Abstract
Polo-like kinase 1 (PLK1) is a critical regulator of cell cycle progression and apoptosis. However, its regulation remains poorly understood. In the present study, we investigated the molecular mechanism underlying the post-transcriptional regulation of PLK1. We observed that heterogeneous nuclear ribonucleoprotein K (hnRNPK) and PLK1 were positively associated in several different cancers and high expression levels of them correlated with poor prognosis in patients with cancer. Knockdown of hnRNPK resulted in reduced expression of PLK1, whereas conversely, PLK1 expression was increased in hnRNPK-overexpressing cells. We found that hnRNPK regulated PLK1 expression through KH1- and KH2-dependent interactions with the 3′UTR of PLK1 mRNA. In addition, microRNA-149-3p (miR-149-3p) and miR-193b-5p suppressed PLK1 expression by targeting the 3′UTR of PLK1 mRNA. MicroRNA-elicited enrichment of PLK1 mRNA in Ago2 immunoprecipitation was altered by the presence or absence of hnRNPK. Furthermore, the deletion of the cytosine (C)-rich sequences of the 3′UTR of PLK1 mRNA abolished the decreased PLK1 expression observed via hnRNPK silencing and administration of miRNAs, a finding that suggests that hnRNPK shares this C-rich motif with miR-149-3p and miR-193b-5p. We also found that downregulation of PLK1 by either silencing hnRNPK or overexpression of miR-149-3p and miR-193b-5p decreased clonogenicity and induced apoptosis. Our findings from this study demonstrate that hnRNPK regulates PLK1 expression by competing with the PLK1-targeting miRNAs, miR-149-3p and miR-193b-5p.
Polo-like kinases (PLKs) are a family of serine/threonine kinases that are highly conserved from yeast to humans and are important regulators of cell cycle progression.1 To date, four isoforms have been identified in mammalian cells: PLK1, PLK2 (also known as serum-inducible kinase, SNK), PLK3 (also known as FNK), and PLK4 (also known as Sak).2, 3, 4 PLK1 is the most commonly known member of the PLK family of kinases and is predominantly localized in the cytoplasm. PLK1 expression is tightly regulated and its regulation is closely associated with the cell cycle. For example, PLK1 is expressed at a very low level at the G1/S transition; its expression increases during the S phase, and reaches a maximum at the G2/M phase.5 It is implicated in the initiation of G2/M transition events by activating cell division cycle 25 (cdc25, known as CDC25C in humans).6 In addition, PLK1 directly phosphorylates components of the anaphase-promoting complex such as cdc27, and therefore, drives mitotic exit.7, 8 PLK1 has a pivotal role in mitotic progression, and in particular, mitosis by activating cdc25C and cdk1/cyclin B1.9, 10 To maintain genomic stability, the cell cycle must be strictly controlled. Since deregulation results in improper chromosome segregation, mitotic progression is properly advanced by cyclin-dependent kinases and tightly regulated by several critical kinases including PLK1.11, 12 There is increasing evidence that overexpression of PLK1 is frequently observed in a number of human cancers and closely correlates with tumor progression. Constitutive expression of PLK1 results in oncogenic transformation, which indicates it is potentially oncogenic.13, 14 In addition, depletion of PLK1 completely abrogated the ability of U2OS cells to form colonies, which provides further support that PLK1 is a key player in cellular proliferation and its overexpression contributes to oncogenic transformation.15 Moreover, several studies have found that PLK1 expression correlates with the metastatic potential of tumors and with prognosis in patients with cancer.16, 17, 18, 19
Heterogeneous nuclear ribonucleoprotein K (hnRNPK) is an evolutionarily conserved RNA-binding protein (RBP).20 It contains three repeats of K homology (KH) domains (KH1, KH2, and KH3), one K-protein-interactive (KI) domain, and one nuclear-cytoplasmic shuttling domain (KNS). As an RNA-recognition motif, KH domains are responsible for recognizing target mRNAs.21 The nuclear-localization signal and KNS confer the ability for the protein to translocate between the cytoplasm and the nucleus.22 hnRNPK has several integrated functions in gene regulation, namely, transcription, pre-mRNA processing, alternative splicing, mRNA export, and translation.23, 24, 25, 26, 27 It was reported that hnRNPK is frequently overexpressed in different types of cancer and is closely associated with poor cancer prognosis in patients.28, 29, 30, 31 In addition to its roles in cancer proliferation, hnRNPK has an important role in metastatic potential such as invasion and migration,32, 33, 34 and therefore, may be a promising therapeutic target for cancer treatment.
Although hnRNPK is a critical regulator of malignancy in cancer cells, its molecular mechanism is not fully understood. Through our continued efforts to identify and characterize important targets of hnRNPK, we identified PLK1 as a critical player in hnRNPK-involved malignancy. In this study, we found that hnRNPK positively regulates PLK1 expression, which is closely associated with cancer prognosis in patients. Using different molecular approaches, we found that hnRNPK interacts with the 3′UTR of PLK1 mRNA in competition with the microRNAs (miRNA, miR), miR-149-3p and miR-193b-5p, which results in inhibition of miRNA-mediated suppression of PLK1.
Results
hnRNPK is involved in the regulation of PLK1 in different types of cancer
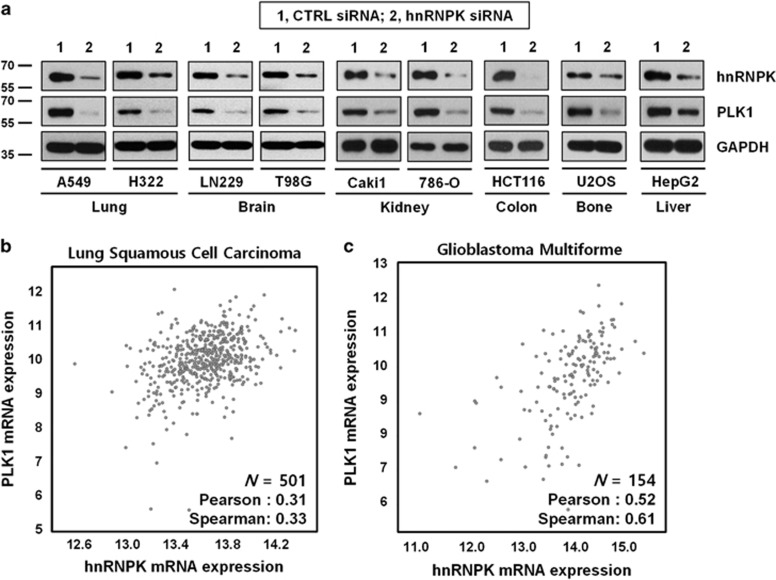
To investigate whether hnRNPK has a role in the regulation of PLK1, we examined the level of PLK1 expression by western blot analysis of hnRNPK-silenced cells from the following cancer cell lines: lung adenocarcinoma (A549 and H322), glioblastoma (LN229 and T98G), renal cell carcinoma (Caki1 and 786-O), colon cancer (HCT116), osteosarcoma (U2OS), and hepatocellular carcinoma (HepG2). In the broad range of cancer cells, knockdown of hnRNPK significantly decreased PLK1 expression, which indicates hnRNPK is closely associated with PLK1, independent of cell context (Figure 1a and Supplementary Figure S2a). To verify this positive relationship between hnRNPK and PLK1, we compared the expression levels of hnRNPK and PLK1 in samples from patients with lung squamous cell carcinoma and glioblastoma multiform (Figures 1b and c, respectively). Consistent with the in vitro data, we found that the level of hnRNPK expression positively correlated with the expression level of PLK1. To determine the association between the survival rate and expression level of hnRNPK and PLK1, Kaplan–Meier analysis of patients with kidney renal clear cell carcinoma and adrenocortical carcinoma (Supplementary Figures S1a and b, respectively) using the expression levels of hnRNPK and PLK1 was performed. In both types of cancer, hnRNPK-high patients had a significantly worse survival rate, compared with those of hnRNPK-low patients (middle panel). In addition, the survival of patients also correlated with PLK1 expression. PLK1-high patients had a worse prognosis than those with PLK1-low (right panel). Furthermore, the patients with high expression of hnRNPK and PLK1 showed a much worse prognosis (left panel).
