Abstract
Background:
Helicobacter pylori is associated with inflammation of different areas, such as the duodenum and stomach, causing gastritis and gastric ulcers leading to lymphoma and cancer. Pathogenic islands are a type of clustered mobile elements ranging from 10-200 Kb contributing to the virulence of the respective pathogen coding for one or more virulence factors. Virulence factors are molecules expressed and secreted by pathogen and are responsible for causing disease in the host. Bacterial genes/virulence factors of the pathogenic islands represent a promising source for identifying novel drug targets.
Objective:
The study aimed at identifying novel drug targets from pathogenic islands in H. pylori.
Material & Methods:
The genome of 23 H. pylori strains were screened for pathogenic islands and bacterial genes/virulence factors to identify drug targets. Protein-protein interactions of drug targets were predicted for identifying interacting partners. Further, host-pathogen interactions of interacting partners were predicted to identify important molecules which are closely associated with gastric cancer.
Results:
Screening the genome of 23 H. pylori strains revealed 642 bacterial genes/virulence factors in 31 pathogenic islands. Further analysis identified 101 genes which were non-homologous to human and essential for the survival of the pathogen, among them 31 are potential drug targets. Protein-protein interactions for 31 drug targets predicted 609 interacting partners. Predicted interacting partners were further subjected to host-pathogen interactions leading to identification of important molecules like TNF receptor associated factor 6, (TRAF6) and MAPKKK7 which are closely associated with gastric cancer.
Conclusion:
These provocative studies enabled us to identify important molecules in H. pylori and their counter interacting molecules in the host leading to gastric cancer and also a pool of novel drug targets for therapeutic intervention of gastric cancer.
Keywords: Pathogenicity, Genomic islands, Virulence factors, Comparative analysis
1. Introduction
Gastric inflammation, ulcer, and cancer are induced by H. pylori infection. H. pylori is also responsible for other disorders like skin, oropharynx, endocrine, respiratory, haemopoietic, central nervous system, eye and reproductive system, etc. [1, 2]. Bacterial virulence factors are important for the development of gastric carcinoma [3, 4]. Stomach cancer is increased by the presence of the Cag Pathogenicity Island (PAI) of which a Cag gene encodes an immunodominant protein called CagA. This belongs to the type IV secretion system along with it VirB proteins. Zanotti and Cendron [5] revealed a large number of copies of CagA and VirB proteins in the type IV secretion system. Once CagA is injected into the host cell, tyrosine is phosphorylated, which interferes with several cancer pathways [5]. Reproduction in male and females is also affected by H. pylori infection [6-8]. These bacterial genes/virulence factors of the pathogenic islands represent a promising source for identifying novel drug targets.
Identification and validation of novel drug targets is a key process for discovery of new compounds. Various methods and approaches are available for discovery and validation of drug targets for infectious diseases [9]. Dutta et al. [10] used subtractive genomics for identification of essential genes in H. pylori strain HpAG1, Hp26695 and J99. Kiranmayi et al. [11] identified essential transporter genes in H. pylori using bioinformatics approaches. Neelapu and Pavani [12] identified 17 novel drug targets in H. pylori strains HpB38, HpP12, HpG27, HpShi470, HpSJM180 using in-silico genome and proteome analysis, whereas in a similar type of analysis carried out in the strain HpAG1 29 novel drug targets were identified [13]. Nammi et al. [14] used comparative genomics, proteomics etc. for 23 H. pylori strains to identify 29 novel drug targets. Mandal and Das [15] used in-silico approach for identifying drug targets in H. pylori. Sarkar et al. [16] used metabolic pathway analysis to identify drug targets in H. pylori. Cai et al. [17] used reverse docking to identify drug targets in H. pylori. However, there are no specific reports to date, on screening of pathogenic islands in H. pylori to identify drug targets. Therefore, the current paper deals with screening of pathogenic islands to identify novel drug targets in 23 strains of H. pylori.
2. Materials and Methods
2.1. Sampling
Genomes of 23 H. pylori strains HpF32 [18], HpF30 [18], Hp2017 [19], Hp2018 [19], Hp26695 [20], Hp35A [21], Hp51 [22], Hp52 [23], HpCuZ20 [24], HpF16 [18], HpF57 [18], HpINDIA7 [25], HpSAT464 [26], HpJ99 [27], HpB8 [28], Hp908 [29], Hp83 [30], HpSJM180 [31], HpAG1 [32], HpShi470 [33], HpG27 [34], HpP12 [35] and HpB38 [36] are sampled based on availability of complete genome, strain history, pathogenicity report of the strains and geographical origin. In our study, identification of novel drug targets for H. pylori has been accomplished for the first time for all the 23 H. pylori strains by using an integrated approach of genome, proteome and primary property analysis followed by protein-protein interactions of genes/proteins and host-pathogen interactions using computational resources.
2.2. Screening of Pathogenic Islands by In silico Genome Analysis for Drug Targets
Islands viewer [37] is used to identify pathogenic islands and the virulence genes in pathogenic islands for 23 H. pylori strains. Islands viewer is an integrated tool with different genomic island prediction methods such as Island Pick, SIGI-HMM and Island Path. These methods identify virulence genes in genomic islands based on three different criteria and methods. Island Pick is used to identify genomic islands and non-genomic islands. SIGI-HMM identifies genomic islands based on sequence composition, GC% and codon usage by implementing Hidden Markov model. Island Path (DIMOB) identifies functionally related mobile genes like transposases, integrases and abnormal sequence composition based on the origin of the genome. Complete genomes of 23 H. pylori strains were submitted to the Islands viewer to screen and identify pathogenic islands in the respective genomes and the virulence genes in pathogenic islands. These virulence genes were screened and confirmed for non homology as per the procedure of Neelapu et al. [13]. Potential drug targets among the pool of catalogued virulence genes were identified as per the procedure of Neelapu et al. [13].
2.3. Prediction of Protein-protein Interactions
Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) at http://string-db.org/ is used to predict the protein interactions for the 31 drug targets [38]. STRING is a database consisting of the known and predicted protein interactions data for more than 2000 organisms. Protein-protein interactions were performed based on amino acid sequence for each drug targets. Amino acid sequence of each drug target was submitted to the STRING database against organism H. pylori.
2.4. Network Analysis
Cytoscape v3.3.0 is a popular bioinformatics tool for biological network visualization and data integration [39]. Cytoscape was used to integrate and visualize the biological network data predicted in STRING. STRING network data consisting of 609 predicted partners for 31 drug targets is imported to Cytoscape. The network data was integrated using option union to predict and visualize the comprehensive network of 609 predicted partners. Network analysis of Network Analyzer of Cytoscape v3.3.0, was used to compute closeness centrality, stress centrality, betweenness centrality, distribution, shortest path length distribution, shared neighbors distribution, node degree distribution, neighbourhood connectivity distribution, node degree distribution, etc., along with other simple parameters of the network such as number of nodes, connected components, network diameter, network radius, network centralization, clustering coefficient, number of selfloops, multi-edge node pairs, shortest paths, characteristic path length, average number of neighbors, network density, network heterogeneity. Data integrated in network was visualized using organic of Y files layout.
2.5. Prediction of Host-Pathogen Interactions
Host-pathogen interactions help us to understand the role and mechanism of infection paving path to understand and identify more efficient strategies to cure or prevent infection. Host-pathogen interaction studies were performed in two ways – first option is by using host-pathogen interaction tools and second option is by text mining of the literature. The following tools were employed to predict the host pathogen interactions: Pathogen-Host interaction search tool (PHISTO) [40], Pathosystems Resource integration Center (PATRIC) [41] and Host Pathogen Interaction Database (HPIDB) [42]. In addition, text mining of literature was performed using drug targets and its predicted interacting partners for data host pathogen interactions.
3. Results
3.1. Potential Drug Targets for H. pylori
Island Pick, SIGI-HMM and Island Path methods of Islands viewer predicted 31 pathogenic islands with genes/virulence factors for 22 H. pylori strains (Table 1). No pathogenic islands were detected in H. pylori strain HpSAT464. The features of the pathogenic islands predicted are mentioned in Table 2. Nearly 642 genes/virulence factors associated with pathogenic islands were identified in 23 H. pylori strains Table 2. Of them 282 were known with known functions and rest 361 were hypothetical proteins (Table 2). Analysis of 642 bacterial genes identified 101 genes which are non-homologous to humans and are essential for pathogen. Gene property analysis of 101 genes identified 31 potential drug targets (Table 3).
Table 1.
