Abstract
Campylobacter jejuni/coli infections are the leading cause of bacterial diarrheal illnesses in humans. Many epidemiological studies indicate that improperly prepared meat from chickens that carry a high load of Campylobacter in their intestinal tracts is the key source of human infections. LAB, mainly members of the Lactococcus and Lactobacillus genera, increasingly have been tested as vehicles for the delivery of heterologous bacterial or viral antigens to animal mucosal immune systems. Thus, the objective of this study was to isolate, identify, and characterize Lactobacillus spp. strains isolated from chickens bred in Poland. Their ability to decrease the level of bird gut colonization by C. jejuni strain was also analyzed. First, the influence of the different chicken rearing systems was evaluated, especially the effect of diets on the Lactobacillus species that colonize the gut of chickens. Next, selected strains were analyzed in terms of their anti‐Campylobacter activity in vitro; potential probiotic traits such as adhesion properties, bile and low pH tolerance; and their ability to grow on a defined carbon source. Given that improperly prepared chicken meat is the main source of human infection by Campylobacter, the selected strains were also assessed for their ability to inhibit Campylobacter colonization in the bird's intestine. These experiments revealed enormous physiological diversity among the Lactobacillus genus strains. Altogether, our results showed that L. plantarum strains isolated from the digestive tracts of chickens bred in Poland displayed some probiotic attributes in vitro and were able to decrease the level of bird gut colonization by C. jejuni strain. This suggests that they can be employed as vectors to deliver Campylobacter immunodominant proteins to the bird's immune system to strengthen the efficacy of in ovo vaccination.
Keywords: Campylobacter, Lactobacillus, poultry, probiotics
1. INTRODUCTION
Microbiota present in human or animal intestinal tracts have enormous impact on the health of the host. Recent advances in technology, in particular progress made in the field of DNA sequencing combined with many bioinformatics methods has replaced culture‐dependent methods with culture‐independent strategies. Data from such analyses reveal details of microbiota diversity and their interactions with the host. Microbial communities of the chicken gastrointestinal tract (GIT), as well as the chicken microbiome, have recently been analyzed by many research group to better understand their influence on bird health and improve poultry industry productivity (for review see [Oakley, Lillehoj, et al., 2014]). The diverse chicken microbiota consists of over 900 bacterial species (Wei, Morrison, & Yu, 2013). Although the microbial communities found in different sections of the chicken GIT vary and their members belong to 13 phyla, the most dominant phyla are Firmicutes, Bacteroides, and Proteobacteria (Pan & Yu, 2014; Stanley, Hughes, & Moore, 2014). Kaakoush et al. (2014), who studied the microbiota of chickens from two farms, assigned them to four enterotypes dominated by varies phyla. Many factors influence the composition of the chicken microbiota; the most substantial factors are the chicken breeds, bird age, and farming practices (Kaakoush et al., 2014; Oakley, Buhr, et al., 2014; Schokker et al., 2015; Thibodeau et al., 2015).
Campylobacteriosis is the most frequently reported foodborne zoonosis worldwide (Kaakoush, Castano‐Rodriguez, Mitchell, & Man, 2015). As chickens harbor a high load of Campylobacter jejuni in their GIT, improperly prepared chicken meat is the main source of human campylobacteriosis (Agunos, Waddell, Leger, & Taboada, 2014; Hermans et al., 2012; Kaakoush et al., 2015; Zambrano, Levy, Menezes, & Freeman, 2014). To date, many interventions to combat the chicken colonization by C. jejuni, such as introducing different hygiene barriers at farms, various nutritional strategies or using bacteriophages, have been examined with varying success (Meunier, Guyard‐Nicodeme, Dory, & Chemaly, 2016; Newell et al., 2011; Wagenaar, French, & Havelaar, 2013). Given that members of the Lactobacillus genus are predominant in the chicken microbiota and that some of them can be classified as probiotics, their anti‐Campylobacter activities have recently been intensively examined using different strategies (Chaveerach, Lipman, & van Knapen, 2004; Robyn, Rasschaert, Messens, Pasmans, & Heyndrickx, 2012; Santini et al., 2010). The aim of these studies was to manipulate the chicken microbiota in order to inhibit chicken digestive tract colonization by Campylobacter (Ghareeb et al., 2012; Santini et al., 2010; Willis & Reid, 2008). The studies showed that only a small percentage of Lactobacillus strains isolated from chicken intestinal tracts or from fecal samples are able to compete with Campylobacter. Although a lot of Lactobacillus isolates have demonstrated high anti‐Campylobacter activity in vitro, many only slightly reduce bird colonization by the pathogen when administered in vivo, which demonstrates the complex mechanisms of interactions among microbiota constituents (Chaveerach et al., 2004; Robyn et al., 2012; Santini et al., 2010). C. jejuni genetic diversity makes the issue even more complicated as inhibition of the pathogen's growth by a specific Lactobacillus isolate is dependent on the C. jejuni strain used in the experiment (Chaveerach et al., 2004; Santini et al., 2010). In some cases, probiotic formulations composed of several bacteria, even belonging to different genera, were proven to be the most effective (Ghareeb et al., 2012; Willis & Reid, 2008). The mechanisms by which Lactobacillus isolates inhibit Campylobacter growth in vivo are diverse and not fully understood. The source of the Lactobacillus strain appears not to be a critical factor, though the data are sometimes contradictory (Chaveerach et al., 2004; Lin, Yu, Jang, & Tsen, 2007; Nishiyama et al., 2014; Santini et al., 2010). Several mechanisms that potentially depend on both the pathogen and the probiotic may be involved in the competitive inhibition of C. jejuni by Lactobacillus. Those that have been proven to play a significant role in inhibition include adhesion to the intestinal epithelium of the host (Nishiyama et al., 2014; Spivey, Dunn‐Horrocks, & Duong, 2014); coaggregation and autoaggregation (Tareb, Bernardeau, Gueguen, & Vernoux, 2013); production of antimicrobial compounds, such as organic acid, bacteriocin (Messaoudi et al., 2013; Neal‐McKinney et al., 2012; Svetoch & Stern, 2010), and hydrogen peroxide (Mota et al., 2006); and immuno‐modulatory effects (Messaoudi et al., 2012).
The objective of this study was to isolate, identify, and characterize the Lactobacillus spp. strains that constitute the chicken intestinal tract microbiota. The first aim of the study was to evaluate the influence of different rearing systems, especially, the dietary treatment of chickens, on Lactobacillus species colonizing their gut. Next, the selected strains were analyzed in terms of their anti‐Campylobacter activity in vitro and their ability to colonize chicken's gut (adhesion properties, bile and low pH tolerance). Their metabolic properties, ability to grow on a defined carbon source, were also evaluated. Given that improperly prepared chicken meat is the main source of human infection by Campylobacter, the ability of the selected strains to inhibit colonization of the bird's intestine by the pathogen was also examined.
2. MATERIALS AND METHODS
2.1. Bacterial strains media and growth conditions
Bacterial strains used in this study are listed in Table 1. A total of 107 Lactobacillus strains were isolated from privately owned “back‐yard” flocks (64 strains) and commercial broiler chicken flocks (43 strains) in Poland. Lactobacillus strains were cultured in MRS broth (de Man, Rogosa, and Sharpe) or MRS agar (solidified with 1.5% agar) medium (Oxoid Ltd., Hampshire, England) at 37°C under microaerobic conditions (85% N2, 10% CO2, 5% O2).
Table 1.
Bacterial strains used in this study
| Strain | Relevant characteristics | Source or reference |
|---|---|---|
| C. jejuni 81–176 | Human clinical isolate | Korlath et al. (1985) |
| C. jejuni 12 | Chicken isolate | Wyszynska et al. (2004) |
| L. salivarius PA14C | Chicken isolate | This study |
| L. salivarius PA17D | Chicken isolate | This study |
| L. salivarius PA18C | Chicken isolate | This study |
| L. reuteri PA12C | Chicken isolate | This study |
| L. reuteri PA8A | Chicken isolate | This study |
| L. reuteri PA19B | Chicken isolate | This study |
| L. crispatus PA11D | Chicken isolate | This study |
| L. plantarum PA11A | Chicken isolate | This study |
| L. plantarum PA18A | Chicken isolate | This study |
| L. plantarum PA20A | Chicken isolate | This study |
| L. agilis PA16B | Chicken isolate | This study |
C. jejuni 12 (PCM 2852, Polish Collection of Microorganisms) and C. jejuni 81–176 (ATCC® BAA‐2151, American Type Culture Collection, USA) were used in C. jejuni inhibition assays in vitro. The C. jejuni 12/2 strain employed in the protection experiment was a broiler‐isolated strain labeled with the pUOA18 plasmid containing a cat gene. Previous experiments have shown that the pUOA18 plasmid is stably maintained in Campylobacter (Wyszynska, Raczko, Lis, & Jagusztyn‐Krynicka, 2004). Campylobacter strains were cultured under microaerobic conditions in Blood Agar (BA) (Oxoid Ltd.) plates supplemented with 5% horse blood and Campylobacter Selective Supplement (Oxoid Ltd. at 37°C. The medium was supplemented with chloramphenicol (15 μg/ml), if necessary.
2.2. Isolation and identification of lactic acid bacteria
Twenty stool samples from back‐yard chickens or cloacal swabs from 25 commercial broiler chickens were streaked onto selective MRS agar and incubated overnight in a microaerobic atmosphere at 37°C. After 24 hr incubation, colonies were randomly picked from the plates and subcultured two or three times on fresh MRS agar plates. The pure LAB cultures were then kept in MRS broth supplemented with 30% (v/v) glycerol and frozen at −80°C until further analysis.
