain, France, Japan, and several other nations (21; 101; 103). Yet despite the founding of the Genome Data Base (an electronic medium for sharing mapping data) in 1993, international coordination of human mapping was disorganized, nucleated around annual meetings and single chromosomes, and complicated by competition and secrecy (18). A 1992 NIH and DOE policy required deposit of data from mapping (to GDB) and sequencing (to GenBank) within six months of generation (18). Aside from the 48-hour sharing policy of the Merck Gene Index Project, this was probably the most specific policy precedent for data sharing in genomics. Standard practice for GenBank, for instance, was to share unpublished sequences concurrently with accompanying journal articles, but not before (9; 133; 137).
The Bermuda Principles extended the 1992 policy, strongly recommending the daily sharing of all HGP-funded DNA sequences of 1 kilobase (Kb) or longer to GenBank, the databank of the EMBL, or the DNA Databank of Japan (DDBJ) (57). For the first time, the HGP had a Project-wide policy, designed to unite all the international contributors and not just those funded by the NIH or the DOE. This facilitated the development of the Human Sequencing and Mapping Index, a website linking laboratory webpages to GenBank and allowing globally distributed centers to “declare” regions for sequencing and avoid duplication (16). Especially in the U.S., moreover, the Principles helped to enforce quality standards (set in 1996 at 99.99% sequence accuracy) and output commitments, providing a means of checking whether the heavily funded centers were delivering on sequencing promises. In 2001, Elliot Marshall called the Principles “community spirit, with teeth,” and for good reason: those centers not producing their sequences, or failing to meet quality standards, could lose their competitive funding and potentially their places within the prestigious HGP (86).
Daily sharing, however, was not necessarily required for these tasks, and the clearest historical test case for this policy was perhaps unlikely source: the C. elegans research community. John Sulston and Robert Waterston, amongst many others in this network, had adopted daily (or as close to daily as possible) sharing in the mapping and early sequencing of the worm genome, beginning in the 1970s and facilitated, by the 1980s, by the rise of networked computing (9; 136; 138). By 1995, Sulston and Waterston (funded by the NIH, the Wellcome, and the U.K. Medical Research Council) had done more large-scale sequencing (first in C. elegans, and later in early human efforts) than anyone, and had become HGP leaders via connections to Collins, Watson, Maynard Olson, and other power players in the field (85; 160). At the Bermuda meeting, Sulston and Waterston co-chaired the final session, on data release policies, which led to the first draft of the Bermuda Principles (57). “It was agreed that all human genomic sequence information,” the statement from that session and a later NCHGR press release read, “generated by centres for large-scale human sequencing, should be freely available and in the public domain in order to encourage further research and development and to maximise its benefit to society” (96).
Daily release, however, was for some a bitter pill to swallow. The adoption of this policy in the HGP was driven by Waterston and Sulston as the C. elegans sequencing leaders, and strongly supported by leaders of the two foremost funders: the NIH and the Wellcome Trust. Debates raged about whether daily release would hurt HGP data quality (3; 15; 128). Perhaps the most significant hurdle for daily data release, however, was its apparent incompatibility with commercialization. The U.S. Bayh-Dole Act allowed universities and businesses the first right to title on inventions funded by government grants, and a German policy allowed HGP investigators three months’ time, before data release, to apply for patents drawing on HGP sequences, including patents on genes (36; 147). In the U.S., daily release did not prevent patents on genes of known function, but it did block patents of the kind HGS and Incyte were seeking. In Europe and other patent jurisdictions, the Bermuda Principles indeed endangered gene patents, because unlike in the U.S., no lag time existed between releases of data into the public domain and when investigators could still apply for patents drawing upon it. (Until the America Invents Act somewhat changed the rules, the grace period in the U.S. was one year (54).) Heidi Williams has since argued convincingly that the patent and database restrictions placed on Celera Genomics’ sequence data led to a 20–30 percent reduction in downstream innovation and development in diagnostics, relative to genes sequenced first by the open and public HGP (158). But in 1996, as today, the HGP policy of daily sharing was just one stance among many, in a spectrum of uncertainty about how best to foster progress in genomics and its applications.
By early 1998, the Bermuda Principles were the official data sharing polices of the HGP, a condition of the large-scale sequencing grants in all participating countries. In 1996, the NIH had put out a statement that while it opposed patents on “raw human genomic DNA sequence, in the absence of additional demonstrated biological information,” because of the Bayh-Dole Act it could only discourage this practice—as it surely did, with the suggestion of removing funding if such patents were filed—but not prevent it outright (96). A series of warning letters from the NIH, the DOE, and the Wellcome Trust helped shift the conflicting policy in Germany and another incompatible policy in Japan, moves which prevented a sharing delay from also materializing in France (12; 28; 29). American and British leaders were flexing their muscles here. The threat of removing other national contributors from the HGP, if they did not agree to daily sharing, held real weight, as NIH and the Wellcome could effectively sequence the genome without collaborators.
The Bermuda Principles continued to exert considerable influence, especially as scientists and administrators amended them to reflect changes in data and practice (21). Two more meetings were convened in Bermuda, in February of 1997 and 1998. Like the 1996 summit, these were intended to address evolving scientific issues in the HGP, but also to revisit data sharing polices and enact policy shifts as attendees saw fit. By the 1998 meeting, the Principles were revised to include a new daily release trigger, of 2,000 (2 Kb) rather than 1,000 (1 Kb) base-pair stretches of DNA, and extended first to mouse and later to all model organism sequences produced under the aegis of the HGP (61; 157). In 2000, the NHGRI extended the Principles again to include several new kinds of data, including those generated during the finishing phase of the draft sequence and through whole genome “shotgun” sequencing (97).
After Bermuda: Broadening Open Science
The Bermuda Principles applied to a small group of laboratories, pulling together to produce a human reference sequence. Even as progress toward this goal accelerated in the late-1990s, it became apparent that studying variations—that is, individuals’ deviations from the reference sequence—and linking genomic data to other kinds of data was essential to making sequences meaningful: in terms of health outcomes, environmental exposures, genealogy, and family history. By 2000, it was clear that linkage amongst databases, through systematic data sharing, would drive the science and its applications.
The data and users were far more heterogeneous than for high-throughput sequencing, yet sharing was still paramount. As microarray technology became pervasive after 1996, identifying and cataloguing single-nucleotide variants (Single Nucleotide Polymorphisms, or SNPs) became possible. Fears of hoarding and patent thickets led to a novel effort to prevent SNP patents, the SNP Consortium (73; 140). This was a public-private partnership that applied for patents on SNPs in profusion, to establish formal legal and scientific priority. It then abandoned the applications, releasing data to the public domain unencumbered by patent rights. Because SNPs were tools in agriculture, pharmaceuticals, biotechnology, and elsewhere, industry supported the Consortium. Academics were involved because they wanted to use the tools, but commercial manufacturers like Affymetrix and Illumina manufactured most SNP chips themselves. These manufacturers, while certainly believing in and holding many patents, nonetheless foresaw a hopelessly dense patent thicket based on DNA sequences and hoped to avoid it. Microarrays routinely included hundreds of thousands of DNA fragments as probes. If SNPs and ESTs were patented, would it not take hundreds of licenses to build a chip? Would anyone be able to afford it? Access to data, including the ability to use DNA molecules that included SNPs, was essential.
Ft. Lauderdale and Beyond
In 2003, as the HGP was nearing completion, the Wellcome Trust convened a meeting in Ft. Lauderdale to define rules for biological infrastructure projects. The meeting produced the first of several statements supporting the open science ethos for “community resource projects,” the likes of which dominate genomics today (98). The meeting focused on the value of prepublication data sharing, but relaxed some of the Bermuda Principles’ provisions. Like at the 1998 Bermuda meeting, Ft. Lauderdale attendees stipulated daily sharing for sequences longer than 2 Kb for any organism. This was very much in the spirit of Bermuda, but their statement also mandated that data generators be credited when data were used. Finally, it acknowledged that hypothesis-driven research did not have the same sharing obligations as those focused on producing “community resources,” like sequencing consortiums, the Merck Gene Index Project, or the SNP Consortium. This recognition, of a need for scientific attribution and credit, grew from the diversity of data-generators and users. Arias, Pham-Kanter and Campbell have beautifully summarized the evolution of data-sharing policies that evolved from the Bermuda Principles (10).
Open science continued to infuse the many projects developed at NHGRI, or in which NHGRI was a partner. These were highly diverse, but many were indeed “community resource” efforts. The HapMap aimed to identify common genomic variations and characterize them from global populations, enabling elucidation of co-inheritance of DNA markers in detail. HapMap policies vigorously discouraged patenting, to a degree that Rebecca Eisenberg questioned their wisdom and enforceability (32; 48; 52). The 1000 Genomes project expanded on the HapMap project, including sequences from a larger sample of populations and intended to identify less common genomic variants (26). Another contemporaneous project, ENCODE, was intended to understand the function of DNA elements, including regulatory sequences, enhancers, promoters and operators (77). Since these elements were by definition functional, however, they might also have practical utility and be proper subjects for patenting. The 2003 ENCODE pilot policy explicitly acknowledged this, another deviation from Bermuda driven by the nature of the work.
