ABSTRACT
Peptides that are synthesized independently of the ribosome in plants, fungi, and bacteria can have clinically relevant anticancer, antihemochromatosis, and antiviral activities, among many other. Despite their natural origin, discovering new natural products is challenging, and there is a need to expand the chemical diversity that is accessible. In this work, we created a novel, compressed synthetic pathway for the heterologous expression and diversification of nonribosomal peptides (NRPs) based on homologs of siderophore pathways from Escherichia coli and Vibrio cholerae. To enhance the likelihood of successful molecule production, we established a selective pressure via the iron-chelating properties of siderophores. By supplementing cells containing our synthetic pathway with different precursors that are incorporated into the pathway independently of NRP enzymes, we generated over 20 predesigned, novel, and structurally diverse NRPs. This engineering approach, where phylogenetically related genes from different organisms are integrated and supplemented with novel precursors, should enable heterologous expression and molecular diversification of NRPs.
KEYWORDS: nonribosomal peptides, pathway engineering, genome engineering, heterologous gene expression, mutasynthesis, polyamines, siderophores
IMPORTANCE
Nonribosomal peptides (NRPs) constitute a source of bioactive molecules with potential therapeutic applications. However, discovering novel NRPs by rational engineering of biosynthetic pathways remains challenging. Here, we show that a synthetic compressed pathway in which we replaced biosynthetic genes with their ancestral homologs and orthologs enabled successful heterologous NRP expression. Polyamines added exogenously were incorporated into nascent NRPs, and molecular production was pressured by growing the host under conditions that make such NRPs beneficial for survival. This multilayered approach resulted in the assembly of over 20 distinct and novel molecules. We envision this strategy being used to enable the production of NRPs from heterologous pathways.
INTRODUCTION
Natural products harvested from plants, fungi, and bacteria have extended human life expectancy and improved quality of life by treating difficult diseases such as cancer and bacterial infections (1). Many of these natural products are assembled by very large enzymes. The biosynthetic enzymes are composed of units (i.e., modules), each of which can be further divided into catalytic domains (2–4). Depending on whether these enzymes catalyze reactions where an amino acid or an α-carboxyacyl coenzyme A (CoA) is activated and condensed into nascent molecules, they generate nonribosomal peptides (NRPs) or polyketides (PKs), respectively. Less commonly, mixed-gene operons or hybrid genes can generate hybrid molecules that have both NRP and PK characteristics. Despite our historic ability to discover natural products with useful therapeutic properties, this process has become much less productive over time. This is in part due to the repeated discovery of molecules that are easily accessible and have already been characterized (5).
Researchers have attempted a multitude of approaches to access new natural products, such as discovering novel producer strains in less-exploited niches, like the human microbiome (6); activating silent gene clusters; and engineering genes, modules, domains, and pathways in heterologous, genetically tractable hosts (7). The first report on the successful assembly of new natural products by combining heterologous and unrelated biosynthetic genes dates back to 1985 (8). In this work, Hopwood et al. cloned genes coding for an assortment of PK antibiotics into several Streptomyces strains and successfully produced new, hybrid molecules. Others have used screening for antibiotic resistance as a means to enrich bacterial libraries for producers of specific groups of antibiotics (9). In addition, by cloning plant genes for substrate synthesis and expressing polyketide synthases and posttranslational modification enzymes, Katsuyama et al. produced plant flavonoids and stilbenes in Escherichia coli when providing the cell with carboxylic acids (10). Nguyen et al. modified daptomycin’s biosynthetic pathway to prevent glycosylation and altered the amino acids to be incorporated into the nascent molecule, resulting in new lipopeptides with various performances (11). Nonetheless, engineering natural product pathways has often yielded poor results, in part due to poor translation of in silico predictions to actual functional pathways and molecules (12, 13).
To tackle the difficulty of generating structural diversity of NRPs, we decided to integrate several distinct approaches in this work. Our goal was to build a single biosynthetic pathway capable of producing a diversity of new and structurally distinct molecules. Specifically, we combined precursor-directed biosynthesis, combinatorial genetics, and heterologous expression of biosynthetic genes to assemble new, unnatural NRPs in a programmable fashion. Precursor-directed biosynthesis enabled us to control the molecules that are made by providing the organism with precursors to be incorporated into the nascent molecule. By using combinatorial genetics, we expanded the diversity of the molecules that could be made through the use of alternative pathways, and by expressing these pathways heterologously, we limited background interference and enabled better control over production.
To increase the likelihood of success, we focused on iron-chelating nonribosomal peptides called siderophores (14). These molecules are key for cell survival under low soluble-iron availability. By linking the production of new molecules to survival, we sought to drive the organism to produce new molecules or otherwise perish. Specifically, we deconstructed the serratiochelin biosynthetic pathway and reconstructed a simple and reduced version incorporating only its biosynthetic genes. We also built an alternative pathway utilizing homologous genes from E. coli and Vibrio cholerae responsible for the biosynthesis of enterobactin (15, 16) and vibriobactin (17), respectively. With this alternative pathway, we explored whether these closely related genes could produce the target molecules, which would yield insights into the evolution of pathways and the exchangeability of homologous enzymes. This synthetic pathway was capable of generating not only natural molecules, such as serratiochelin and enterobactin, but also nonnatural molecules by incorporating exogenously supplied precursors. In summary, we demonstrated the use of heterologous biosynthetic pathways, coupled with lethal selective pressure and distinct precursors, to create an assortment of new and nonnatural NRPs.
RESULTS
Serratiochelins are catechol siderophores produced by Serratia plymuthica V4 (18, 19). These siderophores utilize catechol moieties for iron coordination, obtaining them from the conversion of endogenous chorismate to dihydroxybenzoate (DHB) (19). The chorismate-to-DHB pathway appears to be extremely conserved among catechol siderophores (20). Additional enzymes can then use DHB to form a wide diversity of catechol-based molecules, such as enterobactin, fluvibactin, vibriobactin, photobactin, petrobactin, and vulnibactin (21).
We hypothesized that E. coli, which produces enterobactin (15, 16), could produce serratiochelins. This hypothesis was made based on several assumptions: (i) that the machinery responsible for the import and export of siderophores in this host would recognize serratiochelins and their catechol moieties; (ii) that E. coli could take up polyamines, such as diaminopropane (DAP) (Table 1), which are required for the production of serratiochelins and their nonnatural analogs; (iii) that the genes from the DHB pathway from S. plymuthica would be functional in E. coli, given that they are highly similar to the genes in the DHB pathway from E. coli (4, 19); and (iv) that expressing these pathways in a heterologous organism and under iron-limited conditions would allow us to supplement the medium with different precursors to generate new analogs.
TABLE 1 .
