Abstract
The regulation of chromatin structure is critical for a wide range of essential cellular processes. The Tousled-like kinases, TLK1 and TLK2, regulate ASF1, a histone H3/H4 chaperone, and likely other substrates, and their activity has been implicated in transcription, DNA replication, DNA repair, RNA interference, cell cycle progression, viral latency, chromosome segregation and mitosis. However, little is known about the functions of TLK activity in vivo or the relative functions of the highly similar TLK1 and TLK2 in any cell type. To begin to address this, we have generated Tlk1- and Tlk2-deficient mice. We found that while TLK1 was dispensable for murine viability, TLK2 loss led to late embryonic lethality because of placental failure. TLK2 was required for normal trophoblast differentiation and the phosphorylation of ASF1 was reduced in placentas lacking TLK2. Conditional bypass of the placental phenotype allowed the generation of apparently healthy Tlk2-deficient mice, while only the depletion of both TLK1 and TLK2 led to extensive genomic instability, indicating that both activities contribute to genome maintenance. Our data identifies a specific role for TLK2 in placental function during mammalian development and suggests that TLK1 and TLK2 have largely redundant roles in genome maintenance.
The Tousled kinase (TSL) and Tousled-like kinases (TLKs) are serine–threonine kinases whose activity has been linked to DNA replication, DNA repair, transcription, chromatin structure, chromosome segregation, RNA interference, viral latency and cell cycle checkpoint control in various organisms.1, 2, 3, 4, 5, 6, 7, 8, 9 TSL was identified in Arabidopsis where mutations in the gene led to pleiotropic defects, including delays in flowering time and leaf development.1 In Drosophila and Caenorhabditis elegans, deficiency in the single TLK gene led to severe mitotic defects and lethality during development.8, 10 In Drosophila, these effects were proposed to result from defective chromatin maintenance and could be rescued by the overexpression of the histone H3–H4 chaperone ASF1, a substrate of TLK.3, 10 In C. elegans, the major defects observed following ceTLK-1 downregulation were transcriptional, reflecting reduced phosphorylation of RNA polymerase II, and mitotic, through interactions with Aurora B.7, 8
Vertebrates encode 2 distinct TLK homologs, TLKs 1 and 2 (TLK1 and TLK2), that interact with each other and both ASF1a and ASF1b.3 In Drosophila, TLK phosphorylation of ASF1 controls its stability, while in vertebrates, TLK1-mediated phosphorylation of several sites on the C-terminal tail of ASF1 promote its binding affinity for the H3–H4 heterodimer.11, 12 Following DNA damage, TLK1 is phosphorylated by the checkpoint kinase CHK1, inhibiting its activity, potentially to coordinate global ASF1 histone-binding capacity with the checkpoint response.13, 14 TLK activity has also been linked to cell cycle checkpoint recovery through the phosphorylation of RAD9, a component of the 9-1-1 (RAD9/HUS1/RAD1) alternative clamp loader that has multiple roles in the response to DNA damage, including the regulation of CHK1 activation,15, 16, 17 and through TLK2-ASF1a-mediated transcriptional events.18 Although TLK2 is less well characterized, the amplification of its activity has been specifically linked to breast cancer progression in recent work.19, 20
Despite their identification nearly 15 years ago, the roles of the ubiquitously expressed TLK1 and TLK2 in mammalian physiology remain unexplored. In particular, whether they have distinct, overlapping or entirely redundant roles in particular tissues or cell types remain unknown. Here we describe the generation and characterization of both Tlk1- and Tlk2- deficient mice and cells that reveal distinct and redundant roles for TLK1 and TLK2 activities in development and genome maintenance.
Results
TLK1 is dispensable for embryogenesis and murine viability
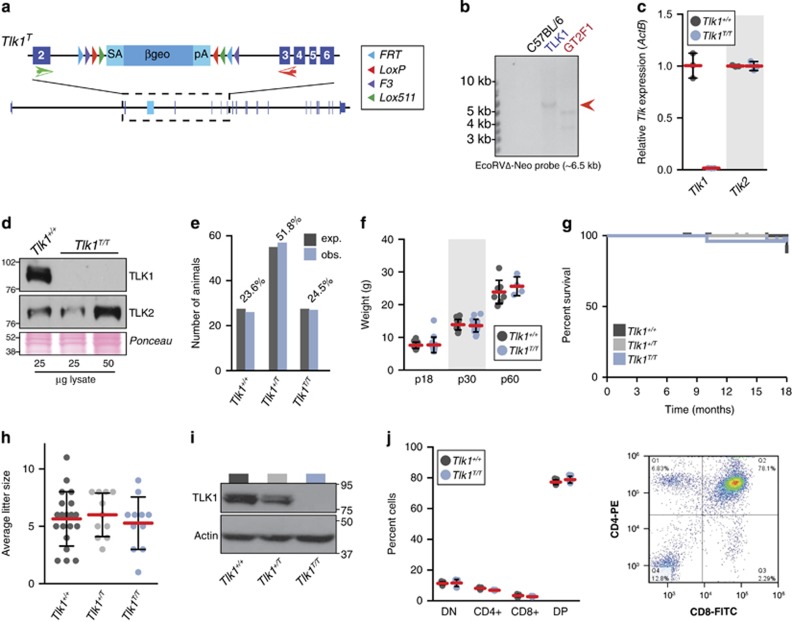
To determine if TLK1 and TLK2 had distinct or redundant roles during development or aging, we generated mice deficient for either Tlk1 or Tlk2. Mice homozygous for a gene-trapped allele of Tlk1 (Tlk1T) (Figures 1a and b) showed efficient reduction of Tlk1 expression (Figure 1c). In addition, no TLK1 protein was detectable in western blot analysis from mouse embryonic fibroblast (MEF) lysates (Figure 1d) and no compensatory changes in Tlk2 mRNA or protein levels were evident (Figures 1c and d). Despite the implication of TLK1 in many critical cellular processes, Tlk1T/T mice were born at normal Mendelian ratios (Figure 1e) and showed no obvious morphological or size differences at birth or during the first months of development (Figure 1f).4, 13, 14, 15, 16, 21, 22, 23 In addition, Tlk1T/T mice did not exhibit any obvious developmental phenotypes or accelerated morbidity over 18 months (Figure 1g). Both male and female Tlk1T/T mice were fertile, indicating that meiotic recombination was largely functional, and Tlk1T/T females produced litter sizes similar to that of Tlk1+/+ and Tlk1+/T littermates (Figure 1h). Additionally, we examined T-cell development that requires the repair of DNA double-strand breaks (DSBs) induced by the RAG recombinase, as previous work has implicated TLK1 in DNA repair.16, 21, 22, 23, 24 Similar to what was observed in MEFs, no TLK1 protein was detectable in T cells from Tlk1T/T mice (Figure 1i) and the distribution of T cells in Tlk1+/+- and Tlk1T/T-deficient mice was indistinguishable (Figure 1j), indicating that non-homologous end-joining-mediated DNA repair in this tissue operates efficiently in the absence of TLK1. Although we cannot rule out more subtle effects on repair processes or developmental programs, these data indicated that TLK1 was dispensable for murine development, aging and multiple physiological DNA repair events.
Figure 1.
TLK1 is dispensable for murine development. (a) Schematic of the Tlk1 genetrap allele (Tlk1T). The position of the Flex-vector genetrap cassette in the Tlk1 locus and approximate positions of primers used for quantitative real-time PCR are shown. (b) The genetrap clone was screened for single-copy insertion using a probe for the neomycin cassette in a Southern blot of EcoRV-digested genomic DNA. A single band of the expected size was observed (red arrow). GTF2 is an unrelated line with a neomycin cassette (positive control) and C57BL6 is wild-type genomic DNA (negative control). (c) Quantitative real-time PCR analysis of Tlk1 and Tlk2 levels in MEF cultures derived from littermates of the indicated gentoype. The mean (red bar) and standard deviation (S.D.) of three replicates are plotted. (d) Western blotting of TLK1 and TLK2 protein levels in MEF cultures of the indicated genotype. Ponceau red-stained blot showed equal loading. (e) Tlk1T/T mice are born at the expected (exp.) Mendelian ratios. Number of pups of the indicated genotype observed (obs.) from 32 litters of Tlk1+/T mice (110 pups total) is indicated. Percentage observed is indicated above the bar graphs. (f) Normal weight and growth of Tlk1T/T mice. Weights of littermate animals at 18 and 30 days postpartum are plotted with the mean (red bars) and S.D. indicated (n=16 for Tlk1+/+ and 15 for Tlk1T/T, respectively). (g) Normal survival of Tlk1T/T mice. Kaplan–Meier plot of animal survival over 18 months (n=25, 41 and 26 for Tlk1+/+, Tlk1+/T and Tlk1T/T, respectively). (h) Normal litter sizes are observed in Tlk1T/T female mice of 2–5 months of age compared with wild type or heterozygous littermates (n=20, 11 and 11 for Tlk1+/+, Tlk1+/T and Tlk1T/T, respectively). The mean (red bars) and S.D. are plotted and statistical analysis using the Wilcoxon's rank-sum test indicated no significant differences (P=0.65;Tlk1+/+ versus Tlk1+/T, 0.75; Tlk1+/+ versus Tlk1T/T and 0.52; Tlk1+/T versus Tlk1T/T). (i) TLK1 protein levels in thymocytes of the indicated genotype are shown. Actin serves as a loading control. (j) Analysis of T-cell subsets in 2-month-old animals of the indicated genotype (n=3 per genotype, each in triplicate). The mean (red bars) and S.D. are shown and representative flow cytometry data showing CD4 and CD8 staining of CD3-positive T cells is shown in the right panel
TLK2 is an essential gene
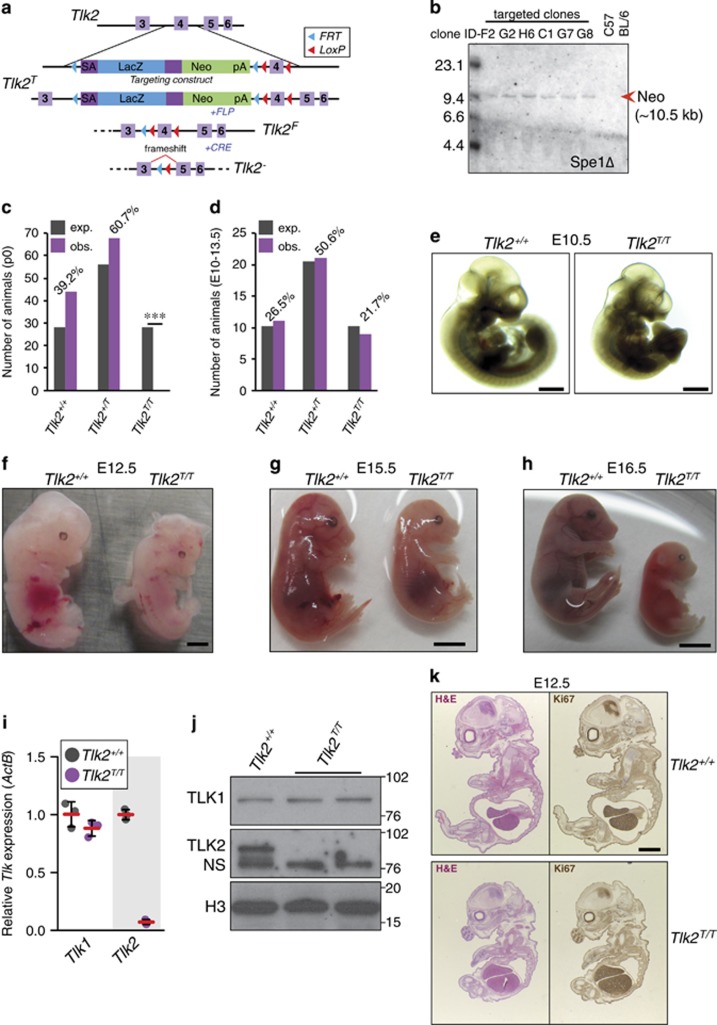
Based on previous data from lower organisms, we expected that loss of TLK1 activity would result in severe phenotypes in the mouse.8, 10 The lack of any clear phenotypes in Tlk1T/T animals suggested potentially redundant activities, with TLK2 being the most likely candidate. To address this, we generated a conditional, gene-trapped allele of Tlk2 (Figures 2a and b and Supplementary Figure S1) and interbred heterozygous mice to generate Tlk2-deficient animals. In sharp contrast to Tlk1, we did not observe any live births of Tlk2T/T mice, indicating embryonic lethality (Figure 2c).
