Abstract
Sulforaphane (SFN) plays an important role in preventing oxidative stress by activating the nuclear factor (erythroid derived 2)-like 2 (Nrf2) signalling pathway. SFN may improve exercise endurance capacity by counteracting oxidative stress-induced damage during exercise. We assessed running ability based on an exhaustive treadmill test (progressive-continuous all-out) and examined the expression of markers for oxidative stress and muscle damage. Twelve- to 13-week-old Male wild-type mice (Nrf2 +/+) and Nrf2-null mice (Nrf2 −/−) on C57BL/6J background were intraperitoneally injected with SFN or vehicle prior to the test. The running distance of SFN-injected Nrf2 +/+ mice was significantly greater compared with that of uninjected mice. Enhanced running capacity was accompanied by upregulation of Nrf2 signalling and downstream genes. Marker of oxidative stress in SFN-injected Nrf2 +/+ mice were lower than those in uninjected mice following the test. SFN produced greater protection against muscle damage during exhaustive exercise conditions in Nrf2 +/+ mice than in Nrf2 −/− mice. SFN-induced Nrf2 upregulation, and its antioxidative effects, might play critical roles in attenuating muscle fatigue via reduction of oxidative stress caused by exhaustive exercise. This in turn leads to enhanced exercise endurance capacity. These results provide new insights into SFN-induced upregulation of Nrf2 and its role in improving exercise performance.
Introduction
The production of reactive oxygen species (ROS) increases during muscle contraction1,2. Although biological systems are able to detoxify ROS and their reactive intermediates, sustained elevation in ROS levels during repetitive or prolonged muscle contractions may produce imbalances in normal redox states, generating oxidative stress3. Exhaustive exercise training in particular promotes bursts in ROS production4. Prolonged exercise causes structural damage and contractile dysfunction in skeletal muscle3,5. In this manner, redox disturbance within skeletal muscles leads to progressive muscular weakness and fatigue (i.e. reduced force capacity and slowed contraction rate). These phenomena manifest as reduced exercise endurance capacity2,3,5–7.
Recent studies highlight the role of transcription factor nuclear factor (erythroid derived 2)-like 2 (Nrf2) in expression of antioxidant response element (ARE)-driven endogenous antioxidant/xenobiotic detoxifying enzymes, and the role of Nrf2 in attenuating oxidative damage8. Modulation of Nrf2-related antioxidant signalling may preserve redox and functional homeostasis in tissues and organs9.
Sulforaphane (SFN), a chemopreventive compound produced by the Brassicaceae family, is a potent activator of Nrf210. Many studies suggest that SFN-induced Nrf2 activation may exert cytoprotective effects on various organs damaged by oxidative stress11. Malaguti et al.12 reported that SFN modulates the redox environment in muscle and attenuates damage to skeletal muscle via the Nrf2-ARE pathway. The antioxidant activity of SFN may contribute to increased exercise endurance capacity by reducing muscle fatigue caused by exercise-induced oxidative stress2,3,5–7.
In this study, we employed SFN to activate the Nrf2-ARE pathway in mice. We assessed effects of Nrf2 activation on exercise endurance capacity by estimating running distances of mice subjected to an exhaustive (progressive-continuous all-out) treadmill test. The expression of markers for oxidative stress and tissue damage were then measured in mouse muscles.
Materials and Methods
Animals and Treatments
The present study was conducted in accordance with the principles and guidelines for international animal care and was approved by the Institutional Animal Care and Use Committees of the University of Tsukuba (Ibaraki, Japan).
Twelve- to 13-week-old (body weight, 25–28 g) male wild-type mice (presence of Nrf2; Nrf2 +/+) and Nrf2-null mice (absence of Nrf2; Nrf2 −/−) on C57BL/6J background were housed in a colony cage under a 12 h:12 h light-dark cycle at 22.5 ± 1.4 °C and 55.6 ± 4.0% relative humidity. Mice were provided with a pellet rodent diet and water ad libitum.
Subject mice of both genotypes (Nrf2 +/+ and Nrf2 −/−) were randomly divided into the following four groups of eight mice each: 1) Nrf2 +/+ mice pretreated with sulforaphane (SFN) (Nrf2 +/+ [SFN]); 2) Nrf2 +/+ mice pretreated with vehicle (Nrf2 +/+ [CON]); 3) Nrf2 −/− mice pretreated with SFN (Nrf2 −/− [SFN]); and 4) Nrf2 −/− mice pretreated with vehicle (Nrf2 −/− [CON]). Mice were injected with SFN or vehicle four times for 3 days (72, 48, 24, and 3 h prior to the exhaustive treadmill tests). The injection frequency and timing of SFN injection was based on results from preliminary experiments to determine optimum Nrf2 activation. Nrf2 +/+ [SFN] and Nrf2 −/− [SFN] mice were intraperitoneally injected with SFN [25 mg/kg12 (MPBio, CA)]. SFN was solubilized in 100% dimethyl sulfoxide (Nacalai Tesque Inc.), and the stock solution was diluted with phosphate-buffered saline. Nrf2 +/+ [CON] and Nrf2 −/− [CON] mice were injected with equal amounts of dimethyl sulfoxide dissolved in phosphate-buffered saline (Fig. 1).
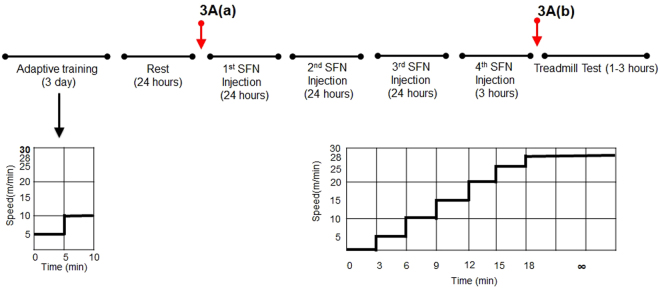
Figure 1.
Experimental protocol. For 3 days, all mice performed 10 min of adaptive training at low speed for 5–10 m/min. Then, all mice were injected with SFN or vehicle four times for 3 days (72, 48, 24, and 3 h prior the exhaustive treadmill tests). On the day of the test, all mice conducted the experimental protocol. Exercise intensity was incrementally increased from running with the treadmill set at an initial speed of 5 m/min to a maximum speed of 28 m/min every 3 min. The maximal velocity was maintained until mice were exhausted. Red arrows indicate the time course of in vivo imaging after SFN or vehicle injection. Results are presented in Fig. 3A[(a)~(b)].
Sample collection
All mice were sacrificed by cervical dislocation under diethylether anaesthesia at baseline, at 50 min following initiation of the exhaustive treadmill test, immediately upon completion of the test, and 18 h following completion of the test. Quadriceps, gastrocnemius, and soleus muscles on the animals’ right side were dissected. Blood was collected from the abdominal aorta and centrifuged (3000 rpm for 20 min at 4 °C) to isolate serum. Samples were frozen in liquid nitrogen and stored at −80 °C pending analysis.
Antibodies and immunoblot analyses
Antibodies were obtained from the following sources: Peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1α; Abcam Cat. #ab54481); AMP-activated protein kinase α (AMPKα; Cell Signaling Cat. # 2532); phosphonate-AMPKα (Cell Signaling Cat. # 2535); Sirtuins 1 (SirT1; Cell Signaling Cat. # 2310); α-tubulin (Cell Signaling Cat. # 3873); and Lamin A/C (Cell Signaling Cat. # 4777).