List of precursors, their reference numbers, final working concentrations, and incorporation in the new molecules
| Polyamine precursor or dipeptide |
Catalog no. (Sigma-Aldrich) or other supplier |
Concn in medium |
Incorporation in molecule |
||
|---|---|---|---|---|---|
| No. | Name | Partial | Full | ||
| Polyamine precursors |
|||||
| 1 | Diaminopropane | D23602 | 8 mM | Yes | Yes |
| 2 | Spermidine | S0266 | 8 mM | Yes | Yes |
| 3 | Spermine | S4264 | 1 mM | NDa | ND |
| 4 | Cadaverine | D22606 | 1 mM | Yes | ND |
| 5 | Putrescine | P5780 | 2.5 mM | Yes | Yes |
| 6 | Norspermidine | I1006 | 10 mM | Yes | Yes |
| 7 | m-Xylylenediamine | X1202 | 2.5 mM | Yes | ND |
| 8 |
N,N′-Bis(2-aminoethyl)-1,3- propanediamine |
333131 | 5 mM | ND | ND |
| 9 | N-Benzylethylenediamine | 462292 | 2.5 mM | Yes | Yes |
| 10 | 4-Aminobenzylamine | 368466 | 2.5 mM | Yes | ND |
| 11 | 4-(2-Aminoethyl)aniline | 123056 | 0.5 mM | Yes | ND |
| 12 | 4,4′-Oxydianiline | 248398 | 0.05 mM | Yes | ND |
| 13 | 4,4′-Diaminodiphenylmethane | 32950 | 0.01 mM | ND | ND |
| 14 | 1,5-Diaminonaphthalene | D21200 | 5 mM | ND | ND |
| 15 | 2,2′-Thiobisacetamide | S365033 | 0.02 mM | ND | ND |
| 16 | Sulfaguanidine | S8751 | 2.5 µM | ND | ND |
| 17 | p-Aminobenzenesulfonamide | S9251 | 0.05 µM | ND | ND |
| 18 | Urea | U5378 | 5 mM | ND | ND |
| 19 | N-Phenylthiourea | P7629 | 5 mM | ND | ND |
| 20 | 3,3′-Diamino-N- methyldipropylamine |
188441 | 5 mM | ND | ND |
| 21 | 1, 8-Diaminooctane | D22401 | 5 mM | Yes | ND |
| Dipeptides | |||||
| 22 | Dipeptide KR | Biomatik USA | 0.01 mM | ND | ND |
| 23 | Dipeptide KK | ND | ND | ||
| 24 | Dipeptide KQ | ND | ND | ||
| 25 | Dipeptide QN | ND | ND | ||
ND, not detected.
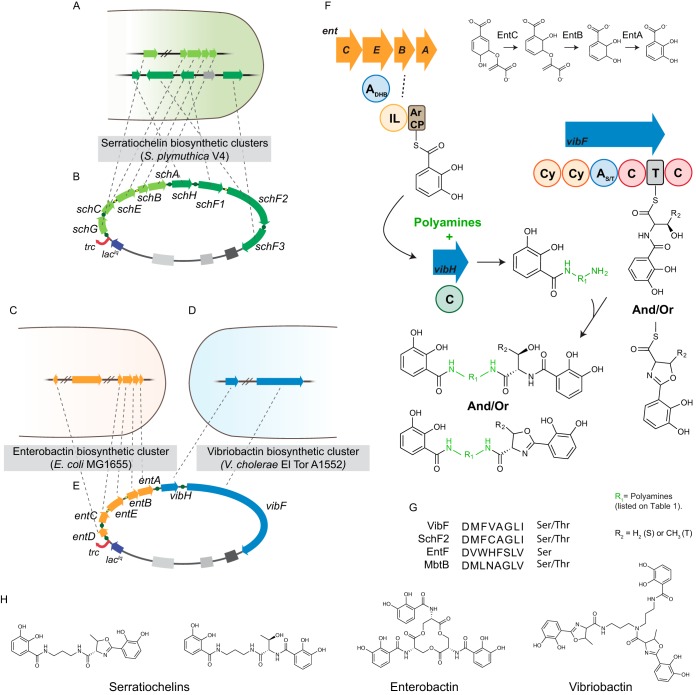
Initially, the S. plymuthica genes involved in the production of serratiochelins were cloned in a single operon into plasmid pDSW204 and driven by its isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible promoter, which is a weaker version of promoter trc99A. This synthetic operon is a compressed version of the two-cluster serratiochelin biosynthetic pathway (Fig. 1A and B). It contained genes schABCEG from S. plymuthica. These genes are homologous to the enterobactin biosynthesis genes entABCDE from E. coli (16, 22, 23), with a protein identity of at least 57%, except for EntD, which shares an identity of 24% with SchG. The synthetic operon also contained schF1F2F3, which are homologous to vibF (51% protein identity), and schH (a vibH homolog; 32% protein identity). Genes vibF and vibH are involved in the biosynthesis of vibriobactin, a siderophore from V. cholerae (24–26). In addition to the large pathway, which we called SP_S, a cos site was also cloned for plasmid stability, and the resulting construct was named pSP_S.
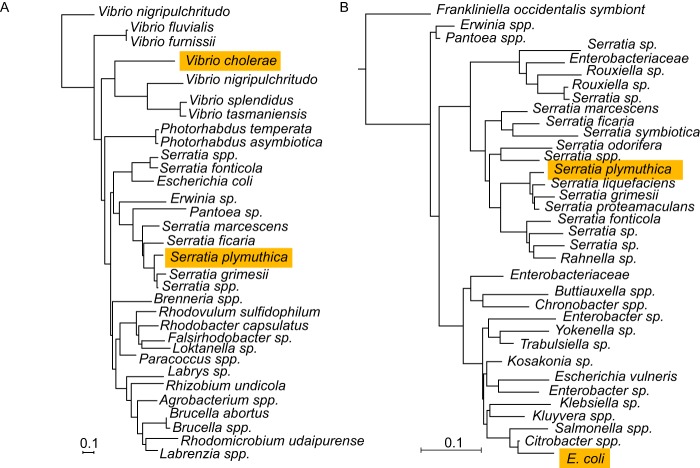
FIG 1 .
Phylogenetic trees displaying the relatedness of SchH and VibH (A) and SchE and EntE (B).
The constructs were transformed into an E. coli Ent− strain in which entABCDEF were deleted. E. coli Ent− carrying pSP_S or the empty vector was grown under iron-deprived conditions at 30°C with agitation, in the presence or absence of DAP. This diamine is naturally produced by S. plymuthica (19). Growth was not observed under either of these conditions. This lack of growth could result from the inability of the E. coli machinery to express S. plymuthica gene clusters or the enzymes being inactive in E. coli. The enzymes responsible for assembling NRPs function in an assembly-line fashion (3), so if a single enzyme is not present or is nonfunctional, the ultimate target molecule will not be made.
To overcome this issue, we hypothesized that homologs of the S. plymuthica genes could produce a functional assembly line capable of synthesizing serratiochelins and new analogs. As noted above, the schABCEG and schF1F2F3 operons are related to E. coli and V. cholerae genes that produce enterobactin and vibriobactin, respectively (Fig. 1). Also, it has been previously reported that holo-EntB, acylated with DHB by EntE, can serve as the substrate for the activity of VibH, similarly to VibB (26) (Fig. 2C and D). Thus, instead of cloning S. plymuthica V4 genes, we used their E. coli orthologs and V. cholerae ancestral homologs (19). Genes entABCDE and vibFH formed the pathway that we named EV_S and were assembled into the same empty pDSW204 backbone, along with a cos site, and introduced into E. coli Ent− (Fig. 2C, D, and E) as a construct called pEV_S.
FIG 2 .
