Abstract
Gene regulatory networks (GRN) are central to developmental processes. They are composed of transcription factors and signaling molecules orchestrating gene expression modules that tightly regulate the development of organisms. The neural crest (NC) is a multipotent cell population that is considered a key innovation of vertebrates. Its derivatives contribute to shaping the astounding morphological diversity of jaws, teeth, head skeleton, or pigmentation. Here, we study the molecular evolution of the NC GRN by analyzing patterns of molecular divergence for a total of 36 genes in 16 species of bony fishes. Analyses of nonsynonymous to synonymous substitution rate ratios (dN/dS) support patterns of variable selective pressures among genes deployed at different stages of NC development, consistent with the developmental hourglass model. Model-based clustering techniques of sequence features support the notion of extreme conservation of NC-genes across the entire network. Our data show that most genes are under strong purifying selection that is maintained throughout ray-finned fish evolution. Late NC development genes reveal a pattern of increased constraints in more recent lineages. Additionally, seven of the NC-genes showed signs of relaxation of purifying selection in the famously species-rich lineage of cichlid fishes. This suggests that NC genes might have played a role in the adaptive radiation of cichlids by granting flexibility in the development of NC-derived traits—suggesting an important role for NC network architecture during the diversification in vertebrates.
Keywords: Actinopterygii, teleost, development, ontogeny, gene regulatory network, cichlid, NC-genes, hourglass model
Introduction
The vertebrate-specific neural crest (NC) is a transient population of migrating multipotent cells that gives rise to various, highly diverse, tissues and cell types (fig. 1A). The NC is considered a key innovation in vertebrate evolution (Gans and Northcutt 1983; Shimeld and Holland 2000). Due to its broad developmental potential, the NC has been even referred to as the fourth germ layer (Hall 2000). Its derivatives provide the developmental basis for the vast morphological and physiological diversification that has occurred during vertebrate evolution (Betancur et al. 2010; Bronner and LeDouarin 2012; Simões-Costa and Bronner 2015). NC cells contribute to evolutionary novelties, such as the cranium, the branchial skeleton, and peripheral nervous system (Shimeld and Holland 2000; Santagati and Rijli 2003). Many of the distinctive phenotypic differences found in bony fishes are largely influenced by the NC cell lineage (Kimmel et al. 2001; Donoghue et al. 2008; Braasch et al. 2009). Most prominently, NC-derived mesenchyme shapes the astonishing diversity of head and jaw morphologies. Also, pigment cells are NC-derived, and populate the whole body axis to generate the extraordinary diversity of coloration and pigmentation patterns found in the most species-rich vertebrate lineage, the ray-finned fish (Actinopterygii) (Sugie et al. 2004; Salzburger et al. 2007; Braasch et al. 2009; Kelsh and Barsh 2011; Renz et al. 2011; Roberts et al. 2011; Powder et al. 2014; Kratochwil et al. 2015). However, up to now, surprisingly little is known about the patterns of molecular evolution of the NC of actinopterygians.
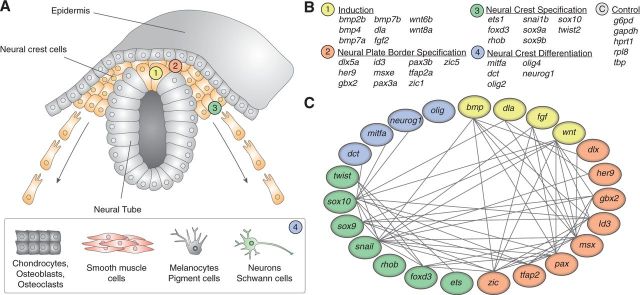
Fig. 1.—
Classification of neural crest-related genes in respect to developmental expression subunits. (A) Thirty-one genes from the NC GRN are classified into four groups depending on their deployment during development, after Betancur et al. (2010): Genes responsible for early induction of future NC (1), genes defining the NPB that lines the neural plate before neurulation (2), specifiers of the NC itself as well as regulators of their migration (3), and genes determining the fate of migrated multipotent NC cells (4). (B) Genes in this study sorted by developmental subunit. Five housekeeping genes (group C) were added to the analyses as a control group. (C) Regulatory connections between network genes without distinction regarding directionality and mode of interaction.
To get a better grasp on the molecular evolution of the NC regulatory network, it is important to know during what ontogenetic phase and how the specific genes of the GRN act and interact. NC genes have been intensively studied in model systems, such as the zebrafish (Betancur et al. 2010; Bronner and LeDouarin 2012; Simões-Costa and Bronner 2015). The aspects of development that are NC-specific are initiated during gastrulation. Cells of the NC first start to appear at the interface of neural plate and surface ectoderm, the so-called neural plate border (NPB) (fig. 1A) (Knecht and Bronner-Fraser 2002; Basch et al. 2006). When the neural tube invaginates and closes during neurulation, the NC is located at its dorsal side (Mayanil 2013). Subsequently, NC cells begin to migrate along predefined paths to colonize various parts of the body (Kulesa and Gammill 2010). Here, the final differentiation is taking place (fig. 1A) (Simões-Costa and Bronner 2015). Induction, specification, and differentiation of NC cells are controlled by a gene regulatory network (GRN) (Sauka-Spengler and Bronner-Fraser 2008). During the last decades, both the genetic components and the regulatory interactions of NC GRN genes have been intensively studied in various vertebrate model organisms (Betancur et al. 2010; Minoux et al. 2013; Simões-Costa and Bronner 2015).
Alterations in GRNs can result in phenotypic and physiological changes (Crombach and Hogeweg 2008; Van Otterloo et al. 2013). To understand the underlying evolutionary dynamics of GRNs, biochemical pathways (Rausher et al. 1999; Lu and Rausher 2003), signal transduction pathways (Riley et al. 2003; Alvarez-Ponce et al. 2009), and developmental pathways (Davila-Velderrain et al. 2014) have been previously analyzed by examining patterns of sequence variation within an evolutionary framework. However, a consensus on general patterns of developmental GRN evolution is still lacking. The topology of GRNs (i.e., the arrangement and connectivity of the GRN elements) has been suggested to influence the rates of molecular evolution within the different members of the network (Cork and Purugganan 2004). Additionally, developmental constraints shape the degree of sequence conservation in developmental GRN. The sequence conservation has been suggested to be higher either during early development (early conservation model or funnel-like model) or at the phylotypic stage, a time point during midembryogenesis when the embryo most closely resembles that of other species (hourglass model)—both models have been controversially discussed in the recent years (Prud’homme and Gompel 2010; Kalinka and Tomancak 2012; Piasecka et al. 2013).
The range of taxa analyzed in this study covers species that diverged between approximately 4 Ma—for the most recently diverged cichlid fishes—and approximately 380 Ma—for the divergence between the coelacanth and ray-finned fish (Amemiya et al. 2013). Furthermore, to study the role of NC genes in cases of extreme phenotypic divergence and adaptive radiations, we included six cichlid species (family Cichlidae) for which genomes have recently become available (Brawand et al. 2014; Elmer et al. 2014). Cichlids have undergone extremely rapid speciation and phenotypic diversification that is often associated with NC-derived traits, such as craniofacial morphology and coloration. This is especially true for cichlid species that arose in the African Great Lakes (Neolamprologus brichardi, Astatotilapia burtoni, Metriaclima zebra, and Pundamilia nyererei) that are one of the best examples for adaptive radiation and fast phenotypic diversification in vertebrates (Turner 2007; Brawand et al. 2014; Henning and Meyer 2014).
The aim of our study was to ascertain whether sequence conservation among the components of the NC GRN is linked to network topology or developmental sequence and how patterns of natural selection might have changed during evolution. Here, we studied the molecular evolution of genes, most of which are transcription factors and guidance molecules (fig. 1) that are part of the NC GRN. We analyzed their molecular divergence in 16 species of bony fishes: 15 actinopterygians (ray-finned fish) and 1 sarcopterygian (lobe-finned fish) that we used as outgroup (fig. 2). Furthermore, we looked for evidence of positive selection or relaxation of purifying selection in these genes among the species-rich and phenotypically diverse clade of African cichlids, where five genomes have become recently available (Brawand et al. 2014).
