Abstract
Background:
The Hoary Edge Skipper (Achalarus lyciades) is an eastern North America endemic butterfly from the Eudaminae subfamily of skippers named for an underside whitish patch near the hindwing edge. Its caterpillars feed on legumes, in contrast to Grass skippers (subfamily Hesperiinae) which feed exclusively on monocots.
Results:
To better understand the evolution and phenotypic diversification of Skippers (family Hesperiidae), we sequenced, assembled and annotated a complete genome draft and transcriptome of a wild-caught specimen of A. lyciades and compared it with the available genome of the Clouded Skipper (Lerema accius) from the Grass skipper subfamily. The genome of A. lyciades is nearly twice the size of L. accius (567 Mbp vs. 298 Mbp), however it encodes a smaller number of proteins (15881 vs. 17411). Gene expansions we identified previously in L. accius apparently did not occur in the genome of A. lyciades. For instance, a family of hypothetical cellulases that diverged from an endochitinase (possibly associated with feeding of L. accius caterpillars on nutrient-poor grasses) is absent in A. lyciades. While L. accius underwent gene expansion in pheromone binding proteins, A. lyciades has more opsins. This difference may be related to the mate recognition mechanisms of the two species: visual cues might be more important for the Eudaminae skippers (which have more variable wing patterns), whereas odor might be more important for Grass skippers (that are hardly distinguishable by their wings). Phylogenetically, A. lyciades is a sister species of L. accius, the only other Hesperiidae with a complete genome.
Conclusions:
A new reference genome of a dicot-feeding skippers, the first from the Eudaminae subfamily, reveals its larger size and suggests hypotheses about phenotypic traits and differences from monocot-feeding skippers.
Keywords: Lepidoptera, Butterflies, Cellulase, Opsin, Catalase, Achalarus lyciades
1. INTRODUCTION
The Hoary Edge (Achalarus lyciades) is a skipper (family Hesperiidae) from the Eudaminae subfamily [1]. It is widely distributed over the entire eastern United States except in south Florida and south Texas [2]. Its English name arises from a large whitish patch underneath the hindwing near its edge (Fig. 1). A combination of large yellowish semi-hyaline spots on forewings with such a patch is unique to this species, making its identification straightforward [2]. The Hoary Edge inhabits open woodlands, forest edges and roadsides [3, 4]. Its caterpillars feed on the leaves of many plants from the legume family (Fabaceae) but are not known to be serious pests to crops [2]. Caterpillars make shelters by tying the leaves of food plants with silk. The skipper has only one brood in the northern parts of its range, but several broods and accompanying facultative larval diapause are characteristic of the southern populations [2, 3].
Fig. (1).
Specimens of Achalarus lyciades. (a, b) left wings of a male voucher NVG-3311 with the reference genome sequenced: USA: Texas, Sabine Co., Sabine National Forest, 1 mi south of Fairmount, near Fox Hunters' Hill, GPS 31.185394, -93.72992, 12-Apr-2015. Specimens reared from eggs: USA: Texas, Wise Co., ca. 10 mi north of Decatur, LBJ National Grassland: (c, d) male, eclosed on 17-Jul-2000, (e, f) female eclosed on 28-Jul-2000. Dorsal (above: a, c, e) and ventral (below: b, d, f) views of each specimen are shown.
Previously, we reported the genome of the Clouded Skipper (Lerema accius) [5], which remains a single genome from the family available to date. The Clouded Skipper belongs to a Grass skipper subfamily Hesperiinae. Its caterpillars feed on grasses, also making shelters from leaves [2]. In addition to mapping phylogenetic diversity of butterflies at the subfamily level with representative genomes, it would be of interest to compare the genomes of two skippers: A. lyciades and L. accius. Their caterpillars have different food plant preferences. While L. accius feeds on nutrient-poor grasses, A. lyciades feeds on leaves of legumes. Uniqueness of the A. lyciades appearance, its wide distribution, the lack of apparent variation in phenotype, and differences in diapause behavior in different parts of the range make it an attractive target for comparative genomics.
Presently, representative genomes are known for five butterfly families: the swallowtails (Papilionidae) [6-8], the Whites and Sulphurs (Pieridae) [9], the Blues (Lycaenidae) [10], the Brushfoots (Nymphalidae) [11-13], and the Skippers (Hesperiidae) [5]. The Brushfoots are currently the best-studied and the most significant efforts have focused on Heliconius and the Monarch (Danaus plexippus) [14, 15]. Skippers traditionally were thought of as a family between butterflies and moths due to their moth-like appearance. However, recent DNA-based evidence suggests that swallowtails are the sister to all other butterflies including skippers [16, 17]. Phylogenetic signal in DNA-based trees is not particularly strong [5] and additional genomes from several phylogenetic lineages of Skippers might help to resolve the phylogenetic uncertainties. Therefore, genomes from different Hesperiidae subfamilies are desirable.
To learn more about skippers and butterflies, we sequenced and annotated the complete genome of Achalarus lyciades from a single specimen. At 567 Mbp, it is one of the largest among known Lepidoptera genomes, and the first from the Eudaminae subfamily. However, phylogenetic analysis with 14 available Lepidoptera genomes remains incongruent with respect to the placement of skippers and swallowtails with the favored tree topology changing depending on the selected evolutionary model. A. lyciades represents a short branch in the phylogenetic tree, suggesting slow evolution. Compared to the Clouded Skipper (L. accius), the only other Hesperiidae (from the Hesperiinae subfamily) with the sequenced genome [5], the larger A. lyciades genome encodes smaller number of proteins. A number of gene duplication events found in L. accius, such as the expansion of hypothetical cellulases and catalases that may be an adaptation to nutrient-poor foodplant and harmful environment are not observed in A. lyciades. Notably, while the L. accius shows expansion in pheromone binding proteins, A. lyciades genome encodes more opsins. Many Grass skippers are similar in wing colors and patterns, and therefore they may rely on pheromones to recognize mates. Eudaminae skippers are frequently characterized by diverse wing patters and visual cues may play more important roles in their mate recognition.
