Abstract
Sediment microbial communities play an important role in lake trophic status. This study determined millions of Illumina reads (16S rRNA gene amplicons) to compare the bacterial communities in moderately eutrophic, lightly eutrophic, and moderately trophic regions using a technically consistent approach. The results indicated that the sediments from moderately eutrophic and trophic lake had the higher bacterial diversity than lightly eutrophic lake. Proteobacteria was the most abundant phylum (22.7%–86.2%) across samples from three regions. The sediments from moderately eutrophic region were enriched with Chloroflexi and Nitrospirae. Alphaproteobacteria, Gammaproteobacteria, and Bacteroidetes were enriched in the sediments from lightly eutrophic lake. The sediments from moderately trophic lake contained a high abundance of Acidobacteria and Deltaproteobacteria because of the low pH of the sediments in this lake. In moderately eutrophic region, Nitrospira held an absolute predominance, while Lysobacter and Flavobacterium were the most predominant genera in lightly eutrophic region. Temperature was the main factor influencing the bacterial community in the three lakes. The bacterial communities in the sediment samples obtained from moderately eutrophic lake were associated with nutrient concentration, whereas organic matter and total nitrogen contents mainly influenced the bacterial communities in sediments obtained from lightly eutrophic lake and moderately trophic lake, respectively.
Keywords: Bacterial community, lake, sediment, sequence, trophic status
1. INTRODUCTION
Lake sediment is one of the main media for nutrient transformation and migration and can act as either nutrient source or pool to the overlying water column; thus, sediments influence the nutrient contents of lakes. Microbial communities play an important role in nutrient cycling in aquatic ecosystems (Cotner & Biddanda, 2002; Jurgens et al., 2000; Ligi et al., 2014). Biogeochemical cycles, especially in sediments, are greatly influenced by microbial communities (Fang et al., 2015; Liu & Liu, 2013; Song, Li, Du, Wang, & Ding, 2012), and changes in the composition of sediment microbial communities can significantly impact the biogeochemical environment of sediments (Haukka et al., 2006; Van der Gucht et al., 2001). Trophic status is the structural and functional quality of water bodies; the trophic status of various aquatic ecosystems is assessed using different indices, such as biological communities and physical, chemical, morphological, and hydrological characteristics (Heiskanen, van de Bund, Cardoso, & Noges, 2004; Molozzi, Feio, Salas, Marques, & Callisto, 2012). The trophic status of aquatic ecosystems was greatly influenced by biological quality elements (Bere & Tundisi, 2010; Kelly et al., 2008; Teixeira et al., 2009). As one of the most critical factors influencing biological quality (Bending, Putland, & Rayns, 2000), microbial community has a significant relationship with trophic status (Duarte, Pascoal, Garabetian, Cassio, & Charcosset, 2009; Kostrzewska‐Szlakowska, 2005). In addition, sediment is a main component of lakes and contains complex microbial communities. Therefore, microbial communities in lake sediments can greatly influence the trophic status of lakes (Dai et al., 2016; Vezzulli & Fabiano, 2006).
Shallow fresh water lakes with different trophic statuses are commonly found in plains. They display different trophic statuses possibly because of various factors, such as amount of aquatic plants in lake (Ginn, 2011), exogenous pollution around the lake (Bertahas, Dimitriou, Karaouzas, Laschou, & Zacharias, 2006), and phytoplankton biomass (Phillips et al., 2013). As the trophic status of lakes changes, bacterial communities in sediments also change. In addition, the composition of bacterial communities can cause variations in nutrient contents of lakes (Molina et al., 2016; Phan et al., 2016). Therefore, trophic status and bacterial community are closely related, and understanding the differences in bacterial community structure of sediments in lake with different trophic statuses is crucial. This knowledge may improve the management and treatment of eutrophic lakes and the prevention of contamination of clean lakes.
In Eastern Plain of China, a large number of shallow fresh water lakes with different trophic statuses exist, and the sediments in these lakes greatly influence the trophic status of these lakes (Berglund, Larsson, Ewald, & Okla, 2001; Trolle, Hamilton, & Pilditch, 2010). Moreover, sediments in lake display higher biomass and microbe taxon richness than the water bodies (Huang & Jiang, 2016), and bacteria in sediments contribute to the decomposition of organic or inorganic compounds, promoting nutrient recycling (Liu et al., 2009; Nealson, 1997); as a result, nutrient contents in lake change, affecting the trophic status of lakes. Gonghu Bay is located in the northeast part of Taihu Lake (T), which is the third largest freshwater lake in China and located in the Eastern Plain of China. Gonghu Bay is an algae‐dominated region in Taihu Lake with an average depth of 2.02 m and an area of 147 km2. Nutrient contents have increased since 2005, and a massive algal bloom has occurred in Gonghu Bay. In 2007, the frequency of algal bloom increased greatly, and the algal bloom affected the security of drinking water in Taihu Lake Basin (Chen et al., 2010). The trophic state index (TSI) (Azevêdo et al., 2015) of Gonghu Bay has recently reached 64.8, and this area is classified as a moderately eutrophic region (Hou et al., 2013). Huangda Lake (HD), which is a shallow fresh water lake, is also located in the Eastern Plain of China. The area of Huangda Lake is approximately 269 km2, and its average depth is 3.64 m. Being a moderately trophic lake, Huangda Lake is a clean lake and its recent TSI value is approximately 45.4. East Dongting Lake (DT) is the largest region (1,328 km2) of Dongting Lake, which is the second largest freshwater lake in China. The East Dongting Lake is 2.1–7.3 m deep, and a large number of shallow areas exist in this region. With the increased anthropogenic activity, the trophic status of East Dongting Lake gradually increased in recent years, and its TSI reached up to 52.6 (in lightly eutrophic region).
This study investigated the bacterial communities in sediments obtained from the three lakes with different trophic statuses (moderately eutrophic, lightly eutrophic, and moderately trophic) using Illumina Miseq high‐throughput sequencing technology. A total of 5.1 million tags of the sediment samples were determined. This work will provide new insights into the nutrient control in lakes.
2. EXPERIMENTAL PROCEDURES
2.1. Sediment sampling
A total of 48 samples were collected from Gonghu Bay of Taihu Lake (T, 12 samples), East Dongting Lake (DT, 16 samples), and Huangda Lake (HD, 20 samples) in August 2016. Figure 1 shows the sampling sites in three regions and the location of these lakes. The composite surface (10 cm) sediment samples (each N = 5) were collected in August 2016, and these samples were assigned as DT, HD, and T, respectively. The weight of the samples collected was sufficient for DNA extraction and for analysis of physicochemical parameters. After sampling, the sediment samples were placed in sealed plastic bags, stored in a portable ice box, transferred into the laboratory within 24 hr, and stored at −80°C before analysis.
Figure 1.
Sampling sites and location of the sampling lakes in China
2.2. Analysis of physicochemical parameters
The temperature of overlying water (Temp.), oxidation reduction potential (ORP) in sediments, and depth were measured in situ using a portable multi‐parameter water quality analyzer (ProPlus, YSI, USA). Total nitrogen (TN) and total phosphorus (TP) in sediments were measured using standardized methods and tests (Institute of Soil Science, Chinese Academy of Science, 1978; Ruban et al., 2001). Organic matter (OM) content in sediments was calculated according to the loss on ignition to constant mass (4 hr) at 550°C (Huang et al., 2015). The pH of the sediment was measured in a 1:2.5 (w/v) mixture of sediment and deionized water (Liao et al., 2014). Table 1 shows the main properties of the sediment samples.
Table 1.