Pathogenic islands identified in 23 H. pylori strains.
| S. No | Strain Name | No of Genomic Islands | Method | Start Position | End Position | Size |
|---|---|---|---|---|---|---|
| 1 | Hp57 | 1 | IslandPath-DIMOB | 284,185 | 324,544 | 40,359 |
| 2 | Hp12 | 1 | IslandPath-DIMOB | 484,957 | 489,788 | 4,831 |
| 3 | HpB8 | 1 | IslandPath-DIMOB | 448,052 | 533,220 | 85,168 |
| 4 | Hp India 7 | 3 | IslandPath-DIMOB | 749,965 | 797,920 | 47,955 |
| __ | __ | __ | IslandPath-DIMOB | 1,217,751 | 1,246,359 | 28,608 |
| __ | __ | __ | IslandPath-DIMOB | 1,616,790 | 1,626,196 | 9,406 |
| 5 | Hp51 | 1 | IslandPath-DIMOB | 992,739 | 1,036,302 | 43,563 |
| 6 | HpF32 | 1 | IslandPath-DIMOB | 1,051,313 | 1,085,082 | 33,769 |
| 7 | HpF16 | 2 | IslandPath-DIMOB | 470,749 | 493,591 | 22,842 |
| __ | __ | __ | IslandPath-DIMOB | 832,543 | 872,347 | 39,804 |
| 8 | HpF30 | 1 | IslandPath-DIMOB | 828,728 | 868,864 | 40,136 |
| 9 | Hp35A | 1 | IslandPath-DIMOB | 1,032,831 | 1,072,644 | 39,813 |
| 10 | HpB38 | 1 | IslandPath-DIMOB | 1,509,346 | 1,522,857 | 13,511 |
| 11 | Hp2018 | 1 | IslandPath-DIMOB | 982,216 | 1,004,243 | 22,027 |
| 12 | Hp908 | 1 | IslandPath-DIMOB | 973,392 | 990,660 | 17,268 |
| 13 | Hp2017 | 2 | IslandPath-DIMOB | 497,212 | 532,400 | 35,188 |
| __ | __ | __ | IslandPath-DIMOB | 974,279 | 996,463 | 22,184 |
| 14 | HpAG1 | 1 | IslandPath-DIMOB | 512,700 | 550,135 | 37,435 |
| 15 | HpSJM180 | 1 | IslandPath-DIMOB | 1,372,341 | 1,416,207 | 43,866 |
| 16 | HpJ99 | 2 | IslandPath-DIMOB | 908,817 | 912,007 | 3,190 |
| __ | __ | __ | IslandPath-DIMOB | 1,010,607 | 1,061,274 | 50,667 |
| 17 | HpShi470 | 1 | IslandPath-DIMOB | 874,701 | 915,726 | 41,025 |
| 18 | HpG27 | 1 | IslandPath-DIMOB | 1,045,375 | 1,082,440 | 37,065 |
| 19 | HpCuz20 | 3 | IslandPick | 205,446 | 215,461 | 10,015 |
| __ | __ | __ | IslandPath-DIMOB | 226,353 | 260,258 | 33,905 |
| __ | __ | __ | IslandPath-DIMOB | 562,530 | 600,842 | 38,312 |
| 20 | Hp26695 | 2 | IslandPath-DIMOB | 449,710 | 479,634 | 29,924 |
| __ | __ | __ | IslandPath-DIMOB | 1,042,255 | 1,070,401 | 28,146 |
| 21 | Hp83 | 2 | IslandPath-DIMOB | 73,583 | 107,449 | 33,866 |
| __ | __ | __ | IslandPath-DIMOB | 852,516 | 892,293 | 39,777 |
| 22 | Hp52 | 1 | IslandPick | 654,123 | 662,394 | 8,271 |
Table 2.
Proteins and drug targets identified in pathogenic islands for 23 H. pylori strains.
| S. No | Strain Name | No. of Pathogenic Islands | No. of Proteins | No. of Hypothetical Proteins | No. of Potential Drug Targets | No. of Drug Targets |
|---|---|---|---|---|---|---|
| 1 | Hp51 | 1 | 9 | 12 | 5 | 3 |
| 2 | HpF32 | 1 | 12 | 25 | 11 | 2 |
| 3 | HpF16 | 2 | 11 | 15 | 10 | 2 |
| 4 | HpF30 | 1 | 28 | 2 | 7 | 1 |
| 5 | Hp35A | 1 | 32 | 4 | 9 | 3 |
| 6 | HpB38 | 1 | 13 | 4 | 3 | 2 |
| 7 | Hp2018 | 1 | 11 | 12 | 2 | 1 |
| 8 | Hp908 | 1 | 6 | 14 | 0 | 0 |
| 9 | Hp2017 | 2 | 28 | 5 | 3 | 2 |
| 10 | HpHPAG1 | 1 | 31 | 0 | 10 | 1 |
| 11 | HpSJM180 | 1 | 9 | 24 | 6 | 3 |
| 12 | HpJ99 | 2 | 2 | 4 | 0 | 0 |
| 13 | HpG27 | 1 | 9 | 16 | 6 | 5 |
| 14 | HpShi470 | 1 | 8 | 27 | 4 | 2 |
| 15 | HpcuZ20 | 3 | 4 | 7 | 0 | 1 |
| 16 | Hp83 | 2 | 13 | 34 | 8 | 2 |
| 17 | Hp26695 | 2 | 9 | 18 | 2 | 0 |
| 18 | HpIndia7 | 3 | 10 | 27 | 7 | 1 |
| 19 | HpB8 | 1 | 21 | 65 | 5 | 3 |
| 20 | Hp12 | 1 | 2 | 0 | 0 | 0 |
| 21 | HpF57 | 1 | 11 | 24 | 3 | 2 |
| 22 | Hp52 | 1 | 4 | 4 | 0 | 0 |
| 23 | HpSAT464 | 0 | 0 | 0 | 0 | 0 |
| Total | 31 | 282 | 361 | 101 | 36 |
Table 3.
Drug targets identified in the 23 H. pylori strains.
| S. No | Drug Target | Metabolic Categories | Gene ID | Strain Name | ||
|---|---|---|---|---|---|---|
| 1 | GTPase | Cellular process | GI:387782588 | Hp51 | ||
| 2 | DNA transfer protein | Cellular process | GI:387782591 | Hp51 | ||
| 3 | Transposase | Cellular process | GI:385224738 | Hp83 | ||
| 4 | Putative IS606 transposase | Cellular process | GI:385223561 | Hp2017 | ||
| 5 | Conjugal transfer protein | Cellular process | GI:317181586 | Hp57 | ||
| 6 | Bacteriophage-related integrase | Cellular process | GI:384892219 | HpcuZ20 | ||
| S. No | Drug Target | Metabolic Categories | Gene ID | Strain Name | ||
| 7 | Cag island DNA transfer proteinCag C | Cellular process | GI:208434927 | HpG27 | ||
| 8 | Cag pathogenicity island protein 5 | Cellular process | GI:384896154 | Hp35A | ||
| 9 | Cag pathogenicity island protein 3 | Cellular process | GI:385223565 | Hp2017 | ||
| 10 | Integrase/recombinase XercD family protein | Cellular process | GI:308185118 | HpSJM180 | ||
| Cellular process | GI:188527674 | HpShi470 | ||||
| 11 | Type IV secretion system protein virB8 | Virulence factors | GI:298355174 | HpB8 | ||
| 12 | Type IV secretion system protein VirB4 | Virulence factors | GI:298355233 | HpB8 | ||
| 13 | Type IV secretion system protein VirB9 | Virulence factors | GI:317009420 | HpIndia7 | ||
| 14 | Periplasmic competence protein-like protein | Virulence factors | GI:308185123 | HpSJM180 | ||
| Virulence factors | GI:208434936 | HpG27 | ||||
| 15 | Poly E-rich protein | Information and storage | GI:385216250 | HpF32 | ||
| 16 | Cag pathogenicity island protein M | Metabolism molecule | GI:384896163 | Hp35A | ||
| 17 | Cag pathogenicity island protein 9 | Metabolism molecule | GI:108562930 | HpHPAG1 | ||
| Metabolism molecule | GI:3848449915 | HpF30 | ||||
| 18 | Cag pathogenicity island protein W | Metabolism molecule | GI:384896173 | Hp35A | ||
| 19 | Hac prophage II protein | Metabolism molecule | GI:254780055 | HpB38 | ||
| 20 | Hac prophage II integrase | Metabolism molecule | GI:254780053 | HpB38 | ||
| 21 | Mechanosensitive channel | Metabolism molecule | GI:385231894 | Hp2018 | ||
| 22 | Relaxase | Metabolism molecule | GI:308185146 | HpSJM180 | ||
| 23 | Putative chromosome partitioning protein | Metabolism molecule | GI:298355190 | HpB8 | ||
| 24 | VirB7 | Metabolism molecule | GI:385224749 | Hp83 | ||
| 25 | Competence protein | Metabolism molecule | GI:208434918 | HpG27 | ||
| 26 | ComB9-like competence protein | Metabolism molecule | GI:208434919 | HpG27 | ||
| 27 | ATPase | Metabolism molecule | GI:387782603 | Hp51 | ||
| 28 | Outer membrane protein HorC | Metabolism molecule | GI:385216248 | HpF32 | ||
| 29 | PARA protein | Metabolism molecule | GI:208434932 | HpG27 | ||
| Metabolism molecule | GI:188527681 | HpShi470 | ||||
| Metabolism molecule | GI:317181592 | Hp57 | ||||
| 30 | Holliday junction resolvase | Metabolism molecule | GI:385217246 | HpF16 | ||
| 31 | Type II adenine specific DNA methyltransferase | Metabolism molecule | GI:385217221 | HpF16 | ||
Literature screening based on the keywords identified that all the drug targets are experimentally validated. Sixteen of the 31 predicted drug targets are critical for the survival of H. pylori. Analysis showed that GTPase, transposase, conjugal transfer protein, cag island DNA transfer protein cag 5, cag pathogenicity island protein Cag 3, cag pathogenicity island protein CagC, type IV secretion system protein virB8, type IV secretion system protein VirB4, type IV secretion system protein VirB9, cag pathogenicity island protein M, cag pathogenicity island protein W/9, relaxase, competence protein comB9-like competence protein, ATPase, Holliday junction resolvase, type II adenine specific DNA methyltransferase might be the critical drug targets for survival of Helicobacter species.
3.2. Protein-protein Interactions
STRING’s reliable algorithms predicted protein-protein interactions for H. pylori in three modes-confidence view, evidence view and action view. Twenty six of the 31 drug targets demonstrated interactions with other proteins, whereas no partners were predicted for rest five of the drug targets (Supplementary Table 1 (236.2KB, pdf) ). The evidence view presents information from different sources such as neighbourhood, coexpression, text mining, homology and gene fusion. The evidence view for the 23 drug targets is presented in Fig. (1). Action view presents interacting information regarding activation, inhibition, binding, phenotype, catalysis, post translation modification and expression whereas confidence view presents score between interaction partners. STRING predicted 609 interacting partners for the 23 drug targets in three modes (Fig. 1; Supplementary Table 1 (236.2KB, pdf) ).