Preliminary identification consisted of Gram staining to assess the morphology of cells by optical microscopy. Next, amplification of the 16S rDNA gene was performed for all gram positive strains. For this, chromosomal DNA was isolated from overnight cultures of selected strains. Pellets were resuspended in TES (25 mmol/L sucrose, 50 mmol/L Tris HCl pH 8.0, 10 mmol/L EDTA) containing lysozyme (20 mg/ml, Sigma) and mutanolysin (500 u/ml, A&A Biotechnology). After incubation for 1 hr at 37°C under constant shaking, chromosomal DNA from the bacterial strains was isolated using a commercial kit (A&A Biotechnology, Poland). Polymerase chain reactions (PCR) were performed with Color optiTaq (EURX) or HotStar HiFidelity Polymerase (Qiagen) under standard conditions. For amplification of the 16S rDNA gene, universal primers F27 (5′‐AGAGTTTGATCCTGGCTCAG‐3′) and 1492 (5′‐GGTTACCTTGTTACGACTT‐3′) were used (McDonald, Kenna, & Murrell, 1995) with an expected PCR product size of 1.5 kb. PCR products were purified using the Clean‐up Concentrator kit (A&A Biotechnology, Poland). Synthetic oligonucleotide synthesis and DNA sequencing of the PCR product was performed by Genomed S.A. (Warsaw, Poland), and then the nucleotide sequences were analyzed using BLAST against the nucleotide database at the NCBI website.
2.3. C. jejuni inhibition assays in vitro
The C. jejuni inhibition assay was performed on BA plates containing 5% horse blood (Oxoid) inoculated with 100 μl of Campylobacter cultures (C. jejuni 12 or C. jejuni 81–176), prepared by suspending the microorganisms in 0.9% NaCl and adjusting the density to 0.5 McFarland standard (~1 × 108 CFU/ml). Plates were incubated for 2 hr at 37°C under microaerobic conditions. Overnight cultures of lactobacilli were centrifuged, and to determine if bacteriocins contributed to inhibition, part of supernatant was neutralized to neutral pH with 6 N NaOH. Untreated supernatants and supernatants neutralized to pH 7 were subsequently filter sterilized (0.22 μm) and spotted onto MRS agar plates inoculated with Campylobacter. Plates were incubated for 24–48 hr at 37°C under microaerobic conditions. Growth inhibition was evaluated as inhibition around the spotted Lactobacillus supernatant.
2.4. Detection of lactic acid production
Detection of lactic acid was carried out as described earlier by Neal‐McKinney et al. (2012). The concentrations of d‐ and l‐lactate in the cell‐free supernatants were measured using stereo‐specific d‐ and l‐lactate assay kits (Megazyme, Wicklow, Ireland). The measurements for selected strains were performed in triplicate for reproducibility. Assays were performed with pure solutions of d‐ and l‐lactate to ensure that the kits were stereo‐specific.
2.5. Tolerance to acid and bile
Tolerance to low pH and bile content was assessed as described by Bujnakova et al. (Bujnakova, Strakova, & Kmet, 2014), with minor modifications. Briefly, 100 μl lactobacilli from overnight cultures (37°C, MRS broth, 5% CO2) diluted to OD660 nm of 1.0 was inoculated into 10 ml of following test solutions: MRS broth control, MRS broth adjusted to pH 2.5, and MRS broth containing 0.5% oxgall (Difco). As a control, broth without inoculation was used. For survival under the different conditions, samples were taken at 6 hr and 100 μl cultures were plated on MRS agar after appropriate dilution. Colonies were counted after 48 hr incubation at 37°C. The survival rate was calculated as the log10 value of CFU/ml.
2.6. Resistance to sodium chloride
To assess effects of osmolarity, 3 μl of o/n culture of each Lactobacillus strain was transferred onto plates with MRS agar (Merck, Germany) and MRS agar containing: 0.34 mol/L (2%), 0.69 mol/L (4%), 1 mol/L (6%), or 1.36 mol/L (8%) (w/v) NaCl (Sigma, USA) and incubated at 37°C for 48 hr. The survival of bacteria was examined visually by comparison of their growth efficiency on MRS agar with their growth on MRS agar with different concentrations of NaCl. Each strain was tested in two independent experiments.
2.7. Sugar fermentation pattern
To determine sugar fermentation profiles of the Lactobacillus spp., API 50 CH kit was used according to the manufacturer's instructions (BioMerieux, France). The resulting fermentation pattern was inspected following anaerobic incubation at 37°C after 48 hr. Fermentation of a carbohydrate was detected by acid production, as demonstrated by a color change in the pH indicator present in the medium. Each strain was tested in three independent experiments.
2.8. Adhesion to bare polystyrene
Adhesion of bacterial cells to polystyrene (PS) was tested on the 96‐well microtiter PS plates (Thermo Fischer Scientific Nunc A/S, Denmark) using a modification of the method already described (Radziwill‐Bienkowska, Zochowska, Bardowski, Mercier‐Bonin, & Kowalczyk, 2014). Briefly, bacteria from overnight cultures diluted to OD660 nm of 1.0 were harvested by centrifugation (103 g; 1 min) and resuspended in an equal volume of PBS (BioShop, Canada). A volume of 100 μl of bacterial suspension was added to each well (eight for each strain). To remove unbound bacteria after 3 hr incubation under static conditions at 37°C, the wells were washed three times with 200 μl of sterile MilliQ‐grade water. Bound cells were stained with crystal violet (Scharlau, Spain) (100 μl per well, RT; 10 min), and excess stain was removed by rinsing three times with water, as above. Finally, stained bacteria were suspended in 200 μl of 96% ethanol and optical density was determined at 583 nm on a Synergy HT Multi‐Detection Reader (BioTek Instruments Inc., USA). The average value of at least six measurements was calculated after rejecting extreme results. Bacterial adhesion was determined in three independent experiments and the results are presented as mean ± SD. Each microtiter plate included blank wells with PBS and control GI (gastrointestinal) strains: high‐adhesive L. rhamnosus GG (Segers & Lebeer, 2014) and nonadhesive L. rhamnosus LOCK 0908 (Aleksandrzak‐Piekarczyk et al., 2016).
To calculate the adherence ratio of bacteria to PS, the value of absorbance of bacteria was divided by the absorbance of the control sample (PBS, only). Bacteria were characterized as strongly adherent (A ≥ 6), moderately adherent (6 > A ≥ 3), weakly adherent (3 > A > 2), and nonadherent (A ≤ 2).
2.9. Campylobacter colonization experiment
Experiments were performed on Hy‐line chickens hatched and reared under controlled conditions from the day of hatch. The chickens were kept under controlled light (L:D 12:12) and temperature (32 ± 2°C during first week and 24 ± 2°C thereafter) conditions, with free access to the standard food and water. Chickens were confirmed to be culture‐negative for Campylobacter by cloacal swabbing. All animal experiments were carried out according to the ethical standards and approval (No. 743/2015) of the Local Ethics Committee No. 1, Warsaw, Poland.
Birds were randomly assigned to six experimental groups (each containing 12 individuals) and housed in an animal facility, with separate cages for each group. On the day of hatch and 4 days post hatch, all groups of chicks were inoculated by oral gavage with 0.1 ml lactobacilli suspension (~108 CFU/ml): L. reuteri PA8A, L. crispatus PA11D, L. salivarius PA14C, L. plantarum PA18A, L. plantarum 20A. A group of birds inoculated with BSG (PBS with 0.1% gelatin) was used as a control. Following vaccination, chickens were observed for development of diarrhea and other potential adverse side effects. At the 14th day of life, birds were orally challenged with ~104 CFU of C. jejuni wild‐type strain 12/2. At 4 and 8 days post challenge (22nd day of life), six birds from each group were killed and samples of cecum were collected. Dilutions of the contents were made in PBS and plated onto BA plates supplemented with 5% horse blood, “Campylobacter Selective Supplement (Blaser‐Wang)” and chloramphenicol (15 μg/ml) for enumeration of C. jejuni. Plates were incubated at 37°C for 48 hr. Plates that were culture‐negative at 48 hr were reincubated for an additional 48 hr. This procedure permits detection of 103 CFU/g of cecal contents.
3. RESULTS
3.1. Isolation and identification of Lactobacillus strains
The Lactobacillus genus includes more than 200 species (http://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=1578, August 2016) that are characterized by diverse physiological properties (Bernardeau, Vernoux, Henri‐Dubernet, & Gueguen, 2008; Lukjancenko, Ussery, & Wassenaar, 2012).
Thus, in this study, we decided to search for Lactobacillus strains with potential probiotic activity among strains isolated from chickens breeding in Poland and we asked whether different chicken rearing systems, especially dietary treatment, have an impact on Lactobacillus species colonizing their gut (Bjerrum et al., 2006; Pan & Yu, 2014).
In total, 107 Lactobacillus strains were isolated: 64 from fecal samples of two privately owned “back‐yard” chicken farms and 43 from cloaca of birds reared in seven commercial poultry farms specializing in production of consumer eggs. The hens (Lohman Brown) from farms were at laying age, 22–50 weeks old. The chickens were fed a commercially available feed for laying hens that consists of raw materials (wheat, triticale, corn, chalk, vegetable oils, vitamins, amino acids, and microelements). This feed does not contain growth‐promoting antibiotics or coccidiostats.
Chicken stool samples were streaked onto selective MRS agar and incubated within an anaerobic atmosphere overnight at 37°C. Pure cultures were obtained. Lactobacillus genus membership was confirmed by molecular analysis (PCR amplification of the intergenic chromosomal DNA region between the 16S rDNA and 23S rDNA and evaluation of the sizes of the DNA fragments). To determine the species of the isolates, the 16S rDNA genes were amplified, and the PCR products were sequenced and analyzed using BLAST against the nucleotide database at the NCBI website. The strains were also characterized by morphological and microscopic observations (Figure S1).
Generally, the most abundant Lactobacillus species was L. salivarius, regardless of chicken breeding methods. L. salivarius (68%) and L. agilis (12%) were more prevalent than other species of Lactobacillus genus obtained from the farm chickens. The species distribution among all the Lactobacillus species isolated from fecal samples of “back‐yard” chickens was more diverse. L. salivarius (29%) was the dominant species, followed by L. reuteri (24%) and L. plantarum (23%). Other Lactobacillus species, such as L. johnsonii (6%), L. oris (3%), L. agilis (5%), and L. kitasatonis (5%), appeared less frequently. The remaining species (L. curvatus, L. crispatus, L. fermentum, and L. ingluviei) were about 1% of all isolates. The detailed data are given in Figure 1.