Physicochemical parameters in sediment and water from the three lakes with different trophic status
| T | DT | HD | ||||
|---|---|---|---|---|---|---|
| Parameter | Mean | Range | Mean | Range | Mean | Range |
| Depth (m) | 2.01 | 1.42–3.11 | 5.28 | 0.31–7.35 | 3.64 | 2.16–5.27 |
| Temp. (°C) | 32.5 | 31.5–33.3 | 29.1 | 27.4–30.7 | 34.0 | 32.9–34.6 |
| OM (%)a | 4.55 | 3.92–6.37 | 9.21 | 7.21–11.02 | 7.51 | 5.06–10.25 |
| ORP (mv) | 35.7 | −5.4–96.4 | 23.8 | −21.3–123.6 | 49.1 | −35.2–171.6 |
| TN (mg kg−1)a | 1708.1 | 1391.2–2170.4 | 912.6 | 611.9–1423.2 | 1969.5 | 1157.3–3333.7 |
| TP (mg kg−1)a | 793.1 | 631.9–891.6 | 823.6 | 602.3–1003.7 | 571.9 | 386.1–797.1 |
| pHa | 7.85 | 7.16–8.31 | 7.15 | 7.11–7.46 | 6.21 | 5.47–7.27 |
Data of T, DT, and HD are obtained from 12, 16, and 20 sites, respectively.
Significant differences exist among the three lakes (p < .05).
2.3. Extraction of genomic DNA
Total genomic DNA was extracted from 250 mg samples using a PowerSoil DNA Isolation Kit (Mobio Laboratories Inc., San Diego, CA, USA) according to the manufacturer's protocol. DNA concentration and quality were determined using a NanoDrop Spectrophotometer. DNA was diluted to 10 ngμl−1 using sterile ultrapure water and stored at −80°C for further analysis.
2.4. PCR amplification of 16S rRNA genes and sequencing
The barcode 515F (5ʹ‐GTGCCAGCMGCCGCGGTAA‐3ʹ) and 926R (5ʹ‐CCGTCAATTCMTTTGAGTTT‐3ʹ) primers were used to amplify the V4–V5 regions of the bacterial 16S rRNA (Liu, DeSantis, Andersen, & Knight, 2008; Wang & Qian, 2009; Xiong et al., 2014). The PCR mixture (25 μl) contained 1 × PCR buffer, 1.5 mmol/L MgCl2, 0.4 μmol/L each of deoxynucleoside triphosphate, 1.0 μmol/L of each primer, 0.5 U of TaKaRa Ex Taq, and 10 ng of template DNA. The PCR amplification program is as follows: initial denaturation at 94°C for 1 min followed by 30 cycles (denaturation at 94°C for 20 s, annealing at 56°C for 30 s, and elongation at 72°C for 45 s) and a final extension at 72°C for 5 min. Three replicates of PCR reactions for each sample were combined. A similar volume of 1 × loading buffer (containing SYB green) was mixed with PCR products and then subjected under electrophoresis on 2% agarose gel for detection. Samples showing bright main strip of 300–500 bp were chosen for further experiments. PCR products were purified using an OMEGA Gel Extraction Kit (Omega Bio‐Tek, USA), and equal molar amounts of the products from different samples were pooled. Sequencing libraries were generated using TruSeq DNA PCR‐Free Sample Prep Kit according to the manufacturer's recommendations, and index codes were added. Library quality was assessed on a Qubit@2.0 Fluorometer (Thermo Scientific) and Agilent Bioanalyzer 2100 system. Finally, the library was subjected under paired‐end sequencing (2 × 250 bp) on an Illumina Miseq apparatus at Rhonin Biosciences Co., Ltd.
2.5. Statistical analysis
After sequencing, Usearch (Edgar, 2010; Edgar, Haas, Clemente, Quince, & Knight, 2011) and QIIME pipeline were used for analysis (Caporaso et al., 2010). Paired‐end reads from the original DNA fragments were merged using FLASH (Magoc & Salzberg, 2011). The sequences were assigned to each sample according to the unique barcode. In this study, we adopted relatively stringent quality controls. We first filtered low‐quality reads (length is < 200 bp, more than two ambiguous base “N,” or average base quality score is < 30) and truncated sequences, wherein quality scores decay (score < 11). After finding the duplicated sequences, we discarded all singletons, which are possibly bad amplicons that lead to overestimation of diversity. Sequences were clustered into operational taxonomic units (OTUs) at 97% identity threshold using UPARSE algorithms (Edgar, 2013). We selected representative sequences and removed potential chimeras using UCHIME algorithm (Edgar et al., 2011). The clean sequences of each sediment sample ranged from 17,770 to 25,9634.
Principal coordinate analysis (PCoA) was employed to explore and visualize the similarities between sediment samples obtained from lakes with different trophic statuses based on Bray–Curtis dissimilarity using package Ape (Paradis, Claude, & Strimmer, 2004). Distance‐based redundancy analysis (db‐RDA) was performed in R environment using the package Vegan. The Pearson correlation method was used to analyze the correlation analysis between genera and environmental variables, and p values were corrected by FDR (False Discovery Rate) method. Then the complete‐linkage clustering of OTU was used to obtain the clustering information which was present using heatmap. The functions Capscale, Anova.cca, and Vif.cca in R package Vegan were used to perform model construction, variables or axes significance permutation test, and variance inflation factor analysis. LEfSe (University of Auckland, Auckland, New Zealand) was used to identify the indicator bacterial groups specific to the sediment samples (Segata et al., 2011). In addition, independent T‐test and permutational multivariate analysis of variance based on Bray–Curtis dissimilarity matrix were used to identify the differences that exist among different groups.
2.6. Accession numbers
All of the sequencing data analyzed in this study can be downloaded from the NCBI's Sequence Read Archive using the accession numbers SRR5131114, SRR531118, and SRR5131121 for samples from Gonghu Bay of Taihu Lake, East Dongting Lake, and Huangda Lake, respectively.
3. RESULTS
3.1. Diversity indices of bacterial community
A total of 48 sediment samples were obtained from moderately eutrophic, lightly eutrophic, and moderately trophic lakes. Usearch and QIIME pipeline were used to analyze the raw sequences. Relatively representative sequences were selected and potential chimeras were removed using UCHIME algorithm. Figure 2 shows the values of diversity (Chao1, ACE, and Shannon) and OTU in sediments obtained from the three lakes. After sequencing and quality control, the observed OTU in East Dongting Lake was 1,229 in average, which was lower than that in Gonghu Bay of Taihu Lake (2,251) and Huangda Lake (2,193). Chao1, ACE, and Shannon indices of the bacterial communities in sediments obtained from Gonghu Bay of Taihu Lake and Huangda Lake showed similar values. The value of Chao1 varied from 2023.4 to 4141.6 (2423.0 in average) and 2255.3 to 4194.9 (3430.7 in average) for Gonghu Bay of Taihu Lake and Huangda Lake, respectively. The value of Chao1 in East Dongting Lake was lower than that in the two other lakes and varied from 1232.5 to 2886.1 (2049.5 in average). Moreover, ACE of bacterial communities in sediments from Gonghu Bay of Taihu Lake and Huangda Lake were higher than that in sediments obtained from East Dongting Lake (p < .05). Shannon index of DT13 in East Dongting Lake showed the lowest value (3.54), and the other positions varied from 4.57 to 5.89 (5.15 in average), which was significantly lower than that in Gonghu Bay of Taihu Lake (6.80 in average) and Huangda Lake (6.57 in average) (p < .05).
Figure 2.
