Abstract
Organ development requires complex signaling by cells in different tissues. Epithelium and mesenchyme interactions are crucial for the development of skin, hair follicles, kidney, lungs, prostate, major glands, and teeth. Despite myriad literature on cell–cell interactions and ligand–receptor binding, the roles of extracellular vesicles in epithelium–mesenchyme interactions during organogenesis are poorly understood. Here, we discovered that ~100 nm exosomes were secreted by the epithelium and mesenchyme of a developing tooth organ and diffused through the basement membrane. Exosomes were entocytosed by epithelium or mesenchyme cells with preference by reciprocal cells rather than self-uptake. Exosomes reciprocally evoked cell differentiation and matrix synthesis: epithelium exosomes induce mesenchyme cells to produce dentin sialoprotein and undergo mineralization, whereas mesenchyme exosomes induce epithelium cells to produce basement membrane components, ameloblastin and amelogenenin. Attenuated exosomal secretion by Rab27a/b knockdown or GW4869 disrupted the basement membrane and reduced enamel and dentin production in organ culture and reduced matrix synthesis and the size of the cervical loop, which harbors epithelium stem cells, in Rab27aash/ash mutant mice. We then profiled exosomal constituents including miRNAs and peptides and further crossed all epithelium exosomal miRNAs with literature-known miRNA Wnt regulators. Epithelium exosome-derived miR135a activated Wnt/β-catenin signaling and escalated mesenchymal production of dentin matrix proteins, partially reversible by Antago-miR135a attenuation. Our results suggest that exosomes may mediate epithelium–mesenchyme crosstalk in organ development, suggesting that these vesicles and/or the molecular contents they are transporting may be interventional targets for treatment of diseases or regeneration of tissues.
Keywords: exosomes, epithelium, mesenchyme, development, miRNA, Wnt, miR135a
Graphical Abstract
Epithelium–mesenchyme crosstalk is required for organogenesis of many mammalian tissues including hair follicles, skin, teeth, mammary glands, prostate, lungs, and kidney.1,2 Extracellular vesicles including exosomes, microvesicles, and apoptotic bodies are intensely studied in cell communication. They have been extensively studied in cancer invasiveness and adaptive immune response,3–5 but little is known of the role of exosomes in mediating epithelium–mesenchyme interactions in organogenesis.
Analogous to the stem cell niche in hair follicles or intestinal mucosa, rodent incisors continually grow throughout life and can be a model for interrogating epithelium and mesenchyme stem cells.6,7 Prenatally, tooth morphogenesis is abolished in the absence of either epithelium or mesenchyme.8 Signaling between epithelium and mesenchyme is transferred through cell junctions, cell-extracellular matrix, and diffused factors. Short- or long-range signaling molecules, including autocrine, paracrine, endocrine, and synaptic signaling, play important roles during development.9 For tooth development, multiple pathways have been demonstrated to be involved in the process including Bmps, Wnt, Shh, and Fgfs at different stages.10,11 For example, a key Wnt–Bmp signaling circuit feedback controls gene expression and signaling dynamics in the interacting epithelial and mesenchymal compartments.12 How the molecules are transported between cells to regulate tooth development remains unclear. For example, tooth differentiation was not disturbed when interposed membrane filters, with pore sizes larger than 0.2 μm, were placed between these two compartments. However, when separated by a filter with pore sizes of 0.1 μm, epithelium and mesenchyme cells fail to polarize or differentiate, suggesting that signaling bodies greater than ~100 nm may be involved.13 Thus, previously well explored signaling modalities, including cell–cell contact and protein signaling, cannot sufficiently account for regulation of epithelium and mesenchyme development.
We discovered these CD63-positive vesicles in the size range of 100–120 nm are present in the epithelium, mesenchyme, and basement membrane. Exosomes are endocytosed reciprocally into epithelium and mesenchyme cells, leading to cellular functions as in the original cells. Attenuation of exosome secretion led to disruption of basement membrane formation and reduced dentinogenesis. Epithelium exosome-derived miR135a not only activated Wnt/β-catenin signaling but also upregulated dentin sialoprotein in mesenchyme cells that lack miR135a. In the future, the manipulation of the exosomal communication between epithelium and mesenchyme cells may control the tissue regeneration process as these cells are involved in organ development.
RESULTS AND DISCUSSION
Presence of Exosomes in Epithelium and Mesenchyme
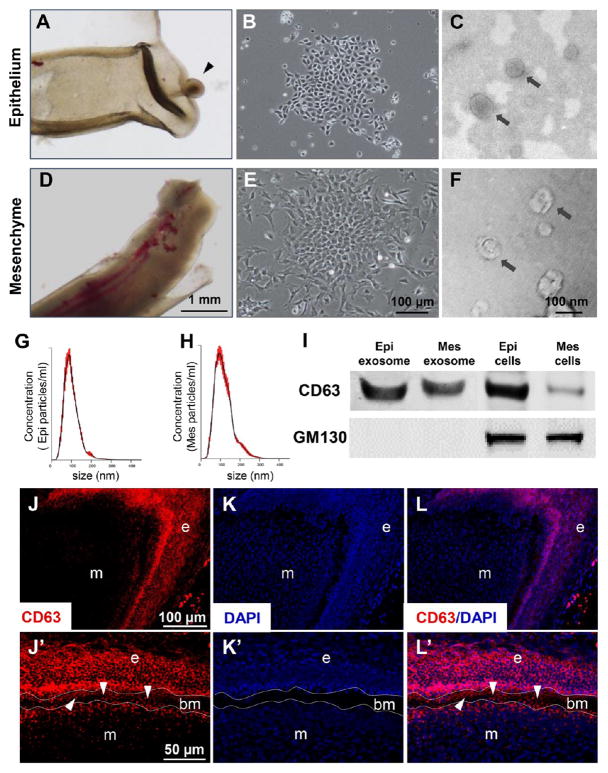
First, we explored whether exosomes were present in the epithelium and mesenchyme. The epithelium was microdissected from the underlying mesenchyme from postnatal 5/6-day-old rat incisors (Figure 1A, D, respectively). The epithelium and mesenchyme stem cells were isolated per our prior methods and cultured in exosome-free media14 and showed typically cobblestone and spindle-like morphologies (Figure 1B, E, respectively). Transmission electron microscopy revealed cell-secreted vesicles in the size range of ~100 nm by epithelium stem cells (Figure 1C) and mesenchyme stem cells (Figure 1F). Nanoparticle tracing revealed 100.1 ± 2.2 nm cellular vesicles in cultured epithelium cell supernatant (Figure 1G) and 116.0 ± 2.3 nm cellular vesicles in cultured mesenchyme cell supernatant (Figure 1H), consistent with known exosomal sizes.15 The isolated epithelium and mesenchyme exosomes were not only CD63 positive but also GM130 negative, suggesting the absence of Golgi or cell contamination (Figure 1I). CD9 and CD81 were negative in the isolated epithelium or mesenchyme cells (data not shown). CD63, a marker for extracellular vesicles, was highly expressed in the epithelium and mesenchyme (marked with e, m in Figure 1J–L) in the cervical loop area. Importantly, CD63-positive vesicles were detected (arrowheads in Figure 1J′–L′) in the acellular basement membrane (bm), suggesting one-way or reciprocal diffusion of extracellular vesicles across the basement membrane. Thus, exosomes are present in epithelium and mesenchyme cells, likely diffuse through the basement membrane, and can be readily isolated.
