To the Editor,
Malignant transformation proceeds as nascent neoplastic cells accumulate oncogenic lesions that confer the ‘hallmarks of cancer’, including self-sufficiency for growth signalling and resistance to anti-growth signals, such as cell senescence and apoptosis.1, 2
Exposure to DNA damaging agents can facilitate neoplastic transformation in mice3 and humans.4, 5 The transcription factor p53 plays a critical role in preventing tumorigenesis, including lymphoma development driven by γ-radiation.6, 7, 8 Apoptosis induction was thought to underpin p53-mediated tumour suppression, however recent findings using various gene targeted mice demonstrated that apoptosis induction (as well as cell cycle arrest and senescence) are dispensable for p53-mediated tumour suppression.9–11 These findings raise the question: can potentially oncogenic DNA lesions induce apoptosis through p53-independent tumour suppression pathways?
We described a role for BIM as a critical inducer of apoptosis (independently of p53) to prevent transformation of T-cell progenitors drive by oncogenic lesions arising from T-cell receptor gene rearrangement.12 To examine whether this BIM-mediated apoptotic pathway might be relevant in the context of tumorigenesis driven by DNA double-strand breaks more broadly, we took advantage of ATM-deficient mice, a model of the human syndrome ataxia telangiectasia, characterised by a high propensity for tumour development.13 This disease is caused by loss of ATM, a DNA damage sensing kinase, acting at the site of DNA lesions to initiate the DNA damage response.14 Interestingly lymphoma development in this model has also been shown to be RAG1/2-dependent.15
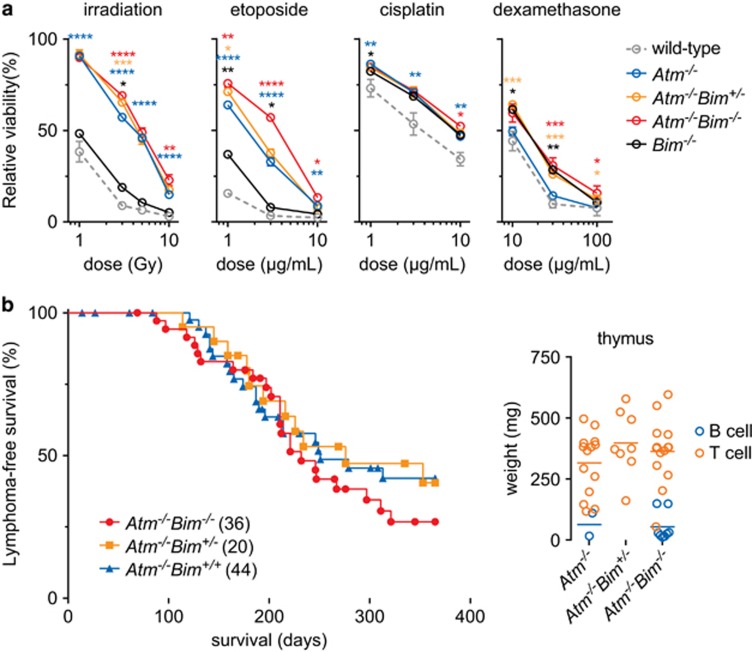
We found that thymocytes deficient for ATM are relatively insensitive to cell death induced by γ-irradiation or etoposide, as reported,16 with the additional loss of BIM conferring additional protection (Figure 1a). Loss of either ATM or BIM conferred resistance to cisplatin; while as expected17,18 only loss of BIM conferred resistance to the glucocorticoid dexamethasone (Figure 1a).
Figure 1.
Loss of BIM augments resistance of Atm−/− thymocytes to DNA damage-induced apoptosis, but does not impact lymphoma development. (a) Thymocytes survival was determined following treatment with the indicated agents relative to untreated cells at 24 h. n=3–9. (b) Mice were reconstituted with HSPCs and lymphoma-free survival was determined (median survival: Atm−/−Bim−/− 232d, Atm−/−Bim+/− 276d and Atm−/−Bim+/+ 251d). Thymus weight from the lymphoma-bearing animals and tumour immunophenotype was determined (donor origin was confirmed with congenic CD45 staining). n=8–22; tumour type mean shown. (a) *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001; black (wild-type vs Bim−/−), blue (wild-type vs Atm−/−), yellow (Atm−/− vs Atm−/−Bim+/−) and red (Atm−/− vs Atm−/−Bim−/−). Mean±S.E.M. depicted
Loss of ATM resulted in reduced circulating T cells,13 and this could be rescued by concomitant loss of BIM (Supplementary Figure 1). As reported,17 loss of BIM resulted in elevated blood T-cell numbers. Loss of neither BIM nor ATM altered erythrocyte numbers, while loss of ATM resulted in an elevation of platelet numbers, which was normalised by concomitant loss of BIM. ATM-deficiency reduced thymus cellularity and concomitant loss of BIM afforded partial rescue (Supplementary Figure 1).