As new genomics tools enabled genome-wide studies, the Genetic Association Information Network (GAIN) was formed (50). GAIN built on tools made possible by the HapMap and SNP chips, probing hundreds of thousands of common variants in large population studies to identify genomic regions associated with diseases and specific traits. It was intended to ensure rigor, facilitate collaboration, forge academic-industry partnerships, encourage sharing, promote publication, and prevent premature intellectual property claims. The Foundation for the NIH, a nonprofit, nongovernment organization, led the effort and invited applications for genome-wide studies. An elaborate peer review process assessed the technical merits of the proposals, but also ensured that study design, computational methods, and data sharing complied with guidelines. GAIN had a set of principles, the first of which was to make results “immediately available for research use by any interested and qualified investigator or organization.” Yet because the data were associated with specific individuals, this was qualified “within the limits of providing appropriate protection of research participants.” The GAIN principles imposed a duty on users to respect the confidentiality of study participants, and also to ensure that uses fell within the terms of their informed consent. Another principle was to acknowledge data sources for any re-use. Finally, the original contributors would have a nine-month period during which they alone could submit abstracts or papers based on their data. This “embargo” period was a marked divergence from Bermuda, driven again by the needs of a different community of producers and users. Labs generating the data were disparate in size, type, geography, and purpose, and those differences needed to be accommodated.
Database structures also had to reflect the new realities. The NCBI’s Database of Genotypes and Phenotypes (dbGaP) was established in 2006 (50). Instead of one unitary bank (i.e., GenBank) for sequences, dbGaP had tiers: a public layer freely available to all, and a large dataset that might contain private or identifiable information, which in turn required a gatekeeper, a Data Access Committee, to make sure users had good reasons for access. They needed to agree to protect privacy and confidentiality and follow rigorous data security practices. Moreover, many studies needed to be approved by an Institutional Review Board to ensure compliance ethics rules. The purpose of dbGaP was to enable research through broad access, but the involvement of possibly identifiable people who had not agreed to post their data on the Internet necessitated new layers of review. NIH’s 2007 policy on Genome-Wide Association Studies (GWAS) mandated deposit of data into dbGaP for NIH-funded studies (99). This was open science, but the database had rules that were not as simple as free and open access.
NIH’s policy for data access was formally modified in 2014 (33; 100). The 2007 policy was only for human GWAS; in 2014 it was generalized to all NIH institutes and all organisms. It permitted a “validation” embargo of the data for several months, to ensure quality, and dbGaP could hold data in a protected private status for 6 months. After six months, however, human data would be freely available, and on animals it would be freely available upon publication. The new policy also made it easier for NIH to continue to tweak the policy in parts in the future, without formal review of the policy in its entirety. Finally, it stipulated that prospectively, studies should get broad consent for deposit and data use, and IRB review for uses of data contributed from past studies, to ensure compliance with the informed consent at the time the data were gathered.
Prepublication data sharing was soon broadened beyond genomics, first to proteomics at a meeting in Amsterdam in 2008, and then to other datasets at a Toronto workshop in 2009 (124; 125; 142). The Toronto statement applied to all datasets that were large-scale, broadly useful, infrastructural (i.e., intended to produce reference datasets), and had community buy-in. This meant respecting the interests of data-generators to publish “first global analyses of their data set[s],” citing the sources of data, and contacting the data-generators if re-uses might “scoop” them.
These iterations and adaptations of data-sharing norms and policies were efforts to keep data open while also respecting the rights and interests of those contributing data (participants) and those generating the data (researchers). Different contexts and uses led to databases with layers of process and rules. Some projects, such as the Personal Genome Project and Open Humans, had an informed consent process that enabled sharing of one’s genome on the Internet with no restrictions, but these were for “information altruists” who did not fear misuse of their own data, or who at least believed the benefits of open access were greater than the risks (7; 8; 25; 108; 127). Most data and studies, however, came from projects that entailed narrower informed consent, which generally promised participants efforts to keep the data secure and prevent identification of individual research participants.
Building a Medical Information Commons: Theory
The Bermuda Principles and its successor statements were practical efforts to set rules and build infrastructure for increasingly diverse research uses. Generally, these rules were crafted while projects were being designed and carried out, and thus generated for and by a community—later including both research and clinical care users—hoping to draw on a “Medical Information Commons.” A 2011 report from the NRC and Institute of Medicine, Toward Precision Medicine, laid out a vision of layers of data oriented around individuals, to enable an evolving health care and public health system based in biomedical research (19; 32; 80; 102; 135; 137; 138). The central recommendations centered on building informational infrastructure, calling for an:
‘Information Commons’ in which data on large populations of patients become broadly available for research use and a ‘Knowledge Network’ that adds value to these data by highlighting their inter-connectedness and integrating them with evolving knowledge of fundamental biological processes. (19; 32; 80; 102; 135; 137; 138)
This language explicitly invoked a “commons,” and thus Garret Hardin’s classic essay from 1968 (63; 64). Hardin’s Presidential address to the American Association for the Advancement of Science, on which this essay was based, addressed problems that had no technical solution, but required collective action to solve. Hardin was mainly concerned with militarization and rising population, but the iconic example he used was drawn from an 1833 tragedy of the commons described by William Forster Lloyd. In a pasture open to many herdsmen, each herdsman would want to have his cattle feed as much as possible, but if each did so, overgrazing would deplete the pasture. Hardin pointed to the inadequacy of the “invisible hand” of a market to solve such problems, which required coercion in order to solve, preferably according to mutually agreed rules. His examples were laws (against bank robbery), taxes, and other exercises of state power. This left the solution options as binary: the market (no rules, free choices, and creation of property rights) or The Leviathan (the state).
Elinor Ostrom and others have also studied the tragedy of the commons. Yet they have expanded the potential solution set, by observing that real-world communities often craft rules preventing the depletion of resources or orchestrating services requiring collective action. Ostrom’s earlier work focused on natural resource depletion (114; 116). She examined how some communities prevented over-fishing, while others tragically depleted fish stocks for lack of a viable commons. She also studied collective interests in urban policing and managing water resources. In her later work, Ostrom and others extended thinking to a knowledge commons, a concept closely aligned to a medical information commons (70). Theoretically, the biggest difference between a natural resource commons and a knowledge (or data) commons is that natural resources, like biorepoistories of samples, can be depleted. Data and knowledge, in contrast, are not depleted no matter how many people use them.
Indeed, data and knowledge achieve network efficiencies that expand dramatically the more users there are, according to Metcalf’s Law (150). Ostrom and her collaborators devoted much of their attention to institutional arrangements that enabled a viable knowledge commons to form, and how to govern it (112; 113; 115; 119). Trust and reciprocity are central themes; and Institutional Analysis and Development (IAD) was the framework she proposed for addressing problems of collective action. In 2010, she concluded that, “rules related to the production of generally accessible data” include:
Who must deposit their data?
How soon after production and authentication of data do researchers have to deposit the data?
How long should the embargo last?
How should conformance to the rules be monitored?
How many researchers are involved in producing and analyzing the particular kind of data?
Should an infraction be made public in order to tarnish the reputation of the infringer? (115)
These questions encompass identifying who is (and is not) a member of the community of contributors and users, how to formulate contributions and use rules, and governance and enforcement procedures. The successive statements about data sharing, beginning with the Bermuda Rules in 1996, were all implicit or explicit responses to questions raised by Ostrom. Contreras’ work on genomics draws directly on this theory; and Madison, Frischmann, and Strandburg are extending theories of the commons into data and knowledge domains (34; 84).
Commons theory is not the only framework for open science, however. The Global Alliance for Genomics and Health appeals to Article 27 of the Universal Declaration of Human Rights, the right “to share in scientific advancement and its benefits” (146). The value of sharing data globally is reiterated in a series of UNESCO declarations, the recommendations of the Council of Europe, and guidelines from the Organization for Economic Cooperation and Development (37; 99; 109–111; 143–145). Much of bioethics has centered on protecting research participants from risk, but the Global Alliance also focuses on the right to benefit, including through data sharing as a means to advance science and improve medicine: the very purpose for which many people contribute samples and information to medical research. Yet the rights to benefit, and implementing it in the real world, are different things; moving from aspiration to realization is a constant struggle. Future statements will no doubt follow as international data sharing continues to take shape.
Emergence of the Global Alliance for Genomics and Health
While the need to link data of many types, housed in many parts of the world, is obvious, the structure of a global commons of genomic and other data faces many practical problems. Some are technical: a need for application interfaces, standard formats, and interoperable data systems. But the legal and social challenges are even more formidable.