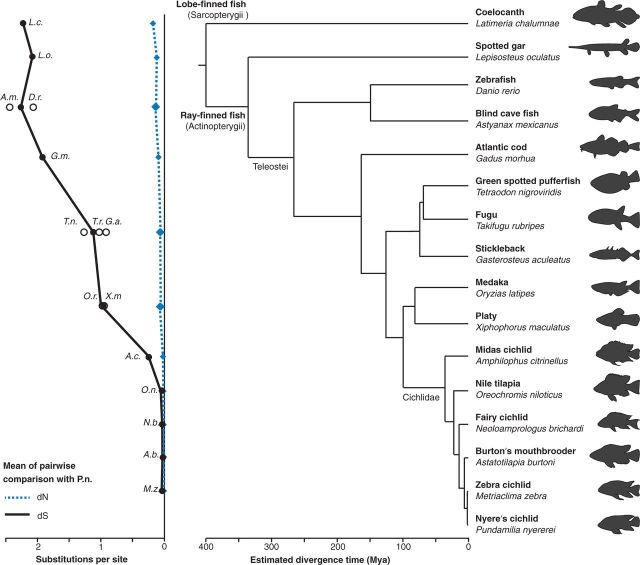
Fig. 2.—
Conservation of the NC GRN during bony fish evolution. Phylogeny of the species used in this study. Clades in dashed boxes share one divergence time to cichlids and are shown as hollow circles in (left). Note the small divergence times within the cichlid lineage leading to small average dN and dS values in corresponding pairwise comparisons. Divergence times are given on the x-axis (Hedges et al. 2006; Genner et al. 2007; Near et al. 2013; Brawand et al. 2014). Average dN (blue) and dS (black) distances in pairwise comparisons against P. nyererei are depicted on the left side (the y-axis does not directly correspond to divergence time). Average values across all genes within the GRN are shown.
Materials and Methods
Data Set
A total of 31 genes associated with the development of the NC in bony fish were included in this study. The genes were chosen based on prior characterizations of NC GRN (Sauka-Spengler and Bronner-Fraser 2008; Betancur et al. 2010; Simões-Costa and Bronner 2015). Only genes that could be found in greater than 80% of our selected species were analyzed. Depending on the time of deployment during development (Sauka-Spengler and Bronner-Fraser 2008; Betancur et al. 2010; Simões-Costa and Bronner 2015), genes were grouped into one of four developmental groups: Induction genes, NPB specification genes, NC specification genes, and NC differentiation genes (fig. 1 and supplementary table S1, Supplementary Material online). Additionally, we analyzed a control group of five housekeeping genes that are not directly related to NC formation, but for which we expected a very high conservation and comparable dN/dS ratios (see below). We downloaded gene sequences of 11 of the 16 species from Ensembl release 75 (Flicek et al. 2014) (fig. 2). After identifying orthologous loci using the EnsemblCompara pipeline (Vilella et al. 2009), we extracted the longest transcript coding sequence (CDS) of each gene. Sequences for cichlid species other than the Nile Tilapia, Oreochromis niloticus, were obtained from the deposited genome assemblies (Brawand et al. 2014; Elmer et al. 2014) (fig. 2). Orthologous loci in cichlid genomes were identified by BLASTN searches using Nile Tilapia sequences as queries and an e-value threshold of 10−4.The integrity of obtained sequences was double-checked by BLASTN searches against available transcriptomes to ensure correct reading frames. Gene orthology was confirmed using phylogenetic analysis (see below). In total, this study contains 483 sequences and 36 genes, each containing at least 13 of the 16 species.
Multiple Sequence Alignment and Phylogeny Reconstruction
Multiple sequence alignments of individual genes were constructed with translatorX (Abascal et al. 2010): After translation, sequences are aligned at the amino acid level using MUSCLE (Edgar 2004), unreliable amino acid positions are removed with Gblocks (Castresana 2000) under the least stringent parameters, and the corresponding nucleotide alignments are created guided by trimmed amino acid alignments. Maximum-likelihood phylogenetic reconstruction was performed with PhyML version 3.1 (Guindon et al. 2010) after best-fit models of evolution were chosen based on AICc (Akaike information criterion with correction) scores (Hurvich and Tsai 1989) calculated in jModeltest version 2.1.5 (Posada 2008). Due to the low sequence divergence among cichlids, single gene alignments contained too few informative sites to permit reconstructing reliable phylogenetic trees. Therefore, we used the previously reported species phylogeny (Brawand et al. 2014) for further analyses of cichlid data.
Saturation of nonsynonymous substitutions in gene alignments could in principle bias dN/dS estimates, even though probabilistic methods have been proven robust in simulation studies (Yang 2006). To exclude this possibility, we built plots of uncorrected p-distances against maximum-likelihood distances for both synonymous and nonsynonymous sites. MEGA version 6 (Tamura et al. 2013) was used to calculate genetic distances, using the parameters estimated by jModeltest for maximum-likelihood distances. Saturation plots did not show noticeable sequence saturation upon visual inspection, supporting the notion that dN/dS calculations are reliable.
Computation of dN/dS Ratios
We estimated the effects of selection on genes by calculating ratios between nonsynonymous and synonymous substitution rates (dN/dS). Using yn00 of the PAML package version 4.7 (Yang 2007) we calculated dN and dS values for pairwise comparisons between all 16 species. Divergence times used for mapping the values onto the phylogeny were taken from the TimeTree database (Hedges et al. 2006).
Using codeml (within the PAML package) we inferred dN/dS values using maximum likelihood. Under neutrality, both synonymous and nonsynonymous changes are expected to accumulate at equal rates (dN/dS = 1). An excess of nonsynonymous over synonymous substitutions is viewed as evidence of positive selection (dN/dS > 1), whereas the opposite is true for negative selection (dN/dS < 1). Measures of dN/dS for each alignment provided information of selective pressures across the different genes and developmental groups (fig. 1), based on the M0 model that averages dN/dS across codons and sequences. Three more sophisticated models were used to examine changes in dN/dS between codon positions (site models), lineages (branch models), and codon positions in particular lineages (branch-site models). 1) Site models can detect specific codons that evolved under positive selection by comparing the M2a and M8 models of positive selection against the nearly neutral models M1a and M7, respectively. The M7/M8 comparison is similar to M1a/M2a, but assumes dN/dS to follow a beta distribution, which can take a variety of shapes (e.g., uniform, linear, exponential, bell) depending on two shape parameters that appear as exponents of the random variable. In this sense, M7 is a more flexible null model than M1a (Yang 2006). 2) In branch models, dN/dS values are calculated for a predefined lineage or lineages (foreground) separately from the rest (background), allowing the examination of changes in selection in specific lineages. In this case, the alternative model assuming different dN/dS across lineages is compared against the M0 model. 3) The branch-site test of positive selection compares model A (that allows dN/dS to vary among sites in predefined branches) and a null model assuming nearly neutral evolution (codons can only have dN/dS ≤ 1). For each of the three model types, the relative fit of nested pairs of models was compared by differences in log-likelihood against a chi-square distribution (i.e., likelihood ratio test), whereas the fit of nonnested models was assessed by differences in AIC scores (Akaike 1974). In both site and branch-site models, positively selected codons were estimated by using a Bayes empirical Bayes (BEB) procedure (Deely and Lindley 2012). Codons under positive selection were only considered, if they had a posterior probability > 0.95. To discard suboptimal results by maximum likelihood getting stuck in local maxima, computations were repeated with different initial dN/dS values (0.05, 0.4, 1.5, and 10) (Bielawski and Yang 2004). All calculations used a F3×4 codon substitution model, where expected codon frequencies are derived from three sets of nucleotide frequencies for the three codon positions. Further scripts used for computational analyses were generated in R (http://www.r-project.org, last accessed October 28, 2015), or Bioconductor (http://www.bioconductor.org, last accessed October 28, 2015), using R packages clusteval, ggbiplot, ggplot2, gplots, grDevices, mclust, and RColorBrewer.