Figure 1.
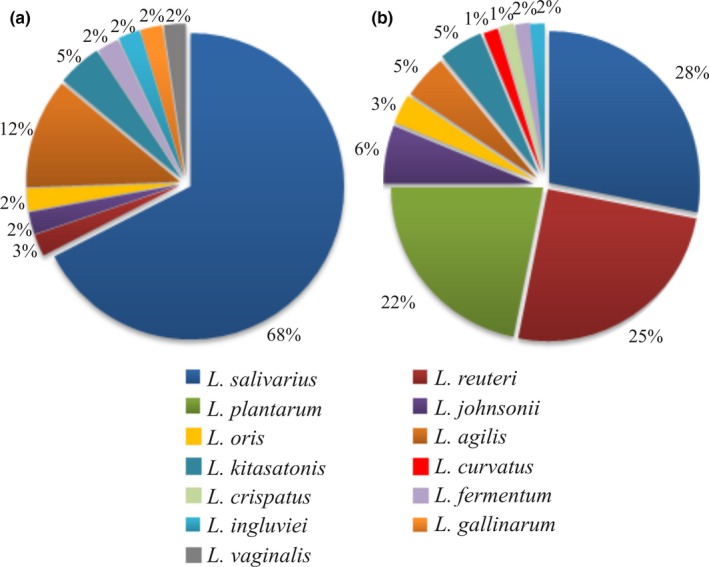
Pie charts represent relative abundance of Lactobacillus species isolated from cloaca of commercial broiler chickens (a) and from stools of backyard chickens (b)
3.2. Preliminary characteristics of isolated strains: anti‐Campylobacter activity in vitro, lactic acid production, and adhesion properties
Subsequent stepwise analysis was performed using Lactobacillus strains isolated from fecal samples of two privately owned “back‐yard” farms, where chickens were allowed access to outdoor areas. The rationale behind this decision was based on two assumptions. First, of all these chickens were never given antibiotics as feed additives at any stage of life. It has been documented that antibiotics may influence the composition of chicken microflora for long periods (Pan & Yu, 2014; Wise & Siragusa, 2007). Second, their food, though difficult to describe in detail, was more diverse than that used on commercial poultry farms.
The preliminary experiments evaluated the inhibitory effect of the Lactobacillus isolates on the growth of Campylobacter using spot titer assays for 64 Lactobacillus strains, where two C. jejuni strains (81176 and 12) of different origins were the target strains. C. jejuni 81176 is human isolate that is widely used in pathogenesis studies, and it has gone through many laboratory passages since it was first isolated (Korlath, Osterholm, Judy, Forfang, & Robinson, 1985). C. jejuni 12 is fresh chicken isolate obtained in our lab. First we checked whether cell‐free supernatants (CFS) of analyzed Lactobacillus strains displayed anti‐Campylobacter activity. The pH of the culture supernatants ranged from 3 to 5 (predominantly 4). We found that anti‐Campylobacter activity was species dependent. Members of L. reuteri species did not exhibit an antagonistic effect on Campylobacter growth. Members of L. curvatus, L. ingluviei, L. kitasatonis, and L. oris also did not inhibit Campylobacter growth, though these results should be treated with caution due to the small number of strains analyzed. Among the strains of L. plantarum and L. salivarius species analyzed, some differences in the size of the inhibition halos were observed (Table S1). Similar results were obtained for both Campylobacter strains.
Next, to find the chemical nature of the CFS component responsible for Campylobacter growth inhibition, the CFSs were neutralized before using them for spot titer assays. We found that none of the neutral supernatants demonstrated an antagonistic effect on Campylobacter, independent on the pathogen strains, suggesting that the inhibition was due to acid production.
To shed more light on the mechanism by which Lactobacillus isolates exert their action, the levels of lactic acid present in the supernatants of the cultures of all Lactobacillus strains were evaluated. In general, we found that L. reuteri strains produced relatively lower amount of lactic acid when compared to L. plantarum or L. salivarius. However, the supernatants of the L. reuteri culture reveal a pH of about 4, suggesting production of the different organic acids by L. reuteri isolates. Of four examined L. johnsonii strains, two generated high levels of lactic acid. In the case of other Lactobacillus species (L. agilis, L. crispatus, L. curvatus, L. ingluviei, L. kitasatonis, and L. oris), too few strains were examined to make a conclusion concerning the ability of a particular species to generate lactic acid. However, in general and similar to the L. reuteri strains, they produced mild level of lactic acid that did not translate into antagonistic effect against Campylobacter.
It is postulated that highly adhesive probiotic bacteria have the greatest beneficial effects on host health and they should, at least transiently, colonize the host gut (Bermudez‐Brito et al., 2012). The adhesion microtiter plate assay is a well‐known and simple method for a preliminary assessment of strain adhesion ability (Radziwill‐Bienkowska et al., 2014), which, as we observed in our previous studies, can correlate with its adhesiveness to biotic surfaces (Aleksandrzak‐Piekarczyk et al., 2016; Radziwill‐Bienkowska et al., 2014, 2016). In this study, we checked the adhesive properties of 64 Lactobacillus strains isolated from chicken gastrointestinal tracts and compared them with highly adhesive or nonadhesive L. rhamnosus strains (GG or LOCK 0908, respectively). The overwhelming majority of bacteria demonstrated adhesion to PS as determined by an adherence ratio higher than 2 (Table S1). The strength of this ability varied between strains and, in some cases, even exceeded the value determined for the positive control strain L. rhamnosus GG (adherence ratio 9.38). Among the three species most widely represented in this test (L. plantarum, L. reuteri, and L. salivarius), L. plantarum seem to have the most consistent adherence since as many as ~60% of all strains efficiently adhered to PS. However, most of them adhered moderately, and only two strains were strongly adherent. Of all L. salivarius strains analyzed, 58% adhered moderately or strongly. L. reuteri strains were slightly less adherent: half adhered either moderately (six strains) or strongly (two strains; Table S1).
On the basis of the preliminary characteristics, 11 strains were chosen for detailed examination in subsequent detailed in vitro experiments. The selection was not obvious as their activity against Campylobacter, lactic acid production and adhesion properties revealed a high level of diversity. The main selection criterion was the strong ability to inhibit Campylobacter growth. The selected strains of L. salivarius and L. plantarum differ in respect to the two other qualities. Two of L. salivarius strains (PA17D and PA18C) generate high levels of lactic acid but each exhibits a different level of adhesion to PS. L. salivarius PA14C, by contrast, is characterized by low production of lactic acid and high adhesion. The differences among the chosen L. plantarum strains were related to their adherence capacity to PS. L. agilis and L. crispatus were added to the list due to their strong ability to inhibit Campylobacter growth. L. reuteri strains were included in consideration of their abundance in the microbiota of “back yard” chickens, although they showed no anti‐Campylobacter activity.
3.3. Examination of the potential probiotic characteristics of selected Lactobacillus strains
3.3.1. Lactic acid production
First, the production of lactic acid by the lactobacilli was assessed in detail (Figure 2). The highest producers were two strains – L. crispatus 11D and L. plantarum 20A, of which the first strain produced lactic acid at a concentration of 255 mmol/L (22.9 g/L), whereas the second one was 244 mmol/L (22.1 g/L). The lowest lactic acid producers were L. reuteri strains – around 100 mmol/L (10 g/L). All the other strains produced 180–200 mmol/L lactic acid concentrations. The ability to produce both isoforms of lactic acid was identified for each of the Lactobacillus strains, and the levels of l‐ and d‐lactate enantiomers (isoforms) produced by the Lactobacillus strains was found to vary (Figure 2). The highest l‐lactate producers are the L. salivarius strains, whereas the L. plantarum strains demonstrated the highest levels of d‐enantiomer. The single L. agilis and L. crispatus strains were very similar in the proportions of l‐ and d‐enantiomers produced, compared to those generated by members of L. salivarius and L. plantarum species, respectively. On the other hand, L. reuteri strains present an intermediate group of bacteria among those tested with respect to lactate generation, since l‐ and d‐isoforms were present in almost equal concentrations.
Figure 2.
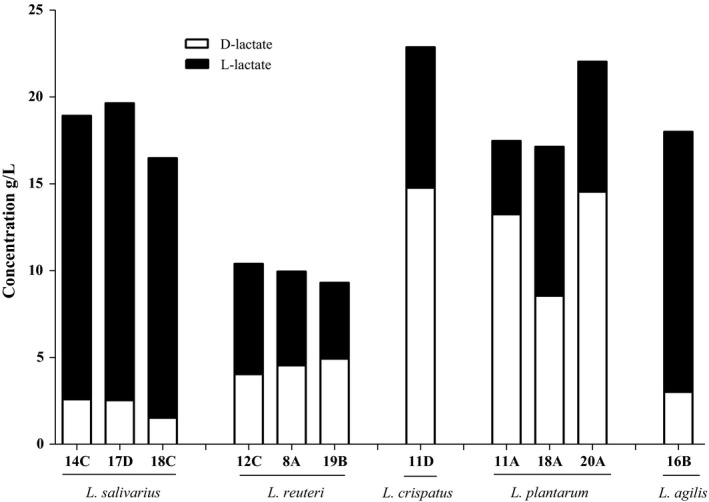
The concentrations (g/L) of d‐ and l‐lactate in the cell‐free supernatants from Lactobacillus culture. The concentration were measured using stereo‐specific d‐ and l‐lactate assay kits (Megazyme, Wicklow, Ireland)
Production of d‐lactate affects synthesis of the cell wall of the bacterial producers by incorporating the d‐lactate in place of d‐alanine, the last molecule of the pentapeptide of peptidoglycan, and thus renders the lactobacilli resistant to the antibiotic vancomycin (Ferain et al., 1996; Goffin et al., 2005). Thus resistance to vancomycin was also checked using M.I.C. Evaluator (Oxoid). We found that all but one (L. crispatus) of the strains were resistant to a high concentration of vancomycin (>256 μg/ml).