Figure 1.
Exosome characterization in epithelium, mesenchyme, and basement membrane. (A) Dental epithelium with cervical loop (arrowhead) was dissected from dental mesenchyme (D). (B, E) Isolated dental epithelial cells and mesenchymal cells. Transmission electron microscopy showing extracellular vesicles with ~100 nm diameters isolated from epithelium (C) and mesenchyme (F). (G, H) Size analysis revealed epithelium vesicles with diameters of 100.1 ± 2.2 nm and mesenchyme vesicles of 116.0 ± 2.3 nm. (I) Total proteins extracted from nanometer vesicles and parent cells probed by anti-CD63 and anti-GM-130 antibodies. (J, J′) CD63 (red); (K, K′) DAPI (blue); (L, L′) overlay. e: epithelium, m: mesenchyme, bm: basement membrane (arrowheads).
Exosomes Are Endocytosed Preferentially by a Reciprocal Cell type
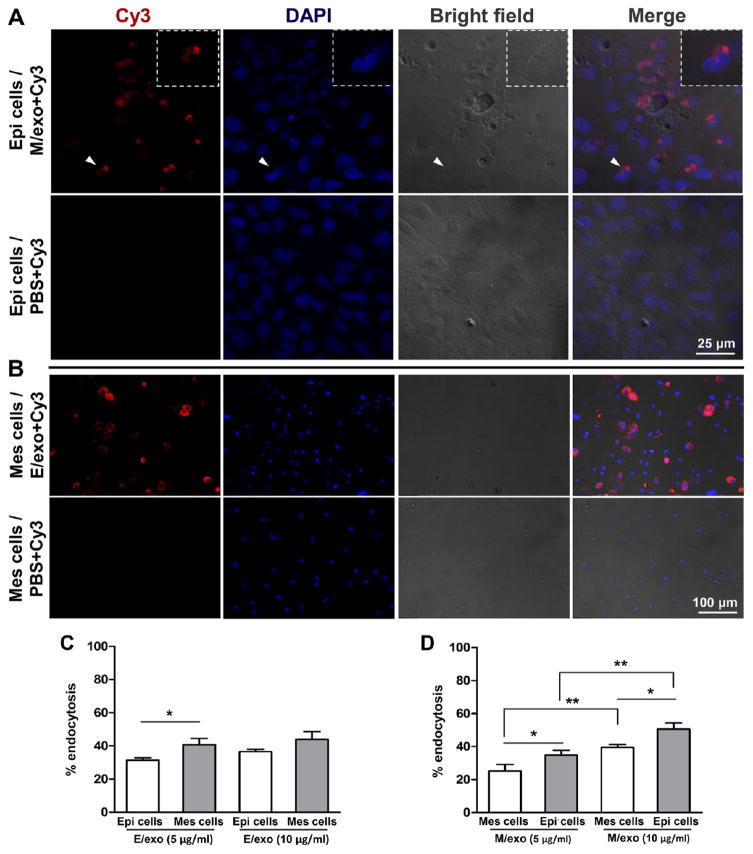
To test exosome endocytosis, we marked exosomes with Cy3-labeled siRNA by electroporation and cross-incubated epithelium and mesenchyme cells in exosome-free media for 12 h. Cy3-labeled mesenchyme exosomes were endocytosed into epithelium cells, relative to controls with the same amount of Cy3-labeled siRNA in medium (Figure 2A). Conversely, Cy3-labeled epithelium exosomes were endocytosed into mesenchyme cells, relative to exosome-free Cy3 in mesenchyme cells (Figure 2B). Remarkably, mesenchyme cells preferentially endocytosed significantly more epithelium exosomes at 5 and 10 μg/mL than donor-matched epithelium cells (Figure 2C). Conversely, epithelium cells endocytosed significantly more mesenchyme exosomes at 5 and 10 μg/mL than donor-matched mesenchyme cells (Figure 2D). These data indicate that epithelium exosomes are preferentially endocytosed into mesenchyme cells, and vice versa, rather than uptake by cells from which exosomes are produced.
Figure 2.
Preferential exosome uptake reciprocally by epithelium and mesenchyme cells. (A) Cy3-labeled siRNA electroporated mesenchyme-derived exosome (M/exo) incubated with epithelium (Epi) cells for 12 h and PBS control. (B) Mesenchyme (Mes) cells incubated with epithelium-derived exosomes (E/exo) for 12 h and PBS control. (C, D) Preferential endocytosis reciprocally by epithelium or mesenchyme cells upon exosome delivery (5.0 or 10.0 μg/mL exosomal protein content) (mean ± SD; six to 10 independent experiments). *P < 0.05, **P < 0.01 (one-way ANOVA and LSD tests).
Multiple pathways can mediate the endocytosis of exosomes.16 To further examine the endocytic pathways involved in dental epithelial and mesenchymal derived exosomes, we labeled exosomes with lipophilic dye and incubated them with inhibitor-pretreated cells reciprocally. As shown in Figure S1A, 10 μM chlorpromazine (CPZ), a cationic amphipathic drug that induces a loss of clathrin,17 caused 75% blockage of epithelial cell-derived exosome endocytosis in mesenchymal cells. Similar results could be observed in the micropinocytosis uptake process. In the presence of LY2940002 (50 μM), exosome uptake was significantly decreased to 34% in mesenchymal cells. Interestingly, mesenchymal cell-derived exosome uptake in epithelial cells was only blocked by nystatin (5 μM) through caveolae-mediated internalization, in contrast to mesenchymal cells (Figure S1B). Statistic results are shown in Figure S1C and D. As a result, the uptake mode of exosomes into epithelial and mesenchymal cells depended on the type of cells and different mechanisms.