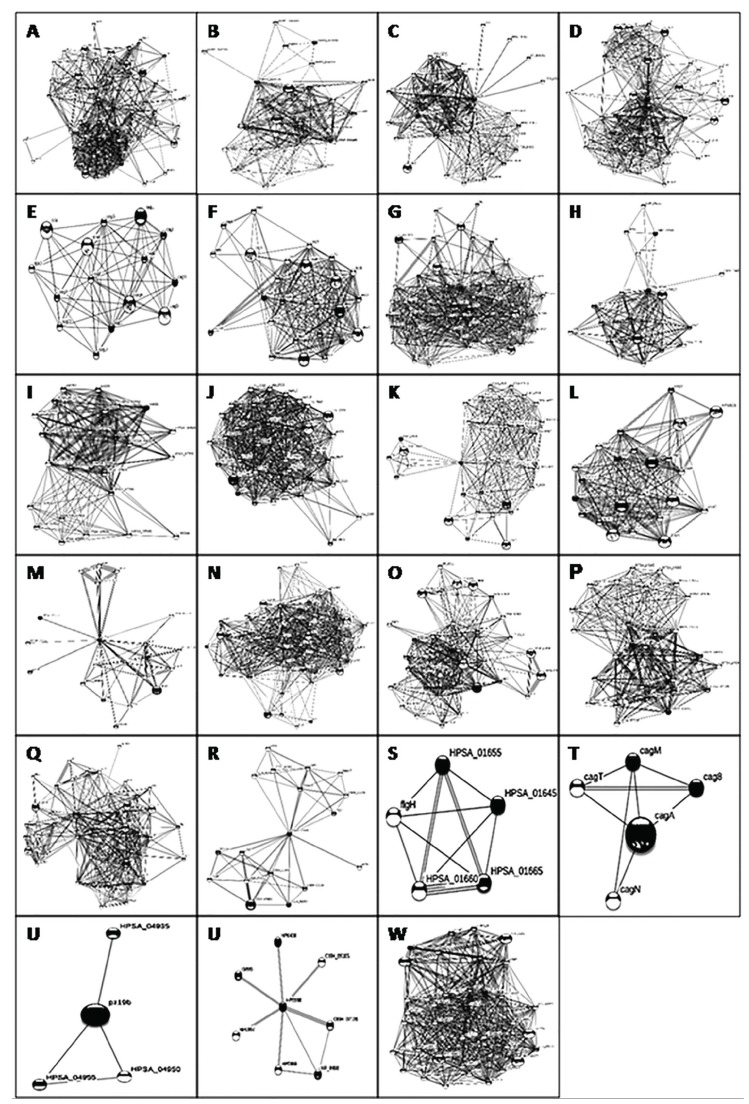
Fig. (1).
Evidence view of protein-protein interactions as predicted in STRING for 23 drug targets identified in H. pylori A). GTPase protein, B). ATPase protein, C). conjugal transfer protein D). Cag 5 protein E). Cag C protein, F). Cag 3 protein, G). integrase/recombinase (XERCD family) protein, H).Vir B4 protein, I). Vir B9 protein, J). Periplasmic competence protein, K). Poly E rich protein, L). Cag 9 protein, M). PARA protein, N). Relaxase protein, O). Competence like protein, P). Com B9 like protein, Q). Holiday junction resolvase protein, R). Type II adenine methyl transferase protein, S). Outer membrane HorC protein, T). Cag M protein, U). Mechanosensitive channel protein, V). Transposase protein, W). Type IV secretion system VirB8 protein.
3.3. Network Analysis
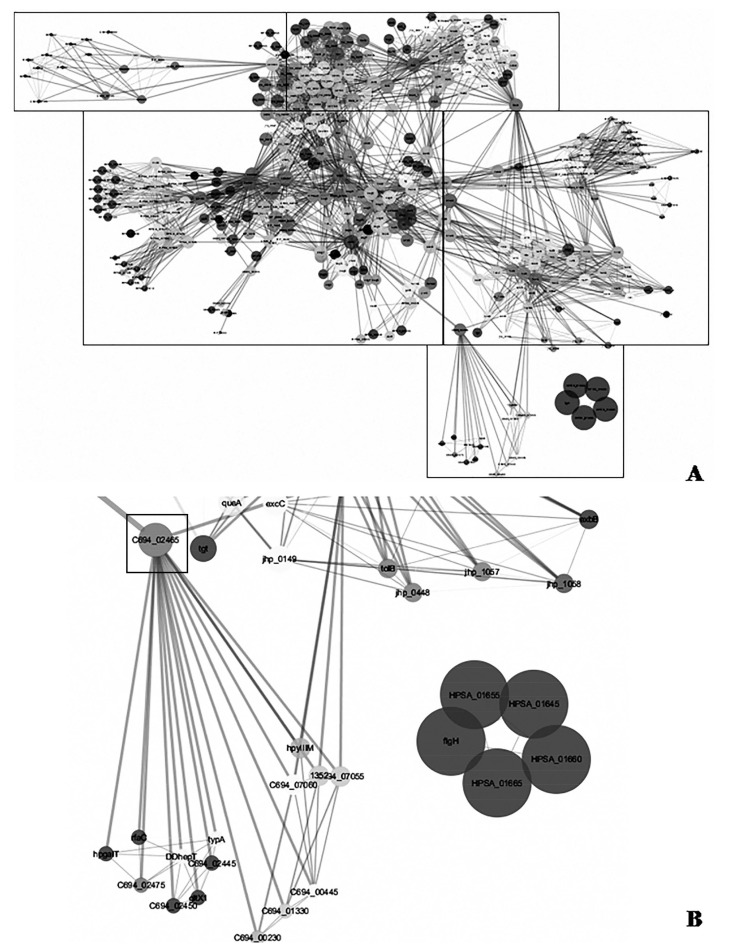
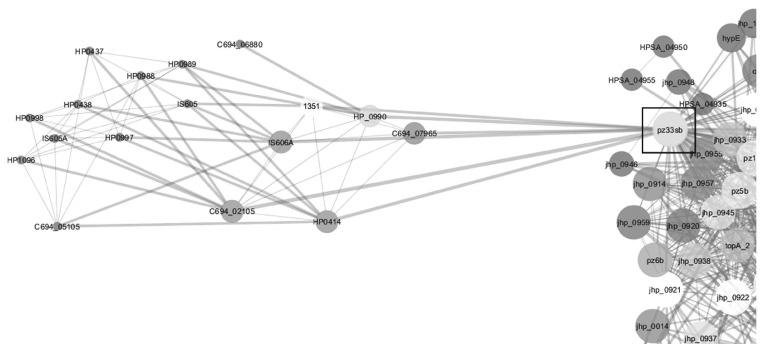
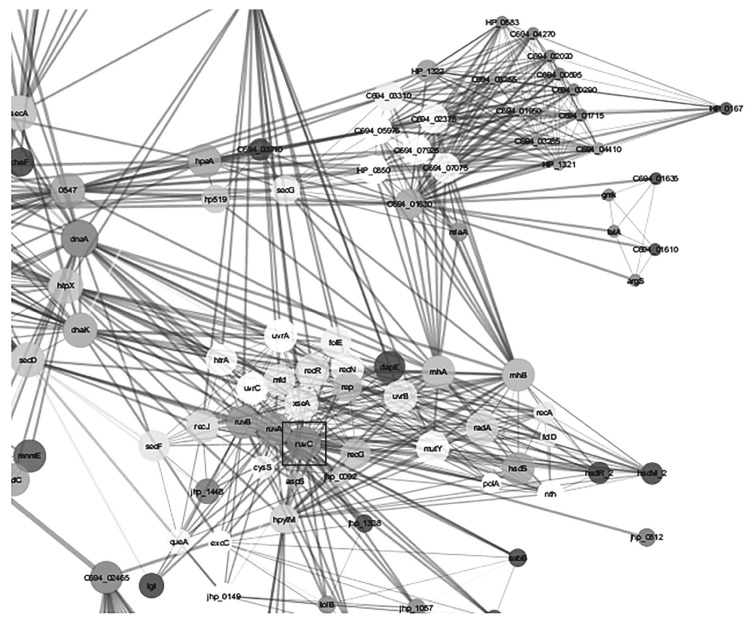
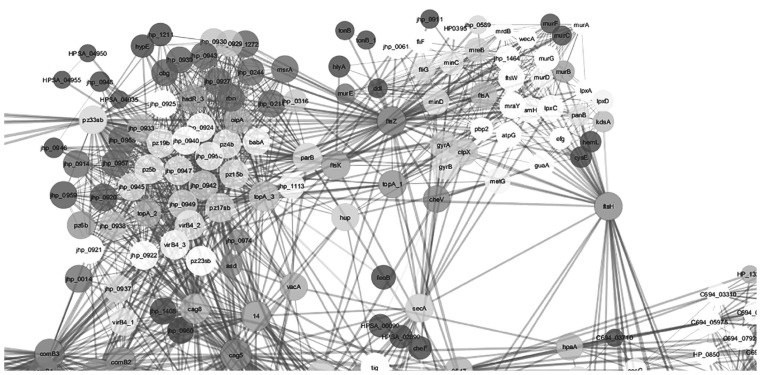
Network analysis on twenty three networks were predicted in STRING when exported and merged in cytoscape. Data visualization and analysis on the merged network demonstrated different protein hubs in the merged network (Figs. 2-6). Protein-protein interactions in bird eye view with 361 nodes and 3146 edges of 609 interacting partners were revealed by cytoscape (Fig. 2A). Very important interactions with specific proteins responsible for gastric cancer were visualized in protein hubs. Protein-protein interactions of drug target C694_01330 (Type II adenine specific DNA methyl tranferase can be visualized in Fig. (2B) by cytoscape. Protein-protein interactions of hub (pz33sb) with IS606A transposase and IS605A transposase were as visualized in Fig. (3). Protein-protein interactions of drug target ruvC (Holiday junction resolvase) with DNA repair system is visualized in Fig. (4). Blocking the drug target/hub of the pathogen with a molecule would lack DNA repairing mechanism affecting survival of the organism.