In January 2013, over fifty individuals from eight countries—roughly comparable to the first Bermuda meeting in size and national representation—met in New York City to develop some standards and articulate the need for infrastructure. The results were summarized in a June 2013 white paper describing a need for collective international action, and to avoid “a hodge-podge of balkanized systems—as developed in the U.S. for electronic medical records—a system that inhibits learning and improving health care” (55). Attendees proposed a global alliance, which came to be known as the Global Alliance for Genomics and Health (GA4GH). By 2016, the Global Alliance comprised 800 people, from 400 organizations, in 70 countries (53). This growth alone indicates how much more complicated it will be to achieve data-sharing among hundreds of diverse stakeholders, as compared to the organizational challenge when HGP leaders met in Bermuda, involving fewer than 100 people in (at the time) only five countries.
Beacon and Matchmaker
As the Global Alliance was being formed, efforts converged on three pilot projects, and in 2016 a fourth was added. The three initial projects were Beacon, Matchmaker, and BRCA Challenge. The Beacon project queried a network of genomic databases to see if they harbored information about particular genomic variants. Those who came upon a variant in research or clinical practice could send a single query, and find out which participating databases had relevant information, anywhere in the world. As of summer 2016, 25 institutions and 250 datasets were participating in Beacon (53). The Matchmaker Exchange was another data-brokering effort for those studying rare disorders, enabling users to find phenotype and genotype data pertinent to a genomic-clinical profile, again allowing a query of participating databases. The October 2015 issue of Human Mutation featured 16 articles from the Matchmaker demonstration project (118). Both Beacon and Matchmaker point to the places where data can be found, while the data themselves remain where they are, and users can seek access.
Case study: Sharing Data about BRCA Variants
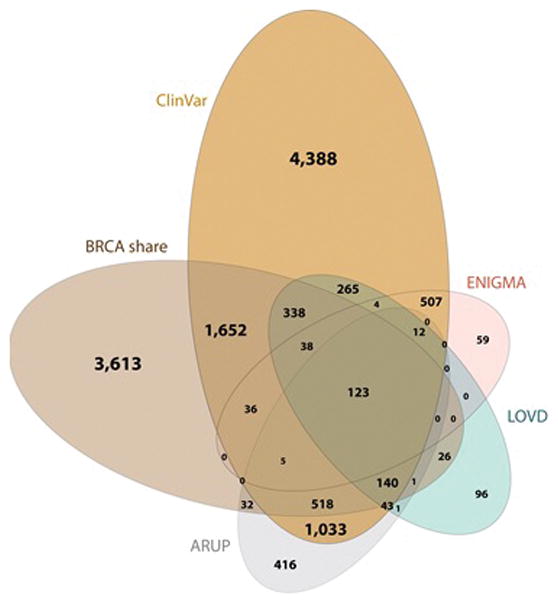
The BRCA Challenge, in contrast to Beacon and Matchmaker, entailed a new database for variants in two of the most studied and clinically significant human genes, BRCA1 and BRCA2, and to pool publicly available data. The intent was to build out from these genes, establishing a precedent for expansion into other genes. The resulting database, BRCA Exchange, has three tiers. The top tier is fully public and lists variants interpreted by the Evidence-based Network for the Interpretation of Germline Mutant Alleles (ENIGMA), an international consortium founded at a May 2009 meeting in Amsterdam (135). ENIGMA is the expert vetting committee for BRCA Exchange variant interpretation. The next layer is a “research” dataset with links to the evidence base, including conflicting interpretations and un-vetted reports, with pointers to other databases containing further information. The third layer, still under construction as of 2016, will contain case-level data that might be linked to identifiable individuals, thus requiring higher levels of security and a gatekeeper to ensure compliance with informed consent and prevent misuse or unauthorized re-identification. BRCA Exchange shares data extensively with ClinVar, LOVD, and other variant databases.
The need for a BRCA Challenge was an anomalous artifact of the history of testing for inherited risk of breast and ovarian cancer. Mary-Claire King’s team found linkage to a putative risk gene in families in 1990 (62). This set off an intense race to identify, clone and sequence the gene likely associated with cancer in high-risk families (6; 10; 40). BRCA1 was identified in 1994 by a team led by Mark Skolnick at the University of Utah, and also associated with the genomic startup Myriad Genetics (58; 93). Linkage to chromosome 13 was found in 1994 by a team in Britain, and BRCA2 was cloned and sequenced in 1995 (161; 162). Two decades later, these remain the two genes most commonly mutated in families with inherited risk of breast and ovarian cancer, although BRCA mutations are found in other cancers, and mutations in another two dozen genes are also associated with carcinomas of the ovaries and breasts (but at much lower frequency).
Both genes were patented, and the story is complicated (24; 36; 58). The first BRCA1 patent was granted to OncorMed in summer 1997, and OncorMed sued Myriad. Myriad countersued the day after it got its first patent that December. The companies settled out of court, with OncorMed agreeing to exit the BRCA testing market and assign its patents to Myriad. There were other BRCA patents, including one granted to the U.K. team that first published on BRCA2, but that sequence was chimeric, and the Myriad team filed a patent application on BRCA2 just a day before the U.K. team published in Nature. Myriad cleared the U.S. market of competitors for commercial BRCA testing, first by sending notification letters to laboratories offering the test, and even suing one such laboratory at the University of Pennsylvania. The laboratory quickly settled and withdrew from testing, beyond for patients in the University of Pennsylvania healthcare system.
From 1998 through 2013 Myriad had a service monopoly on American BRCA testing, yet this did not work anywhere outside the U.S. European patents were opposed and narrowed (88). In Australia, Myriad was forced to license to Genetic Technologies, Ltd. (GTG) to settle a patent infringement suit over use of intervening sequences, and GTG permitted labs in the Australian regional health system to offer testing as a “gift to the people of Australia.” GTG threatened to revoke this gift, yet a firestorm of criticism led to the replacement of the chief executive and a flurry of Senate activity and the “gift” was restored (149). In Canada, Ontario’s Prime and Health Ministers refused to recognize Myriad’s rights, and Myriad never sued in Canada, so most provinces continued to offer BRCA testing (58). In Great Britain, the National Health System (NHS) offered BRCA testing regionally, and largely ignored Myriad’s patents (67; 117). The patent monopoly only held in the U.S., but that was sufficient to give Myriad a dominant position until 2013, by which point it had administered over a million BRCA tests.
The monopoly ended at the U.S. Supreme Court, when Myriad lost an epic patent battle against the American Civil Liberties Union (132). The following month, August 2013, Myriad filed the first of several lawsuits against seven competitors, all of which ended when the Court of Appeals for the Federal Circuit invalidated Myriad’s patents in December of 2014. Myriad had dismissed the last of its suits by February 2015. Several new laboratories entered the BRCA testing market on June 13, 2013, the day of the Supreme Court decision, and several more entered in the following months.
Myriad shared its data on BRCA genetic variants until November 2004, and allowed selective access through its proprietary database to academic collaborators through 2006 (11). But it stopped depositing data at the locus-specific Breast Cancer Information Core at NHGRI, the largest repository of such data. It has since taken further steps to protect its trade secret database through click-through agreements on its website, precluding users from sharing data with third parties and explicitly claiming trade secrecy (data on file with author, R C-D). A decade of testing by the company has revealed thousands of BRCA variants, but Myriad alone knows what these are. While it publishes the names of its interpretive methods, Myriad neither shares them sufficiently for replication nor provides the underlying data, publishing in journals that do not require such disclosure (45; 118).
The BRCA Challenge of GA4GH was intended to address the anomalous situation occasioned by Myriad, which had both a patent monopoly for over a decade and decided to treat data as trade secrets. In order to interpret BRCA variants, the rest of the world had to catch up, since Myriad did not make its data available and did not participate in inter-lab comparisons of variants. The number of BRCA tests administered worldwide is comparable to Myriad’s experience. Yet the company itself had limited access to variants in Africa, Asia, Latin America, and other places where its pricing precludes the use of its tests, different founder mutations have taken root, and rare alleles will continue to crop up. In short, the data have not been shared, stored, curated, or interpreted in ways that can be used for clinical decisions.
BRCA Challenge was announced as ClinVar and ClinGen were getting started. The BRCA Exchange regularly shares data with ClinVar, and ClinVar regularly does variant comparisons with LOVD, ARUP’s BRCA database, and is adding more databases and data as those become available Several of the laboratories that started BRCA testing—Ambry, Invitae, GeneDx, Illumina, and others—promoted the “open science” framework of ClinGen and ClinVar and contributed their data on variants, including different degrees of clinical phenotype information. Another response to Myriad’s data-hoarding policy was an effort, the Sharing Clinical Results Project (SCRP), to secure the laboratory reports that Myriad sent back to ordering laboratories and physicians, or simply to get the same information from individual women who knew their BRCA status through the Genetic Alliance’s “Free the Data” project (14).