Figure 2.
TLK2 is an essential gene. (a) Schematic of the Tlk2 knockout-first allele (Tlk2T). The position of the genetrap cassette, the allelic configurations and their designations following FLP and CRE expression are shown. (b) Southern blot of SpeI-digested genomic DNA with a probe for neomycin to identify clones with single insertions. Six targeted clones and a negative control (C57BL/6) are shown and the expected size indicated. Additional Southern blots confirming correct targeting are shown in Supplementary Figure S1. (c) Tlk2 deficiency is embryonic lethal. The number of animals expected (exp.) from 27 Tlk2+/T breedings (112 pups total) assuming normal Mendelian inheritance is shown compared with the number of observed pups (obs.). Percentage observed is indicated above the bar graphs. Based on the genotypes of a total of 112 pups from 14 independent litters, a P-value of <0.0001 was determined using binomial distribution and indicated by ***. (d) Normal Mendelian distribution of genotypes was observed among 41 E10.5 to E13.5 embryos from six individual Tlk2+/T breedings. Percentage observed is indicated above the bar graphs. (e) Examples of littermate E10.5 embryos (scale bar=1 mm). (f) E12.5 embryos (scale bar=1 mm) (g) E15.5 embryos (scale bar=2 mm) and (h) E16.5 embryos (scale bar=2 mm). (i) Quantitative real-time PCR analysis of Tlk1 and Tlk2 levels in MEFs from littermates of the indicated genotype. The mean (red bars) and standard deviation (S.D.) of three replicates are plotted. (j) Western blotting of TLK1 and TLK2 protein levels in MEFs of the indicated genotype. Histone H3 is shown as a loading control and a nonspecific (NS) band recognized by the TLK2 antibody is indicated. (k) Hematoxylin and eosin (H&E) (left panels) or Ki67 immunohistochemical staining (right panels) of E12.5 embryo sections of the indicated genotype (scale bar=1 mm)
To understand the cause of death, we examined embryos at different developmental stages. The number of Tlk2T/T embryos observed from embryonic days (E) 10.5 to E13.5 was consistent with expected Mendelian ratios (Figure 2d), although embryos were smaller in all cases (Figures 2e–h). Tlk2T/T embryos appeared anemic (Figures 2f and g) and perished by E15.5, as no heartbeat was detectable. By E16.5 extensive tissue autolysis was evident (Figure 2h). Fibroblast cultures from E14.5 embryos confirmed that Tlk2 mRNA and protein levels were reduced and that there was no compensatory increase of TLK1 (Figures 2i and j). The further examination of Tlk2T/T embryos by conventional immunohistochemistry (IHC) (Figure 2k) and lightsheet-based fluorescence macroscopy (Supplementary Figure S2) did not reveal any consistent defects and suggested that embryos were developmentally delayed, but in the majority of cases morphologically normal. Collectively, our data suggested that TLK2, but not TLK1, was essential for embryonic viability.
TLK2 is essential for placental development
As consistent defects in embryonic morphology were not observed, we examined placental function. At E14.5 Tlk2T/T placentas were smaller and there was a lack of blood circulation in the umbilical cord (Supplementary Figures S3A and S3B). To address this phenotype further without potential complications from the genetrap, we excised it by crossing to FlpO-expressing mice, thus rendering exon 4 floxable (Tlk2F) (Figure 2a). Crosses with mice expressing Sox2-Cre, that is active in the embryonic epiblast cells, effectively deleted exon 4 and yielded Tlk2+/− animals that were then interbred to generate Tlk2−/− embryos (Supplementary Figure S3C).
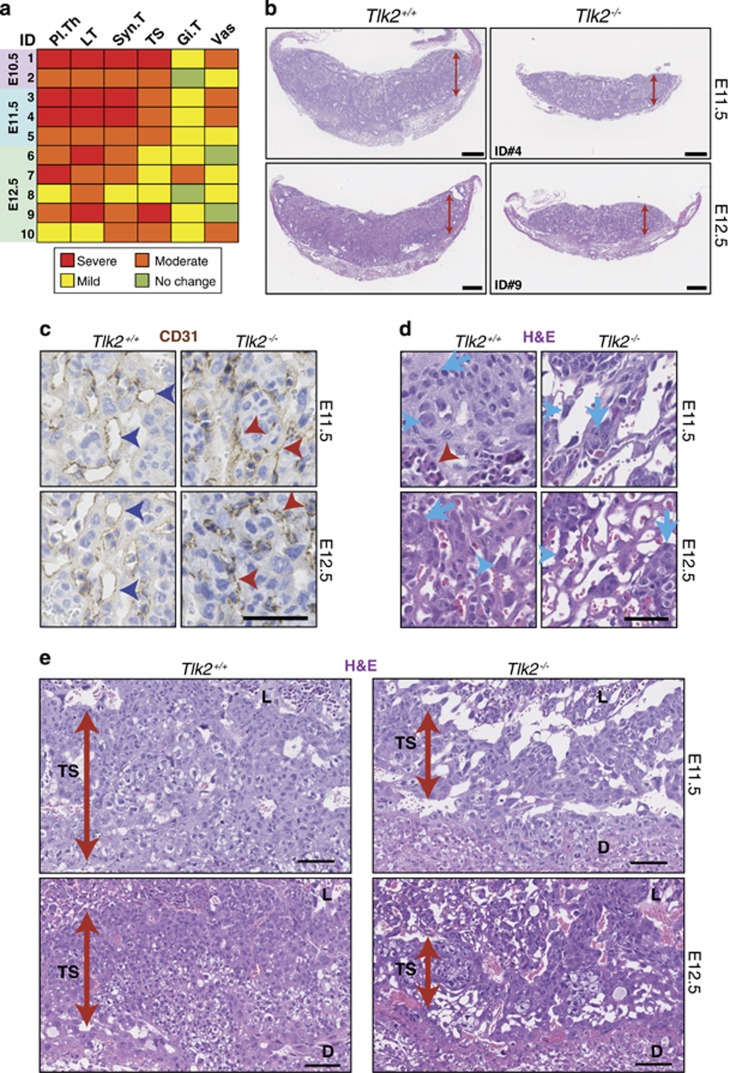
We histologically examined Tlk2−/− placentas from E10.5 to E16.5, and at all stages, Tlk2−/− placentas were moderately to severely smaller, thinner and less cellular compared with Tlk2+/+ or Tlk2+/− littermates, and poorly vascularized (Figures 3a–e and Supplementary Figure S3). At E10.5 through E12.5 the labyrinths of Tlk2−/− placentas were disorganized and composed of less differentiated and less mature trophoblasts (Figure 3d and Supplementary Figure S3). Tlk2−/− placentas had moderately to severely reduced numbers of labyrinth trophoblast, syncytiotrophoblast, spongiotrophoblast (including glycogen cells) and mildly to moderately reduced numbers of trophoblastic giant cells and vasculature (Figures 3c–e and Supplementary Figure S3). These data indicated that TLK2 was required for normal placental development and function, and that its loss led to late embryonic lethality.
Figure 3.
TLK2 is required for placental development. (a) Semiquantitative analysis of histological phenotypes in Tlk2−/− placentas. In each case, Tlk2−/− placentas (n=10) at the indicated age were compared with wild-type littermates. Phenotypes are abbreviated as follows: placental thickness (Pl.Th), numbers/size of Labyrinth trophoblasts (LT), syncytiotrophoblasts (Syn.T), spongiotrophoblasts including glycogen cells (TS), giant cell trophoblasts (Gi.T) and vasculature (Vas). Summary of the grading system used48 was provided in the Materials and Methods. (b) Sections from E11.5 and E12.5 littermate placentas stained with Hematoxylin and eosin (H&E). Tlk2−/− placentas are thinner, less cellular and poorly vascularized compared with Tlk2+/+. Scale bars=500 μm. (c) CD31 staining reveals that the vasculature of Tlk2−/− placentas is miniaturized, collapsed and appears slit-like (red arrowheads) compared with the well-developed, widely opened vasculature of Tlk2+/+ (blue arrowheads). Scale bar=20 μm. (d) H&E staining from the indicated age and genotype showing fewer and smaller trophoblasts (arrows) and syncytiotrophoblasts (blue arrowheads) with an absence of fetal vasculature (red arrowhead) in Tlk2−/− placentas. Scale bar=20 μm. (e) H&E staining of placentas showing a thinner trophospongium (spongiotrophoblasts and glycogen cells) in Tlk2−/− placentas. The trophospongium (TS), labyrinth (L) and decidua (D) are indicated. Scale bar=50 μm
The influence of TLK2 deficiency on proliferation and gene expression in the placenta
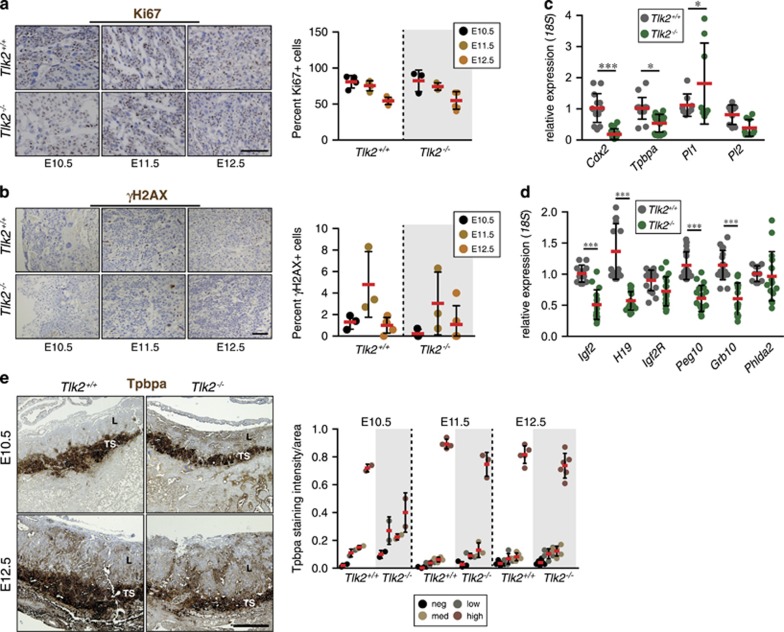
As the regulation of ASF1 by TLK activity has been linked to ongoing DNA replication, we examined trophoblast proliferation by IHC using the marker Ki67.2 By E12.5 most cells of the trophoblastic lineage are differentiated and their rate of proliferation decreases.25, 26 Regardless of genotype, we observed similar percentages of Ki67-positive cells at each stage of development, suggesting that the overall number of proliferating cells was similar (Figure 4a). In addition, we did not observe any significant increase in either the DNA DSB marker γH2AX or the apoptosis marker cleaved caspase-3 (Figure 4b and Supplementary Figure S4A), suggesting that overall levels of proliferation, DNA damage and cell death were similar.