Compressed synthetic pathways for heterologous expression of natural and unnatural nonribosomal peptides. Heterologous expression of serratiochelins in E. coli Ent− was initially attempted by cloning S. plymuthica biosynthetic genes (A) into a single operon, driven by an IPTG-inducible promoter (B). Upon failure to produce serratiochelins heterologously by expressing the S. plymuthica biosynthetic genes in E. coli, genes from E. coli MG1655 (C) and V. cholerae A1552 (D), which are homologous to those involved in the biosynthesis of serratiochelins, were cloned into a single operon (E). The biosynthetic processes for analogs are depicted in panel F. Chorismate is converted to DHB via a series of enzymatic reactions catalyzed by SchC/EntB, SchB/EntB, and SchA/EntA. DHB is then loaded onto the aryl carrier domain of EntB, for incorporation into the nascent molecule. l-Serine or l-threonine is activated by the adenylation domain of VibF and loaded onto the thiolation domain of the same enzyme. The amino acid can be further cyclized, as described elsewhere (19), thus increasing molecular diversity. VibH is responsible for condensing the intermediate molecules with the polyamines. Our results confirm in silico predictions for amino acid activation, where VibF, similarly to serratiochelin’s SchF2 and mycobactin’s MbtB, is predicted to activate l-threonine and l-serine as well, in contrast to enterobactin’s EntF, which activates only l-serine (G). The structures of serratiochelins, enterobactin, and vibriobactin are depicted in panel H.
pEV_S enabled the growth of E. coli Ent− under iron-limited conditions in the presence of DAP. Upon analysis of the Sep-Pak tC18-purified supernatant, we confirmed the production of the serratiochelin precursor (Fig. 3, M1, and see Fig. S1 at https://figshare.com/s/6238bd55b771b7853ff7) and the full-sized serratiochelin as well (Fig. 4, M1Tc, and see Fig. S2 at https://figshare.com/s/4abc2ac1669a6cb52a7b). However, we also observed growth of E. coli Ent− carrying pEV_S in the absence of diaminopropane, suggesting that another siderophore could be assembled by the biosynthetic pathway independently of the polyamine supplemented. We investigated this unexpected observation by analyzing the tC18-purified supernatant, wherein we detected the production of enterobactin (see Fig. S3 at https://figshare.com/s/4e163eaf8f89add99333), as well as linear enterobactin (see Fig. S4 at https://figshare.com/s/c31b4a46cf2a0e73ba76) and its dimers and monomers (see Fig. S5 and S6 at https://figshare.com/s/68fbe7b23716cbc45e2f and https://figshare.com/s/3363ded7d1e0ffc7ef16, respectively). These data indicate that VibF can replace EntF to assemble enterobactin, thus enabling cells to grow even in the absence of the precursor diaminopropane. In addition to analyzing samples with no added amines for enterobactin production, we decided to check whether samples to which amines had been added—thus enabling serratiochelin analogs to be made—also produced enterobactin. Enterobactin was detected in all but two of these samples (see Fig. S7 at https://figshare.com/s/994abf66ce48070d920d). Linear enterobactin and its dimers and monomers were also found in most samples (see Fig. S8d at https://figshare.com/s/0c70fd870d8d66c9fa0d, S9d at https://figshare.com/s/16e58bba259c540abc08, and S10d at https://figshare.com/s/fdf582d9e0addf61666f). The fact that our engineered strains continued to produce enterobactin along with serratiochelin and its analogs suggests that the former molecule is less resource intensive to produce than the latter molecules: enterobactin does not require polyamines (or VibH) for assembly. E. coli Ent−, the control strain that did not contain the heterologous biosynthetic pathway, could not grow under the same conditions tested here.
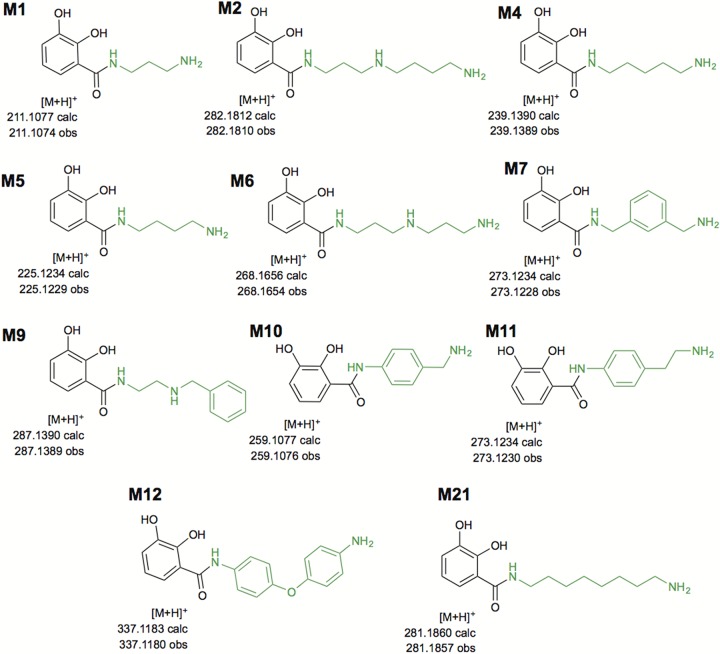
FIG 3 .
Proposed structures for the DHB-polyamine intermediates assembled by the compressed pathway. By adding various polyamines to the growth medium, VibH was found to be able to catalyze the reaction between foreign free polyamines (in green) and the tethered DHB. The [M + H]+ calculated and observed exact mass values for each molecule are also given. Each molecule is identified by the letter “M” and a number, corresponding to the polyamine added to the medium.
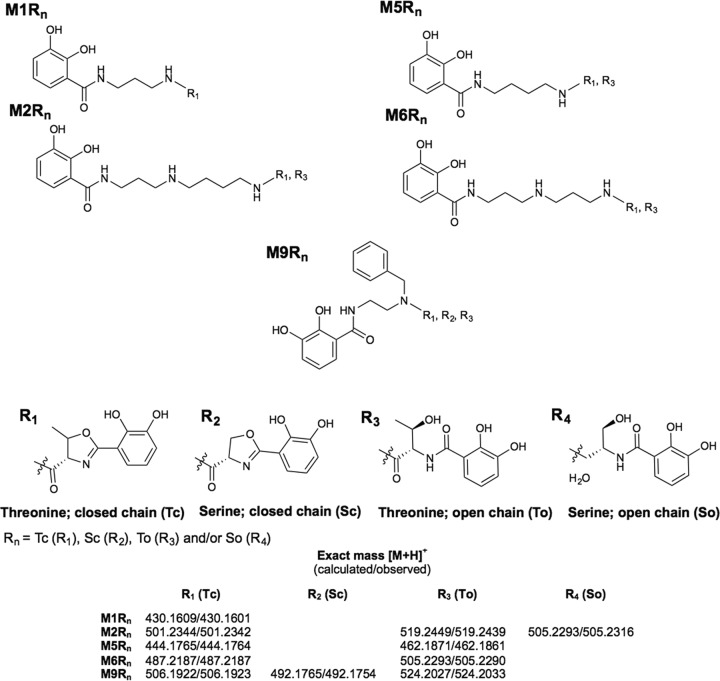
FIG 4 .
Proposed structures for the new serratiochelin analogs. The VibF acylation of the primary amine from the intermediates depicted in Fig. 3 can occur with 2-(2,3-dihydroxyphenyl)-5-methyloxazolinyl (R1 and R3) and/or a 2-(2,3-dihydroxyphenyl)-oxazolinyl (R2 and R4) as well. In some samples, the amino acid incorporated into the intermediate was found not to have gone through an additional cyclization, thus remaining in the open conformation as dihydroxybenzoyl-l-threonine and -serine (R3 and R4). Each molecule is identified by the letter “M” and a number, corresponding to the polyamine added to the medium, as well as the amino acid (S, serine; T, threonine) incorporated and its configuration (c, closed; o, open). Rn indicates the alternative radicals for the structures proposed and detected in the samples.