For western blot analysis, right gastrocnemius muscle lysates were separated by SDS-PAGE and transferred onto nitrocellulose membrane (Bio-Rad, Mississauga, Canada). The membrane was incubated in blocking buffer for one hour (Blocking one-P; Nacalai Tesque Inc.), followed by incubation overnight at 4 °C with the primary antibodies. Then, the blot was incubated with HRP-conjugated secondary antibodies for 2 hours at room temperature and signals were visualized with enhanced chemiluminescence (Chemi-Lumi One Super; Nacalai Tesque Inc.). Target proteins was exposed using by a ChemiDoc XRS + system chemiluminescence imager (Bio-Rad, Hercules, CA, USA). Image Lab software (Bio-Rad) was used for image acquisition and densitometry analysis.
Nuclear protein was extracted from right gastrocnemius muscle using a NE-PER™ Nuclear and Cytoplasmic Extraction Reagent Kit (Thermo Fisher Scientific, MA, USA), according to the kit protocol. Nuclear samples were analysed by SDS-PAGE and western blotting as described above.
Histochemical analysis
Right gastrocnemius muscles were placed in isopentane (C5H12; Nacalai Tesque Inc.) and frozen in liquid nitrogen. A serial cross-section (10 µm) of the muscle was cut transversely using a cryostat (CM 300; Leica Japan, Tokyo, Japan) at −20 °C and stained for myosin-ATPase (preincubation: pH 10.8). Section images were captured using a photomicrographic camera (Olympus AX70; Olympus, Tokyo, Japan) attached to a light microscope (Olympus BX51; Olympus). Images were processed and randomly calculated for percent fibre type (Type I stained light and Type II stained dark) and for cross-sectional areas, using image analysis software (ImageJ; National Institutes of Health, Bethesda, MD, USA)
RNA isolation and reverse transcription
Right gastrocnemius and soleus muscle tissues (20 mg) were homogenized in 1 mL Sepasol-RNA I Super G (Nacalai Tesque Inc.). RNA was isolated via chloroform extraction and precipitated with 2-propanol. RNA was then washed with 75% ethanol and re-dissolved in RNAse-free water. RNA quality and quantity were determined using a spectrophotometer (NanoVue; GE Healthcare, Uppsala, Sweden). First-strand cDNA synthesis was performed using a PrimeScript RT reagent kit (Takara Bio Inc., Shiga, Japan) following the kit protocol.
Mitochondrial DNA Copy Number
Genomic DNA in right gastrocnemius muscle tissues was extracted using a Qiagen DNA Genomic-tip kit according to the kit protocol. Using the DNA, mitochondrial copy number was estimated by real-time quantitative polymerase chain reaction as described below from the respective quantities of nuclear and mitochondrial DNA, which we considered to be the concentration of mitochondria per cell in the tissue.
The following primers (Fasmac, Tokyo, Japan) were used in this assessment: Mitochondrial DNA [mtDNA; F, 5′-CCCTAAAACCCGCC ACATCT-3′, R, 5′-GAGCGATGGTGAGAGC TAAGGT-3′], nuclear DNA [nDNA; F, 5′-CGAGTCGTCTTT CTCCTGATGAT-3′, R, 5′-TTCTGGATTCCAA TGCTTCGA-3′].
Real-time quantitative polymerase chain reaction
Two microliters of the DNAs (diluted to 1:10) were added to Fast SYBR Green Master Mix (Applied Biosystems, Santa Ana, CA). Polymerase chain reaction was performed in 20 µL reaction volumes on a CFX384 Touch Real-Time PCR Detection System (Bio-Rad Lab, CA, US). The following PCR cycling conditions were used for all primer pairs: initial enzyme activation at 95 °C for 20 s followed by 40 cycles of denaturation at 95 °C for 1 s and annealing at 60 °C for 20 s. Fluorescence data were collected at the end of the extension step. After cycling, the melting curve was determined in the range of 65–95 °C with temperature increments of 0.1 °C/s. Each reaction was run in triplicate with appropriate negative controls. Data analysis was performed using the delta-CT-method.
The following primers (Fasmac) were used in this study: haeme oxygenase-1 [HO-1; Forward (F), 5′-CCTCACTGGCAGGAAATCATC-3′, Reverse (R), 5′-CCTCGTGGAGACGCTTTACATA-3′], NAD (P) H: quinone oxidoreductase NQO1; [F, 5′-GGGTCGTCTTGGCAACCA-3′, R, 5′-CAGATGTTGAGGGAGGATCGTAA-3′], γ-glutamylcysteine synthetase [γ-GCS; F, 5′-AGAAGGGGGAGAGGACAAAC-3′, R, 5′-AGTGATGGTGCAGAGAGCCT-3′], and catalase [F, 5′-GGTCACCCACGATATATCACCAGATAC-3′, R, 5′-CGAGGGTCACGAACTGTGTCA-3′], nuclear respiratory factor 1 [NRF-1; F, 5′-GAACGCCACCGATTTCACTGTC-3′, R,5′-CCCTACCACCCACGAATCTGG-3′], transcription factor A, mitochondrial [TFAM; F,5′-CTGATGGGTATGGAGAAGGAGG-3′, R, 5’-CCAACTTCAGCCATCTGCTCTTC-3′], p53R2 [F, 5′-CCAGGTTACCATGGTTGTGG-3′, R, 5′-CCAGTGCACTCAGTAGCTGTG-3′], cytochrome c oxidase [COX IV; F, 5′-ACCAAGCGAATGCTGGACAT-3′, R, 5′-GGCGGAGAAGCCCTGAA-3′], SCO1 [F,5′-CTAGCTTAGCACAATAGCAAGGGCAGGCTAC-3′, R,5′-CCCAGGAATGCAGTTATGACATGACAGCAAAGGCA-3′
SCO2[F, 5′-CAGCCTGTCTTCATCACTGTGGA-3′, R, 5′-GACACTGTGGAAGGCAGCTATGTGCC-3′]. Data were normalized against GAPDH [F, 5′-GTCTTCACCACCATGGAGAAGGCT-3′ and R, 5′-CATGCCAGTGAGCTTCCCGTTCA-3′] and expressed as mean values.
Biochemical analysis
Serum levels of lactate dehydrogenase (LDH) and creatine phosphokinase (CPK) in serum samples were analysed based on the Japan Society of Clinical Chemistry transferable method. Free fatty acids (FFAs) were analysed using the enzymatic method of Kotobiken Co. Ltd. (Tokyo, Japan). Blood samples were obtained by nicking the lateral tail vein using sharp surgical scissors. Blood glucose level was measured using a hand-held glucometer (Ascensia Breeze®2; Bayer Health Care, Osaka, Japan). Blood lactate level was determined using a hand-held lactate analyser (Lactate Pro; Arkray KDK, Kyoto, Japan). Blood was sampled through the tail vein. A commercial enzyme-linked immunosorbent assay kit was used to measure levels of thiobarbituric acid reactive substances (TBARS; Cayman Chemical, MI) from homogenates of gastrocnemius muscles. Reduced (GSH) and oxidized (GSSG) glutathione levels of gastrocnemius muscles were assayed using a GSH/GSSG Ratio Detection Assay Kit (Fluorometric, Abcam, Cambridge, UK).