Figure 4.
Proliferation and gene expression in placentas lacking TLK2. (a) Immunohistochemical staining of the proliferative marker Ki67 or (b) the DNA DSB marker γH2AX in E10.5, E11.5 and E12.5 placentas of the indicated genotype. Similar numbers of proliferative cells are visible in the labyrinth of Tlk2−/− placentas compared with wild type at each stage. Mean percent-positive cells (red bars) and S.D. is graphed in right panels. n=3 and 2 at E10.5, n=3 and 3 at E11.5 and n=5 and 6 at E12.5 for Tlk2+/+and Tlk2−/−, respectively. At least 200 total cells were counted for each individual and scale bar in (a) is equal to 10 μm and in (b) is equal to 100 μm. (c and d) Quantitative real-time PCR analysis of genes expressed in trophoblast lineages from E12.5 placenta samples. Combined data represents the mean of triplicate samples from three individual animals of each genotype. Statistical significance was determined using an upaired t-test (***P<0.0001, **P<0.001, *P<0.05). (e) Immunohistochemical staining for Tpbpa reveals a thinner trophospongium (TS) in the absence of TLK2 at both E10.5 and E12.5. The orientation of the labyrinth (L) is also indicated. Quantification of signal intensity distribution in the TS is shown in the right panel, normalized to area. Scale bar=500 μm
In addition to DNA replication, TLK2 has been implicated in transcriptional regulation through the phosphorylation of ASF1a.18, 27 To address the possibility that transcription may be affected, we examined the mRNA expression levels of several genes required for trophoblast differentiation: Cdx2, Tpbpa, Pl1 and Pl2.28 We observed lower levels of Cdx2, Tpbpa and Pl2 in the Tlk2−/− placentas, reflecting reduced numbers and/or defective differentiation of trophoblast stem cells, spongiotrophoblasts and canal or sinusoidal trophoblast giant cells, respectively (Figure 4c). In contrast, the expression of Pl1, which is a marker for parietal trophoblast giant cells and is normally reduced by E11.5, was slightly upregulated compared with wild-type placenta, although this was more variable among individual samples (Figure 4c).28 In addition, the expression of many imprinted genes that influence placental development, including Igf2, H19, Peg10 and Grb10, was reduced in Tlk2−/− placentas, while others, such as IgfR2 and Phlda2, were expressed at similar levels to that of wild-type (Figure 4d).28, 29 The reduced size of the Tlk2−/− labyrinth and reduced area of Tpbpa-positive cells was further confirmed with IHC (Figure 4e). Taken together, these data suggested that the loss of TLK2 did not cause clear defects in trophoblast proliferation or survival, but reduced the expression of numerous genes important for their normal function and led to impaired differentiation and placental function.
TLK2 interacts with ASF1 and influences its phosphorylation in the placenta
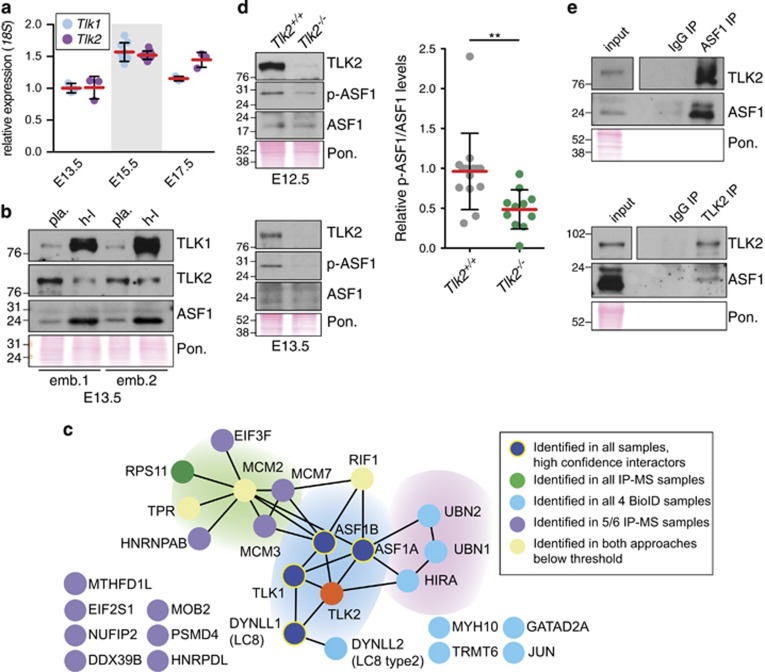
To understand why placental function was dependent on TLK2 and not TLK1, we examined their relative mRNA expression and protein levels. We found similar mRNA expression patterns of both genes in the wild-type placenta at E13.5, E15.5 and E17.5 (Figure 5a). The protein levels of TLK2 were similar in the placental tissue compared with the tissue from the embryo (heart and fetal liver), whereas TLK1 protein levels were strongly increased in the embryonic tissues compared with the placenta, although TLK1 was detectable in placentas (Figure 5b and Supplementary Figure S4B). This suggested that TLK1 was significantly less abundant in the placenta than the embryo, providing a potential explanation as to why placental development was particularly sensitive to Tlk2 deficiency.
Figure 5.
TLK2 interacts with ASF1 and influences its phosphorylation in the placenta. (a) Quantitative real-time PCR of Tlk1 and Tlk2 in wild-type placentas of the indicated age. Mean (red bars) and S.D. of triplicate samples is graphed (n=1, 3 and 1 for each age indicated). (b) Western blotting of TLK1, TLK2 and ASF1 in lysates from E13.5 placenta (Pon.=Ponceau red-stained membrane). TLK2 levels are similar in either tissue (placenta=Pla. and heart/fetal liver=h–l), while TLK1 levels are much higher in embryonic tissue relative to placenta. Samples from two littermate wild-type placentas are shown. Additional examples are shown in Supplementary Figure S4B. (c) Graphic depiction of the combined results from multiple proteomics approaches to identify TLK2-interacting proteins from HEK293T or AD293 cells (IP-MS and BioiD approaches and data are described in further detail in Supplementary Figure S5 and Supplementary Tables S1–S6). TLK1, ASF1a, ASF1b and DYNLL1 (LC8) are the only high confidence interacting proteins identified. LC8 does not appear to be a substrate of TLK2 (Supplementary Figures S5G and S5H). (d) Reduced ASF1-S166 phosphorylation (p-ASF1) in the E12.5 and E13.5 Tlk2−/− placentas. Western blotting of lysates for TLK2, ASF1 and p-ASF1 from littermates of the indicated age. In each case, blots shown are from the same membrane (Pon.=Ponceau red-stained membrane). Quantification of p-ASF1/ASF1 levels in multiple samples of the indicated genotype is shown in the right panel (additional Western blot analysis examples are shown in Supplementary Figure S6A). Statistical significance (P=0.0064) was determined using an unpaired t-test and the mean (red bars) and S.D. are indicated (n=14 and 11, respectively). (e) Co-IP of endogenous TLK2 and ASF1 from wild-type E13.5 placental lysates using antibodies for ASF1 (top panels) or TLK2 (bottom panels). (Pon.=Ponceau red-stained membrane). Additional examples are shown in Supplementary Figure S6B
ASF1a and ASF1b are the most clearly defined targets of TLK1 and TLK2, but other substrates, such as RAD9 and NEK1, have been reported for TLK1.15, 23, 30 To address the potential substrate spectrum of TLK2, we carried out IPs of tagged TLK2 or kinase dead TLK2 followed by quantitative mass spectrometry (IP-MS; Supplementary Figure S5A). In addition, we overexpressed TLK2 fused to the BirA* biotin ligase to identify the proximity interactome of TLK2 using BioID (Supplementary Figure S5B). Only a small number of proteins were consistently identified by both approaches, including ASF1a and ASF1b, TLK1 and LC8 (DYNLL1) (Figure 5c,Supplementary Figure S5 and Supplementary Tables S1–S6). Additional proteins known to be involved in a variety of cellular processes, including DNA replication and chromatin assembly (e.g. MCM helicase components, RIF1 and HIRA/UBN1-2), transcription (e.g. GATAD2A, JUN) and translation (e.g. EIF3F, EIF2S1) were identified in one or both approaches below our statistical thresholds (Figure 5c). We focused on the most high confidence interactions and found that while we could confirm the LC8 interaction, it did not appear to be a substrate of TLK2 activity (Supplementary Figure S5E–H). We therefore focused on determining the level and phosphorylation status of ASF1 in normal and Tlk2-deficient placentas. In Tlk2−/− placentas total ASF1 protein levels were similar to wild-type controls, but the phosphorylation of S166 (p-ASF1), a target site of TLKs, was reduced, suggesting reduced H3/H4 binding potential in the absence of TLK2 (Figure 5d and Supplementary Figure S6A).11 To confirm the interaction of TLK2 with ASF1 in the placenta, we performed co-immunoprecipitations (IPs) of endogenous TLK2 or ASF1 from placental lysates. Following the IP of TLK2, we readily observed both ASF1 and TLK1 and reciprocal IPs with anti-ASF1 antibodies efficiently co-immunoprecipitated TLK2 (Figure 5e and Supplementary Figure S6B). Taken together, these results indicated that despite some levels of detectable TLK1, TLK2 is required for full ASF1 phosphorylation in the placenta, potentially influencing the regulation of genes required for trophoblast differentiation and placental development.