The two largest diagnostic firms in the U.S., Quest and LabCorp, started offering BRCA testing in 2013. In May of 2015, Quest announced it was contributing its data to the Universal Mutation Database (UMD) in Paris, where it would be well curated and interpreted (17; 112). Quest proposed that commercial labs pay for access to the UMD data and contribute to it, while researchers would have free access. LabCorp joined that effort, which came to be known as BRCA Share®. The UMD and ClinVar are now discussing how, and how much, data will flow into freely available databases. ARUP laboratories also established a BRCA database, which is likewise contributing to ClinVar. This spectrum of data-sharing for BRCA variants spans from purely proprietary data-hoarding by Myriad to free and open sharing embodied by ClinVar, SCRP, and Free the Data, with intermediate models of research access and paid commercial storage and curation through BRCA Share®.
Cancer Gene Trust
The Global Alliance’s newest demonstration project is the Cancer Gene Trust, which will first focus on sharing data about somatic genomic cancer variants (53). It is starting with somatic genes in part for simplicity, until privacy issues with potentially identifiable germline data are resolved. One distinctive feature is packets of software that can move amongst linked servers, leaving the large repositories of data in place but allowing analyses and results to be returned to the user. The data stay in place; the analytical software migrates.
The Global Alliance’s projects entail a substantial technical component, and much of the work has centered on developing application programming interfaces (APIs). One group addresses data security, a highly technical domain. Many of the challenges in building a Global Alliance, however, center on ethical issues, law, and policy, the domains of the Regulatory and Ethics Working Group. Indeed, the legal and social impediments to global data sharing are amongst the most challenging obstacles to a medical information commons. One of the early efforts of the working group was to articulate a “Framework for the Responsible Sharing of Genomic Data,” a high-level agreement that has been translated into 13 languages (56). This framework emphasized the need to preserve the scientific value of data where possible, rather than anonymizing it and diminishing the ability to make inferences amongst diverse, individually oriented data types. Major issues include: (a) a need to protect privacy and confidentiality, and to honor informed consent agreements for data generated under diverse national laws; (b) data security; (c) the accommodation of clinical and research uses; and (d) a much more complicated and diverse set of commercial firms involved in genomic research, and incorporating the attendant data into clinical care. We review these briefly in turn.
International Data Sharing Must Abide by National Laws
Privacy and return of results to participants have been the subjects of many articles in Annual Review of Genomics and Human Genetics (4; 59; 82; 91). One of the key findings from legal scholarship is that laws in different nations pertain to activities essential to data sharing. Branum and Wolf recently reviewed the international law on return of results from genomic analysis (20). Another relevant body of law concerns privacy, confidentiality, and informed consent. Many nations have passed laws encouraging the engagement of local researchers with research done in their countries, both to foster economic development and prevent “biocolonialism” and “helicopter research,” wherein foreign researchers extract value but leave little, violating the notion of reciprocity.
A symposium and two recent issues of the Journal of Law, Medicine and Ethics resulted from a massive effort, led by Mark Rothstein and Bartha Knoppers, to review laws pertaining to sharing of samples and data (126; 127). Forty authors surveyed the law in twenty countries. The focus was on biobanks, but included data sharing. Many countries have laws governing data security, privacy and confidentiality. Many require governmental approval and/or sanction from a local ethics review board to export genetic data. Dove notes that a full international harmonization of laws is unlikely, recommending, “foundational responsible data sharing principles in an overarching governance framework” (43). Thorogood and Zawati point out that while incompatible national laws can be impediments to sharing, there is also virtue in pluralism of approaches across nations and cultures (141).
National laws must be respected, and yet they are complicated, and require national, regional, and sometimes local legal expertise to identify and interpret. Rothstein and Knoppers conclude:
[R]elevant laws differ widely among countries engaged in biobank-enabled research in terms of substance, procedure, and underlying public policies. The lack of international regulatory harmonization has been shown to impede data sharing for translational research in genomics and related fields. The daunting task is to identify and characterize the biobank structure and applicable standards in each country and then to devise possible ways to harmonize policies and laws to enable international biobank research while still giving effect to essential privacy protections. (126)
Establishing the infrastructure for international data sharing—to reap the benefits of genomic variants around the globe—confronts drastically greater complexity than did organizing the sequences of mapped DNA segments from anonymized samples, the starting material for the human reference sequence. It is a long voyage from Bermuda.
Scholars are working assiduously to navigate these tumultuous waters. Vanderbilt’s Center for Genetic Privacy and Identity in Community Settings, led by Ellen Wright Clayton and Bradley Malin, is a Center of Excellence for Ethical Legal and Social Implications (ELSI) Research (134). Susan Wolf, Ellen Wright Clayton, and Frances Lawrenz are leading LawSeq, a prodigious accumulation of talent turning its attention to the legal foundations of translating genomics into clinical applications, focusing on U.S. federal law (31). Finally, Amy McGuire and Robert Cook-Deegan are co-directing a grant on “Building the Medical Information Commons,” also centered on American efforts (13).
Clinical Laboratories Generate as Much (or More) Information about Genomic Variants as do Research Laboratories
Since Bermuda, the flow of human genome data has shifted decisively from publicly funded research to commercial laboratory testing, to help individuals make better-informed decisions about medical care, ancestry, or other personal issues. The uses of genomic variation data are likewise embedded in both research and clinical care. The hundreds of databases that nucleated around particular genes (e.g., the CF Transporter Receptor, or the Huntington’s locus) or medical conditions (e.g., epilepsy, Alzheimer’s, or various cancers) generally started from researchers contributing to the literature and depositing their data: sometimes in locus-specific databases, or sometimes in more general databases like the Human Gene Mutation Database in Cardiff, the Universal Mutation Database in Paris, the Leiden Open Variation Database in Belgium, or the many others maintained by the National Center for Biotechnology Information at the National Library of Medicine (e.g., GenBank, dbGaP, RefSeq, OMIM, or ClinVar). These now contain evidence used to make clinical decisions, but (with the exception of ClinVar) were generally not intended for that purpose. The information they contain needs to be validated before clinical use.
Data from commercial testing laboratories is mainly for clinical inference, yet the flow of data into public databases is highly variable, depending in part on business models, history, and how hard it is to get data into them. ClinVar is unique in having been constructed from the beginning for clinical determinations, although it draws heavily from research databases (66). Some of the major contributors to the ClinVar database, through the panel of collaborators that constitute ClinGen, are commercial laboratories(118). This trend towards data flowing primarily from clinical testing laboratories is bound to accelerate as genomic analysis is integrated into healthcare. The shift to “clinical grade” databases, with systematic vetting of variant calls, storage and curation of the data, quality control measures, and participation in proficiency testing, are all part of the nascent infrastructure for clinical genomics.
In most countries, genetic testing is incorporated into laboratory practices in national health systems. Not so in the U.S., where federal regulation of genetic tests has been the subject of debates, books, and several reports from federal advisory committeessince at least 1984 (74; 75; 97; 105; 129). Laboratories in the U.S. are currently regulated by the Clinical Laboratories Improvement Amendments (CLIA) of 1988, through the Centers for Medicare and Medicaid Services. The College of American Pathologists also accredits clinical laboratories. The Food and Drug Administration (FDA) floated draft guidance indicating intent to regulate laboratory-developed genetic tests(LDTs) as medical devices in 2014, proposing to phase-in such regulation over nine years(56). FDA’s entry into regulation caused a kerfuffle, and was opposed by many laboratories, their trade associations, and the Association for Molecular Pathology (121). FDA announced in November 2016 that it would back away from finalizing its guidance (122). But its draft rules on the importance of databases, and the need for independent verification of genomic interpretations used to guide clinical decisions, is nonetheless a clear statement of some of the challenges ahead in building the tools for clinical use, and the need for “regulatory grade” genomic databases(53). Indeed, clinical use requires far more formal oversight and regulation than creating and using data in research. The flow of data from commercial testing laboratories is likely to become the main source of new information about human genomic variation.
The Diversity and Importance of Private Firms is Substantially Greater Than When Generating the Human Reference Sequence
Commercial genomics extends well beyond genetic testing. Even within genetic testing, it includes ancestry testing, personal genomic profiling with chips, exome sequencing, or even whole-genome sequencing, and a panoply of tests ranging from single-gene tests to multi-gene panels. Genomic sequencing has been used to study rare disorders and profile somatic mutations in cancers, to guide choices about treatment and prevention. Other firms specialize in integrating genomic data into medical records or providing bioinformatics tools for genomic data analysis. Some are dedicated to sequencing as all or parts of their business models. In a 2014 editorial, Curnutte documented this diversity of commercial software, hardware, and various services (39). A great deal of expertise in informatics and instrument manufacturing resides in the private sector, in companies of wildly different sizes, ages, financial health, and business models. Many are compatible with data sharing; many are not. The evolving pattern of sharing for BRCA1 and BRCA2, and the complexity of the genomic data commons, illustrate the challenges ahead.
Conclusion
The Bermuda Principles for daily pre-publication data release set a strong foundation for open science. The Principles set a salutary precedent that enabled more rapid progress towards first assembling a human reference sequence and then interpreting the meaning of genomic variants in humans and other organisms. The initial community of fifty people in fewer than a dozen laboratories has broadened to a global endeavor involving hundreds of laboratories, spanning from pure research to clinical use, ancestry, and other applications. As the community of data-contributors and data-users has broadened, the idea of daily sharing has had to adapt, both to comply with the informed consent of people to whom the data pertain, to ensure data security, and to address considerably diversity in international laws governing privacy, confidentiality, and trans-border flow of genetic samples and data. In addition to these challenges, the commercial interests have both intensified and grown far more diverse. The spirit of open science, however, persists. The Bermuda Principles are the taproot from which a global medical information commons will grow.