Fig. (2).
A) Bird eye view of protein-protein interactions with 361 nodes and 3146 edges of 609 interacting partners as revealed by cytoscape. B) Protein-protein interactions of drug target C694_01330 (Type II adenine specific DNA methyl tranferase) highlighted in rectangle box as revealed by cytoscape.
Fig. (6).
Protein-protein interactions of drug targets Cag E, Vir B11, ISO606B transposase with Ure A (urease sububnit aplha) as revealed by cytoscape.
Fig. (3).
Protein-protein interactions from the hub pz33sb highlighted in rectangle box with IS606A transposase and IS605A transposase as revealed by cytoscape.
Fig. (4).
Protein-protein interactions of drug target ruvC (Holiday junction resolvase) highlighted in rectangle box with other DNA repair system as revealed by cytoscape.
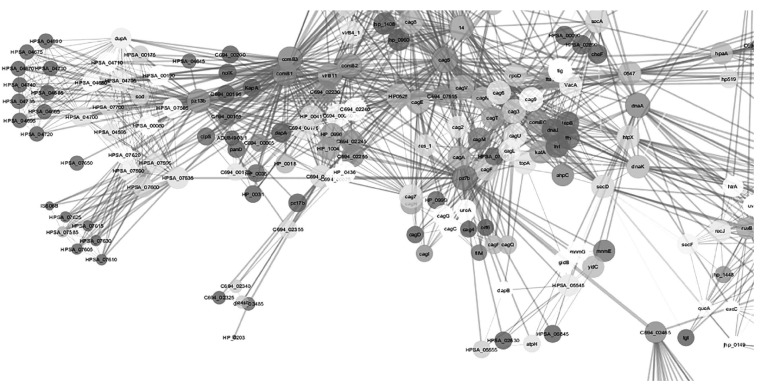
Protein-protein interactions of drug targets Cag 5, Vir B, Vir B8, pz19b (mechanosenstive ion channel protein), ftsz/obg/GTPase, Com9 (DNA transformation competence protein), pz23sb (PARA protein), Vir B4 with Che V (chemotaxis proteins), adhesion proteins like BabA, and toxins like Vac A are visualized in Fig. (5). Interfering with these drug targets which are related to organism movement, adhesion of the organism with the host, and transfer of toxins to host would result in retardation of the growth. Protein-protein interactions of drug targets Cag E, Vir B11, ISO606B transposase with Ure A (urease sububnit aplha) are visualized in Fig. 6. Urease is an important enzyme for neutralizing the acid in the host and lacking this enzyme would be fatal for pathogen.
Fig. (5).
Protein-protein interactions of drug targets Cag 5, Vir B, Vir B8, pz19b (mechanosenstive ion channel protein), ftsz/obg/GTPase, Com9 (DNA transformation competence protein), pz23sb (PARA protein), Vir B4, with Che V(chemotaxis proteins), adhesion proteins like BabA, and toxins like Vac A as revealed by cytoscape.
3.4. Host-pathogen Interactions
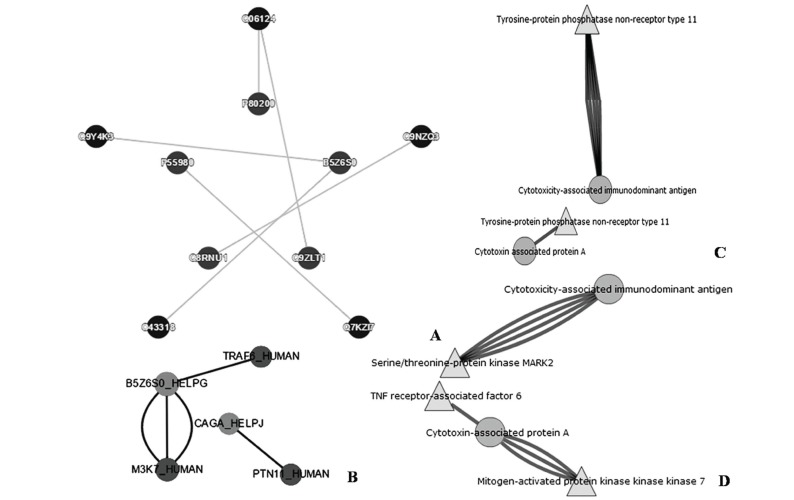
Tools PHISTO, PATRIC and HPIDB predicted host pathogen interactions. PHISTO predicted five interactions between host and pathogen proteins Table 4; (Fig. 7A). Tyrosinase-protein phosphatase non-receptor type II, NCK-interacting protein with SH3 domain, serine/threonine protein kinase MARK2, mitogen-activated protein kinase kinase kinase 7 (MAPKKK7) and TNF receptor associated factor 6 are the proteins from host involved in interactions Table 4; (Fig. 7A). Cytotoxin associated immunodominant antigen (with protein ID's P80200, P55980, B5Z6S0, Q9ZLT1) and vacuolating cytotoxin A (Q8RNUI) are the proteins from pathogen involved in interactions Table 4; (Fig. 7A). Tyrosinase-protein phosphatase non-receptor type II (Q06124), serine/threonine protein kinase MARK2 (Q9NZQ3), mitogen-activated protein kinase kinase kinase 7 (MAPKKK7) (O43318), TNF receptor associated factor 6 (Q9Y4K3) of host interacted with cytotoxin associated immunodominant antigen with protein ID's P80200, P55980, B5Z6S0, Q9ZLT1 of pathogen respectively Table 4; (Fig. 7A). PHISTO also predicted NCK-interacting protein with SH3 domain (Q9NZQ3) of host interacting with vacuolating cytotoxin A (Q8RNUI) of pathogen Table 4; (Fig. 7A).
Table 4.
Host – pathogen interactions as predicted by tools PHISTO, PATRIC and HPIDB.
| S. No | Host ID | Host | Pathogen ID | Pathogen | Interaction Type | Method | Reference |
|---|---|---|---|---|---|---|---|
| PHISTO | |||||||
| 1 | Q06124 | Tyrosine-protein phosphatase non-receptor type 11 | P80200 | Cytotoxicity-associated immunodominant antigen | - | anti bait coimmunoprecipitation | Higashi et al. [43] |
| 2 | Q9NZQ3 | NCK-interacting protein with SH3 domain | Q8RNU1 | Vacuolating cytotoxin A | - | two hybrid/coimmunoprecipitation | de Bernard et al. [44] |
| 3 | Q7KZI7 | Serine/threonine-protein kinase MARK2 | P55980 | Cytotoxicity-associated immunodominant antigen | - | molecular sieving | Nesić et al. [45] |
| 4 | O43318 | Mitogen-activated protein kinase kinase kinase 7 | B5Z6S0 | Cytotoxicity-associated immunodominant antigen | - | anti tag coimmunoprecipitation | Lamb et al. [46] |
| 5 | Q9Y4K3 | TNF receptor-associated factor 6 | Q9ZLT1 | Cytotoxicity-associated immunodominant antigen | - | Other methods | Zhu et al. [47] |
| PATRIC | |||||||
| 1 | Q06124 | Tyrosine-protein phosphatase non-receptor type 11 | B5Z6S0 | Cytotoxicity-associated immunodominant antigen | Physical association | Anti-tagcoimmuniprecption | Higashi et al. [43] |
| 2 | O43318 | Mitogen-activated protein kinase kinase kinase 7 | P80200 | Cytotoxin-associated protein A | Physical association | Anti-tagcoimmuniprecption | Lamb et al. [46] |
| 3 | Q06124 | Tyrosine-protein phosphatase non-receptor type 11 | P80200 | Cytotoxin-associated protein A | Physical association | Anti-tagcoimmuniprecption | Higashi et al. [43] |
| 4 | Q7KZI7 | Serine/threonine-protein kinase MARK2 | P80200 | Cytotoxin-associated protein A | Direct interaction | Molecular seiving | Nesić et al. [45] |
| 5 | Q9Y4K3 | TNF-receptor associated factor 6 | P80200 | Cytotoxin-associated protein A | Direct interaction | Anti-tagcoimmuniprecption | Lamb et al. [46] |
| HPIDB | |||||||
| 1 | Q9NZQ3 | NCK-interacting protein with SH3 domain | P80200 | Cytotoxin-associated protein A | Physical association | colocalization | de Bernard et al. [44] |
| 2 | Q06124 | Tyrosine-protein phosphatase non-receptor type 11 | B5Z6SO | Cytotoxin-associated protein A | Physical association | anti bait coimmunoprecipitation | Higashi et al. [43] |
| 3 | O43318 | Mitogen-activated protein kinase kinase kinase 7 | B5Z6SO | Cytotoxin-associated protein A | Physical association | Anti-tagcoimmuniprecption | Lamb et al. [46] |
Fig. (7).
Host and pathogen interactions as visualized by A) PHISTO B) HPIDB C&D) PATRIC.
HPIDB visualized interactions between 54 proteins from different pathogens with three human proteins (Table 4; Fig. 7B). Proteins 22, 1, 20, 11 were predicted for bacteria, fungi, virus, animal respectively. Among these pool 22 bacterial proteins two proteins from H. pylori were interacting with three human proteins (Table 4). HPIDB predicted three interactions between host and pathogen (Table 4; Fig. 7B). Tyrosinase-protein phosphatase non-receptor type II (Q06124), mitogen-activated protein kinase kinase kinase 7 (MAPKKK7) (O43318) and NCK-interacting protein with SH3 domain (Q9NZQ3) are the proteins from host involved in interactions with cytotoxin associated immunodominant antigen A (with protein ID's P80200; B5Z6S0) of pathogen Table 4; (Fig. 7B).