Figure 1.
Positive correlation between hnRNPK and PLK1 in several types of cancer cells. (a) To examine whether hnRNPK is involved in PLK1 expression, cells from the following cancer cell lines were transfected with hnRNPK-specific siRNA: lung adenocarcinoma (A549 and H322), glioblastoma (LN229 and T98G), renal cell carcinoma (Caki1 and 786-O), colon cancer (HCT116), osteosarcoma (U2OS), and hepatocellular carcinoma (HepG2). The level of hnRNPK and PLK1 was determined by western blot. As a loading control, the level of GAPDH was examined. (b,c) To validate positive correlations, the mRNA level of hnRNPK and PLK1 in lung squamous cell carcinoma (b) and glioblastoma (c) was obtained from the TCGA database
hnRNPK positively regulates the expression of PLK1
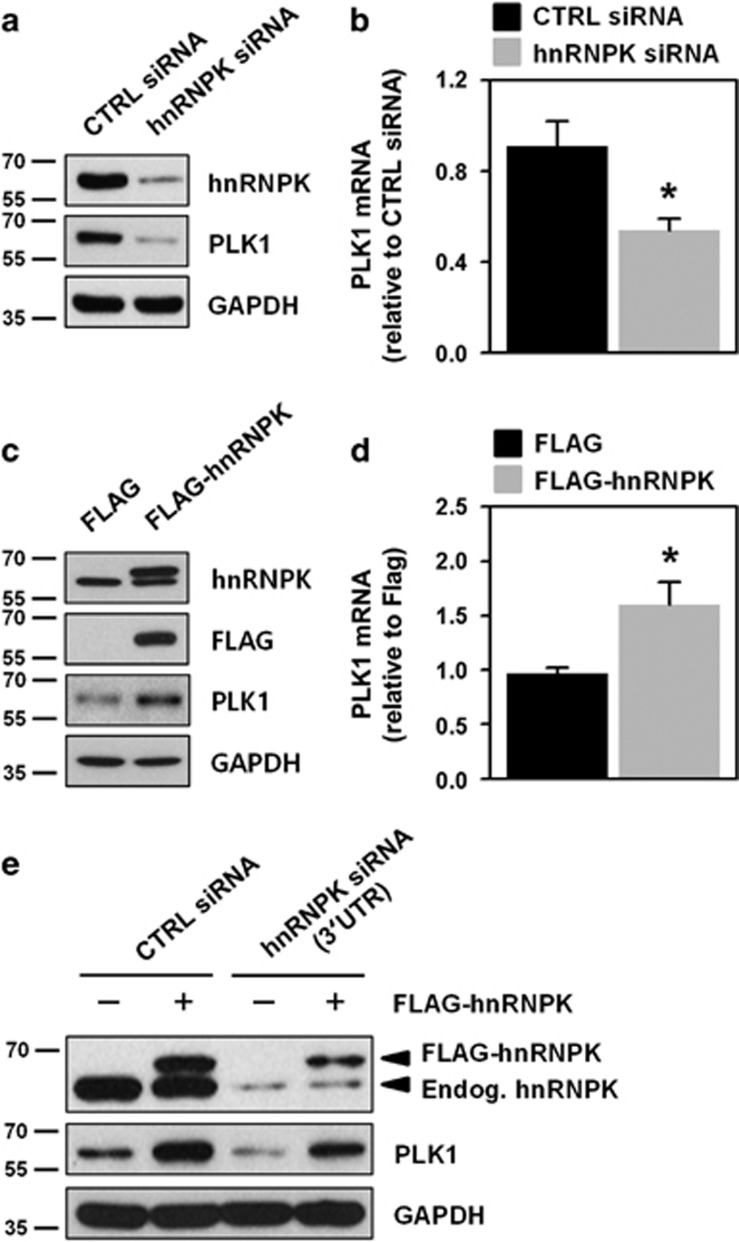
To investigate the molecular mechanism underlying hnRNPK-regulated PLK1 expression, we designed two small interfering RNAs (siRNAs) whose target sites are in different regions of the hnRNPK mRNA. Since administration of either siRNA effectively diminished hnRNPK expression, we used a pooled combination of these siRNAs for further experiments. We found that knockdown of hnRNPK in HeLa cells diminished the expression levels of both PLK1 protein and mRNA (Figures 2a, b and Supplementary Figure S2b). In contrast, we found that the levels of PLK1 protein and mRNA were increased in hnRNPK-overexpressing cells (Figures 2c and d). To confirm our findings, we examined whether ectopic expression of hnRNPK restored PLK1 expression, using siRNA that targeted the 3′UTR of hnRNPK mRNA (Figure 2e). Confirming our previous findings, we found that the level of PLK1 expression was reduced by administration of this 3′UTR-specific siRNA. In addition, we found PLK1 expression was restored through ectopic expression of hnRNPK (FLAG-hnRNPK), a finding that indicates hnRNPK positively regulates PLK1 expression.
Figure 2.
hnRNPK regulates PLK1 expression. (a,b) HeLa cells were transfected with control or hnRNPK-specific siRNA. After 48 h post-transfection, protein and mRNA levels of hnRNPK and PLK1 were determined by western blot and RT–qPCR, respectively. (c,d) Cells were transfected with FLAG or FLAG-hnRNPK vector. Protein and mRNA levels of hnRNPK and PLK1 were assessed as described above. (e) To verify that hnRNPK regulates PLK1, we designed a specific siRNA targeting the 3′UTR of hnRNPK mRNA (Supplementary Table S2). HeLa cells were simultaneously transfected with siRNAs (control or hnRNPK 3′UTR siRNA) and with plasmid DNA (FLAG or FLAG-hnRNPK vector). After 48 h post-transfection, the level of endogenous and ectopic hnRNPK, PLK1, and the loading control, GAPDH, was assessed by western blot. All experiments were performed more than three times and data represent mean±S.D.
hnRNPK directly interacts with the 3′UTR of PLK1 mRNA via the KH1 and KH2 domains
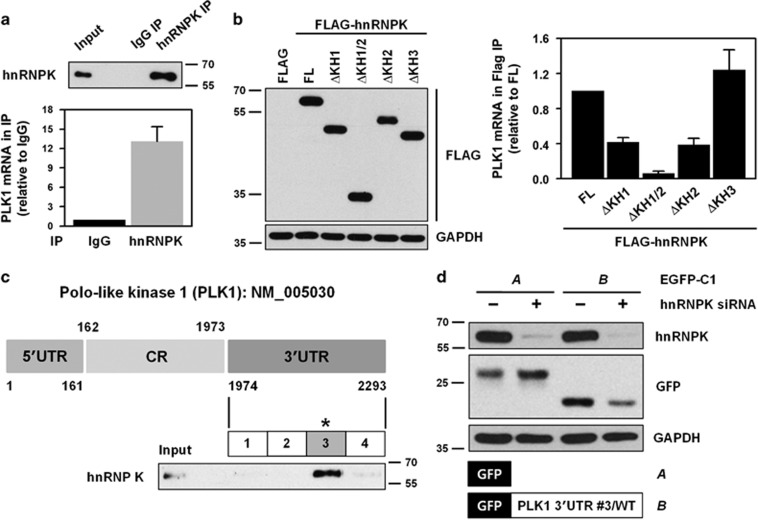
In general, RBPs influence expression of their target genes by directly binding to the 3′UTR of their respective mRNAs. In this regard, we examined whether hnRNPK interacts with the to 3′UTR of PLK1 mRNA by performing immunoprecipitation (IP) with an hnRNPK antibody (RNP-IP experiment). Cytoplasmic extracts of HeLa cells were prepared and incubated with antibody-coated magnetic beads (i.e., either control IgG or hnRNPK antibody). We found greater enrichment of PLK1 mRNA in hnRNPK-IP compared to that found in control IgG-IP, which indicates hnRNPK directly binds to PLK1 mRNA (Figure 3a).