3.3.2. Carbohydrate fermentation profiles of Lactobacillus sp
Utilization of many oligosaccharides from different categories appears to be an ubiquitous feature of lactobacilli (Ganzle & Follador, 2012). We analyzed the ability of selected strains (members of the L. salivarius, plantarum, reuteri, and agilis species) to grow on media containing various sugars as a carbon source, using the API test (Figure 3). The API 50 CH profiles for the strains of lactobacilli demonstrated their phenotypic diversity. Lactobacillus strains within a species showed identical or very similar sugar utilization profiles, but none of the three L. salivarius isolates shared the same phenotypic pattern. Among 49 carbohydrates present in the API 50 CH test, the assimilation of seven sugars (d‐galactose, d‐glucose, d‐lactose, d‐maltose, d‐melibiose, d‐raffinose, and d‐sucrose) was common for all analyzed Lactobacillus spp. (data not shown), whereas the metabolic ability and efficiency for the other 24 carbon sources (amygdaline, arbutin, d‐arabitol, β‐gentiobiose, d‐cellobiose, d‐fructose, d‐mannitol, d‐mannose, d‐melezitose, d‐ribose, d‐sorbitol, d‐trehalose, d‐turanose, esculin, gluconic acid, l‐arabinose, l‐fucose, l‐rhamnose, methyl α‐d‐glucopyranoside, methyl α‐d‐mannopyranoside, N‐acetylglucosamine, salicin, starch, and d‐ melibiose) was found to be species‐dependent. The profiles for each Lactobacillus strain are summarized in Figure 3. Among the species tested, all L. plantarum strains used the widest range of simple and more complex carbohydrates, from 22 different carbohydrates (PA11A and PA20A) up to 26 (PA18A). Among the differentially metabolized sugars (Figure 3), L. plantarum strains were defective in the metabolism of only two l‐form sugars (l‐rhamnose and l‐fucose).
Figure 3.
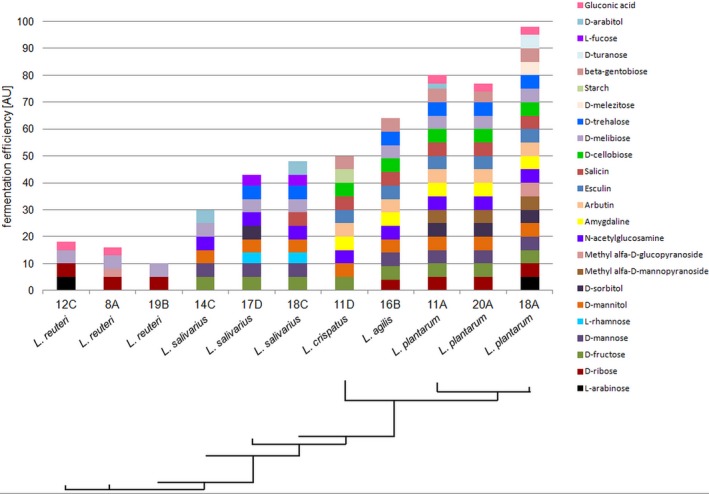
Carbon source assimilation capacities among 11 Lactobacillus strains. Sugar fermentation ability and efficacy is indicated by different color and size tetragons. The assimilation pattern is based on 49 selected carbon sources, excluding invariable sources. The phylogenetic relationships among the analyzed strains phenotypes are shown by a phylogenetic tree. Not used by any of the strains were: 2‐ketogluconate potassium, 5‐ketogluconate potassium, d‐adonitol, d‐arabinose, d‐fucose, d‐xylose, d‐tagatose, dulcitol, l‐xylose, erythritol, glycerol, glycogene, inositol, inuline, l‐arabitol, l‐sorbose, methyl‐α‐d‐mannopyranoside, methyl‐β‐d‐xylopyranoside, and xylitol
Virtually all analyzed strains were able to metabolize maltose, the oligosaccharide from the group of α‐glucans, which are the most omnipresent sugars in the intestinal tract of grain‐eating animals (Ganzle & Follador, 2012). Also, the ability to utilize the α‐galacto‐oligosaccharides‐family (αGOS) d‐melibiose [α‐Gal‐(1 → 6)‐Glu] as well as the raffinose‐family oligosaccharide (RFO) d‐raffinose seems to be a common feature among all Lactobacillus strains tested. On the other hand, the metabolism of a trisaccharide, d‐melezitose (α‐Glu‐(1→3)‐β‐Fru‐(2→1)‐α‐Glu), and the disaccharides d‐trehalose [α‐Glu‐(1 → 1)‐α‐Glu] and d‐turanose (α‐Glu‐(1→3)‐α‐Fru) was species‐ or strain‐specific and restricted mainly to L. plantarum (Figure 3). In contrast to the ubiquitous ability to metabolize oligosaccharides, the majority of the lactobacilli were not amylolytic (Ganzle & Follador, 2012). Indeed, among 11 strains tested, only L. crispatus 11D showed a capacity for starch metabolism (Figure 3), indicating that this strain has a unique ability to produce an extracellular or cell‐bound α‐amylase.
In conclusion, the capacity of individual lactobacilli species for sugar metabolism differed substantially. This metabolic diversity matched with the phylogenetic diversity in the Lactobacillus genus (Figure 3).
3.3.3. Osmotic stress resistance
The ability to resist high salt concentrations is important for probiotic bacteria, since they must survive and grow in the gastrointestinal tract where the environment has an osmolarity equivalent to 0.3 mol/L NaCl (Chowdhury, Sahu, & Das, 1996; West et al., 1996).
In this study, the isolated Lactobacillus spp. showed a variable capacity to survive different concentrations of NaCl. The level of bacterial osmo‐tolerance was more species‐ than strain‐dependent, but none of the isolates could grow in 1.36 mol/L NaCl. The strongest resistance (up to 1 mol/L) was found for all L. plantarum isolates, whereas the resistance of L. salivarius depended on the strain. L. agilis, L. crispatus, and L. reuteri strains showed the weakest resistance, with only faint tolerance to 0.3 mol/L NaCl (Figure 4).
Figure 4.
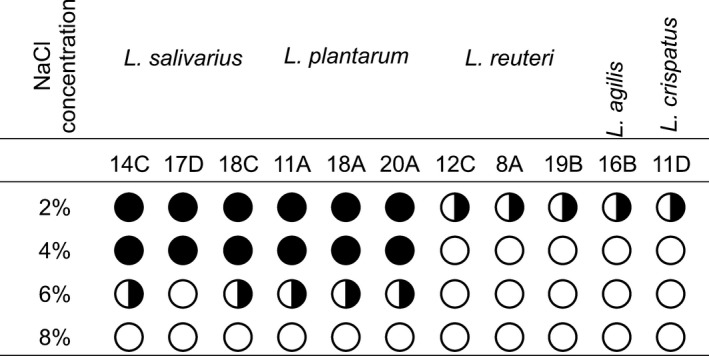
Tolerance of Lactobacillus isolates to different concentration of NaCl. Black circle – full resistance, B&W circle – weak resistance, white circle – lack of resistance
3.3.4. Tolerance to acid and bile salt
The capacity of Lactobacillus strains to act as probiotics is also determined by the ability to survive in the low pH of the stomach and in the high concentration of bile salt of the gastrointestinal tract (Bull, Plummer, Marchesi, & Mahenthiralingam, 2013). In our studies, we decided to check the ability of chosen strains to tolerate pH 2.5 and 0.5% bile salt (Table 2). Only one strain (L. agilis 16B) was unable to grow under such conditions. Other strains employed in the study, members of the species L. plantarum and L. reuteri, showed fairly good survival in low pH and in the presence of bile salt (percentage of the viability ranged from 30 to 87%). Representatives of the species L. salivarius showed rather high tolerance to low pH. At the same time, two of them did not grow in the presence of 0.5% bile salt (14C and 17D), and one exhibited a low percentage of survival (18C). The opposite was observed for L. crispatus 11D – no growth at pH 2.5 and 43.8% of viability in the presence of bile salts.
Table 2.
The tolerance of isolated lactobacilli to low pH and bile salt
| Strain | Control log10 CFU/ml | pH 2.5 | 0.5% of bile salt | ||
|---|---|---|---|---|---|
| Log10 CFU/ml after 6 hr | % of viability | Log10 CFU/ml after 6 hr | % of viability | ||
| L. salivarius PA14C | 8.9 | 4.8 | 49.7 ± 20.4 | 0 | 0 |
| L. salivarius PA17D | 8.9 | 2.6 | 30.2 | 0 | 2.8 ± 5.6 |
| L. salivarius PA18C | 8.9 | 2.9 | 35.9 ± 31.4 | 3.0 | 32.7 ± 17.6 |
| L. crispatus PA11D | 7.4 | 0 | 0 | 3.2 | 43.8 ± 9.2 |
| L. plantarum PA11A | 7.6 | 4.4 | 58.5 ± 34.86 | 6.4 | 87.4 ± 3.5 |
| L. plantarum PA18A | 7.9 | 3.6 | 45.8 ± 3.7 | 6.6 | 83.1 ± 1.2 |
| L. plantarum PA20A | 7.6 | 2.3 | 30.1 ± 9.5 | 5.8 | 76.4 ± 7.0 |
| L. reuteri PA8A | 8.9 | 5.7 | 63.6 ± 3.6 | 4.9 | 55.0 ± 4.6 |
| L. reuteri PA12C | 8.5 | 3.8 | 44.7 ± 18.5 | 5.0 | 59.5 ± 7.4 |
| L. reuteri PA19B | 8.7 | 4.2 | 48.0 ± 6.6 | 5.0 | 57.7 ± 0.6 |
| L. agilis PA16B | 8.6 | 0 | 0 | 0 | 0 |
Values are the mean ± SD from three independent experiments.
