Abstract
Introduction
For the relative quantification of isoform expression, RT-qPCR has been the gold standard for over a decade. More recently, digital PCR is becoming widely implemented, as it is promised to be more accurate, sensitive and less affected by inhibitors, without the need for standard curves. In this study we evaluated RT-qPCR versus RT-droplet digital PCR (ddPCR) for the relative quantification of isoforms in controls and carriers of the splice site mutation BRCA1 c.212+3A>G, associated with increased expression of several isoforms.
Materials and methods
RNA was extracted from EBV cell lines of controls and heterozygous BRCA1 c.212+3A>G carriers. Transcript-specific plasmids were available to determine the efficiency, precision, reproducibility and accuracy of each method.
Results
Both ddPCR and RT-qPCR were able to accurately quantify all targets and showed the same LOB, LOD and LOQ; also precision and reproducibility were similar. Both techniques have the same dynamic range and linearity at biologically relevant template concentrations. However, a significantly higher cost and workload was required for ddPCR experiments.
Conclusions
Our study recognizes the potential and validity of digital PCR but shows the value of a highly optimized qPCR for the relative quantification of isoforms. Cost efficiency and simplicity turned out to be better for RT-qPCR.
Keywords: Reverse transcriptase polymerase chain reaction, Alternative splicing, Droplet digital PCR
1. Introduction
In modern day genetics the concept of alternative splicing is frequently investigated. Alternative splicing is a naturally occurring mechanism that increases the protein coding complexity of the genome. With the formation of several transcripts from one locus the number of proteins that can be formed out of the 20,000 genes, which make out the human protein coding genome, increases tremendously [1]. Besides naturally occurring alternative splicing, alternative (aberrant) transcripts can also arise from mutations in the genome leading to the formation of new splice sites or the removal of existing ones [2], [3]. However, the interpretation of variants modifying alternative transcript ratios in combination with the induction of novel transcripts is less straightforward. Therefore, a first step towards understanding the pathogenicity of a variant, suspected to alter splicing, is accurate quantification of the naturally occurring transcripts together with the discovery and quantification of aberrant transcripts induced by the variant. In case of sufficient expression of normal, functional transcripts, the phenotypic effect of the deleterious variant might be minimal, because the remaining level of the normal transcript can maintain protein functionality [3].
For transcript-specific quantification of RNA RT-qPCR has been the gold standard for several years [4], [5]. Here, quantification with intercalating dyes is the simplest and most cost efficient solution, although this method is prone to detection of non-specific amplification, because the dye can intercalate non-specifically (in amplicons not of interest). Non-specific amplification is expected to be less of a problem when using isoform-specific hydrolysis probes. Here fluorescence is only measured when the probe can anneal to a specific target and subsequently gets cleaved during elongation. Accurate quantification of all targets can however be impaired if one or more transcripts are significantly overrepresented in the sample [5] and the design of the probes in a transcript-specific sequence may not always be feasible for each isoform in combination with the design of transcript-specific primers. This and the added cost of fluorescently labelled probes, are the major limitations for qPCR with probes. Theoretically the number of target amplicons doubles every cycle during a PCR reaction, making it possible to perform relative quantifications (by calculating relative differences in amplification of target between two samples or between targets in a single sample) or absolute quantifications (by calculating the absolute amount of target based on a standard curve of known quantities). To be able to compare quantities between samples, typically several stable reference genes are measured in parallel with the expression of the target for normalization [6], [7].
Recently, droplet digital PCR (ddPCR) is becoming a widely used alternative to qPCR for the quantification of nucleic acids in specific applications such as copy number quantification [8]. In ddPCR thousands of nanoliter scale droplets are generated, each containing none, one or a handful of target molecules. During PCR, each of these droplets acts as a separate reaction volume, amplifying the target. After amplification, each droplet is read out individually and droplets with a higher fluorescence than the threshold are deemed positive. Using Poisson distribution statistics, it is possible to estimate the starting concentration of target in each sample, allowing for both relative and absolute quantification at the same time without the need for a standard curve [9]. Therefore, at first sight, the calculation of target concentration is simpler with ddPCR than with qPCR and accuracy of quantification is not dependent on the accuracy of a standard curve. However, determining the threshold position for ambiguous droplets is a matter of debate and has an impact on the accurate quantification of the target. New ways to tackle this problem have been proposed [10], [11]. Most recently, a data-driven method was developed, that allows for threshold calculation using extreme value theory. According to Trypsteen et al. [12] this method is more accurate than its predecessors because it imposes no assumptions on the distribution of the data and it corrects for the baseline shift between no-template-controls (NTCs) and samples. Using an appropriate method for data analysis, ddPCR is potentially more precise than qPCR [10], [13].
The introduction of digital PCR raises the question if qPCR should remain the gold standard for quantifying alternative transcripts. Here, we make a comparison between qPCR and ddPCR for the relative quantification of such transcripts. Hereto, we use a deleterious BRCA1 mutation (c.212+3A>G) as a model for the evaluation of qPCR and ddPCR-based quantification. BRCA1 c.212+3A>G is a Belgian founder mutation associated with an increased risk for breast and ovarian cancer [14], [15]. The variant was shown to induce a shift in the ratio of naturally occurring isoforms. Three naturally occurring transcripts were identified at this locus. An isoform containing the full-length exon 5 (BRCA1-ex5FL; r.135_212), a transcript with a total skip of exon 5 (BRCA1-Δex5; r.135_212del) and a transcript where the last 22 nucleotides of exon 5 are deleted (BRCA1-Δ22ntex5; r.190_212del) [16]. Several other publications confirmed the expression of these isoforms in relation to this variant [17], [18]. ddPCR was evaluated in comparison to qPCR in terms of accuracy, linearity, dynamic range, precision and reproducibility for the quantification of transcript isoforms.
2. Methods and materials
2.1. Samples
For this study Epstein-Barr Virus (EBV) immortalized B cell lines derived from individuals with a germline BRCA1 c.212+3A>G mutation were used as carrier samples. For the control samples EBV cell lines were used from individuals not carrying a germline BRCA1/2 mutation as determined by screening the entire coding region of both genes. In total 6 carrier and 4 control samples were used. EBV cell lines were made according to Hui-Yuen et al. [19]. Approval for generation and usage of the EBV cell lines for fundamental research purposes was granted by the Gent University Hospital Ethical Committee and by the individuals whom these cell lines were derived from in the form of an informed consent. All experiments were done in accordance with the recommendations and restrictions set by the Gent University Hospital Ethical Committee in compliance with the WMA declaration of Helsinki regarding medical research on human subjects.
