Abstract
Methylenetetrahydrofolate reductase (MTHFR) is a key enzyme of folate pathway. Several polymorphisms were reported in MTHFR gene but C677T polymorphism is most studied and it has been reported to be risk factor for several diseases/disorders. The present study was designed to explore the frequency of MTHFR C677T polymorphism in North Indian healthy population. In addition to this a meta-analysis of published articles was also performed to estimate the global prevalence of MTHFR C677T polymorphism. A total of 1000 unrelated healthy subjects were selected for MTHFR C677T polymorphism analysis. Different databases were searched for eligible articles. Prevalence proportion with 95 % CI was used to determine global prevalence of T allele and TT genotype. Meta-analysis was performed by Open meta-analyst. In 1000 blood samples analyzed, the frequency of T allele and TT genotype was 11 and 1 % respectively. Results of the meta-analysis showed that the global prevalence of T allele and TT genotype were 24.0 % (95 % CI 21.7–26.5) and 7.7 % (95 % CI 6.5–8.9) respectively. In sub-group meta-analysis, the lowest frequency of T allele was found in Africans (10.3 %; 95 % CI 3.8–16.8), and highest in Europeans (34.1 %; 95 % CI 31.9–36.3). The frequency of T allele in the North India is 11 %. The results of the meta-analysis showed that the frequency of the T allele and the TT genotype of C677T is highest in the Caucasian population.
Keywords: MTHFR, C677T, Polymorphism, Meta-analysis, Prevalence proportion, Global frequency
Introduction
Methylenetetrahydrofolate reductase (MTHFR) is a crucial enzyme in folate/homocysteine pathway. It catalyzes the conversion of 5,10-methylenetetrahydrofolate to 5-methylenetetrahydrofolate, which donates methyl group for the conversion of homocysteine to methionine. MTHFR gene is located at chromosome 1p36.3 [1]. A number of single nucleotide polymorphisms (SNPs) are reported in MTHFR gene but C677T mutation (rs1801133) is the most studied and clinically important. C677T polymorphism lies in exon 4, in which cytosine is replaced by thymine at 677th position, which resulted in an alanine to valine substitution at position 222 in protein (A222V) [2, 3]. C677T mutation was shown to render the enzyme thermolabile [2]. MTHFR enzyme functions as dimer or tetramer and Flavin adenine dinucleotide (FAD) is the co-factor. The C677T mutation changes the secondary structure of the peptide and interactions between monomers. The A → V mutation increases the rate of dissociation of FAD and loss of FAD is linked to changes in quaternary structure and enzymatic activity reduces [4–6]. By docking study it is established that the mutant enzyme (222V) has less affinity towards its cofactor FAD than the normal enzyme (222A) [7]. The variant protein loses its cofactor FAD more quickly and has lower stability. C677T polymorphism effect can be suppressed by addition of folate, which causes a higher FAD affinity [8].
MTHFR C677T polymorphism has been reported as a risk factor for several diseases/disorders such as—Down syndrome [9, 10], neural tube defects [11], orofacial clefts [12], type I diabetes [13], cardiovascular diseases [2, 14], male infertility [15], schizophrenia [16], bipolar disorder [16] and cancer [17] etc. The frequency of C677T polymorphism has been described from several world populations. But from India very limited case–control studies were published. So, it is very important to know the frequency of such a clinically important gene polymorphism in the healthy Indian population. Together with this the second objective of present study was to estimate the global prevalence of C677T polymorphism by meta-analysis.
Materials and Methods
Random Sample Analysis
Sample Collection
3 ml blood sample was collected in EDTA coated vials from 1000 unrelated healthy subjects which were domicile of Eastern Uttar Pradesh (UP). The study was approved by the Institutional Ethics Committee of the VBS Purvanchal University, Jaunpur. Blood sample was collected after getting informed written consent. Genomic DNA was extracted by the method of Bartlett and White [18].
Genotyping
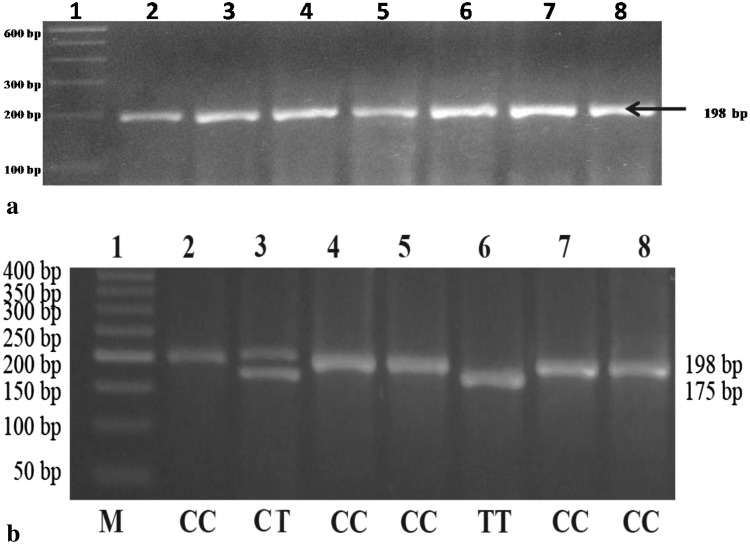
MTHFR C677T genotyping was carried out by PCR–RFLP method of Frosst et al. [2]. Briefly 100 ng of genomic DNA was amplified in a final volume of 15 μl with 4 pM of each of forward and reverse primers, 250 μl of dNTPs mix, 1X Taq DNA polymerase buffer and 1u of Taq DNA polymerase. PCR program was initial denaturation at 94 °C for 4 min followed by 30 cycles of denaturation at 94 °C for 1 min, annealing at 62 °C for 1 min, extension at 72 °C for 1 min and a final extension at 72 °C for 10 min. The amplicons (198 bp) were digested with HinfI as the C677T mutation creates a restriction site for it, and resolved in a 4 % agarose gel. For quality control, 10 % of samples (randomly selected) were re-genotyped and no discrepancy in genotypes were found.
Statistical Analysis
Allele frequencies were calculated by the gene counting method. χ2 test was performed to test the Hardy–Weinberg Equilibrium(HWE). All statistical analysis was performed by DeFinetti program (http://ihg.gsf.de/cgi-bin/hw/hwa1.pl).
Meta-analysis
Searched Strategy and Identification of Studies
For meta-analysis Pubmed, Science Direct, Springer link and Google scholar databases were searched for the suitable articles using keywords “MTHFR”, “methylenetetrahydrofolate reductase”, “C677T”. Large number of articles were retrieved so that only those articles were selected for the inclusion in which C677T polymorphism in healthy subjects were analyzed. The included articles were also hand searched for additional studies which may be included in present meta-analysis.
Inclusion and Exclusion Criteria
The inclusion criteria were as follows: studies should: (1) be original, (2) selected only healthy subjects for C677T analysis and (3) reported MTHFR genotypes. Studies were excluded if they were: (1) case–control, (2) case reports, and (3) review, editorial etc.
Data Extraction
For all the eligible studies following information were extracted: first authors’ family name, year of publication, journal name, country name, population/ethnic group, number of alleles and/or genotypes. If in a study samples were taken from multiple countries or different race/caste then information was abstracted separately for each country/race/caste.
Statistical Analysis
Prevalence proportions (PP) was computed from the number of alleles/genotypes and total number of alleles/sample sizes (N) with the corresponding 95 % confidence interval (CI). A pooled PP was then estimated on the basis of the individual PPs. The PP was estimated either by fixed effect [19] or random effect [20] model depending upon heterogeneity. The heterogeneity between studies was tested using the Q-statistics and was quantified using the I2 statistic [21]. If I2 > 50 % then random effect model was used [22], otherwise fixed effect model was adopted. We have also done sub-group analysis based on geographical area i.e. the region from where the study belonged (Africa, North America, South America, Asia, Australia and Europe). All p values are two tailed with a significance level at <0.05. All statistical analyses were undertaken by Open Meta-analyst [23].
A global C677T frequency map was created by StatPlanet software [24] by pooling the prevalence of MTHFR C677T gene polymorphism in different countries.