3.3.5. Oral administration of selected Lactobacillus strains to chickens – impact on colonization of chickens by Campylobacter
Of the 11 Lactobacillus strains characterized in detail, five were tested in vivo in chicken experiments (Table 3). The aim was to test the effectiveness of lactobacilli application to protect chicken gut against colonization by C. jejuni. Five groups of chickens were administered lactobacilli on day‐of‐hatch and 4 days post hatch. A group of birds inoculated with BSG (PBS with 0.1% gelatin) was used as a control. The details of the procedure are given in Materials and Methods section. At 14 days post hatch chickens were challenged with ~104 CFU of C. jejuni 12/2 strain labeled with the pUOA18 plasmid containing a cat gene. Chickens were killed at 4 and 8 days post challenge and the number of C. jejuni present in the cecum of each chicken was determined. The mean CFU/g of cecal content observed in the untreated control group was about 3 × 107 4 days after challenge and 1 × 108 CFU/g of cecal content 8 days after challenge (Figure 5). Oral inoculation of lactobacilli using strains of L. salivarius, L. crispatus, and L. reuteri, generally did not affect the level of chicken gut colonization by Campylobacter. In contrast, oral inoculation with the L. plantarum strains reduced the number of Campylobacter cells present in the chicken ceca at 4 and 8 days after challenge. The mean level of colonization in the group that received both L. plantarum strains 8 days after challenge was slightly reduced compared to control group (5 × 107 vs. 1 × 108 CFU/g of cecal content). The reduction in C. jejuni cecal content due to L. plantarum application was apparent 4 days after challenge. In the case of L. plantarum PA18A, the median reduction was 1 log10. In the case of L. plantarum PA20A, the mean concentration of C. jejuni cells 4 days after challenge was similar to that observed for control group. However, distinct differences appeared among birds in this group. Two of six chickens were colonized at a low level (about 1 × 105 and 1 × 106 CFU/g of cecal content).
Table 3.
Properties of selected Lactobacillus strains isolated from feces of chickens
| Species | Strain | Anti‐Campylobacter activity | Adhesion to bare polystyrene (adherence ratio)a | l‐lactate (g/L)a | d‐lactate (g/L)a | % of viability at pH 2.5a | % of viability in the presence of 0.5% bile salt |
|---|---|---|---|---|---|---|---|
| L. salivarius | PA14C | + | 7.80 ± 1.34 | 16.4 ± 0.44 | 2.6 ± 0.11 | 49.7 ± 20.4 | 0 |
| PA17D | + | 6.22 ± 1.11 | 17.1 ± 0.40 | 2.5 ± 0.53 | 30.2 | 2.8 ± 5.6 | |
| PA18C | + | 3.50 ± 0.31 | 14.9 ± 1.03 | 1.5 ± 0.14 | 35.9 ± 31.4 | 32.7 ± 17.6 | |
| L. reuteri | PA12C | − | 3.62 ± 0.41 | 6.4 ± 0.45 | 4.0 ± 0.28 | 44.7 ± 18.5 | 59.5 ± 7.4 |
| PA8A | − | 3.66 ± 1.00 | 5.4 ± 0.19 | 4.5 ± 0.35 | 63.6 ± 3.6 | 55.0 ± 4.6 | |
| PA19B | − | 2.79 ± 0.27 | 4.4 ± 0.98 | 4.9 ± 1.19 | 48.0 ± 6.6 | 57.7 ± 0.6 | |
| L. crispatus | PA11D | + | 3.46 ± 1.68 | 8.1 ± 1.09 | 14.8 ± 1.94 | 0 | 43.8 ± 9.2 |
| L. plantarum | PA11A | + | 11.24 ± 0.54 | 4.2 ± 0.46 | 13.2 ± 1.65 | 58.5 ± 34.86 | 87.4 ± 3.5 |
| PA18A | + | 3.48 ± 0.26 | 8.6 ± 0.68 | 8.6 ± 1.04 | 45.8 ± 3.7 | 83.1 ± 1.2 | |
| PA20A | + | 1.10 ± 0.32 | 7.5 ± 0.66 | 14.6 ± 0.83 | 30.1 ± 9.5 | 76.4 ± 7.0 | |
| L. agilis | PA16B | + | 2.20 ± 0.33 | 15.0 ± 1.21 | 3.0 ± 0.45 | 0 | 0 |
The Campylobacter jejuni inhibition assay was performed on Blood Agar plates containing 5% horse blood (Oxoid) inoculated with 100 μl of Campylobacter cultures. Campylobacter growth inhibition by supernatant of Lactobacillus strains is marked as +. Bacterial adhesion was determined using the technique described by Radziwill‐Bienkowska et al. (2014) in three independent experiments. Bacteria were characterized as strongly adherent (A ≥ 6), moderately adherent (6 > A ≥ 3), weakly adherent (3 > A > 2), and nonadherent (A ≤ 2). The adherence ratio for strain L. rhamnosus GG (positive control) was 9.38 and the ratio for nonadhesive L. rhamnosus LOCK 0908 (negative control) was 1.05.
The concentrations (g/L) of d‐ and l‐lactate in the cell‐free supernatants were measured using stereo‐specific d‐ and l‐lactate assay kits (Megazyme, Wicklow, Ireland).
Values are the mean ± SD from three independent experiments.
Figure 5.
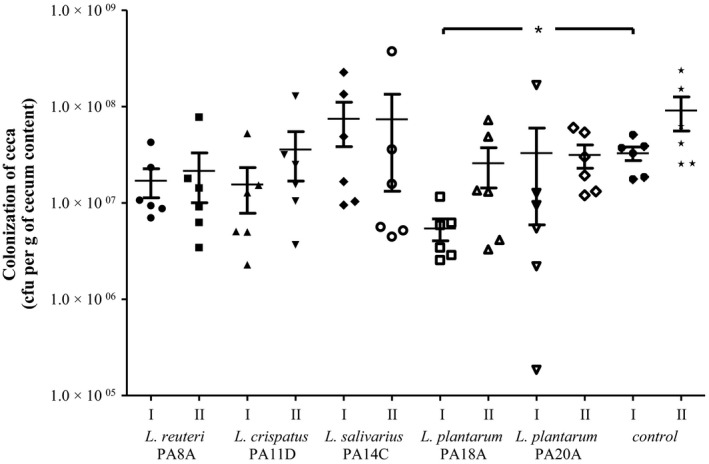
Impact of the Lactobacillus strains on colonization of chickens by Campylobacter. Chickens were orally given Lactobacillus strains (~108 CFU) at day of hatch and 4 days post hatch. Next, birds receiving a Campylobacter jejuni challenge were administered C. jejuni 12/2 (104 CFU) at day 14 post hatch. Control birds were given BSG (PBS with 0.01% gelatin). Chickens were killed at four (I) and eight (II) days post challenge. Cecal contents were serially diluted and plated onto Blood Agar for enumeration of C. jejuni. Bacterial recoveries represent colonization levels of six birds per time interval. The geometric mean for each group is denoted by bars. A statistical analysis was carried out using Kruskal–Wallis test. Asterisks indicate significant differences (p < .05) between analyzed groups and the control group
4. DISCUSSION
A wide array of carriers, both living and nonliving, has been developed to deliver heterologous antigens to the immune system of humans or animals. Among live antigen delivery systems, live attenuated bacterial vectors have been the subject of the most thorough investigations (Curtiss, 2015). LAB bacteria have received considerable attention as biotherapeutic and immunoprophylactic agents due to their GRAS status and because members of this group of microorganisms are autochthonous residents in the human or animal gastrointestinal tract (Bermudez‐Humaran, Kharrat, Chatel, & Langella, 2011; Daniel, Roussel, Kleerebezem, & Pot, 2011; Foligne, Daniel, & Pot, 2013; Wyszynska, Kobierecka, Bardowski, & Jagusztyn‐Krynicka, 2015).
We have shown that in ovo chicken immunization with liposomes containing Campylobacter hybrid protein rCjaAD results in a moderate protection against colonization (Kobierecka, Wyszynska, et al., 2016). The same hybrid protein expressed on the surface of L. lactis also exerted a moderately positive effect, when used for chicken vaccination (Kobierecka, Olech, et al., 2016). We postulate that the efficacy of in ovo immunization with nonliving carriers may be increased by a boost and immunization after hatching with live vectors carrying Campylobacter antigens known to induce a mucosal immune response. The main drawback of the L. lactis delivery vehicle is the lack of long‐lasting colonization of the chick digestive tract. Using Lactobacillus spp. that can consistently colonize, or at least persist for an extended period in the bird intestine and display probiotic properties, should result in more efficient induction of the immune response than that exerted by L. lactis. Lactobacillus strains may be used to decrease the number of Campylobacter colonizing the chicken digestive tract, but at the same time, they can be employed as a vector to deliver Campylobacter immunodominant proteins to the bird immune system. However, members of Lactobacillus spp. constitute an extremely diverse group as regards of their physiological attributes. Thus the main goal of the present work was to identify and characterize Lactobacillus strains isolated from chicken breeds in Poland. First, we compared the composition of Lactobacillus genera colonizing the digestive tract of chickens reared under different conditions. Various species of Lactobacillus genus, such as L. plantarum, L. agilis, L. brevis, L. reuteri, L. salivarius, L. fermentum, and L. crispatus, have been isolated from chicken digestive tracts (Abbas Hilmi, Surakka, Apajalahti, & Saris, 2007; Hammons et al., 2010; Lin et al., 2007; Noohi, Ebrahimipour, Rohani, Talebi, & Pourshafie, 2014). However, studies performed in various countries reported different species as being dominant. The study conducted by Hammons et al. (2010) indicated that the chicken feeding affects the type of L. agilis strains colonizing the birds. Due to enormously discrepant data concerning lactobacilli colonizing chicken gut, we decided to examine lactobacilli strains isolated from birds reared in Poland, as they probably have evolved together with their hosts. The results of this study showed that although the L. salivarius was a dominant species in both “back‐yard” and commercial chickens, the species distribution among the examined Lactobacillus strains isolated from fecal samples of “back‐yard” chickens was more diverse than that among isolates from commercially reared birds. The more diversified diet of privately owned birds may influence the composition of their microbial community and can have an impact on which bacterial strains become dominant in the bird intestine. Also it should be noted that “back‐yard” chickens, in contrast to commercially reared birds, have contact with microorganisms colonizing other animals or present in the environment.
