Abstract
Introduction:
Cardiovascular and neurodegenerative disorders are among the major causes of mortality in the developed countries. Population studies evaluate the genetic risk, i.e. the probability of an individual carrying a specific disease-associated polymorphism. Identification of risk polymorphisms is essential for an accurate diagnosis or prognosis of a number of pathologies.
Aims:
The aim of this study was to characterize the influence of risk polymorphisms associated with lipid metabolism, hypertension, thrombosis, and dementia, in a large population of Spanish individuals affected by a variety of brain and vascular disorders as well as metabolic syndrome.
Material & Method:
We performed a cross-sectional study on 4415 individuals from a widespread regional distribution in Spain (48.15% males and 51.85% females), with mental, neurodegenerative, cerebrovascular, and metabolic disorders. We evaluated polymorphisms in 20 genes involved in obesity, vascular and cardiovascular risk, and dementia in our population and compared it with representative Spanish and European populations. Risk polymorphisms in ACE, AGT(235), IL6(573), PSEN1, and APOE (specially the APOE-ε4 allele) are representative of our population as compared to the reference data of Spanish and European individuals.
Conclusion:
The significantly higher distribution of risk polymorphisms in PSEN1 and APOE-ε4 is characteristic of a representative number of patients with Alzheimer’s disease; whereas polymorphisms in ACE, AGT(235), and IL6(573), are most probably related with the high number of patients with metabolic syndrome or cerebrovascular damage.
Keywords: Dementia, Alzheimer's disease, Hypertension, Metabolic syndrome, APOE, Vascular risk
1. INTRODUCTION
Vascular and neurodegenerative disorders are among the most prevalent causes of death in Western Countries. The knowledge of the human genome allows the detection of modifications in the sequence of certain genes responsible for a number of these diseases. The appropriate genetic counseling relies on the characterization of these risk polymorphisms for accurate individual or family decision making.
Metabolic Syndrome (MS) affects 20-34% of the population [1, 2], primarily in the developed countries. Definition of MS slightly differs depending on the source [3-5], although it is generally defined by the confluence of several medical conditions, including low high-density lipoprotein (HDL) levels, high blood serum triglycerides, high blood pressure, abdominal obesity, and elevated fasting plasma glucose. Individuals with MS are more likely candidates to develop cerebrovascular and cardiovascular disease and diabetes than the general population.
Cerebrovascular disorders and stroke are the third leading cause of death in the US and in Europe with around 200 cases per 100,000 inhabitants per year [6] and almost six million victims every year, according to the World Health Organization [7]. Cerebrovascular disorders are characterized by the blockade of arterial vascularization (atherosclerosis) with the consequent deprivation of oxygen and glucose supply to the affected tissue, leading to cell death and tissue necrosis or ischemia. Depending on the anatomical site of the ischemic insult, cerebral stroke can lead to hemiplegia, aphasia, motor dysfunction, or dementia. Atherosclerosis is a form of chronic inflammation resulting from the interaction of different factors, including enhanced uptake of low-density lipoprotein (LDL) by monocytes and macrophages [8, 9], immune response, and development of plaque deposits in the lumen of the blood vessels [10, 11]. Several genetic risk markers involved in the atherosclerosis-related pathways have been identified [12].
Certain polymorphisms in genes encoding apolipoproteins (APOB, APOC3, APOE) and the cholesterol ester transfer protein (CTEP) are associated with an aberrant increase in LDL-family lipoproteins, cholesterol, and triglyceride levels. Therefore, these gene variants are involved in the risk for atherosclerosis and vascular disorders [13-20]. However, the polymorphism C1421G (rs328) in the LPL gene, coding for the lipoprotein lipase, develops a protective effect by increasing the levels of high density lipoproteins (HDL) and decreasing the triglycerides [13, 21, 22]. One of the signs of the progression from the initial fatty streak to the more complex lesions in the artery wall is the high blood pressure. Specific polymorphisms in NOS3, ACE, and AGT, involved in endothelial function and regulation of blood pressure, are indicative of the progression of the atherosclerotic plaque [13, 23-27]. The role of inflammation in atherosclerosis is well established [11], as well as its implication in thrombosis by enhancing the coagulation process [28]. In this regard, polymorphisms in genes encoding proinflammatory interleukins, especially IL1 and IL6, promote their expression leading to atherothrombosis and lacunar infarction [13, 29-34]. The variant -308G>A (rs1800629) in the tumor necrosis factor gene (TNFA) has also been observed to be associated with migraine, which suggests the role of this polymorphism in the regulation of blood flow throughout the brain [35]. Although the progressive narrowing of the blood vessel during the atherosclerotic process can develop ischemic symptoms, stroke is normally promoted by the rupture of the sclerotic plaque and thrombosis. Polymorphisms in the genes coding the coagulation factors F2 and F5 are associated with the accumulation of prothrombin, increasing the risk of thrombosis [13, 36-38]. The MTHFR gene encodes for the methylentetrahydrofolate reductase which remethylates homocysteine into methionine. The polymorphisms 1298A>C (rs1801131) and 677C>T (rs1801133) in MTHFR result in the accumulation of homocysteine in plasma and increase the risk of a premature cardiovascular disorder up to three times compared to the general population [39, 40].
According to the World Health Organization (WHO), cerebrovascular and neurodegenerative disorders affect one billion people around the world. A number of these disorders are characterized by the onset of dementia. Disability caused by dementia increases dramatically with aging, by affecting 9 per 1000 of the population aged 65-74 years to 83 per 1000 in the population over 85 years old [41]. Alzheimer’s disease (AD) is the major cause of dementia in Western Countries, affecting 45-60% of the population, followed by vascular dementia and mixed dementia with prevalences of 30-40% and 10-20%, respectively [42, 43]. AD is a polygenic and complex disorder characterized by the accumulation of β-amyloid (Aβ) in senile plaques, neurofibrillary tangles, dendritic desarborization, and neuronal loss, which leads to memory deterioration, dementia, and functional decline [42-44].
The PSEN1 and PSEN2 genes, encoding presenilin1 and 2, are important determinants of the β-secretase activity responsible for proteolytic cleavage of the Aβ-precursor protein (APP). Mutations in the PSEN1, PSEN2, and APP confer phenotypes of amyloidogenic pathology and dementia [19, 42, 43, 45]. One of the most prevalent risk genes in AD is the APOE, especially in those individuals harboring the APOE-ε4 allele [13, 19, 44, 46]. Interestingly, the allele APOE-ε2, which is associated with vascular risk, seems to be protective against dementia [13, 19, 44]. The A2M gene, encoding for the alpha-2-macroglobulin (a protease inhibitor), is also localized in amyloid plaques and interacts with Aβ and APOE. The polymorphism 2998G>A (rs669) in homozygosis increases the risk for the onset of AD by 4-fold when compared to the general population [13, 19, 45].
We characterized risk polymorphisms of genes related to obesity, cardiovascular disorders, and dementia in 4415 individuals from Spain. This population included a high rate of individuals diagnosed with vascular-related disorders (25%), and with dementia-related diseases (15%). The other 55% of the population was diagnosed with other pathologies that might be indirectly related to vascular or neurodegenerative disorders. We aimed to characterize which risk polymorphisms were predominant in our population and found that polymorphisms in ACE, AGT(235), IL6(573), PSEN1, and APOE (especially the APOE-ε4 allele) were representative of our population as compared to reference data from Spanish and European individuals. The significantly high rate of the polymorphism +16G>T (rs165932) in the PSEN1 gene, along with the high representation of the APOE-ε4 allele, strongly suggests the presence of a significantly large group of individuals with potential AD disease onset in this population. The large number of individuals analyzed in this study may provide the main representative polymorphisms associated with risk for vascular disorders and dementia in the Spanish population.
2. MATERIALS AND METHODS
2.1. Subjects
Risk polymorphisms were analyzed in a population of 4415 individuals from Spain (72% from Galicia and Northern Spanish regions, and the remaining 18% from a widespread Spanish distribution) attending the outpatient clinic at the EuroEspes Biomedical Research Center from January 1995 to December 2015. The individuals were equally distributed in terms of gender, being 48.15% males and 51.85% females. Table 1 shows the distribution of the population according to age-range and diagnosis. The most representative age ranges were 30-60 years old (43%) and older than 60 (37%), whereas only 20% of individuals were younger than 30 years old. A representative number of those individuals (25%) were diagnosed with disorders associated with vascular damage or vascular risk, such as metabolic syndrome, brain trauma, cerebral stroke, epilepsy, cephalea. A total of 15% of individuals were diagnosed with dementia-related disorders, including AD, and vascular and mixed dementias; these last two being the most representative ones. Among neurodegenerative disorders, besides AD, approximately 4% of individuals were diagnosed with Parkinson’s disease. Over 26% carried psychiatric disorders, including stress-anxiety and depression. The last 30% of patients were diagnosed with peripheral nervous system disorders and other pathologies (Table 1).
