Summary
Hippocampal CA3 neurons form synapses with CA1 neurons in two layers, stratum oriens (SO) and stratum radiatum (SR). Each layer develops unique synaptic properties but molecular mechanisms that mediate these differences are unknown. Here, we show SO synapses normally have significantly more mushroom spines and higher magnitude long-term potentiation (LTP) than SR synapses. Further, we discovered these differences require the Type II classic cadherins, cadherins-6, 9, and 10. Though cadherins typically function via trans-cellular homophilic interactions, our results suggest presynaptic cadherin-9 binds postsynaptic cadherins-6 and 10 to regulate mushroom spine density and high magnitude LTP in the SO layer. Loss of these cadherins has no effect on the lower magnitude LTP typically observed in the SR layer, demonstrating that cadherins-6, 9, and 10 are gatekeepers for high magnitude LTP. Thus, Type II cadherins may uniquely contribute to the specificity and strength of synaptic changes associated with learning and memory.
Introduction
Synapses are broadly classified by the neurotransmitter released and much research has focused on comparing and contrasting glutamatergic versus GABAergic synapses. However, even synapses releasing the same neurotransmitter have unique structural, molecular, and functional properties (Arai et al., 1994; Nicholson et al., 2006; Nicoll and Schmitz, 2005). Whereas unique features of highly unusual synapses like DG-CA3 mossy fiber synapses are well appreciated (Nicoll and Schmitz, 2005), subtle differences between more closely related types of excitatory synapses remain less explored. Here we address this issue in CA1 neurons, which receive glutamatergic excitatory synapses in three distinct layers (Figure 1A). In the stratum lacunosum-moleculare (SLM) layer, CA1 neurons receive input from entorhinal cortex layer III axons. In the stratum oriens (SO) and stratum radiatum (SR) layers, CA1 neurons receive input from CA3 axons (Figure 1A). In addition, about 20% of CA1 SO inputs originate from CA2 axons (Dudek et al., 2016).
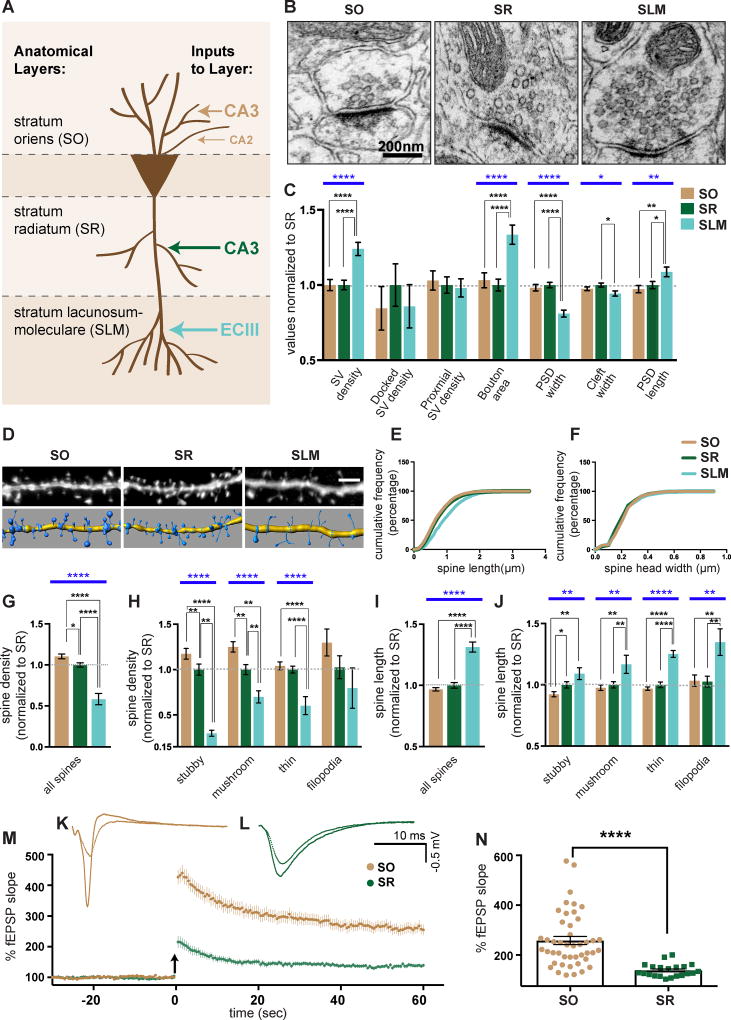
Figure 1. CA1 excitatory synapses have layer-specific properties.
(A) Schematic of excitatory inputs to CA1 neurons. (B) Representative EM images of CA1 synapses. (C) Quantification of EM parameters. All values are normalized to mean SR values. n = 181 (SR), 149 (SO), and 134 (SLM) synapses evenly sampled from 3 mice aged P23. (D) Representative confocal images of Lucifer Yellow filled CA1 dendrites (top) and corresponding 3D models (bottom). (E and F) Cumulative distribution of spine length (E) and spine head width (F) from SO, SR, and SLM spines. Sample sizes: 6028 spines (SO), 7731 spines (SR), 884 spines (SLM). (G–J) Quantification of average spine density (G-H) and spine length (I-J) of indicated spine classes. All values are normalized to mean SR values. n = 49 (SO), 39 (SR), and 11 (SLM) cells from 6, 6, and 3 mice respectively aged P21-P23. (K and L) Representative LTP traces from CA1 SO (K) and CA1 SR (L) layer. (M) Mean LTP time course induced in CA1 SO and SR. Arrow indicates TBS. (N) Mean LTP amplitudes defined as average percentage of fEPSP slope 58.5–60 minutes after TBS in SO and SR layers. n = 45 SO and 21 SR slices from 16 and 9 wildtype mice aged 3–5 months. Statistics for LTP quantification were calculated using the Mann-Whitney test. Statistical differences between SO, SR, and SLM for EM and spine analyses were calculated using one-way ANOVA followed by pairwise Holm-Šidák multiple comparison tests. Blue bars represent one-way ANOVA p-values. Black bars represent pairwise post-test p values. p<0.05, p<0.01, p<0.001, and p<0.0001 is denoted by *, **, ***, and **** respectively, otherwise p>0.05. All data shown as mean ± s.e.m. Data on wildtype mice reported here is a combination of data from Cdh9+/+ and Cdh6+/+;Cdh10+/+ mice introduced in figures 2 ,3, 5, and 6.
Although the major inputs to CA1 SO and SR are from CA3 axons, the two layers have functionally distinct synaptic properties. Notably, several studies identified long-term potentiation (LTP) differences in the CA1 SO and SR (Cavus and Teyler, 1998; Fan, 2013; Kramár and Lynch, 2003; Navakkode et al., 2012; Ramachandran et al., 2014). The most striking difference is that the magnitude of LTP is significantly higher in SO compared to SR in acute slices and in vivo (Arai et al., 1994; Kaibara and Leung, 1993). However, molecular and circuit-wide mechanisms underlying this difference in magnitude are very poorly understood.
One molecular family thought to contribute to specific synapse formation and function are the classic cadherins. Cadherins are calcium-dependent, homophilic cell adhesion molecules. Mice and humans have 18 conserved classic cadherins, which are divided into Type I and Type II cadherins based on sequence similarity in their first extracellular cadherin domain (Nollet et al., 2000). Interestingly, most Type II cadherins are expressed in a cell type-specific manner in the brain. Several studies indicate that differential matching of Type II cadherins provides an adhesive code driving specific synapse formation (Duan et al., 2014; Kuwako et al., 2014; Osterhout et al., 2011; Poskanzer et al., 2003; Redies and Takeichi, 1996; Suzuki et al., 1997; Williams et al., 2011). Moreover, cadherins localize at synapses and regulate many synaptic functions including synaptic vesicle clustering, dendritic spine stabilization, glutamate receptor recruitment, short-term plasticity, and long-term plasticity (Aiga et al., 2010; Bozdagi et al., 2010; Fièvre et al., 2016; Hirano and Takeichi, 2012; Jungling et al., 2006; Mendez et al., 2010; Saglietti et al., 2007a; Tang et al., 1998; Togashi et al., 2002; Vitureira et al., 2011). However, the majority of these functional studies only investigated the role of cadherin-2 (also known as N-cadherin). Cadherin-2 is broadly expressed by neurons and likely affects generic properties common to most synapses rather than conferring synapse-specific properties. Thus, it remains largely untested whether the differentially expressed Type II classic cadherins confer unique properties to specific synapse types.
Here, we investigated whether cadherins confer specific functional properties at CA3-CA1 synapses. First, we used electron microscopy, light microscopy, and electrophysiology to categorize structural and functional differences between excitatory synapses located in different lamina of the CA1 dendritic tree. We demonstrate that, under normal conditions, CA1 SO synapses have significantly more mushroom spines and higher magnitude LTP than CA1 SR synapses. We then identified three Type II cadherins, cadherins-6, 9, and 10, that are selectively required for high magnitude LTP and normal mushroom spine density in the CA1 SO layer. Interestingly, we discovered SR synapses are capable of undergoing high magnitude LTP when inhibition is reduced and this also requires cadherins-6, 9, and 10. Based on expression patterns and binding studies, our results suggest presynaptic cadherin-9 in CA3 neurons binds postsynaptic cadherins-6 and 10 in CA1 neurons. In sum, our results are the first to identify any synaptic molecules required specifically for high magnitude LTP and suggest cadherins-6, 9, and 10 promote high magnitude LTP via trans-synaptic, heterophilic interactions.
Results
CA1 SO synapses exhibit high magnitude synaptic potentiation
To identify layer-specific properties of CA1 excitatory synapses, we first examined presynaptic structure by electron microscopy (EM). We analyzed asymmetric excitatory synapses in CA1 SO (~50–100 µm basal from cell body), SR (~50–100 µm apical from cell body), and SLM (>350 µm apical from cell body). Synaptic vesicles (SVs) were classified as docked if they were touching the active zone membrane and proximal if they were within 30 nm of the active zone but not touching it (Watanabe et al., 2013) (Figure S1A). Results indicate SO and SR synapses have similar structures and vesicle distributions (Figures 1B and 1C). In contrast, SLM synapses are morphologically distinct from SO and SR synapses as indicated by higher SV densities, increased bouton area, and smaller postsynaptic density (PSD) widths (Figures 1B and 1C).
Second, we tested for layer-specific differences in spine shape and density. CA1 neurons were microinjected with Lucifer Yellow dye (Figure 1D) and oblique secondary and tertiary dendritic segments from each layer were analyzed. Spines were classified according to their shape, which reflects the maturity and potentiation state of each synapse (Bourne and Harris, 2007; Harris, 1999; Harris et al., 1992). Absolute spine densities and the relative proportions of spine classes identified by our light microscopic analyses are consistent with those previously observed in EM reconstructions from the SR layer (Figures S1B and S1C) (Harris et al., 1992; Katz et al., 2009).
Results indicate SLM spines are distinct from those in SR and SO. SLM spine densities are significantly lower and spine lengths significantly longer across most spine classes (Figures 1D–J, S1B, and S1C). Interestingly, we also identified significant differences between SR and SO spines. SO has significantly higher densities of stubby and mushroom spines compared to SR (Figures 1H and S1B). We did not observe a significant difference in spine head width among layers (Figure 1F), but this is likely because mushroom spines make up only about 20% of all spines in each layer (Figure S1B). Because mushroom and stubby spines represent the most mature and potentiated spine states (Harris et al., 1992; Holtmaat et al., 2005; Matsuzaki et al., 2004; Roberts et al., 2010; Tønnesen et al., 2014; Xu et al., 2009), we reasoned higher mushroom and stubby spine densities in SO at this basal state may reflect the prior observation that LTP has different properties in SO and SR (Arai et al., 1994). We tested this in our system and found that LTP magnitude induced by theta burst stimulation (TBS) of CA3 axons in acute hippocampal slices is significantly higher in SO compared to SR (Figures 1K–N). We also tested whether short-term plasticity is different between the two layers. We observed no significant difference in paired pulse ratio 30 minutes before or 60 minutes after TBS in either the SO or SR layer (Figures S1D and S1E). This agrees with previous research (Arai et al., 1994) and suggests there is no change in presynaptic release probability following TBS in either layer and that layer-specific differences in LTP are likely mediated via postsynaptic mechanisms.
Together, our results indicate CA1 SO and SR synapses differ in electrophysiological and morphological characteristics. This is particularly interesting because both SO and SR are primarily composed of CA3-CA1 synapses. Thus, we next focused on elucidating molecular mechanisms required for high magnitude potentiation observed in the SO layer. For clarity, throughout the manuscript we use “normal magnitude LTP” to refer to LTP observed in SR layer that is ~150% above baseline and “high magnitude LTP” to refer to LTP observed in SO layer that is >200% above baseline.
Cadherin-9 is required for high magnitude synaptic potentiation in the CA1 SO layer
We previously showed cadherin-9 regulates DG-CA3 synapse formation, functioning presynaptically in DG neurons and postsynaptically in CA3 neurons (Williams et al., 2011). Because cadherin-9 (Cdh9) mRNA is expressed by CA3 neurons (Figure 2A) (Williams et al., 2011) and cadherins generally localize to both pre- and postsynaptic sites, we tested if cadherin-9 also localizes to CA3 axons. Cadherin-9 fused to the high performance epitope tag smFPFLAG (Viswanathan et al., 2015) was expressed in mouse embryos by in utero electroporation of plasmid DNA. Immunostaining at postnatal day (P) 14 revealed that Cdh9-smFPFLAG is found in distinct puncta along CA3 axons (Figure 2B), suggesting cadherin-9 localizes to CA3 presynaptic boutons.