Figure 3.
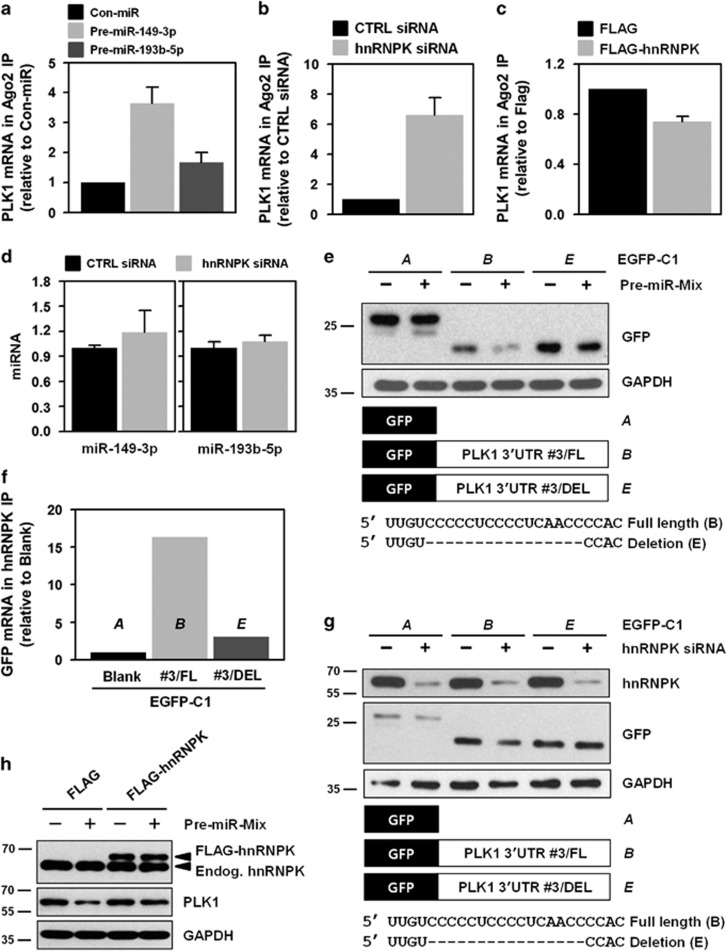
hnRNPK directly interacts with the 3′UTR of PLK1 mRNA through its KH1 and KH2 domains. (a) To determine whether hnRNPK binds to the 3′UTR of PLK1 mRNA, cytoplasmic lysates were prepared as described in the Materials and Methods, and immunoprecipitated (IP) using hnRNPK antibody-coated beads. RNA was isolated from IP materials and the level of PLK1 mRNA was determined by RT–qPCR. Western blot was performed to confirm IP efficiency. (b) As an RNA-binding protein, hnRNPK has three KH domains: KH1, KH2, and KH3. To evaluate which domain or domains are responsible for the interaction between hnRNPK and PLK1 mRNA, four KH deletion mutants were manufactured: ΔKH1, ΔKH1/2, ΔKH2, and ΔKH3 (Supplementary Figure S3). HeLa cells were transfected with FLAG vectors containing the wild-type (full length, FL) and the four deletion mutants. Cytoplasmic lysates were prepared and IP was performed using FLAG antibody. Identical expression levels of ectopic hnRNPK (FLAG-hnRNPK) in input were determined by western blot, and the level of enriched PLK1 mRNA in FLAG-IP was assessed by RT–qPCR. (c) Schematic depiction of the biotinylated RNAs of the 3′UTR of PLK1 mRNA used for biotin pull-down analysis. The level of hnRNPK in biotin pull-down samples was determined by western blot. (d) We constructed vectors expressing chimeric RNAs spanning the GFP (A) and fragment #3 of the 3′UTR PLK1 mRNA (B). HeLa cells were first transfected with control or hnRNPK siRNA. After 24 h post-transfection, cells were resuspended into six-well plates followed by transfection with GFP vectors (blank or fragment #3 of the 3′UTR PLK1 mRNA). The level of hnRNPK, GFP, and the loading control, GAPDH, was assessed by western blot. All experiments were performed more than three times and data represent mean±S.D.
To determine which KH domain or domains were involved in the interaction between hnRNPK and PLK1 mRNA, we generated four KH deletion mutants: KH1 (ΔKH1), KH1/2 (ΔKH1/2), KH2 (ΔKH2), and KH3 (ΔKH3) (Supplementary Figure S3). HeLa cells were transfected with FLAG vectors containing wild type or a deletion mutant of hnRNPK, and RNP-IP was performed from cytoplasmic extracts. Western blot analysis was performed to confirm similar expression levels of ectopic hnRNPK in the input (Figure 3b, left panel) and the level of enriched PLK1 mRNA in FLAG-IP materials were determined by RT–qPCR (Figure 3b, right panel). We found that deletion of the KH1 (ΔKH1) and/or the KH2 (ΔKH2) domain resulted in significantly decreased enrichment of PLK1 mRNA in hnRNPK-IP materials. In contrast, we found no difference in the interaction between hnRNPK and PLK1 mRNA in the absence of the KH3 domain. Our findings indicate that hnRNPK interacts with the 3′UTR of PLK1 mRNA, an interaction that requires the KH1 and KH2 domains of hnRNPK. To identify the region or regions of interaction, biotin pull-down experiments using four biotinylated fragments of the 3′UTR of PLK1 mRNA were performed (Figure 3c and Supplementary Figure S4). Of the four fragments, we found that hnRNPK demonstrated a preferential affinity to the third fragment (#3) of the 3′UTR of PLK1 mRNA. To test whether hnRNPK influenced PLK1 expression by acting on the 3′UTR of PLK1 mRNA, a heterologous reporter construct expressing a chimeric RNA, which spanned the GFP coding region, and fragment #3 of the PLK1 3′UTR was constructed. As shown in Figure 3d, knockdown of hnRNPK decreased GFP expression from the reporter chimeric plasmid pGFP/PLK1 3′UTR #3, but not from pGFP alone, a finding that indicates fragment #3 is responsible for the hnRNPK-mediated regulation of PLK1.