Preferential uptake of epithelial exosomes by mesenchyme cells, and vice versa, suggests cell surface recognition mechanisms that are yet to be understood. Cells internalize exosomes either by fusion or via endocytosis. Several pathways mediate endocytosis including clathrin-mediated endocytosis, caveolae-mediated endocytosis, phagocytosis, and micropinocytosis.18,19 Clathrin-mediated endocytosis is one of the critical pathways, which is inherently active in almost all mammalian cells and inhibited by CPZ.17 Caveolae-mediated endocytosis is another route for exsomal internalization and is blocked by lipid raft disruption, such as that by nystatin.20 The micropinocytosis pathway could be inhibited by a PI3K inhibitor, LY294002. Our finding of mesenchymal cell uptake of epithelial exosomes may be through clathrin and micropinocytosis pathways. On the other hand, mesenchymal exosomes were endocytosed into epithelial cells mainly according to the caveolae pathway. Cells appear to recognize ligands from the exosomal membrane surface and selectively take up exosomes.21 Exosome uptake may be cell-type specific22,23 and can affect cell functions.24
Exosomes Reciprocally Induce Epithelium and Mesenchyme Differentiation and Matrix Synthesis
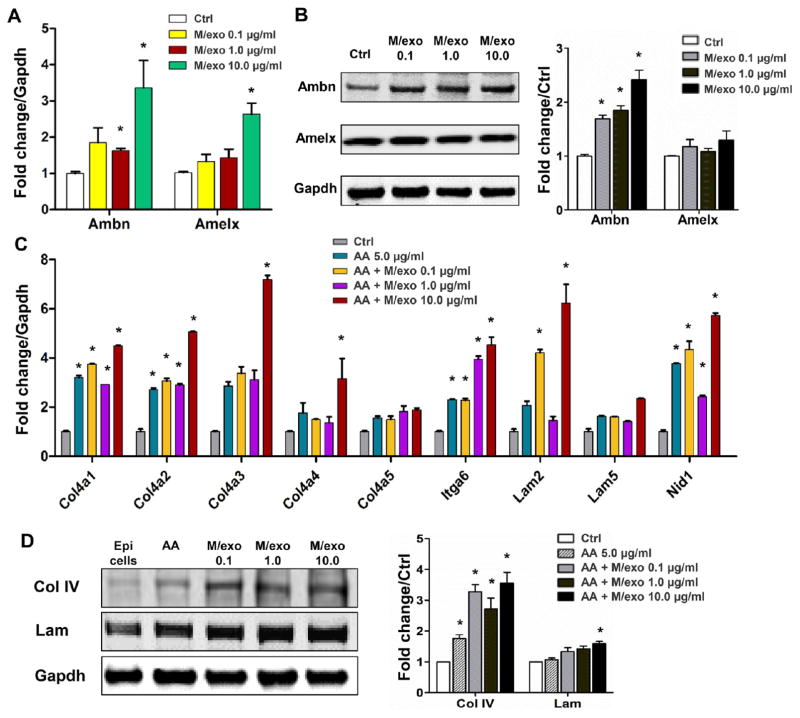
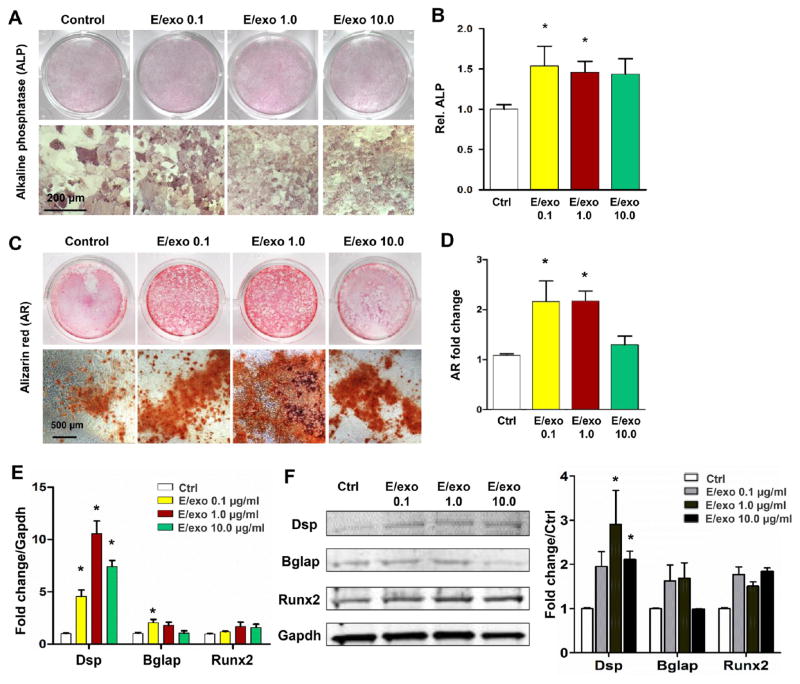
Epithelium cells incubated with mesenchyme exosomes robustly produced amelogenin and ameloblastin mRNAs and proteins (Figure 3A and B), suggesting that mesenchyme exosomes may substitute mesenchyme cells in stimulating the epithelium to produce these two major amelogenesis scaffolding proteins. Basement membrane is an indispensable structure in epithelium and mesenchyme development including enamel and dentin formation in tooth morphogenesis.25 Mesenchyme exosomes stimulated epithelium cells to produce basement membrane components, including collagen type IV (Col IV) and laminin (lam) (Figure 3C and D). Conversely, epithelium exosomes induced mesenchyme cells to elevate alkaline phosphatase production (Figure 4A), an important enzyme in mineralization, with data quantified in Figure 4B, and mineral nodule formation (Figure 4C and D). Epithelium exosomes further stimulated the mesenchyme to produce dentin sialophosphoprotein (Dsp) and osteocalcin (Bglap), two crucial gene and protein products for dentinogenesis (Figure 4E and F). Runx2, a transcriptional factor for osteogenesis that needs to be downregulated during odontoblast differentiation,26 was not effected when epithelium exosomes were incubated with mesenchyme cells (Figure 4E and F). Therefore, epithelium or mesenchyme exosomes may at least partially substitute their parent cells and reciprocally induce cellular differentiation and matrix synthesis.
Figure 3.
Mesenchyme-derived exosomes induced epithelial cell differentiation and matrix synthesis. (A, B) Mesenchyme exosomes stimulated epithelium cells to produce ameloblastin (Ambn) and amelogenin (Amelx) mRNAs and proteins. (C, D) Collagen IV (Col IV) and Laminin (Lam) production by epithelium cells upon stimulation by mesenchyme exosomes at mRNA and protein level (mean ± SD; three to five independent experiments). *P < 0.05 (one-way ANOVA and LSD test).
Figure 4.
Epithelium-derived exosomes induced mesenchymal cell differentiation and mineralization. (A) Epithelial exosomes promoted alkaline phosphatase (ALP) with higher magnification, quantified in B. (C) Alizarin Red (AR)-positive mineral nodule formation was increased with different doses of epithelium exosomes, with higher magnification and quantification (D). (E, F) Epithelium exosomes stimulated mesenchyme cells to produce Dsp at mRNA and protein (mean ± SD; five independent experiments). *P < 0.05 (one-way ANOVA and LSD test).