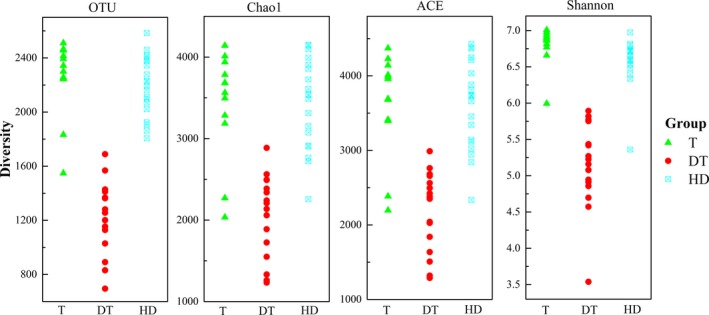
Diversity indices of 16S rRNA gene libraries with significant differences (p < .05)
3.2. Bacterial community composition in sediment samples
Among the filtered sequences, 56 phyla were determined in the sediment samples obtained from the three lakes. Figure 3 shows the dominant groups (relative abundance >0.1%) in each sample. Proteobacteria was the most abundant phylum (22.7%–86.2%) across samples, and the bacterial sequences were affiliated with Acidobacteria (12.9%–34.7%), Chloroflexi (2.5%–19.6%), Bacteroidetes (2.5%–44.4%), and Nitrospirae (1.3%–11.8%). Figure 3a shows the differences in bacterial communities at the phylum level between the sediment samples from three regions. The diversity of Chloroflexi and Nitrospirae was more abundant in the sediment samples obtained from Gonghu Bay of Taihu Lake and Huangda Lake than in the samples obtained from East Dongting Lake, and significant differences (p < .01) were observed among the samples obtained from these lakes. Firmicutes was more abundant in the sediments obtained from East Dongting Lake (e.g., 4.0% in DT9 and 12.3% in DT16) than in the sediments obtained from Gonghu Bay of Taihu Lake and Huangda Lake. Gemmatimonadetes, Actinobacteria, Sporochaetes, Planctomycetes, OP8, NC10, and WS3 were the other phyla which were universally detected in the sediment samples from three regions.
Figure 3.
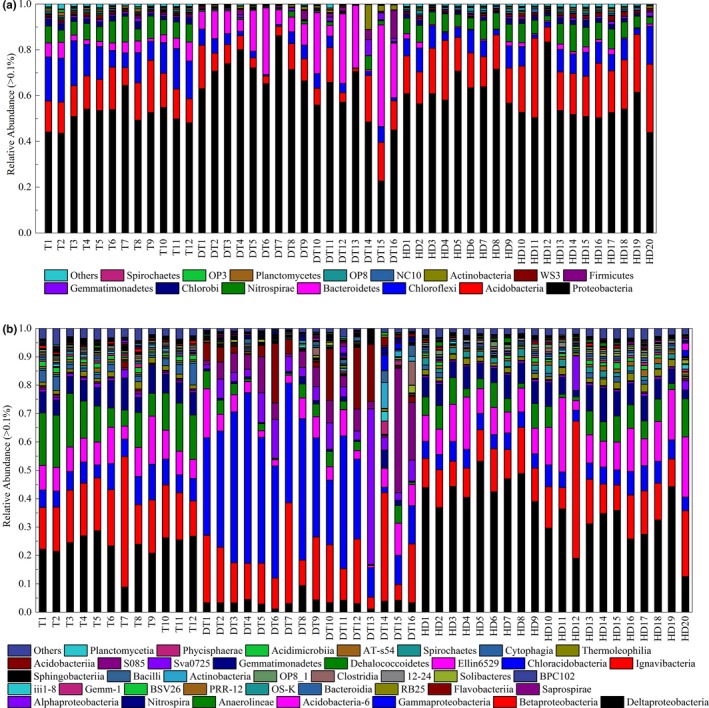
Abundances of different phyla (a) and classes (b) in 48 sediment samples based on pyrosequencing at 0.03 level (12, 20, and 16 samples from Gonghu Bay of Taihu Lake, Huangda Lake, and East Dongting Lake, respectively)
Figure 3b shows the changes in bacterial community composition at the class level. Deltaproteobacteria, Betaproteobacteria, Gammaproteobacteria, Alphaproteobacteria, Anaerolineae, and Nitrospira predominated in the three lakes. The sediment samples obtained from Gonghu Bay of Taihu Lake and Huangda Lake displayed higher abundance of Deltaproteobacteria (23.2% and 36.3%, respectively) than the samples obtained from East Dongting Lake (3.6%); Nitrospira is also more abundant in the Gonghu Bay of Taihu Lake and Huangda Lake (7.5% and 6.9%, respectively) than in East Dongting Lake (1.2%). In addition, Gammaproteobacteria and Flavobacteria were more abundant in the sediment samples obtained from East Dongting Lake than in the samples obtained from Gonghu Bay of Taihu Lake and Huangda Lake (p < .01). Saprospirae, Bacteroidia, Solibacteres, Closstridia, Actinobacteria, Bacilli, Dehalococcoidetes, Gemmatimonadetes, etc. were the other classes which could be detected in the sediment samples from three regions.
The taxonomic composition at the genus level of the bacterial communities in sediment samples from three regions is shown in Figure 4 . Lysobacter and Flavobacterium were the most predominant genera in most of sediment samples from East Dongting Lake (with relative abundance, >7%). The relative abundance of Nitrospira in sediment samples from Gonghu Bay was higher than other two regions (with relative abundance, >5%). HB118, GOUTA19, and Delftia in the sediment samples from Huangda Lake had higher relative abundance than other two regions. In addition, other dominant genera with appreciable relative abundance (average values above 0.1%) in the sediment sample from the three regions included Desulfococcus, Geobacter, Anaerolinea, Methylobacterium, Desulfobulbus, Hydrogenophaga, Sphingomonas, Polaromonas, Rubrivivax, etc.
Figure 4.

Heatmap analysis of bacterial community at genus level
3.3. Principal coordinate analysis
PCoA revealed the differences in microbial community patterns among sediment samples obtained from the three lakes. Figure 5 shows the grouping of the sediment samples according to their bacterial community structure. Two PCoAs explained 43.7% of the total variation in the microbial community structure. Bacterial communities were clearly clustered in the ordination plot according to sediment samples (Figure 5), with the samples from Gonghu Bay of Taihu Lake (T), East Dongting Lake (DT), and Huangda Lake (HD) separated from each other. In addition, the samples from HD20 and HD12 were separated from other samples obtained from Huangda Lake, and the sediment samples of T7 showed similarity with the other samples from Gonghu Bay of Taihu Lake. Independent T‐test indicated that the bacterial communities in sediment samples from the three lakes significantly differed from one another (p < .05).
Figure 5.
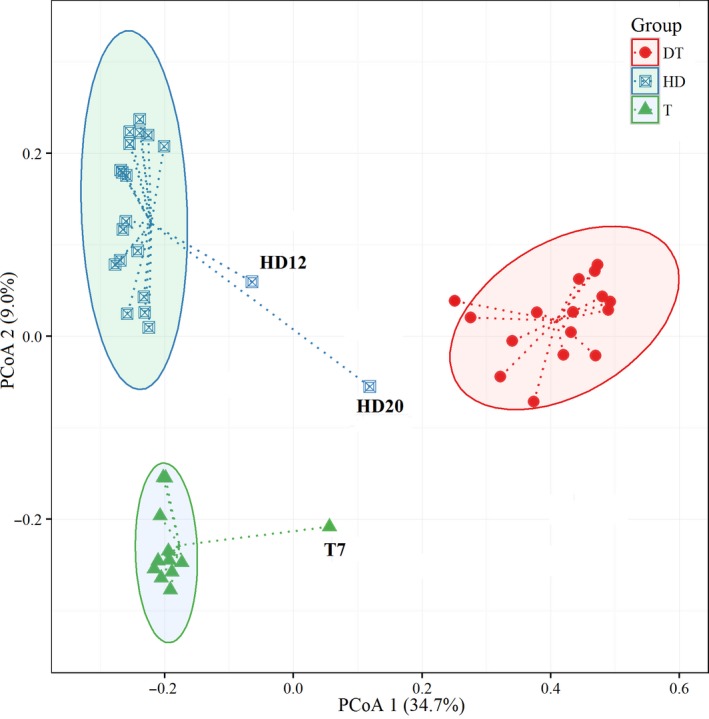
Ordination plot showing the grouping of the sediment samples from the lakes with different trophic statuses according to their bacterial community structure; the principle coordination analysis was based on Bray–Curtis distance matrix
3.4. Distance‐based redundancy analysis (db‐RDA)
db‐RDA was used to partition the variation in beta‐diversity into fractions based on environmental variables (physiochemical parameters). Figure 6a shows the relationship between bacterial community composition and physiochemical parameters. The db‐RDA results suggest that the bacterial community composition of the sediment samples changed with trophic status. The bacterial communities in East Dongting Lake were associated with OM content; by contrast, those in the Huangda Lake were mainly associated with TN concentration, whereas some communities were associated with ORP. In Gonghu Bay of Taihu Lake, multiple physiochemical parameters, such as TN, TP, OM, and temperature, could affect the bacterial community in sediments, resulting in the differences in these communities from those in the two other lakes.