2. RESULTS AND DISCUSSION
2.1. Genome Assembly, Annotation and Comparison to Other Lepidoptera Genomes
We assembled a 567 Mb reference genome of Achalarus lyciades (Aly) from a single specimen. The genome is one of the largest among currently sequenced Lepidoptera genomes [6, 11-13, 18-22]. The scaffold N50 of Aly genome assembly is 558 kb, similar to many other published Lepidoptera genomes. The genome assembly is more complete than many other Lepidoptera genomes measured by the presence of Core Eukaryotic Genes Mapping Approach (CEGMA) genes (supplemental Table S1 (10.1MB, pdf) ) [23], cytoplasmic ribosomal proteins and independently assembled transcripts (Table 1). The genome sequence has been deposited at DDBJ/EMBL/GenBank under the accession MOOZ00000000. The version described in this paper is version MOOZ00000000. In addition, the main results from genome assembly, annotation and analysis can be downloaded at http://prodata.swmed.edu/LepDB/.
Table 1.
Quality and composition of lepidoptera genomes.
| Feature | Aly | Pra | Cce | Lac | Pgl | Dpl | Hme | Mci | Bmo | Pxy | Mse | Ppo | Pse | Pxu |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genome size (Mb) | 567 | 246 | 729 | 298 | 375 | 249 | 274 | 390 | 481 | 394 | 419 | 227 | 406 | 244 |
| Genome size without gap (Mb) | 536 | 243 | 689 | 290 | 361 | 242 | 270 | 361 | 432 | 387 | 400 | 218 | 347 | 238 |
| Heterozygosity (%) | 1.5 | 1.5 | 1.2 | 1.5 | 2.3 | 0.55 | n.a. | n.a. | n.a. | ~2 | n.a. | n.a. | 1.2 | n.a. |
| Scaffold N50 (kb) | 558 | 617 | 233 | 525 | 231 | 716 | 194 | 119 | 3999 | 734 | 664 | 3672 | 257 | 6199 |
| CEGMA (%) | 99.6 | 99.6 | 100 | 99.3 | 99.6 | 99.6 | 98.2 | 98.9 | 99.6 | 98.7 | 99.8 | 99.3 | 99.3 | 99.6 |
| CEGMA coverage by single scaffold (%) | 87.1 | 88.7 | 85.3 | 86.6 | 86.9 | 87.4 | 86.5 | 79.2 | 86.8 | 84.1 | 86.4 | 85.8 | 87.4 | 88.8 |
| Cytoplasmic Ribosomal Proteins (%) | 98.9 | 98.9 | 98.9 | 98.9 | 98.9 | 98.9 | 94.6 | 94.6 | 98.9 | 93.5 | 98.9 | 98.9 | 98.9 | 97.8 |
| De novo assembled transcripts (%) | 98 | 99 | 97 | 98 | 98 | 96 | n.a. | 97 | 98 | 83 | n.a. | n.a. | 97 | n.a. |
| GC content (%) | 35.3 | 32.7 | 37.1 | 34.4 | 35.4 | 31.6 | 32.8 | 32.6 | 37.7 | 38.3 | 35.3 | 34.0 | 39.0 | 33.8 |
| Repeat (%) | 25.0 | 22.7 | 34 | 15.5 | 22.0 | 16.3 | 24.9 | 28.0 | 44.1 | 34.0 | 24.9 | n.a. | 17.2 | n.a. |
| Exon (%) | 3.57 | 7.91 | 3.11 | 6.96 | 5.07 | 8.40 | 6.38 | 6.36 | 4.03 | 6.35 | 5.34 | 5.11 | 6.20 | 8.59 |
| Intron (%) | 28.4 | 33.3 | 24 | 31.6 | 25.6 | 28.1 | 25.4 | 30.7 | 15.9 | 30.7 | 38.3 | 24.8 | 25.5 | 45.5 |
| Number of proteins (thousands) | 15.9 | 13.2 | 16.5 | 17.4 | 15.7 | 15.1 | 12.8 | 16.7 | 14.3 | 18.1 | 15.6 | 15.7 | 16.5 | 13.1 |
n.a. Data not available
Aly: Achalarus lyciades; Pra: Pieris rapae; Cce: Calycopis cecrops; Lac: Lerema accius; Pgl: Pterourus glaucus; Dpl: Danaus plexippus; Hme: Heliconius melpomene; Mci: Melitaea cinxia; Bmo: Bombyx mori; Pxy: Plutella xylostella; Mse: Manduca sexta; Ppo: Papilio polytes; Pse: Phoebis sennae; Pxu: Papilio xuthus.
We assembled the transcriptome of Aly from the same specimen that was used for the genome assembly. Based on the transcriptome, homologs from other Lepidoptera and Drosophila melanogaster, de novo gene predictions, and repeat identification (supplemental Table S2A & 2B), we predicted 15881 protein-coding genes in the Aly genome (supplemental Table S2C). 71.7% of these genes are likely expressed in the adult, as they fully or partially overlap with the transcripts. We annotated the putative functions of the 11778 protein-coding genes (supplemental Table S2D). Although the genome size of Aly is larger than that of most other Lepidoptera genomes, the number of proteins encoded by the
Heterozygosity: Calculated as the percent of heterozygous positions detected by the Genome Analysis Toolkit (GATK) [69] for Pgl, Lac, Cce, Pra and Pse; or taken from information in the literature for Dpl [13]; or estimated based on the histogram of K-mer frequencies for Pxy [19, 43].
genome is comparable to other Lepidoptera and smaller than in the other Skipper Lerema accius. This discrepancy indicates that the increase in size of Aly genome arises from expansion in the non-coding regions and transposons.