Adenosine triphosphate (ATP) levels in right quadriceps muscles were measured using a luciferin-luciferase assay kit (TOYO Ink, Tokyo, Japan.). Glycogen content in the quadriceps muscles was measured using a Glycogen Assay kit (Biovision, CA). The level of cyclic AMP (cAMP) in gastrocnemius muscle was measured using a cAMP Direct Immunoassay Detection Kit (Fluorometric, Abcam, Cambridge, UK).
Exhaustive (progressive-continuous all-out) treadmill test
A motor-driven two-lane treadmill chamber was used during the exhaustive treadmill tests to analyse respiratory parameters such as oxygen (VO2) consumption, carbon dioxide (VCO2) production, and respiratory quotient (RQ) (Muromachi, Tokyo, Japan). Fresh air was pumped into the chamber at a flow rate of 1.5 L/min during the tests.
For 3 days prior to injection with SFN or vehicle, all mice were subjected to 10 min adaptive training at low speed (5–10 m/min) to familiarize them with the treadmill. During the test, all mice were placed on a horizontal treadmill, after which exercise intensity was incrementally increased from an initial running speed of 5 m/min to a maximum speed of 28 m/min every 3 min; maximal velocity was maintained until mice were exhausted (see below for exhaustion criteria) (Fig. 1). All experimental protocols were performed during the mouse dark cycle.
The criterion for exhaustion was defined as the point at which the mice failed to run on the treadmill even after hand prodding using a soft bristle brush and electrical grid and failure of the righting reflex when the mice were laid on their backs.
Indirect calorimetry
Metabolic parameters were measured using an indirect calorimeter (Muromachi) at the Laboratory Animal Resource Center (University of Tsukuba). Calorimetry measurements were acquired during a 2-day acclimation period at baseline. VO2 and VCO2 levels were measured in each chamber every 3 min. The flow rate was set at 0.6 L/min during the measurements. Energy expenditure (EE) was calculated as VO2 × [3.815 + (1.232 × RQ)]13.
Nrf2-Luciferin in vivo imaging and luciferase activity detection
Transgenic OKD48 (Keap1-dependent oxidative stress detector, No-48-luciferase) mice were used for Nrf2 activity detection using in vivo imaging14. We used mice produced by crossing an OKD48 mouse with an Nrf2+/+ (albino-BL/6) or Nrf2−/− (albino-Nrf2−/−) mouse to improve signal detection. After intraperitoneal injection with 150 mg/kg d-luciferin (Promega, Madison, WI, USA) dissolved in PBS, mice were imaged using an In Vivo Imaging System (Vivo Vision IVIS Spectrum; Xenogen, Alameda, CA, USA) under anaesthesia with isoflurane. At 2 min following injection, mice were placed in the In Vivo Imaging System imaging chamber. Data were collected using high sensitivity, 10 min exposure, followed by analysis with Living Image software (Xenogen).
Luciferase reporter assays
We used the Luciferase Assay System (Promega, Madison, WI, USA). Briefly, 20 mg of the right gastrocnemius muscle containing the firefly luciferase gene of the OKD48 mouse was homogenized in Passive Lysis Buffer. After incubation at 4 °C for 3 h, homogenates were centrifuged at 12,000 rpm for 5 min. Supernatants (20 μL) were centrifuged and mixed with 100 μL of luciferase assay reagent, after which total light emission was measured using a TD-20/20 luminometer (Turner Design, CA, USA) according to the manufacturer’s protocol. The total protein in each supernatant was measured using the Pierce BCA protein assay kit (Thermo Fisher Scientific). Luciferase activities were calculated per mg protein in the gastrocnemius muscle.
Statistics
Statistical analysis was performed using SPSS Statistics for Windows, version 23.0 (IBM Corporation, Armonk, NY, USA). Descriptive parameters were expressed as mean ± SE. To compare between groups for all dependent variables, we performed analysis with a one-way analysis of variance. P < 0.05 was considered significant.
Results
Baseline assessment
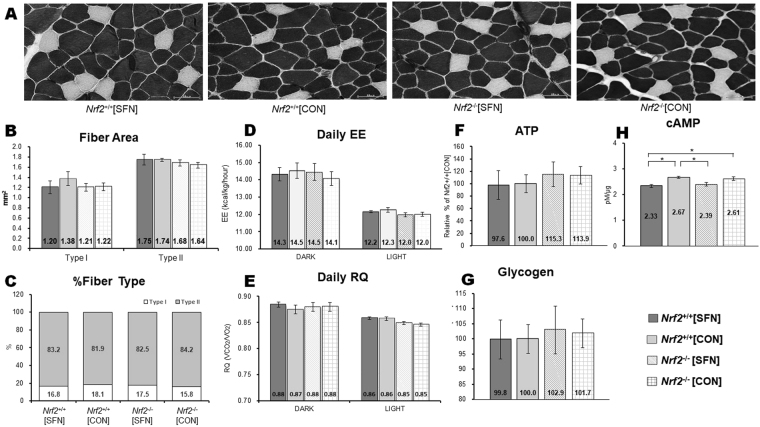
ATPase-stained cross sections of muscle fibres from the central portion of the deep gastrocnemius of the lower right limb were obtained from Nrf2 +/+ and Nrf2 −/− mice injected with SFN or vehicle (Fig. 2A). Diameters of Type I and Type II fibres were not significantly different in Nrf2 +/+ and Nrf2 −/− mice. Fibre diameters did not change following four SFN injections (Fig. 2B). There were no significant differences in the ratio of Type I to Type II muscle fibres between Nrf2 +/+ and Nrf2 −/− mice. These ratios were not significantly altered following four SFN injections (Fig. 2C). There were no significant differences in energy expenditure (Fig. 2D) and respiratory quotient (RQ) (Fig. 2E) for each dark and light cycle among all groups. There were no significant differences in mean levels of ATP (Fig. 2F), glycogen (Fig. 2G), and cAMP (Fig. 2H) among all groups. These data suggest that Nrf2 genotype and SFN injection did not significantly affect muscle fibre morphology, physiology, or metabolism at baseline.
Figure 2.
Fibre types in gastrocnemius muscle obtained by ATPase staining at pH 10.8 following SFN or vehicle injection (A). Fibre area (B) and fibre-type distribution (C) in the gastrocnemius muscle were determined by ATPase staining. Daily EE (D) and RQ (E) values assessed during each dark and light cycle using an indirect calorimeter. ATP (F), Glycogen (G) and cAMP (H) levels were measured in gastrocnemius muscle.