Multivariate Cluster Analysis
The clustering analysis of sequence conservation features was performed as previously described for the floral organ specification GRN (Davila-Velderrain et al. 2014). The four variables used were 1) the coefficient of variation in protein sequence lengths, 2) mean pairwise DNA and 3) protein distances, and 4) dN/dS ratios. Genetic distances were calculated in MEGA 6. A principal component analysis—based on the different sequence conservation features 1–4—was performed in R using the princomp and ggbiplot libraries and normal data ellipses were generated with the standard probability (0.69). The cluster analysis itself was conducted with the package mclust version 4 using the implemented function “Mclust.” Mclust uses the Bayesian Information Criterion (BIC) to identify the best-fit model for constructing covariance matrices that define the clusters within the data set. To validate the clustering results, we calculated the similarity of the clustering decisions to the four a priori conceived developmental subgroups of the NC GRN (fig. 1B) using the Rand index (Rand 1971). Through bootstrap resampling of 100,000 random clusters of similar composition, we determined whether the Rand index of our clustering is significantly higher than the Rand index of randomly generated clusterings with the same number of clusters compared with the four developmental groups.
Results
Changes in Selective Pressure Follow an Hourglass Pattern along Developmental Timing
Genes were grouped into the following four developmental groups: 1) Induction genes, 2) NPB specification genes, 3) NC specification genes, and 4) NC differentiation genes (fig. 1 and supplementary table S1, Supplementary Material online). The four groups (although partially overlapping) act in chronological order within the NC GRN, with the induction genes (1) being the first genes active and NC differentiation genes (4) being active during the last steps of NC development (fig. 1). Since specification genes (3) regulate NC development around the phylotypic stage, they would be expected to exhibit the strongest conservation according to the hourglass model. On the contrary, following the funnel-like model, the early expressed Induction genes (1) should be most conserved (fig. 1A). We added five housekeeping genes as a control group. Here, we expected consistently high conservation throughout all analyzed species thereby serving as comparative base line for highly conserved genes. All analyzed genes—both from NC group and control group—show signs of strong purifying selection, with dN/dS values well beneath what would be considered neutral evolution (dN/dS = 1). For ray-finned fish, ratios among NC-associated genes range from 0.024 in zic1 to 0.150 in bmp7a (mean: 0.072 ± 0.030) (fig. 3B, table 1, and supplementary table S2, Supplementary Material online). Values in the control group of housekeeping genes are consistently low and range from 0.021 to 0.057 (mean: 0.040 ± 0.015) (fig. 3B, table 1, and supplementary table S2, Supplementary Material online).
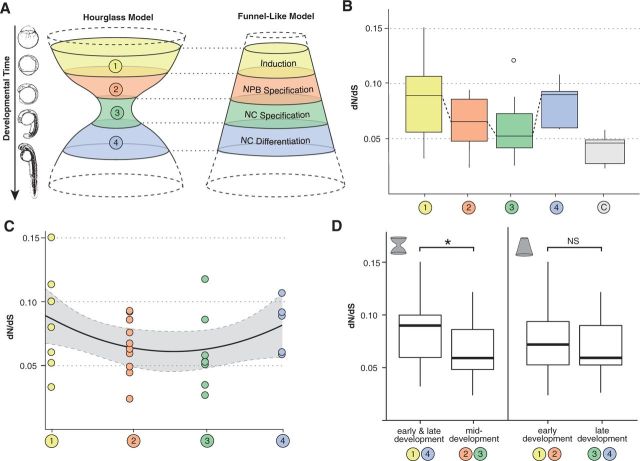
Fig. 3.—
Comparison of dN/dS distribution according to developmental groups. (A) Two hypotheses on GRN evolution. The hourglass model (left) predicts genes in deployed in middevelopment to be under stronger selection than early induction and late differentiation genes. On the other hand, a funnel-like model (right) assumes molecular constraint to be highest in early genes with constraint decreasing as development progresses. (B) Box plot of overall dN/dS ratios (codeml M0 model). Of the four subunits within the NC GRN the neural crest specification genes show the lowest dN/dS values and are most similar in conservation to the housekeeping genes. (C) dN/dS values of individual NC GRN genes and polynomic regression supporting the hourglass model. (D) Group 2/3 has significantly lower dN/dS values than group 1/4. This does not apply for the comparison between group 1/2 and group 3/4.
Table 1.
Summary of dN/dS Values for the Five Gene Groups within Ray-Finned Fishes
| dN/dS | Group 1–4 | Group 1 | Group 2 | Group 3 | Group 4 | Group C |
|---|---|---|---|---|---|---|
| Mean | 0.072 ± 0.030 | 0.086 ± 0.038 | 0.066 ± 0.023 | 0.060 ± 0.031 | 0.082 ± 0.022 | 0.040 ± 0.015 |
| Median | 0.064 | 0.090 | 0.065 | 0.053 | 0.090 | 0.046 |
| Minimum | 0.024 (zic1) | 0.033 (wnt6b) | 0.024 (zic1) | 0.027 (twist2) | 0.059 (neurog1) | 0.021 (rpl8) |
| Maximum | 0.150 (bmp7a) | 0.150 (bmp7a) | 0.094 (msxe) | 0.121 (snai1b) | 0.108 (mitfa) | 0.057 (g6pd) |
In an effort to discover differences in selective regimes between genes deployed at different developmental points, we analyzed the distribution of dN/dS values across the four developmental groups (fig. 1A–C) to determine whether the distribution of molecular constraints follows an hourglass (fig. 3A, left) or a funnel-like distribution (fig. 3A, right). To compare and evaluate the fit of the data to these alternative hypotheses, we performed a linear model fit (fig. 3C and supplementary fig. S1, Supplementary Material online). AIC values for the different linear fit curves favor a quadratic fit over a simple linear model (fig. 3C and supplementary fig. S1, Supplementary Material online). A quadratic fit supports the idea of an hourglass type of conservation across the NC GRN versus a funnel-like or random distribution. In fact, the two central groups (2 and 3) have significantly lower dN/dS values (and therefore higher constraints) than the early and late groups (1 and 4) (t-test; P = 0.0486; fig. 3D), whereas the differences between early (1 and 2) and late groups (3 and 4) are not significant (t-test; P = 0.5973; fig. 3D).
Evolutionary Conservation Features Are Highly Homogeneous across the NC GRN
In an attempt to find groups within the NC GRN that display similar evolutionary patterns, we conducted an unbiased clustering of genes based on different conservation features. This method has previously been reported to yield clear results for another developmental GRN (Davila-Velderrain et al. 2014). An initial clustering of NC GRN genes using four conservation features (dN/dS, DNA and protein sequence distances and coefficient of variation of protein length) resulted in a classification of genes into ten different clusters (supplementary fig. S2 and Supplementary Data and Supplementary Data, Supplementary Material online). Ten clusters is more than any initially anticipated biological classification of genes (such as GRN topology; fig. 1C). The members of the NC GRN are highly conserved at the sequence level, which is supported by the comparison with a previously analyzed developmental GRN that shows more variation and therefore a clearer clustering (Davila-Velderrain et al. 2014) (supplementary fig. S3, Supplementary Material online). We repeated the clustering analyses for modified species-sets for which we removed phylogenetically very distant (spotted gar, coelacanth) or very close (cichlids) species, in order to exclude the possibility that our clustering results could be biased by too many or too few differences, respectively (supplementary tables S5 and Supplementary Data, Supplementary Material online). Clustering analyses on these modified data sets rendered the same clustering results.