TLK2 is dispensable for viability in embryonic and adult tissues
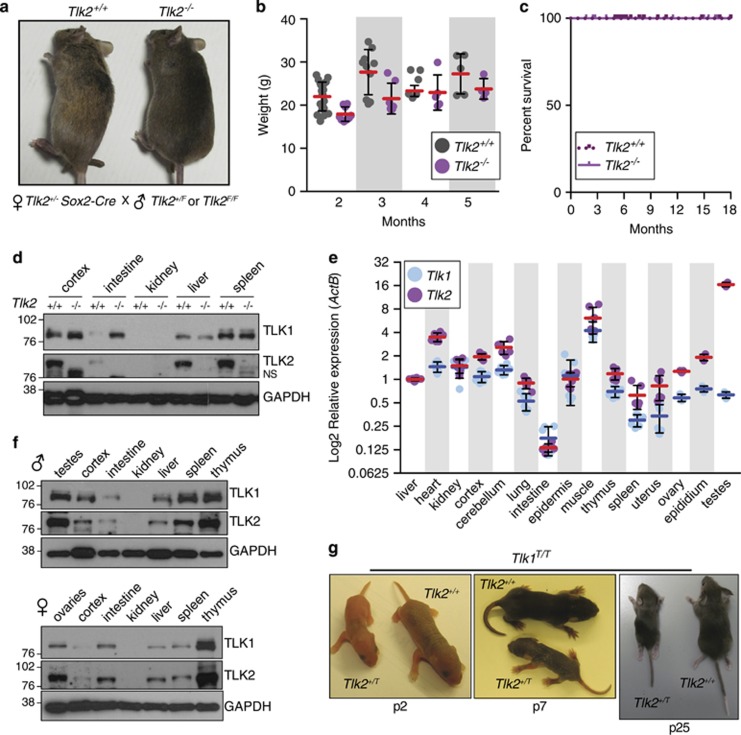
The differential protein levels of TLK1 between the placenta and embryonic tissue provided a potential explanation for placental dependence on TLK2 (Figure 5b). We considered that a similar situation in other cell types in the embryo could also contribute to the embryonic death of Tlk2−/− animals. To address this, we crossed female Tlk2+/− mice carrying the Sox2-Cre transgene, which expresses Cre in the epiblast, with males carrying the floxed conditional allele (Tlk2+/F) to allow the generation of Tlk2−/− embryos, while maintaining at least one functional copy of Tlk2 in the placenta (Figure 6a).31 This strategy bypassed the embryonic lethality and we observed the birth of live, morphologically normal, Tlk2−/− animals (Figure 6a). These animals, although sometimes smaller than littermates, showed a similar growth capacity (Figure 6b) and no increased morbidity has been observed up to 18 months (Figure 6c), although we cannot yet rule out tissue-specific or aging-related phenotypes that are not overtly evident. Importantly, we could not detect TLK2 protein in any tissues examined by western blotting, indicating that Sox2-Cre was effective at eliminating Tlk2 expression (Figure 6d). These data demonstrate that, like TLK1, TLK2 is not required for organismal viability, despite its critical role in the development and function of the placenta.
Figure 6.
TLK2 is dispensable in embryonic and adult tissues. (a) Tlk2+/+ and Tlk2−/− littermates are shown from breedings of female Tlk2+/−, Sox2-Cre+ and male Tlk2+/F mice. (b) Weight of mice lacking TLK2 is slightly reduced but growth is similar to wild type. Mean (red bars) and S.D. are indicated (n=20 and 9 for 1 month, n=10 and 7 for 2 months, n=6 and 5 for 3 months, n=5 and 4 for 4 months for Tlk2+/+ and Tlk2−/−, respectively). (c) Kaplan–Meier survival curve of mice with the indicated genotypes (Tlk2+/−, n=42; Tlk2−/−, n=26). (d) Western blotting of TLK1 and TLK2 using lysates from selected tissues of Tlk2+/+ and Tlk2−/− mice. NS refers to a nonspecific band observed with the TLK2 antibody in many tissues and cell lines. (e) Similar expression patterns of Tlk1 and Tlk2 in adult mice. Taqman real-time PCR analysis of Tlk1 and Tlk2 mRNA levels (log 2-transformed) relative to ActB. Mean (blue bars for Tlk1 and red bars for Tlk2) and S.D. of triplicate reactions from at least two animals are plotted. In each case, levels are normalized to that of the liver (set to 1). (f) Western blotting of TLK1/2 protein levels in 2-month-old male and female wild-type tissues. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) is shown as a loading control. (g) Examples of littermate Tlk1T/T Tlk2+/+ and Tlk1T/T Tlk2+/T animals at different ages. All Tlk1T/T Tlk2+/T animals (n=4) observed were severely runted and one lacked limbs on the left torso. All mice were killed before 1 month following veterinary advice owing to the lack of mobility and trembling. A Kaplan–Meier survival plot for all related genotypes is presented in Supplementary Figure S7
As relative TLK1 and TLK2 protein levels were different between embryonic and placental tissues, we sought to determine if a similar situation existed in other adult tissues. We freshly isolated mRNA and protein from a panel of wild-type tissues at 2 months of age. Using quantitative real-time PCR, we determined the expression levels of Tlk1 and Tlk2 in different tissues (Figure 6e). In general, the expression of Tlk1 and Tlk2 followed a similar pattern across tissues, although the expression of Tlk2 was particularly high in the testes, where the human TLK2 gene was originally identified.6 To determine if the mRNA levels reflected relative protein levels, we performed western blotting from lysates prepared from some of the same tissues of both male and female mice (Figure 6f). These results were generally consistent with the mRNA expression levels and the patterns of TLK1 and TLK2 protein levels were similar relative to tissue type. One exception was the kidney, where both TLK1 and TLK2 protein were undetectable, despite similar levels of mRNA expression as other tissues (Figures 6e and f). These results suggested that the lack of clear phenotypes in adult animals deficient for either TLK1 or TLK2 could be due to their redundancy in most tissues, in contrast to the placenta where TLK2 is required because of the low levels of TLK1. Supporting this possibility, we found that Tlk2 is haploinsufficient on a Tlk1-deficient background, as all Tlk1T/TTlk2+/T pups we observed displayed severe birth defects (Figure 6g and Supplementary Figure S7).
TLK1 and TLK2 cooperate to maintain cell viability and chromosomal stability
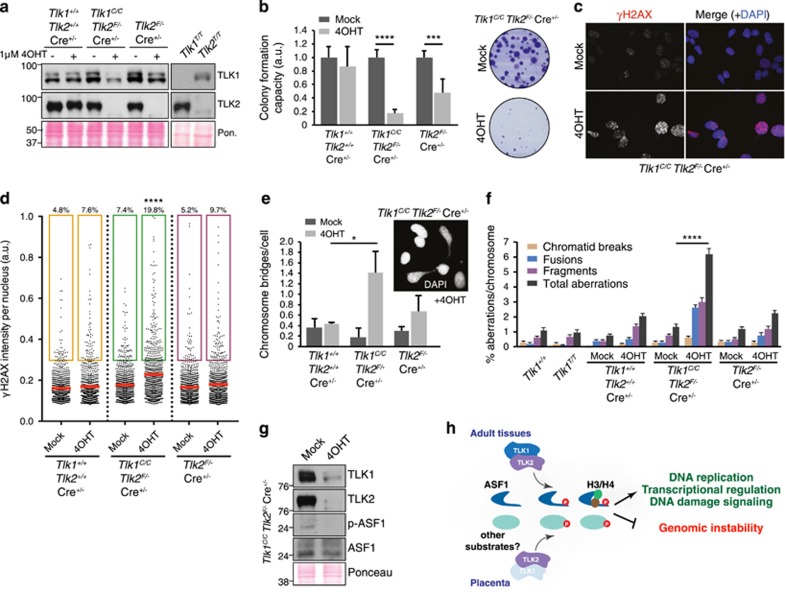
As TLK1- or TLK2-deficient animals appeared phenotypically normal, we wanted to investigate the effects of depleting both activities. To accomplish this, we generated single (Tlk1+/+Tlk2F/−) and double conditional (Tlk1C/CTlk2F/−) transformed MEF lines that allow the 4-hydroxytamoxifen (4OHT)-inducible trapping and deletion of Tlk1 and Tlk2, respectively. Depletion of TLK1 and TLK2 protein levels following 4OHT induced Cre recombinase induction was effective (Figure 7a) and led to reduced colony formation compared with wild-type or single mutants for Tlk2 (Figure 7b).
Figure 7.
TLK1 and TLK2 cooperate to maintain cell viability and chromosome stability. (a) Western blot with antibodies against TLK1 and TLK2 in whole-cell extracts of transformed MEFs of the indicated genotypes mock-treated or treated with 4OHT to induce trapping of TLK1 and/or deletion of TLK2 (Pon.=Ponceau red-stained membrane). (b) Deletion of TLKs reduces colony formation. MEFs were mock-treated or treated with 4OHT for 72 h, washed, plated and cultured for 10–14 days. Relative colony number to mock-treated cells is displayed. Tlk1+/+ Tlk2+/+ Cre+/−, n= 3; Tlk1C/CTlk2F/− Cre+/−, n=6; Tlk2F/− Cre+/−, n=6. Results are shown as mean±S.E.M. Experiment was performed in technical duplicates, at least in biological triplicate. Statistical significance was determined using an upaired t-test (****P<0.0001, ***P<0.001). (c) Representative images of IF experiments staining for γH2AX are shown from Tlk1T/T Tlk2F/− cells that were mock-treated or treated with 4OHT. Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI). (d) Increased DNA DSB formation following the depletion of TLK1 and TLK2. High-throughput microscopy of γH2AX levels per individual nucleus in response to TLK depletion in MEFs mock-treated or treated with 4OHT. At least 800 nuclei were quantified per condition. Mean is shown as a red line; a.u., arbitrary units. Statistical significance was determined using ordinary one-way analysis of variance (ANOVA) Tukey's multiple comparisons test (****P<0.0001). (e) Increased chromosome bridges are evident in 4OHT induced double TLK-deficient cultures relative to mock-treated or Tlk2 deletion alone. A minimum of 200 cells was scored in each independent experiment. Tlk1+/+ Tlk2+/+ Cre+/−, n= 2; Tlk1C/CTlk2F/− Cre+/−, n=5; Tlk2F/− Cre+/−, n=4. Results are shown as mean±S.D. Statistical significance was determined using an upaired t-test (*P<0.05). (f) Plot of chromosomal aberration types scored from multiple transformed MEF cultures depicted as percent aberrations per chromosome. Results are shown as mean±S.E.M. Statistical significance was determined using Fisher’s exact test (****P<0.0001). (g) Western blotting of Tlk1C/CTlk2F/− MEF lysates mock-treated or treated with 4OHT for TLK1, TLK2, ASF1 and p-ASF1. Experiment performed in biological triplicate and a representative example shown. (h) Proposed model for the contribution of TLK1 and TLK2 activity to the placenta and adult tissues. In the placenta, and possibly other cell types or tissues, TLK2 is the prevalent activity and is required to maintain a threshold of signaling required for viability. This likely involves ASF1a/b and potentially other substrates. In most adult tissues, TLK1 and TLK2 are largely redundant and the presence of either is sufficient to provide the necessary activity to support cellular functions required for the maintenance of genome integrity and viability
The simultaneous depletion of TLK1 and TLK2 resulted in a marked increase in DNA DSBs, indicated by increased intensity of γH2AX foci in the nucleus (Figures 7c and d). These cells also displayed mitotic defects, such as chromosome bridges (Figure 7e), as well as increased levels of chromosome aberrations scored in metaphase spreads, particularly chromosome fragments and fusions (Figure 7f). This was accompanied by a reduction in total ASF1 phosphorylation, similar to what was observed in the placenta (Figure 7g). Collectively, our in vivo and in vitro results indicated that TLK1 and TLK2 activities are largely redundant for cell viability and chromosome maintenance, but that TLK2 is critical for mammalian development through its role in supporting placental function (Figure 7h).