Mitochondria biogenesis
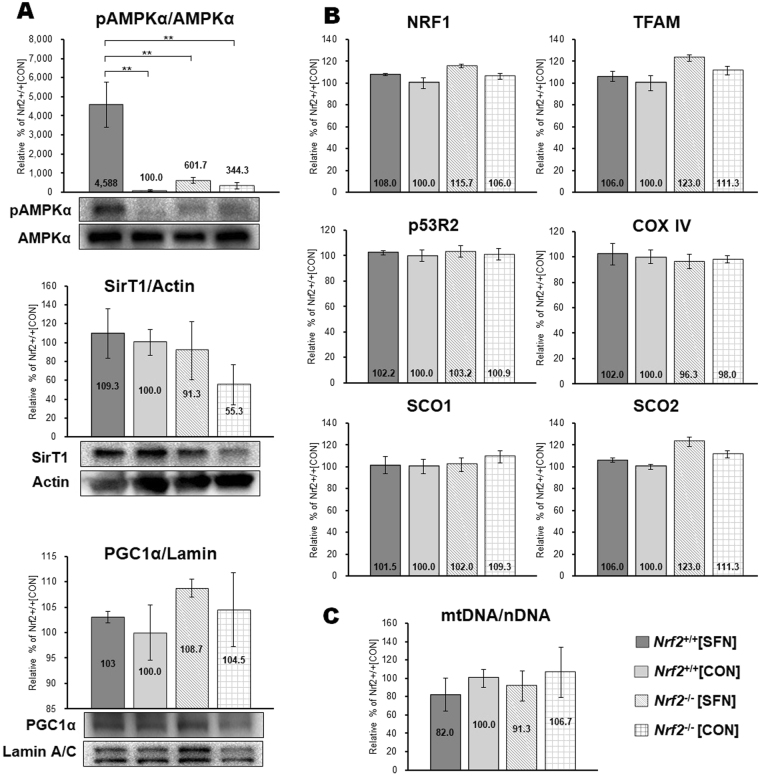
Figure 3 displays biomarkers for mitochondria biogenesis in gastrocnemius homogenates at baseline. Western blot analysis revealed that phosphorylated AMPKα significantly increased in Nrf2 +/+[SFN] mice following four SFN injections (Fig. 3A). However, there were no significant differences in protein expression of SirT1 and PGC1α among all groups. Figure 3B shows mRNA expression of mitochondrial biogenesis markers, including NRF-1, TFAM, p53R2, COX IV, SCO1 and SCO2. No elevated mRNA expression was detected in any group. There was an elevated mtDNA copy number relative to nDNA. This is considered a good marker for mitochondrial biogenesis15. After the fourth SFN injection in Nrf2 +/+ mice and in Nrf2 in Nrf2 −/− groups, mtDNA copy number was not significantly different (Fig. 3C). These data suggest that the connection was weak between activating mitochondria biogenesis signaling and the SFN injection, which was administered only four times.
Figure 3.
Effect of SFN injection. SFN or vehicle was injected four times for 3 days and mitochondrial biogenesis markers were measured in mice gastrocnemius muscle. (A) protein expression levels (western blot strips) of mitochondrial biogenesis markers. The amount of phosphorylated AMPKα was normalized to the amount of AMPKα protein. Protein content of mitochondrial biogenesis markers SirT1 and PGC1α were also measured. The loading volume was normalized by the expression levels of actin and lamin A/C, respectively. (B) mRNA expression of mitochondrial biogenesis markers in AA. mRNA levels of NRF1, TFAM, p53R2, COX IV, SCO1, and SCO2 were determined by real-time PCR. Values were normalized to the level of the housekeeping gene, GAPDH. (C) mtDNA copy number in the gastrocnemius muscle. This number factors into the ratio of nuclear and mtDNA (mtDNA/nDNA); brackets **P < 0.01.
Luminescence activity
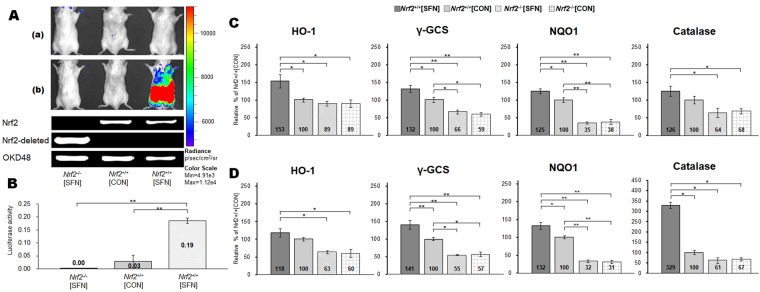
Figure 4A shows real-time in vivo imaging of an Nrf2 response during SFN injection in transgenic OKD48 (Keap1-dependent oxidative stress detector, No-48-luciferase) mice. The OKD48 mice expressed the N-terminal half of Nrf2-Luc fusion protein, driven by the Nrf2-based 3xAre promoter13. No Nrf2-Luc activation was detected in Nrf2 +/+[CON] and Nrf2 −/−[CON] mice. Substantial Nrf2 upregulation was observed in Nrf2 +/+[SFN] mice 3 h following the fourth SFN injection (at 75 h), shortly before the exhaustion treadmill test [Fig. 4A(a) and (b)]. During the SFN injection period, Nrf2-Luc activation gradually increased in Nrf2 +/+[SFN] mice at baseline at 24, 48, 72, and 75 h [Fig. S1(a),(b),(c),(d), and (e)]. Nrf2-Luc was not activated in the Nrf2 +/+[CON] and Nrf2 −/−[CON] groups. Luciferase activity of the gastrocnemius muscle is shown in Fig. 4B. Nrf2-Luc was upregulated in the muscles of Nrf2 +/+[SFN] mice at 75 h. This change was significantly higher than those of the other groups (P < 0.01). These results suggest that SFN pretreatment upregulates Nrf2.
Figure 4.
SFN-induced Nrf2-Luc activity detected using an In Vivo Imaging System and results of genotyping of Nrf2 and OKD48 transgenic mice (A) in the prone position. Time course [(a: baseline) ~ (b: following SFN injections)] of luciferase assay is shown in Fig. 1. Nrf2-Luc activity in gastrocnemius muscle after the fourth SFN injection measured using a luminometer (B). mRNA levels of Nrf2 target genes in gastrocnemius (C) and soleus (D) muscles following four injections of SFN or vehicle. Brackets *P < 0.05, **P < 0.01.
Nrf2 target genes
Figure 4C,D display the mRNA levels of selected Nrf2 target genes: haeme oxygenase-1 (HO-1), NAD (P) H: quinone oxidoreductase A (NQO1), gamma-glutamylcysteine synthetase (γ-GCS), and catalase. These levels were measured in gastrocnemius (Fig. 4C) and soleus (Fig. 4D) tissue homogenates with and without SFN pretreatment. Nrf2 +/+[SFN] mice demonstrated significantly higher expression levels of all target genes in both the gastrocnemius and soleus muscles compared with Nrf2 −/− groups (Nrf2 −/− [SFN] and Nrf2 −/−[CON]). Between the two Nrf2 +/+groups, catalase was not upregulated in the gastrocnemius. HO-1 was not upregulated in the soleus in Nrf2 +/+[SFN] mice than those in Nrf2 +/+[CON] mice. However, expression levels of the other Nrf2 target genes were significantly higher in both tissues in Nrf2 +/+[SFN] mice compared with those in Nrf2 +/+[CON]: NQO1 and γ-GCS expression levels in gastrocnemius and soleus of Nrf2 +/+[CON] mice were significantly higher than those in Nrf2 −/− mice.