2.2. Phylogeny of Lepidoptera
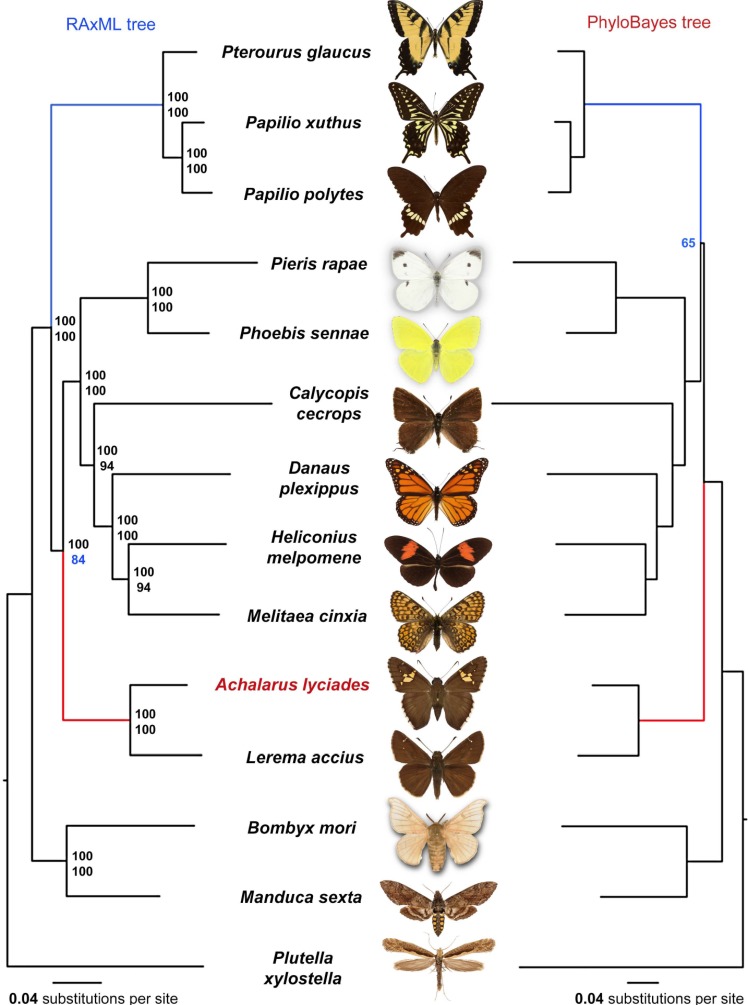
We investigated the position of Achalarus in a phylogenetic tree constructed from protein-coding genes in Lepidoptera with completely sequenced genomes. Analysis of proteins encoded by 14 available Lepidoptera genomes (Achalarus lyciades, Bombyx mori, Manduca sexta, Lerema accius, Pterourus glaucus, Papilio polytes, Papilio xuthus, Phoebis sennae, Melitaea cinxia, Heliconius melpomene, Danaus plexippus, and Plutella xylostella, Calycopis cecrops and Pieris rapae) revealed 4886 universal orthologous groups. Each Lepidoptera species is represented by a single ortholog in 1814 of these groups. Concatenated alignment of such single-copy orthologs resulted in RAxML [24] tree placing Achalarus as the sister to Lerema (Fig. 2 left), the only other member of the Hesperiidae family with sequenced genome. Even with two skipper genomes now included, Papilionidae (and not Hesperiidae) is a sister to all other butterflies in the RAxML tree (Fig. 2 left). Traditional morphology-based view placed Hesperiidae as a sister to all other butterflies [25, 26]. However, starting from Wahlberg [16, 17] the majority of DNA-based phylogenies show this alternative topology with swallowtails being a sister to other butterflies and skippers [5, 27].
Fig. (2).
Phylogenetic trees of the Lepidoptera species with complete genome sequences. Majority-rule consensus tree of the maximal likelihood trees constructed by RAxML on the concatenated alignment of universal single-copy orthologous proteins is shown on the left. Numbers by the nodes refer to bootstrap percentages. The numbers above are obtained from complete alignments, the number below are obtained on 1% of the dataset. The tree shown on the right is constructed by PhyloBayes using the topology constrained to the RAxML topology everywhere except the relative position of Skippers and Swallowtails. The topology shown (Skippers as sister to other butterflies) was recovered in 65% of PhyloBayes runs. The difference between the trees in relative position of Skippers (red) and swallowtails (blue) is indicated. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this paper.)
RAxML gives 100% bootstrap support to all nodes in the tree constructed from a concatenated alignment of all single-copy orthologs. Because bootstrap reflects consistency of phylogenetic signal between different parts of the alignment, very long alignments typically produce 100% support even if the tree topology results from various systematic biases, for instance, nucleotide composition or long branch attraction. To probe weaker nodes in the RAxML tree, we randomly split the concatenated alignment into 50 alignments (5944 positions in each alignment) and obtained a consensus tree of trees built from these shorter alignments. The node placing the skippers within other butterflies receives the lowest support (84%), indicating that sequencing additional representatives of skippers is needed to determine the relative position of skippers and swallowtails.
Interestingly, when we used PhyloBayes [28] with the CAT model [29] (4 categories of evolutionary rates for sites) to analyze the two relative positions of skippers and swallowtails, the traditional topology (skippers as a sister to all butterflies) was supported in 65% of the 50 random samples (Fig. 2 right). Discrepancies between morphology-based and DNA-based trees are commonly seen in phylogenetic analysis [30]. The incongruence between trees constructed with different methods or using different data sets is also frequent [30, 31], and phylogenetic studies of other organisms revealed uncertainties similar to the one we encountered for butterflies [32, 33]. In addition to imperfections in phylogeny-reconstruction methods, this uncertainty in butterfly phylogeny may also result from incomplete lineage sorting [34] or ancient introgression [35]. Finally, the limited number of taxa with complete genomes sequenced may further impede phylogenetic reconstruction. Thus, complete genomes of species involved in deeper branching of each butterfly family could resolve the uncertainty in the phylogenetic tree of butterflies.
2.3. Spread-wing and Grass Skippers: Comparison with Lerema
In Lerema accius genome, we found a unique large expansion into 10 endochitinase-like proteins that arose by duplication and diversification of an endochitinase gene, which is shared among all Lepidoptera and is present as a single copy in genomes other than Lerema accius [5]. The coding genes of these additional endochitinase-like proteins all cluster together in the Lac genome near the locus of the classic endochitinase gene. Their sequences diverged rapidly from the classic endochitinase and lost the chitin-binding domain. We hypothesized that the additional endochitinase-like proteins encoded by the Lac genome could have adopted new functions, possibly cellulase or other sugar food source degrading enzymes, because: (1) Lac feeds on nutrient poor but cellulose-rich grasses; (2) the Lac genome and other Lepidoptera genomes do not encode proteins that belong to the families of known cellulases; (3) endochitinases are homologs of cellulases and they are structurally similar [36]; (4) cellulose and chitin are chemically similar; (5) Lepidoptera genomes, including Lac, encode more than 10 other families of endochitinases and chitinases, and therefore having additional enzymes to hydrolize chitin may not be crucial for them.