To determine the impact of compound loss of BIM and ATM on lymphoma development, a haematopoietic reconstitution approach was adopted to obviate potentially confounding effects caused by ATM loss in other tissues.13 Thus, lethally irradiated wild-type mice were reconstituted with bone marrow from Atm−/−Bim−/−, Atm−/−Bim+/− or Atm−/−Bim+/+ mice, but no differences in the onset, rate or severity of lymphoma development were observed between the different genotypes (CD45 congenic markers were used to confirm donor origin of all lymphomas; Figure 1b).
We conclude that while loss of BIM confers additional resistance to DNA damage-induced apoptosis and can ameliorate certain haematopoietic defects caused by loss of ATM, it does not significantly impact lymphoma development in this disease setting.
Acknowledgments
We thank Drs P Bouillet and M Lavin for gifts of mice; C Gatt, S O’Connor, J Mansheim, K McKenzie and G Siciliano for expert animal care; B Helbert for genotyping; J Corbin and J McManus for automated blood analysis. This work was supported by grants and fellowships from the Cancer Council of Victoria (SG, AD ‘Sydney Parker Smith Postdoctoral Fellowship’), Leukaemia Foundation Australia (SG), the Lady Tata Memorial Trust (SG), Cure Brain Cancer Australia (AS), the National Health and Medical Research Council (Program Grants #1016701 and 1016647, NHMRC Australia Fellowship 1020363 to AS), the Leukemia and Lymphoma Society (SOCR Grant #7001-03 to AS), Melbourne International Research and the Melbourne International Fee Remission Scholarship (University of Melbourne, SG) and Cancer Therapeutics CRC Top-up Scholarship (SG, AD). The estate of Anthony (Toni) Redstone OAM, University of Melbourne International Research and International Fee Remission Scholarships (SG), Australian Postgraduate Award (ARDD), and the operational infrastructure grants through the Australian Government IRIISS and the Victorian State Government OIS.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Differentiation website (http://www.nature.com/cdd).
The authors declare no conflict of interest.
Supplementary Material
References
- Hanahan D et al. Cell 2011; 144(5): 646–74. [DOI] [PubMed]
- Delbridge AR et al. Cold Spring Harb Perspect Biol 2012; 4: a008789. [DOI] [PMC free article] [PubMed]
- Kaplan HS. Natl Cancer Inst Monogr 1964; 14: 207–20. [PubMed]
- Richardson D et al. Radiat Res 2009; 172(3): 368–82. [DOI] [PubMed]
- Krestinina L et al. Radiat Environ Biophys 2010; 49(2): 195–201. [DOI] [PMC free article] [PubMed]
- Kemp CJ et al. Nat Genet 1994; 8: 66–9. [DOI] [PubMed]
- Donehower LA et al. Nature 1992; 356: 215–21. [DOI] [PubMed]
- Jacks T et al. Curr Biol 1994; 4(1): 1–7. [DOI] [PubMed]
- Brady CA et al. Cell 2011; 145(4): 571–583. [DOI] [PMC free article] [PubMed]
- Li T et al. Cell 2012; 149(6): 1269–1283. [DOI] [PMC free article] [PubMed]
- Valente LJ et al. Cell Rep 2013; 3(5): 1339–1345. [DOI] [PubMed]
- Delbridge AR et al. J Exp Med 2016; 213(10): 2039–2048. [DOI] [PMC free article] [PubMed]
- Barlow C et al. Cell 1996; 86(1): 159–171. [DOI] [PubMed]
- Lavin MF et al. Biomolecules 2015; 5(4): 2877–2902. [DOI] [PMC free article] [PubMed]
- Liao M-J et al. Genes Dev 1999; 13(10): 1246–1250. [DOI] [PMC free article] [PubMed]
- Biswas S et al. Cell Death Differ 2013; 20(7): 869–877. [DOI] [PMC free article] [PubMed]
- Bouillet P et al. Science 1999; 286(5445): 1735–1738. [DOI] [PubMed]
- Erlacher M et al. Blood 2005; 106(3): 4131–4138. [DOI] [PMC free article] [PubMed]
- Delbridge AR et al. Blood 2015; 125(21): 3273–3280. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.