Given this observation, we then asked whether this pathway could produce Thr-enterobactin analogs, which would confirm predictions by other groups (27, 28). This question is relevant because based on in silico analysis, the adenylation domain signature of VibF is predicted to activate l-threonine and l-serine, whereas that of EntF is predicted to activate l-serine exclusively (Fig. 2G). The signature in VibF is very similar to that of MbtB (Fig. 2G). MbtB is a part of the pathway that synthesizes the siderophore mycobactin in Mycobacterium tuberculosis (29). In other species of the genus Mycobacterium, this siderophore has been found to contain l-serine or l-threonine via MbtB activation (29–31). Regarding VibF, in vivo incorporation of l-serine was initially thought to be unlikely by some (24) but was subsequently observed in vitro, albeit at a low level (27, 28). Keating et al. (27) made this observation when l-serine was added as a supplement individually to the in vitro reaction and different amino acids were not available to be activated by the adenylation domain of VibF. In our experiments, only the Thr-enterobactin dimer (see Fig. S11 at https://figshare.com/s/3a9f8994f468e552ce09) and monomer (see Fig. S12 at https://figshare.com/s/b73a0a71a913c93f49a7) were detected and only in a reduced number of samples (see Fig. S13 and S14 at https://figshare.com/s/5f2475fd6487f7ff196d and https://figshare.com/s/85e9b17edbb34b86f05f, respectively). We cannot exclude the possibility that Thr-enterobactin was produced but at levels too low to be detected in some of the samples.
To the best of our knowledge, this is the first report of in vivo VibF l-serine activation to generate enterobactin, as well as l-threonine activation to produce a new Thr-enterobactin dimer. This alternative pathway for enterobactin assembly enabled bacterial survival under low-iron conditions and in the absence of externally supplemented polyamines. For this reason, survival was dependent on the biosynthetic pathway but independent of the assembly of serratiochelin analogs and might have led to the production of a lower variety of molecules due to a relief in the strong, lethal selective pressure that results from the unpredicted production of the iron chelator enterobactin. We could potentially engineer a VibF that would not incorporate l-serine but only l-threonine, or other amino acids, to avoid the production of enterobactin. Nonetheless, this would be expected to reduce diversity, as no l-serine molecules would be assembled. To assess whether we can enact selective pressure for the production of polyamine-dependent siderophores, we grew the engineered E. coli Ent− carrying pEV_S in iron-depleted medium with bipyridyl (which chelates the little soluble iron available) and/or DAP (since the absence of DAP should make the cells rely on only enterobactin for iron uptake). We found that DAP enabled a higher growth rate in the presence of bipyridyl (µ = 0.357) compared to cells grown with bipyridyl alone (µ = 0.298, P = 0.02 [Fig. 5]). This suggests that the catechol molecules incorporating DAP are indeed siderophores. In fact, both serratiochelin (which incorporates DAP) and M5Tc/photobactin (which differs from serratiochelin only in its polyamine moiety, which in photobactin is putrescine) are known siderophores (19, 32).
FIG 5 .
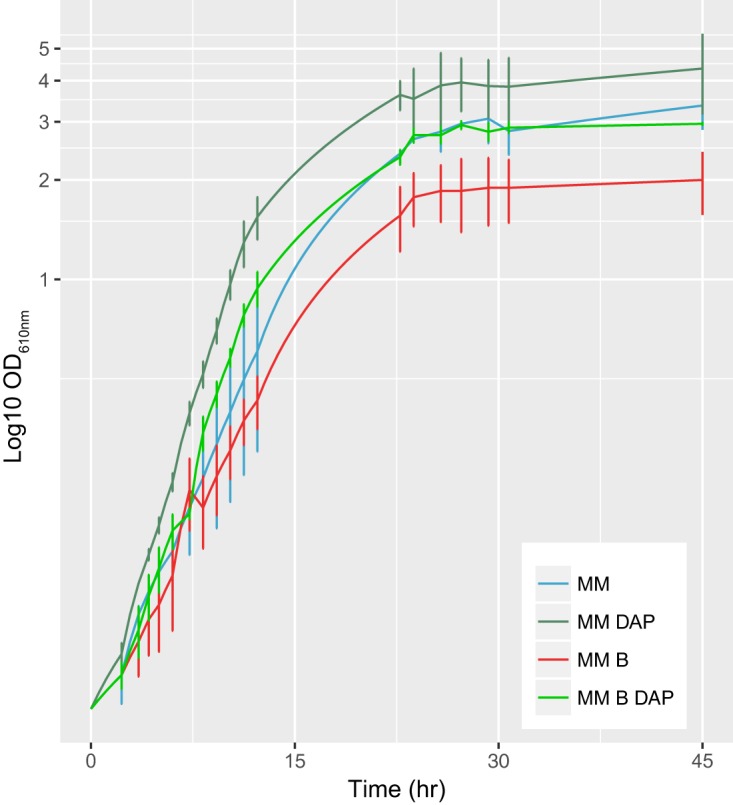
Growth over time of E. coli Ent− pEV_S in minimal medium, in the presence of 0.1% bipyridyl and/or 8 mM DAP, measured as optical density at 610 nm over the course of time.
Having designed and built a hybrid pathway that produces predicted molecules as a function of the precursor added (Fig. 2F), we wanted to determine the extent to which this approach could be adapted to create additional molecules. Polyamines with various numbers of carbons and amine groups, with and without other moieties, as well as four dipeptides, were independently added to the growth medium of E. coli Ent− carrying pEV_S (Table 1). The concentration of polyamines used was determined as the highest concentration that would not inhibit the growth of the producer strain in the absence of iron.
The biosynthetic and programmable pathway was capable of generating several predicted intermediate NRPs in which a polyamine was condensed with DHB (Fig. 3). In fact, VibH condensed linear polyamines and also condensed aromatic ones, such as aminobenzylamine (M10, Fig. 2) and oxydianiline (M21, Fig. 3), forming intermediate molecules that are the base for full-sized analogs. The annotated tandem mass spectrometry (MS/MS) spectra for the proposed structures in Fig. 3 can be found summarized in Table 2 and fully described in the supplementary information at figshare (see Fig. S13 to S34 at https://figshare.com/s/43bb422cac98e3dfeebd). In addition, we observed that the capacity of the pathway to generate full-sized serratiochelin analogs seemed to be restricted mostly to linear polyamines containing up to 4 amine groups and 9 carbons (Table 1, polyamine 2; Fig. 4, M2Rn). Nonetheless, VibF incorporated l-serine or l-threonine into these molecules and cyclized them or not, depending on the polyamine precursor supplemented. VibH was found to be very flexible in the substrate upon which it could act based on transferring activated DHB from EntB to a diversity of acceptor amines (Fig. 3; also Fig. S15 to S34 at https://figshare.com/s/43bb422cac98e3dfeebd).
TABLE 2 .