miRNA-149-3p and miR-193b-5p suppress PLK1 expression
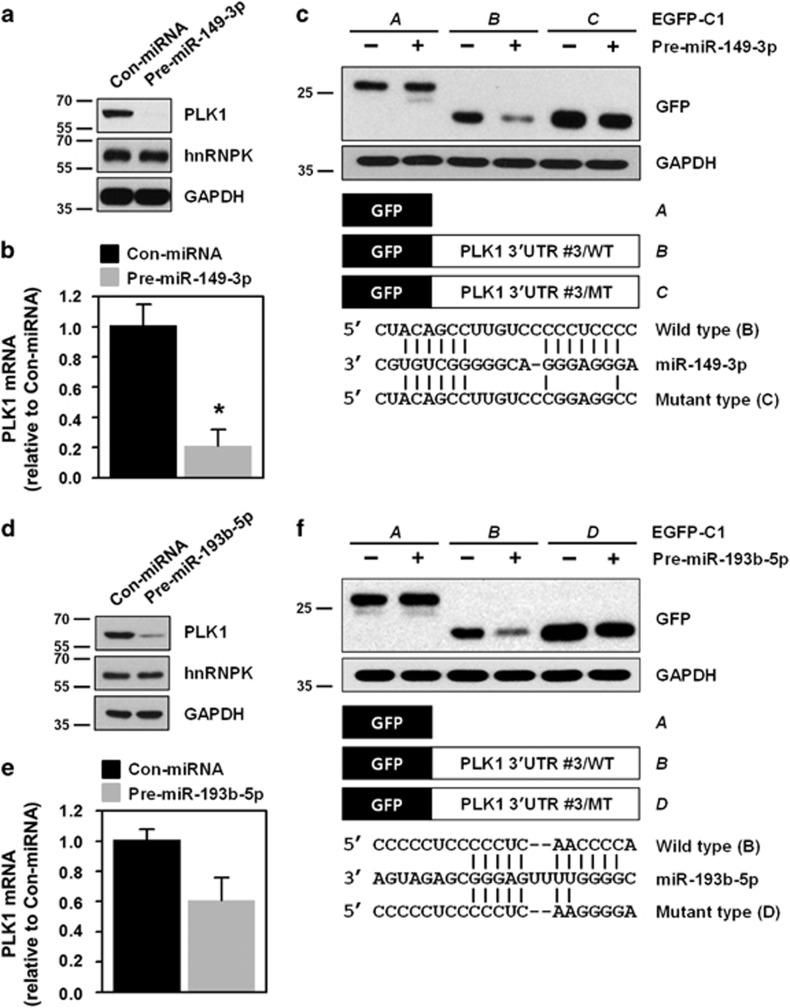
Since it was determined that hnRNPK recognizes and binds to a C-rich region (fragment #3) in the 3′UTR of PLK1 mRNA, we examined fragment #3 for miRNA-binding sites, using several prediction programs such as TargetScan, miRanda, and PicTar. Because of the presence of G-rich sequences in their seed, which provides a high probability of competing with hnRNPK, two miRNAs (miR-149-3p and miR-193b-5p) were selected for further investigation. First, we tested the effect of miR-149-3p on PLK1 expression, and found that transfection of HeLa cells with an miR-149-3p mimic suppressed expression of PLK1 protein and mRNA (Figures 4a and b, respectively). In addition, it reduced the expression level of GFP bearing the wild-type 3′UTR of PLK1 mRNA (#3 fragment), but not that of the mutant (Figure 4c). Consistent with our findings on miR-149-3p, we found decreased expression of PLK1 protein and mRNA in cells transfected with an miR-193b-5p mimic (Figures 4d and e). As expected, we found reduced expression of GFP harboring the wild-type 3′UTR of PLK1 mRNA (#3 fragment), but not that of the mutant (Figure 4f). These findings revealed that miR-149-3p and miR-193b-5p were PLK1-targeting miRNAs.
Figure 4.
miR-149-3p and miR-193b-5p suppress PLK1 expression through direct interactions with the 3′UTR of PLK1 mRNA. (a,b) HeLa cells were transfected with control miRNA or pre-miR-149-3p. After 48 h post-transfection, PLK1 protein and mRNA levels were determined by western blot and RT–qPCR, respectively. hnRNPK and the loading control, GAPHD, were examined by western blot. (c) We constructed two GFP vectors to investigate whether a direct interaction with the 3′UTR PLK1 mRNA is required for downregulation of PLK1 by miR-149-3p. Based on vectors expressing chimeric RNAs spanning the GFP (A) and fragment #3 of the 3′UTR PLK1 mRNA (B), we constructed vectors (C) containing mutated sequences of the miR-149-3p-binding sites in the 3′UTR PLK1 mRNA (fragment #3). After overexpression of miR-149-3p, cells were transfected with the indicated GFP vectors: blank, wild-type (WT), or mutated type (MT). (d–f) Similar to our investigation of miR-149-3p, we tested the effect of miR-193b-5p on PLK1 expression as described above. All experiments were performed more than three times and data represent mean±S.D.
hnRNPK competes with miR-149-3p and miR-193b-5p for the 3′UTR of PLK1 mRNA
As mentioned previously, we found that hnRNPK and two miRNAs (miR-149-3p and miR-193b-5p) were involved in the regulation of PLK1 expression. Interestingly, these trans-regulatory factors may share their binding sites in the 3′UTR of PLK1 mRNA (Supplementary Figure S4). Therefore, we hypothesized that PLK1 is tightly regulated through the interactions between hnRNPK and these two miRNAs (miR-149-3p and miR-193b-5p). To test this hypothesis, we first tested whether the RISC complex, which contains miR-149-3p or miR-193b-5p, interacted with the 3′UTR of PLK1 mRNA. We found that enrichment of PLK1 mRNA in Ago2-IP was increased by miR-149-3p and miR-193b-5p (Figure 5a). Next, the effect of hnRNPK on the interaction between miRNAs and the 3′UTR of PLK1 mRNA was investigated. We found that knockdown of hnRNPK increased enrichment of PLK1 mRNA in Ago2-IP compared to that found in controls (Figure 5b). Conversely, we found that the level of PLK1 mRNA in Ago2-IP was diminished in hnRNPK-overexpressing cells (Figure 5c), which indicated that hnRNPK can disrupt the interaction between miRNA-loaded RISC and PLK1 mRNA. Next, the expression level of these miRNAs was assessed to exclude the possibility that hnRNPK affects enrichment of PLK1 mRNA in Ago2-IP by altering the expression of miR-149-3p and miR-193b-5p. In the hnRNPK-silenced cells, we found that their expression levels were almost identical to those found in controls (Figure 5d). These findings demonstrated that hnRNPK regulates PLK1 expression through a competitive interaction with miR-149-3p and miR-193b-5p at the 3′UTR of PLK1 mRNA.
Figure 5.
Competitive regulation of PLK1 by hnRNPK and miR-149-3p/193b-5p. (a) Cytoplasmic lysates were obtained from miR-149-3p- or miR-193b-5p-overexpressing cells and immunoprecipitated (IP) with Ago2-specific antibody. Enrichment of PLK1 mRNA in Ago2 IP was assessed by RT–qPCR. (b,c) To investigate the effect of hnRNPK on the interaction between PLK1 mRNA and an miRNA-loaded RISC complex, enrichment of PLK1 mRNA in Ago2 IP was examined using cytoplasmic lysates obtained from hnRNPK-silenced (b) or -overexpressing (c) HeLa cells. The level of PLK1 mRNA in Ago2 IP was assessed by RT–qPCR. (d) To test whether hnRNPK affects expression of miR-149-3p and miR-193b-5p, cells were transfected with control or hnRNPK siRNA. After 48 h post-transfection, miRNA expression levels were determined by RT–qPCR. (e) To examine the effect of miRNA mimics on GFP expression, GFP reporters were generated in which GFP was linked to fragment #3 harboring or lacking the binding sequence for hnRNPK and the miRNAs (GFP vector B and E in the schematic). GFP expression was assessed by western blot. (f) The interaction between hnRNPK and GFP chimeric mRNAs was examined. Cells were transfected with the previously described GFP vectors and cytoplasmic lysates were prepared. GFP mRNA enrichment was measured by RNP IP using hnRNPK antibody followed by RT–qPCR. (g) To test whether hnRNPK affects GFP expression in the absence of an miRNA-binding sequence, GFP reporters described in e were used. The expression levels of hnRNPK and GFP were assessed by western blot. (h) To determine whether hnRNPK restored PLK1 expression, cells were transfected with miRNA mimics (pre-miR-149-3p and miR-193b-5p) and an hnRNPK vector (FLAG-hnRNPK). Expression of hnRNPK and PLK1 was assessed by western blot. All experiments were performed more than three times and data represent mean±S.D.