No nonsense mediated decay inhibitors were added to the cell cultures only stable transcripts were of interest in this study. For RNA extraction 1.5 × 106 cells were pelleted and resuspended in 3 mL culture medium (RPMI medium 1640, foetal bovine serum 10%, sodium pyruvate 1.11%, β-mercaptoethanol 0.11%, interleukin 2 0.11%, glutamine 1% and penicillin-streptomycin 0.5%; Thermo Fisher Scientific, Waltham, USA). Cells were counted using a Bürker counting chamber. From 1 mL RNA was extracted right away (0 h samples). 1 mL was left in culture for another 4 h (4 h samples) and another 1 mL for 24 h (24 h samples) before starting RNA extraction. RNA extraction was done using RNeasy mini kit (Qiagen, Hilden, Germany) according to manufacturer’s protocol (without the optional DNase treatment), after which RNA was measured on DropSense96 (Trinean, Gentbrugge, Belgium) and stored at −80 °C. Removal of contaminating gDNA was done prior to RT with Heat & Run DNA removal (ArticZymes, Tromsø, Norway) following the manufacturer’s instructions. For RT, the iScript cDNA Synthesis Kit (Bio-Rad, Temse, Belgium) was used in compliance with the manufacturer’s recommendations. DNase treatment and RT were done as consecutive steps on the same batch of 1 μg of total RNA.
A number of dilution series were constructed. Dilutions were made from either EBV derived samples (controls or BRCA1 c.212+3A>G carriers) or transcript-specific plasmids. In total 11 dilution points were made and a no template control. 5 ng/μL yeast tRNA carrier used in all dilutions. Plasmid series were diluted 1/10, EBV series 1 in 2. Template concentration in all dilution points was calculated from DropSense96 measurements of the undiluted sample. A 424 bp amplicon (spanning exons 2–7 of BRCA1) was amplified using cDNA from a patient heterozygous for BRCA1 c.212+3A>G as described before [16]. This amplicon was cloned in a pCR2.1-TOPO vector (Invitrogen, Carlsbad, USA). Individual clones were investigated through Sanger sequencing; three clones were retained, each containing one of the three transcripts under investigation. In all experiments these plasmids were used as circular molecules without enzymatic linearization.
2.2. Quality control
For both qPCR and ddPCR the same primer sets were used. Specificity of primers for all three assays was verified with qPCR on plasmid constructs, each containing a specific isoform (Supplementary Data A). Furthermore, specificity was also investigated for each individual qPCR reaction with melting curve analysis. All reactions contained only one detectable amplification product.
Quality control (QC) on cDNA and original RNA samples was done via qPCR using an artificial SPUD assay [20], an exon-spanning assay from exon 2–4 in MKNK2 (reference sequence NM_199054), which doesn’t yield a product on gDNA as the involved introns have a combined length of >4 kb, (160 bp, F: 5′-CCAGCCGAACTTCAGGGTTT-3′, R: 5′-CGTCCGGGATGTCAATGGG-3′; forward primer sequence in exon 2, reverse primer sequence in exon 4) and an intronic assay located between exon 30 and 31 in ABCA4 (bp, F: 5′-CCAAGCCTACCTACATGGTGT-3′, R: 5′-AGGGATCCCAAAAGAAGGAC-3′; both primers are entirely located within the same intron; reference sequence NM_000350). Amplification as described in the qPCR section.
2.3. Discovery of all relevant alternative isoforms
For the identification of alternative splicing transcripts at the region of interest a targeted next generation sequencing approach was devised. Primers (F: 5′-CTCCACAAAGTGTGACCACA-3′, R: 5′-CGGTTTCTGTAGCCCATACT-3′) spanning exons 3–7 of the BRCA1 gene were designed to pick up all isoforms in this region. Numbering of the nucleotides and exons according to reference sequence NM_007294.3. cDNA from all controls or carriers was pooled together in two separate pools. These pools provided the input material (10 ng, total RNA equivalents) for discovery of relevant isoforms. After PCR amplification (see qPCR section for details) for 32 cycles on a regular thermocycler, sequencing libraries were constructed using NEXTflex PCR-free DNA Sequencing (Illumina compatible, BiooScientific, Austin, USA) with a customization of the size selection range, exclusion of fragmentation and an additional amplification of the adapter-ligated library (10 cycles). The quality of each library was evaluated on Bioanalyzer (High Sensitivity DNA, Agilent, Santa Clara, USA) and quantification of libraries was done using KAPA Library Quantification Kit (KapaBiosystems, Wilmington, USA). Finally sequencing was done on a MiSeq (Illumina, San Diego, USA) instrument, yielding 150 bp bidirectional reads. Reads were mapped to the hg19 reference genome using TopHat2 (default settings). The sashimi plot function (MISO) integrated in IGV was used to visualise and count the splice junctions present in the gene of interest. RNA integrity was not explicitely investigated, because of the small genomic region under investigation and the small size of the amplicons of interest. The success of this targeted RNA sequencing approach indicates that the RNA integrity is sufficient for the planned PCR experiments.
2.4. qPCR experiments
qPCR reactions were executed using SsoAdvanced universal SYBR Green supermix (Bio-Rad) in 5 μL reactions in a 384-well format, containing 10 ng cDNA (total RNA equivalents) or variable amount for dilution series and 500 nM primer (SPUD template 0.5 pM). Amplification of the target was carried out on LC480 (Roche, Basel, Switzerland), after which melting curve analysis was done of the reaction products on the same machine. Denaturation for 2 min at 95 °C was followed by 45 cycles with 5 s at 95 °C, 60 °C for 30 s and 72 °C for 1 s. For melting curve analysis, temperature increased steadily to 95 °C. All synthetic nucleic acid oligomers in this study were synthesized by IDT (Coralville, USA) and checked for specificity at the design stage by primer-BLAST [21] and secondary structures using Mfold [22]. No isoform-specific probes were used in any of the qPCR reactions. In total three isoform-specific primer sets were used to assess the relative quantification of all three isoforms in each sample: BRCA1-ex5FL (81 bp, F:5′-CATGCTGAAACTTCTCAACCAGAA-3′, R:5′-CTTACTATATTGGTTTTCCTCGGATGT-3′), BRCA1-Δex5 (100 bp, F:5′-GCAGAAAATCTTAGAGTGTCCCATCT-3′, R:5′-AAACGTTCTCGGATGTTCTTTC ATGC-3′) and BRCA1-Δ22ntex5 (74 bp, F:5′-CATGCTGAAACTTCTCAACCAGA-3′, R:5′-AATACTCGGATGTTCTTTCATGCTCTAA-3′).