FIGURE 1. Participants at the first Bermuda meeting, February 1996.
[http://hdl.handle.net/10161/7713, accessed 25 November 2016, photo credit: Richard Myers, HudsonAlpha]
FIGURE 2. The white board on which John Sulston scribbled the Bermuda Principles at the 1996 meeting’s final session. Robert Waterston was leading the discussion, and there was an (informal) vote to adopt the statement.

[http://hdl.handle.net/10161/7721, accessed 25 November 2016, photo credit: Richard Myers.]
FIGURE 3.
Adapted from (17), p. 1323.
Variants in BRCA1/2 genes associated with inherited risk of breast, ovarian and other cancers. Different databases have data on different variants, and none of these databases includes many cases from Africa, Latin America, Asia, or other populations expected to have different founder mutations and population frequency of uncommon alleles.
Contributor Information
Robert Cook-Deegan, Arizona State University.
Rachel A. Ankeny, University of Adelaide
Kathryn Maxson Jones, Princeton University.
Works Cited
- 1.Adams MD, Kelley JM, Heannine D, Gocayne, et al. Complementary DNA Sequencing: Expressed Sequence Tags and the Human Genome Project. Science. 1991;252:1651–6. doi: 10.1126/science.2047873. [DOI] [PubMed] [Google Scholar]
- 2.Adams MD, Sutton GG, Smith HO, Myers EW, Venter JC. The independence of our genome assemblies. Proceedings of the National Academy of Sciences. 2003;100:3025–6. doi: 10.1073/pnas.0637478100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adams MD, Venter JC. Should non-peer-reviewed raw DNA sequence data release be forced on the scientific community? Science. 1996;274:534–6. doi: 10.1126/science.274.5287.534. [DOI] [PubMed] [Google Scholar]
- 4.Anderlik MR, Rothstein MA. Privacy and Confidentiality of Genetic Information: What Rules for the New Science? Annual Review of Genomics and Human Genetics. 2001;2:401–33. doi: 10.1146/annurev.genom.2.1.401. [DOI] [PubMed] [Google Scholar]
- 5.Anderson A. Yeast genome project 300, 000 and counting. Science. 1992;256:462. doi: 10.1126/science.256.5056.462. [DOI] [PubMed] [Google Scholar]
- 6.Angier N. Fierce Competition Marked Fervid Race for Cancer Gene. The New York Times. 1994 Sep 20; [Google Scholar]
- 7.Angrist M. Eyes wide open: the personal genome project, citizen science and veracity in informed consent. Per Med. 2009;6:691–9. doi: 10.2217/pme.09.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Angrist M. Here is a Human Being at the Dawn of Personal Genomics. New York, NY: HarperCollins Publishers; 2010. [Google Scholar]
- 9.Ankeny RA, Leonelli S. Valuing Data in Postgenomic Biology: How Data Donation and Curation Practices Challenge the Scientific Publication System. In: Richardson SS, Stevens H, editors. Postgenomics: Perspectives on Biology after the Genome. Durham, NC and London, UK: Duke University Press; 2015. pp. 126–49.pp. 126–49. [Google Scholar]
- 10.Arias JJ, Pham-Kanter G, Campbell EG. The growth and gaps of genetic data sharing policies in the United States. J Law Biosci. 2015;2:56–68. doi: 10.1093/jlb/lsu032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baldwin AL, Cook-Deegan R. Constructing narratives of heroism and villainy: case study of Myriad’s BRACAnalysis((R)) compared to Genentech’s Herceptin((R)) Genome Med. 2013;5:8. doi: 10.1186/gm412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Balter M. France’s Sequencers Aim to Join the Big League. Science. 1998;280:30–1. [PubMed] [Google Scholar]
- 13.Baylor College of Medicine. Building the Medical Information Information Commons. 2016 https://www.bcm.edu/centers/medical-ethics-and-healthpolicy/research/elsi-research/information-commons.
- 14.Bean LJ, Tinker SW, da Silva C, Hegde MR. Free the data: one laboratory’s approach to knowledge-based genomic variant classification and preparation for EMR integration of genomic data. Hum Mutat. 2013;34:1183–8. doi: 10.1002/humu.22364. [DOI] [PubMed] [Google Scholar]
- 15.Bentley DR. Genomic sequence information should be released immediately and freely in the public domain. Science. 1996;274:533–4. doi: 10.1126/science.274.5287.533. [DOI] [PubMed] [Google Scholar]
- 16.Bentley DR, Pruitt KD, Deloukas P, Schuler GD, Ostell J. Coordination of human genome sequencing via a consensus framework map. Trends Genet. 1998;14:381–4. doi: 10.1016/s0168-9525(98)01591-1. [DOI] [PubMed] [Google Scholar]
- 17.Beroud C, Letovsky SI, Braastad CD, Caputo SM, Beaudoux O, et al. BRCA Share: A Collection of Clinical BRCA Gene Variants. Hum Mutat. 2016;37:1318–28. doi: 10.1002/humu.23113. [DOI] [PubMed] [Google Scholar]
- 18.Brandt KA. The GDB Human Genome Data Base: A Source of Integrated Genetic Mapping and Disease Data. Bulletin of the Medical Library Association. 1993;81:285–92. [PMC free article] [PubMed] [Google Scholar]
- 19.Branscomb E. In: Telephone interview. Cook-Deegan R, Maxson K, editors. Durham, NC: Duke University Libraries; 2011. available at http://hdl.handle.net/10161/7693. [Google Scholar]
- 20.Branum R, Wolf SM. International Policies on Sharing Genomic Research Results with Relatives: Approaches to Balancing Privacy with Access. J Law Med Ethics. 2015;43:576–93. doi: 10.1111/jlme.12301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burris J, Cook-Deegan R, Alberts B. The Human Genome Project after a decade: policy issues. Nat Genet. 1998;20:333–5. doi: 10.1038/3803. [DOI] [PubMed] [Google Scholar]
- 22.Calia KG. Patentability of Expressed Sequence Tags: A Study of the Venter Application. Biotechnology Law Report. 1992;11:540–57. [Google Scholar]
- 23.Cantor CR. Orchestrating the Human Genome Project. Science. 1990;248:49–51. doi: 10.1126/science.2181666. [DOI] [PubMed] [Google Scholar]
- 24.Cheon JY, Mozersky J, Cook-Deegan R. Variants of uncertain significance in BRCA: a harbinger of ethical and policy issues to come? Genome Med. 2014;6:121. doi: 10.1186/s13073-014-0121-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Church GM. The personal genome project. Mol Syst Biol. 2005;1:2005-0030. doi: 10.1038/msb4100040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clarke L, Zheng-Bradley X, Smith R, Kulesha E, Xiao C, et al. The 1000 Genomes Project: data management and community access. Nat Methods. 2012;9:459–62. doi: 10.1038/nmeth.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Collins F. In-person interview. In: Cook-Deegan R, Maxson K, editors. Durham, NC: Duke University Libraries; 2012. available at http://hdl.handle.net/10161/7704. [Google Scholar]
- 28.Collins F, Patrinos A. Center for Public Genomics Research Files. Durham, NC: Duke University Libraries; 1997. Collins (NIH) and Patrinos (DOE) letter to Frank Laplace (BMBF), German Federal Ministry of Education and Research, 21 March 1997. available at http://hdl.handle.net/10161/7727. [Google Scholar]
- 29.Collins F, Patrinos A, Morgan M. Center for Public Genomics Research Files. Durham, NC: Duke University Libraries; 1998. Collins (NIH), Patrinos (DOE), and Morgan (Wellcome) letter to Kenichi Matsubara (Osaka University, Japan), 9 March 1998. In. available at http://hdl.handle.net/10161/7737. [Google Scholar]
- 30.Collins FS, Green ED, Guttmacher AE, Guyer MS. A vision for the future of genomics research. Nature. 2003;422:835–47. doi: 10.1038/nature01626. [DOI] [PubMed] [Google Scholar]
- 31.Consortium on Law and Values in Health Environment & the Life Sciences. LawSeq: Building a Sound Legal Foundation for Translating Genomics into Clinical Application. 2016 https://consortium.umn.edu/research/lawseq-building-sound-legal-foundation-translating-genomics-clinical-application.