Table 1.
Description of the population characterized in this study: 4415 patients distributed throughout Spain (48.15% males and 51.85% females) who attended the outpatient clinic at the EuroEspes Biomedical Research Center (Spain) from January 1995 to December 2015.
| Age Range | Age Rate (%) | Disease | Disease Rate (%) |
|---|---|---|---|
| 0-15 | 7.18 | Cephalea | 9.33 |
| 16-30 | 13.16 | Stress-Anxiety | 18.10 |
| 30-60 | 43.10 | Depression | 8.18 |
| >60 | 36.56 | Dementia (AD,VD,MD) | 14.75 |
| __ | __ | Parkinson's | 3.94 |
| __ | __ | Cerebral Stroke | 9.99 |
| __ | __ | Epilepsy | 4.01 |
| __ | __ | PNS Disorders | 1.18 |
| __ | __ | Brain Trauma | 0.45 |
| __ | __ | Metabolic Syndrome |
0.68 |
| __ | __ | Others | 29.40 |
AD: Alzheimer’s Disease; VD: Vascular Dementia; MD: Mixed Dementia; PNS Disorders: Peripheral Nervous System Disorders.
2.2. Genotype Analysis
Blood was extracted in EDTA-coated tubes and DNA from peripheral blood was extracted using the Qiagen DNA Blood Minikit (Qiagen, Hilden, Germany). A total of 20 different genes were genotyped (Table 2) by Real Time PCR (RT-PCR) using TaqMan assays designed for single nucleotide polymorphisms (SNPs). RT-PCRs were performed in Step One Plus Real Time PCR System (Life Technologies, Waltham, Massachusetts, USA), and TaqMan® Open Array® DNA microchips in Quant StudioTM 12K Flex RT-PCR System (Life Technologies, Waltham, Massachusetts, USA). Open Array® genotyping analyses were performed using the Genotyper software provided by Thermo Fisher Scientific, Waltham, Massachusetts, USA.
Table 2.
Allelic and genotype frequencies of different gene polymorphisms associated with obesity, vascular and cardiovascular risk, and neurodegeneration.
| Panel [Subpanel] | Gene | OMIM | Locus | Polymorphism | Risk SNP | N | Allele Freq | Genotype Freq | H-W | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| -Obesity -Vascular Risk [Lipid Metabolism] | APOB | 107730 | 2p24p23 | rs693 [7545C>T] | 7545T | 931 | C = 0.565 T = 0.435 | CC = 0.319 CT = 0.492 TT = 0.189 | Yes (P=0.65-0.7) | ||||||||||
| -Obesity -Vascular Risk [Lipid Metabolism] | APOC3 | 107720 | 11q23.3 | rs5128 [3175G>C, S1/S2] | 3175G | 932 | C = 0.906 G = 0.094 | CC = 0.821 CG = 0.170 GG = 0.009 | Yes (P=0.09-0.1) | ||||||||||
| -Obesity -Vascular Risk -Neurodegeneration [Lipid Metabolism] | APOE | 107741 | 19q13.2 | rs429358/rs7412 [112T>C/158T>C] E2, E3, E4 | 112T/158T (E2) 112C/158C (E4) | 4377 | ε3 = 0.815 ε2 = 0.05 ε4 = 0.135 | ε 3 ε 3 = 0.668 ε 2 ε 3 = 0.079 ε 2 ε 4 = 0.014 ε 3 ε 4 = 0.212 ε 2 ε 2 = 0.002 ε 4 ε 4 = 0.022 | Yes (P=0.35-0.4) | ||||||||||
| -Obesity -Vascular Risk [Lipid Metabolism] | CETP | 118460 | 16q21 | rs708272 [+279G>A, B1/B2] | +279G (B1) | 2006 | A = 0.377 G = 0.623 | AA = 0.142 AG = 0.47 GG = 0.388 | Yes (P=0.1-0.15) | ||||||||||
| -Obesity -Vascular Risk [Lipid Metabolism] | LPL | 609708 | 8p22 | rs328 [1421C>G, S474X] | 1421G (Protective) | 931 | C = 0.851 G = 0.149 | CC = 0.724 CG = 0.254 GG = 0.022 | No (P<0.001) | ||||||||||
| -Vascular Risk -Neurodegeneration [Endothelial Function] [Hypertension] | NOS3 | 163729 | 7q36 | rs1799983 [894G>T] | 894T | 2711 | G = 0.621 T = 0.379 | GG = 0.386 GT = 0.471 TT = 0.144 | Yes (P=0.25-0.3) | ||||||||||
| -Vascular Risk -Neurodegeneration [Endothelial Function] [Hypertension] | ACE | 106180 | 17q23.3 | rs4332 [547C>T] | 547T | 954 | C = 0.342 T = 0.658 | CC = 0.117 CT = 0.450 TT = 0.433 | No (P<0.001) | ||||||||||
| -Vascular Risk [Endothelial Function] [Hypertension] | AGT-174 | 1906150 | 1q42.2 | rs4762 [9360G>A, T174M] | 9360A | 3453 | G = 0.873 A = 0.127 | GG = 0.762 GA = 0.222 AA = 0.016 | No (P< 0.025) | ||||||||||
| -Vascular Risk [Endothelial Function] [Hypertension] | AGT-235 | 1906150 | 1q42.2 | rs699 [9543A>G, M235T] | 9543G | 3453 | A = 0.504 G = 0.496 | AA = 0.254 AG = 0.500 GG = 0.246 | No (P<0.001) | ||||||||||
| -Vascular Risk [Immune Response] | IL1B | 147720 | 2q14 | rs1143634 [3954C>T] | 3954T | 930 | C = 0.795 T = 0.205 | CC = 0.632 CT = 0.326 TT = 0.042 | No (P<0.025) | ||||||||||
| -Vascular Risk [Immune Response] | IL6 -174 | 147620 | 7p21 | rs1800795 [-174G>C] | -174C | 930 | G = 0.577 C = 0.423 | GG = 0.333 GC = 0.489 CC = 0.179 | No (P< 0.001) | ||||||||||
| -Vascular Risk [Immune Response] | IL6 -573 | 147620 | 7p21 | rs1800796 [-573G>C] | -573C | 930 | G = 0.856 C = 0.144 | GG = 0.733 GC = 0.247 CC = 0.021 | No (P< 0.001) | ||||||||||
| -Vascular Risk [Immune Response] | IL6R | 147880 | 1q21 | rs8192284 [1510A>C] | 1510C | 930 | A = 0.612 C = 0.388 | AA = 0.375 AC = 0.475 CC = 0.151 | Yes (P=0.85-0.9) | ||||||||||
| -Vascular Risk [Immune Response] | TNFA | 191160 | 6p21.33 | rs1800629 [-308G>A] | -308A | 929 | G = 0.854 A = 0.146 | GG = 0.729 GA = 0.249 AA = 0.021 | Yes (P=0.55-0.6) | ||||||||||
| Panel [Subpanel] | Gene | OMIM | Locus | Polymorphism | Risk SNP | N | Allele Freq | Genotype Freq | H-W | ||||||||||
| -Vascular Risk [Thrombosis] | F2 | 17693 | 11p11.2 | rs1799963 [20210G>A] | 20210A | 931 | G = 0.983 A = 0.017 | GG = 0.966 GA = 0.033 AA = 0.0002 | Yes (P=0.6-0.65) | ||||||||||
| -Vascular Risk [Thrombosis] | F5 | 227400 | 1q24.2 | rs6025 [1691G>A] | 1691A | 932 | G = 0.991 A = 0.009 | GG = 0.982 GA = 0.018 AA = 0.00008 | Yes (P=0.75-0.8) | ||||||||||
| -Vascular Risk [Thrombosis] | MTHFR (A/C) | 607093 | 1p36.22 | rs1801131 [1298A>C] | 1298C | 1004 | A = 0.705 C = 0.295 | AA = 0.497 AC = 0.416 CC = 0.087 | Yes (P=0.55-0.6) | ||||||||||
| -Vascular Risk [Thrombosis] | MTHFR (C/T) | 607093 | 1p36.22 | rs1801133 [677C>T] | 677T | 2018 | C = 0.624 T = 0.376 | CC = 0.389 CT = 0.469 TT = 0.141 | Yes (P=0.7-0.75) | ||||||||||
| -Neurodegeneration | PSEN1 | 104311 | 14q24.2 | rs165932 [+16G>T] | +16G | 2087 | T = 0.429 G = 0.571 | TT = 0.184 TG = 0.490 GG = 0.326 | No (P< 0.001) | ||||||||||
| -Neurodegeneration | A2M | 103950 | 12p13.31 | rs669 [2998G>A] | 2998G | 2000 | A = 0.694 G = 0.306 | AA = 0.482 AG = 0.425 GG = 0.094 | Yes (P=0.05-0.1) | ||||||||||
A2M: alpha-2-macroglobulin; ACE: angiotensin I converting enzyme; AGT: angiotensinogen; APOB: apolipoprotein B; APOC3: apolipoprotein CIII; APOE: apolipoprotein E; CETP: cholesteryl ester transfer protein plasma; F2: coagulation factor II; F5: coagulation factor V; IL1B: Interleukin 1 beta; IL6: Interleukin 6; IL6R: Interleukin 6 receptor; LPL: lipoprotein lipase; MTHFR: methylenetetrahydrofolate reductase (NAD(P)H); NOS3: nitric oxide synthase 3; PRNP: prion protein; PSEN1: Presenilin 1; TNFA: tumor necrosis factor A. H-W: Hardy-Weinberg equilibrium. Significant deviation from H-W equilibrium (χ2, P<0.05).