For the calculation of relative expression values (REijl), raw Cq values were first transformed to relative quantities (RQijl) similar to Hellemans et al. [23]. An average Cq () was calculated from the raw Cq values of all m replicates (k) of a sample/assay combination (i/j) on a qPCR plate (l). From here RQijl were calculated by transforming the difference, named △Cqijl, between each and the average Cq over all assays () using the assay specific PCR efficiency (Ej). All assays showed efficiency of nearly 100%, as determined in advance in accordance with Hellemans et al. [23]. The RQijlfor each sample/assay combination was used to determine the relative expression (REijl) of the target to the sum of BRCA1 expression.
| equation | error propagation |
|---|---|
2.5. Droplet digital PCR experiments
All ddPCR reactions were executed according to an optimized and standardized protocol and contained isoform-specific primers and hydrolysis probes for one of the BRCA1 isoforms and RPP30 reference target. Prior to this study the optimal annealing temperature was investigated for every primer-probe combination. BRCA1 primers were the same as for qPCR. BRCA1 probes were labelled with 6-FAM (BRCA1-ex5FL: 5′-/56-FAM/AAAGGGCCT/ZEN/TCACAGTGTCCTTTATGTAA/3IABkFQ/-3′, BRCA1-Δex5: 5′-/56-FAM/AGTTGATCA/ZEN/AGGAACCTGTCTCCACAAAGT/3IABkFQ/-3′ and BRCA1-Δ22ntex5: 5-/56-FAM/AGAAAGGGC/ZEN/CTTCACAGTGTCCT/3IABkFQ/-3′), while the RPP30 probe was HEX labelled (F: 5′-GATTTGGACCTGCGAGCG-3′, probe: 5′-/5HEX/TCTGACCTG/ZEN/AAGGCTCTGCGCG/3IABkFQ/-3′, R: 5′-GCGGCTGTCTCCACAAGT- 3′). On average 14,728 accepted droplets (range 11,842–17,227) were generated (QX100 Droplet Generator, Bio-Rad) from 20 μL mixtures containing 5 ng (or variable amount for dilution series) of cDNA (total RNA equivalents), 250 nM primers, 900 nM probes and 10 μL ddPCR Supermix for Probes (Bio-Rad) and were read out with a QX100 Droplet Reader (Bio-Rad). Mean copies per partition (λ) were dependent on sample-assay combinations, with an average over all ddPCR reactions of 2.35e-2, calculated using positive partitions as described. [24]
The PCR reaction was executed using a regular thermocycler with a 10 min denaturation step at 95 °C followed by repeated cycles consisting of 15 s at 95 °C and one min at 60 °C. After 40 cycles the products were heated to 98 °C for 10 min before cooling to room temperature.
Calling of positive and negative droplets and calculation of the target-concentration was done by Quantasoft software (Bio-Rad) and ddpcRquant webtool (http://www.ddpcrquant.ugent.be) [12]. Normalized concentration values () were obtained by dividing the average concentration of m replicates (k) for a particular sample/assay combination () with average concentration of the corresponding RPP30 measurements. REijl for all targets in a sample were calculated relative to the total expression of BRCA1 in the sample.
| equation | error propagation |
|---|---|
2.6. Statistical analysis
Data analysis, generation of figures and calculation of statistics for qPCR and ddPCR data was executed in the R language and environment for statistical computing [25]. Statistical significance for the comparison of controls and carriers tested by the Mann-Whitney test, while qPCR and ddPCR results were compared using the Wilcoxon signed-rank test. We adopted α = 0.001, making sure that we only claim there is a difference between the compared results with a very small margin of error. Coefficients of variation were calculated for each EBV cell line as the standard deviation between measurements of a cell line over all time points divided by the mean of these measurements. LOB, LOD and LOQ were defined based on Armbruster and Pry. [26]
3. Results
3.1. Quality control on RNA and cDNA samples
Total RNA was obtained from Epstein Barr Virus (EBV) cell lines, derived from both control individuals and BRCA1 c.212+3A>G carriers, and treated with HL-dsDNase to remove remaining genomic DNA (gDNA) before reverse transcription (RT) and quality control. Cells were harvested at three time points: right after (0 h), 4 h and 24 h after culture medium renewal. gDNA contamination was detected in all initial total RNA extracts by an intronic assay, no gDNA was measured in any sample after RT, proving the efficiency of the HL-dsDNase treatment. An exon-spanning assay (not allowing amplification on gDNA) showed no amplification on RNA samples before RT and clear amplification on cDNA samples, implying that RT was successfully executed. Finally all samples showed highly similar amplification with the SPUD assay [20], so it was concluded that none of the samples showed elevated inhibition compared to any of the other samples. Data of all quality control measures can be seen in Supplementary data B.