Discussion
Here we describe for the first time the relative contributions of TLK1 and TLK2 to mammalian embryogenesis. Of the two proteins, TLK1 has been the most studied and has been implicated in many critical cellular processes, such as transcription, DNA repair, DNA replication and mitosis.2, 4, 7, 8, 10, 11, 13, 14, 15, 16, 21, 22, 23, 32, 33, 34, 35, 36 Although it may have a role directly or indirectly in these processes, our results indicate that TLK1 is not required for essential cellular functions when TLK2 is expressed normally. We did however observe that TLK1 becomes essential in Tlk2+/T animals (Figure 6g), and we believe that this likely reflects a need for TLK1 in supporting placental or possibly embryonic function, when Tlk2 is haploinsufficient, but have not investigated this further. Regardless, the loss of TLK1 does not impair the generation of viable, apparently healthy animals, notably impact several physiological repair processes or detectably influence genomic stability.
In contrast to TLK1, TLK2 was strictly required for embryogenesis through its role in supporting placental development. As both Tlk1 and Tlk2 mRNAs are readily detectable in every tissue that we (Figure 6e), and others,37 have examined, this essential role for TLK2 was unexpected. We observed reduced ASF1 phosphorylation, impaired gene expression and histological evidence that indicated a requirement for TLK2 in the proper differentiation of the trophoblast lineages. As we did not observe any indications that proliferation was strongly affected, we believe this is likely through a role in transcriptional regulation.18 We, however, cannot rule out the possibility that loss of TLK2 impairs DNA replication to some extent, leading to either cellular attrition or indirect defects on transcription and that TLK2 may influence cell migration19 or other cellular functions that have yet to be uncovered. A similar differential requirement for ASF1a and ASF1b in development has been reported previously. The depletion of murine ASF1a leads to lethality by E9.5,38 notably earlier than we observed with TLK2 deletion, potentially consistent with the incomplete effect we observed on total ASF1 phosphorylation. In contrast, ASF1b is dispensable for viability, although it affects fertility.39 While mice lacking either TLK1 or TLK2 were fertile, we cannot rule out subtle or age-related differences in fertility from existing data. Future analysis of mice lacking combinations of both TLK and ASF1 alleles may be informative for identifying more specific functions and determining how much of the TLK-deficient phenotype relies on ASF1 regulation.
Although mRNA levels of Tlk1 and Tlk2 were similar from E13.5 to 17.5 of placental development, TLK1 protein levels were much higher in embryonic tissues compared with placenta, whereas TLK2 protein levels were similar regardless of the tissue. We believe this provides the most parsimonious explanation for the essential nature of TLK2 in the placenta, although we cannot rule out that TLK2 is uniquely capable of modifying a cell type-specific substrate or carrying out another unknown, essential role during development. Whether similar unique dependencies of either TLK1 or TLK2 occur in other cell types of embryonic or adult animals remain to be determined, as do the mechanisms that regulate TLK1 or TLK2 protein levels, activity and substrate engagement.
TLK1 activity appears to buffer Tlk2 haploinsufficiency (Figure 6g), but it is insufficient to fully support placental development in the absence of TLK2. This may be because of its lower levels, and presumably lower activity, relative to embryonic tissue, reduced affinity for ASF1a,27 or because of specific interactions that have yet to be identified. However, in other tissues of the embryo and adult animals, the protein levels of TLK1 and TLK2 appear to be sufficient to act interchangeably to support cellular functions, evidenced by the viability of both Tlk1T/T and Tlk2−/− cell cultures and animals (Figures 1,6 and 7).
While our results suggest that there may be a high level of redundancy between TLK1 and TLK2 in many cell types, recent work has specifically connected TLK2 mutations to patients with intellectual disabilities40 and it is tempting to speculate that this could reflect placental defects, although we would expect them to be less marked than what we have observed in mice with a full TLK2 knockout. Moreover, TLK2 has been identified as a regulator of starvation-induced autophagy41 and gammaherpesvirus latency,9 amplifications and hyperphosphorylation of specifically TLK2 have been identified in subtypes of breast cancer and shown to influence cancer progression19, 20, 42, 43 and TLK2 has been shown to have a critical role in cell cycle checkpoint recovery that was not observed following TLK1 depletion.18 These apparently specific roles may reflect differences in their transcriptional or translational regulation and relative ratios, differential control of their activity or distinct substrate specificity. Recent work identified around 150 putative TLK1 interactors30 and we found only seven proteins in common using different experimental approaches for TLK2, including ABLIM1, DNAJC7, EMG1, PDCD6, SERBP1, SNRPC and SRSF1, although these fell below the stringent thresholds we used for selecting interactions to validate (Figure 5c and Supplementary Tables S1–S6). Further investigation in a wider array of cell types will be required to understand the unique and overlapping substrates and functions of TLK1 and TLK2 and their specific roles in development, genome maintenance and other cellular processes.
Materials and methods
Generation of Tlk1- and Tlk2-deficient mice
Embryonic stem (ES) cells with a gene-trapped Tlk1 allele were purchased from the German Genetrap Consortium (clone ID E067A02). A single insertion was confirmed and cells injected into 3.5-day mouse C57B6/j blastocysts (Supplementary Figure S1). A total of 12–15 ES cells were injected into each blastocyst and reimplanted into the oviduct of 2.5-day pseudopregnant foster mice (CD1s). Chimeras were scored and mated with C57B6/j wild-type mice. Agouti offsprings were screened by PCR for the Tlk1 allele and pups were screened for zygosity using PCR or by quantitative real-time PCR (Transnetyx, Cordova, TN, USA). Primer details are available upon request.
The knockout-first Tlk2 targeting construct was designed and constructed in the IRB Mutant Mouse facility using standard recombineering methods.44 The targeting vector was confirmed by sequencing and linearized vector transfected into ES cells for targeting. Cells were selected with neomycin and digested genomic DNA analyzed by Southern blot (performed by service in the Centro Ricerche Biotecnologiche of the Università Cattolica del Sacro Cuore, Milan, Italy) for correct targeting (Supplementary Figure S1). For Southern analysis, three probes were used and correctly targeted clones injected as described for Tlk1. We analyzed the 5′ (5P) and 3′ (3P) integration sites following KpnI digestion (probe primers in Table S7). Pgk1-FlpO (011065) and Sox2-Cre (008454) were purchased from Jackson Laboratories (Bar Harbor, ME, USA) and ERT2-Cre mice were described previously.31, 45, 46 All animals were maintained in strict accordance with the European Community (86/609/EEC) guidelines at the animal facilities in the Barcelona Science Park (PCB). Protocols (CEAA13/0008) were approved by the Animal Care and Use Committee of the PCB (IACUC; CEEA-PCB) in accordance with applicable legislation (Law 5/1995/GC; Order 214/1997/GC; Law 1201/2005/SG). All efforts were made to minimize use and suffering.
Generation of MEFs
MEF cultures were established from E14.5 embryos that were dissected into fresh PBS and kept overnight at 4 °C in trypsin/EDTA (Life Technologies, Carlsbad, CA, USA) solution. Following incubation at 37 °C for 20 min, cells were disaggregated with a serological pipette and supernatant plated for culture in DMEM supplemented with 10% (v/v) fetal bovine serum (FBS; Sigma-Aldrich, St. Louis, MO, USA), 2 mM l-glutamine and 100 U/ml penicillin. Transformed MEFs were generated by transfecting a linearized p129 plasmid, containing an origin-less SV40 genome, using an Amaxa nucleofector with MEF reagent 2 (Lonza, Basel, Switzerland). For the induction of Cre recombinase in ERT2-Cre-expressing cell lines, cells were exposed to 4OHT (Sigma-Aldrich; H7904; 1 μM) for 72 h, washed and plated for experiments.
Immunological analysis
Analysis of thymocytes was performed as described previously.47 Thymi were dissected from 8-week-old animals and a single-cell suspension was obtained using a 45 μm nylon mesh (Thermo Fisher Scientific, Waltham, MA, USA). Apoptosis of thymoctes was assessed by flow cytometry after double staining with FITC-conjugated Annexin V (BD Biosciences, East Rutherford, NJ, USA) and propidium iodide. Thymic sub-populations were quantified by staining with fluorescence-conjugated antibodies for CD3e, CD4 and CD8 (BD Biosciences) and analysis by flow cytometry using the FlowJo Software (Tree Star).