Attenuated Exosome Secretion Evokes Epithelium–Mesenchyme Dysmorphogenesis
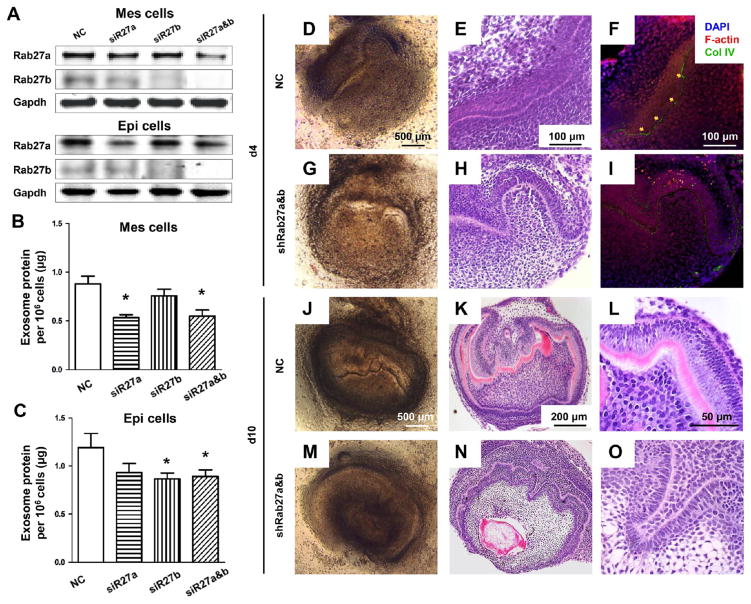
Given that exosomes reciprocally evoke epithelium and mesenchyme functions, we then tested whether attenuated exosomal communication induces dysmorphogenesis. The isolated E16.5 dental epithelium and mesenchyme (Figure S2A), when reconstituted in organ culture (Figure S2B and C), synthesized basement membrane by day 2 (Figure S2D). By day 12, a tooth organ formed (Figure S2E) with polarized ameloblasts and odontoblasts. Using this model, knockdown of rab27a/b, members of the Rab family of GTPase,27 by transfecting with Lipofectamine 2000 into the tooth germ (Figure S3), reduced exosomal secretion by ~20–40% in epithelium and mesenchyme cells (Figure 5A–C). By day 4, compared to control group (NC) (Figure 5D–F), Rab27a/b knockdown disrupted epithelially derived basement membrane component formation such as collagen type IV (Figure 5G–I) and attenuated mesenchymally derived dentinogenesis (Figure 5J–O) by day 10. We also tested GW4869 effects, a small-molecule inhibitor that attenuates exosomal secretion through inhibition of ceramide synthesis.28 Proliferation rates of mesenchyme cells were tested and did not decreased with GW4869 presence in the observed 12 days (Figure S4A). Exosomal protein secretion decreased to ~70% (Figure S4B and C) at 10 μg/mL in dental mesenchyme and epithelium cells. The reconstituted native epithelium and mesenchyme formed a tissue mass, with a basement membrane-like structure that was positive for Col IV (Figure S4D–F). Contrastingly, GW4869 attenuated basement membrane formation with a virtual absence of Col IV (Figure S4G–I). The reconstituted tooth germ continued to develop and had formed a tooth crown-like structure with formation of a dentin-like structure by day 10 (Figure S5J–L). In the GW4869-treated groups, ameloblasts and odontoblasts were well polarized but with no obvious matrix formation (Figure S5M–O).
Figure 5.
Attenuated exosome secretion evokes epithelium–mesenchyme dysmorphogenesis. (A) Rab27a and Rab27b expression in mesenchyme (Mes) and epithelium (Epi) cells following silencing of Rab27a and/or Rab27b. (B, C) Total exosomal proteins from stably transfected Rab27a and/or Rab27b per million Mes and Epi cells (mean ± SD; four independent experiments). *P < 0.05 (one-way ANOVA and LSD test). (D, E, G, H) Epithelium and mesenchyme in E16.5 tooth germs were microdissected and reconstituted/cultured for 4 days with scramble (NC) or Rab27a/b knockdown. (F, I) Fluorescence of F-actin and collagen IV (Col IV) in basement membrane (arrows). (J, M) Reconstituted tooth germs cultured for 10 days with scramble (NC) or Rab27a/b knockdown. Dentin formation in control (K, L) but not in Rab27a/b knockdown group (N, O).
Development Deficiency in Rab27aash/ash Mutation Mouse
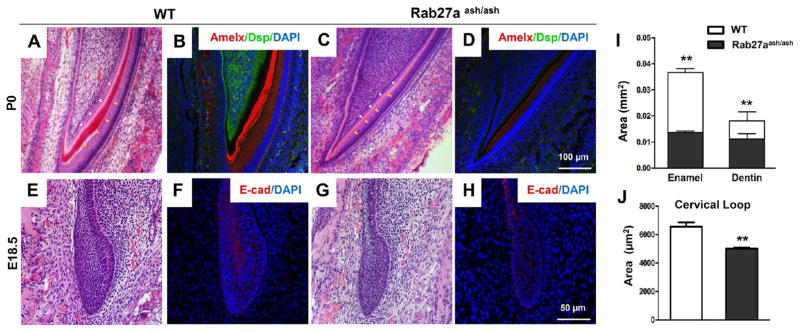
To gain further insight into the exosome function, the Rab27aash/ash mutation mice tooth architecture was investigated. Rab27aash/ash mutant mice, which are known for disrupted cellular vesicle transportation,29,30 showed consistently thinner enamel and dentin (Figure 6A and C), in addition to attenuated amelogenin (Amelx) and Dsp (Figure 6B and D). At E18.5, Rab27aash/ash mutation reduced both the size of the cervical loop (Figure 6E and G), which harbors epithelium stem cells responsible for enamel formation, and E-cadherin (E-cad) expression (Figure 6F and H), which likely account for the underdeveloped enamel and dentin in the reconstituted tooth organ (Figure 5J–O). Thus, exosomes appear to be important, if not indispensable, for epithelium–mesenchyme interactions in organ development.
Figure 6.
Development deficiency in Rab27aash/ash mutation mouse. (A, C) HE staining of postnatal day 0 (P0) incisor of wild-type (WT) and Rab27aash/ash mice. Amelogenin (Amgn) (red) and Dsp (green) expression in WT incisor (B) but with reduced expression in Rab27aash/ash incisor (D), with enamel and dentin area quantified (I). HE staining of the cervical loop of E18.5 incisor tooth organ in WT (E) and Rab27aash/ash (G) mice, with E-cadherin (E-cad) (red) expression in WT (F) and Rab27aash/ash (H) mice, and quantified cervical loop area (J) (mean ± SD; five independent experiments, *P < 0.05, **P < 0.01, independent t test).
Profiling of Exosomal Constituents and miR135a
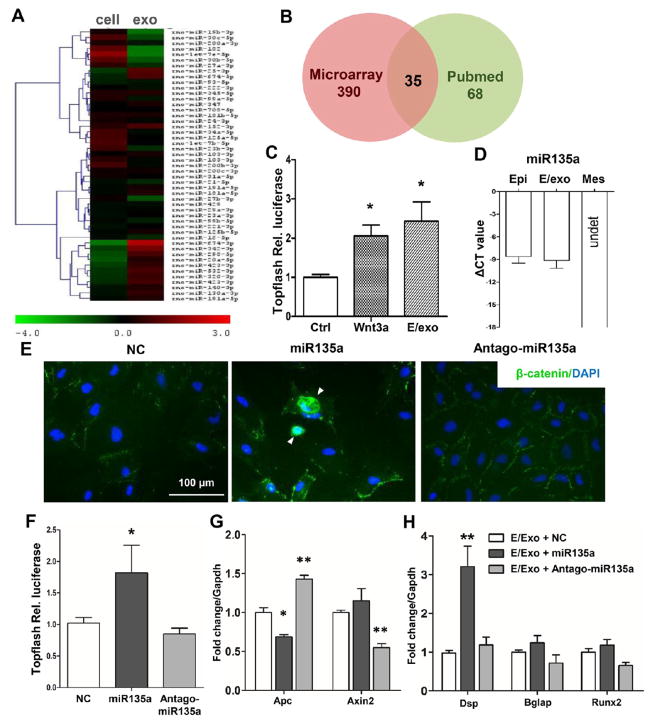
Mass spectrometry revealed 104 and 60 proteins in epithelium and mesenchyme exosomes, respectively (Figure S5 and Table S1). We further profiled exosomal miRNA and found a total of 390 miRNAs in epithelium exosomes (Figure S6A–C) and, as exemplified in Figure 7A, and 385 miRNAs in mesenchyme exosomes (Figure S6D–F) by miRCURY LNA array. To understand pivotal developmental signals that may serve as postnatal diagnostic and/or therapeutic cues, we focused on Wnt signaling given its recently illustrated roles in tooth development.31,32 Mesenchyme cells indeed escalated their Wnt activity (TOPflash luciferase) when treated with epithelium exosomes relative to the control but statistically similar to 50 ng/mL exogenous Wnt3a, a canonical Wnt ligand (Figure 7C). By crossing all 390 epithelium exosome miRNAs with 68 known Wnt-regulating miRNAs in the literature, we identified 35 miRNAs in epithelium exosomes that may regulate Wnt activities (Figure 7B). Accordingly, we performed a bioinformatics analysis and first focused on miR135a. Remarkably, miR135a showed a robust presence in epithelium exosomes, virtually the same as in epithelium cells, but was undetectable in mesenchyme cells (Figure 7D). miR135a transfection in mesenchyme cells induced β-catenin transnucleation (Figure 7E) relative to the control (NC), whereas Antago-miR135a largely abolished β-catenin transnucleation. miR135a further upregulated WNT activity (TOPflash luciferase) in mesenchyme cells, with effects neutralized by Antago-miR135a (Figure 7F). We also tested WNT signaling target gene Axin2 and adenomatous polyposis coli (APC). It showed that Axin2 was upregulated with miR135a stimulation, while APC expression decreased when miR135a was transfected into epithelium-derived exosomes (Figure 7G). Overexpressing miR135a in dental mesenchyme cells upregulated dentin sialoprotein, a key transcriptional factor for dentinogenesis, with the effect neutralized by Antago-miR135a (Figure S7). Additionally, incubation of mesenchyme cells with epithelium-derived exosomes transfected with miR135a or antagonist had virtually the same effects as on DSP expression (Figure 7H) as miR135 treatment. Thus, epithelium exosomes induce the differentiation of mesenchyme tissue, in the absence of epithelium cells, potentially by activating pivotal signaling pathways such as Wnt/β-catenin.