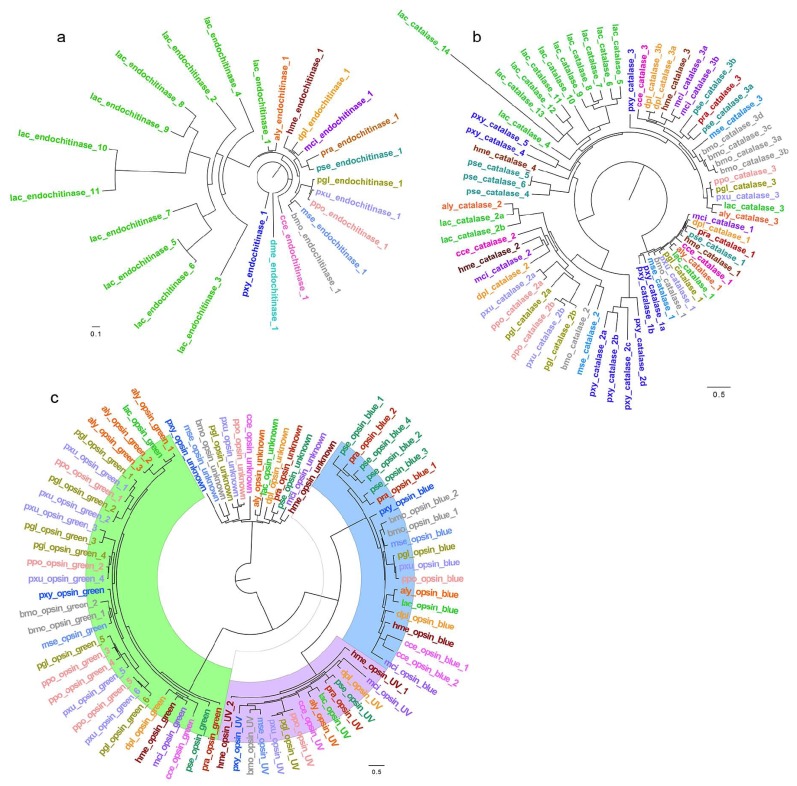
The Aly genome, similarly to the other 13 available Lepidoptera genomes compared in this study, lacks this gene expansion (Fig. 3a). The expansion is apparently unique to Lerema and possibly other Grass skippers, but not to all Hesperiidae, possibly enabling the Grass skipper caterpillars to digest cellulose and thus allowing them to feed on nutrient-poor and cellulose-rich grasses. In contrast, A. lyciades, whose caterpillars feed on nutrient-rich leaves of bean family plants, are not expected to benefit from the ability to digest cellulose more than other non-grass feeding butterflies. The lack of this family expansion in Aly gives further evidence for the hypothesis that the endochitinase-like proteins expanded in Grass skippers may function in cellulose digestion.
Fig. (3).
Gene expansions in Skippers. (a) Expansion of the endochitinase-like proteins coding genes is only found in grass feeding skipper Lerema accius, but not in bean family plants leaves feeding Achalarus lyciades and other Lepidoptera genomes. (b) Catalase is the second family of genes that is expanded in L. accius but not in A. lyciades. (c) Besides one UV-sensing opsin and one blue-light-sensing opsin both L. accius and A. lyciades genome possess, A. lyciades genome encodes two more copies of green-light sensing opsins than L. accius. aly: Achalarus lyciades; pra: Pieris rapae; cce: calycopis cecrops; lac: Lerema accius; pgl: Pterourus glaucus; dpl: Danaus plexippus; hme: Heliconius melpomene; mci: Melitaea cinxia; bmo: Bombyx mori; pxy: Plutella xylostella; mse: Manduca sexta; ppo: Papilio polytes; pse: Phoebis sennae; pxu: Papilio xuthus.
Another large gene expansion detected in Lerema but not in A. lyciades involves catalase (Fig. 3b). Catalases protect against oxidative stress by the decomposition of hydrogen peroxide into water and oxygen. The functional relevance of this gene expansion event is not clear. It is possible that Lerema is exposed to higher oxidative stress due to its food source and environment, and expansion in catalases may be another example of adaptive evolution.
We identified pheromone-binding proteins (PBPs) encoded in all Lepidoptera genomes. Lerema genome encodes the largest number of PBPs among the 14 Lepidoptera species in this study. Lerema has 61 genes encoding for PBPs, while other Lepidoptera genomes encode 36 ± 7 PBPs. Expansion of PBPs suggests a more advanced pheromone sensing system in Lac. Butterflies can select their mates both by using visual cues and by sensing pheromones. Because many Grass skippers are similar in wing colors and patterns, a stronger pheromone system in Lerema should allow for better detection of mates.
Many Spread-wing skippers, as represented by A. lyciades, possess more colorful and diverse wing patterns. Therefore, they may rely to a greater degree on visual cues for mate recognition. The A. lyciades genome encodes 37 PBPs, comparable to most other Lepidoptera species but much less than Lerema. In contrast, A. lyciades genome encodes more opsins than Lerema (Fig. 3c). Lerema and A. lyciades both have one UV-sensing opsin and one blue-light-sensing opsin. However, A. lyciades genome encodes two additional copies of green-light sensing opsins compared to Lerema, suggesting better perception of shorter wavelength light (possibly ultraviolet), and could be an adaptation for better recognition of mates and flowers for feeding, or better vision in dim areas such as forests that it inhabits and at dusk when A. lyciades is still active.
3. METHODS
3.1. Library Preparation and Sequencing
Methods used in this study are generally similar to those we used in our previous genomics work [5, 6, 10, 37, 38]. Parts of the body of a freshly caught Achalarus lyciades specimens (NVG-3311, USA: Texas, Sabine Co., Sabine National Forest, 1 mi south of Fairmount, near Fox Hunters' Hill, GPS 31.185394, -93.72992, 12-Apr-2015), were stored in RNAlater solution and wings preserved to be deposited in the National Museum of Natural History, Smithsonian Institution, Washington, DC, USA (USNM). We used this specimen NVG-3311 to assemble the reference genome. We extracted genomic DNA from its body with the ChargeSwitch gDNA mini tissue kit. 250 bp and 500 bp paired-end libraries were prepared using genomic DNA with enzymes from NEBNext Modules and following the Illumina TruSeq DNA sample preparation guide. 2 kb, 6 kb and 15 kb mate pair libraries were prepared using a protocol similar to previously published Cre-Lox-based method [39]. For the 250 bp, 500 bp, 2 kbp, 6 kbp and 15 kbp libraries, approximately 400 ng, 400 ng, 2 μg, 3 μg and 5 μg of DNA were used, respectively. We quantified the amount of DNA from all the libraries with the KAPA Library Quantification Kit, and mixed 250 bp, 500 bp, 2 kbp, 6 kbp, 15 kbp libraries at relative molar concentration 40:20:8:4:3. The mixed library was sequenced for 150 bp at both ends using Illumina HiSeq 2500 at UT Southwestern Medical Center genomics core facility.