Results
MTHFR C677T Polymorphism Analysis
Total 1000 blood samples were collected from healthy individuals selected from Eastern UP and analyzed for MTHFR C677T polymorphism. The 198 bp long MTHFR amplicon was digested with HinfI and resolved in 2 % agarose gel. The C677T substitution at nucleotide 677 creates a HinfI digestion site. T allele was digested into 2 fragments (175 and 23 bp), whereas C allele remains uncut (Fig. 1). In total 1000 samples the number of CC, CT and TT genotype was 797 (80 %), 192 (19 %) and 11 (1 %) respectively (Table 1). The number of C and T alleles was 1786 (89 %) and 214 (11 %) respectively. The population was in Hardy–Weinberg equilibrium (p = 0.88). Out of 1000 samples, 528 were male and 472 were female. In male samples the number of CC, CT and TT genotype 416 (79 %), 103 (19 %) and 9 (2 %) respectively. The number of C and T alleles was 935 (89 %) and 121 (11 %) respectively (Table 1). In female samples the number of CC, CT and TT genotype 381 (81 %), 89 (18 %) and 2 (0.04 %) respectively. The number of C and T alleles was 851 (90 %) and 93 (10 %) respectively.
Fig. 1.
a MTHFR C677T amplicon with 100 bp DNA ladder in lane 1; b Gel showing HinfI digested different MTHFR C677T genotypes
Table 1.
MTHFR C677T genotype distribution in randomly collected samples and gender wise distribution of samples, number of genotypes, number of alleles, HWE p value
| Sample | Number | Age (mean ± SD) | Genotype | Number of Alleles | HWE p value | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CC | CT | TT | C | T | |||||||||
| No. | Freq | No. | Freq | No. | Freq | No. | Freq | No. | Freq | ||||
| All | |||||||||||||
| Male | 528 | 36.68 ± 16.49 | 416 | 0.79 | 103 | 0.19 | 9 | 0.02 | 935 | 0.89 | 121 | 0.11 | 0.37 |
| Female | 472 | 36.89 ± 17.70 | 381 | 0.81 | 89 | 0.18 | 2 | 0.004 | 851 | 0.90 | 93 | 0.10 | 0.18 |
| Total | 1000 | 36.44 ± 15.03 | 797 | 0.80 | 192 | 0.19 | 11 | 0.01 | 1786 | 0.89 | 214 | 0.11 | 0.88 |
Freq frequency
Meta-analysis
Characteristic of Eligible Studies
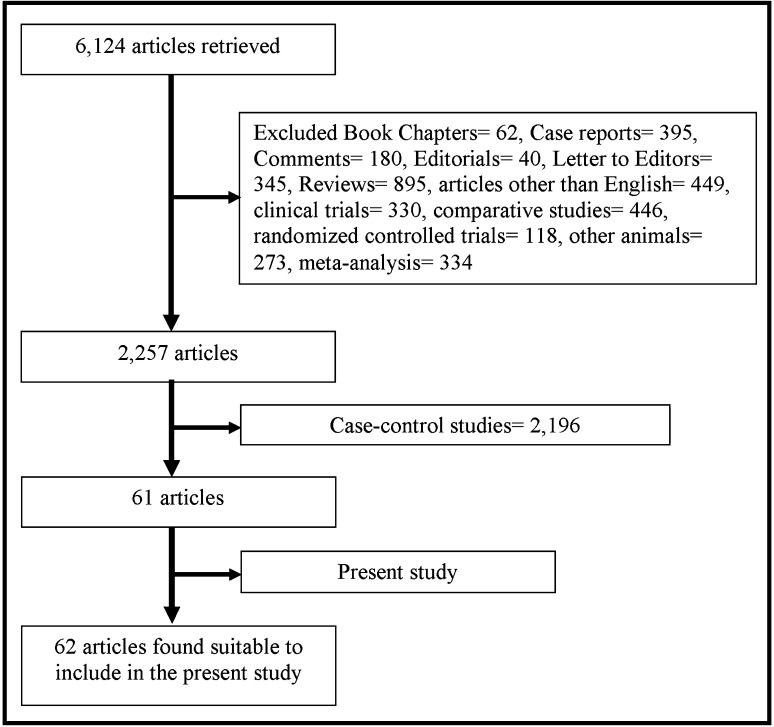
With our initial search strategy, total 6124 articles were retrieved. After exclusion of reviews, meta-analysis, case–control studies, letter to editor, editorials, comments and observational studies etc., 62 articles (investigated 178 populations) with 47,592 samples were found suitable for the inclusion in the present meta-analysis (including present study) (Fig. 2). In included 62 articles—two articles were published by Australian scientists and they analyzed C677T polymorphism in three populations [25, 26] and 3 articles were published from African region, investigated total seven populations [25, 27, 28]. Five articles with twelve populations were published from South America [29–33], and 12 articles with 27 populations were published from North America [25, 26, 34–43], 5 articles with twelve populations from South America [29–33]. From Europe 19 articles were published which investigated 31 populations [25, 26, 44–60]. Highest number of articles were published from Asian continent i.e. 30 studies, in which total 98 populations were investigated [25–28, 61–85, present study]. These studies include 47,592 samples. The highest sample size was 14,405 in the study of Yang et al. [85] and lowest sample size was 20 in the study of Angeline et al. [69] (Table 2).
Fig. 2.
Flow diagram of study search and selection process
Table 2.
Eligible included studies
| Author | Country | No. of population studies | Sample size |
|---|---|---|---|
| Harmon et al. [44] | Ireland | 1 | 625 |
| Stevenson et al. [35] | USA | 2 | 297 |
| Arruda et al. [29] | Brazil | 3 | 327 |
| Bowen et al. [45] | UK | 1 | 300 |
| Franco et al. [30] | Brazil | 5 | 337 |
| Hill and FitzPatrick [46] | UK | 1 | 122 |
| Schneider et al. [25] | Central African Republic; Gambia; Kenya; Madagascar; Australia, UK, USA; French Polynesia; Hong Kong; Mongolia; Indonesia; Sri Lanka; Yemen | 16 | 881 |
| Antoniadi et al. [47] | Greece | 1 | 160 |
| Friedman et al. [61] | Israel | 1 | 401 |
| Mack et al. [34] | USA | 4 | 84 |
| Mutchinick et al. [36] | Mexico | 1 | 250 |
| Zuo et al. [62] | Korea, Japan | 2 | 239 |
| Chango et al. [48] | France | 1 | 169 |
| Pollak et al. [63] | Israel | 8 | 897 |
| Yu et al. [64] | China | 1 | 200 |
| Murakami et al. [65] | Japan | 1 | 816 |
| Mynett-Johnson et al. [49] | Ireland | 1 | 115 |
| Rady et al. [37] | USA | 4 | 507 |
| Rosenberg et al. [27] | Ghana; Israel; Japan | 3 | 444 |
| Sadewa et al. [66] | Indonesia, Japan | 2 | 312 |
| Al-Habboubi et al. [67] | Bahrain, Lebanon | 2 | 560 |
| Bailey et al. [38] | USA | 1 | 185 |
| Chowdary et al. [39] | USA | 1 | 172 |
| Esfahani et al. [40] | USA | 1 | 433 |
| Wilcken et al. [26] | Australia; Italy; Spain; France; Netherland; Finland; Hungary; Russia; Canada; USA; Mexico; China, Israel | 24 | 7130 |
| Zijno et al. [50] | Italy | 1 | 172 |
| Almawi et al. [68] | Lebanon | 7 | 1178 |
| Angeline et al. [69] | India | 1 | 20 |
| Lovricevic et al. [51] | Croatia | 1 | 228 |
| Spiridonova et al. [52] | Yakutia | 3 | 477 |
| Vaughn et al. [41] | USA | 1 | 360 |
| Ameen et al. [28] | Tunisia; Lebanon; Saudi Arabia; Bahrain | 4 | 1234 |
| Golbahar et al. [70] | Iran | 1 | 391 |
| Sazci et al. [53] | Turkey | 1 | 1684 |
| Parle-McDermott et al. [54] | Ireland | 1 | 508 |
| Barbosa et al. [31] | Brazil | 1 | 100 |
| Bhat et al. [71] | India | 1 | 110 |
| Gialeraki et al. [55] | Greece | 1 | 100 |
| Sabbagh et al. [72] | Lebanon | 1 | 205 |
| Saraswathy et al. [73] | India | 1 | 81 |
| Al-Allawi et al. [74] | Iraq | 1 | 150 |
| Algasham et al. [75] | Saudi Arabia | 1 | 270 |
| Mansoor et al. [76] | Pakistan | 14 | 701 |
| Misra et al. [78] | India | 1 | 200 |
| Ozarda et al. [56] | Turkey | 1 | 402 |
| Tsai et al. [42] | USA | 1 | 1689 |
| Zappacosta et al. [57] | Italy | 1 | 104 |
| Bagheri et al. [77] | Iran | 1 | 108 |
| Gra et al. [58] | Russia | 1 | 352 |
| Ghodke et al. [79] | India | 1 | 144 |
| Murry et al. [80] | India | 2 | 112 |
| Ramos et al. [43] | Mexico | 1 | 150 |
| Sukla and Raman [81] | India | 4 | 1426 |
| Sachdeva et al. [82] | India | 1 | 350 |
| Saraswathy et al. [83] | India | 24 | 1142 |
| Stur et al. [32] | Brazil | 2 | 200 |
| Amela et al. [59] | Bosnia | 1 | 206 |
| Dajani et al. [84] | Jordan | 2 | 192 |
| Seremak-Mrozikiewicz et al. [60] | Poland | 1 | 1326 |
| Yang et al. [85] | China | 1 | 14,405 |
| Romero-Sánchez et al. [33] | Colombia | 1 | 152 |
| Present study (2016) | India | 1 | 1000 |
Meta-analysis
The meta-analysis showed that the global frequency of T allele was 24.0 % by adopting random effect model (95 % CI 21.7–26.5, p < 0.001; I2 = 98.74 %) and 26.0 % (95 % CI 25.8–26.3, p < 0.001) by fixed effect model. The frequency of TT genotype was 7.7 % (95 % CI 6.5–8.9, p < 0.001; I2 = 96.25 %) by random effect model and 6.5 (95 % CI 6.3–6.7, p < 0.001) by fixed effect model (Table 3). Between studies heterogeneity was high so random effect model was adopted.