Figure 6.
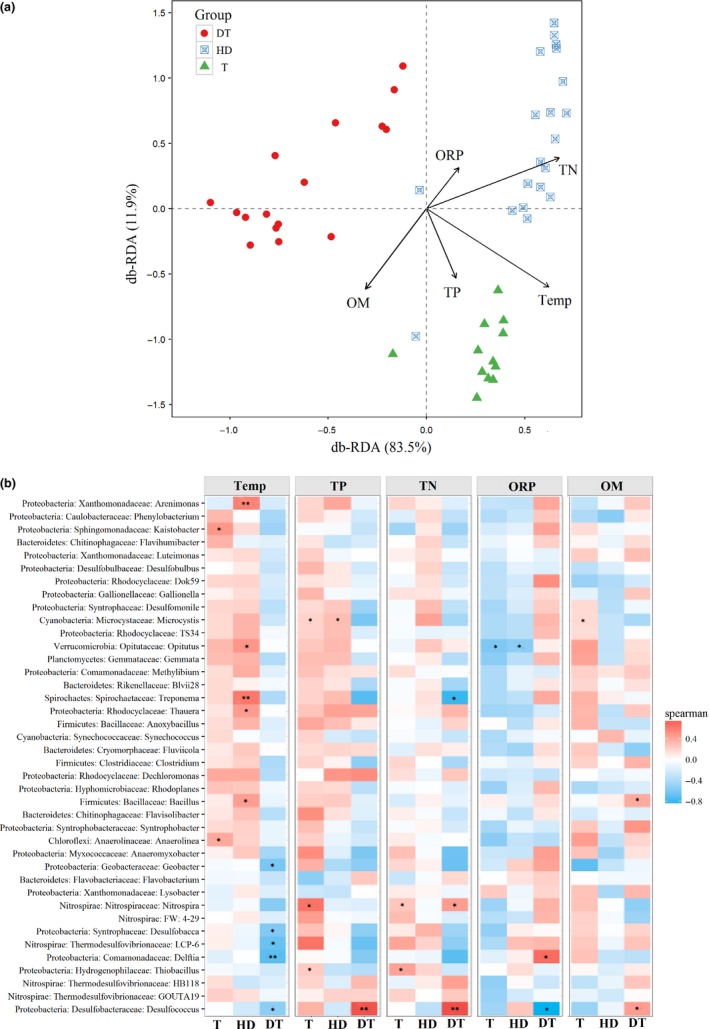
Relationship between bacterial communities and physiochemical parameters in three lakes with different trophic statuses. (a) Structure of bacterial community composition constrained by physiochemical parameters. (b) Correlation analysis between genera and physiochemical parameter. * p < .05, **p < .01
Figure 6b shows the correlation analysis between genera and physicochemical parameters based on db‐RDA. Temperature is the main influencing factor in bacterial community at the genus level in the three lakes. The genus Microcystis (phylum Cyanobacteria) was positively correlated with TP concentration (p < .05) in sediments obtained from Gonghu Bay of Taihu Lake and Huangda Lake, as well as correlated with OM content (p < .05) in sediments obtained from Gonghu Bay of Taihu Lake. Moreover, the db‐RDA results show that in Gonghu Bay of Taihu Lake, Nitrospira and Thiobacillus were associated with TN concentration (p < .05). In the sediments of East Dongting Lake, Desulfococcus (phylum Proteobacteria) was positively correlated with TP, TN (p < .01), and OM content (p < .05). In addition, Bacillus, Treponema, and Opitutus were correlated with OM, temperature, and ORP, respectively.
3.5. LEfSe analysis based on community abundance
Figure 7 shows that the bacterial lineages enriched in sediment samples from Gonghu Bay of Taihu Lake were Chloroflexi, Nitrospirae (from phylum to order), Bacteroidia (the class and order of Bacteroidales), Desulfobacterales (the class and order of Desulfobacteraceae), and Entotheonellales (within Deltaproteobacteria). Within these groups, six fine lineages showed an LDA value of 4 or higher; these lineages are Nitrospirales, GCA004, Entotheonellales, Desulfobacteraceae, envOPS12, and Bacteroidales (Figure 8).
Figure 7.
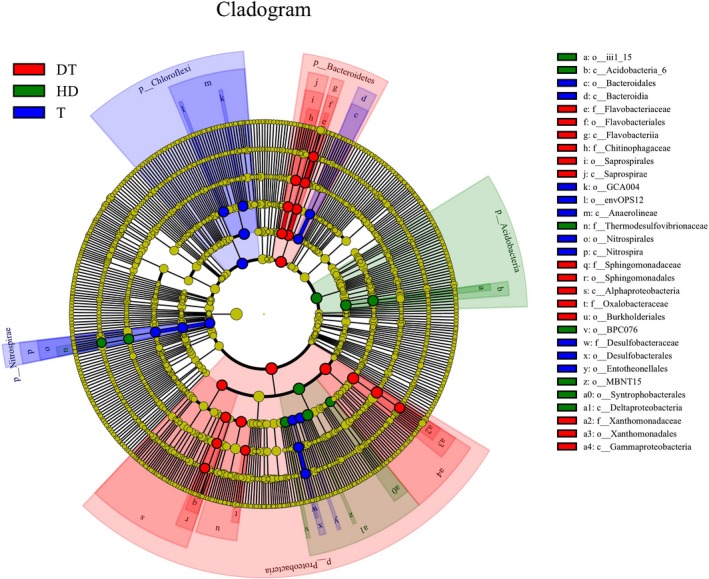
Cladogram showing the phylogenetic distribution of microbial lineages associated with the sediments in lakes with different trophic statuses; lineages with LDA values of 4.0 or higher as determined by LEfSe are shown. Differences are represented by the color of the most abundant class (red indicating East Dongting Lake, green, Huangda Lake, and blue, Gonghu Bay of Taihu Lake; yellow represents insignificant difference). The diameter of each circle is proportional to a taxon's abundance. Circles from inner region to outer region represent the phylogenetic levels from domain to genus
Figure 8.