2.3. Statistical Analysis
Deviation from Hardy-Weinberg equilibrium (HWE) was analyzed by a chi-square test (χ2). Genotype distribution was considered in HWE when observed and theoretical distribution was similar with a P>0.05. The comparison of genotype frequencies in our population with others was analyzed using the Pearson’s chi-square test, considering an equal genotype distribution when the two populations showed a similar distribution with a P>0.05.
2.4. Patient Consent
Written informed consent was obtained from all participants, or from a legal caregiver or representative on their behalf in case of incapability. This study and the consent procedures were approved by the institutional review board at the EuroEspes Medical Center, in line with the ethical code of the World Medical Association (Declaration of Helsinki) and the Recommendations for the Conduct, Reporting, Editing, and Publication of Scholarly Work in Medical Journals of the International Committee of Medical Journals Editors (ICMJE).
3. Results
3.1. Polymorphisms in ACE, AGT-235, IL6-573, and PSEN1 were Significantly More Represented in Our Population Compared to Representative Spanish and European Populations
Polymorphisms in a total of 20 selected genes associated with vascular risk and dementia-related neurodegeneration were analyzed. Gene polymorphisms were selected into panels according to i) previous research [47] and ii) genotyping in over 4000 patients. Genes were then classified into risk panels, defined as groups of genes and gene polymorphisms involved in the different steps (subpanels) which lead to the atherogenic process. In this regard, gene polymorphisms were classified into Obesity, Vascular Risk, and Neurodegeneration panels, and subpanels including Lipid Metabolism, Endothelial Function, Hypertension, Immune Response, and Thrombosis (Table 2). Allele and genotype frequencies were calculated for each gene polymorphism. Risk alleles and their corresponding risk genotypes in homozygosis are in bold Table 2.
A total of 8 out of the 20 polymorphism genotype distributions analyzed were not in Hardy-Weinberg equilibrium (HWE), these including LPL (χ2=34.29; d.f.=2), ACE (χ2=48.20; d.f.=2), AGT174 (χ2=9.37; d.f.=2), AGT235 (χ2=157.41; d.f.=2), IL1B (χ2=4.88; d.f.=2), IL6(174) (χ2=6.18; d.f.=2), IL6(573) (χ2=76.31; d.f.=2), and PSEN1 (χ2=79.56; d.f.=2). Importantly, each polymorphism was analyzed in more than 900 patients (Table 2), which rules out the possibility of a non-representative sample size. This finding suggested that those genes might be biased in our population, most probably due to the variety of vascular risk and dementia phenotypes in our patients.
Fig. (1) plots the risk allele frequencies of these polymorphisms outside HWE in our population (EuroEspes, black bars) with a differential distribution by comparison with other European populations, and especially with other Spanish groups (Mixed European: 1006 individuals; Spanish: 107 individuals; Italians: 107 individuals; British: 91 individuals; Finnish: 99 individuals. These data were extracted from “dbSNP” [48] and “1000 Genomes” [49] databases). In addition, polymorphisms in APOC3 and A2M, which did not deviate from HWE in our population, also showed a differential risk allele frequency distribution when compared to another representative Spanish population (Fig. 1). We used Pearson’s chi-square test to assess whether the genotype distribution of the above mentioned genes in our population was significantly different from other representative European populations which do not deviate from HWE (Table 3). The genes with a significant genotype distribution in our patients compared to representative Spanish populations were ACE (χ2=9.49; P<0.01), AGT-235 (χ2=17.04; P<0.001), IL6-573 (χ2=10.99; P<0.005), and PSEN1 (χ2=13.59; P<0.0025) (Table 3). Indeed, the genotype distribution of IL6-573 and PSEN1 in our population significantly differed from all the other European populations tested (P<0.05), whereas those distributions of AGT-235 and ACE show similarities with a few other European populations (Table 3).
Fig. (1).
Risk allele SNP frequencies with a differential distribution in our population compared to other European populations.
Table 3.
Comparative distribution of genotype frequencies of gene polymorphisms associated with obesity, vascular/cardiovascular risk, and neurodegeneration in our study and other European populations [48, 49].
| __ | __ | Genotype Frequencies | ||||||
|---|---|---|---|---|---|---|---|---|
| Gene | Genotypes | Our Study | N-W Europeans |
Western
Europeans |
Finnish | British | Spanish | Italian |
| A2M | AA AG GG | 0.482 0.425 0.093 | 0.465 0.446 0.089 |
0.625 0.250 0.125 |
0.333 0.465 0.202 | 0.428 0.495 0.077 |
0.551 0.402 0.047 |
0.504 0.421 0.075 |
| χ2 = 0.07 P>0.95 | χ2 = 7.13 P<0.05 | χ2 = 18.31 P<0.001 | χ2 = 0.97 P>0.65 | χ2 = 3.46 P>0.2 | χ2 = 0.44 P=0.8 | |||
| ACE | CC CT TT | 0.117 0.450 0.433 | -- | 0.149 0.474 0.376 |
0.152 0.515 0.333 | 0.263 0.473 0.264 | 0.178 0.486 0.336 |
0.121 0.43 0.449 |
| -- | χ2 = 5.78 P>0.05 | χ2 = 11.22 P<0.005 | χ2 = 16.43 P<0.001 | χ2 = 9.49 P<0.01 | χ2 = 3.29 P>0.15 | |||
| AGT-174 | GG GA AA | 0.762 0.222 0.016 | 0.791 0.200 0.009 |
0.783 0.217 0.000 |
0.627 0.343 0.030 | 0.791 0.209 0.000 | 0.803 0.178 0.019 |
0.757 0.215 0.028 |
| χ2 = 1.97 P>0.4 | χ2 = 1.08 P>0.55 | χ2 = 10.59 P=0.005 | χ2 = 2.07 P>0.35 | χ2 = 0.75 P>0.65 | χ2 = 0.19 P>0.9 | |||
| AGT-235 | AA AG GG | 0.254 0.500 0.246 | 0.371 0.434 0.195 |
0.218 0.652 0.130 |
0.303 0.535 0.162 | 0.384 0.484 0.132 |
0.355 0.439 0.206 | 0.365 0.439 0.196 |
| χ2 = 39.89 P<0.001 | χ2 = 1.21 P>0.5 | χ2 = 6.28 P<0.05 | χ2 = 18.5 P<0.001 | χ2 = 17.04 P<0.001 | χ2 = 18.55 P<0.001 | |||
| APOC3 | CC CG GG | 0.821 0.170 0.009 | 1.000 0.000 0.000 |
0.841 0.159 0.000 |
0.727 0.263 0.010 | 0.770 0.220 0.010 |
0.766 0.206 0.028 |
0.757 0.224 0.019 |
| χ2 = 46.00 P<0.001 | χ2 = 0.63 P>0.7 | χ2 = 6.43 P<0.05 | χ2 = 2.03 P>0.35 | χ2 = 3.05 P>0.2 | χ2 = 3.00 P>0.2 | |||
| IL1B | CC CT TT | 0.632 0.326 0.042 | 0.159 0.310 0.531 |
0.583 0.250 0.167 |
0.566 0.394 0.040 | 0.450 0.473 0.077 | 0.617 0.374 0.009 | 0.533 0.402 0.065 |
| χ2 = 359.5 P<0.001 | χ2 = 10.55 P<0.01 | χ2 = 3.61 P>0.15 | χ2 = 13.41 P<0.0025 | χ2 = 5.62 P>0.05 | χ2 = 5.15 P>0.05 | |||
| IL6-174 | GG GC CC | 0.333 0.489 0.179 | 0.247 0.434 0.319 |
0.208 0.542 0.250 |
0.313 0.465 0.222 | 0.373 0.429 0.198 | 0.411 0.477 0.112 | 0.420 0.449 0.131 |
| χ2 = 17.99 P<0.001 | χ2 = 4.24 P>0.1 | χ2 = 0.7 P>0.65 | χ2 = 0.17 P>0.95 | χ2 = 4.9 P>0.1 | χ2 = 3.52 P>0.15 | |||
| IL6-573 | GG GC CC | 0.733 0.247 0.021 | 0.914 0.086 0.000 |
0.990 0.010 0.000 |
0.899 0.101 0.000 | 0.923 0.077 0.000 | 0.897 0.103 0.000 | 0.897 0.103 0.000 |
| χ2= 26.62 P<0.001 | χ2 = 13.01 P<0.0025 | χ2 = 10.41 P<0.01 | χ2 = 12.41 P<0.025 | χ2 = 10.99 P<0.005 | χ2 = 10.99 P<0.005 | |||
| LPL | CC CG GG | 0.724 0.254 0.022 | 0.766 0.217 0.017 |
0.545 0.364 0.091 |
0.788 0.192 0.020 | 0.791 0.187 0.022 | 0.701 0.271 0.028 | 0.776 0.196 0.028 |
| χ2 = 4.02 P>0.1 | χ2 = 9.87 P<0.01 | χ2 = 1.67 P>0.4 | χ2 = 1.43 P>0.45 | χ2 = 2.97 P>0.2 | χ2 = 0.85 P>0.65 | |||
| PSEN1 | TT TG GG | 0.184 0.490 0.326 | 0.319 0.504 0.177 |
0.435 0.391 0.174 |
0.222 0.505 0.273 | 0.285 0.506 0.209 | 0.261 0.533 0.206 | 0.336 0.486 0.178 |
| χ2 = 54.47 P<0.001 | χ2 = 34.27 P<0.001 | χ2 = 6.05 P<0.05 | χ2 = 16.06 P<0.001 | χ2 = 13.59 P<0.0025 | χ2 = 33.73 P<0.001 | |||
A2M: alpha-2-macroglobulin; ACE: angiotensin I converting enzyme; AGT: angiotensinogen; APOC3: apolipoprotein CIII; CETP: cholesteryl ester transfer protein plasma; IL1B: Interleukin 1 beta; IL6: Interleukin 6; LPL: lipoprotein lipase; PSEN1: Presenilin 1.