- 32.Contreras JL. Bermuda’s legacy: policy, patents. and the design of the genome commons. Minnesota Journal of Law, Science, & Technology. 2011;12:61–125. [Google Scholar]
- 33.Contreras JL. NIH’s genomic data sharing policy: timing and tradeoffs. Trends in Genetics. 2015;31:55–7. doi: 10.1016/j.tig.2014.12.006. [DOI] [PubMed] [Google Scholar]
- 34.Contreras JL. Genetic Property. Georgetown University Law Journal. 2016;105:1–54. [Google Scholar]
- 35.Cook-Deegan R. The Gene Wars: Science, Politics, and the Human Genome. New York: W. W. Norton & Company; 1994. [Google Scholar]
- 36.Cook-Deegan R, Heaney C. Patents in genomics and human genetics. Annu Rev Genomics Hum Genet. 2010;11:383–425. doi: 10.1146/annurev-genom-082509-141811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Council of Europe, Committee of Ministers. Recommendation Rec(2006)4 of the Committee of Ministers to member states on research on biological materials of human origin. 2006 https://search.coe.int/cm/Pages/result_details.aspx?ObjectID=09000016805d84f0.
- 38.Cozzarelli NR. UPSIDE: Uniform Principle for Sharing Integral Data and Materials Expeditiously. Proceedings of the National Academy of Sciences. 2004;101:3721–2. doi: 10.1073/pnas.0400437101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Curnutte MA, Frumovitz KL, Bollinger JM, McGuire AL, Kaufman DJ. Development of the clinical next-generation sequencing industry in a shifting policy climate. Nat Biotechnol. 2014;32:980–2. doi: 10.1038/nbt.3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davies K, White M. Breakthrough: The Race to Find the Breast Cancer Gene. New York, NY: John Wiley & Sons, Inc; 1996. [Google Scholar]
- 41.de Chadarevian S. Designs for Life: Molecular Biology After WWII. Cambridge, UK: Cambridge University Press; 2002. [Google Scholar]
- 42.Dickson D. Merck to back ‘public’ sequencing. Nature. 1994;371:365. [Google Scholar]
- 43.Dove ES. Biobanks, Data Sharing, and the Drive for a Global Privacy Governance Framework. J Law Med Ethics. 2015;43:675–89. doi: 10.1111/jlme.12311. [DOI] [PubMed] [Google Scholar]
- 44.How to get ahead: The success of the $1,000 genome programme offers lessons for fostering innovation. Nature. 2014;507:273–4. Editorial. [Google Scholar]
- 45.Eggington JM, Bowles KR, Moyes K, Manley S, Esterling L, et al. A comprehensive laboratory-based program for classification of variants of uncertain significance in hereditary cancer genes. Clin Genet. 2014;86:229–37. doi: 10.1111/cge.12315. [DOI] [PubMed] [Google Scholar]
- 46.Eisenberg RS. Patenting the Human Genome. Emory Law Journal. 1990;39:721–45. [PubMed] [Google Scholar]
- 47.Eisenberg RS. Intellectual property issues in genomics. Trends in Biotechnology. 1996;14:302–7. doi: 10.1016/0167-7799(96)10040-8. [DOI] [PubMed] [Google Scholar]
- 48.Eisenberg RS. Patents and data-sharing in public science. Industrial and Corporate Change. 2006;15:1013–31. [Google Scholar]
- 49.Eisenberg RS, Merges RP. Opinion Letter as to the Patentability of Certain Inventions Associated with the Identification of Partial cDNA Sequences. AIPLA Quarterly Journal. 1995;23:1–52. [Google Scholar]
- 50.GAIN Collaborative Research Group. Manolio TA, Rodriguez LL, Brooks L, Abecasis G, et al. New models of collaboration in genome-wide association studies: the Genetic Association Information Network. Nat Genet. 2007;39:1045–51. doi: 10.1038/ng2127. [DOI] [PubMed] [Google Scholar]
- 51.Garcia-Sancho M. Biology, Computing, and the History of Molecular Sequencing: From Proteins to DNA, 1945–2000. New York: Palgrave Macmillan; 2015. [2012] [Google Scholar]
- 52.Gibbs RA The International HapMap Consortium. The International HapMap Project. Nature. 2003;426:789–96. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 53.Global Alliance for Genomics and Health (GA4GH) About the Global Alliance. http://genomicsandhealth.org/about-global-alliance.
- 54.Global Alliance for Genomics and Health (GA4GH) Beacon Project. http://ga4gh.org/-/beacon.
- 55.Global Alliance for Genomics and Health (GA4GH) White Paper: Creating a Global Alliance to Enable Responsible Sharing of Genomic and Clinical Data. Global Alliance for Genomics and Health (GA4GH); 2013. available at http://genomicsandhealth.org/white-paper-creating-global-alliance-enableresponsible-sharing-genomic-and-clinical-data-read-online. [Google Scholar]
- 56.Global Alliance for Genomics and Health (GA4GH) Framework for Responsible Sharing of Genomic and Health-Related Data. Global Alliance for Genomics and Health (GA4GH); 2014. available at https://genomicsandhealth.org/about-the-global-alliance/key-documents/framework-responsible-sharing-genomic-and-health-related-data. [Google Scholar]
- 57.Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, et al. Life with 6000 genes. Science. 1996;274:546, 63–7. doi: 10.1126/science.274.5287.546. [DOI] [PubMed] [Google Scholar]
- 58.Gold ER, Carbone J. Myriad Genetics: In the eye of the policy storm. Genetics in medicine: official journal of the American College of Medical Genetics. 2010;12:S39–70. doi: 10.1097/GIM.0b013e3181d72661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Greely HT. The uneasy ethical and legal underpinnings of large-scale genomic biobanks. Annu Rev Genomics Hum Genet. 2007;8:343–64. doi: 10.1146/annurev.genom.7.080505.115721. [DOI] [PubMed] [Google Scholar]
- 60.Green ED, Watson JD, Collins FS. Human Genome Project: Twenty-five years of big biology. Nature. 2015;526:29–31. doi: 10.1038/526029a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guyer M. Statement on the rapid release of genomic DNA sequence. Genome Research. 1998;8:413. doi: 10.1101/gr.8.5.413. [DOI] [PubMed] [Google Scholar]
- 62.Hall JM, Lee MK, Newman B, Morrow JE, Anderson LA, et al. Linkage of early-onset familial breast cancer to chromosome 17q21. Science. 1990;250:1684–9. doi: 10.1126/science.2270482. [DOI] [PubMed] [Google Scholar]
- 63.Hardin G. The Tragedy of the Commons. Science. 1968;162:1243–8. [PubMed] [Google Scholar]
- 64.Hardin G. Extensions of “The Tragedy of the Commons”. Science. 1998;280:682–3. [Google Scholar]
- 65.Harris R, Dixon B. Genome jousting enlivens a sporting lull. Current Biology. 2001;11:R202–R3. doi: 10.1016/s0960-9822(01)00104-x. [DOI] [PubMed] [Google Scholar]
- 66.Harrison SM, Riggs ER, Maglott DR, Lee JM, Azzariti DR, et al. Using ClinVar as a Resource to Support Variant Interpretation. Current Protocols in Human Genetics. 2016;89:1–23. doi: 10.1002/0471142905.hg0816s89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hawkins N. An exception to infringement for genetic testing--addressing patient access and divergence between law and practice. International Review of Intellectual Property and Competition Law. 2012;43:641–61. [Google Scholar]
- 68.Hayden EC. The $1,000 genome. Nature. 2014;507:294–5. doi: 10.1038/507294a. [DOI] [PubMed] [Google Scholar]
- 69.Healy B. Special Report on Gene Patenting. The New England Jounal of Medicine. 1992;327:664–8. doi: 10.1056/NEJM199208273270930. [DOI] [PubMed] [Google Scholar]
- 70.Hess C, Ostrom E, editors. Understanding Knowledge as a Commons: From Theory to Practice. Cambridge, MA: MIT Press; 2007. [Google Scholar]
- 71.Hilgartner S. Private Science: Biotechnology and the Rise of the Molecular Sciences. Philadelphia: University of Pennsylania Press; 1998. Data Access Policy in Genome Research; pp. 202–18. [Google Scholar]
- 72.Hilgartner S, Brandt-Rauf SI. Data Access, Ownership, and Control. Knowledge: Creation, Diffusion, Utilization. 1994;15:355–72. [Google Scholar]
- 73.Holden AL. The SNP Consortium: Summary of a private consortium effort to develop and applied map of the human genome. BioTechniques. 2002;32:S22–6. [PubMed] [Google Scholar]
- 74.Holtzman N. Proceed with caution: predicting genetic risks in the recombinant DNA era. Baltimore, MD: The Johns Hopkins University Press; 1989. [Google Scholar]
- 75.Holtzman NA, Watson MS, editors. Promoting Safe and Effective Genetic Testing in the United States: Final Report of the Task Force on Genetic Testing. Washington, DC: NIH-DOE Working Group on Ethical, Legal and Social Implications of Human Genome Research; 1997. [PubMed] [Google Scholar]