Immunohistochemistry
Paraffin-embedded tissue sections (3 μm) were air dried and further dried at 60 °C overnight. IHC was performed using an Autostainer Plus (Dako, Agilent Technologies, Santa Clara, CA, USA) or manually (Tpbpa). Before IHC, sections were dewaxed using the low pH EnVision FLEX Target Retrieval Solutions for caspase-3 detection (Dako) at 97 °C for 20 min using a PT Link (Dako), or dewaxed and rehydrated and thereafter antigen retrieval was performed using a PT link (Dako) at 97 °C for 20 min with a citrate buffer (pH 6). Quenching of endogenous peroxidase was performed for 10 min with Peroxidase-Blocking Solution (Dako; REAL S2023). Primary antibodies used were as follows: rabbit polyclonal anti-Ki67 (Thermo Fisher Scientific; 1 : 75), rabbit polyclonal anti-CD31 (Abcam, Cambridge, UK; ab28364; 1:500), mouse monoclonal anti-phospho-histone H2A.X-pSer139) (clone JBW301; Millipore, Billerica, MA, USA; 05-636; 1:600), rabbit polyclonal anti-cleaved caspase-3 (Asp175) (Cell Signalling Technology, Danvers, MA, USA; no. 9661; 1:300) and rabbit polyclonal anti-trophoblast-specific protein-α (Abcam; ab104401; 1:1500) were incubated at room temperature for 120 min. BrightVision Poly-HRP-Anti Rabbit IgG Biotin-free (Immunologic, VWR International, Radnor, PA, USA; DPVR-110HRP) was used as a secondary antibody. Antigen–antibody complexes were revealed with 3-3′-diaminobenzidine (Dako; K3468), with the same time exposure per antibody (5 min). Sections were counterstained with hematoxylin (Dako; S202084) and mounted with a mounting medium, toluene-free (CS705; Dako), using a CoverStainer (Dako). Specificity of staining was confirmed by omission of the primary antibody. Slides were imaged using a Olympus DP25 camera (Olympus, Tokyo, Japan) mounted on a Olympus BX45 microscope (Olympus) or using a E600 Nikon epifluorescence microscope (Nikon, Tokyo, Japan) equipped with a x40 NA 0.75 objective lens and a charge-coupled Olympus DP72 device camera (Olympus).
Histopathology
Embryos and placentas were harvested and fixed in 4% paraformaldehyde overnight at room temperature. Representative sections were trimmed, processed, embedded in paraffin, blocked, sectioned and stained with H&E and periodic acid/Schiff reagent. Stained slides were examined microscopically, unbiased by two experienced board-certified pathologists (SAY, AdB). The different types of trophoblastic cell lineage (i.e. labyrinth trophoblast, syncytiotrophoblast, giant cell trophoblast and trophospongium) were identified based on the characteristic histologic features of every cell. The numbers and/or size of a particular cell from the trophoblastic cell lineage were compared between different genotypes and scored by an ordinal semiquantitative system as described,48 where normal=no change in the numbers/size from control tissue, mild=decreased between 10 and 25%, moderate=decreased between 25 and 50% and severe=the number and/or size of a particular cell type is decreased >50% when compared with control tissue.
Isolation of RNA and quantitative real-time PCR
Dissected tissue or primary MEFs were collected on ice, washed in PBS and snap frozen on dry ice. Tissues were disrupted in Tri-Reagent (Sigma-Aldrich) by zirconium beads in a mechanical tissue disruptor (Precellys 24; Bertin Technologies, Montigny-le-Bretonneux, France). RNA was isolated by centrifugation followed by chloroform extraction, isopropanol precipitation, washing two times in 75% ethanol and resuspension in DEPC-treated water. Nucleic acid quantification was performed with a Nanodrop 8000 Instrument (Thermo Fisher Scientific). Reverse transcription reaction was carried out using High Capacity RNA-to-cDNA Kit (Applied Biosystems, Thermo Fisher Scientific, Waltham, MA, USA), following the manufacturer's instructions, in a reaction volume of 20 μl and with the primers contained in the 2 × RT buffer mix, which is composed of dNTPs, random octamers, and oligo(dT)16 and cDNA was stored at −20 °C. qPCR was performed using the comparative CT method and a Step-One-Plus Real-Time PCR Instrument (Applied Biosystems). SYBR Green and Taqman reactions were carried out in technical triplicate in a final volume of 10 μl. For SYBR Green 1 × SYBR Green PCR Master Mix (no. 4364344; Applied Biosystems), forward and reverse primer (Sigma-Aldrich; 100 nM each) and 25 ng of template were used. For TaqMan reactions 1 × TaqMan Universal PCR Master Mix (no. 4324018; Applied Biosystems), 0.5 μl of probe and 25 ng of template were used. Thermocycling parameters used for SYBR Green reactions were: 95 °C 20 s; 40 cycles 95 °C 3 s, 60 °C 30 s; melting curve and for TaqMan reactions were: 95 °C 20 s; 40 cycles 95 °C 1 s, 60 °C 20 s. For TaqMan assays, mACTB probe was used as an endogenous control for normalization and for SYBR Green assays; primers for 18 S were used as endogenous controls for normalization. The 2−ddCT method was used for the analysis of the amplification products. Primer pairs used are indicated in Supplementary Table S7. Data are presented as the means±S.D. of at least three experiments, with at least three animals within each experimental group. Statistical significance was determined using an unpaired t-test (***P⩽0.0001, **P⩽0.001, *P⩽0.05, NS=nonsignificant).
Affinity purification of TLK2 and quantitative mass spectrometry
The human TLK2 cDNA was obtained in a Gateway-compatible pENTR223 vector (Sigma-Aldrich Mission cDNA library) and introduced into the N-SF-TAP-DEST vector (a kind gift from J Gloeckner)49 by recombination reaction (300 ng TLK2-pENTR223, 300 ng N-SF-TAP-DEST, 4 μl Gateway buffer (Invitrogen, Carlsbad, CA, USA) and 4 μl clonase LR (Invitrogen)) and incubated at 25 °C for 1 h. Proteinase K (2 μl) (Sigma-Aldrich) was added and incubated at 37 °C for 10 min to stop the reaction. Competent Escherichia coli DH5α (Sigma-Aldrich) were transformed with the resulting recombination reaction and selected with kanamycin. Plasmids were checked by restriction digests and sequencing (Macrogen, Amsterdam, the Netherlands) and DNA prepared with MaxiPreps (Promega, Madison, WI, USA). The kinase dead mutation (D592V) was introduced using QuickChange Lightning (Agilent Technologies, Santa Clara, CA, USA).
HEK293T cells were grown in DMEM (Life Technologies) supplemented with 10% FBS and penicillin–streptomycin at 37 °C in a 5% CO2 incubator. Transient transfections of 15 μg plasmid per 15 cm plate were carried out using polyethylenimine (PEI) (Polysciences Inc., Warminster, PA, USA). Cells were harvested 48 h after transfection and collected by centrifugation. Pellets were lysed in 1 ml lysis buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% Tween-20, 0.5% NP-40 with 1x protease inhibitor cocktail (Roche, Basel, Switzerland) and 1x phosphatase inhibitor cocktail (Sigma-Aldrich)) on ice for 20 min. Cells were freeze-thawed three times and lysates were cleared by centrifugation at 16,000 × g at 4 °C for 20 min. Proteins were quantified using the DC Protein Assay (Bio-Rad, Hercules, CA, USA) and 100 μl was retained for input. Protein extracts were incubated with 100 μl prewashed streptactin superflow resin (IBA GmbH, Göttingen, Germany) at 4 °C for 2 h using an overhead tumbler. Resin was spun down (7000 × g for 30 s) and transferred to microspin columns (GE Healthcare, Little Chalfont, UK). Resin was washed three times with 500 μl washing buffer (30 mM Tris, pH 7.4, 150 mM NaCl, 0.1% NP-40, protease and phosphatase inhibitors). Fusion proteins were eluted from the Strep-Tactin matrix with 500 μl of desthiobiotin elution buffer (IBA Gmbh) (30 mM Tris, pH 7.4, 150 mM NaCl, 1 mM EDTA, 2 mM desthiobiotin, protease and phosphatase inhibitors) on ice for 10 min. One hundred and fifty microliter was retained for western blot analysis and the remaining protein was concentrated in centrifugal filter units and taken to a final concentration of 6 M urea and 100 mM ammonium bicarbonate. Then they were reduced, alkylated and digested to peptides using sequence-grade trypsin at 1:10 ratio (w : w; enzyme : substrate) at 37 °C overnight. Tryptic peptide mixtures were desalted using a C18 UltraMicroSpin (Thermo Fisher Scientific) column.50
Samples were analyzed in a LTQ-Orbitrap Velos Pro Mass Spectrometer (Thermo Fisher Scientific) coupled to nano-LC (Proxeon, Thermo Fisher Scientific, Waltham, MA, USA) equipped with a reversed-phase chromatography 12-cm column with an inner diameter of 75 μm, packed with 5 μm C18 particles (Nikkyo Technos Co. Ltd, Tokyo, Japan). Chromatographic gradients from 93% buffer A, 7% buffer B to 65% buffer A, 35% buffer B in 60 min at a flow rate of 300 nl/min, in which buffer A: 0.1% formic acid in water and buffer B: 0.1% formic acid in acetonitrile. The instrument was operated in DDA mode and full MS scans with 1 micro-scans at resolution of 60 000 were used over a mass range of m/z 250–2000 with detection in the Orbitrap. Following each survey scan the top 20 most intense ions with multiple charged ions above a threshold ion count of 5000 were selected for fragmentation at normalized collision energy of 35%. Fragment ion spectra produced via collision-induced dissociation (CID) were acquired in the linear ion trap. All data were acquired with Xcalibur Software v.2.2 (Thermo Fisher Scientific, Waltham, MA, USA).
Acquired data were analyzed using the Proteome Discoverer Software suite (v.1.3.0.339; Thermo Fisher Scientific) and the Mascot search engine (v.2.3; Matrix Science, London, UK) were used for peptide identification. Data were searched against an in-house generated database containing all proteins corresponding to human in the SwissProt database plus the most common contaminants, as described previously.51 A precursor ion mass tolerance of 7 p.p.m. at the MS1 level was used, and up to three miscleavages for trypsin were allowed. The fragment ion mass tolerance was set to 0.5 Da. Oxidation of methionine and protein acetylation at the N-terminal were defined as variable modification. Carbamidomethylation on cysteines was set as a fix modification. The identified peptides were filtered using a FDR <1%. For protein quantitation, the top 3 method was used.52 In brief, the average of the peptide areas for the three most intense peptides per protein was used a measure of protein abundance. Additional data is presented in Supplementary Tables S1–S4.