The best-fit model obtained in the clustering analysis (EEV) utilizes covariance matrices of equal shape and volume (BIC = 627.46) (supplementary fig. S2, Supplementary Material online). The ten identified groups show a Rand similarity index of 0.71 when compared with the four initial function-related developmental subgroups. The clustering with Rand = 0.71 has a lower similarity to the predefined developmental groups than 15% of random clusterings of ten clusters. A principal component analysis of all NC genes including all conservation features reveals a rather homogeneous cluster of genes (fig. 4A). However, although the four developmental groups are overlapping, centroids of the central groups (2 and 3) separate from the early and late groups (1 and 4) on the first PC axis (73% of variance) (fig. 4A). The grouping between middevelopment versus early/late groups further supports an hourglass pattern based on more similar evolutionary conservation. In fact, when comparing the two pairs of groups solely on the first principal component (PC1) the difference between groups 1/4 and 2/3 (t-test; P = 0.005), but not between groups 1/2 and 3/4 (t-test; P = 0.654) is statistically significant (fig. 4B).
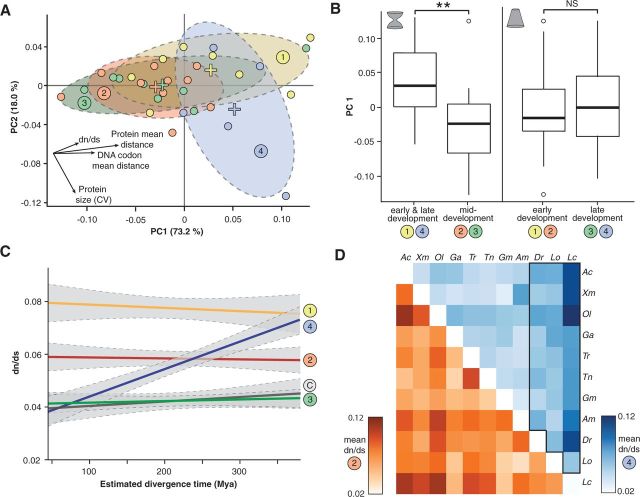
Fig. 4.—
Principal component analysis of the NC gene data set and pairwise comparisons of dN/dS in relation to evolutionary distance. (A) Two-dimensional representation of conservation features after principal component analysis. Developmental subgroups of genes are colored accordingly. Crosses indicate centroids of the respective groups. Normal probability ellipses are shown in dashed lines. (B) Comparison between the principal components (PC1) of groups 1/4 and 2/3 as well as groups 1/2 and 3/4, respectively. PC1 of group 2/3 and group 1/4 are significantly different. This does not apply for the comparison between group 1/2 and group 3/4. (C) Linear regression for the dN/dS of pairwise comparisons for all groups plotted against evolutionary distance of the species pairs. (D) Pairwise-comparisons shown as heatmaps for group 1 (orange) and group 4 (blue). Dark colors indicate high dN/dS, light colors low dN/dS (range 0.02–0.12). Species are referred by the initials of genus and species. Although values are distributed relatively random in group 1, high dN/dS values can be mainly found within comparisons with long evolutionary distance in group 4.
Molecular Constraints of the Majority of NC Genes Are Constant throughout Ray-Finned Fish Evolution
In an effort to gain insight into if and how molecular constraints changed during the evolution of ray-finned fish, we calculated dN/dS for pairwise comparisons between all species and plotted them against their estimated divergence times (Hedges et al. 2006; Genner et al. 2007; Near et al. 2013; Brawand et al. 2014) for both single genes and gene groups (fig. 4C and supplementary figs. S4 and Supplementary Data, Supplementary Material online). Additionally, we performed comparisons centered on single species to emphasize differences of comparisons to particular species (supplementary fig. S6, Supplementary Material online). For stable selective constraints, synonymous and nonsynonymous substitutions are expected to accumulate at constant rates resulting in dN/dS values that do not change with evolutionary time. Indeed, both nonsynonymous (supplementary fig. S7, Supplementary Material online) and synonymous substitution rates (supplementary fig. S8, Supplementary Material online) can be described as a linear function of evolutionary distance (Nei et al. 2010). Consequently, dN/dS values for genes of the NC GRN were almost constant, with only a slight decrease in dN/dS ratios from long to short evolutionary distances (ΔdN/dS = −0.006 per 380 Ma; linear regression; P = 0.015) (fig. 4C and supplementary figs. S4 and Supplementary Data, Supplementary Material online). To account for possible effects due to phylogenetic nonindependence and species selection biases, we repeated the analyses with two differently composed reduced data sets (supplementary figs. S9 and Supplementary Data, Supplementary Material online) and two phylogenetically completely independent data sets of pairwise-comparisons (supplementary figs. S11 and Supplementary Data, Supplementary Material online). The analyses of these data sets suggest that dN/dS values are rather constant over evolutionary time (linear regression; P > 0.05 for all data sets).
Additionally, pairwise comparisons (fig. 4D and supplementary figs. S5–S15, Supplementary Material online) provide further evidence for the particular high selective constraints among NC specification genes. In almost half (48.5%) of all 66 pairwise comparisons (without radiating cichlids) of the group of NC specification genes, dN/dS values of NC specifiers are smaller than the average value of the housekeeping gene group. Furthermore, all 66 (100%) pairwise dN/dS values of group 3 are below the average of the remaining NC groups. This is further evidence for the strong constraints of the NC specification genes that are active around the phylotypic stage and provides additional support for the hourglass model of molecular constraint in the NC GRN.
Selective Constraints of Late NC Genes Increased after the Split between Ray- and Lobe-Finned Fish
To investigate whether there are changes in selective constraints across time within specific groups of the NC GRN, estimated pairwise dN/dS values were plotted as a function of evolutionary time, separately for the five groups of genes (fig. 4C). The NC differentiation genes (group 4) were the only group of genes that showed a significant change in dN/dS values (fig. 4C). This suggests an increase of molecular constraints throughout ray-finned fish evolution within the NC differentiation genes of group 4 (ΔdN/dS = −0.043 per 380 Ma; linear regression; P < 0.0001). We confirmed this result by analyzing the reduced data sets that account for taxon sampling biases (i.e., compensating for the presence of many closely related cichlids or equilibrating lineage representation), which equally showed no significant change for groups 1–3, and significant change for group 4 (supplementary figs. S9 and Supplementary Data, Supplementary Material online). Significant changes in group 4 could be also observed in two phylogenetically independent data sets of species pairs (supplementary figs. S11 and Supplementary Data, Supplementary Material online) as well as for all of the genes of group 4 analyzed individually (supplementary fig. S4, Supplementary Material online). To get a better grasp on the distribution of dN/dS values over all pairwise comparisons, we visualized all developmental groups as heatmaps (fig. 4D and supplementary figs. S13–S15, Supplementary Material online). In group 1, high dN/dS values are distributed rather randomly and apparently without a strong correlation to evolutionary distance of the respective species pair (fig. 4D and supplementary fig. S13, Supplementary Material online). In contrast, for group 4 genes there is a trend of an increase in selective constraint. High dN/dS values are consistently found within the comparisons against coelacanth, spotted gar, and zebrafish. The highest dN/dS values are found among the pairwise comparisons that involve the coelacanth, however the observed trend is still significant, if the coelacanth is removed (ΔdN/dS = −0.022 per 380 Ma; linear regression; P = 0.0001). Interestingly, groups 2 and 3 as well as the control group have no clear trend; however, some species stand out more clearly than in group 1 (coelacanth and pufferfish for group 2, spotted gar and zebrafish for group 3, and blind cave fish for the control group; supplementary fig. S4, Supplementary Material online). The high values for the species pairs with blind cave fish and pufferfish are however due to a small number of genes with high dN/dS values (hprt1 and rpl8 for blind cave fish; tfap2a and id3 for pufferfish) and disappear if one visualizes medians of groups instead of means (supplementary figs. S13–S15, Supplementary Material online).