3.2. Discovery of transcript variants in the BRCA1 c.212+3A>G model
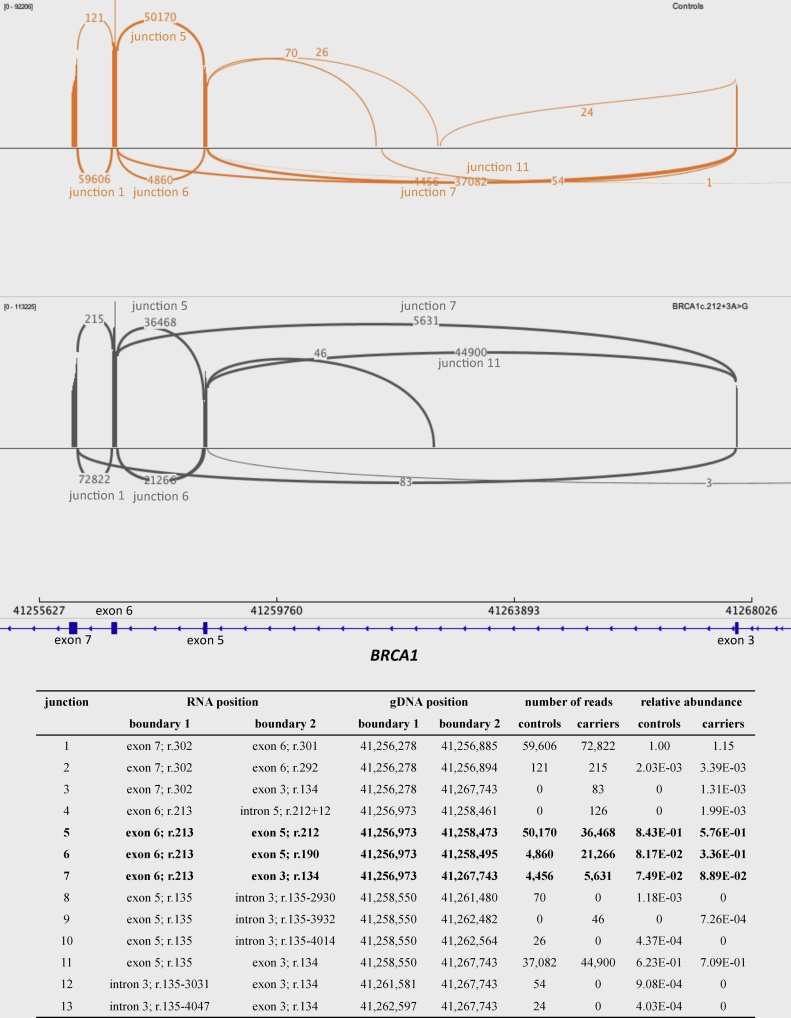
For the relative quantification of isoforms within samples, it is important to have an understanding of all (stably) expressed transcripts. Earlier reports [16], [17], [18] described three isoforms of interest around exon 5 in BRCA1 c.212+3A>G carriers: BRCA1-ex5FL (r.135_212), BRCA1-Δex5 (r.135_212del) and BRCA1-Δ22ntex5 (r.190_212del). Here we verified these using a PCR-based targeted deep sequencing approach, which has never been done before for patient samples with this variant. Fig. 1 summarizes the results of this experiment and visualises them as a sashimi plot.
Fig. 1.
Sashimi plot of targeted RNA sequencing results. For the relative transcript quantification, the relative abundance of the three relevant transcripts was calculated as the number of reads covering the transcript-specific splice site divided by the total number of reads spanning the 3 splice sites (junctions 5–7). RNA positions are based on reference sequence NM_007294.3 starting at the A of the translation initiation codon (ATG) as position +1. Genomic positions are derived from hg19 reference sequence. 13 unique junctions were identified in controls (depicted in orange) and/or BRCA1 c.212+3A>G carriers (grey). At the boundary between exon 6 and 7 (junction 1), 59,606 and 72,822 reads were identified for the controls and the carriers respectively. As expected, three highly abundant transcripts are found on the other end of exon 6, connecting it to either exon 5 (junctions 5 and 6) or exon 3 (junction 7). For junction 5 (BRCA1-ex5FL), junction 6 (BRCA1-Δ22ntex5; c.191_212del22) and junction 7 (BRCA1-Δex5; c.135_212del78) the sum of the number of identified reads for these junctions is 59,486 and 69,525, which equals 99.80% and 87.01% of the number of reads at junction 1 for controls and the carriers respectively. In carriers the 13% difference is not explained by the novel junctions (junctions 3, 4 and 9), therefore we assume that the difference is caused by a percentage of prematurely terminated reads. The extremely low coverage also suggests that all junctions other than 1, 5, 6, 7 and 11 are artefacts, introduced by PCR or bioinformatics, rather than relevant isoforms. In BRCA1 c.212+3A>G carriers there is almost a five-fold relative difference in expression for BRCA1-Δ22ntex5 compared to controls, while BRCA1-Δex5 expression ratios are similar between carriers and controls.
3.3. Relative quantification of BRCA1 c.212+3A>G transcript variants by qPCR and ddPCR
3.3.1. Efficiency
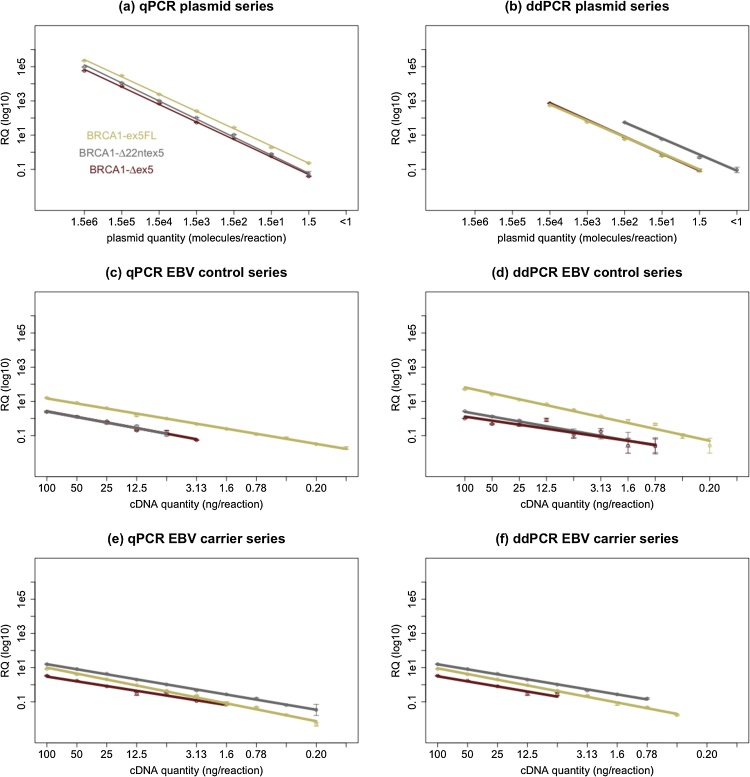
For both qPCR and ddPCR we investigated besides linearity and dynamic range, also limit of blank (LOB), limit of detection (LOD), and limit of quantification (LOQ) [26]. These metrics were evaluated using target quantifications with both methods on dilution series and the data are depicted in Fig. 2.