Table 3.
Summary estimates for the prevalence proportion (PP) of MTHFR C677T in various allele/genotype contrasts, the significance level (p value) of heterogeneity test (Q test), and the I2 metric
| Population | FE estimate (95 % CI) | RE estimate (95 % CI) | I2 | p (Q) |
|---|---|---|---|---|
| All | ||||
| T | 26.0 (25.8–26.3), <0.001 | 24.0 (21.7–26.5), <0.001 | 98.74 | <0.001 |
| C | 74.0 (73.7–74.2), <0.001 | 75.9 (73.5–78.3), <0.001 | 98.74 | <0.001 |
| TT | 6.5 (6.3–6.7), <0.001 | 7.7 (6.5–8.9), <0.001 | 96.25 | <0.001 |
| CT | 33.2 (30.8–35.6), <0.001 | 33.2 (30.8–35.6), <0.001 | 96.49 | <0.001 |
| CC | 50.7 (50.3–51.1), <0.001 | 58.9 (55.4–62.4), <0.001 | 98.41 | <0.001 |
| African | ||||
| T | 10.0 (8.4–11.7), <0.001 | 10.3 (3.8–16.8), 0.002 | 92.79 | <0.001 |
| C | 90.0 (88.3–91.6), <0.001 | 89.7 (83.2–96.2), <0.001 | 92.79 | <0.001 |
| TT | 1.3 (0.3–2.4), 0.01 | 2.4 (–0.0–4.8), <0.001 | 69.43 | 0.006 |
| CT | 18.3 (6.5–30.0), 0.002 | 18.3 (6.5–30.0), <0.001 | 89.03 | <0.001 |
| CC | 77.3 (73.7–80.9), <0.001 | 80.3 (65.0–95.5), <0.001 | 93.57 | <0.001 |
| American (North) | ||||
| T | 28.4 (27.6–29.2), <0.001 | 31.2 (25.7–36.7), <0.001 | 97.58 | <0.001 |
| C | 71.6 (70.8–72.4), <0.001 | 68.8 (63.3–74.3), <0.001 | 97.58 | <0.001 |
| TT | 5.3 (4.8–5.9), <0.001 | 11.9 (8.8–14.9), <0.001 | 95.6 | <0.001 |
| CT | 37.5 (33.5–41.4), <0.001 | 37.5 (33.5–41.4), <0.001 | 87.66 | <0.001 |
| CC | 48.7 (47.5–49.9), <0.001 | 49.8 (41.9–57.7), <0.001 | 97.36 | <0.001 |
| American (South) | ||||
| T | 23.0 (21.3–24.6), <0.001 | 27.8 (19.8–35.8), <0.001 | 95.58 | <0.001 |
| C | 77.0 (75.4–78.7), <0.001 | 72.2 (64.2–80.2), <0.001 | 95.58 | <0.001 |
| TT | 3.4 (2.4–4.4), <0.001 | 7.6 (4.5–10.7), <0.001 | 85.2 | <0.001 |
| CT | 37.9 (29.0–46.9), <0.001 | 37.9 (29.0–46.9), <0.001 | 90.83 | <0.001 |
| CC | 55.0 (52.4–57.7), <0.001 | 53.0 (40.9–65.2), <0.001 | 95.14 | <0.001 |
| Asian | ||||
| T | 24.6 (24.3–25.0), <0.001 | 19.7 (16.0–23.4), <0.001 | 99.13 | <0.001 |
| C | 75.4 (75.0–75.7), <0.001 | 80.3 (76.6–84.0), <0.001 | 99.13 | <0.001 |
| TT | 6.8 (6.5–7.1), <0.001 | 5.5 (3.7–7.2), <0.001 | 97.22 | <0.001 |
| CT | 28.6 (24.8–32.4), <0.001 | 28.6 (24.8–32.4), <0.001 | 97.6 | <0.001 |
| CC | 52.3 (51.7–52.8), <0.001 | 65.9 (60.2–71.6), <0.001 | 98.93 | <0.001 |
| Australian | ||||
| T | 16.3 (14.0–18.6), <0.001 | 20.5 (2.6–38.4), 0.02 | 98.1 | <0.001 |
| C | 83.7 (81.4–86.0), <0.001 | 79.5 (61.6–97.4), <0.001 | 98.1 | <0.001 |
| TT | 2.5 (1.1–3.9), <0.001 | 5.9 (–0.5–12.3), 0.071 | 91.12 | <0.001 |
| CT | 28.8 (6.6–51.0), 0.01 | 28.8 (6.6–51.0), 0.01 | 96.53 | <0.001 |
| CC | 67.2 (63.3–71.2), <0.001 | 65.0 (36.6–93.4), <0.001 | 97.82 | <0.001 |
| European | ||||
| T | 33.0 (32.4–33.6), <0.001 | 34.1 (31.9–36.3), <0.001 | 91.08 | <0.001 |
| C | 67.0 (66.4–67.6), <0.001 | 65.9 (63.7–68.1), <0.001 | 91.08 | <0.001 |
| TT | 9.7 (9.2–10.2), <0.001 | 11.6 (9.9–13.3), <0.001 | 87.33 | <0.001 |
| CT | 44.3 (42.7–45.8), <0.001 | 44.3 (42.7–45.8), <0.001 | 57.83 | <0.001 |
| CC | 44.0 (43.1–45.0), <0.001 | 43.6 (40.8–46.3), <0.001 | 87.77 | <0.001 |
Sub-group Analysis