Figure 7.
Profiling of exosomal constituents. (A) Heat map of microRNA profiles differentially expressed by epithelium cells and exosomes (exo). (B) A total of 35 miRNAs were identified by crossing all 390 miRNAs of our microarray with known miRNAs that regulate Wnt signaling (68). (C) Relative luciferase activity of Topflash reporter with PBS (Ctrl), Wnt3a (50 ng/mL), or 10.0 μg/mL epithelium exosomes (E/exo) (mean ± SD; five independent experiments). (D) miR135a expression in epithelium cells (Epi), epithelium exosomes (E/exo), and mesenchyme cells (Mes). (E) β-Catenin staining (green) of mesenchyme cells transfected with miRNA mimic, miR135a, or Antago-miR135a. Arrowheads indicate β-catenin transnucleation. (F) Relative luciferase activity of the Topflash reporter of mesenchymal cells transfected with miRNA mimic, miR135a, or Antago-miR135a (mean ± SD; seven independent experiments). (G, H) mRNA expression of Apc, Axin2, Dsp, Bglap, and Runx2 of mesenchymal cells incubated with epithelium exosomes (E/exo) that are transfected with miRNA mimic, miR135a, or Antago-miR135a (mean ± SD; four independent experiments). *P < 0.05, **P < 0.01 (one-way ANOVA and LSD test).
Signaling pathways during organogenesis is mediated by miRNAs to affect protein synthesis and/or mRNA stability.33 Exosomal miRNAs regulate several pluripotency factors in tissue repair and oncogenesis. Our presently observed mediation of miR135a, which is derived from epithelium exosomes but virtually absent in mesenchyme exosomes, mediates dentin sialoprotein via Wnt/β-catenin signaling, suggesting that miRNA may affect the target cells and induce them to perform certain cellular functions. This brings an interesting point of whether single exosomal constituents, such as microRNAs or peptides, can induce certain cellular functions in lieu of the entire molecular payload in the exosomes. We show that both exosomes and miR135a act to stimulate mesenchyme matrix synthesis by activating dentin sialoprotein via the Wnt/β-catenin pathway. Recent data showing mediation of salivary epithelial progenitor cell expansion by exosomal microRNA from mesenchyme34 provide additional support of our finding that exosomes from either the epithelium or mesenchyme appear to have broader effects on matrix synthesis and cell differentiation. In the case where a single exosomal constituent can trigger a developmental or regenerative process, exosomal molecules may serve as putative therapeutic targets for correcting diseases or orchestrating tissue regeneration. One of the perceived benefits of exosome delivery, rather than individual molecular ligands, is that exosomes carry a cargo of multiple molecules that may collectively act on effector cells. Extracellular vesicles have been purified and preferentially endocytosed by organ-specific cells such as resident fibroblasts and epithelial and endothelial cells in multiple organs.35,36
CONCLUSIONS
Our findings elucidate not only the presence of exosomes in the epithelium and mesenchyme but also their roles in mediating epithelial and mesenchymal cell functions. Exosomal trafficking between epithelium and mesenchyme through the basement membrane appears purposeful and preferential by reciprocal cell types, leading to specifically induced cell functions including the presently studied cell differentiation and matrix synthesis. Reciprocal exosomal functions, in lieu of epithelium or mesenchyme cells, suggest that exosomes may augment or substitute classic morphogenetic interactions by cell–cell contact and protein–receptor interactions.
METHODS
Animals
Following Columbia University IACUC approval, postnatal 5/6-day-old Sprague–Dawley rats and CD1 mice were used. C3H/HeSn-ash/ash mice were purchased from Jackson Laboratory and propagated.
Exosome Collection Including Nanoparticle Tracing Analysis
Extracellular vehicles were isolated from incisor epithelium and mesenchyme cells of 5/6-day-old Sprague–Dawley rats. Briefly, the epithelium with intact cervical loop was carefully separated from mesenchyme tissue under a dissection microscope per our prior methods.14 Epithelium and mesenchyme stem/progenitor cells were digested and cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% exosome-free serum (System Biosciences, Mountain View, CA, USA) and 1% PS (Gibco). Supernatants were collected after 5–7 days. Upon removing nonadherent cells and debris, extracellular vesicles were further purified by ultracentrifugation or a total exosome isolation kit (Invitrogen, Carlsbad, CA, USA). Ultracentrifugation was performed as the following step.37 Cell culture supernatant was centrifuged at 300g for 10 min. Supernatant was collected and centrifuged at 2000g for 10 min, followed by centrifugeation at 10000g for 60 min. The final supernatant is then ultracentrifuged (Beckman Coulter, USA) at 100000g for 70 min. The pellet was washed in a large volume of phosphate-buffered saline (PBS) to eliminate contamination of proteins and centrifuged at 100000g for 70 min. The collected dental epithelium and mesenchyme vesicles were resuspended in PBS and characterized by NanoSight LM10 (Particle Characterization Laboratories, Novato, CA, USA).
Electron Microscopy
Exosomes were fixed in 2% paraformaldehyde, washed, and loaded onto Formvar-carbon-coated grids. After washing, exosomes were postfixed in 2% glutaraldehyde for 2 min, washed, and contrasted in 2% phosphotungstic acid for 5 min. Samples were washed, dried, and examined by an electron microscope (JEM-1400, Japan).
Histology
Tissues were fixed for 24 h in 4% paraformaldehyde, demineralized in 0.5 M EDTA, dehydrated, embedded in paraffin, and serially sectioned at 4 μm.