Exercise endurance capacity and energy metabolism parameters
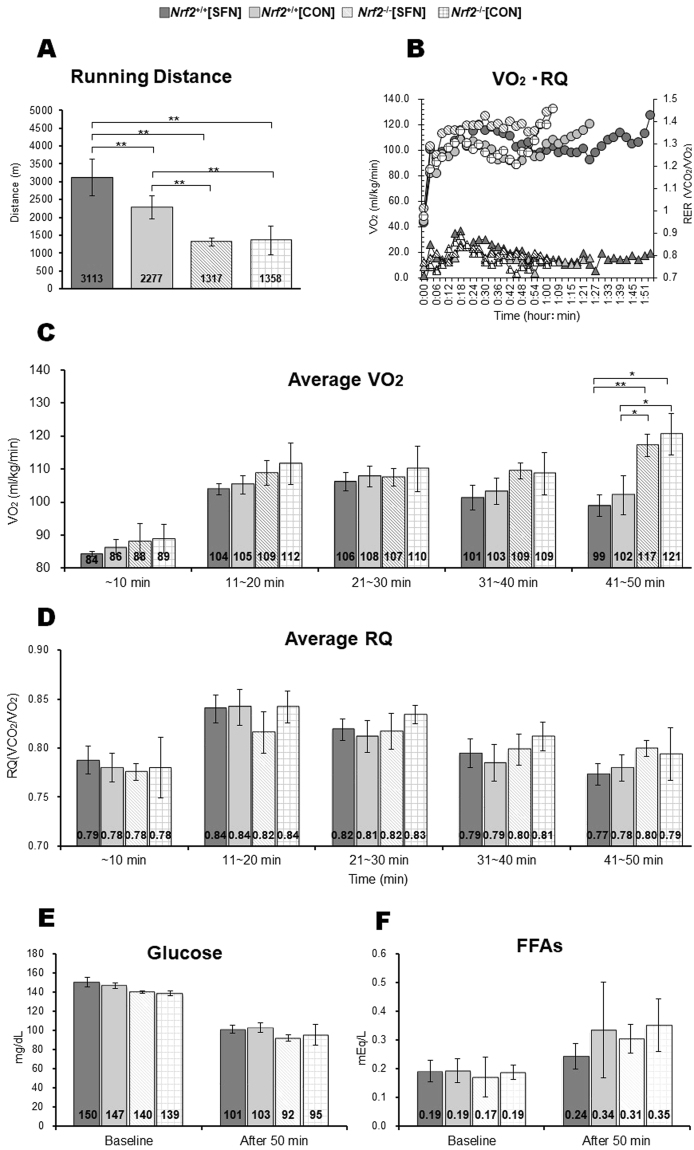
Figure 5A shows the results of the running distance test, which was measured as a proxy for exercise endurance capacity. Nrf2 +/+ mice achieved greater running distances than Nrf2 −/− mice. Among Nrf2 +/+ groups, Nrf2 +/+[SFN] mice achieved greater running distances than those of Nrf2 +/+[CON] mice. Figure 5B shows oxygen (VO2) consumption and RQ per running time, and the recorded median running distance for each group. Figure 5C and D show average VO2 and RQ levels for each group during the 50-min exhaustive treadmill test. There were no significant differences in VO2 levels after 40 min of exercise among all groups. From the period between 41 and 50 min, VO2 levels in the Nrf2 −/− groups were higher than those in the Nrf2 +/+ groups. However, there were no significant differences among the four groups in terms of average RQ per time period. Serum levels of glucose and free fatty acids (FFAs) showed no significant difference among the four groups at baseline and after 50 min (Fig. 5E and F).
Figure 5.
Running distance of each group in exhaustive treadmill test (A) and VO2 (circle) and RQ (triangle) per running time in mouse recorded median running distance in each group (B). Average levels of energy substrates: VO2 (C), RQ (D), glucose (E), and FFA (F) in each group after the 50-min exhaustive treadmill test for comparison among groups at the same time course. Brackets *P < 0.05, **P < 0.01.
Biomarkers for oxidative stress and muscle damage
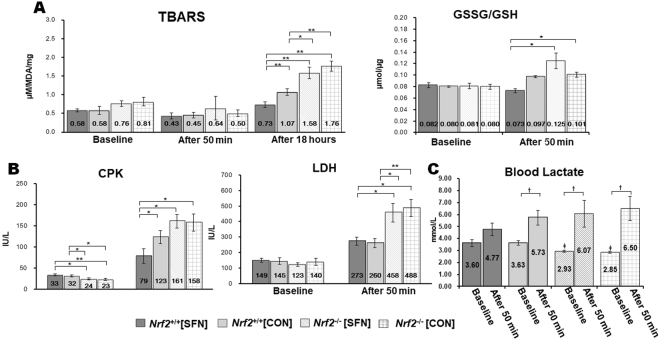
Figure 6A shows levels of thiobarbituric acid reactive substances (TBARS) in gastrocnemius homogenates at baseline, following an exhaustive test at 50 min, and at 18 h following the end of the test. In the baseline and 50-min periods, all groups showed comparable TBARS levels. However, at 18 h following the test, Nrf2 +/+ mice exhibited significantly lower TBARS levels than those of Nrf2 −/− mice. TBARS levels in SFN-injected Nrf2 +/+ mice were lower than those of uninjected Nrf2 +/+ mice. For the GSSG/GSH ratio in baseline, all groups also showed to be comparable. However, at 50-min following the test, SFN injection in Nrf2 +/+ mice showed that the GSSG/GSH ratio was lower than that in Nrf2 −/− groups (Fig. 6A). Figure 6B shows serum levels of muscle damage biomarkers creatine phosphokinase (CPK) and lactate dehydrogenase (LDH) for each group at baseline and following 50 min of treadmill testing. CPK levels were significantly higher in Nrf2 +/+ groups than in Nrf2 −/− groups at baseline. At the 50-min mark of exhaustive treadmill testing, CPK levels in Nrf2 +/+ groups were lower than those in Nrf2 −/− groups. In particular, CPK levels in Nrf2 +/+[SFN] mice were significantly lower compared with other treatment groups. All groups showed comparable LDH levels at baseline. However, LDH levels in Nrf2 +/+ groups were lower than those in Nrf2 −/− groups following the 50-min treadmill test. Blood lactate levels in Nrf2 −/− were significantly lower than those of the Nrf2 +/+ at the baseline. We did not see a significant increase in blood lactate levels in Nrf2 +/+ mice with SFN injection after 50 minutes. However, other groups showed a significant increase (Fig. 6C).
Figure 6.
Oxidative, TBARS and GSSG/GSH (A) markers in the gastrocnemius muscle, and muscle damage, CPK and LDH (B) markers, muscle fatigue, lactate (C) marker in the blood. Brackets *P < 0.05, **P < 0.01, † P < 0.05 vs. after the 50-min exhaustive treadmill test, ‡ P < 0.05 vs. Nrf2+/+ groups.
These data suggest that SFN injection in Nrf2 +/+ mice exerted protective effects in muscle under exhaustive exercise conditions.
Discussion
Elevated oxygen utilization in muscles with increasing exercise intensity may promote production of free radicals and other ROS from spill-over reactions of electrons in the mitochondrial electron transport chain5. Enhanced free radical formation in active muscles regulates various cellular signalling pathways that may result in restricted muscle contraction16. These processes may be caused by impaired central nervous system function, disordered sarcolemmal function, microvascular regulation, calcium regulation, impaired myofilament contraction, and/or altered mitochondrial metabolism17. Although controversial18,19, a growing body of evidence suggests that nutritional supplementation with molecules that enhance the body’s antioxidant defence system, and that act as ROS scavengers, might prevent exercise-induced oxidative stress and reduce muscle damage20–22. Considering the activation of Nrf2, SFN is one such powerful antioxidant11.