Populations: 1301 healthy individuals with a Northern and Western European ancestry extracted from the HapMap-CEU study (N-W Europeans) [48]; 23 healthy individuals from France and Utah residents with European ancestry (Western Europeans) [48]; and 99, 91, 107, and 107 healthy individuals from Finland, Britain, Spain, and Italy, respectively, extracted from the 1000 Genomes database [49].
Populations: 1324 healthy individuals with a Northern and Western European ancestry extracted from the HapMap-CEU and Parc European studies [48]; and 99, 91, 107, and 107 healthy individuals from Finland, Britain, Spain, and Italy, respectively, extracted from the 1000 Genomes database [49].
3.2. The APOE-ε4 Allele is Highly Represented in Our Study Population
When comparing the distribution of all APOE alleles around the world, APOE-ε3 is the most predominant with frequencies of 0.76 ± 0.09, followed by APOE-ε4 (0.13 ± 0.07), and APOE-ε2 (0.06 ± 0.02) [13]. It has been widely reported that the most prevalent risk factor for AD is an increased APOE-ε4 distribution (0.36 ± 0.07) and decreased APOE-ε2 (0.03 ± 0.02) [13, 19, 44, 46]. Allele distribution for APOE in our study population was apparently among the usual in general European populations [13]: APOE-ε3=0.815, APOE-ε4=0.135, and
APOE-ε2=0.050 (Table 2). Indeed, despite the representative number of individuals with vascular risk and dementia in our group, distribution of APOE-ε2 and APOE-ε4 appeared within the range of those of other European and Worldwide populations [50-67] (Fig. 2). It is widely documented that the APOE-ε4 allele rate is lower in Spanish and South European populations [50, 64-67], but the levels of APOE-ε4 in our study population are rather higher than those of South Europeans, including Spanish individuals (Fig. 2), which, once again, might be related to the rate of dementia and atherosclerosis risk in our group. APOE-ε2 distribution is also slightly lower in our population compared to all the other groups, although still far from those with AD (grey bars, Fig. 2).
Fig. (2).
Distribution of the APOE alleles in our study compared to other Spanish, European, and Worldwide populations, including healthy individuals, and patients with Alzheimer’s disease [50-67].
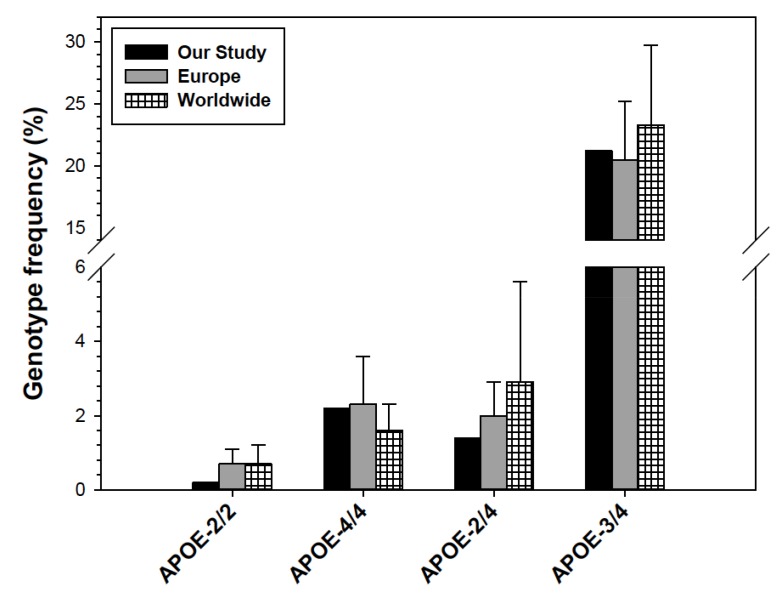
In order to establish the influence of APOE-ε4 and APOE-ε2 distribution in our population, we performed a comparative analysis of the APOE genotype variants with other representative Spanish populations [51, 52, 54, 56, 58, 61, 63-67] (Fig. 3A). APOE genotypes containing the alleles ε2 and ε3 (ε2/ε2, ε2/ε3, and ε3/ε3) did not display large differences between our population and average Spanish groups. However, genotypes containing the ε4 allele were much more evident in our population, especially the homozygous ε4/ε4, with a 2.5-fold increase, followed by the heterozygous ε2/ε4, with a near 2-fold increase, and ε3/ε4 showing an approximately 1.5-fold increase (Fig. 3B). Pearson’s chi-square test provided evidence of a significantly different distribution of the APOE genotypes in our population as compared to five separate representative Spanish populations (P<0.01-0.001) (Table 4), which strongly suggests the high influence of the APOE-ε4 allele in our population. Those particularly higher levels of APOE-ε4 and PSEN1 may be a sign of a representative number of individuals with AD in our group. Nevertheless, the APOE-ε4 rate in our group still far differs from Spanish individuals with AD (P<0.001) Table 4. Although APOE genotypes involving the ε4 allele (ε4/ε4, ε2/ε4, and ε3/ε4) are significantly higher in our population compared to other Spanish groups, it is nevertheless sited within the range values of average European and Worldwide populations, unlike the ε2/ε2 genotype (Fig. 4).
Fig. (3).
Distribution of APOE genotypes in our study and in five other representative Spanish populations [51, 52, 54, 56, 58, 61, 63-67] (A). Genotype distribution ratio between our population (bars) and the other five Spanish populations (dashed line) (B). Spanish groups description: control (healthy) individuals from a broad distribution of Spanish regions (n=1286), and other groups from specific regions, such as Asturias (n=250), Barcelona (n=478), and Navarra (n=188).
Table 4.
Genotype frequencies of APOE polymorphisms in our population and different Spanish populations of healthy individuals (Control) or individuals with Alzheimer’s disease (AD).
| __ | __ | Genotype Frequencies | __ | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Gene | Genotypes | Our Study | Spanish Control | Spanish Control | Spanish Control | Spanish Control | Spanish Control | Spanish AD | Spanish AD | References |
| APOE | ε-2/2 ε-4/4 ε-3/3 ε-2/3 ε-2/4 ε-3/4 | 0.002 0.022 0.668 0.079 0.014 0.212 | 0.005 0.005 0.780 0.100 0.000 0.120 | 0.002 0.008 0.732 0.089 0.009 0.155 | 0.005 0.010 0.672 0.116 0.014 0.164 | 0.002 0.011 0.731 0.070 0.009 0.178 | 0.000 0.010 0.830 0.027 0.005 0.128 | 0.000 0.080 0.580 0.020 0.000 0.320 | 0.001 0.092 0.531 0.051 0.020 0.296 | [51, 52, 54, 56, 58, 61] [63-67] |
| __ | __ | __ | χ2 = 23.37 P<0.001 | χ2 = 34.43 P<0.001 | χ2 = 16.71 P<0.01 | χ2 = 12.23 P<0.01 | χ2 = 22.44 P<0.001 | χ2 = 27.99 P<0.001 | χ2 = 29.43 P<0.001 | __ |
Spanish groups description: control (healthy) individuals from a broad distribution of Spanish regions (n=1286), and other groups from specific regions, such as Asturias (n=250), Barcelona (n=478), and Navarra (n=188). Table also includes two Spanish groups with individuals diagnosed with Alzheimer Disease from Navarra (n=98) and Asturias (n=120).