Immunofluorescence
Tissue sections were incubated with anti-CD63 (1:200, Santa Cruz Biotechnology, Santa Cruz, CA, USA). Tissue sections from Rab27aash/ash mutant mice were incubated with anti-DSP (1:100, Santa Cruz) and anit-amelogenin (1:200, Santa Cruz). Secondary antibodies, Alexa Fluor 647 donkey anti-rabbit IgG (H+L) antibody (1:2000, Invitrogen) or Alexa Fluor 488 donkey anti-goat IgG (H+L) antibody (1:2000, Invitrogen), were applied for 60 min at room temperature. Samples were sealed with Vecta shield mounting medium containing DAPI. Images were taken by using a Nikon A1 Confocal or Leica DMI6000B.
Western Blot
To measure exosome proteins, total proteins extracted from cell lysates and secreted extracellular vesicles were separated by SDS-PAGE, transferred to a nitrocellulose membrane (Millipore), and blotted with anti-CD63 antibodies (1:500, Santa Cruz Biotechnology) and GM130 (1:250, BD Bioscience, San Jose, CA, USA).
To measure endogenous proteins, cell lysate proteins (10 μg) were separated by a 4–12% SDS polyacrylamide gel. The proteins were extracted by RIPA buffer (Thermo Scientific, Rockford, IL, USA), transferred to a nitrocellulose or PVDF membrane (Invitrogen), and immunoblotted using primary antibodies against ameloblastin (1:200, Santa Cruz Biotechnology), amelogenin (1:200, Santa Cruz Biotechnology), Col IV (1:400, Abcam, Cambridge, MA, USA), laminin (1:400, abcam), DSP (1:200, Santa Cruz), Bglap (1:400, Millipore), RunX2 (1:500, abcam), Rab27A (1:200, abcam), Rab27B (1:400, abcam), β-catenin (1:500, abcam), lamin B (1:500, Santa Cruz), and Gapdh (1:500, Santa Cruz).
IRDye 800CW secondary antibodies (1:10 000, LI-COR, Lincoln, NE, USA) were applied for 60 min. The signals were detected by an Odyssey Imaging System (LI-COR).
Transient Transfection and Luciferase Assay
Exosomes were transfected with Cy3-labeled siRNA, mirVana miRNA mimic, miR135a, or miR135 inhibitor (Invitrogen) in electroporation buffer38 at 1200 mV and 20 ms one pulse time. The transfected epithelium and mesenchyme exosomes with Cy3-labeled siRNA were cross-incubated with mesenchyme and epithelium cells for 12 h followed by imaging using a Leica DMI6000B.
For the luciferase assay, mesenchyme cells were transfected by electroporation following the Neon transfection protocol. Briefly, one million cells were resuspended and mixed with 6 μg of Topflash plasmid (Millipore) and 20 ng of reporter plasmid (pGL4). After 24 h, cells transfected with mirVana miRNA mimic, miR135a, or miR135 inhibitor by Lipofectamine RNAiMAX (Invitrogen) were cultured for 24–36 h, lysed, and assayed for reporter activity. Epithelium exosomes with miRNA mimic, miR135a, or miR135 Inhibitors were cocultured with mesenchyme cells for 24–36 h in exosome-free medium. Luciferase was measured using the dual luciferase reporter assay (Promega, Madison, WI, USA).
Endocytosis Inhibition Studies
To study exosome endocytosis pathways, epithelial or mesenchymal cells were preincubated with inhibitors from Sigma-Aldrich.39 CPZ (10 μM) or LY294002 (50 μM) was applied to pretreat cells at 37 °C for 30 min. Nystatin (5 μM) was applied to cells for 120 min and washed excessively prior to exosome addition. The same amount of dimethyl sulfoxide (DMSO) was added into cells as a control. For labeling, the 2.0 μg/mL exosome solution was incubated with PKH67. The unincorporated dyes were neutralized with exosome-free medium and washed in PBS. Labeled exosomes were added into cells after inhibitor treatment and subsequently incubated at 37 °C for 6 h. After being fixed by 2% paraformaldehyde and stained with Dapi for 15 min, cells were imaged by confocal microsopy (Carl Zeiss LSM 710). To quantify the cellular uptake of exosomes, cell numbers with positive-labeled exosome signals were counted and total cell numbers were determined by counting the nuclei.
qRT-PCR
Total RNA was isolated using TRIzol. Complementary DNA (cDNA) was synthesized following the Bio-Rad protocol. PCR amplification was performed using the TaqMan protocol.
Cell Differentiation
For odontogenic/osteogenic differentiation, dental mesenchyme cells were cultured in DMEM with 10% fetal bovine serum (FBS), 5 mM β-glycerophosphate, 50 mM L-ascorbic acid 2-phosphate, 10 nM dexamethasone, and 1% PS. In 7 days, alkaline phosphatase was stained with alkaline phosphatase staining kit II (Stemgent, Cambridge, MA, USA) and quantified by a detection kit (Sigma-Aldrich, St. Louis, MO, USA). In 14 days, cells were fixed and stained with 1% Alizarin red-S (Sigma-Aldrich) to detect mineral nodules. Total RNA and protein were extracted from at least three independent samples by qRT-PCR and Western blot.
Tooth Organ Reconstitution
Mandibular first molar tooth germs from E16.5 (vaginal plug = day 0.5) CD1 mouse embryos were harvested under a dissection microscope. Epithelium and mesenchyme were treated with 50 U/mL Dispase I (BD, Franklin Lakes, NJ, USA) for 10.5 min and mechanically separated.25 The separated epithelium and mesenchyme were reconstituted in a modified Trowell-type organ culture in a 12-well 8.0 μm Transwell (BD Biosciences) and grown at the medium–gas interface, with culture medium consisting of DMEM (Gibco) with 10% heat-inactived serum (Gibco), 1% Gluta-Max (Gibco), and 0.2% PS (Gibco). F12 was used as a 1:1 mixture in culture medium. Ascorbic acid (100 μg/mL, Sigma) was added to the culture medium to allow collagen deposition.40 The culture plates were kept in a humidified incubator at 37 °C in an atmosphere of 5% CO2 for 12 days, with medium change every 2 days.
GW4869 (10 μM) was added and replenished every 2 days. For transfection of RNAi, Cy3-labeled RNAi or siRab27a/b was directly pipetted to the reconstituted tooth germ. The reconstituted tooth germs were harvested at multiple time points.41
Mass Spectrometry
Total proteins were extracted separately from epithelium and mesenchyme exosomes. The lysates were separated using 4–12% SDS-PAGE gel. The gel was stained with a silver stain kit (Biorad, Hercules, CA, USA) and then analyzed by mass spectrometry.
MicroRNA Array and Profiling
Small RNA was isolated per the PureLink miRNA isolation kit (Invitrogen). The quality of total RNA was verified by an Agilent 2100 bioanalyzer profile. The miRNA identification was performed by Exiqon (Exiqon, Woburn, MA, USA). Briefly, all microRNAs were polyadenylated and reverse transcribed into cDNA in a single reaction step. cDNA and SYBR Green Mastermix was transferred to qPCR panels preloaded with primers. Amplification was performed in a Roche Lightcycler480.
Data Analysis and Statistics
All quantitative data were analyzed by using ImageJ (http://rsbweb.nih.gov/ij/download.html). Upon confirmation of normal data distribution, data were treated with one-way ANOVA with LSD tests or independent t tests with an α level of 0.05.