Fig. 2.
Linearity and dynamic range was tested on for all assays with both quantitative PCR methods, using dilution series containing transcript-specific plasmids ((a) and (b)) or cDNA from EBV samples (non-carriers ((c) and (d)) and carriers ((e) and (f)). Measured relative template quantity (RQ) is depicted on the y-axis in function of theoretical template concentration on the x-axis. Almost all assay-sample combinations show excellent linearity with both techniques (R2 > 0.98). Linearity was only worse for BRCA1-Δex5 in dilutions from EBV controls with ddPCR (R2 = 0.81). An important difference is that the dynamic range of qPCR across the dilution series was larger for several sample-target combinations, especially when quantifying transcript-specific plasmids. From these data we also estimated the LOB, LOD and LOQ values. For both methods LOB was equal to 0 (none of the no template controls yielded any amplification) and the LOQ and LOD for both methods reaches the theoretical minimum of 1 copy per reaction in transcript-specific plasmid dilution series. When using EBV derived samples the LOQ for both qPCR and ddPCR was found to range between 100 pg and 6.25 ng of cDNA depending on the abundance of a certain transcript in each sample.
RPP30, the internal reference target for ddPCR, was also found to have a wide dynamic range, excellent linearity within this range and a LOQ similar to BRCA1-ex5FL in all EBV derived samples as can be seen in Supplementary Data C.
3.3.2. Precision and reproducibility
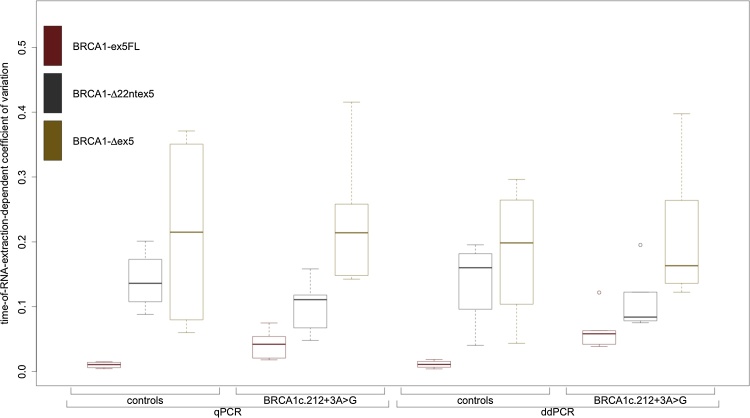
The variation in relative expression for each isoform over all time points of extraction was studied by calculating the coefficient of variation (CV). Boxplots of these CVs, for all isoforms in both sample groups using both techniques, are depicted in Fig. 3.
Fig. 3.
Boxplot of coefficients of variation for all cDNA-assay combinations over all time points of both quantitative PCR techniques. We observed an inverse correlation between the CV and the relative abundance of each isoform; CVs are consistently higher for isoforms with lower expression. CVs for each transcript range within the same order of magnitude and all experiments show very small standard errors, leading us to conclude that precision, reproducibility and repeatability are not an issue with either quantitative PCR technique.
3.3.3. Accuracy
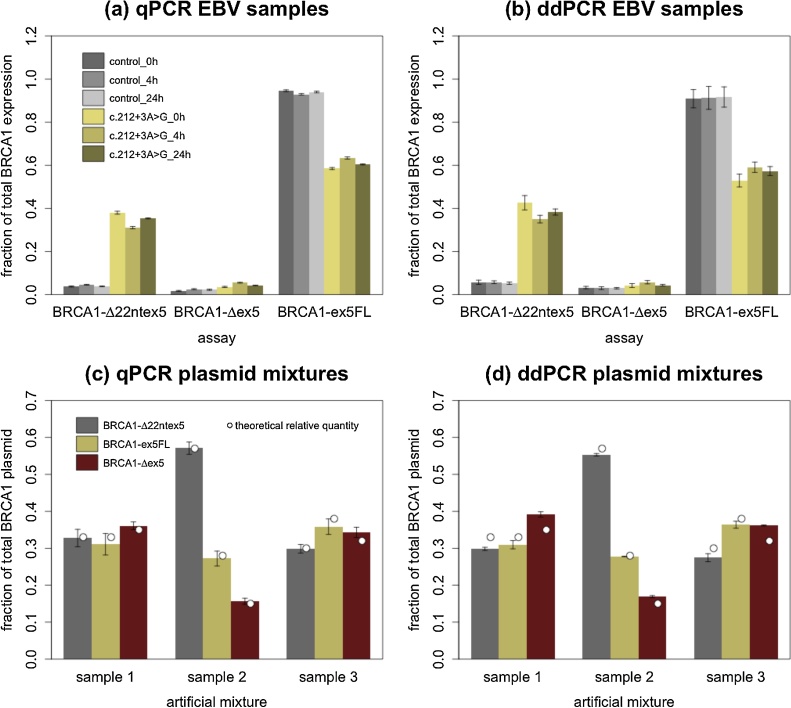
Quantification of the expression of each isoform relative to the sum of all three isoforms is shown in Fig. 4. In BRCA1 c.212+3A>G mutation carriers both qPCR (a) and ddPCR (b) confirmed results that were previously published [16]. There is a diminished relative abundance of BRCA1-ex5FL caused by a statistically significant increase of BRCA1-Δ22ntex5 expression in carriers: a 8.84 ± 1.78 fold increase in relative abundance was measured using qPCR, whereas ddPCR showed a 6.97 ± 0.75 increase (p = 1.16e-8 for both techniques, mean of all three time points). The impact of BRCA1 c.212+3A>G on the BRCA1-Δex5 isoform is less drastic. For BRCA1-Δex5 a 1.95 ± 0.128 fold change is seen with qPCR and a 1.56 ± 0.32 difference with ddPCR (p = 2.31e-8 and 6.14e-5 respectively, mean of all three time points).
Fig. 4.