BioID analysis of proximity interactions
Human TLK2 was amplified with forward primer containing 5′ AscI (TLK2-AscI-F: 5′-AAAAAAGGCGCGCCTATGGAAGAATTGCATAGCC-3′) or reverse primer containing 3′ NotI restriction sites (TLK2-NotI-R: 5′-AAAAAAGCGGCCGCTTAATTAGAAGAACTGTT-3′) using KOD Hot Start DNA Polymerase (Millipore) and cycling conditions recommended from the manufacturer (polymerase activation at 95 °C for 2 min, denaturation at 95 °C for 20 s, annealing at 55 °C for 10 s and extension at 70 °C for 50 s, repeated for 40 cycles). PCR products were purified using the PureLink Quick Gel Extraction Kit (Invitrogen) and cloned into pCR2.1-TOPO vector (Invitrogen). Top 10 competent E. coli cells (Invitrogen) were transformed with pCR2.1-TLK2 and colonies were selected in carbenicillin. Constructs were verified by restriction digestion and sequencing (Macrogen) with primers for the TOPO vector (T7 Promoter-F and M13-R). Afterwards, TLK2 was cut from the pCR2.1-TOPO vector by restriction digest with AscI (New England BioLabs (NEB), Ipswich, MA, USA) and NotI-HF (NEB), purified using the PureLink Quick Gel Extraction Kit (Invitrogen) and ligated into pcDNA5/FRT/TO-N-FLAG-hBirA* (TLK2) using Quick Ligation Kit (NEB). Top 10 competent E. coli cells (Invitrogen) were transformed with pcDNA5/FRT/TO-N-Flag-hBirA*-TLK2 vector and carbenicillin selected. The constructs were confirmed by restriction digestion with AscI (NEB) and NotI-HF (NEB) and sequencing (Macrogen).
AD293 cells were seeded and transfected the next day with either pcDNA5/FRT/TO-N-FLAG-hBirA*-TLK2 or pcDNA5/FRT/TO-N-FLAG-hBirA* using PEI (Sigma-Aldrich)±50 μM biotin (IBA GmbH; 2-1016-002). For mass spectrometry, 5x15 cm2 plates per condition were harvested 24 h after transfection by scraping cells into PBS, washing two times in PBS and snap freezing on dry ice. Cell pellets were lysed in 5 ml modified RIPA buffer (1% TX-100, 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA, 0.1% SDS, 0.5% sodium deoxycholate and protease inhibitors) on ice, treated with 250 U benzonase (Millipore) and biotinylated proteins were isolated using streptavidin-sepharose beads (GE Healthcare). Proteins were washed in ammonium bicarbonate and digested with trypsin. Mass spectrometry was performed as described previously.53 Additional data are presented in Supplementary Tables S5 and S6.
Endogenous co-IP
Dissected placentas were collected on ice and disrupted in Extraction Buffer (300 mM NaCl, 0.5% NP-40, 50 mM Tris, pH 7.8, 0.2 mM EDTA, 5% glycerol, 1x protease (Roche, Basel, Switzerland) and phosphatase inhibitors (Sigma-Aldrich)) by zirconium beads in a mechanical tissue disruptor (Precellys 24; Bertin Technologies). After two centrifugation steps (10 min, max speed, 4 °C and 5 min, max speed, 4 °C), supernatants were collected, precleared and immunoprecipitated (overnight, 4 °C, on rotation) with 10 μg of anti-TLK2 antibody (Bethyl Laboratories, Montgomery, TX, USA; A301-257A), 10 μl of polyclonal ASF1 serum (generated in Anja Groth's Laboratory, BRIC, Copenhagen) or 10 μg of rabbit IgG (Sigma-Aldrich) bound to 50 μl of Dynabeads Protein G (Invitrogen, Carlsbad, CA, USA). Beads were washed five times with cold Wash Buffer (150 mM NaCl, 0.2% NP-40, 50 mM Tris, pH 7.8, 0.2 mM EDTA, 5% glycerol, 1x protease (Roche) and phosphatase inhibitors (Sigma-Aldrich)), resuspended in 30 μl of Laemmli sample buffer and boiled for 20 min.
Protein extraction for western blotting
Dissected tissues or cells were collected on ice, washed in PBS and resuspended in TNG-150 buffer (50 mM Tris-HCl, 150 mM NaCl, 1% Tween-20, 0.5% NP-40, 1x protease (Roche) and phosphatase inhibitors (Sigma-Aldrich)). Tissues were disrupted by zirconium beads in a mechanical tissue disruptor (Precellys 24; Bertin Technologies). Protein concentration was quantified using the DC Protein Assay (Bio-Rad), separated by SDS-PAGE and transferred to PVDF membrane (Millipore). Membranes were probed with antibodies for TLK1 (Cell Signaling; 4125; 1:1000), TLK2 (Bethyl Laboratories; A301-257A; 1:1000), ASF1 (Sigma-Aldrich, A5236, 1:800, for HEK-293 T human cells; Groth Laboratory, rabbit polyclonal, 1:2000, for mouse tissue and MEFs),14 actin (Sigma-Aldrich; A4700; 1:3000), FLAG (Sigma-Aldrich; F3165; 1:1000), LC8 (Abcam; ab51603; 1:1000), GAPDH (Millipore; MAB374; 1:10 000) and p-ASF1-S166 (Groth Laboratory; rabbit polyclonal, 1:30 000).11 Primary antibodies were detected with appropriate secondary antibodies conjugated to HRP and visualized by ECL-Plus (GE Healthcare).
Analysis of genomic instability
For colony formation assays, cells were seeded in duplicate on 6-cm plates and the number of seeded cells were adjusted to the plating efficiency. Cells were incubated at 37 °C for 10–14 days and colonies stained with crystal violet (Sigma-Aldrich) and counted using ImageJ-plugin Trainable Weka Segmentation (NIH, Bethesda, MD, USA). For immunofluorescence and high-throughput microscopy (HTM), MEFs were grown in LabTek II Chamber slides (Nunc, Thermo Fisher Scientific). Cells were pre-extracted for 5 min in 0.2% TX-100/PBS on ice and fixed for 10 min in 4% paraformaldehyde at room temperature. Fixed cells were blocked in 3% BSA with 0.1%Tween/PBS for 1 h and stained using standard procedures. For HTM, 48 images were automatically acquired per well with a Scan^R (Olympus) at × 40 magnification and non-saturating conditions. Images were segmented using the DAPI staining to generate nuclei masks from which the corresponding signals were calculated using an in-house-developed package based on the Cell Profiler. For immunofluorescence analysis the antibody anti-γH2AX (sc-101696; Santa Cruz Biotechnology, Dallas, TX, USA) and the corresponding secondary antibody conjugated with Alexa Fluor 568 were used. For metaphase spread preparations, cells were treated with colcemid (0.1 g/ml for 2 h). Cells were trypsinized, hypotonically swollen in 0.075 M KCl for 20 min at 37 °C and fixed (75% MeOH and 25% acetic acid, ice cold). Metaphase preparations were spread on glass slides and stained with DAPI in Vectashield mounting medium. Images were taken using a Leica DM6000 microscope (Leica, Wetzlar, Germany) and analyzed using Fiji Software (https://fiji.sc/).
Acknowledgments
We are grateful to the Stracker lab, I del Barco, A Igea, J Roig and C Gekas for helpful input; A Nebreda for critical reading of the manuscript; R Medema for sharing unpublished results; S Aivio for animal maintenance; D Reverter and P Gallego Alonso for purified LC8 protein; J Gloeckner for SF-TAP vectors; the IRB Histopathology facility and N Prats, M Aguilera, B Domínguez, E Llonch and N Vasconcelos for assistance; the IRB Advanced Digital Microscopy Core Facility for use of the Lightsheet Macroscope; L Bardia, M Marsal and S Tosi for sample preparation and image analysis; and E Sabido for technical advice and mass spectrometry measurements. Analysis was performed in the CRG/UPF Proteomics Unit, part of the 'Plataforma de Recursos Biomoleculares y Bioinformàticos' (ProteoRed-Instituto de Salud Carlos III, PT13/0001). THS was a Ramon y Cajal scholar and supported by the Ministerio de Economía y Competitividad (MINECO) (BFU2012-39521, BFU2015-68354, Ayudas para incentivar la incorporación estable de doctores (IED) 2015) and institutional funding from MINECO through the Centres of Excellence Severo Ochoa award and from the CERCA Programme of the Catalan Government. AG is an EMBO Young Investigator, and supported by the Danish National Research Foundation to the Center for Epigenetics (DNRF82), the European Commission ITN FP7 'aDDRess', a European Research Council Starting Grant (ERC2011StG, no. 281 765), the Danish Cancer Society, the Danish Medical Research Council, the Novo Nordisk Foundation and the Lundbeck Foundation. SSB and HG were supported by PhD fellowships from Fundacio La Caixa, PAK was supported by an Advanced Postdoc Mobility fellowship from the Swiss National Science Foundation and the Kurt und Senta Herrmann Stiftung and KR was supported by an FPI fellowship (MINECO).
Author contributions
SSB, PAK and HG designed, performed and analyzed most of the experiments; HG and AP carried out IP-MS experiments; PAK, EC and BR carried out BioID experiments; SAY and AdB assessed placental pathology; KR performed experiments; LP and PAK maintained mouse cohorts; JC performed lightsheet imaging; AG, TLR and SF generated and provided critical reagents and experimental advice; and THS designed and analyzed experiments and wrote the manuscript.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd)
Edited by X Lu
The authors declare no conflict of interest.