Part of the specimen NVG-3311 was used to extract RNA using QIAGEN RNeasy Mini Kit. We further isolated mRNA using NEBNext Poly(A) mRNA Magnetic Isolation Module and RNA-seq libraries for both specimens were prepared with NEBNext Ultra Directional RNA Library Prep Kit for Illumina following manufactory’s protocol. The RNA-seq library was sequenced for 150 bp from both ends using Illumina HiSeq 2500.
3.2. Genome and Transcriptome Assembly
We removed sequence reads that did not pass the purity filter and classified the pass-filter reads according to their TruSeq adapter indices to get individual sequencing libraries. Mate pair libraries were processed by the Delox script [39] to remove the loxP sequences and to separate true mate pair from paired-end reads. All reads were processed by mirabait [40] to remove contamination from the TruSeq adapters, an in-house script to remove low quality portions (quality score < 20) at the ends of both reads, by JELLYFISH [41] to obtain k-mer frequencies in all the libraries, and by QUAKE [42] to correct sequencing errors. The data processing resulted in seven libraries that were supplied to Platanus [43] for genome assembly: 250 bp and 500 bp paired-end libraries, 2 kbp, 6kbp, 15kbp true mate pair libraries, a library containing all the paired-end reads from the mate pair libraries, and a single-end library containing all reads whose pairs were removed in the process (Supplemental Table S2A).
We mapped these reads to the initial assembly with Bowtie2 [44] and calculated the coverage of each scaffold with the help of SAMtools [45]. Many short scaffolds in the assembly showed coverage that was about half of the expected
value; they likely came from highly heterozygous regions that were not merged to the equivalent segments in the homologous chromosomes. We removed them if they could be fully aligned to another significantly less covered region (coverage > 90% and uncovered region < 500 bp) in a longer scaffold with high sequence identity (>95%). Similar problems occurred in the Heliconius melpomene, Pterourus glaucus and Lerema accius genome projects, and similar strategies were used to improve the assemblies [5, 6, 12].
The RNA-seq reads were processed using a similar procedure as the genomic DNA reads to remove contamination from TruSeq adapters and the low quality portion of the reads. Afterwards, we applied three methods to assemble the transcriptomes: (1) de novo assembly by Trinity [46], (2) reference-based assembly by TopHat [47] (v2.0.10) and Cufflinks [48] (v2.2.1), and (3) reference-guided assembly by Trinity. The results from all three methods were then integrated by Program to Assemble Spliced Alignment (PASA) [49].
3.3. Identification of Repeats and Gene Annotation
Two approaches were used to identify repeats in the genome: the RepeatModeler [50] pipeline and in-house scripts that extracted regions with coverage 3 times higher than expected. These repeats were submitted to the CENSOR [51] server to assign them to the repeat classification hierarchy. The species-specific repeat library and all repeats classified in RepBase [52] (V18.12) were used to mask repeats in the genome by RepeatMasker [53].
We obtained two sets of transcript-based annotations from two pipelines: TopHat followed by Cufflinks and Trinity followed by PASA. In addition, we obtained six sets of homology-based annotations by aligning protein sets from Drosophila melanogaster [54] and five published Lepidoptera genomes (Bombyx mori, Lerema accius, Papilio xuthus, Heliconius melpomene, and Danaus plexippus) to the Achalarus lyciades genome with exonerate [55]. Proteins from insects in the entire UniRef90 [56] database were used to generate another set of gene predictions by genblastG [57]. We manually curated and selected 1204 confident gene models by integrating the evidence from transcripts and homologs to train de novo gene predictors: AUGUSTUS [58], SNAP [59] and GlimmerHMM [60]. These trained predictors, the self-trained Genemark [61] and a consensus-based pipeline Maker [62], were used to generate another five sets of gene models. Transcript-based and homology-based annotations were supplied to AUGUSTUS, SNAP and Maker to boost their performance. In total, we generated 14 sets of gene predictions and integrated them with EvidenceModeller [49] to generate the final gene models.
We predicted the function of Aly proteins by transferring annotations and GO-terms from the closest BLAST [63] hits (E-value < 10-5) in both the Swissprot [64] database and Flybase [65]. Finally, we performed InterproScan [66] to identify conserved protein domains and functional motifs, to predict coiled coils, transmembrane helices and signal peptides, to detect homologous 3D structures, to assign proteins to protein families and to map them to metabolic pathways.
3.4. Identification of Orthologous Proteins, Gene Expansion and Phylogenetic Tree Construction
We identified the orthologous groups from 14 Lepidoptera genomes using OrthoMCL [67]. If two OrthoMCL-defined orthologous groups overlapped in the Drosophila proteins that they mapped to, we merged them into one family. The function of each family is annotated using GO terms. GO terms that are associated with any gene in a family are considered to be associated with this family. The total number and total length of proteins in a family were used to identify expanded gene families in Achalarus. If the total number and length of Achalarus proteins in a family are more than 1.5 times of the average number and length across other Lepidoptera species, we consider this protein family to have undergone expansion in Achalarus. The enrichment of GO terms associated with these expanded families is identified using a binomial test: m = the number of expanded gene families that were associated with this GO term, N = number of expanded gene families, p = the probability for this GO term to be associated with any gene family.
1814 orthologous groups consisted of single-copy genes from every species, and they were used for phylogenetic analysis. An alignment was built for each universal single-copy orthologous group using both global sequence aligner MAFFT [68] and local sequence aligner BLASTP. Positions that were consistently aligned by both aligners were extracted from each individual alignment and concatenated to obtain an alignment containing 297,210 positions. The concatenated alignment was used to obtain a phylogenetic tree using RAxML [24]. Bootstrap resampling of the aligned positions was performed to assign the confidence level of each node in the tree. In addition, in order to detect the weakest nodes in the tree, we reduced the amount of data by randomly splitting the concatenated alignment into 100 alignments (about 2,972 positions in each alignment) and applied RAxML to each alignment. We obtained a 50% majority rule consensus tree and assigned confidence level to each node based on the percent of individual trees supporting this node.
AUTHORS’ CONTRIBUTIONS
J.S. and Q.C. designed and carried out the experiments, performed the computational analyses and drafted the manuscript. D.B. and Z.O. designed and supervised experimental studies; N.V.G. directed the project and drafted the manuscript. All authors wrote the manuscript.