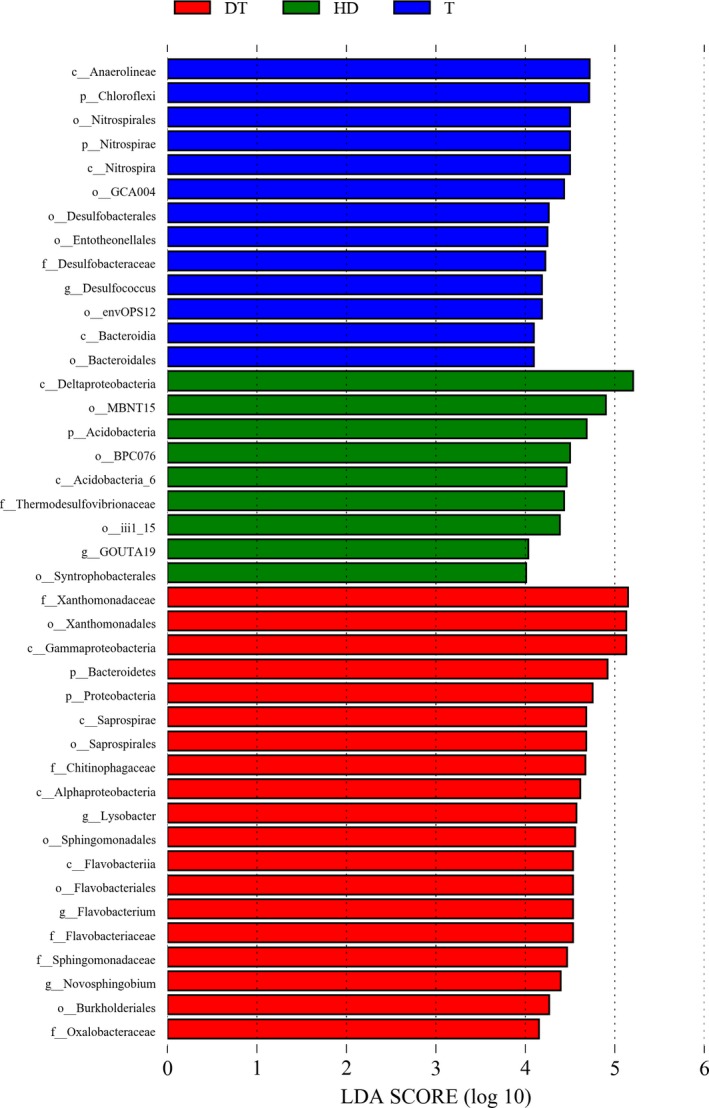
Indicator microbial groups in the three group of sediment samples with LDA values higher than 4.0
Phylum Proteobacteria (Figures 7 and 8), particularly Alphaproteobacteria, one order in Gammaproteobacteria (Xanthomonadales), and two classes in Bacteroidetes (Flavobacteriia and Saprospirae) were enriched in the sediments obtained from East Dongting Lake. At the fine taxonomy levels, Xanthomonadaceae, Chitinophagaceae, Flavobacteriaceae, Sphingomonadaceae, and Oxalobacteraceae found in the sediment samples from East Dongting Lake showed LDA values of higher than 4 (Figure 8).
In the sediment samples from Huangda Lake, Thermodesulfovibrionaceae (a family within Nitrospirae), Acidobacteria, and Deltaproteobacteria (the class and orders of Syntrophobacterales, MBNT15, and BPC076) were enriched in bacterial community (Figure 7). In the sediment samples from Huangda Lake, Thermodesulfovibrionaceae and three groups of Deltaproteobacteria showed LDA values of higher than 4 (Figure 8).
4. DISCUSSION
Proteobacteria is nearly the most abundant phylum in the three districts. Proteobacteria is commonly the most abundant phylum in sediment or soil and that it strongly plays a role in degradation and metabolism in lake sediments (Bai et al., 2012; Chaudhry, Rehman, Mishra, Chauhan, & Nautiyal, 2012). In the Taihu Lake Basin, amounts of exogenous pollutants entered into Taihu Lake, especially in Gonghu Bay, making the latter moderately eutrophic. The sedimentation of organics occurred, and Chloroflexi, which is highly abundant, participated in organic degradation (Daniel, Pozzi, Foresti, & Chinalia, 2009). Therefore, the OM content of sediments was relatively lower in Gonghu Bay of Taihu Lake than in two other lakes (Table 1). In lightly eutrophic lake (East Dongting Lake), Firmicutes predominated in some sediment samples, such as DT9 and DT16. Every August, the water level in Dongting Lake is low, and dry‐rewet cycles occur, especially in East Dongting Lake. Therefore, some sampling sites show high abundance of Firmicutes, which can produce spores that resist dehydration and extreme environmental conditions (Huang & Jiang, 2016).
Studies have indicated that Gammaproteobacteria existed in organic‐rich sediments, such as in the sediments in areas contaminated with agricultural pollution or organic sediments (Beardsley, Moss, Malfatti, & Azam, 2011; Bissett, Bowman, & Burke, 2006; Kunihiro et al., 2011). Anthropogenic activities and agricultural pollution around the basin of East Dongting Lake have recently worsened, and amounts of organic pollutants have entered into East Dongting Lake, worsening the trophic status of the lake. The organic content of sediments increased due to the sedimentation of exogenous organic pollutants. As a result, Gammaproteobacteria in sediment samples is more abundant in East Dongting Lake than in the two other lakes. In addition, Flavobacteria played a significant role in biopolymer degradation in sedimentary organic matter (Bauer et al., 2006; Bissett, Bowman, & Burke, 2008; Gomez‐Pereira et al., 2012) and was more highly abundant in East Dongting Lake than in the two other lakes. Deltaproteobacteria and Nitrospira predominated in the sediment samples from Gonghu Bay of Taihu Lake, which is an algae‐dominated and moderately eutrophic area and where massive algal bloom occurs every August. In Gonghu Bay of Taihu Lake, Deltaproteobacteria and Nitrospira are possibly the important classes for nutrient release in sediments, and the amount of these two classes can serves as potential early warning for the occurrence of algal bloom. The distribution of bacterial community in sediments obtained from Huangda Lake at the class level is similar to that in the sediments obtained from Gonghu Bay of Taihu Lake, indicating that although Huangda Lake is a moderately trophic lake, the nutrient in sediments can be potentially released. Therefore, being a clean lake, the sediments of Huangda Lake must be treated to retain the better water quality.
Sediments from Huangda Lake, which is moderately trophic lake, showed a unique pattern of bacterial diversity (phylum of Acidobacteria) compared with that in East Dongting Lake (lightly eutrophic lake) and Gonghu Bay of Taihu Lake (moderately eutrophic lake). In addition, the bacterial groups enriched in Huangda Lake were mainly limited to Nitrospirae and Deltaproteobacteria (Figures 3a and b). Huangda Lake is a freshwater and moderately trophic lake, and Acidobacteria are ubiquitous and abundant members of bacterial communities in the sediment of freshwater lake. Studies have suggested that abundance of Acidobacteria is significantly correlated with pH, and Acidobacteria prefers an environment with low pH of approximately 5.5 (Jones et al., 2009; Lauber, Hamady, Knight, & Fierer, 2009). The pH of sediments in Huangda Lake is lower than that in sediments of East Dongting Lake and Gonghu Bay of Taihu Lake (Table 1), possibly resulting in higher abundance of Acidobacteria in Huangda Lake than in the two other lakes (Figure 3a). Furthermore, LEfSe analysis showed that subdivisions 6 (Acidobacteria_6) are enriched in the sediments from Huangda Lake (Figure 7), indicating that the distribution of Acidobacteria subdivisions in lakes varies with trophic status (Orcutt, Sylvan, Knab, & Edwards, 2011). The trophic status in East Dongting Lake has recently increased, and the sediments played a significant role in the trophic status of the lake. Several microbes (e.g., Bacteroidetes, Firmicutes, Alphaproteobacteria, and Gammaproteobacteria), which prefer eutrophic conditions, were enriched in sediments obtained from East Dongting Lake, and the bacterial community in the sediment possibly affected the trophic status of the lake. Chloroflexi and Nitrospirae (from phylum to order) were mainly enriched in the sediments obtained from Gonghu Bay of Taihu Lake (Figure 7). Nitrospira from Nitrospirae was the most abundant genus in the sediments obtained from Gonghu Bay of Taihu Lake, and nitrogen is possibly one of the main limiting factors of eutrophication instead of phosphorus in Taihu Lake; Nitrospira is a strong indicator of nitrogen cycle. Chloroflexi is a photoautotrophic microbe and has possibly participated in the degradation of organics, thereby influencing nutrient content, which is one of the main factors affecting trophic status. Therefore, Nitrospirae or Chloroflexi can potentially serve as warning for the occurrence of algal bloom.