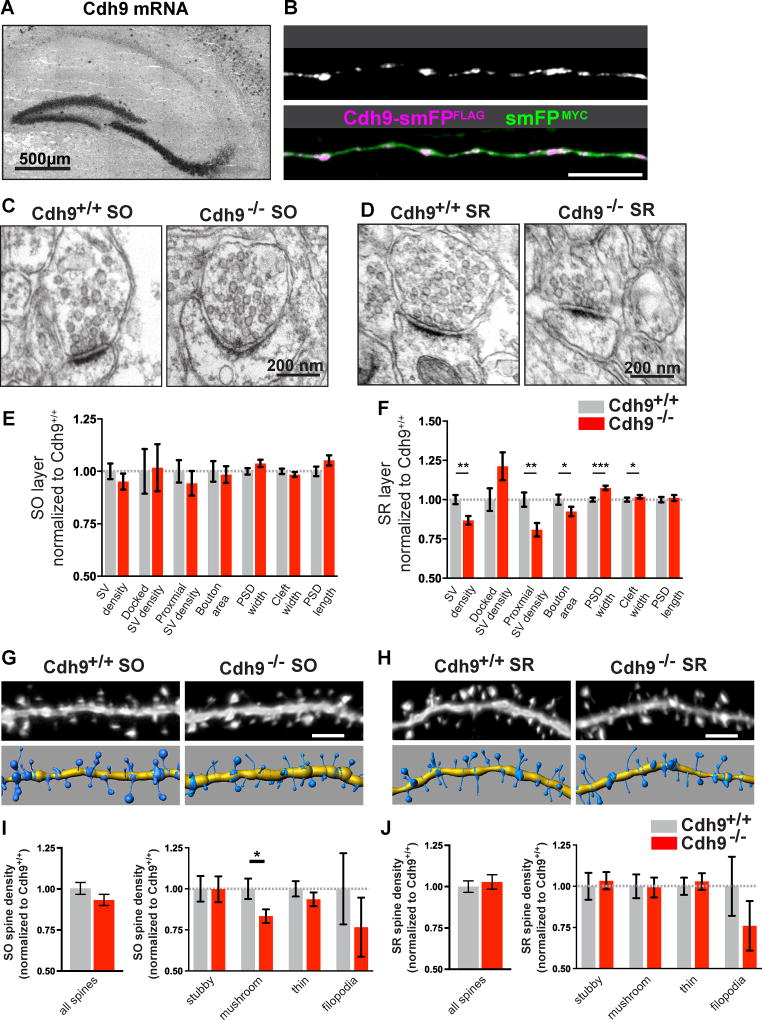
Figure 2. Cadherin-9 regulates mushroom spines in the CA1 SO layer.
(A) In situ hybridization shows Cdh9 mRNA is expressed in DG and CA3 neurons. (B) Immunostaining of a P14 CA3 axon in utero electroporated with Cdh9-smFPFLAG (purple) and smFPMYC (green) at embryonic age 14.5 days. smFPMYC was used to fill the axon. Composite image shown in lower panel. (C–D) Representative EM images of synapses from the SO (C) and SR (D) of Cdh9+/+ and Cdh9−/− mice. (E–F) Quantification of synapses from the SO (E) and SR (F) layers imaged by EM. n = 190 Cdh9+/+ and 170 Cdh9−/− synapses in SO and 314 Cdh9+/+ and 316 Cdh9−/− synapses in SR. All analyses were evenly sampled from 3 mice aged P21 and done blind to genotype. pvalues were calculated using Mann-Whitney test. (G,H) Representative images of SO (G) and SR (H) dendrites analyzed in Cdh9+/+ and Cdh9−/− mice (top) and corresponding 3D models (bottom). (I,J) Quantification of average spine density of total spines (left) and indicated spine classes (right) from the SO (I) and SR (J) layers. All data is normalized to Cdh9+/+. Absolute values are shown in Figure S2. n = 28 Cdh9+/+ and 25 Cdh9−/− cells for SO and 23 Cdh9+/+ and 18 Cdh9−/− cells from SR. All analyses were evenly sampled from 3 mice aged P21-P23 and done blind to genotype. p-values were calculated using students t-test and p<0.05, p<0.01, and p<0.001 is denoted by *, **, and *** respectively, otherwise p>0.05. All data shown as mean ± s.e.m.
Next, we used cadherin-9 knockout (Cdh9−/−) mice (Duan et al., 2014) to test whether cadherin-9 is required for synapse formation or function in CA3-CA1 synapses located in SR or SO. We confirmed knockout mice lack cadherin-9 protein in hippocampal lysates (Figure S2A) and then analyzed synapse morphology in P21 mice by EM (Figures 2C–F). Cdh9−/− mice have no significant changes to synaptic structures in the SO layer compared to wildtype (Figure 2E). In contrast, we identified changes in SV densities, bouton size, and PSD size in the SR layer (Figure 2F). However, we could not identify a functional presynaptic defect associated with these changes as the paired-pulse ratio in the SR layer at numerous inter-stimulus intervals (Figure S2B) is similar between Cdh9+/+ and Cdh9−/− mice.
We next considered the possibility that cadherin-9 acts trans-synaptically at CA3-CA1 synapses. To test this, we analyzed dendritic spines in SO and SR layers of CA1 neurons in young (P24) Cdh9+/+ and Cdh9−/− mice. All spine analyses were conducted blind to genotype. We observed a specific and significant reduction of mushroom spine density in the SO layer of Cdh9−/− animals (17% average reduction compared to Cdh9+/+ animals) although no changes in total spine densities or spine head widths were detected in either layer (Figures 2G–J and S2C–F). Again, this is likely because mushroom spines make up about 20% of all spines and a reduction specifically of mushroom spines may not significantly affect spine population data.
We then tested if the reduction of SO mushroom spines in Cdh9−/− animals correlates with impaired synaptic potentiation specifically in SO by measuring LTP in the SO and SR layers of Cdh9+/+ and Cdh9−/− hippocampal slices. To match the EM and spine analyses, LTP experiments were first conducted in young mice aged P21–P35. LTP is significantly lower in the SO but not the SR of Cdh9−/− mice compared to Cdh9+/+ mice (Figures 3A–D). Interestingly, LTP levels in Cdh9−/− SO are not significantly different than LTP levels observed in Cdh9+/+ SR (Figure S3A). This suggests cadherin-9 is not required for normal magnitude LTP but is required for high magnitude LTP specific to the SO layer. Moreover, analysis of total spine density (Figure 2G–J), input-output curves from field recordings (Figures S3B and C), and spontaneous miniature excitatory postsynaptic currents (mEPSCs) (Figures S3D–F) suggest the LTP defect in Cdh9−/− mice is not due to impaired synapse formation or basic function. In addition, the density of CA3 axons in CA1 SO and SR is similar between Cdh9+/+ and Cdh9−/− mice (Figures 3E–F), suggesting there is no major defect in CA3 axon guidance to either layer in the absence of cadherin-9.
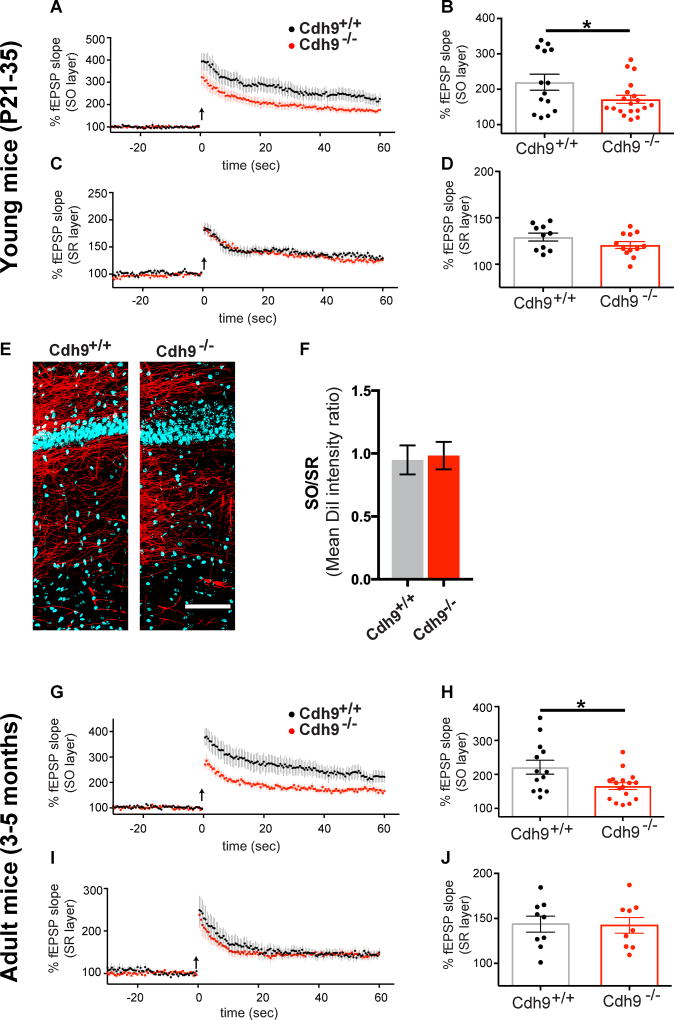
Figure 3. Cadherin-9 regulates high magnitude LTP in CA1 SO.
(A–D) Mean LTP time course (A,C) and amplitudes (B,D) recorded in CA1 SO and SR of Cdh9+/+ and Cdh9−/− hippocampal slices. n = 14 Cdh9+/+ and 19 Cdh9−/− slices from SO and 10 Cdh9+/+ and 12 Cdh9−/− slices from SR, each from 6–7 animals aged P21–35. (E) Representative composite images of DiI labeled CA3 axons (red) projecting to area CA1 and Hoechst (blue). (F) Quantification of mean DiI staining intensity in SO relative to SR layer of Cdh9+/+ and Cdh9−/− hippocampal slices. n = 9–10 slices from 4–5 animals each. All animals aged P21–P35. Student’s t-test indicates no significant differences. (G–J) Same as A–D except experiments were done on adult Cdh9+/+ and Cdh9−/−mice. n = 13 Cdh9+/+ and 17 Cdh9−/− slices for SO and 9 Cdh9+/+ and 9 Cdh9−/− slices for SR, each from 4–5 animals aged 3–5 months. p-values were calculated using students t-test. p<0.05 is represented by *, otherwise p>0.05. All data shown as mean ± s.e.m.
Next, we tested if the LTP phenotype in the SO layer persists in adult animals by conducting LTP experiments in adult (3–5 months old) mice. Similar to young mice, SO LTP is significantly reduced in adult Cdh9−/− compared to Cdh9+/+ mice while SR LTP is unaffected (Figures 3G–J and S3G). As before, input-output curves are normal in adult Cdh9−/− mice (Figures S3H–I) in SO and SR suggesting that reduced SO LTP is not due to impaired basal synaptic transmission of CA3-CA1 synapses. Our results indicate cadherin-9 is specifically required for normal mushroom spine density and high magnitude LTP in the SO layer of CA1 basal dendrites.
Cadherin-6, 9, and 10 heterophilic interactions mediate trans-cellular adhesion
Cadherins typically function via homophilic interaction. However, at CA3-CA1 SO synapses, cadherin-9 is expressed by CA3 but not CA1 neurons (Figure 2A). We therefore reasoned presynaptic cadherin-9 may bind other cadherins expressed in CA1 neurons to carry out the synaptic functions described above. Heterophilic cadherin interactions have been observed in cultured cell lines (Katsamba et al., 2009; Shan et al., 2000; Shimoyama et al., 2000), but have not yet been shown to have functional relevance in the brain.
To begin to test if cadherin-9 functions trans-synaptically via other classic cadherins expressed in CA1, we first identified all classic cadherins expressed in principle neurons of the hippocampus using the Allen Brain Atlas (Lein et al., 2007) and previous reports (Bekirov et al., 2002). We next confirmed hippocampal expression patterns of the identified cadherins by in situ hybridization. Cadherins-2, 8, and 11 are broadly expressed in all principal hippocampal neurons (Figure 4A). In contrast, cadherin-24 is expressed primarily in CA3 neurons and cadherins-6 and 10 are specifically expressed in CA1 neurons (Figure 4A). We also examined the expression pattern of cadherin-10 by genetic labeling. Cadherin-10 knockout mice (Cdh10−/−) were generated by inserting CreERT2 in the first exon of the Cdh10 gene. These mice were crossed to the Credependent Ai3 YFP reporter line to generate heterozygous Cdh10-CreERT2+/−;Ai3+/− mice, which were injected with tamoxifen and immunostained for YFP. Our results indicate cadherin-10 expression is highly restricted to spiny, glutamatergic CA1 pyramidal neurons (Figures 4B and S4A). However, we did occasionally observe CA3 neurons expressing YFP (Figure S4A), suggesting low level expression of cadherin-10 in these cells.
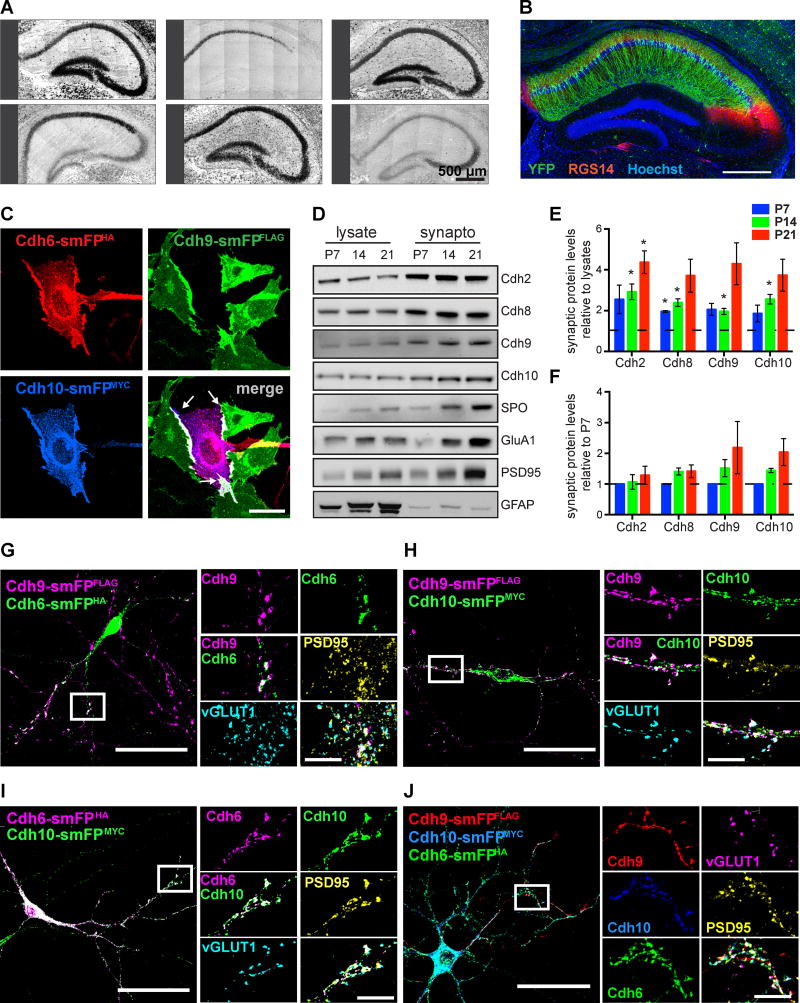
Figure 4. Cadherin-9 mediates trans-cellular adhesion via cadherins-6 and 10.