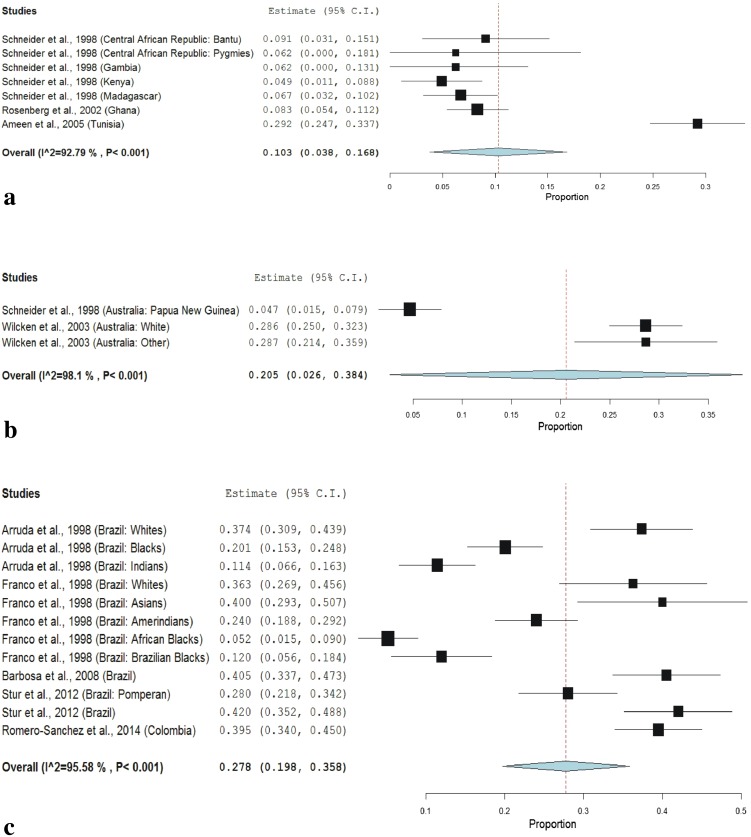
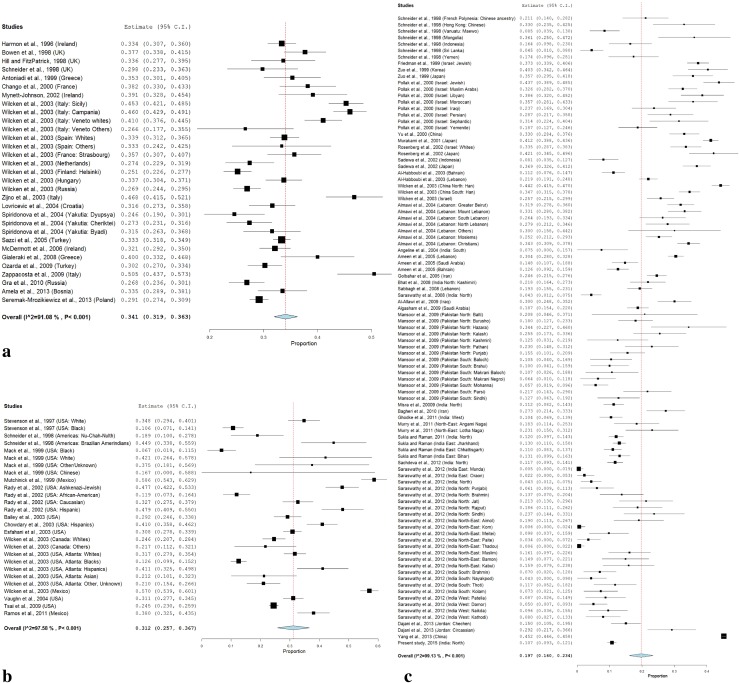
In sub-group meta-analysis, the frequency of T allele was 10.3 % (95 % CI 3.8–16.8, p = 0.002; I2 = 92.79 %) and frequency of TT genotype was 2.4 % (95 % CI −0.00 to 4.8, p < 0.001; I2 = 69.43 %) in Africa (Table 3; Fig. 3a), the frequency of T allele was 20.5 % (95 % CI 2.6–38.4, p = 0.02; I2 = 98.1 %) and frequency of TT genotype was 5.9 % (95 % CI −0.5 to 12.3, p < 0.071; I2 = 91.12 %) in Australia (Table 3; Fig. 3b), the frequency of T allele was 27.8 % (95 % CI 19.8–35.8, p < 0.001; I2 = 95.58 %) and frequency of TT genotype was 7.6 % (95 % CI 4.5–10.7, p < 0.001; I2 = 85.2 %) in South America (Table 3; Fig. 3c), the frequency of T allele was 34.1 % (95 % CI 31.9–36.3, p < 0.001; I2 = 91.08 %) and frequency of TT genotype was 11.6 % (95 % CI 9.9–13.3, p < 0.001; I2 = 87.33 %) in Europe (Table 3; Fig. 4a), the frequency of T allele was 31.2 % (95 % CI 25.7–36.7, p < 0.001; I2 = 97.58 %) and frequency of TT genotype was 11.9 % (95 % CI 8.8–14.9, p < 0.001; I2 = 95.6 %) in North America (Table 3; Fig. 4b), the frequency of T allele was 19.7 % (95 % CI 16.0–23.4, p < 0.001; I2 = 99.13 %) and the frequency of TT genotype was 5.5 % (95 % CI 3.7–7.2, p < 0.001; I2 = 97.22 %) in Asia (Table 3; Fig. 4c). Heterogeneity was high so random effect model was adopted.
Fig. 3.
a Random effect forest plot of T allele in African population; b random effect forest plot of T allele in Australian population; c random effect forest plot of T allele in South American population
Fig. 4.
a Random effect forest plot of T allele in European population; b random effect forest plot of T allele in North American population; c random effect forest plot of T allele in Asian population
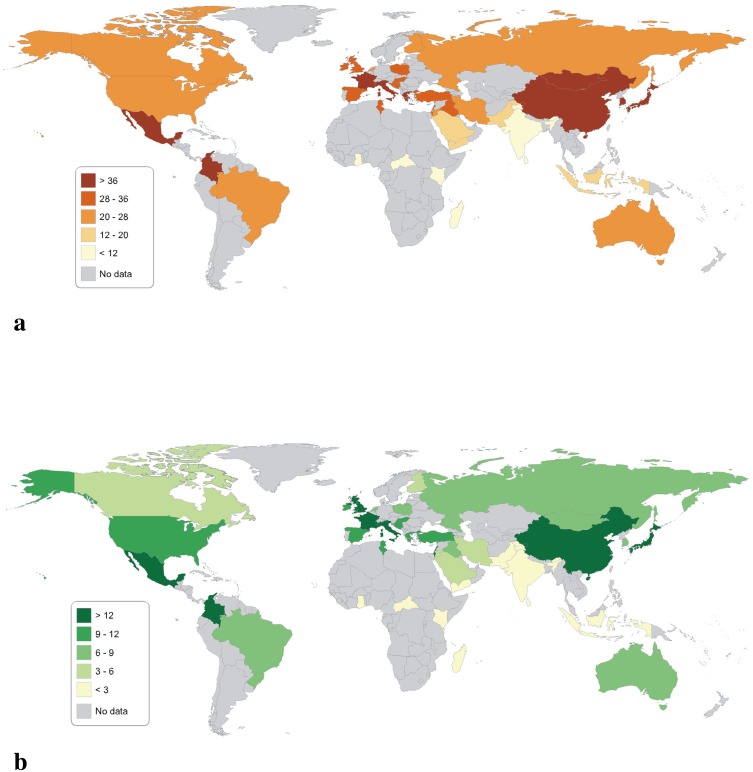
Country-specific prevalence estimates for T allele is presented in Fig. 5a. Visual inspection of the map, revealed the highest prevalence of T allele in China, Mexico and some part of Europe. Same trend are also seen for the TT genotype (Fig. 5b).
Fig. 5.
a Worldwide distribution of T allele; b worldwide distribution of TT genotype
Discussion
The objectives of present study were to determine (1) the frequency of MTHFR C677T polymorphism in Eastern UP population, and (2) the worldwide prevalence of C677T polymorphism by conducting a meta-analysis. MTHFR C677T has a high degree of heterogeneity in its world distribution. T allele is less prevalent in African population, its incidence is intermediate among Asian populations, and it is more prevalent in European and American populations. In present study, the prevalence of the mutant T allele was found as 11 % in the Eastern UP population (1 % TT homozygous) which is similar to that reported earlier from other Indian population. The main strength of this study is that this is the largest study (1000 samples) conducted on healthy individuals from Eastern U P population. The present study has some limitations like-only distribution of genotype in healthy subjects was reported and not presenting an association report.