Our study suggests that SFN pretreatment may increase running distance in exhaustive treadmill tests (Fig. 5A) by suppressing production of TBARS and GSSG-to-GSH ratio in skeletal muscles (Fig. 6A), and by suppressing release of CPK, LDH (Fig. 6B) and lactate (Fig. 6C) in the blood. Increased TBARS levels indicate increased in vivo lipid peroxidation. Serum levels of CPK and LDH are often used as measures of structural damage to muscle cells. The accumulation of lactate causes fatigue in muscles. To the best of our knowledge, our study provides the first direct evidence demonstrating that upregulation of the Nrf2 signalling pathway by SFN, as well as its regulation of downstream phase II and antioxidant genes (HO-1, NQO1, γ-GCS, and catalase), play a crucial role in protecting against exhaustive exercise-mediated oxidative stress and tissue damage in skeletal muscles. SFN-induced Nrf2 activation in skeletal muscles might play a critical role in improving exercise endurance capacity.
Another major goal of the present study is evaluation of in vivo Nrf2 activation via Nrf2-luciferin imaging in whole body and luciferase activity determination in muscles. To date, there have been many unsuccessful attempts at Nrf2 detection in muscle using SDS–PAGE. We had identified Nrf2 in vitro activity via electrical stimulation in C2C12 cells (the size of 95–110 kDa band)23, but detecting Nrf2 activity in mouse muscle has proved difficult. Some studies targeted a non-specific band in the 55–65 kDa region as Nrf224,25. In 2012, we were the first to develop a transgenic mouse (OKD48) that was used to investigate the Keap1-Nrf2 pathway via an imaging technique that measures luminescence activity14. To improve the detection of the Nrf2 signal, we developed a new mouse model by crossing OKD48 with Nrf2 +/+ (albino-BL/6) or Nrf2 −/− (albino-Nrf2 −/−) mice. Using this new model, we demonstrated that SFN activates the Nrf2 signalling pathway in whole body (Fig. 4A), in muscle tissues (Fig. 4B), and strongly protects against oxidative stresses.
TBARS is well-established biological indicators of oxidative stress26. The extent of lipid peroxidation reflects key pathological and toxicological states related to oxidative stress27. A growing body of evidence suggests that TBARS concentrations are increased in both plasma and tissues as a result of short bouts of exhaustive exercise28–30. Glutathione is known as an antioxidant, and has been used as a marker of oxidative stress. Under oxidative stress conditions, there are lower concentrations of GSH and higher concentrations of GSSG, and thus a GSSG:GSH ratio increase31. Many studies have demonstrated that the GSSG:GSH ratio increases in response to exercise training in a manner that is highly correlated with lactate: pyruvate ratios31,32. High levels of oxidants can cause contractile dysfunction, mitochondrial dysfunction, and muscle atrophy, all of which give rise to muscle weakness and fatigue3,16,33. Our data suggest that reduction in TBARS and GSSG:GSH ratio concentrations following the treadmill test suggests protection against muscle damage12, mediated by upregulation of antioxidant and detoxifying genes downstream from SFN-induced Nrf2 activation. Our observation of a relationship between SFN pretreatment and inhibition of oxidants is consistent with recent reports. Malaguti et al.12 reported that in vivo levels of oxidative stress increased after exhaustive treadmill tests but were considerably decreased by SFN pretreatment. Angeloni et al. demonstrated that SFN significantly increases total in vitro antioxidant activity34.
Nrf2 may play crucial roles in lipid and glucose metabolism in addition to attenuating oxidative stress35–37. Zhang and co-workers38 showed that SFN enhances Nrf2 expression and activates the liver kinase B1/AMPK pathway and downstream genes, such as peroxisome proliferator activated receptor γ coactivator-1, phosphorylated acetyl-CoA carboxylase, and carnitine palmitoyl transferase-1. AMPK is known to be regulated in muscles, and acts by facilitating fatty acid uptake and oxidation, glucose uptake, and mitochondrial biogenesis39,40. In addition, Uruno et al. demonstrated that Nrf2 regulates skeletal muscle glycogen metabolism, particularly glycogen branching enzyme (GBE1) in the context of a maximal incremental treadmill protocol41. It may be the case that the observed increase in SFN improves energy metabolism and is responsible for increased running distance in mice. This represents an important subject for further research and requires further experimentation to improve our understanding of the mechanisms by which SFN influences exercise endurance capacity. In this study, VO2 and RQ data were obtained by evaluating energy substrate, carbohydrate, and fat utilization using a treadmill chamber. Dramatic changes in VO2 levels were observed from 41 to 50 min between Nrf2 +/+ and Nrf2 −/− groups (Fig. 5C). This is accompanied by increased LDH and CPK levels in Nrf2 −/− groups (Fig. 6B); however, we detected no significant changes in carbohydrate and fat oxidation and RQ (Fig. 5D). We examined serum levels of glucose and FFAs, which serve as major fuel sources in skeletal muscle42, but we demonstrated no difference among the four groups. Therefore, severe muscle damage observed in Nrf2 −/− groups could be due to lack of Nrf2 expression during the tests rather than by lack of energy supplementation. Strong SFN-mediated induction of total cellular and mitochondrial antioxidants, as well as phase 2 enzymes in Nrf2 +/+ mice might explain the observed enhanced exercise endurance capacity. The change in skeletal muscular oxidative stress levels may account for increased running distance.
Sun et al.43 reported that SFN treatment (2 mg/kg) for 8 weeks improved muscle function and reduced pathology by exerting protective effects against oxidative stress in dystrophic muscles in muscular dystrophy (mdx) mice. These results suggest that SFN injection of Nrf2 +/+ mice and the absence of Nrf2 in Nrf2 −/− groups could affect muscle composition, which in turn could contribute to the observed differences in exercise endurance capacity. However, in our study, injection with SFN over 4 days did not change the muscle fibre types and sizes (Fig. 2A–C). Fibre composition and morphology was not influenced by the presence or absence of Nrf2 genes. Therefore, we propose that SFN-induced improvement in exercise capacity in current exhaustive tests is not mediated by changes in muscle composition.
We displayed markers of mitochondrial biogenesis (Fig. 3) to clarify the cause of the increased exercise capacity. We found significant activation of AMPKα after four SFN injections (Fig. 3A), consistent with a recent study32. However, we did not measure a significant difference in other mitochondria biogenesis markers, including mtDNA copy number among the four groups (Fig. 3A–C). In addition, we assessed changes in cAMP (Fig. 2H), thought to be an effect of mitochondria biogenesis44. However, we did not measure a significant difference in cAMP among the groups. Kitaoka et al.45 demonstrate that the presence of Nrf2 gene does not influence the alterations in mitochondrial morphology. Thus, we assume that the connection was weak between activating mitochondria biogenesis signalling and the improvement in exercise endurance capacity through SFN injection, which was administered only four times.
There are inconsistencies with some research41,46 reported compared with exercise capacity between Nrf2+/+ and Nrf2−/− mice. We recognized that this inconsistency arose from a difference of exercise endurance test protocol. Because the most important goal of this study was to establish a difference in exercise capacity in SFN injection of Nrf2+/+ mice and the absence of Nrf2 in Nrf2−/− groups, we put more value on the exercise endurance test protocol. Through a pilot experiment, we found that the result of exercise endurance capacity can vary widely by the exercise test protocols, among which we have chosen the optimal test protocol to demonstrate our hypothesis. The studies mentioned earlier measured the exercise capacity by incremental increase in speed and/or incline to maximal exercise level on a treadmill. There was the possibility of using anaerobic power rather than aerobic power in exercise endurance capacity tests in this study. Provided that it is based on anaerobic power, the leg’s power of the mouse cannot follow the higher speed load and slope resistance. Therefore, the discordance of results between researches following the test protocols is quite thinkable. However, we recognized maintaining the ~28 m/min to all-out is very appropriate for the assessment of exercise endurance capacity.