- 76.Hudson TJ, Stein LD, Gerety Sebastian S, editors. An STS-Based Map of the Human Genome. Science. 1995;270:1945–54. doi: 10.1126/science.270.5244.1945. [DOI] [PubMed] [Google Scholar]
- 77.International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–45. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 78.Istrail S, Sutton GG, Florea L, et al. Whole-genome shotgun assembly and comparison of human genome assemblies. Proceedings of the National Academy of Sciences. 2004;101:1916–21. doi: 10.1073/pnas.0307971100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jasanoff S. Designs on Nature: Science and Democracy in Europe and the United States. Princeton, NJ: Princeton University Press; 2005. [Google Scholar]
- 80.Jasanoff S, editor. Reframing Rights: Bioconstitutionalism in the Genetic Age. Cambridge, MA and London, UK: The MIT Press; 2011. [Google Scholar]
- 81.Joint Genome Institute, United States Department of Energy. History. 2016 http://jgi.doe.gov/about-us/history/
- 82.Kaye J. The Tension Between Data Sharing and the Protection of Privacy in Genomics Research. Annual Review of Genomics and Human Genetics. 2012;13:415–31. doi: 10.1146/annurev-genom-082410-101454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 84.Madison MJ, Frischmann BM, Strandburg KJ, editors. Governing Medical Knowledge Commons. Cambridge University Press; 2017. forthcoming. [Google Scholar]
- 85.Marshall E. A Strategy for Sequencing the Genome 5 Years Early. Science. 1995;267:783–4. doi: 10.1126/science.7846520. [DOI] [PubMed] [Google Scholar]
- 86.Marshall E. Bermuda Rules: Community Spirit, With Teeth. Science. 2001;291:1192. doi: 10.1126/science.291.5507.1192. [DOI] [PubMed] [Google Scholar]
- 87.Martinell J. Initial rejection of NIH Patent Application 07/837,195 (EST sequences and methods) Biotechnology Law Report. 1992;11:578–96. [Google Scholar]
- 88.Matthijs G. The European Opposition Against the BRCA Gene Patents. Familial Cancer. 2006;5:95–102. doi: 10.1007/s10689-005-2580-6. [DOI] [PubMed] [Google Scholar]
- 89.Maxson Jones K, Ankeny RA, Cook-Deegan R. 2017 forthcoming. [Google Scholar]
- 90.McElheny VK. Drawing the Map of Life: Inside the Human Genome Project. New York: Basic Books; 2010. [Google Scholar]
- 91.McGuire AL, Beskow LM. Informed consent in genomics and genetic research. Annu Rev Genomics Hum Genet. 2010;11:361–81. doi: 10.1146/annurev-genom-082509-141711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.McKusick VA, Ruddle FH. Editorial: A New Discipline, A New Name, A New Journal. Genomics. 1987;1:1–2. [Google Scholar]
- 93.Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, et al. A strong candidate for the breast and ovarian cancer susceptibility gene BRCA1. Science. 1994;266:66–71. doi: 10.1126/science.7545954. [DOI] [PubMed] [Google Scholar]
- 94.Murray JC, Buetow KH, Weber JL, Ludwigsen S, Scherpbier-Heddema T, et al. A comprehensive human linkage map with centimorgan density. Cooperative Human Linkage Center (CHLC) Science. 1994;265:2049–54. doi: 10.1126/science.8091227. [DOI] [PubMed] [Google Scholar]
- 95.Myers EW, Sutton GG, Smith HO, Adams MD, Venter JC. On the sequencing and assembly of the human genome. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:4145–6. doi: 10.1073/pnas.092136699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.National Human Genome Research Institute (NHGRI) Policy on Availability and Patenting of Human Genomic DNA Sequence Produced by NHGRI Pilot Projects (Funded Under RFA HG-95–005) 1996 https://www.genome.gov/10000926/
- 97.National Human Genome Research Institute (NHGRI) NHGRI Policy for Release and Database Deposition of Sequence Data. 2000 https://www.genome.gov/10000910/policy-on-release-of-human-genomic-sequence-data-2000/
- 98.National Human Genome Research Institute (NHGRI) International Consortium Completes Human Genome Project. All Goals Achieved; New Vision for Genome Research Unveiled. 2003 http://www.genome.gov/11006929/
- 99.National Institutes of Health (NIH) Policy for Sharing of Data Obtained in NIH Supported or Conducted Genome-Wide Association Studies (GWAS) 2007 http://grants.nih.gov/grants/guide/notice-files/NOT-OD-07-088.html.
- 100.National Institutes of Health (NIH) NIH Genomic Data Sharing Policy. 2014 http://grants.nih.gov/grants/guide/notice-files/NOT-OD-14-124.html.
- 101.National Research Council. Mapping and Sequencing the Human Genome. National Research Council, National Academy of Sciences; Washington, D.C: 1988. [Google Scholar]
- 102.National Research Council (NRC) Toward Precision Medicine: Building a Knowledge Network for Biomedical Research and a New Taxonomy of Disease. The National Academies Press; Washington, DC: 2011. [PubMed] [Google Scholar]
- 103.Office of Technology Assessment, US Congress. Mapping Our Genes—The Genome Projects: How Big, How Fast? U.S. Congress, Office of Technology Assessment (OTA); Washington, D.C: 1988. [Google Scholar]
- 104.Office of Technology Assessment, US Congress. The Human Genome Project and Patenting DNA Sequences (approved prepublication draft) Office of Technology Assessment, U.S. Congress; Washington, D.C: 1994. [Google Scholar]
- 105.Office of Technology Assessment (OTA), US Congress. Human Gene Therapy-- A Background Paper. Office of Technology Assessment (OTA), U.S. Congress; Washington, DC: 1984. [Google Scholar]
- 106.Olson M. The Human Genome Project: A Player’s Perspective. Journal of Molecular Biology. 2002;319:931–42. doi: 10.1016/S0022-2836(02)00333-9. [DOI] [PubMed] [Google Scholar]
- 107.Olson MV. A time to sequence. Science. 1995;270:394–6. doi: 10.1126/science.270.5235.394. [DOI] [PubMed] [Google Scholar]
- 108.Open Humans. About Open Humans: Contribute to research & citizen science! 2016 https://www.openhumans.org/about/
- 109.Organization for Economic Cooperation and Development (OECD) OECD Guidelines on the Protection of Privacy and Transborder Flows of Personal Data. 1980 https://www.oecd.org/sti/ieconomy/oecdguidelinesontheprotectionofprivacyandtransborderflowsofpersonaldata.htm.
- 110.Organization for Economic Cooperation and Development (OECD) Guidelines for Human Biobanks and Genetic Research Databases (HBGRDs) 2009 www.oecd.org/sti/biotechnology/hbgrd.
- 111.Organization for Economic Cooperation and Development (OECD) OECD work on privacy. 2013 http://oe.cd/privacy.
- 112.Ostrom AL. Service Research Priorities in a Rapidly Changing Context. Journal of Service Research. 2015;18:127–59. [Google Scholar]
- 113.Ostrom E. An Agenda for the Study of Institutions. Public Choice. 1986;48:3–25. [Google Scholar]
- 114.Ostrom E. Governing the Commons: The Evolution of Institutions for Collective Action. Cambridge, U.K: Cambridge University Press; 1990. [Google Scholar]
- 115.Ostrom E. The Institutional Analysis and Development Framework and the Commons. Cornell Law Review. 2010;95:807–16. [Google Scholar]
- 116.Ostrom E, Schroeder L, Wynne S, editors. Institutional incentives and sustainable development: infrastructure policies in perspective. Boulder, CO: Westview Press; 1993. [Google Scholar]
- 117.Parthasarathy S. Building Genetic Medicine: Breast Cancer, Technology, and the Comparative Politics of Health Care. Cambridge, MA: The MIT Press; 2007. [Google Scholar]
- 118.Philippakis AA, Azzariti DR, Beltran S, Brookes AJ, Brownstein CA, et al. The Matchmaker Exchange: a platform for rare disease gene discovery. Hum Mutat. 2015;36:915–21. doi: 10.1002/humu.22858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Polsky MM, Ostrom E. An Institutional Framework for Policy Analysis and Design. 1999 available at https://mason.gmu.edu/~mpolski/documents/PolskiOstromIAD.pdf.
- 120.Qiang B. Human genome research in China. J Mol Med (Berl) 2004;82:214–22. doi: 10.1007/s00109-003-0515-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ray T. FDA Draft LDT Guidance Slated for Release in 60 Days; Industry Readying Recommendations, Objections. GenomeWeb. 2014 available at https://www.genomeweb.com/clinical-genomics/fda-draft-ldt-guidance-slatedrelease-60-days-industry-readying-recommendations.
- 122.Ray T. FDA Holding Off on Finalizing Regulatory Guidance for Lab-Developed Tests. GenomeWeb. 2016 available at https://www.genomeweb.com/moleculardiagnostics/fda-holding-finalizing-regulatory-guidance-lab-developed-tests.