In meta-analysis, global frequency of T allele was found 24.0 %. In subgroup analysis the frequency of T allele was 10.3 % in Africans, 31.2 % in North Americans, 27.8 % in South Americans, 19.7 % in Asians, 20.5 % in Australians and 34.1 % in Europeans. The global frequency of TT genotype was 7.7 %. In subgroup analysis, the frequency of TT genotype was 2.4 % in Africans, 11.9 % in North Americans, 7.6 % in South Americans, 5.5 % in Asians, 5.9 % in Australians and 11.6 % in Europeans. The frequency of both the T allele and TT genotype was lowest in Africans and highest in Europeans. Among Asian studies the frequency of this gene polymorphism was found higher in East Asian countries (44.7 % in China, 40.3 % in Korea, 39.9 % in Japan) than South Asian countries (11.4 % in India, 16 % in Pakistan, 4.5 % in Sri Lanka. The high frequency of T allele in East Asian countries might be due to folate and vitamin B12 rich non-vegetarian food habits of the population. Similarly, in South Asian countries highest frequency of T allele is found in Pakistan where majority of the population is non-vegetarian.
Meta-analysis is a powerful tool for analyzing cumulative data of studies where the individual sample sizes are small. During past decade several meta-analyses were published assessing MTHFR C677T gene polymorphism as risk factor for various diseases/disorders like Down syndrome [86, 87], cleft lip and palate [88], cardiovascular disease [89], diabetes [90], psychiatric disorder [91], neural tube defects [92], cancer [93] etc.
The meta-analysis has a number of strengths—(1) this is the first meta-analysis on the prevalence of MTHFR C677T gene polymorphism and (2) absence of publication bias. However, there are also some limitations—(1) we did not include case–control studies, (2) some studies might have not been included in this meta-analysis due to limitations of searched databases and (3) presence of high heterogeneity.
In conclusion, the frequency of the MTHFR C677T gene polymorphism discriminates the Indian population from other ethnic groups like Africans or Caucasians. Overall, our study showed a varied distribution of MTHFR C677T allele in various ethnic groups residing in diverse geographical regions of the world. The reason for the varied distribution seems to be directed not only by the environmental effect (specially folate and B12 rich/deficient diet) but also due to diversity in the origin and relatedness of various ethnic groups residing in the world. The aim of the study was to show countrywide prevalence of this highly important gene variant. Since MTHFR C677T polymorphism has been found to be associated with different diseases, the data would be very useful in regional health management programs and will facilitate predicting population-based risk factors for a number of congenital and other anomalies associated with MTHFR polymorphism.
Acknowledgments
The authors are grateful to all subjects for their participation in the study. This study was supported by the financial assistance from Department of Biotechnology, New Delhi (No. BT/PR98887/SPD/11/1028/2007) to Vandana Rai to conduct this study.
Compliance with Ethical Standards
Conflict of interest
None.
References
- 1.Goyette P, Pai A, Milos R, Frosst P, Tran P, Chen Z, et al. Gene structure of human and mouse methylenetetrahydrofolate reductase (MTHFR) Mamm Genome. 1998;9:652–656. doi: 10.1007/s003359900838. [DOI] [PubMed] [Google Scholar]
- 2.Frosst P, Blom HJ, Milos R, Goyette P, Sheppard CA, Matthews RG, et al. A candidate genetic risk factor for vascular disease: a common mutation in methylenetetrahydrofolate reductase. Nat Genet. 1995;10:111–113. doi: 10.1038/ng0595-111. [DOI] [PubMed] [Google Scholar]
- 3.Rozen R. Genetic predisposition to hyperhomocysteinemia: deficiency of methylenetetrahydrofolate reductase (MTHFR) Thromb Haemost. 1997;78:523–526. [PubMed] [Google Scholar]
- 4.Guenther BD, Sheppard CA, Tran P, Rozen R, Matthews RG, Ludwig ML. The structure and properties of methylenetetrahydrofolate reductase from Escherichia coli suggest how folate ameliorates human hyperhomocysteinemia. Nat Struct Biol. 1999;6:359–365. doi: 10.1038/7594. [DOI] [PubMed] [Google Scholar]
- 5.Yamada K, Chen Z, Rozen R, Matthews RG. Effects of common polymorphisms on the properties of recombinant human methylenetetrahydrofolate reductase. Proc Natl Acad Sci USA. 2001;98:14853–14858. doi: 10.1073/pnas.261469998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pejchal R, Campbell E, Guenther BD, Lennon BW, Matthews RG, Ludwig ML. Structural perturbations in the Ala → Val polymorphism of methylenetetrahydrofolate reductase: how binding of folates may protect against inactivation. Biochemistry. 2006;45:4808–4818. doi: 10.1021/bi052294c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yadav U, Kumar P, Rai V. Docking study of MTHFR with FAD by Hex. J Comput Intell Bioinform. 2011;4(1):171–178. [Google Scholar]
- 8.Homberger A, Linnebank M, Winter C, Willenbring H, Marquardt T, Harms E, et al. Genomic structure and transcript variants of the human methylenetetrahydrofolate reductase gene. Eur J Hum Genet. 2000;8(9):725–729. doi: 10.1038/sj.ejhg.5200522. [DOI] [PubMed] [Google Scholar]
- 9.Hobbs CA, Sherman SL, Yi P, Hopkins SE, Torfs CP, Hine RJ, et al. Polymorphisms in genes involved in folate metabolism as maternal risk factors for Down syndrome. Am J Hum Genet. 2000;67:623–630. doi: 10.1086/303055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cyril C, Rai P, Chandra N, Gopinath PM, Satyamoorthy K. MTHFR gene variants C677T, A1298C and association with Down syndrome: a case-control study from South India. Indian J Hum Genet. 2009;15:60–64. doi: 10.4103/0971-6866.55217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Godbole K, Gayathri P, Ghule S, Sasirekha BV, Kanitkar-Damle A, Memane N, et al. Maternal one-carbon metabolism, MTHFR and TCN2 genotypes and neural tube defects in India. Birth Defects Res A Clin Mol Teratol. 2011;91(9):848–856. doi: 10.1002/bdra.20841. [DOI] [PubMed] [Google Scholar]
- 12.Mills JL, Molloy AM, Parle-McDermott A, Troendle JF, Brody LC, Conley MR, et al. Folate-related gene polymorphisms as risk factors for cleft lip and cleft palate. Birth Defects Res A Clin Mol Teratol. 2008;82(9):636–643. doi: 10.1002/bdra.20491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Benes P, Kanková K, Muzík J, Groch L, Benedik J. Methylenetetrahydrofolate reductase polymorphism, type II diabetes mellitus, coronary artery disease, and essential hypertension in the Czech population. Mol Genet Metab. 2007;73:188–195. doi: 10.1006/mgme.2001.3188. [DOI] [PubMed] [Google Scholar]
- 14.Lakshmi SV, Naushad SM, Ruparsree Y, Rao DS, Kutala VK. Interactions of 5′-UTR thymidylate synthase polymorphism with 677C→T methylene tetrahydrofolate reductase and 66A→G methyltetrahydrofolate homocysteine methyl-transferase reductase polymorphisms determine susceptibility to coronary artery disease. J Atheroscler Thromb. 2010;18:56–64. doi: 10.5551/jat.5702. [DOI] [PubMed] [Google Scholar]
- 15.Gupta N, Gupta S, Dama M, David A, Khanna G, Khanna A, et al. Strong association of 677 C>T substitution in the MTHFR gene with male infertility-a study on an Indian population and a meta-analysis. PLoS ONE. 2011;6:e22277. doi: 10.1371/journal.pone.0022277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jönsson EG, Larsson K, Vares M, Hansen T, Wang AG, Djurovic S, et al. Two methylenetetrahydrofolate reductase gene (MTHFR) polymorphisms, schizophrenia and bipolar disorder: an association study. Am J Med Genet B Neuropsychiatr Genet. 2008;147B(6):976–982. doi: 10.1002/ajmg.b.30671. [DOI] [PubMed] [Google Scholar]
- 17.Kotsopoulos J, Zhang WW, Zhang S, McCready D, Trudeau M, Zhang P, et al. Polymorphisms in folate metabolizing enzymes and transport proteins and the risk of breast cancer. Breast Cancer Res Treat. 2008;112:585–593. doi: 10.1007/s10549-008-9895-6. [DOI] [PubMed] [Google Scholar]
- 18.Bartlett JMS, White A. Extraction of DNA from whole blood. In: Bartlett JMS, Stirling D, editors. Methods in molecular biology, vol. 226: PCR protocols. 2. Totowa: Humana Press Inc; 2003. pp. 29–31. [DOI] [PubMed] [Google Scholar]
- 19.Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst. 1959;22(4):719–748. [PubMed] [Google Scholar]
- 20.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 21.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 22.Whitehead A. Meta-analysis of controlled clinical trials. West Sussex: Wiley; 2002. [Google Scholar]
- 23.Wallace BC, Dahabreh IJ, Trikalinos TA, Lau J, Trow P, Schmid CH. Closing the gap between methodologists and end-users: R as a computational back-end. J Stat Softw. 2013;49:1–15. [Google Scholar]
- 24.StatPlanet software. http://www.statsilk.com/software/statplanet.