(a) and (b) show relative expression values for all relevant isoforms at all time points of extraction for qPCR and ddPCR. Data points from carrier samples and control samples are shown in yellow and grey respectively. In accordance with the literature and our NGS data, BRCA1-Δ22ntex5 is highly overexpressed in carriers in comparison to controls. (c) and (d) show the quantification of sample 1, 2 and 3. Sample 1 contained 33%, 33% and 34% of plasmid containing BRCA1-Δ22ntex5, BRCA1-ex5FL and BRCA1-Δex5 respectively. For samples 2 and 3 this was 57%/28%/15% and 30%/38%/32% respectively. The theoretical relative amount of each transcript in each sample (depicted with white dots in each bar) was calculated based on the volume of plasmid stock that was added, the measured amount of plasmid (ng/μL) in the stock solution and the theoretical molecular weight of each plasmid (based on the plasmid sequence).
In a correlation plot (Supplementary D, left side panel) we evaluated the correlation between qPCR versus ddPCR results obtained for each isoform relative to the sum of all isoforms, as an estimate of total BRCA1 expression. This graph indicates that there is a small but systematic difference in measurement of the expression as the data are situated primarily on one site of the diagonal. In Supplementary D (right side panel) a Bland-Altman plot clarifies the extent of the difference. This shows that the discrepancy between both methods is small, but clusters to one side of the mean and a perfect agreement between methods cannot be concluded. In control samples (depicted by black data points) the data are almost entirely located to one side of the mean indicating a true difference between both techniques. Underestimation of BRCA1-ex5FL will lead to an overestimation of BRCA1-Δ22ntex5 and BRCA1-Δex5 expression levels, or vice versa, by which the data points shift to opposite sides of the curve. There is a statistically significant difference between the qPCR and ddPCR results for BRCA1-Δ22ntex5 (p = 1.89e-5) and BRCA1-ex5FL (p = 3.79e-6) in controls. In carriers the same effect is observed, although less pronounced and not statistically significant. This explains why the fold-change of BRCA1-Δ22ntex5 expression is roughly 25% higher for qPCR. In Supplementary data E the same analysis can be seen using Quantasoft (Bio-Rad). Here the discrepancies between qPCR and ddPCR data are even bigger, demonstrating the importance of an accurate way to call positive droplets.
To further evaluate the accuracy of both methods we used artificial mixtures, each containing a known relative amount of transcript-specific plasmid. Three such mixtures (hereafter designated as samples 1–3) were made and measured using both methods, to test if consistent overestimation of one target leads to the consistent underestimation of one or multiple other targets. Results of the relative quantification of these mixtures and their accuracy can be seen in Fig. 4(c) and (d). All relative quantifications correlated relatively well to the theoretical relative abundance. qPCR measurements were found to correlate slightly better with the theoretical relative quantities than ddPCR. This effect is mainly explained by the overestimation of BRCA1-Δex5 leading predominately to an underestimation of BRCA1-Δ22ntex5 and BRCA1-ex5FL or vice versa.
3.3.4. Economics
A last consideration when comparing ddPCR and qPCR is the cost, workload and time required to perform analyses. For this study, the cost of digital PCR experiments per reaction was a little over two times greater than that of qPCR. We looked at the cost per reaction carried out as described in the methods section taking into account the four-fold difference in reaction volume. The cost to generate droplets and read out the droplets was not taking into account. This would make the ddPCR experiments 6 times more expensive than the qPCR approach. The number of ddPCR reactions that could be performed in the same timeframe was significantly smaller than with qPCR. In one workday three 384-well plates of qPCR reactions were possible, whereas only two 96-well plates with ddPCR droplets could be executed.
4. Discussion
ddPCR has gained popularity in recent years for nucleic acid quantification and is said to be highly accurate, sensitive and less affected by inhibitors, while measuring target concentrations in a direct manner without the need for standard curves [27]. Studies evaluating the use of ddPCR for pathogen detection, copy number analysis and other applications have been published and some conclude that ddPCR has a lower LOD and is more robust than qPCR in terms of coefficients of variation and resistance to inhibition [8], [27]. Yet, not all sources deem ddPCR to be more accurate and precise than qPCR; both techniques often yield similar results with a high degree of correlation [9], [28]. For relative abundance of expressed transcripts and isoforms, published data are limited to a multiplex ddPCR approach for the quantification of 4 isoforms simultaneously. Also here the acquired expression profile is highly similar to previously reported data generated via qPCR [29]. A multiplex ddPCR setup might not always be feasible for the quantification of multiple transcripts. Design of transcript-specific probes can be challenging and multiple probes for a single locus have the potential to cross-react in a multiplex reaction. Using boundary spanning probes can also interfere with accurate quantification of isoforms, especially in case of a large difference in abundance between transcripts [5].
The aim of this study was to evaluate the performance of digital PCR (dPCR), in the form of the droplet digital PCR platform, compared to qPCR for quantification of the relative abundance of transcript isoforms. We report on linearity and dynamic range, precision and reproducibility, accuracy, cost and labour-intensity for both techniques, using BRCA1 c.212+3A>G as a model splice site mutation.
Here qPCR assays often had a wider dynamic range, as ddPCR could not accurately quantify the upper limit of the plasmid series and the lower limit of the EBV derived series. It seems that at first sight, ddPCR is more easily saturated in some samples and less sensitive in other samples than qPCR, contradicting what has earlier been suggested [27]. The saturation limit of ddPCR reactions is dependent on the number of droplets. The maximal theoretical number of plasmid molecules that could be detected by our ddPCR setup was 15,000 molecules. With an average number of partitions of 14,728 in this study, the ratio of the most concentrated plasmid dilution point that was efficiently quantified is approximately 1.02. All dilution samples with concentrations >15,000 template molecules contained at least 10 x the amount of molecules, demonstrating ddPCR’s inability to quantify samples with a large template to partition ratio. For optimal precision, dPCR is best used at template to partition ratio of 1.59. [24] It is however highly unlikely that biological samples will contain such numbers. In EBV carrier samples all three assays seem to have lower LOD values with qPCR, but in EBV control series ddPCR seems to outperform qPCR for BRCA1-Δ22ntex5 and BRCA1-Δex5 and in plasmid dilutions the LOQ for both techniques is situated at the theoretical minimum. With these discrepancies we should take into account the low amounts of total cDNA that is used as input in these low regions of the EBV series and therefore stochastic effects should be considered. Our conclusion is that ddPCR and qPCR show the same LOB, LOD and LOQ and have the same dynamic range and linearity at biologically relevant template concentrations.