To verify our findings, we prepared a GFP chimeric vector that lacked the sequences recognized by hnRNPK and the miRNAs, and tested the effect of miRNA mimics (miR-149-3p and miR-193b-5p) on GFP expression. Similar to our previous findings, we found that the miRNA mimics suppressed the expression of GFP harboring the binding sequence for hnRNPK and the miRNAs; however, in the case of the deletion mutant, we found that the miRNA mimics did not have an effect (Figure 5e). Next, we hypothesized that hnRNPK and these two miRNAs share their binding sites in the 3′UTR of PLK1 mRNA. To test our hypothesis, we first confirmed the interaction between hnRNPK and the GFP chimeric mRNAs. Concordant with our previous RNP IP findings, we found that hnRNPK was bound to #3 fragment of the 3′UTR of PLK1 mRNA. However, we found that hnRNPK did not interact with GFP chimeric mRNAs that lacked the miRNA-binding sites (Figure 5f). Furthermore, we compared GFP expression in the presence and absence of hnRNPK. While we found that knockdown of hnRNPK decreased GFP expression, it had no effect on expression in the deletion mutant (Figure 5g). Next, we investigated whether hnRNPK reverses miRNA-mediated downregulation of PLK1. Similar to previous findings, we found that overexpression of the miRNAs, miR-149-3p and miR-193b-5p, suppressed PLK1 expression, whereas hnRNPK restored PLK1 expression (Figure 5h). These results indicate that hnRNPK shares a binding sequence with miR-149-3p and miR-193b-5p, and furthermore, reverses the functional effects of these miRNAs by disrupting their interaction with the 3′UTR of PLK1 mRNA.
Decreased expression of PLK1 inhibits clonogenicity and induces apoptotic cell death
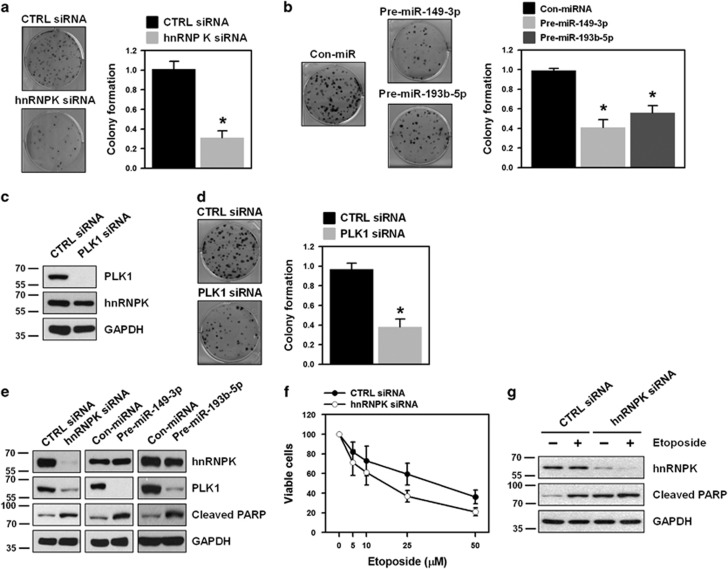
Owing to its critical roles in the cell cycle, knockdown of PLK1 inhibits cellular proliferation and triggers apoptotic cell death in a wide range of cancers. Therefore, we investigated the role of PLK1 in clonogenic abilities. We found that downregulation of PLK1 by hnRNPK silencing or using miRNA mimics significantly decreased the number of colonies (Figures 6a and b, respectively). To test whether downregulation of PLK1 results in the decrease of clonogenicity, the function of PLK1 was investigated. We found that knockdown of PLK1 inhibited clonogenicity of HeLa cells (Figures 6c and d). Two different siRNAs targeting hnRNPK and PLK1 showed similar results (Supplementary Figures S2c and e, respectively). Next, we examined the degree of apoptotic cell death in PLK1-suppressed cells and found a significant increase of PARP cleavage in hnRNPK-silenced cells (Figure 6e). Similarly, we found that overexpression of PLK1-targeting miRNAs (miR-149-3p and miR-193b-5p) by introducing their respective mimic dramatically induced PARP cleavage compared to that found in control miRNA-transfected cells (Figure 6e). Since PLK1 is implicated in the acquisition of resistance against cancer chemotherapeutics, we next examined whether knockdown of hnRNPK sensitized HeLa cells to etoposide treatment. It was found that apoptotic cell death by etoposide was potentiated in hnRNPK-silenced cells (Figure 6f). Furthermore, knockdown of hnRNPK not only triggered apoptotic processes, but also potentiated etoposide-induced cell death (Figure 6g).
Figure 6.
PLK1 regulation through interplay between hnRNPK and miRNAs is implicated in clonogenicity and drug resistance. HeLa cells were transfected with hnRNPK siRNA (a), miRNA mimics (pre-miR-149-3p and miR-193b-5p) (b), or PLK1 siRNA (c–d). After 48 h post-transfection, cells were resuspended into six-well plates and cultured for 2 weeks. Representative images are shown and clonogenic abilities were determined by counting the number of colonies. (e) To determine whether downregulation of PLK1 induces apoptotic cell death, HeLa cells were transfected with hnRNPK siRNA or miRNA mimics (pre-miR-149-3p and miR-193b-5p). The level of hnRNPK, PLK1, and cleaved PARP was assessed by western blot. (f) HeLa cells were transfected with control or hnRNPK-specific siRNA. Transfected cells were resuspended into 96-well plates and treated with different concentrations of etoposide. After incubation for 48 h, cell viability was determined by the MTS cell proliferation assay. (g) To test whether knockdown of hnRNPK sensitized cancer cells to etoposide treatment, HeLa cells were transfected with control or hnRNPK siRNA. Transfected cells were resuspended into 96-well plates and treated with 10 μM etoposide for 48 h. The level of hnRNPK and cleaved PARP was assessed by western blot. All experiments were performed more than three times and data represent mean±S.D.
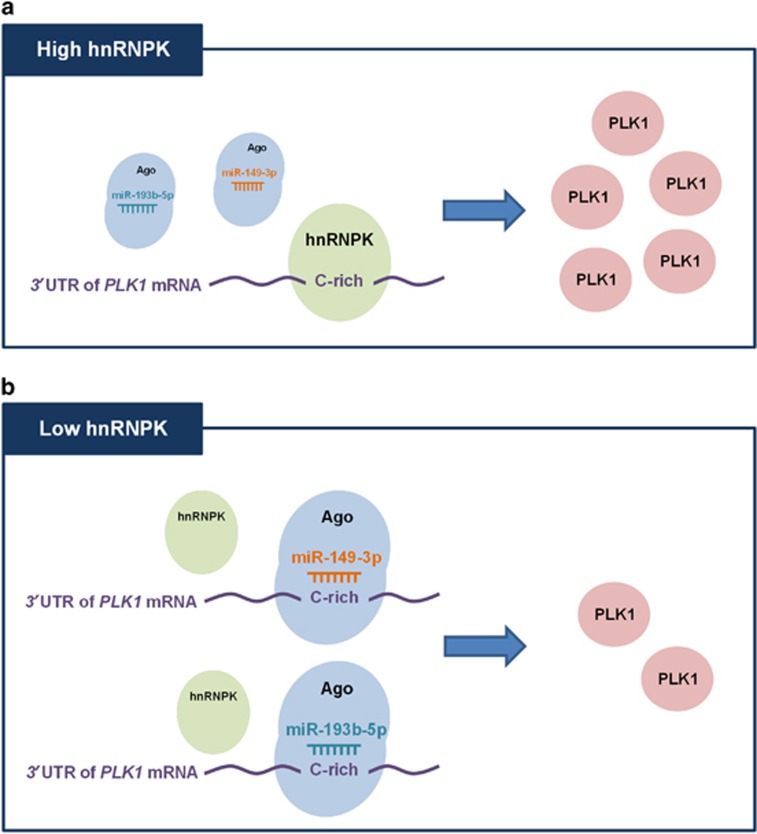
Collectively, our results indicate that hnRNPK and PLK1-tageting miRNAs (miR-149-3p and miR-193b-5p) share their preferred C-rich binding sequence in the 3′UTR of PLK1 mRNA and thus it competes with miRNA-loaded RISC to interact with PLK1 mRNA. Based on our findings, we propose a novel mechanism by which hnRNPK regulates PLK1 expression (Figure 7). The interaction between miRNA-loaded RISC and PLK1 mRNA 3′UTR was disrupted in the condition of high hnRNPK (Figure 7a). Conversely, PLK1 mRNA is very vulnerable to suppression by miR-149-3p and miR-193b-5p in the presence of low hnRNPK level (Figure 7b).