- 25.Schneider JA, Rees DC, Liu YT, Clegg JB. Worldwide distribution of a common methylenetetrahydrofolate reductase mutation. Am J Hum Genet. 1998;62:1258–1260. doi: 10.1086/301836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wilcken B, Bamforth F, Li Z, Zhu H, Ritvanen A, Renlund M, et al. Geographical and ethnic variation of the 677C>T allele of 5,10 methylenetetrahydrofolate reductase (MTHFR): findings from over 7000 newborns from 16 areas world-wide. J Med Genet. 2003;40:619–625. doi: 10.1136/jmg.40.8.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosenberg N, Murata M, Ikeda Y, Opare-Sem O, Zivelin A, Geffen E, et al. The frequent 5,10-methylenetetrahydrofolate reductase C677T polymorphism is associated with a common haplotype in whites, Japanese, and Africans. Am J Hum Genet. 2002;70:758–762. doi: 10.1086/338932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ameen G, Irani-Hakime N, Fawaz NA, Mahjoub T, Almawi WY. An Arab selective gradient in the distribution of factor V G1691A (Leiden), prothrombin G20210A, and methylenetetrahydrofolate reductase (MTHFR) C677T. J Thromb Haemost. 2005;3:2126–2127. doi: 10.1111/j.1538-7836.2005.01546.x. [DOI] [PubMed] [Google Scholar]
- 29.Arruda VR, Siqueira LH, Goncalves MS, von Zuben PM, Soares MC, Menezes R, et al. Prevalence of the mutation C677→T in the methylene tetrahydrofolate reductase gene among distinct ethnic groups in Brazil. Am J Med Genet. 1998;78:332–335. doi: 10.1002/(SICI)1096-8628(19980724)78:4<332::AID-AJMG5>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 30.Franco RF, Araújo AG, Guerreiro JF, Elion J, Zago MA. Analysis of the 677 C→T mutation of the methylenetetrahydrofolate reductase gene in different ethnic groups. Thromb Haemost. 1998;79(1):119–121. [PubMed] [Google Scholar]
- 31.Barbosa PR, Stabler SP, Machado AL, Braga RC, Hirata RD, Hirata MH, et al. Association between decreased vitamin levels and MTHFR, MTR and MTRR gene polymorphisms as determinants for elevated total homocysteine concentrations in pregnant women. Eur J Clin Nutr. 2008;62:1010–1021. doi: 10.1038/sj.ejcn.1602810. [DOI] [PubMed] [Google Scholar]
- 32.Stur E, Silveira AN, Selvatici LS, Alves LN, de Vargas Wolfgramm E, Tovar TT, et al. Polymorphism analysis of MTHFR, factor II, and factor V genes in the Pomeranian population of Espirito Santo, Brazil. Genet Test Mol Biomarkers. 2012;16:219–222. doi: 10.1089/gtmb.2011.0163. [DOI] [PubMed] [Google Scholar]
- 33.Romero-Sánchez C, Gómez-Gutierrez A, Gómez PE, Casas-Gomez MC, Briceño I. C677T (rs1801133) MTHFR gene polymorphism frequency in a Colombian population. Colomb Med (Cali) 2015;46(2):75–79. [PMC free article] [PubMed] [Google Scholar]
- 34.Mack R, Chowdary D, Streck D, Dermody J. Inherited thrombophilia genes in minorities. Genet Test. 1999;3:371–373. doi: 10.1089/gte.1999.3.371. [DOI] [PubMed] [Google Scholar]
- 35.Stevenson RE, Schwartz CE, Du YZ, Adams MJ., Jr Differences in methylenetetrahydrofolate reductase genotype frequencies, between Whites and Blacks. Am J Hum Genet. 1997;60:229–230. [PMC free article] [PubMed] [Google Scholar]
- 36.Mutchinick OM, Lopez MA, Luna L, Waxman J, Babinsky VE. High prevalence of the thermolabile methylenetetrahydrofolate reductase variant in Mexico: a country with a very high prevalence of neural tube defects. Mol Genet Metab. 1999;68:461–467. doi: 10.1006/mgme.1999.2939. [DOI] [PubMed] [Google Scholar]
- 37.Rady PL, Szucs S, Grady J, Hudnall SD, Kellner LH, Nitowsky H, et al. Genetic polymorphisms of methylenetetrahydrofolate reductase (MTHFR) and methionine synthase reductase (MTRR) in ethnic populations in Texas; a report of a novel MTHFR polymorphic site, G1793A. Am J Med Genet. 2002;107:162–168. doi: 10.1002/ajmg.10122. [DOI] [PubMed] [Google Scholar]
- 38.Bailey LB, Duhaney RL, Maneval DR, Kauwell GP, Quinlivan EP, Davis SR, et al. Vitamin B-12 status is inversely associated with plasma homocysteine in young women with C677T and/or A1298C methylenetetrahydrofolate reductase polymorphisms. J Nutr. 2002;132(7):1872–1878. doi: 10.1093/jn/132.7.1872. [DOI] [PubMed] [Google Scholar]
- 39.Chowdary D, Streck D, Schwalb MN, Dermody JJ. High incidence of two methylenetetrahydrofolate reductase mutations (C677T and A1298C) in Hispanics. Genet Test. 2003;7:255–257. doi: 10.1089/109065703322537296. [DOI] [PubMed] [Google Scholar]
- 40.Esfahani ST, Cogger EA, Caudill MA. Heterogeneity in the prevalence of methylenetetrahydrofolate reductase gene polymorphisms in women of different ethnic groups. J Am Diet Assoc. 2003;103:200–207. doi: 10.1053/jada.2003.50030. [DOI] [PubMed] [Google Scholar]
- 41.Vaughn JD, Bailey LB, Shelnutt KP, Dunwoody KM, Maneval DR, Davis SR, et al. Methionine synthase reductase 66A→G polymorphism is associated with increased plasma homocysteine concentration when combined with the homozygous methylenetetrahydrofolate reductase 677C→T variant. J Nutr. 2004;134:2985–2990. doi: 10.1093/jn/134.11.2985. [DOI] [PubMed] [Google Scholar]
- 42.Tsai MY, Loria CM, Cao J, Kim Y, Siscovick DS, Schreiner PJ, et al. Polygenic association with total homocysteine in the post-folic acid fortification era: the CARDIA study. Mol Genet Metab. 2009;98:181–186. doi: 10.1016/j.ymgme.2009.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ramos MA, Mares RE, Avalos ED, Hernandez A, Hernandez R, Lameda R, et al. Pharmacogenetic screening of N-acetyltransferase 2, thiopurine s-methyltransferase, and 5,10-methylene-tetrahydrofolate reductase polymorphisms in Northwestern Mexicans. Genet Test Mol Biomarkers. 2011;15:351–355. doi: 10.1089/gtmb.2010.0216. [DOI] [PubMed] [Google Scholar]
- 44.Harmon DL, Woodside JV, Yarnell JW, McMaster D, Young IS, McCrum EE, et al. The common ‘thermolabile’ variant of methylene tetrahydrofolate reductase is a major determinant of mild hyperhomocysteinaemia. QJM. 1996;89:571–577. doi: 10.1093/qjmed/89.8.571. [DOI] [PubMed] [Google Scholar]
- 45.Bowen DJ, Bowley S, John M, Collins PW. Factor V Leiden (G1691A), the prothrombin 3′-untranslated region variant (G20210A) and thermolabile methylenetetrahydrofolate reductase (C677T): a single genetic test genotypes all three loci-determination of frequencies in the S. Wales population of the UK. Thromb Haemost. 1998;79:949–954. [PubMed] [Google Scholar]
- 46.Hill AE, FitzPatrick DR. MS-PCR assay to detect 677C→T mutation in the 5,10-methylenetetrahydrofolate reductase gene. J Inherit Metab Dis. 1998;21:694–695. doi: 10.1023/A:1005413524070. [DOI] [PubMed] [Google Scholar]