Supplementary Material
Acknowledgments
We thank Q. Guo, P. Ralph-Birkett, and Y. Tse for administrative and technical assistance. This work is supported by Beijing Natural Science Foundation (7174365), National Natural Science Foundation of China (81600820), NIH grants R01DE025643, R01DE023112, R01AR065023, and R34DE026645 to J.J.M., a Guangdong Pioneer Grant (52000-52010002), and the Guangdong Science and Technology Program (2016B030229003).
Footnotes
Author Contributions
N.J. was responsible for the technical design and primary technical undertaking, conducted the experiments, collected and analyzed data, and drafted the manuscript. L.X., L.H., G.Y., J.Z., C.W., Y.Z., S.W., Y.Z., T.-J.S., and S.H.K. performed and/or assisted in molecular assays. J.W. and K.C. assisted in microarray data analysis. P.G.C. and N.T. assisted in histology imaging. J.J.M., Y.Z., and M.C. conceived and designed the experiments, oversaw the collection of results and data interpretation, and wrote the manuscript.
The authors declare no competing financial interest.
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsnano.7b01087.
Supporting data showing endocytosis pathways, organogenesis of E16.5 tooth germs with GW4869 treatment, mass spectrometry, and miRNA arrays (PDF)
References
- 1.Pan BT, Johnstone RM. Fate of the Transferrin Receptor During Maturation of Sheep Reticulocytes in Vitro: Selective Externalization of the Receptor. Cell. 1983;33:967–78. doi: 10.1016/0092-8674(83)90040-5. [DOI] [PubMed] [Google Scholar]
- 2.Record M, Carayon K, Poirot M, Silvente-Poirot S. Exosomes as New Vesicular Lipid Transporters Involved in Cell-Cell Communication and Various Pathophysiologies. Biochim Biophys Acta, Mol Cell Biol Lipids. 2014;1841:108–120. doi: 10.1016/j.bbalip.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 3.Raposo G, Nijman HW, Stoorvogel W, Liejendekker R, Harding CV, Melief CJ, Geuze HJ. B Lymphocytes Secrete Antigen-Presenting Vesicles. J Exp Med. 1996;183:1161–72. doi: 10.1084/jem.183.3.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoshino D, Kirkbride KC, Costello K, Clark ES, Sinha S, Grega-Larson N, Tyska MJ, Weaver AM. Exosome Secretion Is Enhanced by Invadopodia and Drives Invasive Behavior. Cell Rep. 2013;5:1159–1168. doi: 10.1016/j.celrep.2013.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ridder K, Keller S, Dams M, Rupp AK, Schlaudraff J, Del Turco D, Starmann J, Macas J, Karpova D, Devraj K, Depboylu C, Landfried B, Arnold B, Plate KH, Hoglinger G, Sultmann H, Altevogt P, Momma S. Extracellular Vesicle-Mediated Transfer of Genetic Information between the Hematopoietic System and the Brain in Response to Inflammation. PLoS Biol. 2014;12:e1001874. doi: 10.1371/journal.pbio.1001874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tummers M, Thesleff I. Root or Crown: A Developmental Choice Orchestrated by the Differential Regulation of the Epithelial Stem Cell Niche in the Tooth of Two Rodent Species. Development. 2003;130:1049–57. doi: 10.1242/dev.00332. [DOI] [PubMed] [Google Scholar]
- 7.Harada H, Kettunen P, Jung HS, Mustonen T, Wang YA, Thesleff I. Localization of Putative Stem Cells in Dental Epithelium and Their Association with Notch and Fgf Signaling. J Cell Biol. 1999;147:105–120. doi: 10.1083/jcb.147.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Saxen L, Thesleff I. Epithelial-Mesenchymal Interactions in Murine Organogenesis. Ciba Foundation symposium. 1992;165:183–93. doi: 10.1002/9780470514221.ch11. discussion 193–8. [DOI] [PubMed] [Google Scholar]
- 9.Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Molecular Biology of the Cell. 4. Garland Science; New York: 2002. General Principles of Cell Communication. [Google Scholar]
- 10.Klein OD, Lyons DB, Balooch G, Marshall GW, Basson MA, Peterka M, Boran T, Peterkova R, Martin GR. An Fgf Signaling Loop Sustains the Generation of Differentiated Progeny from Stem Cells in Mouse Incisors. Development. 2008;135:377–85. doi: 10.1242/dev.015081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang XP, Suomalainen M, Felszeghy S, Zelarayan LC, Alonso MT, Plikus MV, Maas RL, Chuong CM, Schimmang T, Thesleff I. An Integrated Gene Regulatory Network Controls Stem Cell Proliferation in Teeth. PLoS Biol. 2007;5:e159. doi: 10.1371/journal.pbio.0050159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Connell DJ, Ho JW, Mammoto T, Turbe-Doan A, O’Connell JT, Haseley PS, Koo S, Kamiya N, Ingber DE, Park PJ, Maas RL. A Wnt-Bmp Feedback Circuit Controls Intertissue Signaling Dynamics in Tooth Organogenesis. Sci Signaling. 2012;5:ra4. doi: 10.1126/scisignal.2002414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thesleff I, Lehtonen E, Wartiovaara J, Saxen L. Interference of Tooth Differentiation with Interposed Filters. Dev Biol. 1977;58:197–203. doi: 10.1016/0012-1606(77)90085-9. [DOI] [PubMed] [Google Scholar]
- 14.Jiang N, Zhou J, Chen M, Schiff MD, Lee CH, Kong K, Embree MC, Zhou Y, Mao JJ. Postnatal Epithelium and Mesenchyme Stem/Progenitor Cells in Bioengineered Amelogenesis and Dentinogenesis. Biomaterials. 2014;35:2172–80. doi: 10.1016/j.biomaterials.2013.11.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.SELA, Mager I, Breakefield XO, Wood MJ. Extracellular Vesicles: Biology and Emerging Therapeutic Opportunities. Nat Rev Drug Discovery. 2013;12:347–57. doi: 10.1038/nrd3978. [DOI] [PubMed] [Google Scholar]
- 16.Delenclos M, Trendafilova T, Mahesh D, Baine AM, Moussaud S, Yan IK, Patel T, McLean PJ. Investigation of Endocytic Pathways for the Internalization of Exosome-Associated Oligomeric Alpha-Synuclein. Front Neurosci. 2017;11 doi: 10.3389/fnins.2017.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Watanabe S, Trimbuch T, Camacho-Perez M, Rost BR, Brokowski B, Sohl-Kielczynski B, Felies A, Davis MW, Rosenmund C, Jorgensen EM. Clathrin Regenerates Synaptic Vesicles from Endosomes. Nature. 2014;515:228–33. doi: 10.1038/nature13846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mulcahy LA, Pink RC, Carter DR. Routes and Mechanisms of Extracellular Vesicle Uptake. J Extracell Vesicles. 2014;3:24641. doi: 10.3402/jev.v3.24641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Conner SD, Schmid SL. Regulated Portals of Entry into the Cell. Nature. 2003;422:37–44. doi: 10.1038/nature01451. [DOI] [PubMed] [Google Scholar]