(A) In situ hybridizations of hippocampal cadherins. All images are tiled. (B) Immunostaining against YFP (green) and the CA2 marker RGS14 (red) in hippocampus from Cdh10-CreERT2+/−;Ai3+/− mice injected with tamoxifen. Hoechst (blue) labels all cell nuclei. (C) CHO cells expressing Cdh9-smFPFLAG (green) were mixed with cells co-expressing Cdh6-smFPHA (red) and Cdh10-smFPMYC (blue). Note that cadherins-6, 9, and 10 co-cluster at the interaction interfaces (white arrows in the merged image). (D) Immunoblots show cadherins-9 and 10 are enriched in hippocampal synaptosomes from P7, P14, and P21 mice. Samples were also probed for cadherins-2 and 8, the presynaptic marker synaptoporin (SPO), the postsynaptic markers PSD95 and GluA1, and a nonsynaptic marker GFAP. (E) Quantification of synaptic enrichment of each indicated cadherin relative to their levels in lysate at each time point. For each time point, a one sample t-test was used to determine if the mean enrichment value is significantly different than 1 (depicted as the dotted line), which would denote no enrichment. (F) Quantification of cadherin levels in synaptosomes over time. Each protein is normalized to its level in synaptosomes at P7. Statistical difference between means were calculated using one-way ANOVA (performed on each cadherin separately) followed by pairwise Holm-Šidák multiple comparison tests. For Figures E and F, 3 independent experiments were done for each age with hippocampi from 3–4 animals pooled per experiment. (G–J) Cultured neurons expressing Cdh9-smFPFLAG were plated with neurons expressing either Cdh6-smFPHA, Cdh10-smFPMYC, or both Cdh6-smFPHA and Cdh10-smFPMYC. Neurons were immunostained for epitope tags to label cadherins and vGLUT1 and PSD95 to label pre- and postsynaptic sites. Boxed regions are shown magnified at right and arrowheads indicate points of co-localization at synapses. All data shown as mean ± s.e.m. All fluorescent images are composite and each channel is indicated.
To determine if cadherin-9 can bind in trans to other cadherins expressed in the hippocampus, we conducted cell aggregation assays (Takeichi and Nakagawa, 2001). CHO cells, which express no endogenous cadherins (Figure S4B) (Ginsberg et al., 1991), were transfected with cadherins fused to GFP or mCherry and cell suspensions were mixed. If the two cadherins interact in trans, mixed red and green aggregates form (Figure S4C, middle). If the two cadherins do not interact heterophilically, separate red and green aggregates form because all cadherins undergo homophilic binding (Figure S4C, right). Due to poor expression in CHO cells cadherin-24 was omitted from the interaction screen. An aggregation index was calculated for each cadherin pair tested (see Supplemental Experimental Procedures). Consistent with a previous report (Shimoyama et al., 2000), we identified 4 heterophilic cadherin pairs; cadherins-6/9, 9/10, 10/6, and 8/11 (Figures S4C–D). As expected, all cadherins tested showed homophilic binding while cells expressing GFP and mCherry alone showed no binding (Figures S4C–D). All homophilic and heterophilic binding is calcium dependent as it was completely prevented in the presence of EDTA (Figure S4C). Though we could not test cadherin-24, it is evolutionarily more distant (Nollet et al., 2000) and therefore not predicted to interact with other hippocampal cadherins based on sequence analysis.
Cadherins-6, 9, and 10 accumulate at cell-cell junctions and synapses
Because cadherins-6, 9, and 10 interact with one another, we next investigated whether they are co-recruited to cell-cell junctions in CHO cells. Each cadherin was tagged with a different high performance spaghetti monster epitope tag (smFP) (Viswanathan et al., 2015). We then expressed Cdh9-smFPFLAG in one set of CHO cells to simulate CA3 neurons and plated them with a second set of CHO cells expressing either Cdh6-smFPHA, Cdh10-smFPMYC, or Cdh6-smFPHA and Cdh10-smFPMYC (Figure 4C) to simulate CA1 neurons. Immunostaining shows that cadherins localize at the interaction interfaces in all combinations tested (Figures 4C and S4E).
For cadherin-6, 9, 10 heterophilic interactions to be biologically relevant, the binding partners need to be expressed at the same place and time in hippocampal synapses. To test this, we purified hippocampal synaptosomes from P7, P14, and P21 mice and immunoblotted for cadherins and synaptic markers (Figure 4D). All cadherins tested were enriched in the synaptosome fraction relative to lysates by P14 (Figures 4E). Moreover, the levels of cadherins-9 and 10 in the synaptosome fraction tended to increase with age but the differences were not statistically significant (Figures 4F).
Next, we asked if epitope tagged cadherins-6, 9, and 10 co-localize at synaptic sites in hippocampal neurons in vitro. We co-cultured neurons expressing Cdh9-smFPFLAG with neurons expressing either Cdh6-smFPHA, Cdh10-smFPMYC, or both Cdh6-smFPHA and Cdh10-smFPMYC and immunostained for epitope tags and the synaptic markers vGLUT1 and PSD95. We observed co-localization of every pairwise combination of cadherins at synapses (Figures 4G–I). Moreover, five color labeling reveals that all three cadherins simultaneously co-localize at synaptic sites marked by juxtaposed vGLUT and PSD95 puncta (Figure 4J). To rule out the possibility that co-localization is an over-expression artifact, we repeated the experiment by mixing neurons expressing Cdh9-smFPFLAG with neurons expressing cadherins that do not bind cadherin-9, namely Cdh2-smFPHA and Cdh11-smFPMYC. In this case, we did not observe colocalization among the cadherins (Figure S4F). Together, our results indicate cadherins-6, 9 and 10 are selectively enriched at cell junctions and synapses in vitro.
Cadherins-6 and 10 are required for normal mushroom spine density and high magnitude synaptic potentiation in the CA1 SO layer
Our results suggest cadherins-6 and 10 are postsynaptic binding partners of cadherin-9 at CA3-CA1 synapses. If so, mice lacking these cadherins should have reduced mushroom spines and LTP in the SO layer similar to Cdh9−/− mice. We first examined the Cdh10−/− mouse line. We confirmed Cdh10−/− mice lack cadherin-10 protein in hippocampal lysates and synaptosomes while expressing normal levels of cadherin-9 (Figure S5A).
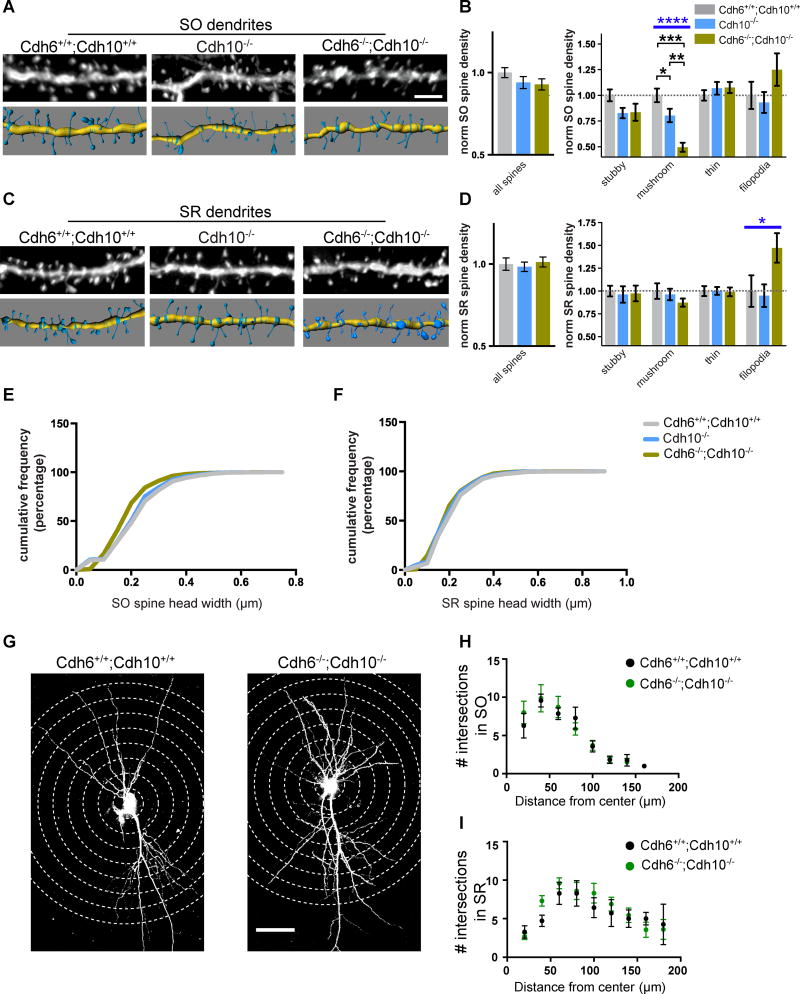
Spine analysis shows Cdh10−/− mice have a significantly reduced density of mushroom spines specifically in the SO layer compared to wildtype mice (Figures 5A–D and S5H–I), resembling the spine phenotype observed in Cdh9−/− mice (Figure 2). However, LTP in both the SO and SR layers of adult Cdh10−/− mice is unchanged compared to wildtype mice (Figures S5B–E). We reasoned that the lack of an LTP phenotype may be due to the redundant function of cadherin-6, which is still present in CA1 neurons of these mice. To test this, we analyzed double knockout mice that lack expression of both cadherins-6 and 10 (Figures S5F–G). Spine analysis indicates Cdh6−/−;Cdh10−/− double knockout mice have a significantly more severe spine defect than loss of cadherin-10. CA1 neurons lacking both cadherins-6 and 10 have a 50% reduction of mushroom spines in the SO layer compared to wildtype (Figures 5A–D and S5H–I). The more severe reduction in mushroom spines results in an overall reduction of spine head width in Cdh6−/−;Cdh10−/− double knockout SO spines (Figures 5E–F). Despite the spine defect, Sholl analysis indicates that the overall dendritic morphology of CA1 neurons in Cdh6−/−;Cdh10−/− mice is normal (Figures 5G–I).
Figure 5. Cadherins-6 and 10 regulate mushroom spine formation in CA1 SO.
(A,C) Representative images of SO (A) and SR (C) dendrites analyzed in wildtype (Cdh6+/+;Cdh10+/+), cadherin-10 knockout (Cdh10−/−), and cadherin-6/10 double knockout (Cdh6−/−;Cdh10−/−) mice (top), and their 3D models (bottom). (B,D) Quantification of average density of total spines (left) and indicated spine classes (right). All measurements are normalized to mean wildtype values. n = 16–21 cells from 3 mice aged P21–P23 for each layer and genotype. Statistical differences calculated using one-way ANOVA followed by pairwise Holm-Šidák multiple comparison tests. (E–F) Cumulative distribution of spine head width from SO (E) and SR (F) layers from Cdh6+/+;Cdh10+/+, Cdh10−/−, and Cdh6−/−;Cdh10−/− mice. n= >1000 spines for each. All spine analyses were evenly sampled from 3 mice aged P21–P23 and conducted blind to genotype. (G) Representative images of Lucifer yellow filled CA1 neurons from Cdh6+/+;Cdh10+/+ and Cdh6−/− ;Cdh10−/− mice for Sholl analysis. Concentric circles used to quantify dendritic branching are indicated by dotted lines. (H, I) Quantification of intersection points of SO (H) and SR (I) dendrites from Cdh6+/+;Cdh10+/+ and Cdh6−/−;Cdh10−/− mice. n= 7–10 neurons from 3 animals for each layer and genotype. p-values were calculated using Holm-Šidák multiple comparison test and no significant differences were found. Blue bars represent ANOVA p-values. Black bars represent post-test p values. p<0.05, p<0.01, p<0.001, and p<0.0001 is denoted by *, **, ***, and **** respectively, otherwise p>0.05. All data shown as mean ± s.e.m.