PATRIC predicted five interactions between host and pathogen Table 4; (Fig. 7C, D). Tyrosinase-protein phosphatase non-receptor type II, mitogen-activated protein kinase kinase kinase 7 (MAPKKK7), serine/threonine protein kinase MARK2 and TNF receptor associated factor 6 are the proteins from host involved in interactions Table 4; (Fig. 7C, D). Cytotoxin associated immunodominant antigen (with protein ID B5Z6S0) and Cytotoxin associated immunodominant antigen A (with protein ID P80200) are the proteins from pathogen involved in interactions Table 4; (Fig. 7C, D). Serine/threonine protein kinase MARK2 (Q9NZQ3), mitogen-activated protein kinase kinase kinase 7 (MAPKKK7) (O43318), TNF receptor associated factor 6 (Q9Y4K3) and NCK-interacting protein with SH3 domain (Q9NZQ3) of host interacted with cytotoxin associated immunodominant antigen A (P80200) of pathogen respectively Table 4; (Fig. 7C, D). PATRIC also predicted tyrosinase-protein phosphatase non-receptor type II (Q06124) of host interacting with cytotoxin associated immunodominant antigen (B5Z6S0) of pathogen respectively Table 4; (Fig. 7C, D). In addition text mining of literature showed that eight proteins of H. pylori are interacting with 14 human proteins Table 5.
Table 5.
Host – pathogen interactions revealed from the text mining of the literature.
| S. No | Host Pathogen Interactions | Interactions Causes | Reference | |
|---|---|---|---|---|
| Pathogen Proteins | Human Proteins | |||
| 1 | CagA | E-cadherin | Gastric cancer | Murata-Kamiya et al. [48] |
| Erk mitogen-activated protein kinase | Gastric cancer | Zhu et al. [47] | ||
| Transforming growth factor-b-activated kinase 1 (TAK1) | Gastric cancer | Lamb et al. [46] | ||
| Src family kinases | Gastric cancer | Higashi et al. [49] | ||
| human kinase PAR1b/MARK2 | Gastric cancer | Nesić et al. [45] | ||
| 2 | CagE type IV secretion system | Dendritic cells | MALT and Gastric cancer | Donald et al. [50] |
| NF- B activator. TAK1, TRAF6, and MyD88 |
Intestinal metaplasia and Gastric cancer | Hirata et al. [51] | ||
| 3 | Holliday junctions resolves | Mus81 | block DNA replication | Chen et al. [52] |
| 4 | Mechanosensitive ion channel protein | integrin-b-catenin | human articular chondrocyte (HAC) responses | Lee et al. [53] |
| Focal Adhesion Kinase pp125FAK | osteoblast activation | Rezzonico et al. [54] | ||
| 5 | Type IV secretion system | protein kinase B, PKB | Gastric cancer | King et al. [55] |
| 6 | GTPase | Guanine Nucleotide Exchange FactorSec7 Domain |
IL-8 expression | Mossessova et al. [56] |
| 7 | Transpoase | RNA-proteins | To regulate the RNA-proteins network | Kelley et al. [57] |
| 8 | VacA | VIP54 | Infection/Gastric cancer | de Bernard et al. [44] |
4. Discussion
Discovery, identification and validation of drug targets have been a debate from long time. Recent advances on discovery and validation of drug targets for infectious diseases focused on disease understanding and mechanism. Previously subtractive genomics [11-13] was implemented by our group to identify novel drug targets for 23 H. pylori strains. Different methods like essential gene identification [10, 11], metabolic pathway analysis [16] and reverse docking [17] were implemented by other groups to identify novel drug targets for H. pylori. Though, these methods were successful in identifying novel drug targets for H. pylori we foresee pathogenic islands as the potential source for novel drug targets.
The current study was the first report till date to employ successful systematic insilico analysis for identification of potential and novel drug target candidates from pathogenic islands of 23 H. pylori strains. Systematic in silico analysis included five steps - the first two steps were used to identify drug targets and the next three steps were used to characterize the drug targets. The initial step in the systematic analysis is to screen the genome of H. pylori strains to identify pathogenic islands. Screening the genome of H. pylori strains using islands viewer [37] identified 31 pathogenic islands Table 1. The second step is to analyzae the pathogenic islands to identify the potential drug targets for
H. pylori. Analysis of the pathogenic islands resulted in identification of 642 virulence factors (bacterial genes) in 31 pathogenic islands Table 2. The analysis of the 642 virulence factors identified 101 genes which were non-homologous to human and are essential for the survival of the pathogen. Further, analysis of 101 genes for gene property identified 31 novel and potential drug targets for H. pylori Table 3. The third step in the systematic analysis is to implement protein-protein interactions to identify the interacting partners for the potential drug targets. STRING was used to study the protein-protein interactions and predicted 609 interacting partners for the 23 drug targets (Fig. 1); Supplementary Table 1 (236.2KB, pdf) . The fourth step is to accomplish network analysis on the interacting partners associated in the protein-protein interactions. Data of twenty three networks was exported and merged in cytoscape to perform network analysis. Data visualization and analysis on the merged network demonstrated bird eye view of different protein hubs in the merged network with 361 nodes and 3146 edges of 609 interacting partners (Fig. 2A). And the fifth and final step in the systematic analysis is to proceed with the host-pathogen interactions based on tools and literature mining. PHISTO, PATRIC and Host Pathogen Interaction Database were used to predict the host pathogen interactions for predicted interacting partners. Host-pathogen interactions identified important molecules which are closely associated with gastric cancer. These studies persuaded us to ascertain key molecules in H. pylori and their counter interacting molecules in the host leading to gastric cancer.
Data on protein-protein interactions, network analysis and host-pathogenic interactions provided few insights and understanding of H. pylori associated gastric cancer. As revealed by the data in the present study H. pylori uses cytokines, gastrin and toxin VacA to weaken the gastric mucosal barrier and colonize in the submucous. After the gastric mucosal barrier is weakened BabA facilitates H. pylori in adhering to the epithelial lining of the stomach [58-62]. T4SS system coded by cytotoxin associated gene pathogenicity island (cag PAI) injects CagA, peptidoglycan and VacA into the host to establish interaction with the host leading to inflammation, a condition known as gastritis.
Cag A changes the expression of host cells; induces elongation of cell, loss of cell polarity and cell proliferation; decreases acid secretion; and degrade cell-cell junctions [63]. CagA is phosphorylated and activated by src/Lyn kinase disturbing mitogen-activated protein kinase (MAPK) signaling in host cells through NCK-interacting with SH3/SH2 domain to modify cellular responses [64]. Cell focal adhesions are disrupted by CagA by binding and activating SHP2 phosphates/Tyrosine protein phosphatase non receptor type 11 [64]. Normal epithelial architecture is disrupted when polarity regulator PAR1b/MARK2 kinase is inhibited by CagA leading to loss of polarity in epithelial cells [64]. Another surface receptor protein in H. pylori Toll like receptor (TLR)-2 disrupts adherin junctions within gastricepithelial cells. TLR-2 activates protease calpain cleaving E-cadherin and allows increased β-catenin signaling to disrupt adherin junctions [65]. CagA-dependent, TRAF6-mediated Lys 63-ubiquitination and activation of TAK1 activate transcription factor NF-κβ, resulting in chronic inflammation and cancer when is constitutively expressed [66-68]. CagA and COX-2 were known for cell proliferation, prostaglandin biosynthesis and angiogenesis. Cag A induces proteasome mediated degradation directly by inactivating gastric tumor suppressor gene RUNX3 [69, 70] or indirectly p53 to modulate ASPP2 tumor suppressor genes [71].
Vacuolating cytotoxin (Vac) A induce ROS at the site of infection damaging mitochondrial DNA of gastric epithelial cells [44]. Vac A interact with a number of host surface receptors to trigger responses such as poreformation, cell vacuolation, endolysomal functions modification, immune inhibition and apoptosis [72-74]. VacA along with other virulence factors such as γ-glutamyl transpeptidase, and cholesterol α-glucosides modulate responses of T cells. Cag A and Vac A induce ROS and NF-κβ, along with cytokines, and chemokines.
Cytokines (IL-1, 6, 8), chemokines (CXCL8, CCL3, 4), metalloproteinases (MMPs), prostaglandin E2 (PGE2) and reactive oxygen nitrogen species (RONS) prolong inflammation inducing G cells to secrete the hormone gastrin in turn stimulating loads of acid damaging duodenum a condition known as ulcers [75, 76]. NF-κβ and β-catenin signaling pathways induce double stranded breaks, defective mitotic checkpoints, deregulate HR pathway of DSB repair and DNA repair enzymes leading to genetic diversification randomly heading towards activation of oncogenes and inactivation of tumor suppressor genes leading to gastric cancer [77].
CONCLUSION
Pathogenic islands are the good source for drug targets. Analysis of genomes in 23 H. pylori strains identified 31 pathogenic islands of them 29 bacterial genes which are nonhomologous to humans and are essential for pathogen. All the drug targets were found to be critical for the species and are already experimentally validated lending credence to our approach. PHISTO, HPIDB, PATRIC tools visualized host-pathogen interactions directly or indirectly predicting the role of certain pathogen molecules (drug targets) in gastric cancer. These novel drug targets may have possible therapeutic implications for gastric cancer.
Consent for Publication
Not applicable.
ACKNOWLEDGEMENTS
DN, AMCP, NSY and NNRR are thankful to the GITAM University, Visakhapatnam, India for providing the facility and support. NNRR, DN and AMCP are thankful to University Grants Commission, New Delhi for the project funding [UGC Project F.No.42-636/2013 (SR) letter dated 25-03-2013]. DN is thankful for the Project Fellowship sponsored by UGC, New Delhi, India. The authors are also thankful to Prof. I. Bhaskar Reddy and Prof. Malla Rama Rao for constant support throughout the research work. We generously thank Dr Ch. Surekha, GITAM University, Visakhapatnam, India for critical comments and reviewing of the manuscript.