Fig. (4).
Genotype frequencies of the APOE-ε2/2 (A), APOE-ε4/4 (B), APOE-ε2/4 (C), and APOE-ε3/4 (D) in our population, compared to representative European and Worldwide populations [13, 50, 51, 61]. Error bars correspond to STD.
4. DISCUSSION
This study evaluates the impact of 20 gene polymorphisms associated with vascular risk and dementia (Table 2) in a broad population of 4415 individuals from Spain carrying a variety of disorders involving metabolic syndrome, vascular damage, and impaired cognitive decline leading to dementia, among others Table 1. A total of approximately 40% of patients were diagnosed with disorders involving, directly or indirectly, vascular damage or cognitive decline leading to dementia. A significant part of those patients with dementia have a cerebrovascular component. Some of the remaining 60% of pathologies (psychiatric disorders, peripheral nervous system (PNS) impairment, or others) might also lead to vascular damage or dementia, depending on the disease progression or genetic predisposition of the individuals.
Genotype distributions in 8 out of the 20 polymorphisms analyzed (LPL, ACE, AGT-174, AGT-235, IL1B, IL6-174, IL6-573, PSEN1) deviate from HWE Table 2. In this case, the number of individuals analyzed was large enough to be representative. Therefore, those gene polymorphisms were good candidates to distinguish our biased group from average populations. However, when the genotype distribution of these genes in our group was compared with other representative Spanish or European populations, we found that only the polymorphisms ACE (547C>T, rs4332), AGT (9543A>G, M235T, rs699), IL6 (573G>C, rs1800796), and PSEN1 (16G>T, rs165932) were representative in our population (Fig. 1; Table 3).
Hypertension is one of the leading risks (50-70%) for cardiovascular disorders and cerebral stroke, which are considered to be among the top three causes of death in the US and Europe [6, 7]. Polymorphisms in genes involved in blood pressure regulation are markedly represented in our population. Genes encoding for angiotensin I-converting enzyme (ACE) and the angiotensinogen (AGT) are involved in the renin-angiotensin system pathway that regulates blood pressure. Polymorphisms in those genes have been reported to be associated with hypertension [13, 24-27]. Polymorphisms in inflammation-related genes such as IL6 are also involved in hypertension and atherosclerosis [11, 13, 28, 31, 33], although the physiological mechanisms are not well characterized.
Apparently, hypertension-related polymorphisms, rather than lipid metabolism-related variations, are the major genetic cause for vascular disorders in our population. Indeed, correlation studies performed in Spanish populations demonstrate that regions with a higher total fat consumption presented a lower coronary-leading mortality, and vice versa [68, 69]. This finding suggests (i) the strong influence of genetic predisposition over environmental factors; (ii) the lower importance of variations in genes involved in lipid metabolism (APOB, APOC3, CETP) compared to those involving blood pressure regulation in Spanish populations.
The only gene involved in lipid metabolism with a representative influence in our population was APOE, and particularly, the APOE-ε4 allele (Figs. 2-4, Table 4). It is widely reported that the increased risk of developing an ischemic cardiopathy, which is directly proportional to the APOE-ε4 allele frequency, follows a north-south gradient throughout Europe, being more frequent in northern than southern countries [50, 64-67, 70, 71]. Although the APOE-ε4 allele frequency in our population fitted within the range of European individuals, it was nevertheless higher than the average Spanish and South European populations (Fig. 2). In addition, all genotype variants involving APOE-ε4 were predominant in our population compared to other Spanish groups (Fig. 3, Table 4), but not to other Europeans or Worldwide populations (Fig. 4).
APOE-ε4, and especially the APOE-ε4/ε4 genotype, is one of the most prevalent risk factors associated with neurological hallmarks of AD [13, 19, 44, 46]. It has been reported that APOE-ε4 may influence AD by interacting with APP metabolism and Aβ accumulation, enhancing the hyperphosphorylation of tau protein, and starting a chain reaction involving oxidative processes, modification of the neuroimmunotrophic activity, altering lipid metabolism and transport, and membrane biosynthesis in sprouting and synaptic remodeling, and inducing apoptosis [13, 19, 44, 72-74].
However, other different reasons may also explain the high rate of APOE-ε4, and the APOE-ε4/ε4 genotype in our population. The APOE-ε4 allele is highly associated with an aberrant increase in LDL-family lipoproteins and cholesterol, which promotes the risk of developing atherosclerosis and vascular disorders [13-20], which may be explained by the significant rate of disorders with a vascular component represented in our population. Some of the individuals belonging to the 15% carrying dementia-related disorders were diagnosed with AD, although a high proportion of them had a vascular component rather than neurodegenerative. In addition, APOE-related pathogenic mechanisms are also associated with brain aging [75], and over 35% of individuals in our population were older than 60.
The presence of APOE-ε4 is a hallmark but not enough to cause AD. Nevertheless, besides the prevalence of APOE-ε4, there are also other good reasons to suggest that our population may have a higher rate of individuals with a potential onset of AD than previously expected: (i) Polymorphism +16G>T (rs165932) in the PSEN1 gene is also prevalent in our population compared to other representative Spanish and European populations (Table 3). This polymorphism is a hallmark of AD, since it affects β-secretase activity leading to accumulation of Aβ [19, 42, 43, 45]. (ii) Contrary to APOE-ε4, carriers of APOE-ε2 may be protected against dementia, and in fact, APOE-ε2 allele distribution is usually lower in individuals with AD [13, 19, 44]. Our population displays a higher rate of APOE-ε4 and lower APOE-ε2 than other Spanish and South European populations (Figs. 2,3). (iii) Several reports associate metabolic syndrome and diabetes as promoters of future AD onset [76-78], which would suggest a prognosis for a potential number of individuals in our population. However, some of those reports define as AD cases of dementia associated with a vascular component turning into neurodegeneration, which is not entirely correct.
CONCLUSION
The objective of this study was to characterize the influence of gene polymorphisms related to vascular risk and dementia on a broad population of Spanish individuals diagnosed with a variety of disorders concerning impaired lipid metabolism, hypertension, atherosclerosis, and dementia, among others. Polymorphisms in ACE, AGT(235), IL6(573), PSEN1, and APOE (especially the APOE-ε4 allele) were representative of our study population as compared to reference data of Spanish and European individuals. Results obtained in this work may yield the following conclusions: (i) the large number of individuals analyzed in this study might provide a hallmark of cardiovascular and dementia risk polymorphisms in the Spanish population; (ii) polymorphisms involving hypercholesterolemia are more pronounced in our population, rather than those related to lipid metabolism; (iii) the prevalence of APOE-ε4 and +16G>T (rs165932) variation in the PSEN1 gene suggests a higher potential rate of AD onset in our population. Thus, AD diagnosis might be underestimated in our population, or not yet diagnosed since the classic symptoms had not yet shown up, although they might already be genetically imprinted.
Identification of risk polymorphisms provides information about the susceptibility of an individual for the onset of certain diseases, compared to the average population. A proper risk assessment requires the analysis of variations in multiple potential interacting genes at individual, family, and ethnical levels. Identification and proper analysis of risk polymorphisms will ensure more reliable, accurate, and affordable treatments.
ACKNOWLEDGEMENTS
This work was supported by the EuroEspes Biomedical Research Center, Institute of Medical Science and Genomic Medicine, and the International Agency for Brain Research and Aging, Corunna, Spain.