The bacterial communities in sediment samples from Gonghu Bay were influenced by TP, TN, temperature, and OM. Studies have indicated that TP and temperature are the main factors influencing bacterial communities, and phosphorus is a significant limiting factor of eutrophication in Taihu Lake (Shao et al., 2011; Song et al., 2012; Tang et al., 2009). In addition, Gonghu Bay is an algae‐dominated area in Taihu Lake, and the concentration of nutrients, such as TN and TP, should stabilize the bacterial community in the sediments. Therefore, TP and temperature were the main environmental factors affecting the bacterial communities. Gonghu Bay of Taihu Lake, which is a moderately eutrophic lake, suffers from serious eutrophication every August, and the growth and metabolism of algae is significantly related to the sediments. The genus Microcystis (phylum Cyanobacteria) is positively related to TP concentration, demonstrating that P remains one of the most significant limiting factors of eutrophication in Gonghu Bay of Taihu Lake, and the sediments of the algae‐dominated area is dominated by Microcystis (Shao et al., 2011; Wu, Chen, Xu, Liu, & Hahn, 2007). In addition, Microcystis is positively correlated with TP concentration in sediments from Huangda Lake. Huangda Lake is a clean lake and has a moderately trophic status with TSI of 45.4, and the average TP concentration in sediments is 571.9 mg kg−1 (Table 1). However, the TP concentration in sediments or water has recently increased due to fish farming and mining of phosphate near Huangda Lake; these anthropogenic activities possibly promote the activity of Microcystis (phylum Cyanobacteria) community, increasing the risk of eutrophication.
Although P is a limiting factor of eutrophication for many years in Taihu Lake (Shao et al., 2011; Song et al., 2012; Zeng et al., 2009), P is no longer the only limiting factor of eutrophication eventually. In addition, the genera Thiobacillus (phylum Proteobacteria) and Nitrospira (phylum Nitrospirae) were relatively abundant in Gonghu Bay of Taihu Lake, and they were positively correlated with TN concentration (Figure 6a). N forms were affected by Nitrospirae, and NO3‐N concentration greatly changed in Gonghu Bay of Taihu Lake. Furthermore, Thiobacillus can directly participate in NO3‐N metabolism, thereby influencing TN concentration (Bruckner et al., 2013; Li, Zhao, & Wang, 2011). Therefore, Nitrospirae and Thiobacillus are possibly among the main reasons for the change in limiting factors in Gonghu Bay of Taihu Lake. East Dongting Lake is a moderately eutrophic lake, and its eutrophic conditions have recently worsened because of anthropogenic activities and agricultural pollution around its basin. Large amounts of nitrogen have entered into the lake, and nitrogen deposition resulted in relatively high N content in sediments and abundant bacterial communities, such as Nitrospirae.
Correlation analysis between genera and physiochemical parameters in sediments showed that the genus Desulfococcus (family Desulfobacteraceae) was significantly positively correlated with TN and TP concentrations in sediments obtained from East Dongting Lake. In East Dongting Lake, OM content was relatively high, and the transformation of nutrients from inorganic to organic forms occurs frequently, resulting in the association between Desulfobacteraceae and organic N or P. Moreover, Figure 6b shows that OM content is positively correlated with the genus Desulfococcus in the sediments obtained from East Dongting Lake, and a study has estimated that sulfate reduction can account for as much as 50% of OM degradation (Jorgensen, 1982). Therefore, the high OM content in sediments obtained from East Dongting Lake can ensure the activity of sulfate reducers (Pallud & Van Cappellen, 2006).
In summary, obvious differences in bacterial communities in the three lakes with different trophic statuses were observed, and the bacterial communities were significantly correlated with the trophic status of the lakes. This study comprehensively compared the three lakes with different trophic statuses (moderately eutrophic, lightly eutrophic, and moderately trophic lake) using high‐throughput sequencing method. Proteobacteria was the most abundant phylum in these lakes. The sediments from moderately eutrophic lake (Gonghu Bay of Taihu Lake) were enriched with Chloroflexi and Nitrospirae. The sediment from lightly eutrophic region (East Dongting Lake) showed a low diversity and was enriched with Alphaproteobacteria, Gammaproteobacteria, and Bacteroidetes. The sediments from Huangda Lake, which is a moderately trophic lake, displayed a high abundance of Acidobacteria and Deltaproteobacteria because of the low pH in sediments in the lake. Temperature could influence the sediment bacterial communities in these lakes. Nutrient concentration was the main factor influencing bacterial communities in Gonghu Bay of Taihu Lake. In East Donging Lake, the organic matter contents could affect the bacterial communities in the sediments, and the bacterial communities in the sediment obtained from Huangda Lake were associated with TN concentration.
CONFLICT OF INTEREST
None declared.
ACKNOWLEDGMENT
This study was supported by the National Basic Research Program of China (No. 2012CB417004), the Major Science and Technology Program for Water Pollution Control and Treatment (No. 2012ZX07101‐013), and State Key Laboratory of Environmental Criteria and Risk Assessment, Chinese Research Academy of Environmental Sciences .
Huang W, Chen X, Jiang X, Zheng B. Characterization of sediment bacterial communities in plain lakes with different trophic statuses. MicrobiologyOpen. 2017;6:e503 https://doi.org/10.1002/mbo3.503
REFERENCES
- Azevêdo, D. J. S. , Barbosa, J. E. L. , Gomes, W. I. A. , Porto, D. E. , Marques, J. C. , & Molozzi, J. (2015). Diversity measures in macroinvertebrate and zooplankton communities related to the trophic status of subtropical reservoirs: Contradictory or complementary responses? Ecological Indicators, 50, 135–149. [Google Scholar]
- Bai, Y. , Shi, Q. , Wen, D. , Li, Z. , Jefferson, W. A. , Feng, C. , & Tang, X. (2012). Bacterial communities in the sediments of Dianchi Lake, a partitioned eutrophic waterbody in China. PLoS ONE, 7, e37796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer, M. , Kube, M. , Teeling, H. , Richter, M. , Lombardot, T. , Allers, E. , … Glockner, F. O. (2006). Whole genome analysis of the marine Bacteroidetes ‘Gramella forsetii’ reveals adaptations to degradation of polymeric organic matter. Environmental Microbiology, 8, 2201–2213. [DOI] [PubMed] [Google Scholar]
- Beardsley, C. , Moss, S. , Malfatti, F. , & Azam, F. (2011). Quantitative role of shrimp fecal bacteria in organic matter fluxes in a recirculating shrimp aquaculture system. FEMS Microbiology Ecology, 77, 134–145. [DOI] [PubMed] [Google Scholar]
- Bending, G. D. , Putland, C. , & Rayns, F. (2000). Changes in microbial community metabolism and labile organic matter fractions as early indicators of the impact of management on soil biological quality. Biology and Fertility of Soils, 31, 78–84. [Google Scholar]
- Bere, T. , & Tundisi, J. G. (2010). Biological monitoring of lotic ecosystems: The role of diatoms. Brazilian Journal of Biology, 70, 493–502. [DOI] [PubMed] [Google Scholar]