ACKNOWLEDGEMENTS
We acknowledge Texas Parks and Wildlife Department (Natural Resources Program Director David H. Riskind) for the permit #08-02Rev that makes research based on material collected in Texas State Parks possible. We thank R. Dustin Schaeffer for critical suggestions and proofreading of the manuscript; Qian Cong is a Howard Hughes Medical Institute International Student Research fellow. This work was supported in part by the National Institutes of Health (GM094575 to NVG) and the Welch Foundation (I-1505 to NVG).
SUPPLEMENTARY MATERIAL
Supplementary material is available on the publisher’s web site along with the published article.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
REFERENCES
- 1.Warren A.D., Ogawa J.R., Brower A.V. Revised classification of the family Hesperiidae (Lepidoptera: Hesperioidea) based on combined molecular and morphological data. Syst. Entomol. 2009;34(3):467–523. [Google Scholar]
- 2.Scott J.A. The butterflies of north america: A natural history and field guide. Stanford University Press; 1986. [Google Scholar]
- 3.Opler P.A., Krizek G.O. Butterflies east of the Great Plains: an illustrated natural history. Baltimore: Johns Hopkins University Press; 1984. [Google Scholar]
- 4.Cech R., Tudor G. Butterflies of the East Coast: An Observer’s Guide. New Jersey: Princeton University Press; 2005. [Google Scholar]
- 5.Cong Q., Borek D., Otwinowski Z., Grishin N.V. Skipper genome sheds light on unique phenotypic traits and phylogeny. BMC Genomics. 2015;16:639. doi: 10.1186/s12864-015-1846-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cong Q., Borek D., Otwinowski Z., Grishin N.V. Tiger swallowtail genome reveals mechanisms for speciation and caterpillar chemical defense. Cell Rep. 2015. [DOI] [PMC free article] [PubMed]
- 7.Mallet J. New genomes clarify mimicry evolution. Nat. Genet. 2015;47(4):306–307. doi: 10.1038/ng.3260. [DOI] [PubMed] [Google Scholar]
- 8.Li X., Fan D., Zhang W., Liu G., Zhang L., Zhao L., Fang X., Chen L., Dong Y., Chen Y., Ding Y., Zhao R., Feng M., Zhu Y., Feng Y., Jiang X., Zhu D., Xiang H., Feng X., Li S., Wang J., Zhang G., Kronforst M.R., Wang W. Outbred genome sequencing and CRISPR/Cas9 gene editing in butterflies. Nat. Commun. 2015;6:8212. doi: 10.1038/ncomms9212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cong Q., Shen J., Warren A.D., Borek D., Otwinowski Z., Grishin N.V. Speciation in cloudless sulphurs gleaned from complete genomes. Genome Biol. Evol. 2016;8(3):915–931. doi: 10.1093/gbe/evw045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cong Q., Shen J., Borek D., Robbins R.K., Otwinowski Z., Grishin N.V. Complete genomes of Hairstreak butterflies, their speciation, and nucleo-mitochondrial incongruence. Sci. Rep. 2016;6:24863. doi: 10.1038/srep24863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ahola V., Lehtonen R., Somervuo P., Salmela L., Koskinen P., Rastas P., Valimaki N., Paulin L., Kvist J., Wahlberg N., Tanskanen J., Hornett E.A., Ferguson L.C., Luo S., Cao Z., de Jong M.A., Duplouy A., Smolander O.P., Vogel H., McCoy R.C., Qian K., Chong W.S., Zhang Q., Ahmad F., Haukka J.K., Joshi A., Salojarvi J., Wheat C.W., Grosse-Wilde E., Hughes D., Katainen R., Pitkanen E., Ylinen J., Waterhouse R.M., Turunen M., Vaharautio A., Ojanen S.P., Schulman A.H., Taipale M., Lawson D., Ukkonen E., Makinen V., Goldsmith M.R., Holm L., Auvinen P., Frilander M.J., Hanski I. The Glanville fritillary genome retains an ancient karyotype and reveals selective chromosomal fusions in Lepidoptera. Nat. Commun. 2014;5:4737. doi: 10.1038/ncomms5737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heliconius Genome C. Butterfly genome reveals promiscuous exchange of mimicry adaptations among species. Nature. 2012;487(7405):94–98. doi: 10.1038/nature11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhan S., Merlin C., Boore J.L., Reppert S.M. The monarch butterfly genome yields insights into long-distance migration. Cell. 2011;147(5):1171–1185. doi: 10.1016/j.cell.2011.09.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhan S., Zhang W., Niitepold K., Hsu J., Haeger J.F., Zalucki M.P., Altizer S., de Roode J.C., Reppert S.M., Kronforst M.R. The genetics of monarch butterfly migration and warning colouration. Nature. 2014;514(7522):317–321. doi: 10.1038/nature13812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nadeau N.J., Ruiz M., Salazar P., Counterman B., Medina J.A., Ortiz-Zuazaga H., Morrison A., McMillan W.O., Jiggins C.D., Papa R. Population genomics of parallel hybrid zones in the mimetic butterflies, H. melpomene and H. erato. Genome Res. 2014;24(8):1316–1333. doi: 10.1101/gr.169292.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mutanen M., Wahlberg N., Kaila L. Comprehensive gene and taxon coverage elucidates radiation patterns in moths and butterflies. Proc. Biol. Sci. 1695;2010(277):2839–2848. doi: 10.1098/rspb.2010.0392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heikkila M., Kaila L., Mutanen M., Pena C., Wahlberg N. Cretaceous origin and repeated tertiary diversification of the redefined butterflies. Proc. Biol. Sci. 1731;2012(279):1093–1099. doi: 10.1098/rspb.2011.1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.International Silkworm Genome C. The genome of a lepidopteran model insect, the silkworm Bombyx mori. Insect Biochem. Mol. Biol. 2008;38(12):1036–1045. doi: 10.1016/j.ibmb.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 19.You M., Yue Z., He W., Yang X., Yang G., Xie M., Zhan D., Baxter S.W., Vasseur L., Gurr G.M., Douglas C.J., Bai J., Wang P., Cui K., Huang S., Li X., Zhou Q., Wu Z., Chen Q., Liu C., Wang B., Li X., Xu X., Lu C., Hu M., Davey J.W., Smith S.M., Chen M., Xia X., Tang W., Ke F., Zheng D., Hu Y., Song F., You Y., Ma X., Peng L., Zheng Y., Liang Y., Chen Y., Yu L., Zhang Y., Liu Y., Li G., Fang L., Li J., Zhou X., Luo Y., Gou C., Wang J., Wang J., Yang H., Wang J. A heterozygous moth genome provides insights into herbivory and detoxification. Nat. Genet. 2013;45(2):220–225. doi: 10.1038/ng.2524. [DOI] [PubMed] [Google Scholar]