- 20.Khalil IA, Kogure K, Akita H, Harashima H. Uptake Pathways and Subsequent Intracellular Trafficking in Nonviral Gene Delivery. Pharmacol Rev. 2006;58:32–45. doi: 10.1124/pr.58.1.8. [DOI] [PubMed] [Google Scholar]
- 21.Rana S, Yue S, Stadel D, Zoller M. Toward Tailored Exosomes: The Exosomal Tetraspanin Web Contributes to Target Cell Selection. Int J Biochem Cell Biol. 2012;44:1574–84. doi: 10.1016/j.biocel.2012.06.018. [DOI] [PubMed] [Google Scholar]
- 22.Feng D, Zhao WL, Ye YY, Bai XC, Liu RQ, Chang LF, Zhou Q, Sui SF. Cellular Internalization of Exosomes Occurs through Phagocytosis. Traffic. 2010;11:675–687. doi: 10.1111/j.1600-0854.2010.01041.x. [DOI] [PubMed] [Google Scholar]
- 23.Fitzner D, Schnaars M, van Rossum D, Krishnamoorthy G, Dibaj P, Bakhti M, Regen T, Hanisch UK, Simons M. Selective Transfer of Exosomes from Oligodendrocytes to Microglia by Macropinocytosis. J Cell Sci. 2011;124:447–458. doi: 10.1242/jcs.074088. [DOI] [PubMed] [Google Scholar]
- 24.Abels ER, Breakefield XO. Introduction to Extracellular Vesicles: Biogenesis, Rna Cargo Selection, Content, Release, and Uptake. Cell Mol Neurobiol. 2016;36:301–12. doi: 10.1007/s10571-016-0366-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thesleff I, Lehtonen E, Saxen L. Basement Membrane Formation in Transfilter Tooth Culture and Its Relation to Odontoblast Differentiation. Differentiation. 1978;10:71–9. doi: 10.1111/j.1432-0436.1978.tb00948.x. [DOI] [PubMed] [Google Scholar]
- 26.Miyazaki T, Kanatani N, Rokutanda S, Yoshida C, Toyosawa S, Nakamura R, Takada S, Komori T. Inhibition of the Terminal Differentiation of Odontoblasts and Their Transdifferentiation into Osteoblasts in Runx2 Transgenic Mice. Arch Histol Cytol. 2008;71:131–46. doi: 10.1679/aohc.71.131. [DOI] [PubMed] [Google Scholar]
- 27.Ostrowski M, Carmo NB, Krumeich S, Fanget I, Raposo G, Savina A, Moita CF, Schauer K, Hume AN, Freitas RP, Goud B, Benaroch P, Hacohen N, Fukuda M, Desnos C, Seabra MC, Darchen F, Amigorena S, Moita LF, Thery C. Rab27a and Rab27b Control Different Steps of the Exosome Secretion Pathway. Nat Cell Biol. 2010;12:19–U61. doi: 10.1038/ncb2000. [DOI] [PubMed] [Google Scholar]
- 28.Trajkovic K, Hsu C, Chiantia S, Rajendran L, Wenzel D, Wieland F, Schwille P, Brugger B, Simons M. Ceramide Triggers Budding of Exosome Vesicles into Multivesicular Endosomes. Science. 2008;319:1244–1247. doi: 10.1126/science.1153124. [DOI] [PubMed] [Google Scholar]
- 29.Tolmachova T, Anders R, Stinchcombe J, Bossi G, Griffiths GM, Huxley C, Seabra MC. A General Role for Rab27a in Secretory Cells. Molecular biology of the cell. 2004;15:332–44. doi: 10.1091/mbc.E03-07-0452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilson SM, Yip R, Swing DA, O’Sullivan TN, Zhang Y, Novak EK, Swank RT, Russell LB, Copeland NG, Jenkins NA. A Mutation in Rab27a Causes the Vesicle Transport Defects Observed in Ashen Mice. Proc Natl Acad Sci U S A. 2000;97:7933–8. doi: 10.1073/pnas.140212797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lim WH, Liu B, Cheng D, Hunter DJ, Zhong Z, Ramos DM, Williams BO, Sharpe PT, Bardet C, Mah SJ, Helms JA. Wnt Signaling Regulates Pulp Volume and Dentin Thickness. J Bone Miner Res. 2014;29:892–901. doi: 10.1002/jbmr.2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suomalainen M, Thesleff I. Patterns of Wnt Pathway Activity in the Mouse Incisor Indicate Absence of Wnt/Beta-Catenin Signaling in the Epithelial Stem Cells. Dev Dyn. 2010;239:364–72. doi: 10.1002/dvdy.22106. [DOI] [PubMed] [Google Scholar]
- 33.Eulalio A, Huntzinger E, Izaurralde E. Getting to the Root of Mirna-Mediated Gene Silencing. Cell. 2008;132:9–14. doi: 10.1016/j.cell.2007.12.024. [DOI] [PubMed] [Google Scholar]
- 34.Hayashi T, Lombaert IM, Hauser BR, Patel VN, Hoffman MP. Exosomal Microrna Transport from Salivary Mesenchyme Regulates Epithelial Progenitor Expansion During Organogenesis. Dev Cell. 2017;40:95–103. doi: 10.1016/j.devcel.2016.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zitvogel L, Regnault A, Lozier A, Wolfers J, Flament C, Tenza D, Ricciardi-Castagnoli P, Raposo G, Amigorena S. Eradication of Established Murine Tumors Using a Novel Cell-Free Vaccine: Dendritic Cell-Derived Exosomes. Nat Med. 1998;4:594–600. doi: 10.1038/nm0598-594. [DOI] [PubMed] [Google Scholar]
- 36.Faure J, Lachenal G, Court M, Hirrlinger J, Chatellard-Causse C, Blot B, Grange J, Schoehn G, Goldberg Y, Boyer V, Kirchhoff F, Raposo G, Garin J, Sadoul R. Exosomes Are Released by Cultured Cortical Neurones. Mol Cell Neurosci. 2006;31:642–8. doi: 10.1016/j.mcn.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 37.Thery C, Amigorena S, Raposo G, Clayton A. Isolation and Characterization of Exosomes from Cell Culture Supernatants and Biological Fluids. Current Protocols in Cell Biology. 2006;Chapter 3(Unit 3):22. doi: 10.1002/0471143030.cb0322s30. [DOI] [PubMed] [Google Scholar]
- 38.El-Andaloussi S, Lee Y, Lakhal-Littleton S, Li J, Seow Y, Gardiner C, Alvarez-Erviti L, Sargent IL, Wood MJ. Exosome-Mediated Delivery of Sirna in Vitro and in Vivo. Nat Protoc. 2012;7:2112–26. doi: 10.1038/nprot.2012.131. [DOI] [PubMed] [Google Scholar]
- 39.Tian T, Zhu YL, Zhou YY, Liang GF, Wang YY, Hu FH, Xiao ZD. Exosome Uptake through Clathrin-Mediated Endocytosis and Macropinocytosis and Mediating Mir-21 Delivery. J Biol Chem. 2014;289:22258–22267. doi: 10.1074/jbc.M114.588046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Munne PM, Narhi K, Michon F. Analysis of Tissue Interactions in Ectodermal Organ Culture. Methods Mol Biol. 2012;945:401–16. doi: 10.1007/978-1-62703-125-7_24. [DOI] [PubMed] [Google Scholar]
- 41.Narhi K, Thesleff I. Explant Culture of Embryonic Craniofacial Tissues: Analyzing Effects of Signaling Molecules on Gene Expression. Methods Mol Biol. 2010;666:253–67. doi: 10.1007/978-1-60761-820-1_16. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.