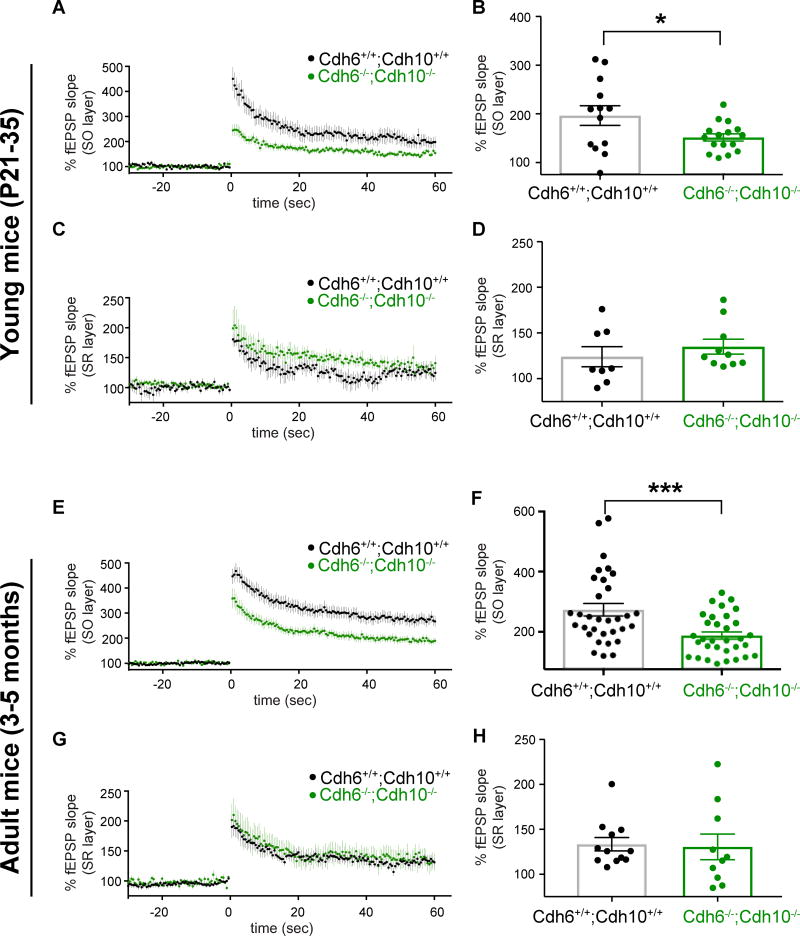
Next we tested if LTP is affected in Cdh6−/−;Cdh10−/− mice. Our results indicate Cdh6−/− ;Cdh10−/− mice have significantly reduced SO LTP in young and adult mice, while SR LTP remains unchanged compared to wildtype mice (Figure 6). Input/output curves suggest baseline synaptic transmission in SO and SR layer is normal in the Cdh6−/−;Cdh10−/− double knockout mice (Figures S6B–C and S6E–F). Similar to loss of cadherin-9, the loss of cadherins-6 and 10 reduces SO high magnitude LTP to relatively normal magnitude levels (Figures S6A and S6D). This suggests that, like cadherin-9, cadherins-6 and 10 are required for normal mushroom spine formation and high magnitude LTP.
Figure 6. Cadherins-6 and 10 are required for high magnitude LTP in CA1 SO.
(A–B) Mean LTP time course (A) and amplitudes (B) recorded in CA1 SO layer of Cdh6+/+;Cdh10+/+ and Cdh6−/−;Cdh10−/− hippocampal slices. (C–D) Same as figures 6A–B except data from SR layer is shown. n= 13 Cdh6+/+;Cdh10+/+ and 16 Cdh6−/−;Cdh10−/− slices for SO and 8 Cdh6+/+;Cdh10+/+ and 10 Cdh6−/−;Cdh10−/− slices for SR. Data collected from 4 mice aged P21–35. (E–H) Same as in figures 4A–D except recordings were performed in adult mice. n= 32 Cdh6+/+;Cdh10+/+ and 32 Cdh6−/−;Cdh10−/− slices for SO and 12 Cdh6+/+;Cdh10+/+ and 10 Cdh6−/−;Cdh10−/− slices for SR, each from 5–11 animals aged 3–5 months. p-values were calculated using students t-test. *=p<0.05 and ***=p<0.001, otherwise p>0.05. All data shown as mean ± s.e.m.
SR synapses undergo cadherin-dependent high magnitude LTP when inhibition is reduced
Why might cadherins-6, 9, and 10 affect synaptic potentiation specifically in CA1 SO but not SR? First, we tested if these cadherins specifically localize in the SO but not SR layer. We immunoblotted tissue from CA1 SO and SR layers and found cadherins-9 and 10 are expressed at similar levels in SO and SR (Figure S7A). We verified our dissection technique using myelin, which is high in SO and low in SR (Gil et al., 2010) (Figure S7A). It is still possible these cadherins and/or cadherin-6 are preferentially enriched at active synaptic sites in SO compared to SR but overall layer-specific localization of cadherins-9 and 10 does not explain their specific role in SO.
Second, because CA2 neurons project preferentially to CA1 SO compared to SR (Hitti and Siegelbaum, 2014), we considered the possibility that reduced SO LTP in cadherin knockout animals could reflect disruption of CA2-CA1 synapses instead of CA3-CA1 synapses. However, we find that neither cadherins-6, 9, nor 10 are expressed in CA2 neurons. Using a CA2 specific marker on sections from Cdh10-CreERT2+/−;Ai3+/− mice, we demonstrate cadherin-10 expression is limited to CA1 neurons and a few scattered CA3 neurons but no CA2 neurons (Figures 4B and S4A). Moreover, double in situ hybridization of cadherins-9/10 and cadherins-9/6 indicates there is consistently a gap in the signal for these probes in the CA2 region (Figure S7B–C). Thus, it is unlikely CA2-CA1 synapses are primarily affected in these knockout mice.
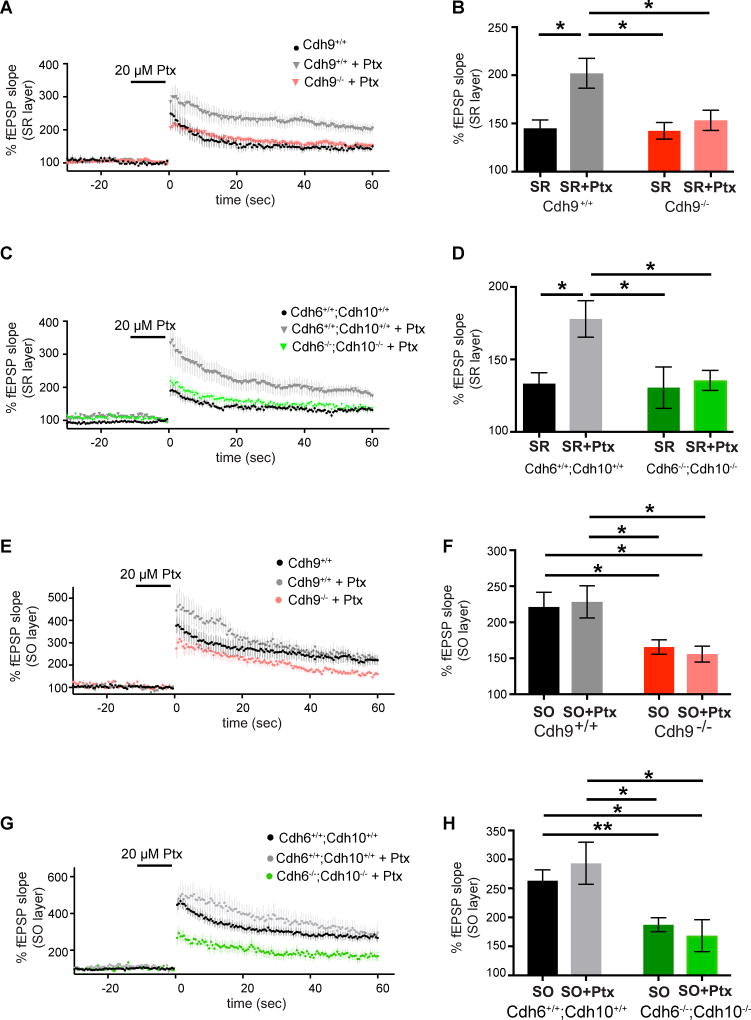
Next, we reasoned that cadherins-6, 9, and 10 may not function only in the SO layer but instead, may function specifically in high magnitude LTP, which happens to occur specifically in the SO layer under normal conditions. To test this, we determined if cadherins-6, 9, and 10 function in the SR layer when SR LTP is artificially forced to undergo high magnitude LTP. Blocking inhibition with 20 µM picrotoxin for 10 min before and during TBS increases SR LTP to high magnitude LTP levels in wildtype slices (Figures 7A–D). However, SR LTP in Cdh9−/− and Cdh6−/−;Cdh10−/− double knockout slices cannot reach high magnitude LTP levels even in the presence of picrotoxin (Figures 7A–D), indicating that cadherins-6, 9, and 10 are specifically required for high magnitude LTP in both the SO and SR layers.
Figure 7. Cadherins-6, 9, and 10 are required for picrotoxin-induced high magnitude LTP in SR.
(A–B) Time course (A) and mean LTP magnitude comparison (B) of SR LTP recorded from Cdh9 wildtype and knockout mice with and without 20 µM picrotoxin (Ptx). n=9–16 slices, each from 3–5 animals aged 3–5 months. (C–D) Time course (C) and mean LTP magnitude comparison (D) of SR LTP recorded from Cdh6/Cdh10 wildtype and double knockout mice with and without 20 µM picrotoxin (Ptx). n= 10–15 slices, each from 3–6 animals aged 3–5 months. (E–F) Time course (E) and mean LTP magnitude comparison (F) of SO LTP recorded from Cdh9 wildtype and knockout mice with and without 20 µM picrotoxin (Ptx). n=8–17 slices, each from 3–5 animals aged 3–5 months. (G–H) Time course (G) and mean LTP magnitude comparison (H) of SO LTP recorded from Cdh6/Cdh10 wildtype and double knockout mice with and without 20 µM picrotoxin (Ptx). n=8–32 slices, each from 3–11 animals aged 3–5 months. Statistical differences were measured using two-way ANOVA followed by pairwise p-value calculation using Holm-Šidák multiple comparison test. Two-way ANOVA p-values are reported in supplementary table 2. p<0.05, p<0.01, p<0.001, and p<0.0001 is denoted by *, **, ***, and **** respectively, otherwise p>0.05. All data shown as mean ± s.e.m. Note: untreated wildtype and knockout data without Ptx is the same data reported in previous figures and shown here again for comparison with Ptx treatment.
Picrotoxin likely induces high magnitude LTP in SR by reducing feed forward inhibition, allowing the same TBS stimulation to depolarize postsynaptic neurons to a greater extent. Regardless of mechanism, these results indicate that levels of local inhibition affect LTP magnitude. Therefore, we next considered two possibilities to explain how loss of cadherins-6, 9, and 10 decreases SO LTP magnitude. The cadherins could either act directly in postsynaptic CA1 neurons or indirectly by increasing inhibition in the SO layer. We reasoned if loss of these cadherins works indirectly by increasing inhibition in the SO layer, then picrotoxin should rescue the attenuation of SO LTP magnitude in Cdh9−/− and Cdh6−/−;Cdh10−/− double knockout mice. However, our results indicate recording in picrotoxin does not rescue LTP defects in the SO layer of either Cdh9−/− or Cdh6−/−;Cdh10−/− double knockout mice (Figures 7E–H). Taken together, our results indicate that LTP in the SO and SR layers of Cdh9−/− and Cdh6−/−;Cdh10−/− double knockout mice are limited to normal magnitude LTP and suggest that cadherins-6, 9, and 10 specifically regulate high magnitude LTP by acting directly in CA1 synapses.
Discussion
Deciphering the function of the brain at the cellular level requires identifying specific connections, understanding molecular mechanisms regulating those connections, and using that molecular and cellular knowledge to manipulate those specific connections to determine their function. Here we identified several unique structural and functional properties of different CA1 synapses. We demonstrate that CA1 spines in SO and SR have distinct synaptic potentiation properties despite consisting primarily of similar CA3 inputs. Subsequently, we identified three Type II cadherins, cadherins-6, 9, and 10, specifically required for mushroom spine formation and high magnitude LTP characteristic of CA1 basal dendrites in the hippocampal SO.
Identifying layer-specific synaptic properties of hippocampal CA1 neurons
We initially conducted a thorough characterization of the structure of excitatory synapses in three CA1 synaptic layers using electron microscopy and 3D light microscopy. Our results from these layer-specific analyses of wildtype mice support two main conclusions. First, EC-CA1 synapses in the SLM layer have significantly different pre- and postsynaptic structures than CA3-CA1 synapses in SR or SO. Specifically, SLM excitatory synapses tend to be larger and less dense than SR or SO synapses. The fact that SLM synapses differ from SR and SO synapses is not entirely surprising given that SLM synapses are located on the thinnest, most distal dendrites and receive inputs from entorhinal cortex. Second, and more surprising because they primarily originate from the same class of input neuron, we identified significant differences between synapses in SR and SO. Our results indicate the SO has a significantly higher density of mushroom spines than SR and the magnitude of LTP is significantly higher in SO versus SR.
What is high magnitude LTP?
LTP is well established as the key molecular mechanism underlying learning and memory but few studies have considered how different modes of LTP acting on one neuron or cell type may contribute to different aspects of learning. High magnitude LTP has been observed in the CA1 SO of the intact hippocampus in vivo (Kaibara and Leung, 1993) but its function remains unknown. A deeper understanding of the role of high magnitude LTP in hippocampal function requires the ability to specifically manipulate high versus normal magnitude LTP. Here, we show cadherins-6, 9, and 10 are specifically required for high but not normal magnitude LTP. Therefore, the knockout mice analyzed here and other tools targeting cadherins-6, 9, and 10 should provide important access to directly test the function of high magnitude CA1 SO LTP in hippocampal-dependent behaviors and circuit activity.
At the mechanistic level, SO LTP magnitude is significantly higher than SR at all time points tested following TBS from 0.5 min to 60 min and beyond (Arai et al., 1994; Fan, 2013). This suggests that both the induction and maintenance of SO LTP are significantly different from SR LTP. Therefore, it is possible that the cellular mechanisms underlying high magnitude LTP have both a pre and postsynaptic component. However, results from our lab and others suggest high magnitude LTP is primarily mediated via a postsynaptic mechanism. High magnitude LTP in the SO is NMDA receptor dependent (Arai et al., 1994; Cavus and Teyler, 1998) and no changes in paired pulse ratio, a classic measure of presynaptic release probability, are observed between SR and SO before or after TBS (Arai et al., 1994). These results suggest mechanisms underlying high magnitude LTP overlap with mechanisms mediating normal magnitude NMDA receptor-dependent LTP (Herring and Nicoll, 2016). However, although some molecular components between normal and high magnitude LTP are likely shared, our new results indicate there are clear distinctions as loss of cadherins-6, 9, and 10 specifically prevents high magnitude LTP but not normal magnitude LTP.