- 47.Antoniadi T, Hatzis T, Kroupis C, Economou-Petersen E, Petersen MB. Prevalence of factor V Leiden, prothrombin G20210A, and MTHFR C677T mutations in a Greek population of blood donors. Am J Hematol. 1999;61:265–267. doi: 10.1002/(SICI)1096-8652(199908)61:4<265::AID-AJH8>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 48.Chango A, Boisson F, Barbe F, Quilliot D, Droesch S, Pfister M, et al. The effect of 677C→T and 1298A→C mutations on plasma homocysteine and 5,10-methylenetetrahydrofolate reductase activity in healthy subjects. Br J Nutr. 2000;83:593–596. doi: 10.1017/S0007114500000751. [DOI] [PubMed] [Google Scholar]
- 49.Mynett-Johnson LA, Keenan C, Black IL, Livingstone WJ, Lawler M, Roche HM, et al. Thermolabile methylenetetrahydrofolate reductase (C677T): frequency in the Irish population. Ir J Med Sci. 2002;171:37–39. doi: 10.1007/BF03168940. [DOI] [PubMed] [Google Scholar]
- 50.Zijno A, Andreoli C, Leopardi P, Marcon F, Rossi S, Caiola S, et al. Folate status, metabolic genotype, and biomarkers of genotoxicity in healthy subjects. Carcinogenesis. 2003;24:1097–1103. doi: 10.1093/carcin/bgg064. [DOI] [PubMed] [Google Scholar]
- 51.Lovricevic I, Franjic BD, Tomicic M, Vrkic N, De Syo D, Hudorovic N, et al. 5,10-Methylenetetrahydrofolate reductase (MTHFR) 677 C→T genetic polymorphism in 228 Croatian volunteers. Coll Antropol. 2004;28:647–654. [PubMed] [Google Scholar]
- 52.Spiridonova MG, Stepanov VA, Maksimova NR, Puzyrev VP. Population study of frequency of methylenetetrahydrofolate reductase C677T gene polymorphism in Yakutia. Genetika. 2004;40:704–708. [PubMed] [Google Scholar]
- 53.Sazci A, Ergul E, Kaya G, Kara I. Genotype and allele frequencies of the polymorphic methylenetetrahydrofolate reductase gene in Turkey. Cell Biochem Funct. 2005;23:51–54. doi: 10.1002/cbf.1132. [DOI] [PubMed] [Google Scholar]
- 54.Parle-McDermott A, Mills JL, Molloy AM, Carroll N, Kirke PN, Cox C, et al. The MTHFR 1298CC and 677TT genotypes have opposite associations with red cell folate levels. Mol Genet Metab. 2006;88:290–294. doi: 10.1016/j.ymgme.2006.02.011. [DOI] [PubMed] [Google Scholar]
- 55.Gialeraki A, Politou M, Rallidis L, Merkouri E, Markatos C, Kremastinos D, et al. Prevalence of prothrombotic polymorphisms in Greece. Genet Test. 2008;12:541–547. doi: 10.1089/gte.2008.0060. [DOI] [PubMed] [Google Scholar]
- 56.Ozarda Y, Sucu DK, Hizli B, Aslan D. Rate of T alleles and TT genotype at MTHFR 677C→T locus or C alleles and CC genotype at MTHFR 1298A→C locus among healthy subjects in Turkey: impact on homocysteine and folic acid status and reference intervals. Cell Biochem Funct. 2009;27:568–577. doi: 10.1002/cbf.1610. [DOI] [PubMed] [Google Scholar]
- 57.Zappacosta B, Romano L, Persichilli S, Cutrone LA, Graziano M, Vittani A, et al. Genotype prevalence and allele frequencies of 5,10-methylenetetrahydrofolate reductase (MTHFR) C677T and A1298C polymorphisms in Italian newborns. Labmed. 2009;40(12):732–736. [Google Scholar]
- 58.Gra O, Mityaeva O, Berdichevets I, Kozhekbaeva Z, Fesenko D, Kurbatova O, et al. Microarray-based detection of CYP1A1, CYP2C9, CYP2C19, CYP2D6, GSTT1, GSTM1, MTHFR, MTRR, NQO1, NAT2, HLA-DQA1, and ABO allele frequencies in native Russians. Genet Test Mol Biomarkers. 2010;14:329–342. doi: 10.1089/gtmb.2009.0158. [DOI] [PubMed] [Google Scholar]
- 59.Amela K, Rifet T, Zoran J, Jasminka MM. The frequency of C677T methylenetetrahydrofolate reductase (MTHFR) polymorphism in Southern East Bosnian population. J Biomet Biostat. 2013;4:169. doi: 10.4172/2155-6180.1000169. [DOI] [Google Scholar]
- 60.Seremak-Mrozikiewicz A, Barlik M, Borowczak P, Kurzawinska G, Krasnik W, Nowocien G, et al. The frequency of 677C→T polymorphism of MTHFR gene in the Polish population. Arch Perinat Med. 2013;19(1):12–18. [Google Scholar]
- 61.Friedman G, Goldschmidt N, Friedlander Y, Ben-Yehuda A, Selhub J, Babaey S, et al. A common mutation A1298C in human methylenetetrahydrofolate reductase gene: association with plasma total homocysteine and folate concentrations. J Nutr. 1999;129:1656–1661. doi: 10.1093/jn/129.9.1656. [DOI] [PubMed] [Google Scholar]
- 62.Zuo M, Lee MJ, Kim MH, Wu Y, Ayaki H, Nishio H, et al. C677T mutation of the methylenetetrahydrofolate reductase gene among the Korean infants in Seoul city. Kobe J Med Sci. 1999;45:271–279. [PubMed] [Google Scholar]
- 63.Pollak RD, Friedlander Y, Pollak A, Idelson M, Bejarano Achache I, Blumenfeld A. Ethnic differences in the frequency of the C677T mutation in the methylenetetrahydrofolate reductase (MTHFR) gene in healthy Israeli populations. Genet Test. 2000;4:309–311. doi: 10.1089/10906570050501560. [DOI] [PubMed] [Google Scholar]
- 64.Yu J, Chen B, Zhang G, Fu S, Li P. The 677 C→T mutation in the methylenetetrahydrofolate reductase (MTHFR) gene in five Chinese ethnic groups. Hum Hered. 2000;50:268–270. doi: 10.1159/000022929. [DOI] [PubMed] [Google Scholar]
- 65.Murakami S, Matsubara N, Saitoh M, Miyakaw S, Shoji M, Kubo T. The relation between plasma homocysteine concentration and methylenetetrahydrofolate reductase gene polymorphism in pregnant women. J Obstet Gynaecol Res. 2001;27:349–352. doi: 10.1111/j.1447-0756.2001.tb01284.x. [DOI] [PubMed] [Google Scholar]
- 66.Sadewa AH, Sunarti Sutomo R, Hayashi C, Lee MJ, Ayaki H, et al. The C677T mutation in the methylenetetrahydrofolate reductase gene among the Indonesian Javanese population. Kobe J Med Sci. 2002;48:137–144. [PubMed] [Google Scholar]
- 67.Al-Habboubi H, Tamim H, Ameen G, Almawi WY. A common mutation in 5,10-methylenetetrahydrofolate reductase (MTHFR) gene in two Arab communities. J Thromb Haemost. 2003;1:2246–2248. doi: 10.1046/j.1538-7836.2003.00390.x. [DOI] [PubMed] [Google Scholar]
- 68.Almawi WY, Ameen G, Tamim H, Finan RR, Irani-Hakime N. Factor V G1691A, prothrombin G20210A, and methylenetetrahydrofolate reductase [MTHFR] C677T gene polymorphism in angiographically documented coronary artery disease. J Thromb Thrombolysis. 2004;17:199–205. doi: 10.1023/B:THRO.0000040489.86029.27. [DOI] [PubMed] [Google Scholar]
- 69.Angeline T, Jeyaraj N, Granito S, Tsongalis GJ. Prevalence of MTHFR gene polymorphisms (C677T and A1298C) among Tamilians. Exp Mol Pathol. 2004;77:85–88. doi: 10.1016/j.yexmp.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 70.Golbahar J, Fathi Z, Tamadon M. Distribution of 5,10-methylenetetrahydrofolate reductase (C667T) polymorphism and its association with red blood cell 5-methyltetrahydrofolate in the healthy Iranians. Clin Nutr. 2005;24:83–87. doi: 10.1016/j.clnu.2004.07.019. [DOI] [PubMed] [Google Scholar]