Lactobacillus plantarum species were only isolated from intestines of “back yards” chickens. Analysis of the sugars utilized by each strain showed that three L. plantarum strains were able to utilize an enormously wide spectrum of sugars, from 22 different carbohydrates up to 26. Among all the sugars analyzed, the L. plantarum strains were unable to metabolize only two l‐form sugars (l‐rhamnose and l‐fucose) and starch. This fact suggests the high versatility of this species. Indeed, L. plantarum is commonly found in many different ecological niches, such as vegetables, meat, fish, dairy products as well as in the gastrointestinal tracts (Siezen & van Hylckama Vlieg, 2011). This broad repertoire of sugar metabolism corresponds with the more than 3 Mb L. plantarum genome, which is among the largest Lactobacillus genomes reported to date (http://www.ncbi.nlm.nih.gov/genome/). Similar to L. plantarum, a slightly narrower carbohydrate utilization profile was observed for L. agilis. In contrast, the observed spectrum of metabolic activity of all the L. salivarius and L. reuteri isolates was low and, among the differentially metabolized sugars, limited to only a few carbon sources. Neither L. salivarius nor L. reuteri was able to metabolize any of the β‐glucosides, and additionally, all L. reuteri strains were defective in metabolism of α‐glucosides and more simple sugars, such as d‐arabitol, d‐fructose, d‐mannitol, d‐mannose, l‐rhamnose, and d‐sorbitol metabolism. The significant reduction in the range of sugars metabolized by L. salivarius and L. reuteri, as compared to L. plantarum, may result from the considerably smaller genome size of both species (http://www.ncbi.nlm.nih.gov/genome/), and that genome size may be subject to an ongoing process of degradation for niche adaptation, as was found in the case of L. salivarius (Raftis, Salvetti, Torriani, Felis, & O'Toole, 2011). L. salivarius and L. reuteri are reported to be highly specialized, and their primary habitats are limited to niches such as human and animal gastrointestinal tracts (Casas & Dobrogosz, 2000; Raftis et al., 2011).
Acid and bile tolerance and the ability to overcome high concentration of NaCl are essential natural attributes of probiotic strains since they have to survive in the conditions present in the intestinal tract (stomach and small intestine) of their host. Thus, we checked whether the selected strains display these properties. In this study, the isolated Lactobacillus sp. showed a variable capacity to survive in different concentrations of NaCl. According to Mohd Adnan and Tan (2007), high osmotolerance could be a desirable feature for potentially commercial LAB strains, because lactic acid production, would result in pumping alkali outside to prevent an excessive reduction in pH, and converting the free acid to its salt form, finally elevating the osmotic pressure on the bacterial cells (Mohd Adnan & Tan, 2007). Our results suggest that all the analyzed strains could resist the NaCl osmolarity of the animal GI tract, but only L. plantarum and some L. salivarius strains could resist technological processing, as they were the most tolerant to high NaCl concentrations that can be present in pelleted or dried animal food. Moreover, it has been reported that Lactobacillus strains adapted to NaCl exhibited up to 16‐fold higher tolerance to lethal temperatures used during spray drying technological process (Desmond, Stanton, Fitzgerald, Collins, & Ross, 2001).
One possible mechanism by which lactobacilli restrict the growth of pathogenic microorganisms is their ability to produce inhibitory metabolites such as organic acids. Lactic acid is widely involved in various branches of industry, for example, food and feed production, medical and pharmaceutical use, and also in cosmetics. The biologically active isoform of lactate is l‐lactate (Martinez et al., 2013), which unlike d‐lactate, can be metabolized by humans. Lactic acid impairs growth of various microorganisms, including pathogenic and spoilage bacteria (Neal‐McKinney et al., 2012).
Members of the Lactobacillus genus are l‐ and d‐lactate producers, though the ratio of two isomers differs considerably and is recognized as species dependent (Manome, Okada, Uchimura, & Komagata, 1998). Our results indicate that this feature of lactobacilli may be not only species‐ but also strain‐dependent. The L. crispatus strain characterized by Neal‐McKinney et al. (2012) produces similar levels of two lactate isomers, whereas L. crispatus analyzed in this work produces mainly d‐lactate. Although high l‐lactate concentrations were observed only for the L. salivarius and L. agilis strains, strains with high d‐lactate levels also impaired growth of the Campylobacter pathogen. It is thus possible, that a certain level of total lactate concentration (both l‐ and d‐isoforms) is important to hamper the growth of pathogenic bacteria. Production of d‐lactate, mediated by either d‐lactate dehydrogenase or l‐lactate racemase, affects synthesis of the cell wall of the producer bacteria that incorporate it in place of d‐alanine, the last molecule of the pentapeptide of peptidoglycan, which renders the lactobacilli resistant to the high concentrations of antibiotic vancomycin (Ferain et al., 1996; Goffin et al., 2005). This phenotype is seen mostly in L. plantarum and some other heterofermentative lactobacilli. Vancomycin resistance of homofermentative L. salivarius species is controversial and potentially is strain‐dependent (Ammor, Florez, & Mayo, 2007; Ammor et al., 2008; EFSA, 2012). Our observations corroborate these data, since all the strains tested were found to be resistant to vancomycin (intrinsic resistance, nonspecific, nontransferable). The only strain sensitive to vancomycin was L. crispatus, which similar to L. plantarum produces high levels of d‐lactate and belongs to the facultatively heterofermentative group of lactobacilli. Explanation of this vancomycin sensitivity requires more research, as the mechanism that determines vancomycin resistance was studied in detail only in L. plantarum (Goffin et al., 2005).
Anti‐Campylobacter activity expressed in in vitro experiments has been documented for some Lactobacillus strains of different origins (Ghareeb et al., 2012; Robyn et al., 2012; Santini et al., 2010). Although many Lactobacillus strains have been examined for their anti‐Campylobacter activity in vitro, only limited studies to date have been undertaken to confirm this effect in vivo. Our knowledge about interaction among constituents of intestinal microbiota is rather limited. Thus, the data arising from in vitro experiments, although they provide useful information about strain physiology, have to be treated with caution and only as indication for planning further experiments. To evaluate anti‐Campylobacter activities in vivo, potential probiotic Lactobacillus species have been administered as mono‐ or multi‐species feed additives. The studies have differed in their experimental designs and the data obtained through in vitro examination were not always consistent with those provided by animal studies (Robyn, Rasschaert, Hermans, Pasmans, & Heyndrickx, 2013; Robyn et al., 2012; Santini et al., 2010). The main differences among in vivo experiments concern the length of time of formula administration and C. jejuni strains used (Neal‐McKinney et al., 2012; Nishiyama et al., 2014; Santini et al., 2010). The procedures most often used daily administration of the probiotic formula. Although most of Lactobacillus strains only transiently colonize chicken gastrointestinal chicken tracts, three Lactobacillus strains (L. agilis, L. crispatus, and L. vaginalis) that were able to persist in the chicken intestine for the entire production period after a single inoculation (day 0) have been isolated (Spivey et al., 2014; Stephenson, Moore, & Allison, 2010). Generally the clearest efficacy was achieved by daily applying a multispecies probiotic product (Poultry Star sol) (Ghareeb et al., 2012; Guyard‐Nicodeme et al., 2016). In the present work, we employed a schema of chicken experiments identical as that used by Neal‐McKinney et al. (2012), but some elements differed, mainly the C. jejuni strain used for challenge and the Lactobacillus strains administered. The one common Lactobacillus strain used by us and Neal‐McKinney et al. was a member of the L. crispatus species, but the two strains appear to differ in terms of their physiology, as based on type and amount of lactate production. Of five species tested in this work, lactobacilli strains that are members of L. plantarum appeared to be the most effective. Other strains did not display anti‐Campylobacter activity, though it is not known whether a more frequent and longer application period would improve their efficacy.
Overall, the present data show that L. plantarum strains isolated from digestive tracts of chicken bred in Poland displayed some probiotic attributes in vitro and were able to decrease the level of bird gut colonization by C. jejuni strain. However, further experiments are needed to understand details concerning their interaction with C. jejuni, as well as with the host, which should help to improve the schema of its application. Additionally, recent experiments in our lab show that L. plantarum strains are susceptible to genetic manipulation, so they may be used as potential delivery vectors for C. jejuni antigens.
CONFLICT OF INTEREST
None declared.
Supporting information
ACKNOWLEDGMENTS
We thank Dr. Jeffrey Hansen for critical reading of the manuscript. The work was supported by grants from the National Science Center, Poland (grants No 2011/03/B/NZ1/00592, 2014/15/N/NZ2/00380) and the Ministry of Science and Higher Education through the Faculty of Biology, University of Warsaw intramural grant DSM 110120.