Positive Selection on rhob and dct Genes
After assessing the evolution of the NC GRN as a whole, we investigated how selection regimes in single genes changed during ray-finned fish evolution. Codeml site models found clear evidence for positive selection in two genes. In rhob (Ras homolog gene family member b), likelihood ratio tests were significant for the codeml models M1a/M2a (LR = 16.48, P = 2.6 × 10−4) and M7/M8 (LR = 31.41, P = 1.51 × 10−7) comparisons (supplementary table S7, Supplementary Material online). In both cases, BEB analyses identified amino acid position 186 (based on the Nile Tilapia sequence) as positively selected. Position 186 in Nile Tilapia is a proline located close to the C-terminal end of the protein. It is a site within a highly variable region at the C-terminus of the protein among the six cichlid species. Amino acid position 186 is three amino acids away from a highly conserved C-terminal motif (CVNCCKVL in the coelacanth/CFKCCVLM in the African cichlids) that is involved in the localization and degradation of the protein (Pérez-Sala et al. 2009). The second gene with positively selected codons was dct (dopachrome tautomerase), where the test comparing M7/M8 models was significant (LR = 16.68, P = 2.3 10−4) and BEB analysis showed that amino acid position 7 (in Nile Tilapia) has evolved under positive selection (supplementary table S7, Supplementary Material online). The amino acid at position 7 is a glycine in all cichlids. This glycine is shared with the platyfish Xiphophorus maculatus, but it is otherwise variable among the rest of species. The N-terminus of the encoded enzyme, the dopachrome tautomerase (also TRP-2 for tyrosine related protein 2), is involved in its catalytic reaction (Hiroshi Sugimoto et al. 1999). However, both sites appear to lie within regions of strong variability of their respective amino acid sequences.
Differences in Selective Pressures Arose during Cichlid Evolution
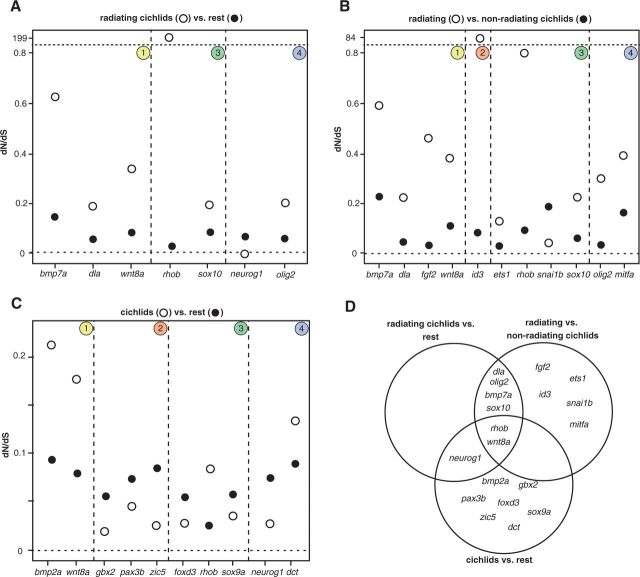
To test whether the NC GRN evolved under different selective regimes in lineages that are highly diverse in NC-derived traits, we analyzed the family of cichlid fishes (fig. 2). Cichlids underwent exceedingly rapid speciation often associated with astonishing diversification of NC-derived traits (Albertson and Kocher 2006). This is especially true for species that arose in the African Great Lakes (Stiassny and Meyer 1999; Albertson and Kocher 2006; Renz et al. 2011; Albertson et al. 2014; Powder et al. 2014). Therefore, we grouped the five cichlid species into two separate groups: One including cichlids associated with adaptive radiations (N. brichardi, A. burtoni, M. zebra, and P. nyererei) and the other including the rest of species (O. niloticus and Amphilophus citrinellus). Using branch models in codeml, we found significant differences in dN/dS ratios between these two groups in 7 of 31 genes (fig. 5A and supplementary table S8, Supplementary Material online). Most estimated dN/dS ratios were lower than 1, implying prevalence of purifying selection in both groups of species. Six of these genes showed a dN/dS that was at least 2.3 times higher in radiating cichlids than in nonradiating cichlid lineages: bmp7a, dla, olig2, rhob, sox10, and wnt8a (fig. 5A and supplementary table S8, Supplementary Material online). In addition, rhob had a dN/dS >1 in radiating cichlids. This gene had already shown evidences of positive selection in site model analyses (see above). Interestingly, neurog1 was the only gene that showed the reverse pattern of a lower dN/dS in radiating cichlids (fig. 5A and supplementary table S8, Supplementary Material online).
Fig. 5.—
Differential conservation of NC GRN genes during cichlid evolution. (A) Seven genes with different selective pressures between radiation-associated cichlids (N. brichardi, A. burtoni, M. zebra, and P. nyererei) and all other 12 analyzed species based on codeml branch models. (B) Eleven genes showed differential selective pressures when radiating cichlids are compared only with the other two cichlid species (Am. citrinellus and O. niloticus). (C) Comparison of all six cichlids against the rest of the species. Eleven of 31 genes show significant differences in dN/dS ratios. (D) Overview over which genes are shared between the comparisons.
To maximize information content in the comparisons between cichlids associated or not with evolutionary radiations, all branch models were repeated on alignments containing only the six cichlid species. The removal of more divergent ray-finned fish allowed establishing more reliable codon homologies within cichlids. Additionally, resulting alignments were longer after Gblocks filtering, thus including more informative codon sites. In this case, the four radiating African cichlids were compared with the Nile Tilapia (O. niloticus) and the Neotropical Midas cichlid (Am. citrinellus). Results are consistent with already reported analyses: Six of the genes from the earlier analysis show significantly higher dN/dS in radiating cichlids (fig. 5B and D and supplementary table S9, Supplementary Material online). Five additional genes display significant differences in dN/dS between radiating and nonradiating cichlids: ets1, fgf2, id3, mitfa, and snai1b (fig. 5B and D and supplementary table S9, Supplementary Material online). Except for snai1b, all of them have an at least 2.3-fold higher dN/dS in radiating cichlids.
To ascertain whether the changes in dN/dS inferred for radiation-associated cichlids were specific to these species or rather characteristic of the whole cichlid lineage, we further studied differences in dN/dS using branch models comparing all cichlids (as a foreground group) and all other fish species (as background) (fig. 5C and supplementary table S10, Supplementary Material online). In these tests rhob and wnt8a, two of the genes that appeared in the earlier analyses, also showed a higher dN/dS in cichlids (fig. 5C and supplementary table S10, Supplementary Material online). The comparison of the relevant models by AIC suggests that the increase in dN/dS in wnt8 most likely occurred in radiating cichlids rather than in the whole cichlid lineage (ΔAIC = 11.03). Aside from that, we observed significant changes in selective pressure in eight additional genes in cichlids. Of these, two show an increase of dN/dS in cichlids (bmp2b and dct), whereas six show a decrease (foxd3, gbx2, pax3a, neurog1, sox9a, and zic5) (fig. 5C and supplementary table S10, Supplementary Material online).
Natural selection is expected to act on particular amino acid positions of functional relevance, and during specific time periods after which selection turns into purifying to maintain adaptations (Zhang et al. 2005). Branch models average dN/dS over the alignment, and thus can have low power to detect positive selection if it has occurred in only a few localized positions (Yang 2006). Therefore, we used branch-site models that allow detecting positive selection in specific codons in particular branches. Based on the same comparisons between species groups as described above, branch-site models detected position 186 in rhob to have evolved under positive selection in radiating cichlids (supplementary table S11, Supplementary Material online), as was already found by site models (supplementary table S7, Supplementary Material online). The comparison of the relevant models favored the hypothesis that position 186 in rhob is under positive selection on all studied fish species rather than on cichlids specifically (best was M8 site model, with ΔAIC > 25.9 to M2a, branch and branch-site models assuming dN/dS changes in either radiating or all cichlids).
Discussion
The NC GRN Is under Strong Selective Constraint
The NC is a transient developmental unit of migrating multipotent cells that contributes to many different tissues within the vertebrate body. NC development is directed by an intricate GRN of transcription factors and signaling molecules. Here, we analyzed the molecular evolution of the CDSs of involved genes to gain insights on how the genes within this network change over evolutionary times and look for patterns of changes in selective pressures. Our results show that almost all genes of the NC GRN are under strong selective constraint across ray-finned fish, as evidenced by the values of dN/dS <1 (Yang 2006). These overall low dN/dS values found across the whole NC GRN are typical for a regulatory module that is active in such early developmental stages (Garfield and Wray 2009). Additionally, pleiotropic effects that affect the development of other tissues and cell types that changes in these genes might create, are an explanation for the strong purifying selection they evolve under (Wagner and Zhang 2011).