Cadherins-6, 9, and 10 are specifically required for high magnitude LTP
Our initial observation that cadherins-6, 9, and 10 function in high magnitude SO LTP could be explained by two possibilities. One is that cadherins-6, 9, and 10 selectively function at SO but not SR synapses, possibly via differential localization. The other is that they function selectively in high magnitude LTP, which happens to only occur normally in the SO. To distinguish these possibilities, we developed a protocol to generate high-magnitude LTP in SR synapses by recording from picrotoxin-treated slices. We then demonstrated that this effect requires cadherins-6, 9, and 10 because it is abolished in cadherin-9 knockout and cadherin-6/10 double knockout mice. Additionally, immunoblotting indicates cadherins-9 and 10 are found in similar amounts in SO and SR. Taken together, these results strongly suggest cadherins-6, 9, and 10 are present in both layers but are only required during high magnitude LTP, which is normally only observed in SO.
Our analyses of germ line knockout mice clearly indicate cadherins-6, 9, and 10 are required for high magnitude synaptic plasticity at SO synapses. Because we used germ line knockout mice, we cannot distinguish between the possibilities that these cadherins regulate LTP by functioning directly in CA1 neurons or indirectly via other network defects. However, we provide several pieces of experimental evidence against an indirect role on the network. First, picrotoxin is unable to rescue the reduction in LTP magnitude in the SO layer of cadherin-9 knockout and cadherin-6/10 double knockout mice suggesting that cadherin loss does not reduce high magnitude LTP by increasing inhibition. Second, cadherin-9 knockout and cadherin-6/10 double knockout mice have a similar reduction in SO LTP magnitude despite the fact that the three cadherins are expressed by different populations of neurons throughout the brain (Bekirov et al., 2002; Lein et al., 2007). Moreover, in situs and genetic reporters indicate these cadherins-6 and 10 are expressed in CA1 principal neurons but not inhibitory neurons. Third, in all cases, SR spine morphology, normal magnitude LTP, paired-pulse ratio, mEPSCs, and input-output curves are unaffected, providing internal controls for largely normal hippocampal network form and function. Fourth, LTP defects are similar in young P21-P35 and adult 3–5 months old mice in all mouse lines, suggesting no change as hippocampal function matures. Together, these results favor the model that the critical function of pre-synaptic cadherin-9 and postsynaptic cadherins-6 and 10 is to regulate high magnitude LTP in CA1 postsynaptic neurons but, importantly, the cell autonomous nature of cadherin-6 and 10 function remains to be directly tested.
The mechanism by which cadherins-6, 9, and 10 regulate high magnitude LTP is still unknown. One model posits that cadherin-9 is present throughout CA3 axons and cadherins-6 and 10 are present throughout CA1 dendrites. Then, if the local neural activity reaches a critical threshold, presynaptic cadherin-9 and postsynaptic cadherins-6 and 10 (or a critical interacting molecule) may be locally recruited to active synapses where they may form a trans-synaptic complex that stabilizes mushroom spines and triggers intracellular events that uniquely contribute to high magnitude LTP. Cadherins are generally known to mediate cellular processes active during LTP including actin reorganization (Herring and Nicoll, 2016) and AMPA receptor recruitment (Patterson et al., 2010; Saglietti et al., 2007b). Though an increase in spine head size often accompanies LTP (Bosch and Hayashi, 2012), the results presented here cannot determine whether cadherins-6, 9, and 10 regulate mushroom spine density and high magnitude LTP via a singular mechanism. It is possible these cadherins use multiple pathways to mediate different cellular events. Future studies are needed to test these open questions by analyzing the precise synaptic localization of each cadherin before and after LTP induction and determining specific intracellular binding partners.
Heterophilic interactions of Type II cadherins
Thus far, most biological functions of cadherins are attributed to trans-cellular homophilic interactions. However, some cadherins engage in heterophilic interactions in cultured cell lines (Shan et al., 2000; Shimoyama et al., 2000). It was suggested that cadherins may use heterophilic interactions in vivo (Duan et al., 2014), but this idea had not yet been investigated directly. Our new results provide strong evidence that trans-cellular heterophilic interactions between pre-synaptic cadherin-9 and postsynaptic cadherins-6 and 10 regulate high magnitude synaptic potentiation in CA3-CA1 synapses.
Cadherins interact laterally in cis as well as in trans (Wu et al., 2010). Similar to trans interactions, most attention has been paid to homophilic cis interactions (Harrison et al., 2011). However, heterophilic cis interactions may be particularly important in the nervous system where most neurons express multiple cadherins. Here, we show that cadherins-6 and 10 expressed in CA1 neurons co-localize at synapses when exogenously co-expressed in cultured neurons. Due to a lack of suitable reagents, it will be difficult to determine if cadherins-6, 9, and 10 directly interact in vivo. However, our results support a model whereby cadherins-6, 9, and 10 use heterophilic trans (between cadherins-9 and 6 and cadherins-9 and 10) and cis interactions (between cadherins-6 and 10) to form dimeric and trimeric complexes regulating mushroom spine formation and high magnitude LTP.
Cadherins-6, 9, and 10 are highly similar genes that likely arose through gene duplication (Nollet et al., 2000). The three genes are linked on the same chromosome and have 85% similarity and 72% identity to one another. Given this, it is not surprising these cadherins interact with one another and they are likely to act as redundant molecules when expressed in the same neuron. If so, we predicted cadherin-9 knockout mice would closely phenocopy cadherin-6/10 double knockout mice, while loss of cadherin-10 alone should have no or little effect because cadherin-6 is present to preserve function. In support, our data suggest cadherins-6 and 10 act redundantly in CA1 neurons during high magnitude LTP. Interestingly, the role of these cadherins in spine morphology is more complex. Here, cadherin-9 and 10 single knockout mice have similar reductions in mushroom spine density compared to wildtype but the cadherin-6/10 double knockout mice are more severe. This suggests that there is a low level of cadherins-6 and/or 10 expressed in presynaptic CA3 neurons that can substitute for cadherin-9 or cadherins-6 and 10 have a postsynaptic function that does not fully depend on trans-synaptic binding with cadherin-9. These alternatives are not mutually exclusive but, in support of the first possibility, we occasionally observe YFP labeled CA3 neurons in the cadherin-10-reporter mice. While the reduction of mushroom spines and decrease in LTP magnitude are both regulated by cadherins-6, 9, and 10, they may utilize distinct mechanisms. Because CA1 neurons also express cadherins-2, 8, and 11, it will be interesting to determine in future studies how adhesion, spine shape, and LTP is affected by even more complex combinations of synaptic cadherins.
The role of cadherin diversity in the brain
Understanding the true function of the classic cadherins in the brain has been challenging. Most of the 18 classic cadherins are expressed in the brain and thus, as we demonstrate here, many likely have overlapping functions that mask defects in single gene gain and loss of function experiments. Most classic cadherins are persistently expressed through brain development and maturity. Thus, they may take on new functions as the animal develops. Consistent with this, cadherins function in diverse processes including neural tube formation (Hirano and Takeichi, 2012), axon targeting (Duan et al., 2014; Kuwako et al., 2014; Osterhout et al., 2011; Poskanzer et al., 2003), synapse formation (Togashi et al., 2002; Williams et al., 2011), synapse pruning (Bian et al., 2015), and synapse function (Bozdagi et al., 2010; Fièvre et al., 2016; Jungling et al., 2006; Mendez et al., 2010; Tang et al., 1998; Vitureira et al., 2011). Moreover, there is an overwhelming focus on the study and function of the broadly expressed N-cadherin/cadherin-2 and β-catenin, an intracellular binding partner common to all classic cadherins. This has led many to assume that all cadherins function in the same manner with little attention on the differences between cadherin family members.
By analyzing input-specific excitatory synapses in different layers of the CA1 dendritic tree, we discovered that cadherins-6, 9, and 10 function uniquely in high magnitude but not normal magnitude LTP. In contrast, work from others has shown that blocking cadherin-2 function or deleting the gene results in impaired normal magnitude LTP in CA1 SR synapses (Bozdagi et al., 2010; Tang et al., 1998). Moreover, loss of cadherin-2 does not alter the initial rise of synaptic strength following LTP stimulation but it is required for the sustained persistence of LTP after spines enlarge (Bozdagi et al., 2010; Tang et al., 1998). In contrast, cadherins-6, 9, and 10 are required for the high magnitude potentiation observed in CA1 SO layer starting 30 seconds post TBS. This highlights how, even in the same neurons, different cadherins mediate distinct functions. Further, it was shown that cadherin-8 but not cadherin-2 levels are reduced following LTP induction in medial perforant path-DG synapses (Huntley et al., 2010) and loss of cadherin-11 causes increased CA1 SR LTP (Manabe et al., 2000). Thus, it is becoming clear that different cadherins have complex regulatory roles on synaptic potentiation and the relative levels of diverse cadherins may govern synapse dynamics.
In summary, our study provides new mechanistic insight into the little-studied phenomenon of high magnitude LTP. We identified three heterophilic Type II cadherins required specifically for high magnitude but not normal magnitude LTP in the hippocampus. This study lays a critical framework for understanding the role of high magnitude LTP in hippocampal-dependent learning and memory behaviors and other circuits across the brain.
STAR METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Megan E. Williams (megan.williams@neuro.utah.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
All animals and experiments were maintained and conducted in accordance with the NIH guidelines on the care and use of animals and approved by the University of Utah and Harvard University IACUC committees. All mouse lines (except CD1) were maintained on the c57/Bl6 background and group housed in a dedicated animal facility with a standard 12-hour light/dark cycle. Mice were fed ad libitum and their health status routinely monitored. For all experiments, naïve (no previous procedures and tests) and virgin male and female mice were used. For all wildtype versus knockout animal experiments an approximately equal proportion of male and female mice were used for both genotypes. Wildtype animals and their knockout counterparts were not always littermates but were age matched. Occasionally, the same wildtype data is presented in multiple figures and this is also noted in the figure legends. The ages of animals used in this study vary and are noted in the figure legends.
The cadherin-9 knockout mouse line was generated by inserting a lacZ-containing cassette in exon 5 of the cadherin-9 gene and described previously (Duan et al., 2014). Generation of the cadherin-10 single knockout and cadherin-6/10 double knockout mice will be detailed in an upcoming manuscript (Duan, Sanes et al, submitted). Briefly, the cadherin-10 targeting vector was obtained via lambda phage-mediated recombineering and 6XMyc-CreERT was inserted into the first coding exon (exon 2) of the cadherin-10 gene by homologous recombination in mouse B6/129J embryonic stem (ES) cells. Multiple chimeric mice with the targeted embryonic stem cells were generated and two lines with germ line transmissions were mated to produce stable knockout lines. Knockout of the cadherin-10 protein was confirmed biochemically and immunohistochemically. The cadherin-6 and 10 genes are closely linked on mouse chromosome 15. Therefore, to obtain cadherin-6/10 double knockout mice, first cadherin-6/10 trans-heterozygotes were made by breeding cadherin-6 (Kay et al., 2011) and cadherin-10 single knockouts. Subsequently the trans-heterozygotes were mated with wildtype mice to obtain cadherin-6/10 cis-heterozygotes. One cis-heterozygote was obtained out of 323 pups. This particular heterozygote was mated back to C57/BL/6J mice over multiple generations and progenies were mated to obtain homozygous cadherin-6/10 double knockout mice.
Cdh10-CreERT2+/−;Ai3+/− mice were generated by breeding homozygous cadherin-10 single knockouts with homozygous Ai3−/− mice purchased from the Jackson Laboratory (full name: Gt(ROSA)26Sortm3(CAG-EYFP)Hze; Stock# 007903). C57/BL/6J mice were purchased from the Jackson Laboratory (Stock#000664, RRID: IMSR_JAX:000664). CD1 mice were purchased from Charles River (Strain#022). Sprague Dawley rats were purchased from Charles River (Strain#400, RRID: RGD_734476).
Cell lines and primary cultures
Cultured cell lines used in this paper are: CHO-K1 cells (ATCC, Cat# CCL-61, RRID: CVCL_0214, female), HEK293 cells (ATCC, Cat# CRL-3216, RRID: CVCL_0063, female), and L929 cells (ATCC, Cat#CCL-1, RRID: CVCL_0462, male) were utilized. These cell lines were not authenticated. Cultured cells were kept in a humidified incubator maintained at 37°C and 5% CO2. Primary neuron cultures were prepared from P0 CD1 mouse (Strain#022, RRID: IMSR_CRL:22) or Sprague Dawley rat (Strain#400, RRID: RGD_734476) pups of both sexes and maintained in a humidified incubator at 37°C and 5% CO2.
METHOD DETAILS
Plasmids
A codon optimized version of mouse cadherin-9 cDNA was synthesized (Genscript) and all other mouse cadherin cDNAs were obtained from Open Biosystems (GE Healthcare) (Williams et al., 2011). All cadherins were subsequently cloned using DH5α bacteria (Invitrogen, Cat#18265-017) into the mammalian expression vector pCAG using standard restriction digestion and ligation procedures. The pCAG vector backbone was obtained from GFP pCAG (addgene Plasmid# 11150, RRID:SCR_002037). Spaghetti monster fluorescent proteins (smFPs) (Viswanathan et al., 2015), GFP, and mCherry tags were PCR cloned from pCAG constructs and inserted in frame at the C-terminus of all cadherin constructs to generate fusion proteins. mCherry pCAG was obtained from addgene (plasmid# 41583, RRID:SCR_002037).