Exact mass and electrospray ionization-MS/MS fragmentation pattern for all molecules assembleda
| Molecule | Exact mass (calc/obs) | Fragmentation (calc/obs) |
|---|---|---|
| 1 | 211.1077/211.1073 | 137.0239/137.0231 |
| 194.0817/194.0809 | ||
| 1Tc | 430.1609/430.1601 | 137.0239/137.0230 |
| 194.0817/194.0811 | ||
| 277.1175/277.1188 | ||
| 294.1443/294.1457 | ||
| 2 | 282.1812/282.1810 | 72.0813/72.0813 |
| 129.1392/129.1388 | ||
| 194.0817/194.0810 | ||
| 265.1552/265.1542 | ||
| 2Tc | 501.2344/501.2342 | 194.0817/194.0812 |
| 265.1552/265.1540 | ||
| 308.1610/308.1595 | ||
| 365.2178/365.2173 | ||
| 2So | 505.2293/505.2316 | 137.0239/137.0237 |
| 209.0926/209.0924 | ||
| 224.0559/224.0555 | ||
| 2To | 519.2449/519.2439 | 137.0239/137.0232 |
| 194.0817/194.0809 | ||
| 210.0766/210.0758 | ||
| 265.1552/265.1542 | ||
| 282.1807/282.1809 | ||
| 383.2283/383.2294 | ||
| 4 | 239.1390/239.1389 | 86.0970/86.0970 |
| 103.1229/103.1235 | ||
| 137.0239/137.0232 | ||
| 222.1130/222.1132 | ||
| 5 | 225.1234/225.1229 | 72.0813/72.0815 |
| 89.1068/89.1080 | ||
| 137.0239/137.0234 | ||
| 208.0974/208.0968 | ||
| 5Tc | 444.1765/444.1764 | 137.0239/137.0229 |
| 192.0661/192.0661 | ||
| 208.0974/208.0964 | ||
| 225.1228/225.1237 | ||
| 308.1599/308.1617 | ||
| 5To | 462.1871/462.1861 | 137.0239/137.0230 |
| 208.0974/208.0964 | ||
| 225.1228/225.1237 | ||
| 238.0715/238.0717 | ||
| 210.0766/210.0755 | ||
| 6 | 268.1656/268.1654 | 137.0239/137.0232 |
| 194.0817/194.0810 | ||
| 251.1396/251.1381 | ||
| 443.1925/443.1907 | ||
| 6Tc | 487.2187/487.2187 | 137.0239/137.0232 |
| 194.0817/194.0810 | ||
| 277.1188/277.1188 | ||
| 351.2021/351.2016 | ||
| 443.1925/443.1907 | ||
| 6To | 505.2293/505.2290 | 137.0239/137.0232 |
| 194.0817/194.0810 | ||
| 210.0766/210.0758 | ||
| 369.2127/369.2130 | ||
| 7 | 273.1234/273.1228 | 120.0813/120.0811 |
| 137.0239/137.0232 | ||
| 256.0974/256.0964 | ||
| 9 | 287.1390/287.1389 | 91.0548/91.0545 |
| 180.0661/180.0651 | ||
| 9Tc | 506.1922/506.1923 | 91.0548/91.0546 |
| 180.0661/180.0652 | ||
| 287.1385/287.1381 | ||
| 9Sc | 492.1765/492.1754 | 91.0548/91.0548 |
| 287.1385/287.1385 | ||
| 9To | 524.2027/524.2033 | 91.0548/91.0548 |
| 345.1439/345.1460 | ||
| 389.1934/389.1956 | ||
| 10 | 259.1077/259.1076 | 106.0657/106.0656 |
| 137.0239/137.0237 | ||
| 154.0493/154.0497 | ||
| 11 | 273.1234/273.1230 | 120.0813/120.0813 |
| 137.0239/137.0234 | ||
| 256.0974/256.0963 | ||
| 12 | 337.1183/337.1180 | 108.0449/108.0446 |
| 137.0239/137.0232 | ||
| 201.1017/201.1024 | ||
| 21 | 281.1860/281.1857 | 128.1439/128.1435 |
| 137.0239/137.0231 | ||
| 145.1694/145.1699 | ||
| 264.1600/264.1595 | ||
| Ent | 670.1515/670.1509 | 137.0239/137.02134 |
| 206.0459/206.0452 | ||
| 224.0553/224.0554 | ||
| 447.1034/447.1029 | ||
| Ent trimer | 688.1621/688.1613 | 137.0239/137.0230 |
| 224.0559/224.0555 | ||
| 447.1040/447.1018 | ||
| Ent dimer | 465.1140/465.1133 | 137.02139/137.0231 |
| 196.0610/196.0609 | ||
| 224.0559/224.0557 | ||
| Ent monomer | 242.0659/242.0653 | 106.0493/106.0503 |
| 137.0239/137.0234 | ||
| Thr-Ent dimer | 493.1453/493.1448 | 137.0239/137.02131 |
calc/obs, calculated/observed; Ent, enterobactin.
We sought to determine whether the pathway could be used to assemble vibriobactin by adding norspermidine (Table 1, polyamine 6) to the medium. This molecule was not detected in the supernatant, but its intermediate with only the primary amines acylated was indeed detected (Fig. 4, M6Tc, and see Fig. S24 at https://figshare.com/s/f8395aa5e8853c1e70cb), as well as an additional analog, M6To (Fig. 4; see also Fig. S25 at https://figshare.com/s/fbfcb15ad60f1daf0ecd). This was not surprising, as others had already unsuccessfully tried to assemble vibriobactin in vitro (26).
One of the factors limiting the diversity of analogs generated appeared to be VibF, not VibH. Given that we detected several of the serratiochelin mutasynthon intermediates in the supernatant, VibF seems incapable of condensing its dihydroxyphenyl-5-methoxyxazoline (and l-serine-containing derivative) with the polyamine-containing intermediate. Several approaches could potentially enhance the performance of this synthetic pathway in terms of assembling full-sized molecules. For example, VibF could be subjected to directed evolution and other VibF/SchF1F2F3 homologs could be tested. It is also possible that the molecules were indeed assembled but could not be exported to the extracellular space that was assayed.
Interestingly, we observed what seemed to be a preferential orientation for condensing asymmetrical polyamines with the EntB-tethered DHB. For molecules M9 (Fig. 3; see also Fig. S25 at https://figshare.com/s/fbfcb15ad60f1daf0ecd) and M9Tc (Fig. 4; see also Fig. S26 at https://figshare.com/s/bb5ed59c88ba93b4aa59), a single fragmentation pattern, corresponding to that of a single orientation, was found. Nonetheless, we cannot exclude that the alternative conformation could exist at lower levels.
M5 corresponds to aminochelin (see Fig. S20 at https://figshare.com/s/49a77eddc280d6cc457f), a molecule that itself can act as a siderophore but can be incorporated into a large one, azotochelin, both produced by Azotobacter vinelandii (33, 34). We thus anticipate that the novel intermediate generated via this programmable pathway could possess metal-chelating abilities, similarly to its larger counterparts.
DISCUSSION
Microorganisms display an extraordinary ability to synthesize molecules that have been used to target cancer cells, parasites, iron overload, and bacterial infections (35, 36). They have evolved sets of very large enzymes that interact to assemble acyl-CoA- or peptide-based molecules, PKs or NRPs, respectively. The genes encoding these enzymes are modular, and each module is responsible for the incorporation of one unit (3, 37). Productively tapping these pathways has nonetheless posed a great challenge to researchers. In addition to difficulties in cultivating some producer organisms, it can be challenging to find conditions that lead to novel molecule production or to engineer native or heterologous pathways for inducible or constitutive expression (38). Often, attempts to alter these pathways for the production of new molecules or to express them heterologously result in a complete shutdown of production (12, 39, 40).