- 123.Roberts L. Genome patent fight erupts. Science. 1991;254:184–6. doi: 10.1126/science.254.5029.184. [DOI] [PubMed] [Google Scholar]
- 124.Rodriguez H. International summit on proteomics data release and sharing policy. Journal of Proteome Research. 2008;7:4609. doi: 10.1021/pr800779q. [DOI] [PubMed] [Google Scholar]
- 125.Rodriguez H, Snyder M, Uhlen M, Andrews P, Beavis R, et al. Recommendations from the 2008 international summit on proteomics data release and sharing policy: the Amsterdam principles. Journal of Proteome Research. 2009;8:3689–92. doi: 10.1021/pr900023z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Rothstein MA, Knoppers BM. INTRODUCTION: Harmonizing Privacy Laws to Enable International Biobank Research. J Law Med Ethics. 2015;43:673–4. doi: 10.1111/jlme.12310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Saulnier KM, Joly Y. Locating Biobanks in the Canadian Privacy Maze. J Law Med Ethics. 2016;44:7–19. doi: 10.1177/1073110516644185. [DOI] [PubMed] [Google Scholar]
- 128.Schuler GD, Boguski MS, Stewart EA, Stein LD, Gyapay G, et al. A gene map of the human genome. Science. 1996;274:540–6. [PubMed] [Google Scholar]
- 129.Secretary’s Advisory Committee on Genetics Health and Society (SACGHS) U.S. System of Oversight of Genetic Testing. A Response to the Charge of the Secretary of Health and Human Services, US Department of Health and Human Services, Secretary’s Advisory Committee on Genetics, Health, and Society (SACGHS); Bethesda, MD: 2008. [Google Scholar]
- 130.Sherkow JS, Greely HT. The History of Patenting Genetic Material. Annual Review of Genetics. 2015;49:161–82. doi: 10.1146/annurev-genet-112414-054731. [DOI] [PubMed] [Google Scholar]
- 131.Shreeve J. The Genome War: How Craig Venter Tried to Capture the Code of Life and Save the World. New York: Ballantine Books; 2004. [PubMed] [Google Scholar]
- 132.Simoncelli T, Park S. Making the Case Against Gene Patents. Perspectives on Science. 2015;23:106–45. [Google Scholar]
- 133.Smith TF. The history of the genetic sequence databases. Genomics. 1990;6:701–7. doi: 10.1016/0888-7543(90)90509-s. [DOI] [PubMed] [Google Scholar]
- 134.Snyder B. New center to study genomic privacy concerns. 2016 https://news.vanderbilt.edu/2016/05/19/new-center-to-study-genomic-privacyconcerns/
- 135.Spurdle AB, Healey S, Devereau A, Hogervorst FB, Monteiro AN, et al. ENIGMA- -evidence-based network for the interpretation of germline mutant alleles: an international initiative to evaluate risk and clinical significance associated with sequence variation in BRCA1 and BRCA2 genes. Hum Mutat. 2012;33:2–7. doi: 10.1002/humu.21628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Stevens H. Networking Biology: The Origins of Sequence-Sharing Practices in Genomics. Technology and Culture. 2015;56:839–67. doi: 10.1353/tech.2015.0119. [DOI] [PubMed] [Google Scholar]
- 137.Strasser BJ. The Experimenter’s Museum: GenBank, Natural History, and the Moral Economies of Biomedicine. Isis. 2011;102:60–9. doi: 10.1086/658657. [DOI] [PubMed] [Google Scholar]
- 138.Sulston J, Waterston R. In: In-person interview. Cook-Deegan R, Maxson K, Ankeny R, editors. Durham, NC: Duke University Libraries; 2011. available at http://hdl.handle.net/10161/7692. [Google Scholar]
- 139.Thompson L. Healy and Collins strike a deal. Science. 1993;259:22–3. [PubMed] [Google Scholar]
- 140.Thorisson GA, Stein LD. The SNP Consortium website: past, present and future. Nucleic Acids Research. 2003;31:124–7. doi: 10.1093/nar/gkg052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Thorogood A, Zawati MH. International Guidelines for Privacy in Genomic Biobanking (or the Unexpected Virtue of Pluralism) J Law Med Ethics. 2015;43:690–702. doi: 10.1111/jlme.12312. [DOI] [PubMed] [Google Scholar]
- 142.Toronto International Data Release Workshop A. Birney E, Hudson TJ, Green ED, Gunter C, et al. Prepublication data sharing. Nature. 2009;461:168–70. doi: 10.1038/461168a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.United Nations Educational Scientific and Cultural Organization (UNESCO) Universal Declaration on the Human Genome and Human Rights. 1997 http://portal.unesco.org/en/ev.php-URL_ID=13177&URL_DO=DO_TOPIC&URL_SECTION=201.html.
- 144.United Nations Educational Scientific and Cultural Organization (UNESCO) International Declaration on Human Genetic Data. 2003 http://portal.unesco.org/en/ev.php-URL_ID=17720&URL_DO=DO_TOPIC&URL_SECTION=201.html.
- 145.United Nations Educational Scientific and Cultural Organization (UNESCO) Universal Declaration on Bioethics and Human Rights. 2005 http://portal.unesco.org/en/ev.php-URL_ID=31058&URL_DO=DO_TOPIC&URL_SECTION=201.html.
- 146.United Nations General Assembly. The Universal Declaration of Human Rights. 1948 http://www.un.org/en/universal-declaration-human-rights/
- 147.Unterhuber R. Germany gets warning on access to sequence data. Nature. 1997;387:111. doi: 10.1038/387111a0. [DOI] [PubMed] [Google Scholar]
- 148.US Department of Energy (DOE), US National Institutes of Health. Understanding Our Genetic Inheritance: The U.S. Human Genome Project: The First Five Years: Fiscal Years 1991–1995. U.S. Department of Health and Human Services, Public Health Service, National Institutes of Health; U.S. Department of Energy, Office of Energy Research, Office of Health and Environmental Research; Washington, D.C: 1990. [Google Scholar]
- 149.Van Zimmeren E, Nicol D, Gold R, Carbone J, Chandrasekharan S, et al. The BRCA Patent Controversies: An international review of patent disputes. In: Gibbon S, Joseph G, Mozersky J, zur Nieden A, Palfner S, editors. Breast Cancer Gene Research and Medical Practices: Transnational perspectives in the time of BRCA. New York, NY: Routledge; 2014. pp. 151–74. [Google Scholar]
- 150.Varian HR, Farrell J, Shapiro C. The Economics of Information Technology: An Introduction. New York, NY and Cambridge, U.K: Cambridge University Press; 2005. [Google Scholar]
- 151.Venter JC, Adams MD, Sutton GG, Kerlavage AR, Smith HO, Hunkapiller M. Shotgun Sequencing of the Human Genome. Science. 1998;280:1540–2. doi: 10.1126/science.280.5369.1540. [DOI] [PubMed] [Google Scholar]
- 152.Venter JC, et al. The Sequence of the Human Genome. Science. 2001;291:1304–51. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 153.Waterston R, Sulston JE. The Human Genome Project: Reaching the Finish Line. Science. 1998;282:53–4. doi: 10.1126/science.282.5386.53. [DOI] [PubMed] [Google Scholar]
- 154.Waterston RH, Lander ES, Sulston JE. On the sequencing of the human genome. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:3712–6. doi: 10.1073/pnas.042692499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Watson JD. The human genome project: past, present, and future. Science. 1990;248:44–9. doi: 10.1126/science.2181665. [DOI] [PubMed] [Google Scholar]
- 156.Watson JD, Crick FH. The structure of DNA. Cold Spring Harbor symposia on quantitative biology. 1953;18:123–31. doi: 10.1101/sqb.1953.018.01.020. [DOI] [PubMed] [Google Scholar]
- 157.Wellcome Trust, US National Institutes of Health, US Department of Energy, 164th Genome Technology Committee, Japan Society for Promotion of Science, UK Medical Research Council. Center for Public Genomics Research Files; Third International Strategy Meeting on Human Genome Sequencing Hamilton Princess Hotel; Bermuda. 27th February – 1st March 1998; Durham, NC: Duke University Libraries; 1998. available at http://hdl.handle.net/10161/7745. [Google Scholar]
- 158.Williams H. Intellectual Property Rights and Innovation: Evidence from the Human Genome. Journal of Political Economy. 2013;121:1–27. doi: 10.1086/669706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Williamson AR. The Merck Gene Index Project. Drug Discovery Today. 1999;4:115–22. doi: 10.1016/s1359-6446(99)01303-3. [DOI] [PubMed] [Google Scholar]
- 160.Wilson R. How the worm was won: the C. elegans genome sequencing project. Trends Genet. 1999;15:51–8. doi: 10.1016/s0168-9525(98)01666-7. [DOI] [PubMed] [Google Scholar]
- 161.Wooster R, Bignell G, Lancaster J, Swift S, Seal S, et al. Identification of the breast cancer susceptibility gene BRCA2. Nature. 1995;378:789–92. doi: 10.1038/378789a0. [DOI] [PubMed] [Google Scholar]
- 162.Wooster R, Neuhausen SL, Mangion J, Quirk Y, Ford D, et al. Localization of a breast cancer susceptibility gene, BRCA2, to chromosome 13q12–13. Science. 1994;265:2088–90. doi: 10.1126/science.8091231. [DOI] [PubMed] [Google Scholar]