CONFLICT OF INTEREST
The authors are supported by the EuroEspes Biomedical Research Center, Institute of Medical Science and Genomic Medicine, and the International Agency for Brain Research and Aging, Corunna, Spain. The authors have no other relevant affiliations or financial involvement with any organization or entity in financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
REFERENCES
- 1.Ford E.S., Giles W.H., Dietz W.H. Prevalence of the metabolic syndrome among US adults: findings from the third National Health and Nutrition Examination Survey. JAMA. 2002;287(3):356–359. doi: 10.1001/jama.287.3.356. [DOI] [PubMed] [Google Scholar]
- 2.Mozumdar A., Liguori G. Persistent increase of prevalence of metabolic syndrome among U.S. adults: NHANES III to NHANES 1999-2006. Diabetes Care. 2011;34(1):216–219. doi: 10.2337/dc10-0879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alberti K.G., Zimmet P.Z. Definition, diagnosis and classification of diabetes mellitus and its complications. Part 1: diagnosis and classification of diabetes mellitus provisional report of a WHO consultation. Diabet. Med. 1998;15(7):539–553. doi: 10.1002/(SICI)1096-9136(199807)15:7<539::AID-DIA668>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 4. The IDF consensus worldwide definition of the metabolic syndrome. https://www.idf.org/webdata/docs/MetS_def_update2006.pdf
- 5.Grundy S.M., Cleeman J.I., Daniels S.R., Donato K.A., Eckel R.H., Franklin B.A., Gordon D.J., Krauss R.M., Savage P.J., Smith S.C., Jr, Spertus J.A., Costa F., American Heart Association. National Heart, Lung, and Blood Institute Diagnosis and management of the metabolic syndrome: an American Heart Association/National Heart, Lung, and Blood Institute Scientific Statement. Circulation. 2005;112(17):2735–2752. doi: 10.1161/CIRCULATIONAHA.105.169404. [DOI] [PubMed] [Google Scholar]
- 6.Barrett K.M., Meschia J.F. Acute ischemic stroke management: medical management. Semin. Neurol. 2010;30(5):461–468. doi: 10.1055/s-0030-1268859. [DOI] [PubMed] [Google Scholar]
- 7.Lloyd-Jones D., Adams R.J., Brown T.M., Carnethon M., Dai S., De Simone G., Ferguson T.B., Ford E., Furie K., Gillespie C., Go A., Greenlund K., Haase N., Hailpern S., Ho P.M., Howard V., Kissela B., Kittner S., Lackland D., Lisabeth L., Marelli A., McDermott M.M., Meigs J., Mozaffarian D., Mussolino M., Nichol G., Roger V.L., Rosamond W., Sacco R., Sorlie P., Roger V.L., Thom T., Wasserthiel-Smoller S., Wong N.D., Wylie-Rosett J., WRITING GROUP MEMBERS. American Heart Association Statistics Committee and Stroke Statistics Subcommittee Heart disease and stroke statistics--2010 update: a report from the American Heart Association. Circulation. 2010;121(7):e46–e215. doi: 10.1161/CIRCULATIONAHA.109.192667. [DOI] [PubMed] [Google Scholar]
- 8.Navab M., Berliner J.A., Watson A.D., Hama S.Y., Territo M.C., Lusis A.J., Shih D.M., Van Lenten B.J., Frank J.S., Demer L.L., Edwards P.A., Fogelman A.M. The Yin and Yang of oxidation in the development of the fatty streak. A review based on the 1994 George Lyman Duff Memorial Lecture. Arterioscler. Thromb. Vasc. Biol. 1996;16(7):831–842. doi: 10.1161/01.ATV.16.7.831. [DOI] [PubMed] [Google Scholar]
- 9.Steinberg D., Parthasarathy S., Carew T.E., Khoo J.C., Witztum J.L. Beyond cholesterol. Modifications of low-density lipoprotein that increase its atherogenicity. N. Engl. J. Med. 1989;320(14):915–924. doi: 10.1056/NEJM198904063201407. [DOI] [PubMed] [Google Scholar]
- 10.Glass C.K., Witztum J.L. Atherosclerosis. the road ahead. Cell. 2001;104(4):503–516. doi: 10.1016/S0092-8674(01)00238-0. [DOI] [PubMed] [Google Scholar]
- 11.Libby P. Inflammation in atherosclerosis. Nature. 2002;420(6917):868–874. doi: 10.1038/nature01323. [DOI] [PubMed] [Google Scholar]
- 12.Lusis A.J., Fogelman A.M., Fonarow G.C. Genetic basis of atherosclerosis: part I: new genes and pathways. Circulation. 2004;110(13):1868–1873. doi: 10.1161/01.CIR.0000143041.58692.CC. [DOI] [PubMed] [Google Scholar]
- 13.Cacabelos R. World Guide for Drug Use and Pharmacogenomics. 1st ed. Corunna, Spain: EuroEspes Publishing; 2012. [Google Scholar]
- 14.Law A., Wallis S.C., Powell L.M., Pease R.J., Brunt H., Priestley L.M., Knott T.J., Scott J., Altman D.G., Miller G.J., et al. Common DNA polymorphism within coding sequence of apolipoprotein B gene associated with altered lipid levels. Lancet. 1986;1(8493):1301–1303. doi: 10.1016/S0140-6736(86)91222-5. [DOI] [PubMed] [Google Scholar]
- 15.Hegele R.A., Huang L.S., Herbert P.N., Blum C.B., Buring J.E., Hennekens C.H., Breslow J.L. Apolipoprotein B-gene DNA polymorphisms associated with myocardial infarction. N. Engl. J. Med. 1986;315(24):1509–1515. doi: 10.1056/NEJM198612113152403. [DOI] [PubMed] [Google Scholar]
- 16.Paré G., Serre D., Brisson D., Anand S.S., Montpetit A., Tremblay G., Engert J.C., Hudson T.J., Gaudet D. Genetic analysis of 103 candidate genes for coronary artery disease and associated phenotypes in a founder population reveals a new association between endothelin-1 and high-density lipoprotein cholesterol. Am. J. Hum. Genet. 2007;80(4):673–682. doi: 10.1086/513286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Breslow J.L., McPherson J., Nussbaum A.L., Williams H.W., Lofquist-Kahl F., Karathanasis S.K., Zannis V.I. Identification and DNA sequence of a human apolipoprotein E cDNA clone. J. Biol. Chem. 1982;257(24):14639–14641. [PubMed] [Google Scholar]
- 18.Pedro-Botet J., Sentí M., Nogués X., Rubiés-Prat J., Roquer J., D’Olhaberriague L., Olivé J. Lipoprotein and apolipoprotein profile in men with ischemic stroke. Role of lipoprotein(a), triglyceride-rich lipoproteins, and apolipoprotein E polymorphism. Stroke. 1992;23(11):1556–1562. doi: 10.1161/01.STR.23.11.1556. [DOI] [PubMed] [Google Scholar]
- 19.Cacabelos R., Martínez R., Fernández-Novoa L., Carril J.C., Lombardi V., Carrera I., Corzo L., Tellado I., Leszek J., McKay A., Takeda M. Genomics of dementia: APOE- and CYP2D6-related pharmacogenetics. Int. J. Alzheimers Dis. 2012. [DOI] [PMC free article] [PubMed]
- 20.Ordovas J.M., Cupples L.A., Corella D., Otvos J.D., Osgood D., Martínez A., Lahoz C., Coltell O., Wilson P.W., Schaefer E.J. Association of cholesteryl ester transfer protein-TaqIB polymorphism with variations in lipoprotein subclasses and coronary heart disease risk: the Framingham study. Arterioscler. Thromb. Vasc. Biol. 2000;20(5):1323–1329. doi: 10.1161/01.ATV.20.5.1323. [DOI] [PubMed] [Google Scholar]
- 21.Eckel R.H. Lipoprotein lipase. A multifunctional enzyme relevant to common metabolic diseases. N. Engl. J. Med. 1989;320(16):1060–1068. doi: 10.1056/NEJM198904203201607. [DOI] [PubMed] [Google Scholar]
- 22.Stocks J., Thorn J.A., Galton D.J. Lipoprotein lipase genotypes for a common premature termination codon mutation detected by PCR-mediated site-directed mutagenesis and restriction digestion. J. Lipid Res. 1992;33(6):853–857. [PubMed] [Google Scholar]
- 23.Rankinen T., Rice T., Pérusse L., Chagnon Y.C., Gagnon J., Leon A.S., Skinner J.S., Wilmore J.H., Rao D.C., Bouchard C., Bouchard C. NOS3 Glu298Asp genotype and blood pressure response to endurance training: the HERITAGE family study. Hypertension. 2000;36(5):885–889. doi: 10.1161/01.HYP.36.5.885. [DOI] [PubMed] [Google Scholar]
- 24.Slowik A., Turaj W., Dziedzic T., Haefele A., Pera J., Malecki M.T., Glodzik-Sobanska L., Szermer P., Figlewicz D.A., Szczudlik A. DD genotype of ACE gene is a risk factor for intracerebral hemorrhage. Neurology. 2004;63(2):359–361. doi: 10.1212/01.WNL.0000130200.12993.0C. [DOI] [PubMed] [Google Scholar]