- 71.Bhat TA, Mir MR, Qasim I, Misra SS, Kirmani MA. Genetic polymorphism of 5,10-methylenetetrahydrofolate reductase C677T in Kashmiri population. Biotechnology. 2008;7:822–825. doi: 10.3923/biotech.2008.822.825. [DOI] [Google Scholar]
- 72.Sabbagh AS, Mahfoud Z, Taher A, Zaatari G, Daher R, Mahfouz RA. High prevalence of MTHFR gene A1298C polymorphism in Lebanon. Genet Test. 2008;12:75–80. doi: 10.1089/gte.2007.0064. [DOI] [PubMed] [Google Scholar]
- 73.Saraswathy KN, Mukhopadhyay R, Sinha E, Aggarwal S, Sachdeva MP, Kalla AK. MTHFR C677T polymorphisms among the Ahirs and Jats of Haryana (India) Am J Hum Biol. 2008;20:116–117. doi: 10.1002/ajhb.20682. [DOI] [PubMed] [Google Scholar]
- 74.Al-Allawi NA, Avo AS, Jubrael JM. Methylenetetrahydrofolate reductase C677T polymorphism in Iraqi patients with ischemic stroke. Neurol India. 2009;57:631–635. doi: 10.4103/0028-3886.57821. [DOI] [PubMed] [Google Scholar]
- 75.Algasham A, Ismail H, Dewaidar M, Settin AA. Methylenetetrahydrofolate reductase and angiotensin-converting enzyme gene polymorphisms among Saudi population from Qassim region. Genet Test Mol Biomarkers. 2009;13:817–820. doi: 10.1089/gtmb.2009.0019. [DOI] [PubMed] [Google Scholar]
- 76.Mansoor A, Mazhar K, Ali L, Muazzam AG, Siddiqi S, Usman S. Prevalence of the C677T single-nucleotide polymorphism in the methylenetetrahydrofolate reductase gene among Pakistani ethnic groups. Genet Test Mol Biomarkers. 2009;13:521–526. doi: 10.1089/gtmb.2009.0012. [DOI] [PubMed] [Google Scholar]
- 77.Bagheri M, Abdi Rad I. Frequency of the Methylenetetrahydrofolate reductase 677CT and 1298AC mutations in an Iranian Turkish female population. Maedica (Buchar) 2010;5:171–177. [PMC free article] [PubMed] [Google Scholar]
- 78.Misra UK, Kalita J, Srivastava AK, Agarwal S. MTHFR gene polymorphism and its relationship with plasma homocysteine and folate in a North Indian population. Biochem Genet. 2010;48:229–235. doi: 10.1007/s10528-009-9312-9. [DOI] [PubMed] [Google Scholar]
- 79.Ghodke Y, Chopra A, Shintre P, Puranik A, Joshi K, Patwardhan B. Profiling single nucleotide polymorphisms (SNPs) across intracellular folate metabolic pathway in healthy Indians. Indian J Med Res. 2011;133:274–279. [PMC free article] [PubMed] [Google Scholar]
- 80.Murry B, Vakha N, Achoubi N, Sachdeva MP, Saraswathy KN. APOE, MTHFR, LDLR and ACE polymorphisms among Angami and Lotha Naga populations of Nagaland, India. J Community Health. 2011;36:975–985. doi: 10.1007/s10900-011-9397-z. [DOI] [PubMed] [Google Scholar]
- 81.Sukla KK, Raman R. Association of MTHFR and RFC1 gene polymorphism with hyperhomocysteinemia and its modulation by vitamin B12 and folic acid in an Indian population. Eur J Clin Nutr. 2012;66:111–118. doi: 10.1038/ejcn.2011.152. [DOI] [PubMed] [Google Scholar]
- 82.Sachdeva S, Saraswathy KN, Gulabani M, Kaushik S, Sachdeva MP, Puri M, et al. MTHFR C677T polymorphism among three Mendelian populations: a study from North India. Biochem Genet. 2012;50:893–897. doi: 10.1007/s10528-012-9529-x. [DOI] [PubMed] [Google Scholar]
- 83.Saraswathy KN, Asghar M, Samtani R, Murry B, Mondal PR, Ghosh PK, et al. Spectrum of MTHFR gene SNPs C677T and A1298C: a study among 23 population groups of India. Mol Biol Rep. 2012;39:5025–5031. doi: 10.1007/s11033-011-1299-8. [DOI] [PubMed] [Google Scholar]
- 84.Dajani R, Fathallah R, Arafat A, AbdulQader ME, Hakooz N, Al-Motassem Y, et al. Prevalence of MTHFR C677T single nucleotide polymorphism in genetically isolated populations in Jordan. Biochem Genet. 2013;51:780–788. doi: 10.1007/s10528-013-9606-9. [DOI] [PubMed] [Google Scholar]
- 85.Yang B, Liu Y, Li Y, Fan S, Zhi X, Lu X, et al. Geographical distribution of MTHFR C677T, A1298C and MTRR A66G gene polymorphisms in China: findings from 15357 adults of Han nationality. PLoS ONE. 2013;8:e57917. doi: 10.1371/journal.pone.0057917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wu X, Wang X, Chan Y, Jia S, Luo Y, Tang W. Folate metabolism gene polymorphisms MTHFR C677T and A1298C and risk for down syndrome offspring: a meta-analysis. Eur J Obstet Gynecol Reprod Biol. 2013;167(2):154–159. doi: 10.1016/j.ejogrb.2012.11.022. [DOI] [PubMed] [Google Scholar]
- 87.Rai V, Yadav U, Kumar P, Yadav SK, Mishra OP. Maternal methylenetetrahydrofolate reductase C677T polymorphism and Down syndrome risk: a meta-analysis from 34 studies. PLoS ONE. 2014;9:e108552. doi: 10.1371/journal.pone.0108552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Verkleij-Hagoort A, Bliek J, Sayed-Tabatabaei F, Ursem N, Steegers E, Steegers-Theunissen R. Hyperhomocysteinemia and MTHFR polymorphisms in association with orofacial clefts and congenital heart defects: a meta-analysis. Am J Med Genet A. 2007;143A:952–960. doi: 10.1002/ajmg.a.31684. [DOI] [PubMed] [Google Scholar]
- 89.Xuan C, Bai XY, Gao G, Yang Q, He GW. Association between polymorphism of methylenetetrahydrofolate reductase (MTHFR) C677T and risk of myocardial infarction: a meta-analysis for 8,140 cases and 10,522 controls. Arch Med Res. 2011;42:677–685. doi: 10.1016/j.arcmed.2011.11.009. [DOI] [PubMed] [Google Scholar]
- 90.Yang S, Zhang J, Feng C, Huang G. MTHFR 677T variant contributes to diabetic nephropathy risk in Caucasian individuals with type 2 diabetes: a meta-analysis. Metabolism. 2013;62(4):586–594. doi: 10.1016/j.metabol.2012.10.004. [DOI] [PubMed] [Google Scholar]
- 91.Peerbooms OL, van Os J, Drukker M, Kenis G, Hoogveld L, MTHFR in Psychiatry Group et al. Meta-analysis of MTHFR gene variants in schizophrenia, bipolar disorder and unipolar depressive disorder: evidence for a common genetic vulnerability? Brain Behav Immun. 2011;25(8):1530–1543. doi: 10.1016/j.bbi.2010.12.006. [DOI] [PubMed] [Google Scholar]
- 92.Yadav U, Kumar P, Yadav SK, Mishra OP, Rai V. Polymorphisms in folate metabolism genes as maternal risk factor for neural tube defects: an updated meta-analysis. Metab Brain Dis. 2015;30:7–24. doi: 10.1007/s11011-014-9575-7. [DOI] [PubMed] [Google Scholar]
- 93.Rai V. The methylenetetrahydrofolate reductase C677T polymorphism and breast cancer risk in Asian populations. Asian Pac J Cancer Prev. 2014;15(14):5853–5860. doi: 10.7314/APJCP.2014.15.14.5853. [DOI] [PubMed] [Google Scholar]