- 20.Tang W., Yu L., He W., Yang G., Ke F., Baxter S.W., You S., Douglas C.J., You M. DBM-DB: the diamondback moth genome database. 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhan S., Reppert S.M. MonarchBase: the monarch butterfly genome database. Nucleic Acids Res. 2013;41(Database issue):D758–D763. doi: 10.1093/nar/gks1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Duan J., Li R., Cheng D., Fan W., Zha X., Cheng T., Wu Y., Wang J., Mita K., Xiang Z., Xia Q. SilkDB v2.0: a platform for silkworm (Bombyx mori) genome biology. Nucleic Acids Res. 2010;38(Database issue):D453–D456. doi: 10.1093/nar/gkp801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parra G., Bradnam K., Korf I. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics. 2007;23(9):1061–1067. doi: 10.1093/bioinformatics/btm071. [DOI] [PubMed] [Google Scholar]
- 24.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30(9):1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ackery P.R., de Jong R., Vane-Wright R.I. The butterflies: Hedyloidea, Hesperioidea, and Papilionoidea. in Lepidoptera, Moths and Butterflies. Volume 1: Evolution, Systematics, and Biogeography. Handbuch der Zoologie. Handbook of Zoology. Band / Volume IV Arthropoda: Insecta Teilband / Part 35. de Gruyter: Berlin, New York, 1999. 1999. [Google Scholar]
- 26.Wahlberg N., Braby M.F., Brower A.V., de Jong R., Lee M.M., Nylin S., Pierce N.E., Sperling F.A., Vila R., Warren A.D., Zakharov E. Synergistic effects of combining morphological and molecular data in resolving the phylogeny of butterflies and skippers. . Proc. Biol. Sci. 2005;272(1572):1577–1586. doi: 10.1098/rspb.2005.3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kawahara A.Y., Breinholt J.W. Phylogenomics provides strong evidence for relationships of butterflies and moths. Proc. Biol. Sci. 1788;2014(281):20140970. doi: 10.1098/rspb.2014.0970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lartillot N., Lepage T., Blanquart S. PhyloBayes 3: a Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics. 2009;25(17):2286–2288. doi: 10.1093/bioinformatics/btp368. [DOI] [PubMed] [Google Scholar]
- 29.Lartillot N., Philippe H. A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol. Biol. Evol. 2004;21(6):1095–1109. doi: 10.1093/molbev/msh112. [DOI] [PubMed] [Google Scholar]
- 30.Patterson C., Williams D.M., Humpries C.J. Congruence between molecular and morphological phylogenies. Annu. Rev. Ecol. Syst. 1993;24:153–188. [Google Scholar]
- 31.Pisani D., Benton M.J., Wilkinson M. Congruence of morphological and molecular phylogenies. Acta Biotheor. 2007;55(3):269–281. doi: 10.1007/s10441-007-9015-8. [DOI] [PubMed] [Google Scholar]
- 32.Jékely G., Paps J., Nielsen C. The phylogenetic position of ctenophores and the origin(s) of nervous systems. Evodevo. 2015;6(1):1–9. doi: 10.1186/2041-9139-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Talavera G., Vila R. What is the phylogenetic signal limit from mitogenomes? The reconciliation between mitochondrial and nuclear data in the Insecta class phylogeny. BMC Evol. Biol. 2011;11(1):1–15. doi: 10.1186/1471-2148-11-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jarvis E.D., Mirarab S., Aberer A.J., Li B., Houde P., Li C., Ho S.Y., Faircloth B.C., Nabholz B., Howard J.T., Suh A., Weber C.C., da Fonseca R.R., Li J., Zhang F., Li H., Zhou L., Narula N., Liu L., Ganapathy G., Boussau B., Bayzid M.S., Zavidovych V., Subramanian S., Gabaldon T., Capella-Gutierrez S., Huerta-Cepas J., Rekepalli B., Munch K., Schierup M., Lindow B., Warren W.C., Ray D., Green R.E., Bruford M.W., Zhan X., Dixon A., Li S., Li N., Huang Y., Derryberry E.P., Bertelsen M.F., Sheldon F.H., Brumfield R.T., Mello C.V., Lovell P.V., Wirthlin M., Schneider M.P., Prosdocimi F., Samaniego J.A., Vargas Velazquez A.M., Alfaro-Nunez A., Campos P.F., Petersen B., Sicheritz-Ponten T., Pas A., Bailey T., Scofield P., Bunce M., Lambert D.M., Zhou Q., Perelman P., Driskell A.C., Shapiro B., Xiong Z., Zeng Y., Liu S., Li Z., Liu B., Wu K., Xiao J., Yinqi X., Zheng Q., Zhang Y., Yang H., Wang J., Smeds L., Rheindt F.E., Braun M., Fjeldsa J., Orlando L., Barker F.K., Jonsson K.A., Johnson W., Koepfli K.P., O’Brien S., Haussler D., Ryder O.A., Rahbek C., Willerslev E., Graves G.R., Glenn T.C., McCormack J., Burt D., Ellegren H., Alstrom P., Edwards S.V., Stamatakis A., Mindell D.P., Cracraft J., Braun E.L., Warnow T., Jun W., Gilbert M.T., Zhang G. Whole-genome analyses resolve early branches in the tree of life of modern birds. Science. 2014;346(6215):1320–1331. doi: 10.1126/science.1253451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mallet J., Besansky N., Hahn M.W. How reticulated are species? BioEssays. 2016;38(2):140–149. doi: 10.1002/bies.201500149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheng H., Schaeffer R.D., Liao Y., Kinch L.N., Pei J., Shi S., Kim B.H., Grishin N.V. ECOD: an evolutionary classification of protein domains. PLOS Comput. Biol. 2014;10(12):e1003926. doi: 10.1371/journal.pcbi.1003926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cong Q., Shen J., Warren A.D., Borek D., Otwinowski Z., Grishin N.V. Speciation in Cloudless Sulphurs Gleaned from Complete Genomes. Genome Biol. Evol. 2016;8(3):915–931. doi: 10.1093/gbe/evw045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shen J., Cong Q., Kinch L.N., Borek D., Otwinowski Z., Grishin N.V. Complete genome of Pieris rapae, a resilient alien, a cabbage pest, and a source of anti-cancer proteins. F1000 Res. 2016;5:2631. doi: 10.12688/f1000research.9765.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van Nieuwerburgh F., Thompson R.C., Ledesma J., Deforce D., Gaasterland T., Ordoukhanian P., Head S.R. Illumina mate-paired DNA sequencing-library preparation using Cre-Lox recombination. Nucleic Acids Res. 2012;40(3):e24. doi: 10.1093/nar/gkr1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chevreux B., Wetter T., Suhai S. Genome Sequence Assembly Using Trace Signals and Additional Sequence Information. Comput. Sci. Biol. 1999;99:45–56. [Google Scholar]