Molecular Evolution of NC GRN Modules Supports the Developmental Hourglass Model
Our data show a distribution of dN/dS values across the four developmental groups that support an hourglass model of conservation, where intermediate stages (groups 2 and especially 3) are under stronger purifying selection than earlier and latter stages (groups 1 and 4) (fig. 3C). This result is in line with previous studies that support the hourglass model (Hazkani Covo et al. 2005; Roux and Robinson-Rechavi 2008; Piasecka et al. 2013). There is increasing evidence that the phylotypic stage (i.e., the time point in development when phylogenetically diverse species resemble most closely to each other) can be indeed visualized at the genomic and transcriptomic level by increased constraints (Hazkani Covo et al. 2005; Roux and Robinson-Rechavi 2008; Levin et al. 2012; Piasecka et al. 2013; Irie and Kuratani 2014). A recent comparison of dN/dS ratios of genes expressed at different developmental stages in zebrafish (Piasecka et al. 2013) did not show significant differences between the studied stages—however, the distribution of means suggested that the highest degree of sequence conservation occurred around the phylotypic stage.
It has been hypothesized that the mechanisms behind the phenomenon of high selective constraint during middevelopment (phylotypic stage) is their strong internal recursive wiring within these central nodes of the regulatory network (Davidson and Erwin 2006) (fig. 1C). This is valid for many of the highly conserved genes that can be found among the NC and plate border specifiers (fig. 1B) including msxe, pax3a/b, tfap2, zic1/5, foxd3, snail1b, and sox9/10 (Betancur et al. 2010; Simões-Costa and Bronner 2015) and would explain the occurrence of stronger purifying selection in this subset of genes. The relatively relaxed selective pressure in differentiation genes is expected under all the above models of GRN evolution. Differentiation genes are most divergent in pairwise dN/dS comparisons over evolutionary time and have some of the highest overall dN/dS ratios (codeml M0 model). This agrees with predictions that genes that are more downstream would also experience less severe selective constraints. In this regard, it is interesting to note that group 4 shows lower dN/dS values in pairwise dN/dS comparisons between more recently diverged species (supplementary fig. S12, Supplementary Material online). An increase of genes depending on the NC differentiation genes (i.e., further downstream in different GRNs) might have led to a gradual increase in selective pressure throughout ray-finned fish evolution.
Clustering Does Not Link Conservation Features to Developmental Groups or Network Topology
Clustering the NC GRN genes without prior hypotheses resulted in a classification into ten groups. This number of clusters is much higher than any anticipated biological grouping of the genes. Also, the principal component analysis of gene conservation features did not reveal a clear aggregation pattern of the genes into one of the preconceived classifications (supplementary fig. S2, Supplementary Material online). However, comparing the PC1 of the central groups (2 and 3) with early and late groups (1 and 4) reveals a significant difference (P = 0.005; fig. 4B), further supporting an hourglass pattern of conservation. Additionally, the results reveal that the variation in protein size is a minor contributing factor in the variation of conservation in NC GRN genes, as it is the variable that constitutes most of the second principal component (fig. 4A)
A comparison of the first two principal components from our data set combined with a previous study of another developmental GRN (Davila-Velderrain et al. 2014) reveals that the NC GRN is more conserved along the two axes (supplementary fig. S3, Supplementary Material online). Clustering techniques based on molecular conservation features might thus not be a viable method for GRNs that are extremely conserved at the sequence level.
Specific Codons Are under Positive Selection in rhoB and dct
Positively selected codons across the studied species could be detected in two genes: rhob and dct. Interestingly, both rhob and dct are deployed rather late in development. Rhob is grouped within the NC specifiers, but it is associated with the later steps of this phase including delamination and migration (Liu and Jessell 1998). The dct gene is directly regulated by mitfa, that is a direct part of the enzymatic pathway for melanin production (Potterf et al. 2001). The fact that positive selection was only found in late genes and not in any of the earlier genes shows that gene deployed earlier during development are under stronger selective constraints. This observation, taken together with the pattern of dN/dS values between developmental groups, lends further credence to the assertion that the evolution of the NC GRN follows an hourglass model.
Release of Constraint in the Radiating African Cichlid Lineages
Periods of increased or decreased selective pressure can be detected through statistical testing of branch models. Our results show that six genes (bmp7a, dla, olig2, rhob, sox10, and wnt8a) showed higher dN/dS ratios in the radiation-associated and phenotypically diverse cichlids, both when compared with nonradiating cichlids or with all other fish. This suggests that selective pressure acting on these genes was relaxed in cichlids associated with evolutionary radiations (fig. 5A, C, and D). Five more genes (fgf2, id3, ets1, snai1b, and mitfa) showed significantly different dN/dS between radiating and nonradiating cichlids (fig. 5B and D). Overall, there is statistical evidence for relaxation of purifying selection in 10 of 31 genes in the NC GRN. One might therefore speculate that this release of constraint in cichlids might be in part responsible for the enormous diversity in NC-derived traits displayed by this family, in particular a number of trophic and phenotypic adaptations that enabled the astonishing cichlid radiations (Trainor et al. 2003; Albertson and Kocher 2006; Clabaut et al. 2007). For two of the above mentioned genes (id3 and rhob), a dN/dS >1 is inferred for the lineage of radiating cichlids, which is evidence for positive selection (fig. 5A and B). The fact that these two genes show such high dN/dS values in cichlids is attributable to the very low amount of synonymous substitutions, which are outnumbered by nonsynonymous changes. Surprisingly, even though we found a dN/dS >1 for id3 in cichlids associated with adaptive radiations, no specific codon sites could be identified to have undergone positive selection (estimated in either the site test or the branch-site test).
Tests using branch models were also used to compare all cichlids against the rest of fish species. Only three of the genes that already appeared in the above analysis also showed significantly different dN/dS values in this case: neurog1, rhob, and wnt8a (fig. 5D). For rhob and wnt8, the comparison of AIC scores between relevant branch models favored the scenario of relaxation of purifying selection in the lineage or radiating cichlids rather than in the whole cichlid lineage (ΔAIC = 2.7 and ΔAIC = 11.03, respectively), whereas for rhob both scenarios had a similar fit (ΔAIC = 0.8). Notably, the other nine genes that displayed higher dN/dS in radiating cichlids did not show significant differences in the comparisons with all cichlids, which is further evidence that these changes in purifying selection have indeed arisen in the radiating clade. It may also be of note that all genes showing higher dN/dS in cichlids are either genes that are deployed very early (bmp2b and wnt8a) or rather late (rhob and dct) in development whereas the other genes are more central and have lower dN/dS in cichlids. Nevertheless, the remaining genes that do not show significant differences in dN/dS do not follow a congruent pattern with this observation and thus this conclusion should be taken with caution. The interpretation of lower dN/dS found in cichlids compared with all other lineages must be taken with precaution, as it might be due to other important changes in selective regions of genes in species outside of the cichlid lineage.
Positive Selection at Specific Sites in Radiating Cichlids
Site model tests can detect the effect of selection at specific sites and ignore among-branch differences, whereas branch models allow detecting periodic changes in selective pressure and ignore among-codon differences. Overall, both types of models are highly conservative (Yang 2006). Branch-site models account for both effects and are in principle more sensitive than branch or site models alone. Tests of branch-site models have proven to yield insightful results in other studies, even involving some of the species we have included in this study (Fan et al. 2011; Amemiya et al. 2013). For the NC GRN, branch-site tests of positive selection applied to the comparisons with either radiating or all cichlids as foreground branches only detected positive selection rhob. The comparison among different scenarios of dN/dS change for rhob suggests that the positively selected site 186 is most likely not exclusive of cichlids, but rather shared with other ray-finned fish.
