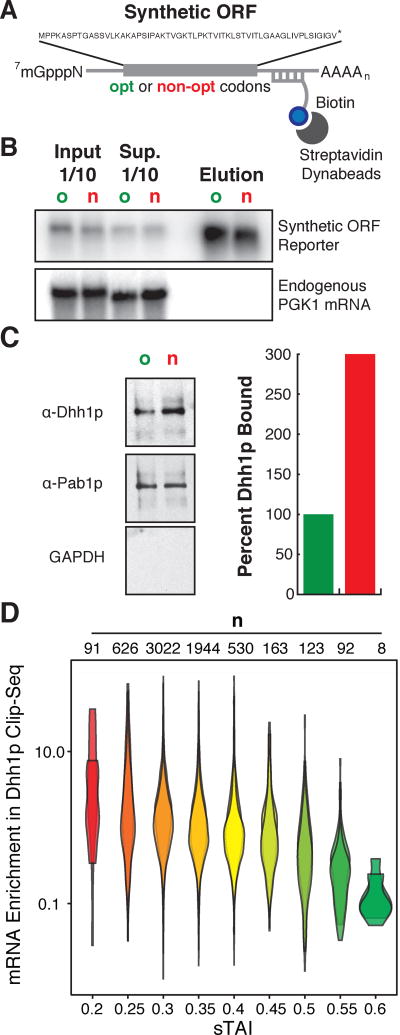
Figure 3. Dhh1p preferentially binds to mRNAs with low codon optimality.
(A) Representation of the reporters and experimental design used for mRNA pulldown. A tag sequence was inserted in the 3'UTR of the SYN reporters for pulldown. (B) Northern blot for the SYN mRNAs pull-downs. PGK1 mRNA was probed as a control of specificity. o: optimal, n: non-optimal. (C) Western blot showing the amount of Dhh1p, Pab1p and GAPDH pulled down by the SYN mRNAs. Quantitations of Dhh1p were normalized to mRNA levels from eluates in b. (D) Reanalysis of previously performed CLIP-Seq on Dhh1p calculating enrichment of mRNA transcripts bound to Dhh1p relative to WT conditions, where transcripts are binned by sTAI. Shown are two biological replicates. A two-tailed Mann-Whitney test shows that low optimality mRNAs (sTAI = 0.25, Med. = 2.02) are preferentially bound to Dhh1p relative to high optimality mRNAs (sTAI = 0.55, Med. = 0.32), U = 304, p = 7.1×10−9.