Here, we report an approach for the effective production and structural diversification of molecules produced by a nonribosomal peptide synthesis pathway. We successfully used ancestral or ortholog biosynthetic genes, rather than original pathways, for the heterologous and programmable production of serratiochelins and new analogs. Given that most NRP synthetases (NRPSs) responsible for the assembly of a given molecule have evolved from preexisting NRPSs (20, 41, 42), we anticipate that this approach can be applied to other biosynthetic pathways for which ancestral or ortholog genes exist. We deconstructed the enterobactin and vibriobactin biosynthetic pathways, which are organized in multiple operons, and then reconstructed them into a single and hybrid pathway comprised only of the biosynthetic genes. Specifically, we cloned entABCDE and vibFH (24, 25) in a single operon whose expression was driven by an IPTG-inducible promoter into an enterobactin-deficient E. coli strain lacking entABCDEF (E. coli Ent−).
An assortment of structurally diverse NRPs was produced by supplementing the iron-deprived growth medium with different small-molecule precursors as the substrates for VibH. These molecules were analogs of serratiochelin and its intermediate. Through this approach, new molecules were generated by adding precursors to the medium. If there is a specific moiety that one desires to include in the nascent molecule, the corresponding precursor can be supplied to the medium for incorporation, as long as it contains at least one amine group and the biosynthetic enzymes can use the precursor as a substrate for activity. Additional structural diversity was generated due to the capacity of VibF to activate not only l-threonine but l-serine as well for incorporation into the nascent molecule. To the best of our knowledge, this is the first report of in vivo VibF activation of l-serine. Nonetheless, not all precursors could serve as a substrate to VibF or VibH, and even when they could, there seemed to be a slight preference for l-threonine over l-serine activation (Fig. 4). Nearly half of the precursors tested were incorporated into the nascent molecule, and over half of these led to additional new structures, as detected and recognized by liquid chromatography (LC)-MS/MS. This methodology has been routinely used for this particular type of application (11, 43–47). The MS/MS pattern of each new molecule is known because the biosynthetic process is well characterized for serratiochelins (19).
We found that our synthetic pathway successfully assembled the cyclic and linear versions of enterobactin, as well as its monomer and its dimer. It also assembled a new version of linear enterobactin, dimer, and monomers, containing not l-serine but l-threonine. The latter observation is in line with the theory of the evolution of gene collectives (20), which are suggested to be sets of genes that coevolved quickly to lead to new molecules with minimal effort. The fact that multiple siderophores are assembled, at least in part, by biosynthetic pathways that are phylogenetically related supports this theory. Here, we show that combining genes from independent pathways can lead to new molecules as well as known molecules that are assembled by different enzymes. Moreover, we also show how ancestral genes and genes can allow for heterologous expression of NRPs when the original genes do not. In addition, it was interesting to observe how the programmable pathway SP_S containing the serratiochelin biosynthetic genes was not capable of synthesizing iron chelators in E. coli, despite it being related to pathway pEV_S. The fact that their ancestors could make both serratiochelin and enterobactin (though not vibriobactin, but its intermediates), and additional derivatives, shows how using ancestor genes instead of the original ones can lead to the successful heterologous expression of molecules.
Algorithm-based analyses of these molecules suggest that they may have relevant properties with clinical applications (see Tables S1 to S3 at https://figshare.com/s/43bb422cac98e3dfeebd). For example, all molecules were predicted to be potentially drug-like (see Table S2 at the URL mentioned above) and well absorbed (violations, <2; see Table S3 at the URL mentioned above). These algorithms suggest that some of the molecules generated could potentially target G-protein-coupled receptors (GPCRs) and ion channel modulators, among others (see Table S2 at the URL mentioned above). Future cellular and in vivo work will be needed to characterize the therapeutic potential of these molecules and whether these predicted activities are real. Siderophores may also have nonclinical applications, such as bioremediation, given their capacity to bind divalent cations (e.g., Cd2+, Cu2+, Ni2+, Pb2+, and Zn2+), trivalent cations (Mn3+, Co3+, and Al3+) and actinides (e.g., Th4+, U4+, and Pu4+) (48). Thus, the new analogs of siderophores could be tested in vitro and assayed for their metal binding affinities and specificities.
In addition to biological testing of the newly assembled molecules, future work should focus on screening amide synthases for their substrate tolerance in order to try to further increase the diversity of molecules. Furthermore, in order to block the assembly of enterobactin, one could mutate the adenylation domain of VibF, specifically the 10-amino-acid signature of this domain that is responsible for amino acid activation (49–51). Given that these 10 amino acids are dispersed throughout the domain, we would expect that most of the mutants generated would result in complete loss of function instead of a threonine-only activator.
In summary, we envision that using ancestor genes for molecular assembly along with precursor supplementation could be used for the diversification of chemical entities of biological interest in tractable, heterologous organisms. This approach could be applied to other nonribosomal pathways for which heterologous expression has posed a problem and improved with in silico molecule design as well as high-throughput strategies for pathway optimization and precursor application.
MATERIALS AND METHODS
Strains and general growth media.
All E. coli strains and S. plymuthica were maintained on lysogeny broth (Miller; Lab Express) supplemented with 1.5% agar and appropriate antibiotics as required. V. cholerae was maintained on agar plates prepared with marine broth 2216 (BD Diagnostics). Saccharomyces cerevisiae was maintained on complete supplement mixture medium (CSM; Sunrise Science Products) or CSM-tryptophan dropout medium for selection and maintenance of the yeast artificial chromosome (YAC; pYES-1L) carrying the assembled pathways.
Construction of an E. coli strain for heterologous expression of serratiochelins.
The genes responsible for serratiochelin production are distributed between two gene clusters in S. plymuthica. One of these clusters contains two genes homologous to those involved in the production of vibriobactin (in V. cholerae). The other contains 6 genes that are homologs to those involved in the production of enterobactin (19). These 6 homologs were removed from the chromosome of E. coli, given that our goal was to express serratiochelin and its analogs in this organism without their interference. The new E. coli strain was called E. coli Ent−. This was done so that the extrachromosomal synthetic pathway was responsible for molecular biosynthesis. Gene deletion was achieved using the Lambda Red recombination system (52); entD, entCEBA, and entF were replaced with chloramphenicol, kanamycin, and gentamicin resistance, respectively. The removal of the enterobactin biosynthetic genes disables this organism’s capacity to assemble this siderophore and grow under iron-limited conditions (16, 53, 54).
Construction of compressed synthetic pathways for the assembly of serratiochelin analogs.
The serratiochelin biosynthetic genes schCEBA (GenBank accession numbers AHY08568.1, Sch_19080, AHY08567.1, and AHY08566.1), schF1F2F3 (GenBank accession numbers AHY05890.1, AHY05889.1, and AHY05892.1), schG (GenBank accession number AHY08579.1), and schH (GenBank accession number AHY05888.1) were cloned from S. plymuthica V4 (identifier [ID] CP007439.1), which produces serratiochelins. The enterobactin genes entCEBA (IDs 945511, 947426, 946178, and 945284) and entD (ID 945194) are from E. coli MG1655 (ID NC_000913.3). vibF (ID 2614958) and vibH (ID 2615318) are from V. cholerae El Tor A1552 (ID N16961) (Table 3). Genes schCEBA, entCEBA, and schF1F2 were cloned as open reading frames and operons. The E. coli ribosome binding site (RBS) GAGGAGA was placed upstream of genes schC, entC, schF1, schH, vibH, schF3, vibF, schG, and entD.
TABLE 3 .