SUPPLEMENTARY MATERIAL
Supplementary material is available on the publisher’s web site along with the published article.
CONFLICT OF INTEREST
The authors declare no conflict of interest, financial or otherwise.
References
- 1.Figura N., Franceschi F., Santucci A., Bernardini G., Gasbarrini G., Gasbarrini A. Extragastric manifestations of Helicobacter pylori infection. Helicobacter. 2010;15(1):60–68. doi: 10.1111/j.1523-5378.2010.00778.x. [DOI] [PubMed] [Google Scholar]
- 2.Roubaud Baudron C., Franceschi F., Salles N., Gasbarrini A. Extragastric diseases and Helicobacter pylori. Helicobacter. 2013;18(1):44–51. doi: 10.1111/hel.12077. [DOI] [PubMed] [Google Scholar]
- 3.Blaser M.J. Gastric Campylobacter-like organisms, gastritis, and peptic ulcer disease. Gastroenterology. 1987;93(2):371–383. doi: 10.1016/0016-5085(87)91028-6. [DOI] [PubMed] [Google Scholar]
- 4.Wotherspoon A.C., Ortiz-Hidalgo C., Falzon M.R., Isaacson P.G. Helicobacter pylori associated gastritis and primary B-cell gastric lymphoma. Lancet. 1991;338(8776):1175–1176. doi: 10.1016/0140-6736(91)92035-z. [DOI] [PubMed] [Google Scholar]
- 5.Zanotti G., Cendron L. Structural and functional aspects of the Helicobacter pylori secretome. World J. Gastroenterol. 2014;20(6):1402–1423. doi: 10.3748/wjg.v20.i6.1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kurotsuchi S., Ando H., Iwase A., Ishida Y., Hamajima N., Kikkawa F. The plausibility of H. pylori related infertility in Japan. Fertil. Steril. 2008;90:866–868. doi: 10.1016/j.fertnstert.2007.06.097. [DOI] [PubMed] [Google Scholar]
- 7.Ambrosini G., Andrisani A., Fiore C., Faggian D., D’Antona D., Ragazzi E., Plebani M., Armanini D. Anti-Helicobacter pylori antibodies in cervical mucus: A new cause of infertility. Eur. J. Obstet. Gynecol. Reprod. Biol. 2011;155(2):157–160. doi: 10.1016/j.ejogrb.2010.12.001. [DOI] [PubMed] [Google Scholar]
- 8.Repaci A., Gambineri A., Pagotto U., Pasquali R. Ghrelin and reproductive disorders. Mol. Cell. Endocrinol. 2011;340:70–79. doi: 10.1016/j.mce.2011.02.022. [DOI] [PubMed] [Google Scholar]
- 9.Neelapu N.R., Nammi D., Pasupuleti A.C., Surekha Ch. New weapons to control bacterial growth. Springer, Hiedelberg, 2016. 2016. Targets against Helicobacter pylori and other tumor-producing bacteria. [Google Scholar]
- 10.Dutta A., Singh S.K., Ghosh P., Mukherjee R., Mitter S., Bandyopadhyay D. In silico identification of potential therapeutic targets in the human pathogen Helicobacter pylori. In Silico Biol. 2006;6:43–47. [PubMed] [Google Scholar]
- 11.Kiranmayi P., Swathi S.V., Neelapu N.R. Comparative analysis of metal transportomes in Helicobacter species: Modelling the molecular structures of nickel transporters. In: Gaillard B., Damien M., editors. Biometals: Molecular structures, binding properties and applications. New York: Nova Sci Pub Inc; 2009. [Google Scholar]
- 12.Neelapu N.R., Pavani T. Identification of novel drug targets in HpB38, HpP12, HpG27, Hpshi470, HpSJM180 strains of Helicobacter pylori: an in silico approach for therapeutic intervention. Curr. Drug Targets. 2013;14:601–611. doi: 10.2174/1389450111314050009. [DOI] [PubMed] [Google Scholar]
- 13.Neelapu N.R., Naresh M.V., Srinivas A. Identification of potential drug targets for Helicobacter pylori strain HPAG1 by in silico genome analysis. Infect. Disord. Drug Targets. 2015;15:106–117. doi: 10.2174/1871526515666150724111528. [DOI] [PubMed] [Google Scholar]
- 14.Nammi D., Srimath-Tirumala-Peddinti R.C., Neelapu N.R. Identification of drug targets in Helicobacter pylori by in silico analysis: possible therapeutic implications for gastric cancer. Curr. Cancer Drug Targets. 2015;16:79–98. doi: 10.2174/1568009615666150602143239. [DOI] [PubMed] [Google Scholar]
- 15.Mandal R.S., Das S. In silico approach towards identification of potential inhibitors of Helicobacter pylori DapE. J. Biomol. Struct. Dyn. 9:1–14. doi: 10.1080/07391102.2014.954272. [DOI] [PubMed] [Google Scholar]
- 16.Sarkar M., Maganti L., Ghoshal N., Dutta C. In silico quest for putative drug targets in Helicobacter pylori HPAG1: Molecular modeling of candidate enzymes from lipopolysaccharide biosynthesis pathway. J. Mol. Model. 2012;18:1855–1866. doi: 10.1007/s00894-011-1204-3. [DOI] [PubMed] [Google Scholar]
- 17.Cai J., Han C., Hu T., Zhang J., Wu D., Wang F., Liu Y., Ding J., Chen K., Yue J., Shen X., Jiang H. Peptide deformylase is a potential target for anti-Helicobacter pylori drugs: Reverse docking, enzymatic assay, and X-ray crystallography validation. Protein Sci. 2006;15:2071–2081. doi: 10.1110/ps.062238406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Furuta Y., Kawai M., Yahara K., Takahashi N., Handa N., Tsuru T., Oshima K., Yoshida M., Azuma T., Hattori M., Uchiyama I., Kobayashi I. Birth and death of genes linked to chromosomal inversion. Proc. Natl. Acad. Sci. USA. 2011;108:1501–1506. doi: 10.1073/pnas.1012579108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Avasthi T.S., Devi S.H., Taylor T.D., Taylor T.D., Kumar N., Baddam R., Kondo S., Suzuki Y., Lamouliatte H., Mégraud F., Ahmed N. Genomes of two chronological isolates (Helicobacter pylori 2017 and 2018) of the West African Helicobacter pylori strain 908 obtained from a single patient. J. Bacteriol. 2011;193:3385–3386. doi: 10.1128/JB.05006-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manolov A., Prihodko E., Larin A., Karpova I., Semashko T., Alexeev D., Kostrjukova E., Govorun V. Direct submission by bioinformatics, research institute for physico-chemical medicine, malaya pirogovskaya 1A, moscow 119992, Russia. Available from: http://www.ncbi.nlm.nih.gov/nuccore/GI:410024832
- 21.Muzny D., Qin X., Buhay C., Dugan-Rocha S., Ding Y., Chen G., Hawes A., Holder M., Jhangiani S., Johnson A., Khan Z., Li Z., Liu W., Liu X., Perez L., Shen H., Wang Q., Watt J., Xi L., Xin Y., Zhou J., Deng J., Jiang H., Liu Y., Qu J., Song X-Z., Zhang L., Villasana D., Johnson A., Liu J., Liyanage D., Lorensuhewa L., Robinson T., Song A., Song B-B., Dinh H., Thornton R., Coyle M., Francisco L., Jackson L., Javaid M., Korchina V., Kovar C., Mata R., Mathew T., Ngo R., Nguyen L., Nguyen N., Okwuonu G., Ongeri F., Pham C., Simmons D., Wilczek-Boney K., Hale W., Jakkamsetti A., Pham P., Ruth R., San Lucas F., Warren J., Zhang J., Zhao Z., Zhou C., Zhu D., Lee S., Bess C., Blankenburg K., Forbes L., Fu Q., Gubbala S., Hirani K., Jayaseelan J.C., Lara F., Munidasa M., Palculict T., Patil S., Pu L-L., Saada N., Tang L., Weissenberger G., Zhu Y., Hemphill L., Shang Y., Youmans B., Ayvaz T., Ross M., Santibanez J., Aqrawi P., Gross S., Joshi V., Fowler G. Human Genome Sequencing Center, Baylor College of Medicine, One Baylor Plaza, Houston, TX 77030, USA. Available from: . http://www.ncbi.nlm.nih.gov/nuccore/GI:384895178
- 22.Kim S., Lee W.K., Choi S.H., Kang S., Park H.S., Kim Y.S., Lee S.G., Byun E.Y., Jeon J.E., Park Y.H., Lee E.J., Kim J.S., Ryu B.D., Lee Y.S., Hahn Y., Yeom Y.I., Park S.G., Youn H.S., Ko G.H., Choi M.B., Park C.H., Lim J.Y., Bae D.W., Song J.Y., Park J.U., Kang H.L., Baik S.C., Cho M.J., Yoo H.S., Rhee K.H. Direct submission by KRIBB, 52, Oun-dong, Yusonggu, Daejeon, 305-333, Korea. Available from: http://www.ncbi. nlm.nih.gov/nuccore/GI:387781698
- 23.Kim S., Lee W.K., Choi S.H., Kang S., Park H.S., Kim Y.S., Lee S.G., Byun E.Y., Jeong J.E., Park Y.H., Lee E.J., Kim J.S., Ryu B.D., Lee Y.S., Hahn Y., Yeom Y.I., Park S.G., Youn H.S., Ko G.H., Choi M.B., Park C.H., Lim J.Y., Bae D.W., Song J.Y., Park J.U., Kang H.L., Baik S.C., Cho M.J., Yoo H.S., Rhee K.H. Direct submission by KRIBB, 111 Gwahangno, Yuseong-gu, Daejeon 305-806, Korea. . Available from: http://www.ncbi.nlm.nih.gov/nuccore/GI:384887043
- 24.Kersulyte D., Herrera P., Gilman R.H., Berg D.E. Direct Submission by Molecular Microbiology, Washington University Medical School, 4940 Parkview Place, Saint Louis, MO 63110, USA. Available from: http://www.ncbi.nlm.nih.gov/nuccore/GI:38489. 2008