The variation between two RNA samples extracted from the same culture at different time points does not correlate between qPCR and ddPCR, which indicated that the variation, between replicates extracted at different time points, is due to technical variation rather than biological. This is further supported by the fact that the magnitude of CV values is inversely correlated to the expression level of each isoform and that the means of these CV values do correlate between both methods. Mestdagh et al. showed coefficients of variations that range from <10%, for RNA triplicates with Cq values between 15 and 30, till 50% for replicates with Cq > 30 [30]. Cq values for BRCA1-Δ22ntex5 and BRCA1-Δex5 were situated in the latter range, while BRCA1-ex5FL reactions were situated in the former. Therefore, the majority of the variation is related to the stochastic effect inherent with analysis of low expression isoforms. Coefficients of variation are within the same range for both quantitative PCR methods and this holds true for all three isoforms under investigation, therefore both methods are deemed equally precise and reproducible.
For analysis of the ddPCR data we applied both ddpcRquant and the standard Quantasoft software. Using ddpcRquant reduced time-of-RNA-extraction-dependent variation, which is considered to be technical variation, and made the relative expression levels more similar to qPCR results, thereby enhancing the correlation between both technologies. Standard errors also decreased when using ddpcRquant, as the variation between technical replicates became smaller. A possible explanation might be that Quantasoft assumes a normal distribution of the fluorescent amplitudes or the threshold is placed too close to the negative droplet population, both of which will result in inaccurate positive droplet calling. The ddpcRquant algorithm stays clear of any distribution assumptions as well as accounts for shifts in baseline fluorescence [12].
Considering ddpcRquant as the analysis method of choice for ddPCR data, relative expression values for all three isoforms yielded similar results with both methods (Fig. 4(a) and (b)), although the correlation and Bland-Altman plot (Supplementary Data D) show that data points are not nicely distributed along the diagonal and the mean difference line for all sample/assay combinations. The same holds true for the artificially created samples, were qPCR is slightly more accurate in quantifying the relative abundance of each transcript (Fig. 4(c) and (d)).
Aside from these considerations, an overall better performance cannot be concluded for either technique, considering the similarity of the quantitative results and the fact that here we only investigated a single splice site mutation as a model for alternative splicing. Both methods were able to accurately quantify several targets over a range of dilutions, showed accurate relative quantification on both human-derived samples and artificially created samples and had similar LOB, LOD and LOQ values. Our results also show similarity between the relative quantification results obtained with both quantitative PCR techniques and next generation sequencing, although it has to be noted that the experimental design of NGS was devised for discovery purposes and not as a validation method for relative quantification.
Cost comparisons seem entirely application and setup-dependent as other studies have reported both worse and better cost ratios for ddPCR compared to qPCR [9], [28]. The cost of ddPCR could go down by introducing a dsDNA intercalating fluorophore instead of target-specific probes, but this would make the use of an internal reference target (RPP30 in this case) impossible. Calculating relative expression values with unnormalized concentrations measured by ddPCR, gave larger standard errors as a result of more variation between replicates. Therefore the use of at least one (possibly more) highly stable internal reference target seems advisable. Performing ddPCR also took more hands-on manipulations such as the addition of probes and droplet generation/calling. Of note, the advantage of ddPCR for absolute quantification through direct quantification without the need for a standard curve is of no use when doing relative quantification. Data-analysis for both methods requires some finesse, as qPCR data need to be converted to relative quantities in order to obtain linear relative expression values and ddpcRquant requires careful consideration of how the positive droplets are called. It should be noted that relative abundance results, calculated in the discovery phase using NGS, are similar to the results of both PCR methods without the need for isoform-specific primer and/or probe design.
Based on the data collected here, ddPCR appears to be an accurate and valid alternative to qPCR for the evaluation of the relative abundance of alternative transcription. For BRCA1 c.212+3A>G carriers and controls both techniques gave similar results, although we admit that these results reflect the performance of qPCR and ddPCR on just one splice site mutation. Although literature promises a whole new generation of PCR, data show here that qPCR should not be cast aside just yet, at least not for relative quantification of splice variants, provided that a carefully optimized qPCR standard operating procedure is used. The arrival of this next generation quantitative PCR method should be welcomed as an addition to qPCR, rather than a replacement of, especially for relative quantification of gene/isoform expression.