Figure 7.
Proposed action mechanism underlying hnRNPK-mediated PLK1 regulation. Under the condition of high hnRNPK, the interaction between PLK1 mRNA 3′UTR and miRNA-loaded RISC is disrupted by hnRNPK, which results in increase of PLK1 expression. Conversely, in the presence of low hnRNPK, miRNA-loaded RISC easily interacts with the 3′UTR of PLK1 mRNA, in turn lowering the PLK1 expression
Discussion
Post-transcriptional gene regulation (PTGR) is involved in the precise control of many oncogenes and tumor-suppressing genes.35 As the trans-acting factors, RBPs and miRNAs have key roles in PTGR mainly through interactions with the 3′UTR of target mRNAs (cis-elements).36 RBPs have been classified into two groups that are defined by the characteristics of their binding sequences: one group consists of the AU-rich element (ARE)-binding proteins and the other consists of the pyrimidine (C/CU)-rich element-binding proteins. As a poly-r(C)-binding RBP, hnRNPK is highly expressed in a number of different cancers and is implicated in both cancer development and gain of metastatic potential.32 Compared to other hnRNP members, hnRNPK has different characteristics, including its KH domains and DNA-binding affinity. For these reasons, hnRNPK has a significant role in various cellular processes in both the nucleus and the cytoplasm. First, it directly interacts with the RNA polymerase machinery through its association with the TATA-box binding protein to activate the transcription of target genes.37 In addition, hnRNPK is involved in alternative splicing processes by interacting with RNA splicing factors; approximately 50% of total alternative splicing in apoptosis-associated genes was affected by hnRNPK.38 Through a direct interaction with elongation factor-1α, hnRNPK affects translation of target genes.39 Depending on cell context and UTR sequence, the translational efficacy of target genes is differentially affected by hnRNPK. For example, hnRNPK increases translation of the androgen receptor by binding to the 5'UTR of its mRNA,40 but it inhibits the translation of p21 through interaction with the 3′UTR of its mRNA.41 In this report, PLK1 mRNA is identified as a novel target of hnRNPK. Through IP experiments using deletion mutants, we found that KH1 and KH2 domains of hnRNPK are required for the recognition of C-rich sequence in the 3′UTR of PLK1 mRNA that is denoted by #3 fragment.
Since many trans-elements have shared binding sequences, they cooperatively or competitively influence the expression of their target genes. In the destabilization of mRNA, tristetraprolin (TTP) was reported to recruit miR-16-loaded Ago2 into ARE-containing mRNAs such as tumor necrosis factor-α and cyclooxygenase-2 (COX-2).42 Human antigen R (HuR) acts in collaboration to regulate translationally the expression of c-myc43 and RhoB44 with let-7a and miR-19, respectively. In addition to its cooperative effects, HuR protects target mRNAs from miRNA-mediated degradation or translational suppression.45 In stressful conditions such as amino acid deprivation, HuR abolishes miR-122-mediated translational suppression of CAT-1 mRNA46 and miR-16-mediated degradation of COX-2 mRNA.47 To date, the most well-known PLK1-targeting miRNA is miR-100-5p. It was reported to repress PLK1 expression in nasopharyngeal cancer, hepatocellular carcinoma, non-small cell lung cancer, and cervical cancer.48, 49, 50, 51, 52 In addition, miR-100-5p is closely associated with cancer prognosis in patients and is considered a potential molecular marker of non-small cell lung cancer. In this study, we also tested whether hnRNPK can influence the function of miR-100-5p. Consistently with our miRNAs, miR-149-3p and miR-193b-5p, PLK1 was downregulated by miR-100-5p of which binding site is located near C-rich sequences recognized by miR-149-3p and miR-193b-5p (denoted as #3 fragment in this report). Similar to the observation of miR-149-3p and miR-193b-5p, we found that hnRNPK can also influence the function of another PLK1-targeting miR-100-5p (Supplementary Figure S6). Collectively, we demonstrate that hnRNPK competitively controls the expression of PLK1 with PLK1-targeting miRNAs for binding to 3′UTR of its target mRNA. We observed that the competitive relation between hnRNPK and PLK1-targeting miRNAs is dependent on the expression levels of miRNAs. Whereas both miR-149-3p and miR-193b-5p suppressed the expression of PLK1 in HeLa and H322 cells, we found that only one miRNA mimic showed the decrease in PLK1 expression: miR-149-3p in T98G cells and miR-193b-5p in HCT116 and HepG2 cells. Similar to what we observed in HeLa cells, the expression level of PLK1 was restored in hnRNPK-overexpressed cells in both cells. (Supplementary Figure S5). These results are possibly due to two aspects: (1) the different levels of miR-149-3p and miR-193b-5p; (2) the different levels of RBPs which can influence the interaction of miRNAs to the 3′UTR of target mRNA. Taken together, we demonstrate that PLK1 expression is governed through a dynamic post-transcriptional regulatory mechanism, depending on the levels of trans-acting factors such as miRNAs and RBPs.
PLK1 is overexpressed in several different types of cancer and is closely associated with cancer prognosis in patients. It has key roles in the control of the cell cycle, and therefore, regulates cellular proliferation.53 As one of the most attractive targets for the development of anticancer drugs, PLK1 has been the subject of extensive research. It was found that inhibition of PLK1 by siRNA or small molecules decreases proliferation and triggers the apoptotic process in both in vitro and in preclinical studies. In this study, it was first reported that miR-149-3p and miR-193b-5p are novel PLK1-targeting mRNAs and hnRNPK is able to regulate PLK1 expression by binding to the 3′UTR of PLK1 mRNA in competition with miR-149-3p and miR-193b-5p. Based on our findings, we propose a novel mechanism underlying hnRNPK-mediated PLK1 regulation (Figure 7). Briefly, hnRNPK can disrupt the interaction between PLK1 mRNA 3′UTR and miRNA-loaded RISC, resulting in the increase of PLK1 expression. On the contrary, miRNA-loaded RISC easily interacts with the 3′UTR of PLK1 mRNA, in turn lowering the PLK1 expression in the presence of low hnRNPK. We demonstrate that hnRNPK which is highly expressed in cancer plays an important role in the post-transcriptional regulation of PLK1 in competition with PLK1-targeting miRNAs.
Materials and methods
Cell culture and transfection
Cancer cells from lung adenocarcinoma (A549 and H322), glioblastoma (LN229 and T98G), renal cell carcinoma (Caki1 and 786-O), colorectal carcinoma (HCT116), osteosarcoma (U2OS), hepatocellular carcinoma (HepG2), and cervical carcinoma (HeLa) cell lines were maintained in Dulbecco’s modified Eagle’s medium (Hyclone, Logan, UT, USA) supplemented with 10% fetal bovine serum and 1% antibiotic-antimycotic solution (GIBCO-BRL, Grand Island, NY, USA).
Complementary DNA (cDNA) from HeLa cells were used to generate constructs of the full-length and deletion mutants of FLAG-hnRNPK (Supplementary Figure S3 and Supplementary Table S1). For knockdown of hnRNPK, we designed two specific siRNAs targeting the coding region or the 3′UTR of hnRNPK mRNA. To reduce PLK1 expression, two specific siRNAs targeting the coding region of PLK1 mRNA were synthesized (ST Pharm, Seoul, South Korea). The sequences of these siRNAs are provided in Supplementary Table S2. Synthesized miRNA mimics (pre-miR-149-3p and pre-miR-193b-5p) were purchased from Ambion (Austin, TX, USA) and used for miRNA overexpression. For transient transfection, cells were plated at an appropriate confluency and transfected with the indicated siRNAs or plasmids using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol.