- 41.Marcais G., Kingsford C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics. 2011;27(6):764–770. doi: 10.1093/bioinformatics/btr011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kelley D.R., Schatz M.C., Salzberg S.L. Quake: quality-aware detection and correction of sequencing errors. Genome Biol. 2010;11(11):R116. doi: 10.1186/gb-2010-11-11-r116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kajitani R., Toshimoto K., Noguchi H., Toyoda A., Ogura Y., Okuno M., Yabana M., Harada M., Nagayasu E., Maruyama H., Kohara Y., Fujiyama A., Hayashi T., Itoh T. Efficient de novo assembly of highly heterozygous genomes from whole-genome shotgun short reads. Genome Res. 2014;24(8):1384–1395. doi: 10.1101/gr.170720.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. Genome Project Data Processing, S. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Haas B.J., Papanicolaou A., Yassour M., Grabherr M., Blood P.D., Bowden J., Couger M.B., Eccles D., Li B., Lieber M., Macmanes M.D., Ott M., Orvis J., Pochet N., Strozzi F., Weeks N., Westerman R., William T., Dewey C.N., Henschel R., Leduc R.D., Friedman N., Regev A. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013;8(8):1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14(4):R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roberts A., Pimentel H., Trapnell C., Pachter L. Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics. 2011;27(17):2325–2329. doi: 10.1093/bioinformatics/btr355. [DOI] [PubMed] [Google Scholar]
- 49.Haas B.J., Salzberg S.L., Zhu W., Pertea M., Allen J.E., Orvis J., White O., Buell C.R., Wortman J.R. Automated eukaryotic gene structure annotation using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome Biol. 2008;9(1):R7. doi: 10.1186/gb-2008-9-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Smit A.F., Hubley R. http://www.repeatmasker.org
- 51.Jurka J., Klonowski P., Dagman V., Pelton P. CENSOR--a program for identification and elimination of repetitive elements from DNA sequences. Comput. Chem. 1996;20(1):119–121. doi: 10.1016/s0097-8485(96)80013-1. [DOI] [PubMed] [Google Scholar]
- 52.Jurka J., Kapitonov V.V., Pavlicek A., Klonowski P., Kohany O., Walichiewicz J. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 2005;110(1-4):462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- 53.Smit A.F., Hubley R., Green P. http://www.repeatmasker.org
- 54.Misra S., Crosby M. A., Mungall C. J., Matthews B. B., Campbell K.S., Hradecky P., Huang Y., Kaminker J.S., Millburn G. H., Prochnik S.E., Smith C.D., Tupy J.L., Whitfied E.J., Bayraktaroglu L., Berman B.P., Bettencourt B.R., Celniker S.E., de Grey A.D., Drysdale R.A., Harris N.L., Richter J., Russo S., Schroeder A.J., Shu S.Q., Stapleton M., Yamada C., Ashburner M., Gelbart W.M., Rubin G.M., Lewis S.E. Annotation of the Drosophila melanogaster euchromatic genome: a systematic review. Genome Biol. 2002 doi: 10.1186/gb-2002-3-12-research0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Slater G.S., Birney E. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics. 2005;6:31. doi: 10.1186/1471-2105-6-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Suzek B.E., Huang H., McGarvey P., Mazumder R., Wu C.H. UniRef: comprehensive and non-redundant UniProt reference clusters. Bioinformatics. 2007;23(10):1282–1288. doi: 10.1093/bioinformatics/btm098. [DOI] [PubMed] [Google Scholar]
- 57.She R., Chu J.S., Uyar B., Wang J., Wang K., Chen N. genBlastG: using BLAST searches to build homologous gene models. Bioinformatics. 2011;27(15):2141–2143. doi: 10.1093/bioinformatics/btr342. [DOI] [PubMed] [Google Scholar]
- 58.Stanke M., Schoffmann O., Morgenstern B., Waack S. Gene prediction in eukaryotes with a generalized hidden Markov model that uses hints from external sources. BMC Bioinformatics. 2006;7:62. doi: 10.1186/1471-2105-7-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Korf I. Gene finding in novel genomes. BMC Bioinformatics. 2004;5:59. doi: 10.1186/1471-2105-5-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Majoros W.H., Pertea M., Salzberg S.L. TigrScan and GlimmerHMM: two open source ab initio eukaryotic gene-finders. Bioinformatics. 2004;20(16):2878–2879. doi: 10.1093/bioinformatics/bth315. [DOI] [PubMed] [Google Scholar]
- 61.Besemer J., Borodovsky M. GeneMark: web software for gene finding in prokaryotes, eukaryotes and viruses. Nucleic Acids Res., 2005 doi: 10.1093/nar/gki487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cantarel B.L., Korf I., Robb S.M., Parra G., Ross E., Moore B., Holt C., Sanchez A.A., Yandell M. MAKER: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 2008;18(1):188–196. doi: 10.1101/gr.6743907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 64.UniProt C. Activities at the Universal Protein Resource (UniProt). Nucleic Acids Res. 2014;42(Database issue):D191–D198. doi: 10.1093/nar/gkt1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.St Pierre S.E., Ponting L., Stefancsik R., McQuilton P. FlyBase, C. FlyBase 102--advanced approaches to interrogating FlyBase. Nucleic Acids Res. 2014;42(Database issue):D780–D788. doi: 10.1093/nar/gkt1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jones P., Binns D., Chang H.Y., Fraser M., Li W., McAnulla C., McWilliam H., Maslen J., Mitchell A., Nuka G., Pesseat S., Quinn A.F., Sangrador-Vegas A., Scheremetjew M., Yong S.Y., Lopez R., Hunter S. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30(9):1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li L., Stoeckert C.J., Jr, Roos D.S. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13(9):2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30(4):772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., DePristo M.A. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20(9):1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material is available on the publisher’s web site along with the published article.