In situ hybridization
Antisense mRNA probes were in vitro transcribed and DIG labeled from linearized full length cadherin cDNAs obtained from OpenBiosystems. In vitro transcription was conducted using SP6 or T7 enzymes and DIG RNA labeling mix (Roche, Cat#11277073910). Full length probes were subject to alkyline hydrolosis in 33mM NaHCO3 and 50mM Na2CO3 at 60°C to generate smaller probes to facilitate tissue penetration. 20 µm thick coronal cryosections of mouse brain tissue were post-fixed in 4% PFA for 20 min, permeabilized in 1 µg/mL RNase-free Proteinase K for 20 min, acetylated in 0.25% acetic anhydride in triethanolamine for 10 min, and incubated in hybridization buffer (50% formamide, 750 mM NaCl, 75 mM Na Citrate, 5xDenhardt’s solution (Invitrogen), 0.25mg/mL yeast tRNA, and 0.5mg/mL salmon sperm DNA) without probe at room temperature for 2–6 hours. Sections were then incubated with 200–800ng/mL DIG-labeled probe in hybridization buffer at 65°C overnight. Sections were washed and immunolabeled with alkaline phosphatase conjugated anti-DIG antibody (Roche, Cat#11093274910, RRID: AB_514497), and detected using NBT/BCIP stock solution (Roche, Cat#11383213001 and Cat#11383221001). All solutions prior to and including RNA probe hybridization steps were done with DEPC-treated water.
DiI labelling
Mice were perfused with 4% PFA and the brains were post-fixed in PFA for 30 minutes. Subsequently 350 µm thick sagittal sections of the hippocampus were obtained and a microscopic DiI (Invitrogen, D282) crystal was placed on the basal side of CA3a region using an insect pin. Slices were immersed in PBS and incubated for 48 hours in 37°C. Next, slices were placed under coverslips and imaged within one day. For analysis, DiI intensity was measured in the entire CA1 SO and SR layer (per slice) followed by mean intensity per pixel calculation for each layer. Subsequently the ratio of mean pixel intensity of SO/SR was calculated per slice.
Synaptosome preparation
Synaptosomes were purified as described previously (Jones and Matus, 1974). Briefly, hippocampi were dissected from mice aged P7, P14, or P21. Tissue was homogenized with a Dounce homogenizer in ice-cold 0.32 M sucrose + 20 mM HEPES, pH 7.4 (20% w/v) supplemented with protease inhibitors (1mM PMSF, 1ug/ml Leupeptin, 1ug/ml Aprotinin) and phosphatase inhibitors (1mM NaVO3 and 1mM NaF). Homogenates were cleared by spinning at 1000 × g for 10 minutes at 4°C. The supernatant was then spun at 17000 × g for 15 minutes. The pellet containing crude synaptosomes was resuspended in 0.32 M sucrose + 20 mM HEPES and layered at the top of a sucrose gradient (made of 4 mls of 1.2 M, 4 mls of 1 M, and 3 mls of 0.8 M sucrose in 20 mM HEPES) in a Beckman Coulter #331372 centrifuge tube and centrifuged at 82000 × g for 2 hours in a Beckman Coulter SW 41 Ti rotor. Purified synaptosomes were collected at the interface between 1.2 M and 1 M sucrose. 20 µg protein was loaded per lane for detection of cadherins while 5 µg of protein per lane was loaded for all other proteins.
Cell aggregation assay
CHO cells were transfected with cadherins fused to GFP or mCherry. Transfections were performed using a transfection mix comprising 2–5 µg DNA, 5 µg of Polyethylenimine (PEI) per µg of DNA, and 500µl of OPTIMEM (Gibco). 48 hours post transfection cells were washed with HEPES based calcium and magnesium free buffer (HCMF, 137 mM NaCl, 5.4 mM KCl, 0.34 mM Na2HPO4, 10mM HEPES, 5.55 mM Glucose) and dissociated with 0.01% Trypsin (Gibco) in HCMF+1 mM CaCl2. Cells were subsequently spun down and resuspended in HCMF and kept on ice. 50,000 cells expressing a GFP tagged cadherin were mixed with 50,000 cells expressing a mCherry tagged cadherin. The cell mixture was supplemented to obtain final concentrations of 4 mM CaCl2, 20 µg/ml DNAse I, and 1 mM MgCl2 and brought to a final volume of 500 µl. Cells were then shaken in a nutating shaker for 90 minutes and subsequently fixed with addition of 500 µl of 8% PFA in PBS pH 7.4 supplemented with 1:10000 dilution of Hoechst to label all cell nuclei and kept at 4°C. 12 hours later cells were transferred to 96 well glass bottom dishes and imaged using a Zeiss LSM 710 confocal microscope with a 10X magnification lens. To calculate the aggregation index (AI), the entire well was imaged and cellular clusters bigger than 900µm2 (based on clusters identified in the HOECST channel) were defined as aggregates. For every aggregate the net GFP (g) and mCherry (m) fluorescence signal was quantified. These values were normalized to the total GFP (G) and mCherry (M) signal in the well to obtain Gn and Mn respectively (i.e. Gn=g/G, Mn=m/M). Next, a heterophilic score (S) for an aggregate was calculated using the formula S = (Gn+Mn)*sine(π*Gn/(Gn+Mn)). This function quantified the ‘heterophilicity’ of an aggregate. Subsequently, the AI for the entire well was calculated as AI = ΣSi /(G+M), where Si is the heterophilic score of the ith aggregate. Image analysis was done using Fiji (NIH).
Immunoblotting and immunostaining
For immunoblotting, protein concentrations from synaptosome preparations were quantified with a Pierce BCA Protein Assay Kit (Thermo Scientific, Cat# 23225). 5µg (or 20µg) of synaptosome or cleared lysate proteins was loaded per lane for Western blot analysis. Proteins were run on Bis-Tris gradient acrylamide gels and transferred to nitrocellulose membranes. Membranes were incubated in blocking solution (50 mM Tris pH7.5, 300 mM NaCl, 3% w/v dry milk powder, and 0.05% tween-20) for 10 minutes, incubated in primary antibody overnight at 4°C, washed, incubated in HRP-conjugated secondary antibodies for 1 hour at room temperature and detected using the BioRad Clarity ECL kit (Cat# 1705061) on a BioRad ChemiDoc XRS+ imaging system. Hippocampal lysates were prepared by homogenizing 100 mg of hippocampal tissue in 1 ml of reducing sample buffer.
For immunostaining, cultured cells were fixed in 4% PFA for 10 minutes, washed with PBS, and incubated in blocking solution (PBS with 3% bovine serum albumin and 0.1% Triton-X 100) for 30 minutes. Cells were incubated in primary antibodies (diluted in blocking solution) for 1–2 hours. After 3 washes, secondary antibody was added for 45 minutes, washed, and cells were mounted for imaging using Fluoromount-G (Southern Biotech, Cat# 0100-01). For tissue sections, mice were transcardially perfused with 4% PFA in PBS. Brains were post-fixed in PFA overnight and 100 µm vibratome sections were cut. Sections were incubated in blocking solution (PBS, 3% BSA, 0.3% Triton-X 100) for 2 hours and incubated in primary antibody (diluted in PBS, 3% BSA, 0.2% Triton-X 100) at 4°C overnight. Secondary antibody (diluted in PBS, 3% BSA, 0.2% Triton-X 100) incubation was performed at room temperature for 1 hour. Sections were mounted in Fluoromount-G for imaging.
The following primary antibodies (along with their concentrations) were used: mouse anti-FLAG M2 1:3000 (Sigma, Cat# F1804, RRID:AB_262044), rabbit anti-Myc 1:1000 (Sigma, Cat# C3956, RRID:AB_439680), rat anti-HA 1:1000 (Roche, Cat# 11867423001, RRID:AB_10094468), goat anti-GFP 1:3000 (Abcam, Cat# ab6673, RRID:AB_305643), chicken anti-MAP2 1:5000 (Abcam, Cat# ab5392, RRID:AB_2138153), rabbit anti-GABA 1:1500 (Sigma, Cat# A2052, RRID:AB_477652), rabbit anti-synaptoporin 1:1000 (Synaptic Systems, Cat# 102002, RRID:AB_887841), mouse anti-PSD95 1:1000 (Neuromab, Cat# 75-348, RRID:AB_2315909), mouse anti-GFAP 1:1000 (EMD Millipore, Cat# MAB360, RRID:AB_11212597), rabbit anti-GluA1 1:1000 (Millipore, Cat# AB1504, RRID:AB_2113602), guinea pig anti-vGLUT1 1:2000 (Millipore, Cat# AB5905, RRID:AB_2301751), mouse anti-GAPDH 1:3000 (Millipore, Cat# AB2302, RRID:AB_11211911), rabbit anti-Myelin basic protein 1:1000 (Abcam, Cat# 40390, RRID:AB_1141521), rabbit anti-cadherin-9 1:500 (gift from Dr. Gerd Klein, University of Tuebingen), mouse anti-cadherin-2 1:1000 (BD Biosciences, Cat# 610920), rabbit anti pancadherin antibody 1:200 (Sigma, Cat# C3678, RRID:AB_258851), mouse anti-RGS14 1:500 (NeuroMab, Cat# 75-372, RRID: AB_2179931) and mouse-anti cadherin-8 1:50 (Developmental studies hybridoma bank, Cat# CAD8-1, RRID:AB_2078272). The rabbit anti-cadherin-10 was used at 1:500 and was generated for this study. A peptide corresponding to part of the intracellular domain of cadherin-10 (QNTIHLRVLESSPV) was synthesized (Selleckchem.com) and used for inoculation (Cocalico Biologicals). Anti-cadherin-10 containing sera was affinity purified and specificity was determined by Western blot analysis of brain lysates from wildtype and cadherin-10 knockout animals (this paper).
Fluorescent dye conjugated secondary antibodies were used for immunostaining at a concentration of 1:1000. HRP conjugated secondary antibodies were used for immunoblotting at a concentration 1:3000. The following secondary antibodies were used from Jackson ImmunoResearch: donkey anti-guinea pig-DyLight 405 (Cat# 706-475-148, RRID:AB_2340470), donkey anti-goat-Alexa 488 (Cat# 705-545-147, RRID:AB_2336933), donkey anti-rat-Cy3 (Cat# 712-165-153, RRID:AB_2340667), donkey anti-mouse-Cy3 (Cat# 715-165-150, RRID:AB_2340813), donkey anti-rabbit-Alexa 647 (Cat#711-605-152, RRID:AB_2492288), donkey anti-mouse-Alexa 647 (Cat# 715-650-150), donkey anti-chicken-DyLight 405 (Cat# 703-475-155, RRID:AB_2340373), goat anti-rabbit-HRP (Cat# 111-036-003, RRID:AB_2337942), goat anti-mouse-HRP (Cat#115-035-003, RRID:AB_10015289). The following secondary antibodies were used from Invitrogen: donkey anti-mouse-Alexa 488 (Cat# A21202, RRID:AB_141607), donkey anti-rabbit-Alexa 594 (Cat# A21207, RRID:AB_141637), donkey anti-rabbit-Alexa 488 (Cat# A21206, RRID:AB_141708). Other: goat anti-guinea pig-HRP (Santa Cruz Biotechnology, Cat# SC2438, RRID:AB_650492). Imaging was done using a Zeiss LSM710 confocal microscope.
Cell culture
Cell culture was carried out as described previously (Martin et al., 2015). For neuron cultures, P2 rat cortical glia were cultured on coverslips coated with 0.032 mg/mL Purecol (Advanced BioMatrix, 5005-100ML) to form a monolayer. One week later, hippocampi from P0 CD1 mouse pups were dissected in cold HEPES-buffered saline solution, incubated in papain for 30 minutes, dissociated, and plated to rat astroglial monolayers at 1×105 cells/ml. Glia media: DMEM (Gibco), 10% FBS (Gibco), 75 mM glucose, and 100 U/ml Penicillin, 100 g/ml Streptomycin (Mixture of Penicillin and Streptomycin purchased from Gibco). Neuron plating media: MEM (Gibco), 10% horse serum (Gibco), 50 mM glucose, 0.250 mM sodium pyruvate, 2 mM Glutamax (Gibco), 100 U/ml Penicillin, 100 g/ml Streptomycin. Neuron feeding media: Neurobasal A (Gibco), B27 (Invitrogen), 30 mM glucose, 0.5 mM Glutamax, 20 U/ml Penicillin, 20 g/ml Streptomycin. Cells were kept in an oxygenated incubator maintained at 37°C. Neurons were transfected by electroporation using a ECM830 model (BTX, Harvard Apparatus, item# 45-0002). Neurons were extracted from animals both sexes.
For CHO cell cultures, cells were maintained in CHO media: F12K media (Corning Inc.), 10% FBS, 100 U/ml Penicillin, and 100 g/ml Streptomycin. For HEK293 and L-929 cell cultures, cells were maintained DMEM, 10%FBS, 100 U/ml Penicillin, and 100 g/ml Streptomycin.