List of bacterial strains and their genotype and/or phenotype and source, as well as plasmids used and built, and their characteristics and source
| Strain/plasmid | Genotype/phenotype/description | Source or reference |
|---|---|---|
| Strains | ||
| E. coli Top10 | Large plasmid cloning strain, F−
mcrA Δ(mrr-hsdRMS-mcrBC) Φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara leu)7697 galU galK rpsL (Strr) endA1 nupG |
GeneArt Life Technologies, Inc. |
| E. coli DH5α | Cloning strain, F− Φ80lacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17 (rK− mK+) phoA supE44 λ− thi-1 gyrA96 relA1 |
Laboratory collection |
| E. coli K-12 MG1655 | Wild type and enterobactin producer, F− λ− ilvG rfb-50 rph-1 | Laboratory collection |
| E. coli Ent− | MG1655 ΔentD::Camr ΔentCEBA::Kanr ΔentF::Gentr | This study |
| Serratia plymuthica V4 | Serratiochelin producer (ZK4911) | 19, 71 |
| Vibrio cholerae O1 El Tor A1552 | Wild type, O1 El Tor Inaba, vibriobactin producer | Laboratory collection |
| Saccharomyces cerevisiae MaV203 |
MATα leu2-3,112 trp1-901 his3Δ200 ade2-101 gal4Δ gal80Δ SPAL10::URA3 GAL1::lacZ HIS3UAS GAL1::HIS3@LYS2 can1R cyh2R |
GeneArt Life Technologies, Inc. |
| Plasmids | ||
| pYES-1L | Yeast artificial chromosome, S. cerevisiae-E. coli shuttle vector, Trp− Specr |
GeneArt Life Technologies, Inc. |
| pDSW204 |
E. coli replicative expression vector with a medium-strength promoter, IPTG inducible |
72 |
| pWEB-TNC | E. coli cosmid, donor of the cos site | Epicentre |
| pEV_S | pDSW204 carrying pathway EV_S, with genes entABCDE and vibHF and a cos site |
This study (Addgene plasmid #100266) |
| pSP_S | pDSW204 carrying pathway SP_S, with genes schABCEF1F2F3GH and a cos site |
This study (Addgene plasmid #100270) |
In addition to building the synthetic sch-based compressed pathway, containing the S. plymuthica genes schABCEF1F2F3GH, we also assembled a pathway from the E. coli enterobactin and V. cholerae vibriobactin genes. Given the homology between these two clusters of genes and those of the serratiochelin biosynthetic pathway, it is of interest to determine whether, when together, these genes can assemble serratiochelin. The degree of similarity between homologous proteins was assessed utilizing the BLAST blastp suite from the National Center for Biotechnology Information (55–57) and has also been addressed elsewhere (19).
The genes or open reading frames were PCR amplified with overhangs homologous to the genes to be located up- and downstream, in the compressed pathway. SpeI sites were added at both ends of each pathway, to allow the pathways to be released by restriction enzyme digestion from the cloning vector, a yeast artificial chromosome. The amplicons were transformed, assembled into full pathways in S. cerevisiae using the GeneArt high-order genetic assembly kit (Life Technologies, Inc.), and checked for proper assembly by PCR. The compressed pathways (Fig. 2B and E) were released from pYES-1L by restriction digestion with SpeI. Each of the inserts was cloned into pDSW204, to which a cos site (for large construct stability) and a SpeI site had been added. After being checked for proper assembly, the constructs were moved to E. coli Ent− for production of serratiochelin and its analogs. The constructs are available on Addgene for distribution.
Selection of exogenously supplied precursors.
To test the substrate limits for VibH, we offered several amine-containing small molecules as DHB acceptors. We selected precursors with the goal of generating a wide diversity of molecules with a range of chemical properties. All polyamine precursors were purchased from Sigma-Aldrich; dipeptides were synthesized by Biomatik. All precursor polyamines were either primary or secondary amines. The product references, names, and concentrations used are listed in Table 1.
Both the compressed and the hybrid pathways, introduced into E. coli Ent−, were tested for production of serratiochelin and vibriobactin. To this end, diaminopropane (incorporated in Fig. 3, M1, and Fig. 4, M1Tc) and norspermidine (incorporated in Fig. 3, M6, and Fig. 4, M6Tc/To) were added to the medium, respectively. To test whether other analogs could be generated, we selected other polyamines to be added to the growth medium. We chose a variety of molecules with up to 12 carbons and 4 amine groups [cadaverine, incorporated in Fig. 3, M4; putrescine, incorporated in Fig. 3, M5, and Fig. 4, M5Tc/To; spermidine, incorporated in Fig. 3, M2, and Fig. 4, M2Tc; 1,8-diaminooctane (incorporated in Fig. 3, M21), 3,3′-diamino-N-methyldipropylamine, and N,N′-bis(2-aminoethyl)-1,3-propanediamine], because they are similar to diaminopropane and norspermidine. 2,2′-Thiobisacetamide has two amides, which could potentially contribute for metal chelation, in addition to being a substrate for condensation by VibH. Furthermore, we selected two molecules, sulfaguanidine and p-aminobenzenesulfonamide, with structural similarity to synthetic sulfonamide antibiotics. Siderophores are taken up by cells using specialized transporters (14), and antibiotics that have evolved to structurally resemble siderophores (58) can take advantage of this mechanism to kill bacterial cells. Urea and N-phenylthiourea were also tested.
Putrescine, spermidine, cadaverine, and aminopropylcadaverine are polyamines naturally occurring in E. coli (59–63); although their molecular functions are not fully understood (64), there is evidence for a role in mRNA translation (63). We supplied these compounds exogenously in hope that they would be incorporated into unnatural nonribosomal peptides (NRPs). We expected that endogenous levels would be too low to contribute to molecule assembly (65).
Additional precursors were chosen with the potential to enhance fluorescence (66, 67) of the products. m-Xylylenediamine, N-benzylethylenediamine, 4-aminobenzylamine (incorporated in Fig. 3, M10), 4-(2-aminoethyl)aniline (incorporated in Fig. 3, M11), 4,4′-oxydianiline (incorporated in Fig. 3, M12), 4,4′-diaminodiphenylmethane, and 1,5-diaminonaphthalene contain one or two benzene rings in addition to the required amine groups. Fluorescent molecules can be tracked as they move into and out of the cell and could be used, for example, as an Fe2+ sensor in the medium, since bacteria will secrete the iron chelator only under low-soluble-iron conditions (68).
The antimicrobial activity of short peptides, such as KR-12, has been established against some bacteria (69). Efficient antimicrobial activity is correlated with inclusion of positively charged amino acids. To explore the potential for synthesizing antimicrobial peptides using one of our pathways, four dipeptides—lysine-lysine (KK), lysine-arginine (KR), lysine-glutamine (KQ), and glutamine-asparagine (QN)—were selected for testing as precursors.
Production and purification of hybrid unnatural NRPs.
Minimal medium optimized for the production of serratiochelins (19) was used for molecule production by the synthetic pathways introduced into E. coli Ent−. It was composed of Na2HPO4 (5.96 g/liter), K2HPO4 (3.0 g/liter), NH4Cl (1.0 g/liter), NaCl (0.5 g/liter), MgSO4 (0.058 g/liter), C6H12O6 (5.0 g/liter), and IPTG (1 mM), pH 7.0. Precursors were added to final concentrations of 0.05 µM to 10 mM (Table 1). Siderophore production and related machinery were further induced by adding the iron chelator 2,2′-bipyridyl (Sigma-Aldrich catalog no. D216305) to a final concentration of 0.1 mM to the growth medium. In wild-type E. coli, bipyridyl activates the PentC promoter, which drives the expression of entCEBA (70). Cultures were grown to an optical density (600 nm) of ~2.8 (glucose depletion), for up to 7 days at 30°C with 250 rpm shaking.