Conclusions
Ray-finned fish exhibit an enormous diversity in NC-derived traits, and yet the NC GRNs and most of its genes have so far not been analyzed from an evolutionary perspective. Analyses of protein CDSs support an hourglass model of protein CDS conservation in the different subunits of the GRN. The common morphological developmental pattern of an “hourglass” is reflected and possibly caused not or not only by developmental constraints but rather by constraints at the underlying genetic or genomic, in the sense of gene network interactions, levels. Cluster analysis revealed the extreme degree of sequence conservation throughout the NC GRN, illustrating its deep roots within vertebrate evolution (Gans and Northcutt 1983; Shimeld and Holland 2000). Values of dN/dS are constant throughout long evolutionary time spans demonstrating the strong, obligatory integration of the NC GRN within ray-finned fish. Interestingly, NC differentiation genes, the last group to be deployed in development, is an exception to this pattern, as it shows increasingly stronger purifying selection in ray-finned fish after the split from lobe-finned fish. The genes identified to have released selective constraint in radiating cichlids (bmp7a, dla, ets1, fgf2, id3, mitfa, olig2, and sox10) could be considered for future studies involving NC-derived traits. Further analyses of the molecular evolution of regulatory interactions will continue to shed light on the evolution of the NC network (Kratochwil and Meyer 2015a, 2015b, 2015c).
Supplementary Material
Acknowledgments
This work was supported by the Swiss National Science Foundation (P2BSP3_148629) and the EU FP7 Marie Curie Zukunftskolleg Incoming Fellowship Program, University of Konstanz (grant no. 291784) to C.F.K.; the Alexander von Humboldt Foundation (application 1150725) and the European Molecular Biology Organization (ALTF 440-2013) to I.I.; several grants of the Deutsche Forschungsgemeinschaft (DFG) and advanced grant 297300 “Genadapt” by the European Research Council to A.M. The authors thank Jose Davila-Velderrain for helpful discussions and further information on R scripts he used for GRN analysis. They appreciate the efforts and constructive input of two anonymous reviewers and thank Darrin Hulsey and Joost Woltering for discussions of this work and comments on the manuscript.
Literature Cited
- Abascal F, Zardoya R, Telford MJ. 2010. TranslatorX: multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res. 38:W7–W13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akaike H. 1974. A new look at the statistical model identification. IEEE Trans Automat Contr. 19:716–723. [Google Scholar]
- Albertson RC, et al. 2014. Genetic basis of continuous variation in the levels and modular inheritance of pigmentation in cichlid fishes. Mol Ecol. 23:5135–5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertson RC, Kocher TD. 2006. Genetic and developmental basis of cichlid trophic diversity. Heredity 97:211–221. [DOI] [PubMed] [Google Scholar]
- Alvarez-Ponce D, Aguade M, Rozas J. 2009. Network-level molecular evolutionary analysis of the insulin/TOR signal transduction pathway across 12 Drosophila genomes. Genome Res. 19:234–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amemiya CT, et al. 2013. The African coelacanth genome provides insights into tetrapod evolution. Nature 496:311–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basch ML, Bronner-Fraser M, García-Castro MI. 2006. Specification of the neural crest occurs during gastrulation and requires Pax7. Nature 441:218–222. [DOI] [PubMed] [Google Scholar]
- Betancur P, Bronner-Fraser M, Sauka-Spengler T. 2010. Assembling neural crest regulatory circuits into a gene regulatory network. Annu Rev Cell Dev Biol. 26:581–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielawski JP, Yang Z. 2004. A maximum likelihood method for detecting functional divergence at individual codon sites, with application to gene family evolution. J Mol Evol. 59:121–132. [DOI] [PubMed] [Google Scholar]
- Braasch I, Brunet F, Volff J-N, Schartl M. 2009. Pigmentation pathway evolution after whole-genome duplication in fish. Genome Biol Evol. 1:479–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brawand D, et al. 2014. The genomic substrate for adaptive radiation in African cichlid fish. Nature 513:375–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronner ME, LeDouarin NM. 2012. Development and evolution of the neural crest: an overview. Dev Biol. 366:2–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 17:540–552. [DOI] [PubMed] [Google Scholar]
- Clabaut C, Bunje PME, Salzburger W, Meyer A. 2007. Comparative phylogenetic analyses of the adaptive radiation of Lake Tanganyika cichlid fish: nuclear sequences are less homoplasious but also less informative than mitochondrial DNA. Evolution 61:560–578. [DOI] [PubMed] [Google Scholar]
- Cork JM, Purugganan MD. 2004. The evolution of molecular genetic pathways and networks. Bioessays 26:479–484. [DOI] [PubMed] [Google Scholar]
- Crombach A, Hogeweg P. 2008. Evolution of evolvability in gene regulatory networks. PLoS Comput Biol. 4:e1000112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson EH, Erwin DH. 2006. Gene regulatory networks and the evolution of animal body plans. Science 311:796–800. [DOI] [PubMed] [Google Scholar]
- Davila-Velderrain J, Servin-Marquez A, Alvarez-Buylla ER. 2014. Molecular evolution constraints in the floral organ specification gene regulatory network module across 18 angiosperm genomes. Mol Biol Evol. 31:560–573. [DOI] [PubMed] [Google Scholar]
- Deely JJ, Lindley DV. 2012. Bayes empirical Bayes. J Am Stat Assoc. 76:833–841. [Google Scholar]
- Donoghue PCJ, Graham A, Kelsh RN. 2008. The origin and evolution of the neural crest. Bioessays 30:530–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmer KR, et al. 2014. Parallel evolution of Nicaraguan crater lake cichlid fishes via non-parallel routes. Nat Commun. 5:5168. [DOI] [PubMed] [Google Scholar]
- Fan S, Elmer KR, Meyer A. 2011. Positive Darwinian selection drives the evolution of the morphology-related gene, EPCAM, in particularly species-rich lineages of African cichlid fishes. J Mol Evol. 73:1–9. [DOI] [PubMed] [Google Scholar]
- Flicek P, et al. 2014. Ensembl 2014. Nucleic Acids Res. 42:D749–D755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gans C, Northcutt RG. 1983. Neural crest and the origin of vertebrates: a new head. Science 220:268–273. [DOI] [PubMed] [Google Scholar]
- Garfield DA, Wray GA. 2009. Comparative embryology without a microscope: using genomic approaches to understand the evolution of development. J Biol. 8:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genner MJ, et al. 2007. Age of cichlids: new dates for ancient lake fish radiations. Mol Biol Evol. 24:1269–1282. [DOI] [PubMed] [Google Scholar]
- Guindon S, et al. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 59:307–321. [DOI] [PubMed] [Google Scholar]
- Hall BK. 2000. The neural crest as a fourth germ layer and vertebrates as quadroblastic not triploblastic. Evol Dev. 2:3–5. [DOI] [PubMed] [Google Scholar]
- Hazkani Covo E, Wool D, Graur D. 2005. In search of the vertebrate phylotypic stage: a molecular examination of the developmental hourglass model and von Baer's third law. J Exp Zool B Mol Dev Evol. 304B:150–158. [DOI] [PubMed] [Google Scholar]
- Hedges SB, Dudley J, Kumar S. 2006. TimeTree: a public knowledge-base of divergence times among organisms. Bioinformatics 22:2971–2972. [DOI] [PubMed] [Google Scholar]
- Henning F, Meyer A. 2014. The evolutionary genomics of cichlid fishes: explosive speciation and adaptation in the postgenomic era. Annu Rev Genomics Hum Genet. 15:417–441. [DOI] [PubMed] [Google Scholar]
- Hurvich CM, Tsai C-L. 1989. Regression and time series model selection in small samples. Biometrika 76:297–307. [Google Scholar]