Recently, there have been important reports18,19 related to ROS and exercise adaptation in muscle. That is, antioxidant supplementation might hamper exercise training-induced adaptations in skeletal muscle. Therefore, we should be cautious drawing conclusions regarding long-term SFN treatment. However, even though an antioxidant has beneficial effects, it can have toxic side effects according to type, duration, frequency, and dosage. Therefore, we believe more scientific evidence behind antioxidant functions is needed. We do not consider sulforaphane to be merely an antioxidant. Of course, it is obvious that the main role of Nrf2 activation by sulforaphane treatment is to indirectly express various transcription factors with antioxidant properties. However, it is difficult to state that sulforaphane directly affects other antioxidants to induce cellular signalling for muscle adaptation. Therefore, more experiments are necessary to answer this question to determine long-term negative impacts of SFN.
Our results suggest that SFN-induced Nrf2 upregulation and SFN-mediated antioxidative effects might play crucial roles in attenuating muscle fatigue by reducing oxidative stress caused by exhaustive exercise. These effects may give rise to improved exercise endurance capacity. We believe this study provides new insights into the role of Nrf2 in improving exercise performance.
Electronic supplementary material
Acknowledgements
This work was supported in part by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology, Japan (nos 17H02174, 17K19887, 17K19888, 16H03255, 16K15188, 26670109, 24659104, 17H02174 and 14F04307).
Author Contributions
Substantial contributions to study conception and design: Sechang Oh, Junichi Shoda Kazunori Ishige, Yuji Mizokami, Keisuke Kuga; data acquisition: Sechang Oh, Shoichi Komine, Eiji Warabi, Kentaro Akiyama, Masaki Horie, Akiko Ishii, Yoshihiro Miwa; data analysis: Sechang Oh, Shoichi Komine, Masaki Horie, Kentaro Akiyama; data interpretation: Sechang Oh, Shoichi Komine, Yoshihiro Miwa, Takao Iwawaki, Masayuki Yamamoto and Junichi Shoda; drafted the article: Sechang Oh and Junichi Shoda; revised the article critically for important intellectual content: Kazunori Ishige, Yuji Mizokami, Keisuke Kuga, Akiko Ishii, Yoshihiro Miwa, Takao Iwawaki and Masayuki Yamamoto.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Sechang Oh and Shoichi Komine contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-017-12926-y.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Jackson MJ. Control of reactive oxygen species production in contracting skeletal muscle. Antioxid Redox Signal. 2011;15:2477–2486. doi: 10.1089/ars.2011.3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reid MB. Invited Review: Redox modulation of skeletal muscle contraction: what we know and what we don’t. J Appl Physiol. 2001;90:724–731. doi: 10.1063/1.1381002. [DOI] [PubMed] [Google Scholar]
- 3.Powers SK, Jackson MJ. Exercise-induced oxidative stress: cellular mechanisms and impact on muscle force production. Physiol Rev. 2008;88:1243–1276. doi: 10.1152/physrev.00031.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goto C, et al. Effect of different intensities of exercise on endothelium-dependent vasodilation in humans: role of endothelium-dependent nitric oxide and oxidative stress. Circulation. 2003;108:530–535. doi: 10.1161/01.CIR.0000080893.55729.28. [DOI] [PubMed] [Google Scholar]
- 5.Jackson MJ. Free radicals generated by contracting muscle: by-products of metabolism or key regulators of muscle function? Free Radic Biol Med. 2008;44:132–141. doi: 10.1016/j.freeradbiomed.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 6.Davies KJA, Quintanilha AT, Brooks GA, Packer L. Free radicals and tissue damage produced by exercise. Biochem Biophys Res Commun. 1982;107:1198–1205. doi: 10.1016/S0006-291X(82)80124-1. [DOI] [PubMed] [Google Scholar]
- 7.Reid MB. Reactive oxygen species as agents of fatigue. Med Sci Sports Exerc. 2016;48:2239–2246. doi: 10.1249/MSS.0000000000001006. [DOI] [PubMed] [Google Scholar]
- 8.Kobayashi M, et al. Identification of the interactive interface and phylogenic conservation of the Nrf2-Keap1 system. Genes Cells. 2002;7:807–820. doi: 10.1046/j.1365-2443.2002.00561.x. [DOI] [PubMed] [Google Scholar]
- 9.Itoh K, Mimura J, Yamamoto M. Discovery of the negative regulator of Nrf2, Keap1: a historical overview. Antioxid Redox Signal. 2010;13:1665–1678. doi: 10.1089/ars.2010.3222. [DOI] [PubMed] [Google Scholar]
- 10.Thimmulappa RK, et al. Identification of Nrf2-regulated genes induced by the chemopreventive agent sulforaphane by oligonucleotide microarray. Cancer Res. 2002;62:5196–5203. [PubMed] [Google Scholar]
- 11.Guerrero-Beltrán CE, Calderón-Oliver M, Pedraza-Chaverri J, Chirino YI. Protective effect of sulforaphane against oxidative stress: Recent advances. Exp Toxicol Pathol. 2012;64:503–508. doi: 10.1016/j.etp.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 12.Malaguti M, et al. Sulforaphane treatment protects skeletal muscle against damage induced by exhaustive exercise in rats. J Appl Physiol. 2009;107:1028–1036. doi: 10.1152/japplphysiol.00293.2009. [DOI] [PubMed] [Google Scholar]
- 13.Lusk, G. The elements of the science of nutrition. (4th ed.) WB Saunders, Philadelphia, (1928).