- 25.Kersulyte D., Mukhopadhyay A., Choudhury A., Nair G.B., Berg D.E. Direct submission by Molecular Microbiology, Washington University Medical School, 4940 Parkview Place, Saint Louis, MO 63110, USA. Available from: http://www.ncbi.nlm. nih.gov/nuccore /GI:385219873
- 26.Kersulyte D., Jahuira A.H., Gilman R.H., Berg D.E. Direct submission by Molecular Microbiology, Washington University Medical School, 4940 Parkview Place, Saint Louis, MO 63110, USA. Available from: http://www.ncbi.nlm.nih.gov/nuccore/GI
- 27.Merrell D.S., Thompson L.J., Kim C.C., Mitchell H., Tompkins L.S., Lee A., Falkow S. Growth phase-dependent response of Helicobacter pylori to iron starvation. Infect. Immun. 2003;71:6510–6525. doi: 10.1128/IAI.71.11.6510-6525.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Farnbacher M., Jahns T., Willrodt D., Daniel R., Haas R., Goesmann A., Kurtz S., Rieder G. Sequencing, annotation, and comparative genome analysis of the gerbil-adapted Helicobacter pylori strain B8. BMC Genomics. 2010;11:335. doi: 10.1186/1471-2164-11-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Devi S.H., Taylor T.D., Avasthi T.S., Kondo S., Suzuki Y., Megraud F., Ahmed N. Genome of Helicobacter pylori Strain 908. J. Bacteriol. 2010;192:6488–6489. doi: 10.1128/JB.01110-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muzny D., Qin X., Deng J., Jiang H., Liu Y., Qu J., Song X-Z., Zhang L., Thornton R., Coyle M., Francisco L., Jackson L., Javaid M., Korchina V., Kovar C., Mata R., Mathew T., Ngo R., Nguyen L., Nguyen N., Okwuonu G., Ongeri F., Pham C., Simmons D., Wilczek-Boney K., Hale W., Jakkamsetti A., Pham P., Ruth R., San Lucas F., Warren J., Zhang J., Zhao Z., Zhou C., Zhu D., Lee S., Bess C., Blankenburg K., Forbes L., Fu Q., Gubbala S., Hirani K., Jayaseelan J.C., Lara F., Munidasa M., Palculict T., Patil S., Pu L-L., Saada N., Tang L., Weissenberger G., Zhu Y., Hemphill L., Shang Y., Youmans B., Ayvaz T., Ross M., Santibanez J., Aqrawi P., Gross S., Joshi V., Fowler G., Nazareth L., Reid J., Worley K., Petrosino J., Highlander S., Gibbs R., Gibbs R. Direct submission by Human Genome Sequencing Center, Baylor College of Medicine, One Baylor Plaza, Houston, TX 77030, USA. Available from: http://www.ncbi.nlm.nih.gov/nuccore/GI:385224642
- 31.Kersulyte D., Velapatino B., Gilman R.H., Berg D.E. Direct submission by Molecular Microbiology, Washington University Medical School, 4940 Parkview Place, Saint Louis, MO 63110, USA. . Available from: http://www.ncbi.nlm.nih.gov/nuccore/GI
- 32.Oh J.D., Kling-Backhed H., Giannakis M., Xu J., Fulton R.S., Fulton L.A., Cordum H.S., Wang C., Elliott G., Edwards J., Mardis E.R., Engstrand L.G., Gordon J.I. The complete genome sequence of a chronic atrophic gastritis Helicobacter pylori strain: evolution during disease progression. Proc. Natl. Acad. Sci. USA. 2006;103:9999–10004. doi: 10.1073/pnas.0603784103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kersulyte D., Kalia A., Gilman R.H., Mendez M., Herrera P., Cabrera L., Velapatino B., Balqui J., Paredes Puente de la Vega F., Rodriguez Ulloa C.A., Cok J., Hooper C.C., Dailide G., Tamma S., Berg D.E. Helicobacter pylori from Peruvian amerindians: traces of human migrations in strains from remote Amazon, and genome sequence of an Amerind strain. PLoS One. 2010;5:e15076. doi: 10.1371/journal.pone.0015076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baltrus D.A., Amieva M.R., Covacci A., Lowe T.M., Merrell D.S., Ottemann K.M., Stein M., Salama N.R., Guillemin K. The complete genome sequence of Helicobacter pylori strain G27. J. Bacteriol. 2009;191:447–448. doi: 10.1128/JB.01416-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fischer W., Windhager L., Rohrer S., Zeiller M., Karnholz A., Hoffmann R., Zimmer R., Haas R. Strain-specific genes of Helicobacter pylori: genome evolution driven by a novel type IV secretion system and genomic island transfer. Nucleic Acids Res. 2010;38:6089–6101. doi: 10.1093/nar/gkq378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thiberge J.M., Boursaux Eude C., Lehours P., Dillies M.A., Creno S., Coppée J.Y., Rouy Z., Lajus A., Ma L., Burucoa C., Ruskoné-Foumestraux A., Courillon-Mallet A., De Reuse H., Boneca I.G., Lamarque D., Mégraud F., Delchier J.C., Médigue C., Bouchier C., Labigne A., Raymond J. From array-based hybridization of Helicobacter pylori isolates to the complete genome sequence of an isolate associated with MALT lymphoma. BMC Genomics. 2010;11:368. doi: 10.1186/1471-2164-11-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dhillon B.K., Chiu T.A., Laird M.R., Langille M.G., Brinkman F.S. IslandViewer update: improved genomic island discovery and visualization. Nucleic Acids Res. 2013;41:W129-132. doi: 10.1093/nar/gkt394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J., Simonovic M., Roth A., Santos A., Tsafou K.P., Kuhn M., Bork P., Jensen L.J., von Mering C. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Durmuş T.S., Çakır T., Ardiç E., Sayılırbaş A.S., Konuk G., Konuk M., Sarıyer H., Uğurlu A., Karadeniz İ., Özgür A., Sevilgen F.E., Ülgen K.Ö. PHISTO: pathogen-host interaction search tool. Bioinformatics. 2013;29(10):1357–1358. doi: 10.1093/bioinformatics/btt137. [DOI] [PubMed] [Google Scholar]
- 41.Wattam A.R., Abraham D., Dalay O., Disz T.L., Driscoll T., Gabbard J.L., Gillespie J.J., Gough R., Hix D., Kenyon R., Machi D., Mao C., Nordberg E.K., Olson R., Overbeek R., Pusch G.D., Shukla M., Schulman J., Stevens R.L., Sullivan D.E., Vonstein V., Warren A., Will R., Wilson M.J., Yoo H.S., Zhang C., Zhang Y., Sobral B.W. PATRIC, the bacterial bioinformatics database and analysis resource. Nucleic Acids Res. 2014;42(D1):D581–D591. doi: 10.1093/nar/gkt1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kumar R. Nanduri. B. HPIDB - a unified resource for host-pathogen interactions. BMC Bioinformatics. 2010;11(16) doi: 10.1186/1471-2105-11-S6-S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Higashi H., Tsutsumi R., Muto S., Sugiyama T., Azuma T., Asaka M., Hatakeyama M. SHP-2 tyrosine phosphatase as an intracellular target of Helicobacter pylori Cag. Protein Sci. 2002;295(5555):683–686. doi: 10.1126/science.1067147. [DOI] [PubMed] [Google Scholar]
- 44.de Bernard M., Moschioni M., Napolitani G., Rappuoli R., Montecucco C. The VacA toxin of Helicobacter pylori identifies a new intermediate filament-interacting protein. EMBO J. 2000;9(1):48–56. doi: 10.1093/emboj/19.1.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nesić D., Miller M.C., Quinkert Z.T., Stein M., Chait B.T., Stebbins C.E. Helicobacter pylori CagA inhibits PAR1-MARK family kinases by mimicking host substrates. Nat. Struct. Mol. Biol. 2010;17(1):30–32. doi: 10.1038/nsmb.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lamb A., Yang X.D., Tsang Y.H., Li J.D., Higashi H., Hatakeyama M., Peek R.M., Blanke S.R., Chen L.F. Helicobacter pylori CagA activates NF-kappaB by targeting TAK1 for TRAF6-mediated Lys 63 ubiquitination. EMBO Rep. 2009;10(11):1242–1249. doi: 10.1038/embor.2009.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu Y., Zhong X., Zheng S., Du Q., Xu W. Transformed immortalized gastric epithelial cells by virulence factor CagA of Helicobacter pylori through Erk mitogen-activated protein kinase pathway. Oncogene. 2005;24(24):3886–3895. doi: 10.1038/sj.onc.1208551. [DOI] [PubMed] [Google Scholar]
- 48.Murata-Kamiya N., Kurashima Y., Teishikata Y., Yamahashi Y., Saito Y., Higashi H., Aburatani H., Akiyama T., Peek R.M., Jr, Azuma T., Hatakeyama M. Helicobacter pylori CagA interacts with E-cadherin and deregulates the beta-catenin signal that promotes intestinal transdifferentiation in gastric epithelial cells. Oncogene. 2007;26(32):4617–4626. doi: 10.1038/sj.onc.1210251. [DOI] [PubMed] [Google Scholar]
- 49.Higashi H., Nakaya A., Tsutsumi R., Yokoyama K., Fujii Y., Ishikawa S., Higuchi M., Takahashi A., Kurashima Y., Teishikata Y., Tanaka S., Azuma T., Hatakeyama M. Helicobacter pylori CagA induces Ras-independent morphogenetic response through SHP-2 recruitment and activation. J. Biol. Chem. 2004;279(17):17205–17216. doi: 10.1074/jbc.M309964200. [DOI] [PubMed] [Google Scholar]