Handled by Justin O’Grady
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.bdq.2017.09.001.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- 1.de Klerk E., ’t Hoen P.A. Alternative mRNA transcription, processing, and translation: insights from RNA sequencing. Trends Genet. 2015;31(3):128–139. doi: 10.1016/j.tig.2015.01.001. Elsevier Ltd. [DOI] [PubMed] [Google Scholar]
- 2.Xiong H.Y., Alipanahi B., Lee L.J., Bretschneider H., Merico D., Yuen R.K.C. The human splicing code reveals new insights into the genetic determinants of disease. Science (80-) 2015;347(6218):1254806. doi: 10.1126/science.1254806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ward A.J., Cooper T.A. The pathobiology of splicing. J. Pathol. 2010;220(2):152–163. doi: 10.1002/path.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Derveaux S., Vandesompele J., Hellemans J. How to do successful gene expression analysis using real-time PCR. Methods. 2010;50(4):227–230. doi: 10.1016/j.ymeth.2009.11.001. Elsevier Inc. [DOI] [PubMed] [Google Scholar]
- 5.Vandenbroucke I.I., Vandesompele J., Paepe A.D., Messiaen L. Quantification of splice variants using real-time PCR. Nucleic Acids Res. 2001;29(13):E68. doi: 10.1093/nar/29.13.e68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M. The MIQE guidelines:Minimum Information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009;55(4):611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 7.Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3(7) doi: 10.1186/gb-2002-3-7-research0034. RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hindson B.J., Ness K.D., Masquelier D.A., Belgrader P., Heredia N.J., Makarewicz A.J. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal. Chem. 2011;83(22):8604–8610. doi: 10.1021/ac202028g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang R., Paparini A., Monis P., Ryan U. Comparison of next-generation droplet digital PCR (ddPCR) with quantitative PCR (qPCR) for enumeration of Cryptosporidium oocysts in faecal samples. Int. J. Parasitol. 2014;44(14):1105–1113. doi: 10.1016/j.ijpara.2014.08.004. Australian Society for Parasitology Inc. [DOI] [PubMed] [Google Scholar]
- 10.Strain M.C., Lada S.M., Luong T., Rought S.E., Gianella S., Terry V.H. Highly precise measurement of HIV DNA by droplet digital PCR. PLoS One. 2013;8(4):2–9. doi: 10.1371/journal.pone.0055943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones M., Williams J., Gärtner K., Phillips R., Hurst J., Frater J. Low copy target detection by Droplet Digital PCR through application of a novel open access bioinformatic pipeline, definetherain. J. Virol. Methods. 2014;202(100):46–53. doi: 10.1016/j.jviromet.2014.02.020. Elsevier/North-Holland Biomedical Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trypsteen W., Vynck M., De Neve J., Bonczkowski P., Kiselinova M., Malatinkova E. ddpcRquant: threshold determination for single channel droplet digital PCR experiments. Anal. Bioanal. Chem. 2015;407(19):5827–5834. doi: 10.1007/s00216-015-8773-4. [DOI] [PubMed] [Google Scholar]
- 13.Whale A.S., Huggett J.F., Cowen S., Speirs V., Shaw J., Ellison S. Comparison of microfluidic digital PCR and conventional quantitative PCR for measuring copy number variation. Nucleic Acids Res. 2012;40(11):e82. doi: 10.1093/nar/gks203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Landrum M.J., Lee J.M., Riley G.R., Jang W., Rubinstein W.S., Church D.M. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014;42(D1):D980–5. doi: 10.1093/nar/gkt1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Claes K., Machackova E., De Vos M., Mortier G., De Paepe A., Messiaen L. Mutation analysis of the BRCA1 and BRCA2 genes results in the identification of novel and recurrent mutations in 6/16 Flemish families with breast and/or ovarian cancer but not in 12 sporadic patients with early-onset disease. Hum. Mutat. 1999;13(3):256. doi: 10.1002/(SICI)1098-1004(1999)13:3<256::AID-HUMU12>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 16.Claes K., Vandesompele J., Poppe B., Dahan K., Coene I., De Paepe A. Pathological splice mutations outside the invariant AG/GT splice sites of BRCA1 exon 5 increase alternative transcript levels in the 5 (end of the BRCA1 gene. Oncogene. 2002;21(26):4171–4175. doi: 10.1038/sj.onc.1205520. [DOI] [PubMed] [Google Scholar]
- 17.Sanz D.J., Acedo A., Infante M., Durán M., Pérez-Cabornero L., Esteban-Cardeñosa E. A high proportion of DNA variants of BRCA1 and BRCA2 is associated with aberrant splicing in breast/ovarian cancer patients. Clin. Cancer Res. 2010;16:1957–1967. doi: 10.1158/1078-0432.CCR-09-2564. [DOI] [PubMed] [Google Scholar]
- 18.Théry J.C., Krieger S., Gaildrat P., Révillion F., Buisine M.-P., Killian A. Contribution of bioinformatics predictions and functional splicing assays to the interpretation of unclassified variants of the BRCA genes. Eur. J. Hum. Genet. 2011;19(10):1052–1058. doi: 10.1038/ejhg.2011.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hui-Yuen J., Mcallister S., Koganti S., Hill E., Bhaduri-Mcintosh S. Establishment of Epstein-Barr virus growth-transformed lymphoblastoid cell lines. J. Vis. Exp. 2011;57(10) doi: 10.3791/3321. 33213791–3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nolan T., Hands R.E., Ogunkolade W. SPUD. A quantitative PCR assay for the detection of inhibitors in nucleic acid preparations. Anal. Biochem. 2006;351(2):308–310. doi: 10.1016/j.ab.2006.01.051. [DOI] [PubMed] [Google Scholar]
- 21.Ye J., Coulouris G., Zaretskaya I., Cutcutache I., Rozen S., Madden T.L. Primer-BLAST. A tool to design target-specific primers for polymerase chain reaction. BMC Bioinf. 2012;13(1):134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31(13):3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hellemans J., Mortier G., De Paepe A., Speleman F., Vandesompele J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 2007;8(2):R19. doi: 10.1186/gb-2007-8-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huggett J.F., Foy C.A., Benes V., Emslie K., Garson J.A., Haynes R. The digital MIQE guidelines: minimum information for publication of quantitative digital PCR experiments. Clin. Chem. 2013:902. doi: 10.1373/clinchem.2013.206375. [DOI] [PubMed] [Google Scholar]
- 25.R Core Team . R Foundation for Statistical Computing; Vienna, Austria: 2014. R: A Language and Environment for Statistical Computing. [Google Scholar]
- 26.Armbruster D.A., Pry T. Limit of blank, limit of detection and limit of quantitation. Clin Biochem Rev. 2008;29(Suppl 1 (August)):S49–S52. [PMC free article] [PubMed] [Google Scholar]
- 27.Hindson C.M., Chevillet J.R., Briggs H.A., Gallichotte E.N., Ruf I.K., Hindson B.J. Absolute quantification by droplet digital PCR versus analog real-time PCR. Nat. Methods. 2013;10(10):1003–1005. doi: 10.1038/nmeth.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hayden R.T., Gu Z., Ingersoll J., Abdul-Ali D., Shi L., Pounds S. Comparison of droplet digital PCR to real-time PCR for quantitative detection of cytomegalovirus. J. Clin. Microbiol. 2013;51(2):540–546. doi: 10.1128/JCM.02620-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun B., Tao L., Zheng Y.L. Simultaneous quantification of alternatively spliced transcripts in a single droplet digital PCR reaction. Biotechniques. 2014;56(6):319–325. doi: 10.2144/000114179. [DOI] [PubMed] [Google Scholar]
- 30.Mestdagh P., Feys T., Bernard N., Guenther S., Chen C., Speleman F. High-throughput stem-loop RT-qPCR miRNA expression profiling using minute amounts of input RNA. Nucleic Acids Res. 2008;36(December (21)):e143. doi: 10.1093/nar/gkn725. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.