Whole cell recordings
Mice were rapidly decapitated and their brains carefully removed and kept in iced, artificial cerebrospinal fluid (aCSF) with sucrose (200 mM sucrose, 3 mM KCl, 1.4 mM Na2PO4, 3mM MgSO4, 26 mM NaHCO3, 10 mM glucose and 0.5 mM CaCl2). 300 µm thick transverse slices were cut on a Leica vibratome (Leica VT1200) and left at room temperature in the holding chamber until recording. Cells were visualized by oblique illumination using a bright light source (Olympus BX51WI microscope, Hitachi color CCD camera KP-D20BU). Slices kept in the patching chamber were continuously superfused with aCSF containing 126 mM NaCl, 26 mM NaHCO3, 3 mM KCl, 1.4 mM NaH2PO4, 2 mM CaCl2, 1.5 mM MgSO4, and 10 mM D-glucose, bubbled with 95% O2–5% CO2. The intracellular pipette solution contained 80 mM Cesium methylsulfonate, 60 mM CsCl, 10 mM HEPES, 1mM EGTA (adjusted with CsOH), 0.5 mM CaCl2, 10 mM Glucose and 5 mM QX-314, adjusted to 290–300 mOsm/Lt at pH 7.3. For mEPSC experiments Tetrodotoxin (Ttx) (Tocris Biosciences, Cat# 1078) was used at working concentration of 0.5 µM. Somatic whole cell recordings were performed with Axon Multiclamp 700B amplifies (Molecular Devices) in voltage clamp mode at 34 ± 1°C bath temperature for mEPSC experiments and room temperature for paired pulse ratio experiments. Data acquisition was performed via an Axon Digidata 1550 (Molecular Devices), connected to a Windows 7 computer, running pClamp (Version 10, Molecular Devices). Current signals were sampled at 1 kHz and filtered with a 2 kHz Bessel filter. Patch pipettes with a tip resistance of 6 – 8 MΩ were pulled with a Flaming/Brown micropipette puller P-97 (Sutter Instruments and Co.) using borosilicate glass capillaries with filaments (1B150F-4, World Precision Instruments). All cells patched were held in voltage clamp at -70mV. Acquired traces were analyzed with pClamp (Version 10, Molecular Devices). mEPSC event detection was performed with a template match search.
Field recordings
Mice were anesthetized with sodium pentobarbital (60 mg/kg, i.p.), and brains rapidly removed and placed in ice-cold (4°C) oxygenated sucrose based aCSF solution (bubbled with 95% O2/5% CO2) containing 200 mM Sucrose, 3 mM KCl, 1.4 mM Na2HPO4, 3 mM MgSO4, 26 mM NaHCO3, 10 mM glucose, and 0.5 mM CaCl2. Subsequently the brain was sectioned horizontally into 350 µm thick sections. Slices were incubated for 2 hours in a chamber containing oxygenated aCSF containing 126 mM NaCl, 3 mM KCl, 1.4 mM Na2HPO4, 1 mM MgSO4, 26 mM NaHCO3, 10 mM glucose, and 2.5 mM CaCl2. The pH (7.30–7.40) and osmolarity (290–300) mOsm of the ACSF were verified prior to each experiment. Extracellular field excitatory postsynaptic potentials (fEPSPs) were recorded using a Slicemaster highthroughput brain slice recording system (Scientifica). Slices were continuously perfused with oxygenated aCSF (2.5 ml/minute). Recordings were performed at 30–31°C. Concentric bipolar stimulating electrodes (MCE-100; Rhodes Medical Instrument) were placed in either the stratum oriens or stratum radiatum of the CA2-CA1 junction region. Recording microelectrodes (2–3 MΩ resistance) were filled with aCSF and placed within 250–500 µm of the stimulating electrodes. Data were acquired using pClamp 10 interfaced to a Digidata 1440A data acquisition board at a sampling rate of 10 kHz, low-pass filtered at 1 kHz, and high-pass filtered at 3 Hz. 100 µs stimuli ranging from 1 to 40 V were used to evoke fEPSPs. Input-output curves were generated and the stimulation strength was set so that the fEPSP amplitude was half that of the smallest fEPSP accompanied by a population spike. Slices were then stimulated every 30 seconds for a 30 minute baseline period. LTP was induced using theta burst stimulation (TBS, five trains of four pulses at 100 Hz separated by 200 msec and repeated once with a 20 second interval). Low frequency stimulation was resumed for 60 minutes post TBS at which point LTP was quantified relative to baseline. fEPSP slope was calculated from 25%–85% of the rising phase of the fEPSP. For picrotoxin experiments, 20 µM picrotoxin (Sigma, Cat# P1675) was perfused in the chamber 10 minutes prior and during the TBS and removed immediately after.
Transmission Electron Microscopy
Mice were transcardially perfused with cold phosphate buffered saline pH 7.4 (PBS) for 1 minute followed by cold fixative (1% paraformaldehyde (PFA) and 2.5% glutaraldehyde in 0.1 M sodium cacodylate buffer pH 7.2) for 7 minutes. Brains were removed, soaked in fixative for two days, and sectioned into 150 µm thick coronal sections. The CA1 region was cut out, washed with 0.1 M sodium cacodylate buffer, fixed with 1% osmium tetroxide and 1.5% potassium ferrocyanide for 1 hour and stained with 1% uranyl acetate for 1 hour. The tissue was subsequently dehydrated in increasing concentrations of ethanol, embedded in epon resin, and cured at 60°C for 48 hours. Next, 40 nm sections were cut using a Leica UC6 ultramicrotome and stained using lead citrate. Images were acquired using a JEM-1400Plus TEM (JEOL) at 10000X magnification and analyzed using Fiji. Only synapses with distinct pre and postsynaptic boundaries were analyzed. Presynaptic boutons were en passant or terminal and were defined as axon swellings containing a cluster of SVs adjacent to a PSD. Bouton boundaries were delineated by the presynaptic membrane and a virtual line was drawn as an extension of the curvature where the axon swells to accommodate the synapse (see figure S1A). Cadherin-9 wildtype and knockout electron microscopy analysis was done blind to genotype.
Microiontophoresis, spine, and Sholl analysis
Lucifer yellow (LY) microiontophoresis was performed as described previously (Dumitriu et al., 2011). Mice were transcardially perfused with a fixative comprising 4% PFA (w/v), 0.125% Glutaraldehyde (v/v) in Phosphate buffer (PB) pH 7.4. Brains were quickly extracted and post-fixed for 30 minutes in the fixative after which it was transferred to PB and sectioned into 200 µm thick slices. CA1 neuron cell bodies were impaled with a sharp (150–250 MΩ) glass electrode containing 100 mM LY (Invitrogen, Cat# L453) dissolved in 200 mM KCl filled until the tips of distal dendrites appeared bright. Slices were post-fixed in 4% PFA in PBS for 15 minutes and dendrites were imaged using a Zeiss LSM 710 confocal microscope using a 63X oil immersion lens (N.A.=1.4). Images were deconvolved using AutoQuant X3 (Bitplane, RRID:SCR_002465) and spines were modeled using Imaris (Bitplane, RRID:SCR_007370) software. All spine analysis was done blind to genotype. Any dendritic protrusions completely within the x, y, and z-planes of the image were counted as spines. Spine parameters like head width (H), mean neck width (N), and length (L) were calculated and used for further classification. Spines were classified into thin (H > 1.2*N and 0.15 µm < H < 0.3 µm), mushroom (H > 1.2*N and H > 0.3 µm), stubby (H < 1.2*N and L < 0.5), and filopodia (H < 0.15 µm and N < 0.15 µm). The rare spine not satisfying any of these conditions was deemed unclassified. The spine head width cutoff of 0.3 µm resulted in a mushroom to thin spine ratio of 0.3 which is close to the value defined previously (Harris et al., 1992). For Sholl analysis we ensured that dendrites in either SO and/or SR layer were completely filled. Dendrites were imaged and analyzed using the Sholl analysis plugin within Fiji using a Sholl radius step size of 20 µm.
RT-PCR analysis
Mice were sacrificed using CO2 mediated asphyxiation and their brains were immediately removed and the hippocampi were dissected out. Total hippocampal RNA was extracted using TRIzol reagent (Invitrogen, Cat# 15596026). Subsequently, cDNA was synthesized from 200 ng RNA using SuperScript VILO cDNA synthesis kit (Invitrogen, Cat# 11754050). The following primers were used: Cadherin-6 forward primer 5’-ACGTGGGCAAGTTACATTCA-3’, Cadherin-6 reverse primer 5’-CCTGTATGTCGCCTGTGTTC-3’, GAPDH forward primer 5’-GAAACCTGCCAAGTATGATGAC-3’, and GAPDH reverse primer 5’-AAGTCGCAGGAGACAACCTG-3’.
QUANTIFICATION AND STATISTICAL ANALYSIS
All statistical analyses were performed using Prism (GraphPad Software, RRID:SCR_002798). To compare between the means of two distributions we first checked if the distributions were Gaussian. For Gaussian distributions, we calculated p-values using student’s t-test while for non-Gaussian distributions we used Mann-Whitney test. When comparing among more than two distributions, we used one-way ANOVA for one dimensional data (for example comparing mushroom spine densities among SO, SR, and SLM layers) followed by post-hoc p-value calculation by Hölm-Sidàk’s method owing to its high statistical power. For two-dimensional data, we used two-way ANOVA followed by post-hoc p-value calculation by Hölm-Sidàk’s method. The two-way ANOVA interaction, row, and column p-values are reported in Supplementary tables 1 and 2. p-values below 0.05 were defined as statistically significant. In the figures the following symbols were used to show different levels of significance: p<0.05, p<0.01, p<0.001, and p<0.0001 is denoted by *, **, ***, and **** respectively. Statistical outliers were removed using Grubbs outlier test (https://graphpad.com/quickcalcs/Grubbs1.cfm) using a p=0.05 as cutoff. All statistical tests and sample sizes for each experiment are listed in the figure legends.
Supplementary Material
Highlights.
CA1 neurons have layer-specific differences in LTP magnitude
Cadherin-9, expressed in CA3 neurons, binds cadherins-6 and 10, expressed in CA1 neurons
Cadherins-6, 9, and 10 are specifically required for high magnitude LTP
Acknowledgments
We thank Dr. Gerd Klein (University of Tuebingen) for cadherin-9 antibody, Dr. Alan R. White (University of Utah) for advice on microiontophoresis, Jennifer Hunter, Keegan Wade Teeter, and Zhirong Wang for technical help, and Dimitri Tränkner, Jason Shepherd, and the entire Williams lab for comments on the manuscript. X.D. was supported by a Life Science Research Foundation fellowship and Research to Prevent Blindness Career Development award. This work was funded by grants from the Alfred P. Sloan, Whitehall, and Edward Mallinckrodt Jr. Foundations (to M.E.W) and N.I.H grant EY022073 (to J.R.S).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
Conceptualization, R.B. and M.E.W; Methodology, R.B., X.D., and J.R.S; Investigation, R.B, M.R.T, E.A.M, S.M., Y.W., L.G-W, S.C.D, M.Y. and M.E.W; Resources X.D., P.J.W., and J.R.S; Writing-Original draft, R.B. and M.E.W; Writing-Review and Editing, R.B., M.R.T, E.A.M, L.G-W, P.J.W., J.R.S, and M.E.W.; Funding Acquisition, J.R.S, and M.E.W.; Supervision; P.J.W., J.R.S., and M.E.W.