Supplementary Material
References
- Roe JL, Rivin CJ, Sessions RA, Feldmann KA, Zambryski PC. The Tousled gene in A. thaliana encodes a protein kinase homolog that is required for leaf and flower development. Cell 1993; 75: 939–950. [DOI] [PubMed] [Google Scholar]
- Sillje HH, Takahashi K, Tanaka K, Van Houwe G, Nigg EA. Mammalian homologues of the plant Tousled gene code for cell-cycle-regulated kinases with maximal activities linked to ongoing DNA replication. EMBO J 1999; 18: 5691–5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sillje HH, Nigg EA. Identification of human Asf1 chromatin assembly factors as substrates of Tousled-like kinases. Curr Biol 2001; 11: 1068–1073. [DOI] [PubMed] [Google Scholar]
- Li Y, DeFatta R, Anthony C, Sunavala G, De Benedetti A. A translationally regulated Tousled kinase phosphorylates histone H3 and confers radioresistance when overexpressed. Oncogene 2001; 20: 726–738. [DOI] [PubMed] [Google Scholar]
- Delacroix S, Wagner JM, Kobayashi M, Yamamoto K, Karnitz LM. The Rad9-Hus1-Rad1 (9-1-1) clamp activates checkpoint signaling via TopBP1. Genes Dev 2007; 21: 1472–1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalom S, Don J. Tlk, a novel evolutionarily conserved murine serine threonine kinase, encodes multiple testis transcripts. Mol Reprod Dev 1999; 52: 392–405. [DOI] [PubMed] [Google Scholar]
- Han Z, Riefler GM, Saam JR, Mango SE, Schumacher JM. The C. elegans Tousled-like kinase contributes to chromosome segregation as a substrate and regulator of the Aurora B kinase. Curr Biol 2005; 15: 894–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Z, Saam JR, Adams HP, Mango SE, Schumacher JM. The C. elegans Tousled-like kinase (TLK-1) has an essential role in transcription. Curr Biol 2003; 13: 1921–1929. [DOI] [PubMed] [Google Scholar]
- Dillon PJ, Gregory SM, Tamburro K, Sanders MK, Johnson GL, Raab-Traub N et al. Tousled-like kinases modulate reactivation of gammaherpesviruses from latency. Cell Host Microbe 2013; 13: 204–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrera P, Moshkin YM, Gronke S, Sillje HH, Nigg EA, Jackle H et al. Tousled-like kinase functions with the chromatin assembly pathway regulating nuclear divisions. Genes Dev 2003; 17: 2578–2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimovskaia IM, Young C, Stromme CB, Menard P, Jasencakova Z, Mejlvang J et al. Tousled-like kinases phosphorylate Asf1 to promote histone supply during DNA replication. Nat Commun 2014; 5: 3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilyugin M, Demmers J, Verrijzer CP, Karch F, Moshkin YM. Phosphorylation-mediated control of histone chaperone ASF1 levels by Tousled-like kinases. PLoS ONE 2009; 4: e8328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groth A, Lukas J, Nigg EA, Sillje HH, Wernstedt C, Bartek J et al. Human Tousled like kinases are targeted by an ATM- and Chk1-dependent DNA damage checkpoint. EMBO J 2003; 22: 1676–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause DR, Jonnalagadda JC, Gatei MH, Sillje HH, Zhou BB, Nigg EA et al. Suppression of Tousled-like kinase activity after DNA damage or replication block requires ATM, NBS1 and Chk1. Oncogene 2003; 22: 5927–5937. [DOI] [PubMed] [Google Scholar]
- Kelly R, Davey SK. Tousled-like kinase-dependent phosphorylation of Rad9 plays a role in cell cycle progression and G2/M checkpoint exit. PLoS ONE 2013; 8: e85859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunavala-Dossabhoy G, De Benedetti A. Tousled homolog, TLK1, binds and phosphorylates Rad9; TLK1 acts as a molecular chaperone in DNA repair. DNA Repair (Amst) 2009; 8: 87–102. [DOI] [PubMed] [Google Scholar]
- Abreu CM, Kumar R, Hamilton D, Dawdy AW, Creavin K, Eivers S et al. Site-specific phosphorylation of the DNA damage response mediator rad9 by cyclin-dependent kinases regulates activation of checkpoint kinase 1. PLoS Genet 2013; 9: e1003310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruinsma W, van den Berg J, Aprelia M, Medema RH. Tousled-like kinase 2 regulates recovery from a DNA damage-induced G2 arrest. EMBO Rep 2016; 17: 659–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JA, Tan Y, Wang X, Cao X, Veeraraghavan J, Liang Y et al. Comprehensive functional analysis of the tousled-like kinase 2 frequently amplified in aggressive luminal breast cancers. Nat Commun 2016; 7: 12991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JA, Anurag M, Veeraraghavan J, Schiff R, Li K, Wang X. Amplification of TLK2 induces genomic instability via impairing the G2/M checkpoint. Mol Cancer Res 2016; 14: 920–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunavala-Dossabhoy G, Balakrishnan SK, Sen S, Nuthalapaty S, De Benedetti A. The radioresistance kinase TLK1B protects the cells by promoting repair of double strand breaks. BMC. Mol Biol 2005; 6: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen SP, De Benedetti A. TLK1B promotes repair of UV-damaged DNA through chromatin remodeling by Asf1. BMC. Mol Biol 2006; 7: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canfield C, Rains J, De Benedetti A. TLK1B promotes repair of DSBs via its interaction with Rad9 and Asf1. BMC. Mol Biol 2009; 10: 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronald S, Sunavala-Dossabhoy G, Adams L, Williams B, De Benedetti A. The expression of Tousled kinases in CaP cell lines and its relation to radiation response and DSB repair. Prostate 2011; 71: 1367–1373. [DOI] [PubMed] [Google Scholar]
- Cross JC. Genetic insights into trophoblast differentiation and placental morphogenesis. Semin Cell Dev Biol 2000; 11: 105–113. [DOI] [PubMed] [Google Scholar]
- Simmons DG, Cross JC. Determinants of trophoblast lineage and cell subtype specification in the mouse placenta. Dev Biol 2005; 284: 12–24. [DOI] [PubMed] [Google Scholar]
- Abascal F, Corpet A, Gurard-Levin ZA, Juan D, Ochsenbein F, Rico D et al. Subfunctionalization via adaptive evolution influenced by genomic context: the case of histone chaperones ASF1a and ASF1b. Mol Biol Evol 2013; 30: 1853–1866. [DOI] [PubMed] [Google Scholar]
- Simmons DG, Fortier AL, Cross JC. Diverse subtypes and developmental origins of trophoblast giant cells in the mouse placenta. Dev Biol 2007; 304: 567–578. [DOI] [PubMed] [Google Scholar]
- Cleaton MA, Edwards CA, Ferguson-Smith AC. Phenotypic outcomes of imprinted gene models in mice: elucidation of pre- and postnatal functions of imprinted genes. Annu Rev Genomics Hum Genet 2014; 15: 93–126. [DOI] [PubMed] [Google Scholar]
- Singh V, Connelly ZM, Shen X, De Benedetti A. Identification of the proteome complement of human TLK1 reveals it binds and phosphorylates NEK1 regulating its activity. Cell Cycle 2017; 16: 915–926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi S, Lewis P, Pevny L, McMahon AP. Efficient gene modulation in mouse epiblast using a Sox2Cre transgenic mouse strain. Mech Dev 2002; 119(Suppl 1): S97–S101. [DOI] [PubMed] [Google Scholar]
- Kodym R, Mayerhofer T, Ortmann E. Purification and identification of a protein kinase activity modulated by ionizing radiation. Biochem Biophys Res Commun 2004; 313: 97–103. [DOI] [PubMed] [Google Scholar]
- Sunavala-Dossabhoy G, Li Y, Williams B, De Benedetti A. A dominant negative mutant of TLK1 causes chromosome missegregation and aneuploidy in normal breast epithelial cells. BMC Cell Biol 2003; 4: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunavala-Dossabhoy G, Fowler M, De Benedetti A. Translation of the radioresistance kinase TLK1B is induced by gamma-irradiation through activation of mTOR and phosphorylation of 4E-BP1. BMC. Mol Biol 2004; 5: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Benedetti A. Tousled kinase TLK1B mediates chromatin assembly in conjunction with Asf1 regardless of its kinase activity. BMC Res Notes 2010; 3: 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Gourguechon S, Wang CC. Tousled-like kinase in a microbial eukaryote regulates spindle assembly and S-phase progression by interacting with Aurora kinase and chromatin assembly factors. J Cell Sci 2007; 120(Part 21): 3883–3894. [DOI] [PubMed] [Google Scholar]
- Petryszak R, Keays M, Tang YA, Fonseca NA, Barrera E, Burdett T et al. Expression Atlas update – an integrated database of gene and protein expression in humans, animals and plants. Nucleic Acids Res 2016; 44: D746–D752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartford SA, Luo Y, Southard TL, Min IM, Lis JT, Schimenti JC. Minichromosome maintenance helicase paralog MCM9 is dispensible for DNA replication but functions in germ-line stem cells and tumor suppression. Proc Natl Acad Sci USA 2011; 108: 17702–17707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messiaen S, Guiard J, Aigueperse C, Fliniaux I, Tourpin S, Barroca V et al. Loss of the histone chaperone ASF1B reduces female reproductive capacity in mice. Reproduction 2016; 151: 477–489. [DOI] [PubMed] [Google Scholar]
- Lelieveld SH, Reijnders MR, Pfundt R, Yntema HG, Kamsteeg EJ, de Vries P et al. Meta-analysis of 2,104 trios provides support for 10 new genes for intellectual disability. Nat Neurosci 2016; 19: 1194–1196. [DOI] [PubMed] [Google Scholar]
- McKnight NC, Jefferies HB, Alemu EA, Saunders RE, Howell M, Johansen T et al. Genome-wide siRNA screen reveals amino acid starvation-induced autophagy requires SCOC and WAC. EMBO J 2012; 31: 1931–1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertins P, Mani DR, Ruggles KV, Gillette MA, Clauser KR, Wang P et al. Proteogenomics connects somatic mutations to signalling in breast cancer. Nature 2016; 534: 55–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srihari S, Singla J, Wong L, Ragan MA. Inferring synthetic lethal interactions from mutual exclusivity of genetic events in cancer. Biol Direct 2015; 10: 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copeland NG, Jenkins NA, Court DL. Recombineering: a powerful new tool for mouse functional genomics. Nat Rev Genet 2001; 2: 769–779. [DOI] [PubMed] [Google Scholar]
- Wu Y, Wang C, Sun H, LeRoith D, Yakar S. High-efficient FLPo deleter mice in C57BL/6J background. PLoS ONE 2009; 4: e8054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzankina Y, Pinzon-Guzman C, Asare A, Ong T, Pontano L, Cotsarelis G et al. Deletion of the developmentally essential gene ATR in adult mice leads to age-related phenotypes and stem cell loss. Cell Stem Cell 2017; 1: 113–126.. [DOI] [PMC free article] [PubMed]
- Knobel PA, Belotserkovskaya R, Galanty Y, Schmidt CK, Jackson SP, Stracker TH. USP28 is recruited to sites of DNA damage by the tandem BRCT domains of 53BP1 but plays a minor role in double-strand break metabolism. Mol Cell Biol 2014; 34: 2062–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson-Corley KN, Olivier AK, Meyerholz DK. Principles for valid histopathologic scoring in research. Vet Pathol 2013; 50: 1007–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gloeckner CJ, Boldt K, Schumacher A, Roepman R, Ueffing M. A novel tandem affinity purification strategy for the efficient isolation and characterisation of native protein complexes. Proteomics 2007; 7: 4228–4234. [DOI] [PubMed] [Google Scholar]
- Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc 2007; 2: 1896–1906. [DOI] [PubMed] [Google Scholar]
- Bunkenborg J, Garcia GE, Paz MI, Andersen JS, Molina H. The minotaur proteome: avoiding cross-species identifications deriving from bovine serum in cell culture models. Proteomics 2010; 10: 3040–3044. [DOI] [PubMed] [Google Scholar]
- Silva JC, Gorenstein MV, Li GZ, Vissers JP, Geromanos SJ. Absolute quantification of proteins by LCMSE: a virtue of parallel MS acquisition. Mol Cell Proteomics 2006; 5: 144–156. [DOI] [PubMed] [Google Scholar]
- Coyaud E, Mis M, Laurent EM, Dunham WH, Couzens AL, Robitaille M et al. BioID-based identification of Skp Cullin F-box (SCF)beta-TrCP1/2 E3 ligase substrates. Mol Cell Proteomics 2015; 14: 1781–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.