- Irie N, Kuratani S. 2014. The developmental hourglass model: a predictor of the basic body plan? Development 141:4649–4655. [DOI] [PubMed] [Google Scholar]
- Kalinka AT, Tomancak P. 2012. The evolution of early animal embryos: conservation or divergence? Trends Ecol Evol. 27:385–393. [DOI] [PubMed] [Google Scholar]
- Kelsh RN, Barsh GS. 2011. A nervous origin for fish stripes. PLoS Genet. 7:e1002081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Miller CT, Keynes RJ. 2001. Neural crest patterning and the evolution of the jaw. J Anat. 199:105–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knecht AK, Bronner-Fraser M. 2002. Induction of the neural crest: a multigene process. Nat Rev Genet. 3:453–461. [DOI] [PubMed] [Google Scholar]
- Kratochwil CF, Meyer A. 2015a. Closing the genotype-phenotype gap: emerging technologies for evolutionary genetics in ecological model vertebrate systems. Bioessays 37:213–226. [DOI] [PubMed] [Google Scholar]
- Kratochwil CF, Meyer A. 2015b. Evolution: tinkering within gene regulatory landscapes. Curr Biol. 25:R285–R288. [DOI] [PubMed] [Google Scholar]
- Kratochwil CF, Meyer A. 2015c. Mapping active promoters by ChIP-seq profiling of H3K4me3 in cichlid fish—a first step to uncover cis-regulatory elements in ecological model teleosts. Mol Ecol Resour. 15:761–771. [DOI] [PubMed] [Google Scholar]
- Kratochwil CF, Sefton MM, Meyer A. 2015. Embryonic and larval development in the Midas cichlid fish species flock (Amphilophus spp.): a new evo-devo model for the investigation of adaptive novelties and species differences. BMC Dev Biol. 15:277–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulesa PM, Gammill LS. 2010. Neural crest migration: patterns, phases and signals. Dev Biol. 344:566–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin M, Hashimshony T, Wagner F, Yanai I. 2012. Developmental milestones punctuate gene expression in the Caenorhabditis embryo. Dev Cell. 22:1101–1108. [DOI] [PubMed] [Google Scholar]
- Liu JP, Jessell TM. 1998. A role for rhoB in the delamination of neural crest cells from the dorsal neural tube. Development 125:5055–5067. [DOI] [PubMed] [Google Scholar]
- Lu Y, Rausher MD. 2003. Evolutionary rate variation in anthocyanin pathway genes. Mol Biol Evol. 20:1844–1853. [DOI] [PubMed] [Google Scholar]
- Mayanil CS. 2013. Transcriptional and epigenetic regulation of neural crest induction during neurulation. Dev Neurosci. 35:361–372. [DOI] [PubMed] [Google Scholar]
- Minoux M, et al. 2013. Mouse Hoxa2 mutations provide a model for microtia and auricle duplication. Development 140:4386–4397. [DOI] [PubMed] [Google Scholar]
- Near TJ, et al. 2013. Phylogeny and tempo of diversification in the superradiation of spiny-rayed fishes. Proc Natl Acad Sci U S A. 110:12738–12743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Suzuki Y, Nozawa M. 2010. The neutral theory of molecular evolution in the genomic era. Annu Rev Genomics Hum Genet. 11:265–289. [DOI] [PubMed] [Google Scholar]
- Pérez-Sala D, Boya P, Ramos I, Herrera M, Stamatakis K. 2009. The C-terminal sequence of RhoB directs protein degradation through an endo-lysosomal pathway. PLoS One 4:e8117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piasecka B, Lichocki P, Moretti S, Bergmann S, Robinson-Rechavi M. 2013. The hourglass and the early conservation models—co-existing patterns of developmental constraints in vertebrates. PLoS Genet. 9:e1003476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D. 2008. jModelTest: phylogenetic model averaging. Mol Biol Evol. 25:1253–1256. [DOI] [PubMed] [Google Scholar]
- Potterf SB, et al. 2001. Analysis of SOX10 function in neural crest-derived melanocyte development: SOX10-dependent transcriptional control of dopachrome tautomerase. Dev Biol. 237:245–257. [DOI] [PubMed] [Google Scholar]
- Powder KE, Cousin H, McLinden GP, Craig Albertson R. 2014. A nonsynonymous mutation in the transcriptional regulator lbh is associated with cichlid craniofacial adaptation and neural crest cell development. Mol Biol Evol. 31:3113–3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prud’homme B, Gompel N. 2010. Evolutionary biology: genomic hourglass. Nature 468:768–769. [DOI] [PubMed] [Google Scholar]
- Rand WM. 1971. Objective criteria for the evaluation of clustering methods. J. Am. Stat. Assoc. 66:846–850. [Google Scholar]
- Rausher MD, Miller RE, Tiffin P. 1999. Patterns of evolutionary rate variation among genes of the anthocyanin biosynthetic pathway. Mol Biol Evol. 16:266–274. [DOI] [PubMed] [Google Scholar]
- Renz AJ, Gunter HM, Qiu H, Meyer A, Kuraku S. 2011. Ancestral and derived attributes of the dlx gene repertoire, cluster structure and expression patterns in an African cichlid fish. Evodevo 2:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley RM, Jin W, Gibson G. 2003. Contrasting selection pressures on components of the Ras-mediated signal transduction pathway in Drosophila. Mol Ecol. 12:1315–1323. [DOI] [PubMed] [Google Scholar]
- Roberts RB, Hu Y, Albertson RC, Kocher TD. 2011. Craniofacial divergence and ongoing adaptation via the hedgehog pathway. Proc Natl Acad Sci U S A. 108:13194–13199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux J, Robinson-Rechavi M. 2008. Developmental constraints on vertebrate genome evolution. PLoS Genet. 4:e1000311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzburger W, Braasch I, Meyer A. 2007. Adaptive sequence evolution in a color gene involved in the formation of the characteristic egg-dummies of male haplochromine cichlid fishes. BMC Biol. 5:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santagati F, Rijli FM. 2003. Cranial neural crest and the building of the vertebrate head. Nat Rev Neurosci. 4:806–818. [DOI] [PubMed] [Google Scholar]
- Sauka-Spengler T, Bronner-Fraser M. 2008. A gene regulatory network orchestrates neural crest formation. Nat Rev Mol Cell Biol. 9:557–568. [DOI] [PubMed] [Google Scholar]
- Shimeld SM, Holland PW. 2000. Vertebrate innovations. Proc Natl Acad Sci U S A. 97:4449–4452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simões-Costa M, Bronner ME. 2015. Establishing neural crest identity: a gene regulatory recipe. Development 142:242–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiassny ML, Meyer A. 1999. Cichlids of the rift lakes. Sci Am. 280:64–69. [Google Scholar]
- Sugie A, Terai Y, Ota R, Okada N. 2004. The evolution of genes for pigmentation in African cichlid fishes. Gene 343:337–346. [DOI] [PubMed] [Google Scholar]
- Sugimoto H, et al. 1999. Crystal structure of human d-dopachrome tautomerase, a homologue of macrophage migration inhibitory factor, at 1.54 Å resolution. Biochemistry 38:3268–3279. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol. 30:2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trainor PA, Melton KR, Manzanares M. 2003. Origins and plasticity of neural crest cells and their roles in jaw and craniofacial evolution. Int J Dev Biol. 47:541–553. [PubMed] [Google Scholar]
- Turner GF. 2007. Adaptive radiation of cichlid fish. Curr Biol. 17:R827–R831. [DOI] [PubMed] [Google Scholar]
- Van Otterloo E, Cornell RA, Medeiros DM, Garnett AT. 2013. Gene regulatory evolution and the origin of macroevolutionary novelties: insights from the neural crest. Genesis 51:457–470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilella AJ, et al. 2009. EnsemblCompara GeneTrees: complete, duplication-aware phylogenetic trees in vertebrates. Genome Res. 19:327–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner GP, Zhang J. 2011. The pleiotropic structure of the genotype–phenotype map: the evolvability of complex organisms. Nat Rev Genet. 12:204–213. [DOI] [PubMed] [Google Scholar]
- Yang Z. 2006. Computational molecular evolution. Oxford: Oxford University Press. [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24:1586–1591. [DOI] [PubMed] [Google Scholar]
- Zhang J, Nielsen R, Yang Z. 2005. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol. 22:2472–2479 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.