Western blot analysis
Cells were lysed using radioimmunoprecipitation (RIPA) buffer containing protease and phosphatase inhibitors (Roche, Basel, Switzerland). Equal amounts of cell lysate were separated by SDS–polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membranes (Millipore, Billerica, MA, USA). After blocking, membranes were incubated with the respective primary antibody, washed, and incubated with the appropriate secondary antibody. The antibodies used in this study are provided in Supplementary Table S3.
RT–qPCR analysis
Total RNA was isolated using TRIzol (Invitrogen) according to the manufacturer’s instructions and used as a template to synthesize cDNA, using the SuperScript III First-Strand Synthesis System (Invitrogen). The amount of mRNA was quantified by RT–qPCR with an ABI Prism 7900 using Power SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA, USA). Primer sequences are provided in Supplementary Table S4.
Immunoprecipitation of ribonucleoprotein (RNP-IP)
To assess direct interactions between hnRNPK and its target mRNA, RNP-IP was performed using Dynabeads Protein A (ThermoFisher Scientific, Rockford, IL, USA) coated with control IgG (Santa Cruz, CA, USA) or hnRNPK antibody (Abcam, Cambridge, UK). Cytoplasmic lysates were prepared using PEB lysis buffer containing protease inhibitor, phosphatase inhibitor, and RNaseOUT (Invitrogen), and were incubated with Dynabeads (ThermoFisher Scientific). Following treatment with DNase I and protease K, RNA was isolated and used to determine enrichment of PLK1 mRNA in IP materials. Enrichment of PLK1 mRNA in Ago2-IP or FLAG-IP was determined by following the procedure described above using Dynabeads coated with Ago2 or FLAG antibody, respectively.
Biotin pull-down assay
To synthesize biotinylated transcripts, PCR fragments were prepared using primers shown in Supplementary Table S5. After biotinylated transcripts were synthesized using the MaxiScript T7 kit (Ambion), whole-cell lysates were incubated with purified biotinylated transcripts for 4 h at room temperature and complexes were isolated using streptavidin-coupled Dynabeads (ThermoFisher Scientific). The level of hnRNPK in the pull-down material was determined by western blot analysis.
Reporter assay
To determine the miRNA recognition elements in the 3′UTR of PLK1 mRNA, we prepared five GFP reporter constructs: (A) blank pEGFP-C1 vector, (B) wild-type (WT) 3′UTR of PLK1 mRNA fragment #3, (C) five point mutations in the miR-149-3p seed region, (D) four point mutations in the miR-193b-5p seed region, and (E) a deletion mutant that does not have a seed region of either miRNA. The effects of these mutations on GFP expression were assessed by western blot analysis.
Colony forming assay
To determine clonogenicity, a colony-forming assay was performed. Briefly, transfected cells were seeded in triplicate in six-well plates and cultured for 2 weeks. Cells were fixed with 4% paraformaldehyde and stained with 0.2% crystal violet. Clonogenic abilities were determined by counting the number of colonies.
Gene expression correlation and survival analysis
The correlation of gene expression and the significance for clinical outcome were plotted using The Cancer Genome Atlas (TCGA) data set.
Acknowledgments
This study was supported by a grant from the Mid-career Research Program through the National Research Foundation (NRF) funded by the Ministry of Science, ICT, and Future Planning, Republic of Korea (2014R1A2A1A11053130).
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by M Blagosklonny
The authors declare no conflict of interest.
Supplementary Material
References
- Hamanaka R, Maloid S, Smith MR, O'Connell CD, Longo DL, Ferris DK. Cloning and characterization of human and murine homologues of the Drosophila polo serine-threonine kinase. Cell growth differ 1994; 5: 249–257. [PubMed] [Google Scholar]
- Clay FJ, McEwen SJ, Bertoncello I, Wilks AF, Dunn AR. Identification and cloning of a protein kinase-encoding mouse gene, Plk, related to the polo gene of Drosophila. Proc Natl Acad Sci USA 1993; 90: 4882–4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lake RJ, Jelinek WR. Cell cycle- and terminal differentiation-associated regulation of the mouse mRNA encoding a conserved mitotic protein kinase. Mol cell biol 1993; 13: 7793–7801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons DL, Neel BG, Stevens R, Evett G, Erikson RL. Identification of an early-growth-response gene encoding a novel putative protein kinase. Mol cell biol 1992; 12: 4164–4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai N, Hamanaka R, Yoshimatsu J, Miyakawa I. Polo-like kinases (Plks) and cancer. Oncogene 2005; 24: 287–291. [DOI] [PubMed] [Google Scholar]
- Kumagai A, Dunphy WG. Purification and molecular cloning of Plx1, a Cdc25-regulatory kinase from Xenopus egg extracts. Science 1996; 273: 1377–1380. [DOI] [PubMed] [Google Scholar]
- Kotani S, Tugendreich S, Fujii M, Jorgensen PM, Watanabe N, Hoog C et al. PKA and MPF-activated polo-like kinase regulate anaphase-promoting complex activity and mitosis progression. Mol cell 1998; 1: 371–380. [DOI] [PubMed] [Google Scholar]
- Descombes P, Nigg EA. The polo-like kinase Plx1 is required for M phase exit and destruction of mitotic regulators in Xenopus egg extracts. EMBO j 1998; 17: 1328–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover DM, Hagan IM, Tavares AA. Polo-like kinases: a team that plays throughout mitosis. Genes dev 1998; 12: 3777–3787. [DOI] [PubMed] [Google Scholar]
- Nigg EA. Mitotic kinases as regulators of cell division and its checkpoints. Nat rev Mol cell biol 2001; 2: 21–32. [DOI] [PubMed] [Google Scholar]
- Sullivan M, Morgan DO. Finishing mitosis, one step at a time. Nat rev Mol cell biol 2007; 8: 894–903. [DOI] [PubMed] [Google Scholar]
- Ferrari S. Protein kinases controlling the onset of mitosis. Cell mol life sci 2006; 63: 781–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y, Miyoshi E, Sasaki N, Kakudo K, Yoshida H, Tomoda C et al. Polo-like kinase 1 overexpression is an early event in the progression of papillary carcinoma. Br j cancer 2004; 90: 414–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studach LL, Rakotomalala L, Wang WH, Hullinger RL, Cairo S, Buendia MA et al. Polo-like kinase 1 inhibition suppresses hepatitis B virus X protein-induced transformation in an in vitro model of liver cancer progression. Hepatology 2009; 50: 414–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Vugt MA, Smits VA, Klompmaker R, Medema RH. Inhibition of Polo-like kinase-1 by DNA damage occurs in an ATM- or ATR-dependent fashion. J biol chem 2001; 276: 41656–41660. [DOI] [PubMed] [Google Scholar]
- Kneisel L, Strebhardt K, Bernd A, Wolter M, Binder A, Kaufmann R. Expression of polo-like kinase (PLK1) in thin melanomas: a novel marker of metastatic disease. J cutan pathol 2002; 29: 354–358. [DOI] [PubMed] [Google Scholar]
- Takai N, Miyazaki T, Fujisawa K, Nasu K, Hamanaka R, Miyakawa I. Polo-like kinase (PLK) expression in endometrial carcinoma. Cancer lett 2001; 169: 41–49. [DOI] [PubMed] [Google Scholar]
- Takai N, Miyazaki T, Fujisawa K, Nasu K, Hamanaka R, Miyakawa I. Expression of polo-like kinase in ovarian cancer is associated with histological grade and clinical stage. Cancer lett 2001; 164: 41–49. [DOI] [PubMed] [Google Scholar]
- Wolf G, Elez R, Doermer A, Holtrich U, Ackermann H, Stutte HJ et al. Prognostic significance of polo-like kinase (PLK) expression in non-small cell lung cancer. Oncogene 1997; 14: 543–549. [DOI] [PubMed] [Google Scholar]