- Berglund, O. , Larsson, P. , Ewald, G. , & Okla, L. (2001). Influence of trophic status on PCB distribution in lake sediments and biota. Environmental Pollution, 113, 199–210. [DOI] [PubMed] [Google Scholar]
- Bertahas, I. , Dimitriou, E. , Karaouzas, I. , Laschou, S. , & Zacharias, I. (2006). Climate change and agricultural pollution effects on the trophic status of a Mediterranean lake. Acta Hydrochimica et Hydrobiologica, 34, 349–359. [Google Scholar]
- Bissett, A. , Bowman, J. , & Burke, C. (2006). Bacterial diversity in organically‐enriched fish farm sediments. FEMS Microbiology Ecology, 55, 48–56. [DOI] [PubMed] [Google Scholar]
- Bissett, A. , Bowman, J. P. , & Burke, C. M. (2008). Flavobacterial response to organic pollution. Aquatic Microbial Ecology, 51, 31–43. [Google Scholar]
- Bruckner, C. G. , Mammitzsch, K. , Jost, G. , Wendt, J. , Labrenz, M. , & Jurgens, K. (2013). Chemolithoautotrophic denitrification of epsilonproteobacteria in marine pelagic redox gradients. Environmental Microbiology, 15, 1505–1513. [DOI] [PubMed] [Google Scholar]
- Caporaso, J. G. , Kuczynski, J. , Stombaugh, J. , Bittinger, K. , Bushman, F. D. , Costello, E. K. , … Knight, R. (2010). QIIME allows analysis of high‐throughput community sequencing data. Nature Methods, 7, 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry, V. , Rehman, A. , Mishra, A. , Chauhan, P. S. , & Nautiyal, C. S. (2012). Changes in bacterial community structure of agricultural land due to long‐term organic and chemical amendments. Microbial Ecology, 64, 450–460. [DOI] [PubMed] [Google Scholar]
- Chen, J. , Xie, P. , Ma, Z. M. , Niu, Y. A. , Tao, M. , Deng, X. W. , & Wang, Q. (2010). A systematic study on spatial and seasonal patterns of eight taste and odor compounds with relation to various biotic and abiotic parameters in Gonghu Bay of Lake Taihu, China. Science of the Total Environment, 409, 314–325. [DOI] [PubMed] [Google Scholar]
- Cotner, J. B. , & Biddanda, B. A. (2002). Small players, large role: Microbial influence on biogeochemical processes in pelagic aquatic ecosystems. Ecosystems, 5, 105–121. [Google Scholar]
- Dai, Y. , Yang, Y. Y. , Wu, Z. , Feng, Q. Y. , Xie, S. G. , & Liu, Y. (2016). Spatiotemporal variation of planktonic and sediment bacterial assemblages in two plateau freshwater lakes at different trophic status. Applied Microbiology and Biotechnology, 100, 4161–4175. [DOI] [PubMed] [Google Scholar]
- Daniel, L. M. C. , Pozzi, E. , Foresti, E. , & Chinalia, F. A. (2009). Removal of ammonium via simultaneous nitrification‐denitrification nitrite‐shortcut in a single packed‐bed batch reactor. Bioresource Technology, 100, 1100–1107. [DOI] [PubMed] [Google Scholar]
- Duarte, S. , Pascoal, C. , Garabetian, F. , Cassio, F. , & Charcosset, J. Y. (2009). Microbial decomposer communities are mainly structured by trophic status in circumneutral and alkaline streams. Applied and Environmental Microbiology, 75, 6211–6221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics, 26, 2460–2461. [DOI] [PubMed] [Google Scholar]
- Edgar, R. C. (2013). UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nature Methods, 10, 996–998. [DOI] [PubMed] [Google Scholar]
- Edgar, R. C. , Haas, B. J. , Clemente, J. C. , Quince, C. , & Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics, 27, 2194–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang, L. , Chen, L. , Liu, Y. , Tao, W. , Zhang, Z. , Liu, H. , & Tang, Y. (2015). Planktonic and sedimentary bacterial diversity of Lake Sayram in summer. MicrobiologyOpen, 4, 814–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginn, B. K. (2011). Distribution and limnological drivers of submerged aquatic plant communities in Lake Simcoe (Ontario, Canada): Utility of macrophytes as bioindicators of lake trophic status. Journal of Great Lakes Research, 37, 83–89. [Google Scholar]
- Gomez‐Pereira, P. R. , Schuler, M. , Fuchs, B. M. , Bennke, C. , Teeling, H. , Waldmann, J. , … Amann, R. (2012). Genomic content of uncultured bacteroidetes from contrasting oceanic provinces in the North Atlantic Ocean. Environmental Microbiology, 14, 52–66. [DOI] [PubMed] [Google Scholar]
- Haukka, K. , Kolmonen, E. , Hyder, R. , Hietala, J. , Vakkilainen, K. , Kairesalo, T. , … Sivonen, K. (2006). Effect of nutrient loading on bacterioplankton community composition in lake mesocosms. Microbial Ecology, 51, 137–146. [DOI] [PubMed] [Google Scholar]
- Heiskanen, A. S. , van de Bund, W. , Cardoso, A. C. , & Noges, P. (2004). Towards good ecological status of surface waters in Europe ‐ interpretation and harmonisation of the concept. Water Science and Technology, 49, 169–177. [PubMed] [Google Scholar]
- Hou, D. , He, J. , Lu, C. , Sun, Y. , Zhang, F. , & Otgonbayar, K. (2013). Effects of environmental factors on nutrients release at sediment‐water interface and assessment of trophic status for a typical shallow lake, northwest China. Scientific World Journal, 2013, 716342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, W. , & Jiang, X. (2016). Profiling of sediment microbial community in dongting lake before and after impoundment of the three gorges dam. International Journal of Environmental Research and Public Health, 13, 617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, W. , Lu, Y. , Li, J. H. , Zheng, Z. , Zhang, J. B. , & Jiang, X. (2015). Effect of ionic strength on phosphorus sorption in different sediments from a eutrophic plateau lake. RSC Advances, 5, 79607–79615. [Google Scholar]
- Institute of Soil Science, Chinese Acadmy of Science . (1978). Soil Physics Chemical Analysis. Shanghai: Shanghai Science and Technology Press. [Google Scholar]
- Jones, R. T. , Robeson, M. S. , Lauber, C. L. , Hamady, M. , Knight, R. , & Fierer, N. (2009). A comprehensive survey of soil acidobacterial diversity using pyrosequencing and clone library analyses. Isme Journal, 3, 442–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen, B. B. (1982). Mineralization of organic‐matter in the sea bed‐the role of sulfate reduction. Nature, 296, 643–645. [Google Scholar]
- Jurgens, G. , Glockner, F. O. , Amann, R. , Saano, A. , Montonen, L. , Likolammi, M. , & Munster, U. (2000). Identification of novel Archaea in bacterioplankton of a boreal forest lake by phylogenetic analysis and fluorescent in situ hybridization. FEMS Microbiology Ecology, 34, 45–56. [DOI] [PubMed] [Google Scholar]
- Kelly, M. , Juggins, S. , Guthrie, R. , Pritchard, S. , Jamieson, J. , Rippey, B. , … Yallop, M. (2008). Assessment of ecological status in UK rivers using diatoms. Freshwater Biology, 53, 403–422. [Google Scholar]
- Kostrzewska‐Szlakowska, I. (2005). Surface microlayer in lakes of different trophic status: Dissolved organic matter and microbial community. Polish Journal of Ecology, 53, 343–351. [Google Scholar]
- Kunihiro, T. , Takasu, H. , Miyazaki, T. , Uramoto, Y. , Kinoshita, K. , Yodnarasri, S. , … Tsutsumi, H. (2011). Increase in Alphaproteobacteria in association with a polychaete, Capitella sp I, in the organically enriched sediment. Isme Journal, 5, 1818–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauber, C. L. , Hamady, M. , Knight, R. , & Fierer, N. (2009). Pyrosequencing‐based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Applied and Environmental Microbiology, 75, 5111–5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. H. , Zhao, S. H. , & Wang, Y. Q. (2011). Microbial desulfurization of ground tire rubber by Thiobacillus ferrooxidans. Polymer Degradation and Stability, 96, 1662–1668. [Google Scholar]