- 50.Guiney D.G., Hasegawa P., Cole S.P. Helicobacter pylori preferentially induces interleukin 12 (IL-12) rather than IL-6 or IL-10 in human dendritic cells. Infect. Immun. 2003;71(7):4163–4166. doi: 10.1128/IAI.71.7.4163-4166.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hirata Y., Ohmae T., Shibata W., Maeda S., Ogura K., Yoshida H., Kawabe T., Omata M. MyD88 and TNF receptor-associated factor 6 are critical signal transducers in Helicobacter pylori-infected human epithelial cells. J. Immunol. 2006;176(6):3796–3803. doi: 10.4049/jimmunol.176.6.3796. [DOI] [PubMed] [Google Scholar]
- 52.Chen X.B., Melchionna R., Denis C.M., Gaillard P.H., Blasina A., Van de Weyer I., Boddy M.N., Russell P., Vialard J., McGowan C.H. Human Mus81-associated endonuclease cleaves Holliday junctions in vitro. Mol. Cell. 2001;8(5):1117–1127. doi: 10.1016/s1097-2765(01)00375-6. [DOI] [PubMed] [Google Scholar]
- 53.Lee H.S., Millward-Sadler S.J., Wright M.O., Nuki G., Salter D.M. Integrin and mechanosensitive ion channel-dependent tyrosine phosphorylation of focal adhesion proteins and beta-catenin in human articular chondrocytes after mechanical stimulation. J. Bone Miner. Res. 2000;15(8):1501–1509. doi: 10.1359/jbmr.2000.15.8.1501. [DOI] [PubMed] [Google Scholar]
- 54.Rezzonico R., Cayatte C., Bourget-Ponzio I., Romey G., Belhacene N., Loubat A., Rocchi S., Van Obberghen E., Girault J.A., Rossi B., Schmid-Antomarchi H. Focal adhesion kinase pp125FAK interacts with the large conductance calcium-activated hSlo potassium channel in human osteoblasts: potential role in mechanotransduction. J. Bone Miner. Res. 2003;18(10):1863–1871. doi: 10.1359/jbmr.2003.18.10.1863. [DOI] [PubMed] [Google Scholar]
- 55.King C.C., Obonyo M. Helicobacter pylori modulates host cell survival regulation through the serine-threonine kinase, 3-phosphoinositide dependent kinase 1 (PDK-1). BMC Microbiol. 2015;15:222. doi: 10.1186/s12866-015-0543-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mossessova E., Gulbis J.M., Goldberg J. Structure of the guanine nucleotide exchange factor Sec7 domain of human arno and analysis of the interaction with ARF GTPase. Cell. 1998;92(3):415–423. doi: 10.1016/s0092-8674(00)80933-2. [DOI] [PubMed] [Google Scholar]
- 57.Kelley D.R., Hendrickson D.G., Tenen D., Rinn J.L. Transposable elements modulate human RNA abundance and splicing via specific RNA-protein interactions. Genome Biol. 2014;15(12):537. doi: 10.1186/s13059-014-0537-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Neelapu N.R., Nammi D., Pasupuleti A.C., Surekha C. Helicobacter pylori induced gastric inflammation, ulcer, and cancer: A pathogenesis perspective. Interdiscip. J. Microinflammation. 2014;1:113. doi: 10.4172/ijm.1000113. [DOI] [Google Scholar]
- 59.Petersen A.M., Krogfelt K.A. Helicobacter pylori: an invading microorganism? A review. FEMS Immunol. Med. Microbiol. 2003;36:117–126. doi: 10.1016/S0928-8244(03)00020-8. [DOI] [PubMed] [Google Scholar]
- 60.Ilver D., Arnqvist A., Ogren J., Frick I.M., Kersulyte D., Incecik E.T., Berg D.E., Covacci A., Engstrand L., Borén T. Helicobacter pylori adhesin binding fucosylated histo-blood group antigens revealed by retagging. Science. 1998;279:373–377. doi: 10.1126/science.279.5349.373. [DOI] [PubMed] [Google Scholar]
- 61.Mahdavi J., Sondén B., Hurtig M., Olfat F.O., Forsberg L., Roche N., Angstrom J., Larsson T., Teneberg S., Karlsson K.A., Altraja S., Wadström T., Kersulyte D., Berg D.E., Dubois A., Petersson C., Magnusson K.E., Norberg T., Lindh F., Lundskog B.B., Arnqvist A., Hammarström L., Borén T. Helicobacter pylori SabA adhesin in persistent infection and chronic inflammation. Science. 2002;297:573–578. doi: 10.1126/science.1069076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moodley Y., Linz B., Yamaoka Y., Windsor H.M., Breurec S., Wu J.Y., Maady A., Bernhöft S., Thiberge J.M., Phuanukoonnon S., Jobb G., Siba P., Graham D.Y., Marshall B.J., Achtman M. The peopling of the Pacific from a bacterial perspective. Science. 2009;323:527–530. doi: 10.1126/science.1166083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yamaoka Y. Mechanisms of disease: Helicobacter pylori virulence factors. Nat. Rev. Gastroenterol. Hepatol. 2010;7:629–641. doi: 10.1038/nrgastro.2010.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hatakeyama M. SagA of CagA in Helicobacter pylori pathogenesis. Curr. Opin. Microbiol. 2008;11(1):30–37. doi: 10.1016/j.mib.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 65.O’Connor P.M., Lapointe T.K., Jackson S., Beck P.L., Jones N.L., Buret A.G. Helicobacter pylori activates calpain via toll-like receptor 2 to disrupt adherens junctions in human gastric epithelial cells. Infect. Immun. 2011;79(10):3887–3894. doi: 10.1128/IAI.05109-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Toback F.G., Walsh-Reitz M.M., Musch M.W., Chang E.B., Del Valle J., Ren H., Huang E., Martin T.E. Peptide fragments of AMP-18, a novel secreted gastric antrum mucosal protein, are mitogenic and motogenic. Am. J. Physiol. Gastrointest. Liver Physiol. 2003;285(2):G344–G353. doi: 10.1152/ajpgi.00455.2002. [DOI] [PubMed] [Google Scholar]
- 67.Walsh-Reitz M.M., Huang E.F., Musch M.W., Chang E.B., Martin T.E., Kartha S., Toback F.G. AMP-18 protects barrier function of colonic epithelial cells: Role of tight junction proteins. Am. J. Physiol. Gastrointest. Liver Physiol. 2005;289(1):G163–G171. doi: 10.1152/ajpgi.00013.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jones N.L., Shannon P.T., Cutz E., Yeger H., Sherman P.M. Increase in proliferation and apoptosis of gastric epithelial cells early in the natural history of Helicobacter pylori infection. Am. J. Pathol. 2005;151:1695–1703. [PMC free article] [PubMed] [Google Scholar]
- 69.Tsang Y.H., Lamb A., Romero-Gallo J., Huang B., Ito. K., Peek R.M., Ito Y., Chen L.F. Helicobacter pylori CagA targets gastric tumor suppressor RUNX3 for proteasome-mediated degradation. Oncogene. 2010;29:5643–5650. doi: 10.1038/onc.2010.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tsang Y.H., Lamb A., Chen L.F. New insights into the inactivation of gastric tumor suppressor RUNX3: The role of H. pylori infection. J. Cell. Biochem. 2011;112:381–386. doi: 10.1002/jcb.22964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Buti L., Spooner E., Van der Veen A.G., Rappuoli R., Covacci A., Ploegh H.L. Helicobacter pylori cytotoxin-associated gene A (CagA) subverts the apoptosis-stimulating protein of p53 (ASPP2) tumor suppressor pathway of the host. Proc. Natl. Acad. Sci. USA. 2011;108(22):9238–9243. doi: 10.1073/pnas.1106200108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lancellotti M., Brocchi M., da Silveira W.D. Bacteria-induced apoptosis: an approach to bacterial pathogenesis. Braz. J. Morphol. Sci. 2006;23(1):75–86. [Google Scholar]
- 73.Fan X., Gunasena H., Cheng Z., Espejo R., Crowe S.E., Ernst P.B., Reyes V.E. Helicobacter pylori urease binds to class II MHC on gastric epithelial cells and induces their apoptosis. J. Immunol. 2000;165(4):1918–1924. doi: 10.4049/jimmunol.165.4.1918. [DOI] [PubMed] [Google Scholar]
- 74.Caulfield A.J., Lathem W.W. Disruption of fas-fas ligand signaling, apoptosis, and innate immunity by bacterial pathogens. PLoS Pathog. 2014;10(8):e1004252. doi: 10.1371/journal.ppat.1004252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Blaser M.J., Atherton J.C. Helicobacter pylori persistence: Biology and disease. J. Clin. Invest. 2004;113(3):321–233. doi: 10.1172/JCI20925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Schubert M.L., Peura D.A. Control of gastric acid secretion in health and disease. Gastroenterology. 2008;134(7):1842–1860. doi: 10.1053/j.gastro.2008.05.021. [DOI] [PubMed] [Google Scholar]
- 77.Colotta F., Allavena P., Sica A., Garlanda C., Mantovani A. Cancer-related inflammation, the seventh hallmark of cancer: Links to genetic instability. Carcinogenesis. 2009;30(7):1073–1081. doi: 10.1093/carcin/bgp127. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material is available on the publisher’s web site along with the published article.