- 25.Slowik A., Dziedzic T., Pera J., Wloch D., Kopec G., Szczudlik A. ACE genotype, risk and causal relationship to stroke: implications for treatment. Curr. Treat. Options Cardiovasc. Med. 2007;9(3):198–204. doi: 10.1007/s11936-007-0013-6. [DOI] [PubMed] [Google Scholar]
- 26.Jeunemaitre X., Soubrier F., Kotelevtsev Y.V., Lifton R.P., Williams C.S., Charru A., Hunt S.C., Hopkins P.N., Williams R.R., Lalouel J.M., Corvol P. Molecular basis of human hypertension: role of angiotensinogen. Cell. 1992;71(1):169–180. doi: 10.1016/0092-8674(92)90275-H. [DOI] [PubMed] [Google Scholar]
- 27.Katsuya T., Koike G., Yee T.W., Sharpe N., Jackson R., Norton R., Horiuchi M., Pratt R.E., Dzau V.J., MacMahon S. Association of angiotensinogen gene T235 variant with increased risk of coronary heart disease. Lancet. 1995;345(8965):1600–1603. doi: 10.1016/S0140-6736(95)90115-9. [DOI] [PubMed] [Google Scholar]
- 28.Luther T., Mackman N. Tissue factor in the heart. Multiple roles in hemostasis, thrombosis, and inflammation. Trends Cardiovasc. Med. 2001;11(8):307–312. doi: 10.1016/S1050-1738(01)00129-3. [DOI] [PubMed] [Google Scholar]
- 29.Danesh J., Appleby P. Persistent infection and vascular disease: a systematic review. Expert Opin. Investig. Drugs. 1998;7(5):691–713. doi: 10.1517/13543784.7.5.691. [DOI] [PubMed] [Google Scholar]
- 30.Fernández-Real J.M., Broch M., Vendrell J., Richart C., Ricart W. Interleukin-6 gene polymorphism and lipid abnormalities in healthy subjects. J. Clin. Endocrinol. Metab. 2000;85(3):1334–1339. doi: 10.1210/jcem.85.3.6555. [DOI] [PubMed] [Google Scholar]
- 31.Revilla M., Obach V., Cervera A., Dávalos A., Castillo J., Chamorro A. A -174G/C polymorphism of the interleukin-6 gene in patients with lacunar infarction. Neurosci. Lett. 2002;324(1):29–32. doi: 10.1016/S0304-3940(02)00169-6. [DOI] [PubMed] [Google Scholar]
- 32.Iacoviello L., Di Castelnuovo A., Gattone M., Pezzini A., Assanelli D., Lorenzet R., Del Zotto E., Colombo M., Napoleone E., Amore C., D’Orazio A., Padovani A., de Gaetano G., Giannuzzi P., Donati M.B. IGIGI Investigators. Polymorphisms of the interleukin-1ß gene affect the risk of myocardial infarction and ischemic stroke at young age and the response of mononuclear cells to stimulation in vitro. Arterioscler. Thromb. Vasc. Biol. 2005;25:222–227. doi: 10.1161/01.ATV.0000150039.60906.02. [DOI] [PubMed] [Google Scholar]
- 33.Yamada Y., Metoki N., Yoshida H., Satoh K., Ichihara S., Kato K., Kameyama T., Yokoi K., Matsuo H., Segawa T., Watanabe S., Nozawa Y. Genetic risk for ischemic and hemorrhagic stroke. Arterioscler. Thromb. Vasc. Biol. 2006;26(8):1920–1925. doi: 10.1161/01.ATV.0000229694.97827.38. [DOI] [PubMed] [Google Scholar]
- 34.Reich D., Patterson N., Ramesh V., De Jager P.L., McDonald G.J., Tandon A., Choy E., Hu D., Tamraz B., Pawlikowska L., Wassel-Fyr C., Huntsman S., Waliszewska A., Rossin E., Li R., Garcia M., Reiner A., Ferrell R., Cummings S., Kwok P.Y., Harris T., Zmuda J.M., Ziv E., Health, Aging and Body Composition (Health ABC) Study Admixture mapping of an allele affecting interleukin 6 soluble receptor and interleukin 6 levels. Am. J. Hum. Genet. 2007;80(4):716–726. doi: 10.1086/513206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rainero I., Grimaldi L.M., Salani G., Valfrè W., Rivoiro C., Savi L., Pinessi L. Association between the tumor necrosis factor-alpha -308 G/A gene polymorphism and migraine. Neurology. 2004;62(1):141–143. doi: 10.1212/01.WNL.0000101717.16799.8F. [DOI] [PubMed] [Google Scholar]
- 36.De Stefano V., Martinelli I., Mannucci P.M., Paciaroni K., Chiusolo P., Casorelli I., Rossi E., Leone G. The risk of recurrent deep venous thrombosis among heterozygous carriers of both factor V Leiden and the G20210A prothrombin mutation. N. Engl. J. Med. 1999;341(11):801–806. doi: 10.1056/NEJM199909093411104. [DOI] [PubMed] [Google Scholar]
- 37.Gerhardt A., Scharf R.E., Beckmann M.W., Struve S., Bender H.G., Pillny M., Sandmann W., Zotz R.B. Prothrombin and factor V mutations in women with a history of thrombosis during pregnancy and the puerperium. N. Engl. J. Med. 2000;342(6):374–380. doi: 10.1056/NEJM200002103420602. [DOI] [PubMed] [Google Scholar]
- 38.Casas J.P., Hingorani A.D., Bautista L.E., Sharma P. Meta-analysis of genetic studies in ischemic stroke: thirty-two genes involving approximately 18,000 cases and 58,000 controls. Arch. Neurol. 2004;61(11):1652–1661. doi: 10.1001/archneur.61.11.1652. [DOI] [PubMed] [Google Scholar]
- 39.Bethke L., Webb E., Murray A., Schoemaker M., Feychting M., Lönn S., Ahlbom A., Malmer B., Henriksson R., Auvinen A., Kiuru A., Salminen T., Johansen C., Christensen H.C., Muir K., McKinney P., Hepworth S., Dimitropoulou P., Lophatananon A., Swerdlow A., Houlston R. Functional polymorphisms in folate metabolism genes influence the risk of meningioma and glioma. Cancer Epidemiol. Biomarkers Prev. 2008;17(5):1195–1202. doi: 10.1158/1055-9965.EPI-07-2733. [DOI] [PubMed] [Google Scholar]
- 40.Wald D.S., Law M., Morris J.K. Homocysteine and cardiovascular disease: evidence on causality from a meta-analysis. BMJ. 2002;325(7374):1202–1207. doi: 10.1136/bmj.325.7374.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Centers for Disease Control and Prevention. . National Center for Health Statistics (NCHS). Atlanta, GA, USA http://www.cdc.gov/ nchs/
- 42.Cacabelos R. Pharmacogenomics in Alzheimer’s disease. Methods Mol. Biol. 2008;448:213–357. doi: 10.1007/978-1-59745-205-2_10. [DOI] [PubMed] [Google Scholar]
- 43.Cacabelos R., Torrellas C. Epigenetic drug discovery for Alzheimer’s disease. Expert Opin. Drug Discov. 2014;9(9):1059–1086. doi: 10.1517/17460441.2014.930124. [DOI] [PubMed] [Google Scholar]
- 44.Cacabelos R., Fernandez-Novoa L., Lombardi V., Kubota Y., Takeda M. Molecular genetics of Alzheimer’s disease and aging. Methods Find. Exp. Clin. Pharmacol. 2005;27(Suppl. A):1–573. [PubMed] [Google Scholar]
- 45.Cacabelos R., Fernández-Novoa L., Martínez-Bouza R., McKay A., Carril J.C., Lombardi V., Corzo L., Carrera I., Tellado I., Nebril L., Alcaraz M., Rodríguez S., Casas A., Couceiro V., Álvarez A. Future trends in the pharmacogenomics of brain disorders and dementia: Influence of APOE and CYP2D6 variants. Pharmaceuticals. 2010;3:3040–3100. doi: 10.3390/ph3103040. [DOI] [Google Scholar]
- 46.Strittmatter W.J., Weisgraber K.H., Huang D.Y., Dong L.M., Salvesen G.S., Pericak-Vance M., Schmechel D., Saunders A.M., Goldgaber D., Roses A.D. Binding of human apolipoprotein E to synthetic amyloid beta peptide: isoform-specific effects and implications for late-onset Alzheimer disease. Proc. Natl. Acad. Sci. USA. 1993;90(17):8098–8102. doi: 10.1073/pnas.90.17.8098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Carril J.C. Genomics of the cerebrovascular pathology. GenT. 2009;4:70–84. [Google Scholar]
- 48. dbSNP Short Genetic Variations. . National Center for Biotechnology Information (NCBI). http://www.ncbi.nlm.nih.gov/projects/ SNP/
- 49. 1000 Genomes. A Deep Catalog of Human Genetic Variation. http://www.1000genomes.org/
- 50.Tiret L., de Knijff P., Menzel H-J., Ehnholm C., Nicaud V., Havekes L.M., EARS group ApoE polymorphism and predisposition to coronary heart disease in youths of different European populations. The EARS Study. European Atherosclerosis Research Study. Arterioscler. Thromb. 1994;14(10):1617–1624. doi: 10.1161/01.ATV.14.10.1617. [DOI] [PubMed] [Google Scholar]