- Liao, X. , Zhang, C. , Yao, L. , Li, J. , Liu, M. , Xu, L. , & Evalde, M. (2014). Sorption behavior of nonylphenol (NP) on sewage‐irrigated soil: Kinetic and thermodynamic studies. Science of the Total Environment, 473–474, 530–536. [DOI] [PubMed] [Google Scholar]
- Ligi, T. , Oopkaup, K. , Truu, M. , Preem, J.‐K. , Nõlvak, H. , Mitsch, W. J. , … Truu, J. (2014). Characterization of bacterial communities in soil and sediment of a created riverine wetland complex using high‐throughput 16S rRNA amplicon sequencing. Ecological Engineering, 72, 56–66. [Google Scholar]
- Liu, Z. Z. , DeSantis, T. Z. , Andersen, G. L. , & Knight, R. (2008). Accurate taxonomy assignments from 16S rRNA sequences produced by highly parallel pyrosequencers. Nucleic Acids Research, 36, e120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, F. H. , Lin, G. H. , Gao, G. , Qin, B. Q. , Zhang, J. S. , Zhao, G. P. , … Shen, J. H. (2009). Bacterial and archaeal assemblages in sediments of a large shallow freshwater lake, Lake Taihu, as revealed by denaturing gradient gel electrophoresis. Journal of Applied Microbiology, 106, 1022–1032. [DOI] [PubMed] [Google Scholar]
- Liu, Z. F. , & Liu, J. Q. (2013). Evaluating bacterial community structures in oil collected from the sea surface and sediment in the northern Gulf of Mexico after the Deepwater Horizon oil spill. MicrobiologyOpen, 2, 492–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magoc, T. , & Salzberg, S. L. (2011). FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics, 27, 2957–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina, V. , Hernandez, K. , Dorador, C. , Eissler, Y. , Hengst, M. , Perez, V. , & Harrod, C. (2016). Bacterial active community cycling in response to solar radiation and their influence on nutrient changes in a high‐altitude wetland. Frontiers in Microbiology, 7, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molozzi, J. , Feio, M. J. , Salas, F. , Marques, J. C. , & Callisto, M. (2012). Development and test of a statistical model for the ecological assessment of tropical reservoirs based on benthic macroinvertebrates. Ecological Indicators, 23, 155–165. [Google Scholar]
- Nealson, K. H. (1997). Sediment bacteria: Who's there, what are they doing, and what's new? Annual Review of Earth and Planetary Sciences, 25, 403–434. [DOI] [PubMed] [Google Scholar]
- Orcutt, B. N. , Sylvan, J. B. , Knab, N. J. , & Edwards, K. J. (2011). Microbial ecology of the dark ocean above, at, and below the seafloor. Microbiology and Molecular Biology Reviews, 75, 361–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallud, C. , & Van Cappellen, P. (2006). Kinetics of microbial sulfate reduction in estuarine sediments. Geochimica et Cosmochimica Acta, 70, 1148–1162. [Google Scholar]
- Paradis, E. , Claude, J. , & Strimmer, K. (2004). APE: Analyses of phylogenetics and evolution in R language. Bioinformatics, 20, 289–290. [DOI] [PubMed] [Google Scholar]
- Phan, H. V. , Hai, F. I. , Zhang, R. , Kang, J. , Price, W. E. , & Nghiem, L. D. (2016). Bacterial community dynamics in an anoxic‐aerobic membrane bioreactor ‐ Impact on nutrient and trace organic contaminant removal. International Biodeterioration & Biodegradation, 109, 61–72. [Google Scholar]
- Phillips, G. , Lyche‐Solheim, A. , Skjelbred, B. , Mischke, U. , Drakare, S. , Free, G. , … Carvalho, L. (2013). A phytoplankton trophic index to assess the status of lakes for the Water Framework Directive. Hydrobiologia, 704, 75–95. [Google Scholar]
- Ruban, V. , Lopez‐Sanchez, J. F. , Pardo, P. , Rauret, G. , Muntau, H. , & Quevauviller, P. (2001). Development of a harmonised phosphorus extraction procedure and certification of a sediment reference material. Journal of Environmental Monitoring, 3, 121–125. [DOI] [PubMed] [Google Scholar]
- Segata, N. , Izard, J. , Waldron, L. , Gevers, D. , Miropolsky, L. , Garrett, W. S. , & Huttenhower, C. (2011). Metagenomic biomarker discovery and explanation. Genome Biology, 12, R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao, K. Q. , Gao, G. , Qin, B. Q. , Tang, X. M. , Wang, Y. P. , Chi, K. X. , & Dai, J. Y. (2011). Comparing sediment bacterial communities in the macrophyte‐dominated and algae‐dominated areas of eutrophic Lake Taihu, China. Canadian Journal of Microbiology, 57, 263–272. [DOI] [PubMed] [Google Scholar]
- Song, H. , Li, Z. , Du, B. , Wang, G. , & Ding, Y. (2012). Bacterial communities in sediments of the shallow Lake Dongping in China. Journal of Applied Microbiology, 112, 79–89. [DOI] [PubMed] [Google Scholar]
- Tang, X. , Gao, G. , Qin, B. , Zhu, L. , Chao, J. , Wang, J. , & Yang, G. (2009). Characterization of bacterial communities associated with organic aggregates in a large, shallow, eutrophic freshwater lake (Lake Taihu, China). Microbial Ecology, 58, 307–322. [DOI] [PubMed] [Google Scholar]
- Teixeira, H. , Neto, J. M. , Patricio, J. , Verissimo, H. , Pinto, R. , Salas, F. , & Marques, J. C. (2009). Quality assessment of benthic macroinvertebrates under the scope of WFD using BAT, the Benthic Assessment Tool. Marine Pollution Bullet, 58, 1477–1486. [DOI] [PubMed] [Google Scholar]
- Trolle, D. , Hamilton, D. P. , & Pilditch, C. A. (2010). Evaluating the influence of lake morphology, trophic status and diagenesis on geochemical profiles in lake sediments. Applied Geochemistry, 25, 621–632. [Google Scholar]
- Van der Gucht, K. , Sabbe, K. , De Meester, L. , Vloemans, N. , Zwart, G. , Gillis, M. , & Vyverman, W. (2001). Contrasting bacterioplankton community composition and seasonal dynamics in two neighbouring hypertrophic freshwater lakes. Environmental Microbiology, 3, 680–690. [DOI] [PubMed] [Google Scholar]
- Vezzulli, L. , & Fabiano, M. (2006). Sediment biochemical and microbial variables for the evaluation of trophic status along the Italian and Albanian continental shelves. Journal of the Marine Biological Association of the United Kingdom, 86, 27–37. [Google Scholar]
- Wang, Y. , & Qian, P. Y. (2009). Conservative Fragments in Bacterial 16S rRNA Genes and Primer Design for 16S Ribosomal DNA Amplicons in Metagenomic Studies. PLoS ONE, 4, e7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Q. L. L. , Chen, Y. W. , Xu, K. D. , Liu, Z. W. , & Hahn, M. W. (2007). Intra‐habitat heterogeneity of microbial food web structure under the regime of eutrophication and sediment resuspension in the large subtropical shallow Lake Taihu, China. Hydrobiologia, 581, 241–254. [Google Scholar]
- Xiong, J. B. , Ye, X. S. , Wang, K. , Chen, H. P. , Hu, C. J. , Zhu, J. L. , & Zhang, D. M. (2014). Biogeography of the Sediment Bacterial Community Responds to a Nitrogen Pollution Gradient in the East China Sea. Applied and Environmental Microbiology, 80, 1919–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, J. , Yang, L. Y. , Li, J. Y. , Liang, Y. , Xiao, L. , Jiang, L. J. , & Zhao, D. Y. (2009). Vertical distribution of bacterial community structure in the sediments of two eutrophic lakes revealed by denaturing gradient gel electrophoresis (DGGE) and multivariate analysis techniques. World Journal of Microbiology & Biotechnology, 25, 225–233. [Google Scholar]