- 14.Oikawa D, Akai R, Tokuda M, Iwawaki T. A transgenic mouse model for monitoring oxidative stress. Sci Rep. 2012;2:229. doi: 10.1038/srep00229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chowanadisai W, et al. Pyrroloquinoline Quinone Stimulates Mitochondrial Biogenesis through cAMP Response Element-binding Protein Phosphorylation and Increased PGC-1α Expression. J Biol Chem. 2010;285:142–152. doi: 10.1074/jbc.M109.030130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Steinbacher P, Eckl P. Impact of oxidative stress on exercising skeletal muscle. Biomolecules. 2015;5:356–377. doi: 10.3390/biom5020356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reid MB. Free radicals and muscle fatigue: Of ROS, canaries, and the IOC. Free Radic Biol Med. 2008;44:169–179. doi: 10.1016/j.freeradbiomed.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 18.Strobel NA, et al. Antioxidant supplementation reduces skeletal muscle mitochondrial biogenesis. Med Sci Sports Exerc. 2011;43:1017–1024. doi: 10.1249/MSS.0b013e318203afa3. [DOI] [PubMed] [Google Scholar]
- 19.Gomez-Cabrera MC, et al. Oral administration of vitamin C decreases muscle mitochondrial biogenesis and hampers training-induced adaptations in endurance performance. Am J Clin Nutr. 2008;87:142–149. doi: 10.1093/ajcn/87.1.142. [DOI] [PubMed] [Google Scholar]
- 20.Boz I, Belviranli M, Okudan N. Curcumin modulates muscle damage but not oxidative stress and antioxidant defense following eccentric exercise in rats. Int J Vitam Nutr Res. 2014;84:163–172. doi: 10.1024/0300-9831/a000203. [DOI] [PubMed] [Google Scholar]
- 21.Deminice R, Jordao AA. Creatine supplementation decreases plasma lipid peroxidation markers and enhances anaerobic performance in rats. Redox Rep. 2016;21:31–36. doi: 10.1179/1351000215Y.0000000020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meier P, Renga M, Hoppeler H, Baum O. The impact of antioxidant supplements and endurance exercise on genes of the carbohydrate and lipid metabolism in skeletal muscle of mice. Cell Biochem Funct. 2013;31:51–59. doi: 10.1002/cbf.2859. [DOI] [PubMed] [Google Scholar]
- 23.Horie M, Warabi E, Komine S, Oh S, Shoda J. Cytoprotective role of nrf2 in electrical pulse stimulated C2C12 myotube. PLoS One. 2015;10:e0144835. doi: 10.1371/journal.pone.0144835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taguchi K, Motohashi H, Yamamoto M. Molecular mechanisms of the Keap1–Nrf2 pathway in stress response and cancer evolution. Genes Cells. 2011;16:123–140. doi: 10.1111/j.1365-2443.2010.01473.x. [DOI] [PubMed] [Google Scholar]
- 25.Lau A, Tian W, Whitman SA, Zhang DD. The predicted molecular weight of nrf2: it is what it is not. Antioxid Redox Signal. 2013;18:91–93. doi: 10.1089/ars.2012.4754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Armstrong D, Browne R. The analysis of free radicals, lipid peroxides, antioxidant enzymes and compounds related to oxidative stress as applied to the clinical chemistry laboratory. Adv Exp Med Biol. 1994;366:43–58. doi: 10.1007/978-1-4615-1833-4_4. [DOI] [PubMed] [Google Scholar]
- 27.Grotto D, et al. Importance of the lipid peroxidation biomarkers and methodological aspects for malondialdehyde quantification. Quím Nova. 2009;32:169–174. doi: 10.1590/S0100-40422009000100032. [DOI] [Google Scholar]
- 28.Qiao D, Hou L, Liu X. Influence of intermittent anaerobic exercise on mouse physical endurance and antioxidant components. Br J Sports Med. 2006;40:214–218. doi: 10.1136/bjsm.2005.020099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Venditti P, Di Meo S. Antioxidants, tissue damage, and endurance in trained and untrained young male rats. Arch Biochem Biophys. 1996;331:63–68. doi: 10.1006/abbi.1996.0283. [DOI] [PubMed] [Google Scholar]
- 30.Frankiewicz-Jozko A, Faff J, Sieradzan-Gabelska B. Changes in concentrations of tissue free radical marker and serum creatine kinase during the post-exercise period in rats. Eur J Appl Physiol Occup Physiol. 1996;74:470–474. doi: 10.1007/BF02337728. [DOI] [PubMed] [Google Scholar]
- 31.Urso ML, Clarkson PM. Oxidative stress, exercise, and antioxidant supplementation. Toxicology. 2003;189:41–54. doi: 10.1016/S0300-483X(03)00151-3. [DOI] [PubMed] [Google Scholar]
- 32.Laaksonen DE, et al. Blood Glutathione homeostasis as a determinant of resting and exercise-induced oxidative stress in young men. Redox Rep. 1999;4:53–59. doi: 10.1179/135100099101534648. [DOI] [PubMed] [Google Scholar]
- 33.Navarro A, Gomez C, Lopez-Cepero JM, Boveris A. Beneficial effects of moderate exercise on mice aging: survival, behavior, oxidative stress, and mitochondrial electron transfer. Am J Physiol Regul Integr Comp Physiol. 2004;286:R505–R511. doi: 10.1152/ajpregu.00208.2003. [DOI] [PubMed] [Google Scholar]
- 34.Angeloni C, et al. Novel targets of sulforaphane in primary cardiomyocytes identified by proteomic analysis. PLoS One. 2013;8:e83283. doi: 10.1371/journal.pone.0083283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Slocum SL, et al. Keap1/Nrf2 pathway activation leads to a repressed hepatic gluconeogenic and lipogenic program in mice on a high-fat diet. Arch Biochem Biophys. 2016;591:57–65. doi: 10.1016/j.abb.2015.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barroso, E. et al. PPARbeta/delta ameliorates fructose-induced insulin resistance in adipocytes by preventing Nrf2 activation. Biochim Biophys Acta1852, 1049−1058 (2015). [DOI] [PubMed]
- 37.Tebay LE, et al. Mechanisms of activation of the transcription factor Nrf2 by redox stressors, nutrient cues, and energy status and the pathways through which it attenuates degenerative disease. Free Radic Biol Med. 2015;88:108–146. doi: 10.1016/j.freeradbiomed.2015.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Z, et al. Sulforaphane prevents the development of cardiomyopathy in type 2 diabetic mice probably by reversing oxidative stress-induced inhibition of LKB1/AMPK pathway. J Mol Cell Cardiol. 2014;77:42–52. doi: 10.1016/j.yjmcc.2014.09.022. [DOI] [PubMed] [Google Scholar]
- 39.Hardie DG, Sakamoto K. AMPK: a key sensor of fuel and energy status in skeletal muscle. Physiology. 2006;21:48–60. doi: 10.1152/physiol.00044.2005. [DOI] [PubMed] [Google Scholar]
- 40.Garcia D, Shaw RJ. AMPK: Mechanisms of Cellular Energy Sensing and Restoration of Metabolic Balance. Mol Cell. 2017;66:789–800. doi: 10.1016/j.molcel.2017.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Uruno A, et al. Nrf2-Mediated Regulation of Skeletal Muscle Glycogen Metabolism. Mol Cell Biol. 2016;36:1655–1672. doi: 10.1128/MCB.01095-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Egan B, Zierath Juleen R. Exercise metabolism and the molecular regulation of skeletal muscle adaptation. Cell Metab. 2013;17:162–184. doi: 10.1016/j.cmet.2012.12.012. [DOI] [PubMed] [Google Scholar]
- 43.Sun C, Li S, Li D. Sulforaphane mitigates muscle fibrosis in mdx mice via Nrf2-mediated inhibition of TGF-beta/Smad signaling. J Appl Physiol. 2016;120:377–390. doi: 10.1152/japplphysiol.00721.2015. [DOI] [PubMed] [Google Scholar]
- 44.Ray Hamidie, R. D., Yamada, T., Ishizawa, R., Saito, Y. & Masuda, K. Curcumin treatment enhances the effect of exercise on mitochondrial biogenesis in skeletal muscle by increasing cAMP levels. Metabolism64, 1334–1347 (2015). [DOI] [PubMed]
- 45.Kitaoka Y, et al. Nrf2 deficiency does not affect denervation-induced alterations in mitochondrial fission and fusion proteins in skeletal muscle. Physiol Rep. 2016;4:e13064. doi: 10.14814/phy2.13064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Merry TL, Ristow M. Nuclear factor erythroid-derived 2-like 2 (NFE2L2, Nrf2) mediates exercise-induced mitochondrial biogenesis and the anti-oxidant response in mice. J Physiol. 2016;594:5195–5207. doi: 10.1113/JP271957. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.