- 51.Corbo R.M., Scacchi R., Apolipoprotein E. Apolipoprotein E (APOE) allele distribution in the world. Is APOE*4 a ‘thrifty’ allele? Ann. Hum. Genet. 1999;63(Pt 4):301–310. doi: 10.1046/j.1469-1809.1999.6340301.x. [DOI] [PubMed] [Google Scholar]
- 52.Valveny N., Esteban E., Kandil M., Moral P. APO E polymorphism in Spanish and Moroccan populations. Clin. Genet. 1997;51(5):354–356. doi: 10.1111/j.1399-0004.1997.tb02488.x. [DOI] [PubMed] [Google Scholar]
- 53.Corbo R.M., Scacchi R., Mureddu L., Mulas G., Alfano G. Apolipoprotein E polymorphism in Italy investigated in native plasma by a simple polyacrylamide gel isoelectric focusing technique. Comparison with frequency data of other European populations. Ann. Hum. Genet. 1995;59(Pt 2):197–209. doi: 10.1111/j.1469-1809.1995.tb00741.x. [DOI] [PubMed] [Google Scholar]
- 54.Gerdes L.U., Klausen I.C., Sihm I., Faergeman O. Apolipoprotein E polymorphism in a Danish population compared to findings in 45 other study populations around the world. Genet. Epidemiol. 1992;9(3):155–167. doi: 10.1002/gepi.1370090302. [DOI] [PubMed] [Google Scholar]
- 55.Braeckman L., De Bacquer D., Rosseneu M., De Backer G. Apolipoprotein E polymorphism in middle-aged Belgian men: phenotype distribution and relation to serum lipids and lipoproteins. Atherosclerosis. 1996;120(1-2):67–73. doi: 10.1016/0021-9150(95)05681-5. [DOI] [PubMed] [Google Scholar]
- 56.Lucotte G., Loirat F., Hazout S. Pattern of gradient of apolipoprotein E allele *4 frequencies in western Europe. Hum. Biol. 1997;69(2):253–262. [PubMed] [Google Scholar]
- 57.Kowalska A., Wiechmann I., Walter H. Genetic variability of apolipoprotein E in a Polish population. Hum. Biol. 1998;70(6):1093–1099. [PubMed] [Google Scholar]
- 58.Muros M., Rodríguez-Ferrer C. Apolipoprotein E polymorphism influence on lipids, apolipoproteins and Lp(a) in a Spanish population underexpressing apo E4. Atherosclerosis. 1996;121(1):13–21. doi: 10.1016/0021-9150(95)06643-8. [DOI] [PubMed] [Google Scholar]
- 59.Cariolou M.A., Kokkofitou A., Manoli P., Christou S., Karagrigoriou A., Middleton L. Underexpression of the apolipoprotein E2 and E4 alleles in the Greek Cypriot population of Cyprus. Genet. Epidemiol. 1995;12(5):489–497. doi: 10.1002/gepi.1370120506. [DOI] [PubMed] [Google Scholar]
- 60.Sklavounou E., Economou-Petersen E., Karadima G., Panas M., Avramopoulos D., Varsou A., Vassilopoulos D., Petersen M.B. Apolipoprotein E polymorphism in the Greek population. Clin. Genet. 1997;52(4):216–218. doi: 10.1111/j.1399-0004.1997.tb02550.x. [DOI] [PubMed] [Google Scholar]
- 61.Marca V., Acosta O., Cornejo-Olivas M., Ortega O., Huerta D., Mazzetti P. [Genetic polymorphism of apolipoprotein E in a Peruvian population]. Rev. Peru. Med. Exp. Salud Publica. 2011;28(4):589–594. [PubMed] [Google Scholar]
- 62.Singh P.P., Singh M., Mastana S.S. APOE distribution in world populations with new data from India and the UK. Ann. Hum. Biol. 2006;33(3):279–308. doi: 10.1080/03014460600594513. [DOI] [PubMed] [Google Scholar]
- 63.Haddy N., De Bacquer D., Chemaly M.M., Maurice M., Ehnholm C., Evans A., Sans S., Do Carmo Martins M., De Backer G., Siest G., Visvikis S. The importance of plasma apolipoprotein E concentration in addition to its common polymorphism on inter-individual variation in lipid levels: results from Apo Europe. Eur. J. Hum. Genet. 2002;10(12):841–850. doi: 10.1038/sj.ejhg.5200864. [DOI] [PubMed] [Google Scholar]
- 64.Alvarez V., Alvarez R., Peña J., Lahoz C.H., Martínez C., Menéndez-Guisasola L., Salas-Puig J., Morís G., Uría D., Menes B.B., Ribacoba R., Vidal J.A., Coto E. [Frequency of the APOE-4 allele in Alzheimer’s disease and its variation with age in Asturias (Spain)]. Med. Clin. (Barc.) 1999;113(12):441–443. [PubMed] [Google Scholar]
- 65.Setó-Salvia N., Clarimón J. Genética en la enfermedad de Alzheimer. Rev. Neurol. 2010;50(6):360–364. [PubMed] [Google Scholar]
- 66.Colsa Gutiérrez P., Cagigas J.C., Ruiz J.L., Hernández-Estefanía R., Cagigas P., Lamuño D., Ovejero V., Escalante A., Ingelmo A. Apolipoproteínas E y CIII en pacientes obesos relacionadas con el Fenotipo IMC tras cirugía bariátrica. BMI. 2014;4(5):564–568. [Google Scholar]
- 67.Hernández-Charro B., Moreno S., Valiente A., Manubens J.M., Villar M.D., Ramos-Arroyo M.A. [ACT/AA polymorphism could duplicate the APOE*epsilon4-associated Alzheimer’s disease risk]. Med. Clin. (Barc.) 2004;123(7):251–254. doi: 10.1016/s0025-7753(04)74479-6. [DOI] [PubMed] [Google Scholar]
- 68.The World Health Organization MONICA Project Ecological analysis of the association between mortality and major risk factors of cardiovascular disease. Int. J. Epidemiol. 1994;23(3):505–516. doi: 10.1093/ije/23.3.505. [DOI] [PubMed] [Google Scholar]
- 69.Menotti A., Blackburn H., Kromhout D., Nissinen A., Karvonen M., Aravanis C., Dontas A., Fidanza F., Giampaoli S. The inverse relation of average population blood pressure and stroke mortality rates in the seven countries study: a paradox. Eur. J. Epidemiol. 1997;13(4):379–386. doi: 10.1023/A:1007326624702. [DOI] [PubMed] [Google Scholar]
- 70.Lucotte G., Loirat F., Hazout S. Pattern of gradient of apolipoprotein E allele *4 frequencies in western Europe. Hum. Biol. 1997;69(2):253–262. [PubMed] [Google Scholar]
- 71.Gerdes L.U., Klausen I.C., Sihm I., Faergeman O. Apolipoprotein E polymorphism in a Danish population compared to findings in 45 other study populations around the world. Genet. Epidemiol. 1992;9(3):155–167. doi: 10.1002/gepi.1370090302. [DOI] [PubMed] [Google Scholar]
- 72.Strittmatter W.J., Saunders A.M., Schmechel D., Pericak-Vance M., Enghild J., Salvesen G.S., Roses A.D. Apolipoprotein E: high-avidity binding to β-amyloid and increased frequency of type 4 allele in late-onset familial Alzheimer disease. Proc. Natl. Acad. Sci. USA. 1993;90(5):1977–1981. doi: 10.1073/pnas.90.5.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Corder E.H., Saunders A.M., Strittmatter W.J., Schmechel D.E., Gaskell P.C., Small G.W., Roses A.D., Haines J.L., Pericak-Vance M.A. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 1993;261(5123):921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- 74.Roses A.D. Pharmacogenetics and drug development: the path to safer and more effective drugs. Nat. Rev. Genet. 2004;5(9):645–656. doi: 10.1038/nrg1432. [DOI] [PubMed] [Google Scholar]
- 75.Filippini N., Ebmeier K.P., MacIntosh B.J., Trachtenberg A.J., Frisoni G.B., Wilcock G.K., Beckmann C.F., Smith S.M., Matthews P.M., Mackay C.E. Differential effects of the APOE genotype on brain function across the lifespan. Neuroimage. 2011;54(1):602–610. doi: 10.1016/j.neuroimage.2010.08.009. [DOI] [PubMed] [Google Scholar]
- 76.Marquer C., Laine J., Dauphinot L., Hanbouch L., Lemercier-Neuillet C., Pierrot N., Bossers K., Le M., Corlier F., Benstaali C., Saudou F., Thinakaran G., Cartier N., Octave J-N., Duyckaerts C., Potier M-C. Increasing membrane cholesterol of neurons in culture recapitulates Alzheimer’s disease early phenotypes. Mol. Neurodegener. 2014;9:60. doi: 10.1186/1750-1326-9-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Akram A., Schmeidler J., Katsel P., Hof P.R., Haroutunian V. Increased expression of RXRα in dementia: an early harbinger for the cholesterol dyshomeostasis? Mol. Neurodegener. 2010;5:36. doi: 10.1186/1750-1326-5-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bourdel-Marchasson I., Mouries A., Helmer C. Hyperglycaemia, microangiopathy, diabetes and dementia risk. Diabetes Metab. 2010;36(Suppl. 3):S112–S118. doi: 10.1016/S1262-3636(10)70477-8. [DOI] [PubMed] [Google Scholar]