Kobierecka PA, Wyszyńska AK, Aleksandrzak‐Piekarczyk T, et al. In vitro characteristics of Lactobacillus spp. strains isolated from the chicken digestive tract and their role in the inhibition of Campylobacter colonization. MicrobiologyOpen. 2017;6:e512 https://doi.org/10.1002/mbo3.512
REFERENCES
- Abbas Hilmi, H. T. , Surakka, A. , Apajalahti, J. , & Saris, P. E. (2007). Identification of the most abundant lactobacillus species in the crop of 1‐ and 5‐week‐old broiler chickens. Applied and Environmental Microbiology, 73, 7867–7873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agunos, A. , Waddell, L. , Leger, D. , & Taboada, E. (2014). A systematic review characterizing on‐farm sources of Campylobacter spp. for broiler chickens. PLoS ONE, 9, e104905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aleksandrzak‐Piekarczyk, T. , Koryszewska‐Baginska, A. , Grynberg, M. , Nowak, A. , Cukrowska, B. , Kozakova, H. , & Bardowski, J. (2016). Genomic and functional characterization of the unusual pLOCK 0919 plasmid harboring the spaCBA pili cluster in Lactobacillus casei LOCK 0919. Genome Biology and Evolution, 8, 202–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammor, M. S. , Florez, A. B. , & Mayo, B. (2007). Antibiotic resistance in non‐enterococcal lactic acid bacteria and bifidobacteria. Food Microbiology, 24, 559–570. [DOI] [PubMed] [Google Scholar]
- Ammor, M. S. , Florez, A. B. , van Hoek, A. H. , de Los Reyes‐Gavilan, C. G. , Aarts, H. J. , Margolles, A. , & Mayo, B. (2008). Molecular characterization of intrinsic and acquired antibiotic resistance in lactic acid bacteria and bifidobacteria. Journal of Molecular Microbiology and Biotechnology, 14, 6–15. [DOI] [PubMed] [Google Scholar]
- Bermudez‐Brito, M. , Plaza‐Díaz, J. , Muñoz‐Quezada, S. , Gómez‐Llorente, C. , & Gil, A. (2012). Probiotic Mechanisms of Action. Annals of Nutrition & Metabolism, 61, 160–174. [DOI] [PubMed] [Google Scholar]
- Bermudez‐Humaran, L. G. , Kharrat, P. , Chatel, J. M. , & Langella, P. (2011). Lactococci and lactobacilli as mucosal delivery vectors for therapeutic proteins and DNA vaccines. Microbial Cell Factories, 10(Suppl 1), S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardeau, M. , Vernoux, J. P. , Henri‐Dubernet, S. , & Gueguen, M. (2008). Safety assessment of dairy microorganisms: The Lactobacillus genus. International Journal of Food Microbiology, 126, 278–285. [DOI] [PubMed] [Google Scholar]
- Bjerrum, L. , Engberg, R. M. , Leser, T. D. , Jensen, B. B. , Finster, K. , & Pedersen, K. (2006). Microbial community composition of the ileum and cecum of broiler chickens as revealed by molecular and culture‐based techniques. Poultry Science, 85, 1151–1164. [DOI] [PubMed] [Google Scholar]
- Bujnakova, D. , Strakova, E. , & Kmet, V. (2014). In vitro evaluation of the safety and probiotic properties of Lactobacilli isolated from chicken and calves. Anaerobe, 29, 118–127. [DOI] [PubMed] [Google Scholar]
- Bull, M. , Plummer, S. , Marchesi, J. , & Mahenthiralingam, E. (2013). The life history of Lactobacillus acidophilus as a probiotic: A tale of revisionary taxonomy, misidentification and commercial success. FEMS Microbiology Letters, 349, 77–87. [DOI] [PubMed] [Google Scholar]
- Casas, I. , & Dobrogosz, W. (2000). Validation of the probiotic concept: Lactobacillus reuteri confers broad‐spectrum protection against disease in humans and animals. Microbial Ecology in Health and Disease, 12, 247–285. [Google Scholar]
- Chaveerach, P. , Lipman, L. J. , & van Knapen, F. (2004). Antagonistic activities of several bacteria on in vitro growth of 10 strains of Campylobacter jejuni/coli . International Journal of Food Microbiology, 90, 43–50. [DOI] [PubMed] [Google Scholar]
- Chowdhury, R. , Sahu, G. K. , & Das, D. (1996). Stress response in pathogenic bacteria. Journal of Biosciences, 21, 149–160. [Google Scholar]
- Curtiss, R. (2015). Antigen delivery system II: Development of live attenuated bacterial vectors In Mestecky J., Strober W., Russell M., Cheroutre H., Lambrecht B., & Kelsall B. (Eds.), Mucosal immunology, 4th ed. (pp. 1233–1269). London: Elsevier Academic Press. [Google Scholar]
- Daniel, C. , Roussel, Y. , Kleerebezem, M. , & Pot, B. (2011). Recombinant lactic acid bacteria as mucosal biotherapeutic agents. Trends in Biotechnology, 29, 499–508. [DOI] [PubMed] [Google Scholar]
- Desmond, C. , Stanton, C. , Fitzgerald, G. , Collins, K. , & Ross, R. (2001). Environmental adaptation of probiotic lactobacilli towards improvement of performance during spray drying. International Dairy Journal, 11, 801–808. [Google Scholar]
- EFSA Panel on Additives and Products or Substances used in Animal Feed (2012). Guidance on the assessment of bacteria susceptibility to antimicrobials of human and veterinary importance. EFSA Journal, 10(6), 2740. [Google Scholar]
- Ferain, T. , Hobbs, J. N. Jr , Richardson, J. , Bernard, N. , Garmyn, D. , Hols, P. , … Delcour, J. (1996). Knockout of the two ldh genes has a major impact on peptidoglycan precursor synthesis in Lactobacillus plantarum . Journal of Bacteriology, 178, 5431–5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foligne, B. , Daniel, C. , & Pot, B. (2013). Probiotics from research to market: The possibilities, risks and challenges. Current Opinion in Microbiology, 16, 284–292. [DOI] [PubMed] [Google Scholar]
- Ganzle, M. G. , & Follador, R. (2012). Metabolism of oligosaccharides and starch in lactobacilli: A review. Frontiers in Microbiology, 3, 340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghareeb, K. , Awad, W. A. , Mohnl, M. , Porta, R. , Biarnes, M. , Bohm, J. , & Schatzmayr, G. (2012). Evaluating the efficacy of an avian‐specific probiotic to reduce the colonization of Campylobacter jejuni in broiler chickens. Poultry Science, 91, 1825–1832. [DOI] [PubMed] [Google Scholar]
- Goffin, P. , Deghorain, M. , Mainardi, J. L. , Tytgat, I. , Champomier‐Verges, M. C. , Kleerebezem, M. , & Hols, P. (2005). Lactate racemization as a rescue pathway for supplying D‐lactate to the cell wall biosynthesis machinery in Lactobacillus plantarum . Journal of Bacteriology, 187, 6750–6761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guyard‐Nicodeme, M. , Keita, A. , Quesne, S. , Amelot, M. , Poezevara, T. , Le Berre, B. , … Chemaly, M. (2016). Efficacy of feed additives against Campylobacter in live broilers during the entire rearing period. Poultry Science, 95, 298–305. [DOI] [PubMed] [Google Scholar]
- Hammons, S. , Oh, P. L. , Martinez, I. , Clark, K. , Schlegel, V. L. , Sitorius, E. , … Walter, J. (2010). A small variation in diet influences the Lactobacillus strain composition in the crop of broiler chickens. Systematic and Applied Microbiology, 33, 275–281. [DOI] [PubMed] [Google Scholar]
- Hermans, D. , Pasmans, F. , Messens, W. , Martel, A. , Van Immerseel, F. , Rasschaert, G. , … Haesebrouck, F. (2012). Poultry as a host for the zoonotic pathogen Campylobacter jejuni . Vector Borne and Zoonotic Diseases, 12, 89–98. [DOI] [PubMed] [Google Scholar]
- Kaakoush, N. O. , Castano‐Rodriguez, N. , Mitchell, H. M. , & Man, S. M. (2015). Global epidemiology of Campylobacter infection. Clinical Microbiology Reviews, 28, 687–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaakoush, N. O. , Sodhi, N. , Chenu, J. W. , Cox, J. M. , Riordan, S. M. , & Mitchell, H. M. (2014). The interplay between Campylobacter and Helicobacter species and other gastrointestinal microbiota of commercial broiler chickens. Gut Pathogens, 6, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobierecka, P. A. , Olech, B. , Ksiazek, M. , Derlatka, K. , Adamska, I. , Majewski, P. M. , … Wyszynska, A. K. (2016). Cell wall anchoring of the Campylobacter Antigens to Lactococcus lactis . Frontiers in Microbiology, 7, 165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobierecka, P. A. , Wyszynska, A. K. , Gubernator, J. , Kuczkowski, M. , Wisniewski, O. , Maruszewska, M. , … Jagusztyn‐Krynicka, E. K. (2016). Chicken anti‐Campylobacter vaccine ‐ comparison of various carriers and routes of immunization. Frontiers in Microbiology, 7, 740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korlath, J. A. , Osterholm, M. T. , Judy, L. A. , Forfang, J. C. , & Robinson, R. A. (1985). A point‐source outbreak of campylobacteriosis associated with consumption of raw milk. The Journal of Infectious Diseases, 152, 592–596. [DOI] [PubMed] [Google Scholar]
- Lin, W. H. , Yu, B. , Jang, S. H. , & Tsen, H. Y. (2007). Different probiotic properties for Lactobacillus fermentum strains isolated from swine and poultry. Anaerobe, 13, 107–113. [DOI] [PubMed] [Google Scholar]
- Lukjancenko, O. , Ussery, D. W. , & Wassenaar, T. M. (2012). Comparative genomics of Bifidobacterium, Lactobacillus and related probiotic genera. Microbial Ecology, 63, 651–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manome, A. , Okada, S. , Uchimura, T. , & Komagata, K. (1998). The ratio of L‐form to D‐form of lactic acid as a criteria for the identification of lactic acid bacteria. The Journal of General and Applied Microbiology, 44, 371–374. [DOI] [PubMed] [Google Scholar]
- Martinez, F. , Balciunas, F. , Salgadb, J. , González, J. , Convertic, A. , & de Souza Oliveira, R. (2013). Lactic acid properties, applications and production: A review. Trends in Food Science & Technology, 30, 70–83. [Google Scholar]