- Barboro P, Ferrari N, Balbi C. Emerging roles of heterogeneous nuclear ribonucleoprotein K (hnRNP K) in cancer progression. Cancer lett 2014; 352: 152–159. [DOI] [PubMed] [Google Scholar]
- Siomi H, Matunis MJ, Michael WM, Dreyfuss G. The pre-mRNA binding K protein contains a novel evolutionarily conserved motif. Nucleic acids res 1993; 21: 1193–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael WM, Eder PS, Dreyfuss G. The K nuclear shuttling domain: a novel signal for nuclear import and nuclear export in the hnRNP K protein. EMBO j 1997; 16: 3587–3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan JY, Huang SM, Liu ST, Huang CH. The transactivation domain of heterogeneous nuclear ribonucleoprotein K overlaps its nuclear shuttling domain. int j biochem cell biol 2008; 40: 2078–2089. [DOI] [PubMed] [Google Scholar]
- Chen Y, Zhou X, Liu N, Wang C, Zhang L, Mo W et al. Arginine methylation of hnRNP K enhances p53 transcriptional activity. FEBS lett 2008; 582: 1761–1765. [DOI] [PubMed] [Google Scholar]
- Lee MH, Mori S, Raychaudhuri P. trans-Activation by the hnRNP K protein involves an increase in RNA synthesis from the reporter genes. J biol chem 1996; 271: 3420–3427. [DOI] [PubMed] [Google Scholar]
- Matunis MJ, Michael WM, Dreyfuss G. Characterization and primary structure of the poly(C)-binding heterogeneous nuclear ribonucleoprotein complex K protein. Mol cell biol 1992; 12: 164–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelotti EF, Michelotti GA, Aronsohn AI, Levens D. Heterogeneous nuclear ribonucleoprotein K is a transcription factor. Mol cell biol 1996; 16: 2350–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal M, Vadlamudi R, Nguyen D, Wang RA, Costa L, Bagheri-Yarmand R et al. Growth factors regulate heterogeneous nuclear ribonucleoprotein K expression and function. J biol chem 2001; 276: 9699–9704. [DOI] [PubMed] [Google Scholar]
- Roychoudhury P, Chaudhuri K. Evidence for heterogeneous nuclear ribonucleoprotein K overexpression in oral squamous cell carcinoma. Br j cancer 2007; 97: 574–575 author reply 576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F, Zhang P, Shi C, Yang Y, Qin H. Immunohistochemical detection of HSP27 and hnRNP K as prognostic and predictive biomarkers for colorectal cancer. Med oncol 2012; 29: 1780–1788. [DOI] [PubMed] [Google Scholar]
- Yang R, Zeng Y, Xu H, Chen Z, Xiang M, Fu Y et al. Heterogeneous nuclear ribonucleoprotein K is overexpressed and associated with poor prognosis in gastric cancer. Oncol rep 2016; 36: 929–935. [DOI] [PubMed] [Google Scholar]
- Gao R, Yu Y, Inoue A, Widodo N, Kaul SC, Wadhwa R. Heterogeneous nuclear ribonucleoprotein K (hnRNP-K) promotes tumor metastasis by induction of genes involved in extracellular matrix, cell movement, and angiogenesis. J biol chem 2013; 288: 15046–15056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope NR, Murray GI. The expression profile of RNA-binding proteins in primary and metastatic colorectal cancer: relationship of heterogeneous nuclear ribonucleoproteins with prognosis. Hum pathol 2011; 42: 393–402. [DOI] [PubMed] [Google Scholar]
- Inoue A, Sawata SY, Taira K, Wadhwa R. Loss-of-function screening by randomized intracellular antibodies: identification of hnRNP-K as a potential target for metastasis. Proc Natl Acad Sci USA 2007; 104: 8983–8988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audic Y, Hartley RS. Post-transcriptional regulation in cancer. Biol cell 2004; 96: 479–498. [DOI] [PubMed] [Google Scholar]
- Li J, Lu X. The emerging roles of 3' untranslated regions in cancer. Cancer lett 2013; 337: 22–25. [DOI] [PubMed] [Google Scholar]
- Choi HS, Hwang CK, Song KY, Law PY, Wei LN, Loh HH. Poly(C)-binding proteins as transcriptional regulators of gene expression. Biochem biophys res commun 2009; 380: 431–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venables JP, Koh CS, Froehlich U, Lapointe E, Couture S, Inkel L et al. Multiple and specific mRNA processing targets for the major human hnRNP proteins. Mol cell biol 2008; 28: 6033–6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomsztyk K, Van Seuningen I, Suzuki H, Denisenko O, Ostrowski J. Diverse molecular interactions of the hnRNP K protein. FEBS lett 1997; 403: 113–115. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay NK, Kim J, Cinar B, Ramachandran A, Hager MH, Di Vizio D et al. Heterogeneous nuclear ribonucleoprotein K is a novel regulator of androgen receptor translation. Cancer res 2009; 69: 2210–2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano M, Okano HJ, Okano H. Involvement of Hu and heterogeneous nuclear ribonucleoprotein K in neuronal differentiation through p21 mRNA post-transcriptional regulation. J biol chem 2005; 280: 12690–12699. [DOI] [PubMed] [Google Scholar]
- Jing Q, Huang S, Guth S, Zarubin T, Motoyama A, Chen J et al. Involvement of microRNA in AU-rich element-mediated mRNA instability. Cell 2005; 120: 623–634. [DOI] [PubMed] [Google Scholar]
- Kim HH, Kuwano Y, Srikantan S, Lee EK, Martindale JL, Gorospe M. HuR recruits let-7/RISC to repress c-Myc expression. Genes dev 2009; 23: 1743–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glorian V, Maillot G, Polès S, Iacovoni JS, Favre G, Vagner S. HuR-dependent loading of miRNA RISC to the mRNA encoding the Ras-related small GTPase RhoB controls its translation during UV-induced apoptosis. Cell death differ 2011; 18: 1692–1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu P, Fabian MR, Sonenberg N, Bhattacharyya SN, Filipowicz W. HuR protein attenuates miRNA-mediated repression by promoting miRISC dissociation from the target RNA. Nucleic acids res 2012; 40: 5088–5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell 2006; 125: 1111–1124. [DOI] [PubMed] [Google Scholar]
- Young LE, Moore AE, Sokol L, Meisner-Kober N, Dixon DA. The mRNA stability factor HuR inhibits microRNA-16 targeting of COX-2. Mol cancer res 2012; 10: 167–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W, Alajez NM, Bastianutto C, Hui AB, Mocanu JD, Ito E et al. Significance of Plk1 regulation by miR-100 in human nasopharyngeal cancer. Int j cancer 1262010: 2036–2048. [DOI] [PubMed] [Google Scholar]
- Chen P, Zhao X, Ma L. Downregulation of microRNA-100 correlates with tumor progression and poor prognosis in hepatocellular carcinoma. Mol cell biochem 2013; 383: 49–58. [DOI] [PubMed] [Google Scholar]
- Petrelli A, Perra A, Schernhuber K, Cargnelutti M, Salvi A, Migliore C et al. Sequential analysis of multistage hepatocarcinogenesis reveals that miR-100 and PLK1 dysregulation is an early event maintained along tumor progression. Oncogene 312012: 4517–4526. [DOI] [PubMed] [Google Scholar]
- Liu J, Lu KH, Liu ZL, Sun M, De W, Wang ZX. MicroRNA-100 is a potential molecular marker of non-small cell lung cancer and functions as a tumor suppressor by targeting polo-like kinase 1. BMC cancer 2012; 12: 519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li BH, Zhou JS, Ye F, Cheng XD, Zhou CY, Lu WG et al. Reduced miR-100 expression in cervical cancer and precursors and its carcinogenic effect through targeting PLK1 protein. Eur j cancer 2011; 47: 2166–2174. [DOI] [PubMed] [Google Scholar]
- Lee KS, Burke TR Jr, Park JE, Bang JK, Lee E. Recent advances and new strategies in targeting Plk1 for anticancer therapy. Trends pharmacol sci 2015; 36: 858–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.