References
- Aiga M, Levinson JN, Bamji SX. N-cadherin and Neuroligins Cooperate to Regulate Synapse Formation in Hippocampal Cultures. J. Biol. Chem. 2010;286:851–858. doi: 10.1074/jbc.M110.176305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai A, Black J, Lynch G. Origins of the variations in long-term potentiation between synapses in the basal versus apical dendrites of hippocampal neurons. Hippocampus. 1994;4:1–9. doi: 10.1002/hipo.450040103. [DOI] [PubMed] [Google Scholar]
- Bekirov IH, Needleman LA, Zhang W, Benson DL. Identification and localization of multiple classic cadherins in developing rat limbic system. Neuroscience. 2002;115:213–227. doi: 10.1016/s0306-4522(02)00375-5. [DOI] [PubMed] [Google Scholar]
- Bian W-J, Miao W-Y, He S-J, Qiu Z, Yu X. Coordinated Spine Pruning and Maturation Mediated by Inter-Spine Competition for Cadherin/Catenin Complexes. Cell. 2015:1–39. doi: 10.1016/j.cell.2015.07.018. [DOI] [PubMed] [Google Scholar]
- Bosch M, Hayashi Y. Structural plasticity of dendritic spines. Curr. Opin. Neurobiol. 2012;22:383–388. doi: 10.1016/j.conb.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne J, Harris KM. Do thin spines learn to be mushroom spines that remember? Curr. Opin. Neurobiol. 2007;17:381–386. doi: 10.1016/j.conb.2007.04.009. [DOI] [PubMed] [Google Scholar]
- Bozdagi O, Wang XB, Nikitczuk JS, Anderson TR, Bloss EB, Radice GL, Zhou Q, Benson DL, Huntley GW. Persistence of Coordinated Long-Term Potentiation and Dendritic Spine Enlargement at Mature Hippocampal CA1 Synapses Requires N-Cadherin. J. Neurosci. 2010;30:9984–9989. doi: 10.1523/JNEUROSCI.1223-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavus I, Teyler TJ. NMDA receptor-independent LTP in basal versus apical dendrites of CA1 pyramidal cells in rat hippocampal slice. Hippocampus. 1998;8:373–379. doi: 10.1002/(SICI)1098-1063(1998)8:4<373::AID-HIPO5>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Duan X, Krishnaswamy A, la Huerta De I, Sanes JR. Type II Cadherins Guide Assembly of a Direction-Selective Retinal Circuit. Cell. 2014;158:793–807. doi: 10.1016/j.cell.2014.06.047. [DOI] [PubMed] [Google Scholar]
- Dudek SM, Alexander GM, Farris S. Rediscovering area CA2: unique properties and functions. Nat. Rev. Neurosci. 2016;17:89–102. doi: 10.1038/nrn.2015.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumitriu D, Rodriguez A, Morrison JH. High-throughput, detailed, cell-specific neuroanatomy of dendritic spines using microinjection and confocal microscopy. Nat. Prot. 2011;6:1391–1411. doi: 10.1038/nprot.2011.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan W. Group I metabotropic glutamate receptors modulate late phase long-term potentiation in hippocampal CA1 pyramidal neurons: comparison of apical and basal dendrites. Neurosci. Lett. 2013;553:132–137. doi: 10.1016/j.neulet.2013.08.030. [DOI] [PubMed] [Google Scholar]
- Fièvre S, Carta M, Chamma I, Labrousse V, Thoumine O, Mulle C. Molecular determinants for the strictly compartmentalized expression of kainate receptors in CA3 pyramidal cells. Nat. Commun. 2016;7:12738. doi: 10.1038/ncomms12738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil V, Bichler Z, Lee JK, Seira O, Llorens F, Bribian A, Morales R, Claverol-Tinture E, Soriano E, Sumoy L, et al. Developmental Expression of the Oligodendrocyte Myelin Glycoprotein in the Mouse Telencephalon. Cereb. Cortex. 2010;20:1769–1779. doi: 10.1093/cercor/bhp246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginsberg D, DeSIMONE D, Geiger B. Expression of a novel cadherin (EP-cadherin) in unfertilized eggs and early Xenopus embryos. Development. 1991;111:315–325. doi: 10.1242/dev.111.2.315. [DOI] [PubMed] [Google Scholar]
- Harris KM. Structure, development, and plasticity of dendritic spines. Curr. Opin. Neurobiol. 1999;9:343–348. doi: 10.1016/s0959-4388(99)80050-6. [DOI] [PubMed] [Google Scholar]
- Harris KM, Jensen FE, Tsao B. Three-dimensional structure of dendritic spines and synapses in rat hippocampus (CA1) at postnatal day 15 and adult ages: implications for the maturation of synaptic physiology and long-term potentiation. J. Neurosci. 1992;12:2685–2705. doi: 10.1523/JNEUROSCI.12-07-02685.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison OJ, Jin X, Hong S, Bahna F, Ahlsen G, Brasch J, Wu Y, Vendome J, Felsovalyi K, Hampton CM, et al. The Extracellular Architecture of Adherens Junctions Revealed by Crystal Structures of Type I Cadherins. Structure. 2011;19:244–256. doi: 10.1016/j.str.2010.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herring BE, Nicoll RA. Long-Term Potentiation: From CaMKII to AMPA Receptor Trafficking. Annu. Rev. Physiol. 2016;78:351–365. doi: 10.1146/annurev-physiol-021014-071753. [DOI] [PubMed] [Google Scholar]
- Hirano S, Takeichi M. Cadherins in brain morphogenesis and wiring. Physiol. Rev. 2012;92:597–634. doi: 10.1152/physrev.00014.2011. [DOI] [PubMed] [Google Scholar]
- Hitti FL, Siegelbaum SA. The hippocampal CA2 region is essential for social memory. Nature. 2014;508:88–92. doi: 10.1038/nature13028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtmaat AJGD, Trachtenberg JT, Wilbrecht L, Shepherd GM, Zhang X, Knott GW, Svoboda K. Transient and persistent dendritic spines in the neocortex in vivo. Neuron. 2005;45:279–291. doi: 10.1016/j.neuron.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Huntley GW, Elste AM, Patil SB, Bozdagi O, Benson DL, Steward O. Synaptic loss and retention of different classic cadherins with LTP-associated synaptic structural remodeling in vivo. Hippocampus. 2010;22:17–28. doi: 10.1002/hipo.20859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DH, Matus AI. Isolation of synaptic plasma membrane from brain by combined flotation-sedimentation density gradient centrifugation. Biochimica Et Biophysica Acta (BBA) - Biomembranes. 1974;356:276–287. doi: 10.1016/0005-2736(74)90268-5. [DOI] [PubMed] [Google Scholar]
- Jungling K, Eulenburg V, Moore R, Kemler R, Lessmann V, Gottmann K. NCadherin Transsynaptically Regulates Short-Term Plasticity at Glutamatergic Synapses in Embryonic Stem Cell-Derived Neurons. J. Neurosci. 2006;26:6968–6978. doi: 10.1523/JNEUROSCI.1013-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaibara T, Leung LS. Basal versus apical dendritic long-term potentiation of commissural afferents to hippocampal CA1: a current-source density study. J. Neurosci. 1993;13:2391–2404. doi: 10.1523/JNEUROSCI.13-06-02391.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsamba P, Carroll K, Ahlsen G, Bahna F, Vendome J, Posy S, Rajebhosale M, Price S, Jessell TM, Ben-Shaul A, et al. Linking molecular affinity and cellular specificity in cadherin-mediated adhesion. Proc. Natl. Acad. Sci. USA. 2009;106:11594–11599. doi: 10.1073/pnas.0905349106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz Y, Menon V, Nicholson DA, Geinisman Y, Kath WL, Spruston N. Synapse Distribution Suggests a Two-Stage Model of Dendritic Integration in CA1 Pyramidal Neurons. Neuron. 2009;63:171–177. doi: 10.1016/j.neuron.2009.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay JN, la Huerta De I, Kim IJ, Zhang Y, Yamagata M, Chu MW, Meister M, Sanes JR. Retinal Ganglion Cells with Distinct Directional Preferences Differ in Molecular Identity, Structure, and Central Projections. J. Neurosci. 2011;31:7753–7762. doi: 10.1523/JNEUROSCI.0907-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramár EA, Lynch G. Developmental and regional differences in the consolidation of long-term potentiation. Neuroscience. 2003;118:387–398. doi: 10.1016/s0306-4522(02)00916-8. [DOI] [PubMed] [Google Scholar]
- Kuwako K-I, Nishimoto Y, Kawase S, Okano HJ, Okano H. Cadherin-7 Regulates Mossy Fiber Connectivity in the Cerebellum. Cell Rep. 2014:1–35. doi: 10.1016/j.celrep.2014.08.063. [DOI] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Manabe T, Togashi H, Uchida N, Suzuki SC, Hayakawa Y, Yamamoto M, Yoda H, Miyakawa T, Takeichi M, Chisaka O. Loss of Cadherin-11 Adhesion Receptor Enhances Plastic Changes in Hippocampal Synapses and Modifies Behavioral Responses. Mol. Cell. Neurosci. 2000;15:534–546. doi: 10.1006/mcne.2000.0849. [DOI] [PubMed] [Google Scholar]
- Martin EA, Muralidhar S, Wang Z, Cervantes DC, Basu R, Taylor MR, Hunter J, Cutforth T, Wilke SA, Ghosh A, et al. The intellectual disability gene Kirrel3 regulates target-specific mossy fiber synapse development in the hippocampus. Elife. 2015;4:e09395. doi: 10.7554/eLife.09395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki M, Honkura N, Ellis-Davies GCR, Kasai H. Structural basis of long-term potentiation in single dendritic spines. Nature. 2004;429:761–766. doi: 10.1038/nature02617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez P, De Roo M, Poglia L, Klauser P, Muller D. N-cadherin mediates plasticity-induced long-term spine stabilization. J. Cell. Biol. 2010;189:589–600. doi: 10.1083/jcb.201003007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navakkode S, Sajikumar S, Korte M, Soong TW. Dopamine induces LTP differentially in apical and basal dendrites through BDNF and voltage-dependent calcium channels. Learn. Mem. 2012;19:294–299. doi: 10.1101/lm.026203.112. [DOI] [PubMed] [Google Scholar]
- Nicholson DA, Trana R, Katz Y, Kath WL, Spruston N, Geinisman Y. Distance-dependent differences in synapse number and AMPA receptor expression in hippocampal CA1 pyramidal neurons. Neuron. 2006;50:431–442. doi: 10.1016/j.neuron.2006.03.022. [DOI] [PubMed] [Google Scholar]
- Nicoll RA, Schmitz D. Synaptic plasticity at hippocampal mossy fibre synapses. Nat. Rev. Neurosci. 2005;6:863–876. doi: 10.1038/nrn1786. [DOI] [PubMed] [Google Scholar]
- Nollet F, Kools P, van Roy F. Phylogenetic analysis of the cadherin superfamily allows identification of six major subfamilies besides several solitary members. J. Mol. Biol. 2000;299:551–572. doi: 10.1006/jmbi.2000.3777. [DOI] [PubMed] [Google Scholar]
- Osterhout JA, Josten N, Yamada J, Pan F, Wu S-W, Nguyen PL, Panagiotakos G, Inoue YU, Egusa SF, Volgyi B, et al. Cadherin-6 Mediates Axon-Target Matching in a Non-Image-Forming Visual Circuit. Neuron. 2011;71:632–639. doi: 10.1016/j.neuron.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson MA, Szatmari EM, Yasuda R. AMPA receptors are exocytosed in stimulated spines and adjacent dendrites in a Ras-ERK-dependent manner during long-term potentiation. Proc. Natl. Acad. Sci. USA. 2010;107:15951–15956. doi: 10.1073/pnas.0913875107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poskanzer K, Needleman LA, Bozdagi O, Huntley GW. N-cadherin regulates ingrowth and laminar targeting of thalamocortical axons. J. Neurosci. 2003;23:2294–2305. doi: 10.1523/JNEUROSCI.23-06-02294.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran B, Ahmed S, Zafar N, Dean C. Ethanol inhibits long-term potentiation in hippocampal CA1 neurons, irrespective of lamina and stimulus strength, through neurosteroidogenesis. Hippocampus. 2014;25:106–118. doi: 10.1002/hipo.22356. [DOI] [PubMed] [Google Scholar]
- Redies C, Takeichi M. Cadherins in the Developing Central Nervous System: An Adhesive Code for Segmental and Functional Subdivisions. Dev. Biol. 1996;180:413–423. doi: 10.1006/dbio.1996.0315. [DOI] [PubMed] [Google Scholar]
- Roberts TF, Tschida KA, Klein ME, Mooney R. Rapid spine stabilization and synaptic enhancement at the onset of behavioural learning. Nature. 2010;463:948–952. doi: 10.1038/nature08759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saglietti L, Dequidt C, Kamieniarz K, Rousset M-C, Valnegri P, Thoumine O, Beretta F, Fagni L, Choquet D, Sala C, et al. Extracellular Interactions between GluR2 and N-Cadherin in Spine Regulation. Neuron. 2007a;54:461–477. doi: 10.1016/j.neuron.2007.04.012. [DOI] [PubMed] [Google Scholar]
- Saglietti L, Dequidt C, Kamieniarz K, Rousset M-C, Valnegri P, Thoumine O, Beretta F, Fagni L, Choquet D, Sala C, et al. Extracellular Interactions between GluR2 and N-Cadherin in Spine Regulation. Neuron. 2007b;54:461–477. doi: 10.1016/j.neuron.2007.04.012. [DOI] [PubMed] [Google Scholar]
- Shan WS, Tanaka H, Phillips GR, Arndt K, Yoshida M, Colman DR, Shapiro L. Functional cis-heterodimers of N- and R-cadherins. J. Cell. Biol. 2000;148:579–590. doi: 10.1083/jcb.148.3.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoyama Y, Tsujimoto G, Kitajima M, Natori M. Identification of three human type-II classic cadherins and frequent heterophilic interactions between different subclasses of type-II classic cadherins. Biochem. J. 2000;349:159–167. doi: 10.1042/0264-6021:3490159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki SC, Inoue T, Kimura Y, Tanaka T, Takeichi M. Neuronal circuits are subdivided by differential expression of type-II classic cadherins in postnatal mouse brains. Mol. Cell. Neurosci. 1997;9:433–447. doi: 10.1006/mcne.1997.0626. [DOI] [PubMed] [Google Scholar]
- Takeichi M, Nakagawa S. Cadherin-dependent cell-cell adhesion. Curr. Protoc. Cell. Biol. 2001 doi: 10.1002/0471143030.cb0903s00. Chapter 9, Unit 9.3–9.3.15. [DOI] [PubMed] [Google Scholar]
- Tang L, Hung CP, Schuman EM. A role for the cadherin family of cell adhesion molecules in hippocampal long-term potentiation. Neuron. 1998;20:1165–1175. doi: 10.1016/s0896-6273(00)80497-3. [DOI] [PubMed] [Google Scholar]
- Thedieck C, Kalbacher H, Kuczyk M, Müller GA, Müller CA, Klein G. Cadherin-9 Is a Novel Cell Surface Marker for the Heterogeneous Pool of Renal Fibroblasts. PLoS ONE. 2007;2:e657. doi: 10.1371/journal.pone.0000657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Togashi H, Abe K, Mizoguchi A, Takaoka K, Chisaka O, Takeichi M. Cadherin regulates dendritic spine morphogenesis. Neuron. 2002;35:77–89. doi: 10.1016/s0896-6273(02)00748-1. [DOI] [PubMed] [Google Scholar]
- Tønnesen J, Katona G, Rózsa B, Nägerl UV. Spine neck plasticity regulates compartmentalization of synapses. Nat. Neurosci. 2014;17:678–685. doi: 10.1038/nn.3682. [DOI] [PubMed] [Google Scholar]
- Viswanathan S, Williams ME, Bloss EB, Stasevich TJ, Speer CM, Nern A, Pfeiffer BD, Hooks BM, Li W-P, English BP, et al. High-performance probes for light and electron microscopy. Nat. Meth. 2015;12:568–576. doi: 10.1038/nmeth.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitureira N, Letellier M, White IJ, Goda Y. Differential control of presynaptic efficacy by postsynaptic N-cadherin and β-catenin. Nat. Neurosci. 2011;15:81–89. doi: 10.1038/nn.2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S, Rost BR, Camacho-Pérez M, Davis MW, Söhl-Kielczynski B, Rosenmund C, Jorgensen EM. Ultrafast endocytosis at mouse hippocampal synapses. Nature. 2013;504:242–247. doi: 10.1038/nature12809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams ME, Wilke SA, Daggett A, Davis E, Otto S, Ravi D, Ripley B, Bushong EA, Ellisman MH, Klein G, et al. Cadherin-9 Regulates Synapse-Specific Differentiation in the Developing Hippocampus. Neuron. 2011;71:640–655. doi: 10.1016/j.neuron.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Jin X, Harrison O, Shapiro L. Cooperativity between trans and cis interactions in cadherin-mediated junction formation. Proc. Natl. Acad. Sci. USA. 2010;107:17592–17597. doi: 10.1073/pnas.1011247107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Yu X, Perlik AJ, Tobin WF, Zweig JA, Tennant K, Jones T, Zuo Y. Rapid formation and selective stabilization of synapses for enduring motor memories. Nature. 2009;462:915–919. doi: 10.1038/nature08389. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.