- McDonald, I. R. , Kenna, E. M. , & Murrell, J. C. (1995). Detection of methanotrophic bacteria in environmental samples with the PCR. Applied and Environmental Microbiology, 61, 116–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messaoudi, S. , Madi, A. , Prevost, H. , Feuilloley, M. , Manai, M. , Dousset, X. , & Connil, N. (2012). In vitro evaluation of the probiotic potential of Lactobacillus salivarius SMXD51. Anaerobe, 18, 584–589. [DOI] [PubMed] [Google Scholar]
- Messaoudi, S. , Manai, M. , Kergourlay, G. , Prevost, H. , Connil, N. , Chobert, J. M. , & Dousset, X. (2013). Lactobacillus salivarius: Bacteriocin and probiotic activity. Food Microbiology, 36, 296–304. [DOI] [PubMed] [Google Scholar]
- Meunier, M. , Guyard‐Nicodeme, M. , Dory, D. , & Chemaly, M. (2016). Control strategies against Campylobacter at the poultry production level: Biosecurity measures, feed additives and vaccination. Journal of Applied Microbiology, 120(5), 1139–1173. [DOI] [PubMed] [Google Scholar]
- Mohd Adnan, A. F. , & Tan, I. K. (2007). Isolation of lactic acid bacteria from Malaysian foods and assessment of the isolates for industrial potential. Bioresource Technology, 98, 1380–1385. [DOI] [PubMed] [Google Scholar]
- Mota, R. M. , Moreira, J. L. , Souza, M. R. , Horta, M. F. , Teixeira, S. M. , Neumann, E. , … Nunes, A. C. (2006). Genetic transformation of novel isolates of chicken Lactobacillus bearing probiotic features for expression of heterologous proteins: A tool to develop live oral vaccines. BMC Biotechnology, 6, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neal‐McKinney, J. M. , Lu, X. , Duong, T. , Larson, C. L. , Call, D. R. , Shah, D. H. , & Konkel, M. E. (2012). Production of organic acids by probiotic lactobacilli can be used to reduce pathogen load in poultry. PLoS ONE, 7, e43928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newell, D. G. , Elvers, K. T. , Dopfer, D. , Hansson, I. , Jones, P. , James, S. , … Allen, V. M. (2011). Biosecurity‐based interventions and strategies to reduce Campylobacter spp. on poultry farms. Applied and Environmental Microbiology, 77, 8605–8614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama, K. , Seto, Y. , Yoshioka, K. , Kakuda, T. , Takai, S. , Yamamoto, Y. , & Mukai, T. (2014). Lactobacillus gasseri SBT2055 reduces infection by and colonization of Campylobacter jejuni . PLoS ONE, 9, e108827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noohi, N. , Ebrahimipour, G. , Rohani, M. , Talebi, M. , & Pourshafie, M. R. (2014). Phenotypic characteristics and probiotic potentials of Lactobacillus spp. isolated from poultry. Jundishapur Journal of Microbiology, 7, e17824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley, B. B. , Buhr, R. J. , Ritz, C. W. , Kiepper, B. H. , Berrang, M. E. , Seal, B. S. , & Cox, N. A. (2014). Successional changes in the chicken cecal microbiome during 42 days of growth are independent of organic acid feed additives. BMC Veterinary Research, 10, 282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley, B. B. , Lillehoj, H. S. , Kogut, M. H. , Kim, W. K. , Maurer, J. J. , Pedroso, A. , … Cox, N. A. (2014). The chicken gastrointestinal microbiome. FEMS Microbiology Letters, 360, 100–112. [DOI] [PubMed] [Google Scholar]
- Pan, D. , & Yu, Z. (2014). Intestinal microbiome of poultry and its interaction with host and diet. Gut Microbes, 5, 108–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radziwill‐Bienkowska, J. M. , Le, D. T. L. , Szczesny, P. , Duviau, M.‐P. , Aleksandrzak‐Piekarczyk, T. , Loubière, P. , … Kowalczyk, M. (2016). Adhesion of the genome‐sequenced Lactococcus lactis subsp. cremoris IBB477 strain is mediated by specific molecular determinants. Applied Microbiology and Biotechnology, 100, 9605–9617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radziwill‐Bienkowska, J. M. , Zochowska, D. , Bardowski, J. , Mercier‐Bonin, M. , & Kowalczyk, M. (2014). Lactococcus lactis IBB477 presenting adhesive and muco‐adhesive properties as a candidate carrier strain for oral vaccination against influenza virus. Acta Biochimica Polonica, 61, 603–607. [PubMed] [Google Scholar]
- Raftis, E. J. , Salvetti, E. , Torriani, S. , Felis, G. E. , & O'Toole, P. W. (2011). Genomic diversity of Lactobacillus salivarius . Applied and Environmental Microbiology, 77, 954–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robyn, J. , Rasschaert, G. , Hermans, D. , Pasmans, F. , & Heyndrickx, M. (2013). In vivo broiler experiments to assess anti‐Campylobacter jejuni activity of a live Enterococcus faecalis strain. Poultry Science, 92, 265–271. [DOI] [PubMed] [Google Scholar]
- Robyn, J. , Rasschaert, G. , Messens, W. , Pasmans, F. , & Heyndrickx, M. (2012). Screening for lactic acid bacteria capable of inhibiting Campylobacter jejuni in in vitro simulations of the broiler chicken caecal environment. Beneficial Microbes, 3, 299–308. [DOI] [PubMed] [Google Scholar]
- Santini, C. , Baffoni, L. , Gaggia, F. , Granata, M. , Gasbarri, R. , Di Gioia, D. , & Biavati, B. (2010). Characterization of probiotic strains: An application as feed additives in poultry against Campylobacter jejuni . International Journal of Food Microbiology, 141(Suppl 1), S98–S108. [DOI] [PubMed] [Google Scholar]
- Schokker, D. , Veninga, G. , Vastenhouw, S. A. , Bossers, A. , de Bree, F. M. , Kaal‐Lansbergen, L. M. , … Smits, M. A. (2015). Early life microbial colonization of the gut and intestinal development differ between genetically divergent broiler lines. BMC Genomics, 16, 418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segers, M. E. , & Lebeer, S. (2014). Towards a better understanding of Lactobacillus rhamnosus GG–host interactions. Microbial Cell Factories, 13(Suppl 1), S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siezen, R. J. , & van Hylckama Vlieg, J. E. (2011). Genomic diversity and versatility of Lactobacillus plantarum, a natural metabolic engineer. Microbial Cell Factories, 10(Suppl 1), S3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spivey, M. A. , Dunn‐Horrocks, S. L. , & Duong, T. (2014). Epithelial cell adhesion and gastrointestinal colonization of Lactobacillus in poultry. Poultry Science, 93, 2910–2919. [DOI] [PubMed] [Google Scholar]
- Stanley, D. , Hughes, R. J. , & Moore, R. J. (2014). Microbiota of the chicken gastrointestinal tract: Influence on health, productivity and disease. Applied Microbiology and Biotechnology, 98, 4301–4310. [DOI] [PubMed] [Google Scholar]
- Stephenson, D. P. , Moore, R. J. , & Allison, G. E. (2010). Lactobacillus strain ecology and persistence within broiler chickens fed different diets: Identification of persistent strains. Applied and Environmental Microbiology, 76, 6494–6503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svetoch, E. A. , & Stern, N. J. (2010). Bacteriocins to control Campylobacter spp. in poultry–A review. Poultry Science, 89, 1763–1768. [DOI] [PubMed] [Google Scholar]
- Tareb, R. , Bernardeau, M. , Gueguen, M. , & Vernoux, J. P. (2013). In vitro characterization of aggregation and adhesion properties of viable and heat‐killed forms of two probiotic Lactobacillus strains and interaction with foodborne zoonotic bacteria, especially Campylobacter jejuni . Journal of Medical Microbiology, 62, 637–649. [DOI] [PubMed] [Google Scholar]
- Thibodeau, A. , Fravalo, P. , Yergeau, E. , Arsenault, J. , Lahaye, L. , & Letellier, A. (2015). Chicken caecal microbiome modifications induced by Campylobacter jejuni colonization and by a non‐antibiotic feed additive. PLoS ONE, 10, e0131978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagenaar, J. A. , French, N. P. , & Havelaar, A. H. (2013). Preventing Campylobacter at the source: Why is it so difficult? Clinical Infectious Diseases, 57, 1600–1606. [DOI] [PubMed] [Google Scholar]
- Wei, S. , Morrison, M. , & Yu, Z. (2013). Bacterial census of poultry intestinal microbiome. Poultry Science, 92, 671–683. [DOI] [PubMed] [Google Scholar]
- West, J. L. , Chowdhury, S. M. , Sawhney, A. S. , Pathak, C. P. , Dunn, R. C. , & Hubbell, J. A. (1996). Efficacy of adhesion barriers. Resorbable hydrogel, oxidized regenerated cellulose and hyaluronic acid. The Journal of Reproductive Medicine, 41, 149–154. [PubMed] [Google Scholar]
- Willis, W. L. , & Reid, L. (2008). Investigating the effects of dietary probiotic feeding regimens on broiler chicken production and Campylobacter jejuni presence. Poultry Science, 87, 606–611. [DOI] [PubMed] [Google Scholar]
- Wise, M. G. , & Siragusa, G. R. (2007). Quantitative analysis of the intestinal bacterial community in one‐ to three‐week‐old commercially reared broiler chickens fed conventional or antibiotic‐free vegetable‐based diets. Journal of Applied Microbiology, 102, 1138–1149. [DOI] [PubMed] [Google Scholar]
- Wyszynska, A. , Kobierecka, P. , Bardowski, J. , & Jagusztyn‐Krynicka, E. K. (2015). Lactic acid bacteria–20 years exploring their potential as live vectors for mucosal vaccination. Applied Microbiology and Biotechnology, 99, 2967–2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyszynska, A. , Raczko, A. , Lis, M. , & Jagusztyn‐Krynicka, E. K. (2004). Oral immunization of chickens with avirulent Salmonella vaccine strain carrying C. jejuni 72Dz/92 cjaA gene elicits specific humoral immune response associated with protection against challenge with wild‐type Campylobacter . Vaccine, 22, 1379–1389. [DOI] [PubMed] [Google Scholar]
- Zambrano, L. D. , Levy, K. , Menezes, N. P. , & Freeman, M. C. (2014). Human diarrhea infections associated with domestic animal husbandry: A systematic review and meta‐analysis. Transactions of the Royal Society of Tropical Medicine and Hygiene, 108, 313–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