After growth, cells were spun down and the supernatant was filter sterilized. Cell-free supernatant was loaded onto Sep-Pak tC18 (5 g) reversed-phase columns (Waters). The columns were washed with water, and the adherent molecules were eluted with 100% acetonitrile.
Liquid chromatography and tandem mass spectrometry (LC-MS/MS) sample analysis were performed at the Small Molecule Mass Spectrometry core facilities at Harvard University. Two-hundred-fifty-microliter aliquots of each sample were injected into a high-resolution, accurate mass Q Exactive Plus Orbitrap, with positive ionization and mass scan ranging from 66.7 to 1,000.0 m/z (resolution 70,000 FWHM [full width at half maximum]), and operated over the course of 30 min at a flow rate of 0.8 ml/min, with a gradient of 10% acetonitrile (ACN) in H2O to 100% ACN. Molecules displaying masses matching the expected ones were fragmented (35,000 FWHM), and the respective fragmentation patterns were compared against those of the predicted structures.
Structure predictions were performed based on previous knowledge of the NRP-based assembly of serratiochelins (19). We anticipated that, instead of condensing diaminopropane, VibH (much like SchH) would potentially incorporate the polyamines provided and VibF (like SchF1F2F3) would finalize the assembly of serratiochelin analogs.
Predicted structures and exact mass values, for the natural and unnatural molecules potentially assembled, were drawn and calculated using ChemDraw Professional 10 (PerkinElmer).
Bacterial growth rate determination.
We determined how the producer strain E. coli Ent− pEV_S grew in the presence of bipyridyl with and without DAP. For this, three independent experiments were performed, where 50 ml of minimal medium containing 0.1 mM bipyridyl only or 0.1 mM bipyridyl plus 8 mM DAP were inoculated with overnight cultures under the same conditions to an optical density (610 nm) of 0.05. Growth was followed until the cultures reached stationary phase. The growth rate for each growth curve was calculated using the R package growthrates.
SchE and SchH phylogenetic trees.
Despite having already been discussed in an earlier work (19), we queried the NCBI database for 500 homologs of SchE and SchH, in order to show the shared origin of these enzymes and EntE and VibH, respectively. The Newick trees (neighbor joining, 0.85 maximum sequence difference, Grishin distance) were generated and downloaded from NCBI.
Data availability.
The supplemental materials and methods, as well as supplemental data, can be accessed at figshare (https://figshare.com/s/43bb422cac98e3dfeebd).
ACKNOWLEDGMENTS
We thank Sunia Trauger for the helpful discussions and for analyzing the samples via high-performance liquid chromatography tandem mass spectrometry.
This project was funded by the National Institutes of Health (1P50GM098792), the Defense Threat Reduction Agency (HDTRA1-15-1-0050), and the Singapore-MIT Alliance for Research and Technology. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Citation Cleto S, Lu TK. 2017. An engineered synthetic pathway for discovering nonnatural nonribosomal peptides in Escherichia coli. mBio 8:e01474-17. https://doi.org/10.1128/mBio.01474-17.
REFERENCES
- 1.Bérdy J. 2005. Bioactive microbial metabolites. J Antibiot 58:1–26. doi: 10.1038/ja.2005.1. [DOI] [PubMed] [Google Scholar]
- 2.Walsh CT. 2004. Polyketide and nonribosomal peptide antibiotics: modularity and versatility. Science 303:1805–1810. doi: 10.1126/science.1094318. [DOI] [PubMed] [Google Scholar]
- 3.Fischbach MA, Walsh CT. 2006. Assembly-line enzymology for polyketide and nonribosomal peptide antibiotics: logic, machinery, and mechanisms. Chem Rev 106:3468–3496. doi: 10.1021/cr0503097. [DOI] [PubMed] [Google Scholar]
- 4.Walsh CT. 2016. Insights into the chemical logic and enzymatic machinery of NRPS assembly lines. Nat Prod Rep 33:127–135. doi: 10.1039/c5np00035a. [DOI] [PubMed] [Google Scholar]
- 5.Silver LL. 2011. Challenges of antibacterial discovery. Clin Microbiol Rev 24:71–109. doi: 10.1128/CMR.00030-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davenport M. 2014. Mining the microbiome for therapeutics. Chem Eng News 92:36. [Google Scholar]
- 7.Ando H, Citorik R, Cleto S, Lemire S, Mimee M, Lu T. 2015. Synthetic biology and therapies for infectious diseases. In Phoenix DA, Harris F, Dennison SR (ed), Novel antimicrobial agents and strategies. Wiley-VCH Verlag GmbH & Co. KGaA, Weinheim, Germany. [Google Scholar]
- 8.Hopwood DA, Malpartida F, Kieser HM, Ikeda H, Duncan J, Fujii I, Rudd BA, Floss HG, Omura S. 1985. Production of “hybrid” antibiotics by genetic engineering. Nature 314:642–644. doi: 10.1038/314642a0. [DOI] [PubMed] [Google Scholar]
- 9.Thaker MN, Wang W, Spanogiannopoulos P, Waglechner N, King AM, Medina R, Wright GD. 2013. Identifying producers of antibacterial compounds by screening for antibiotic resistance. Nat Biotechnol 31:922–927. doi: 10.1038/nbt.2685. [DOI] [PubMed] [Google Scholar]
- 10.Katsuyama Y, Funa N, Miyahisa I, Horinouchi S. 2007. Synthesis of unnatural flavonoids and stilbenes by exploiting the plant biosynthetic pathway in Escherichia coli. Chem Biol 14:613–621. doi: 10.1016/j.chembiol.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 11.Nguyen KT, He X, Alexander DC, Li C, Gu JQ, Mascio C, Van Praagh A, Mortin L, Chu M, Silverman JA, Brian P, Baltz RH. 2010. Genetically engineered lipopeptide antibiotics related to A54145 and daptomycin with improved properties. Antimicrob Agents Chemother 54:1404–1413. doi: 10.1128/AAC.01307-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weissman KJ. 2015. The structural biology of biosynthetic megaenzymes. Nat Chem Biol 11:660–670. doi: 10.1038/nchembio.1883. [DOI] [PubMed] [Google Scholar]
- 13.Zucko J, Cullum J, Hranueli D, Long PF. 2010. Evolutionary dynamics of modular polyketide synthases, with implications for protein design and engineering. J Antibiot 64:89–92. doi: 10.1038/ja.2010.141. [DOI] [PubMed] [Google Scholar]
- 14.Chu BC, Garcia-Herrero A, Johanson TH, Krewulak KD, Lau CK, Peacock RS, Slavinskaya Z, Vogel HJ. 2010. Siderophore uptake in bacteria and the battle for iron with the host; a bird’s eye view. Biometals 23:601–611. doi: 10.1007/s10534-010-9361-x. [DOI] [PubMed] [Google Scholar]
- 15.Brot N, Goodwin J, Fales H. 1966. In vivo and in vitro formation of 2,3-dihydroxybenzoylserine by Escherichia coli K12. Biochem Biophys Res Commun 25:454–461. doi: 10.1016/0006-291X(66)90227-0. [DOI] [PubMed] [Google Scholar]
- 16.Gehring AM, Mori I, Walsh CT. 1998. Reconstitution and characterization of the Escherichia coli enterobactin synthetase from EntB, EntE, and EntF. Biochemistry 37:2648–2659. doi: 10.1021/bi9726584. [DOI] [PubMed] [Google Scholar]
- 17.Griffiths GL, Sigel SP, Payne SM, Neilands JB. 1984. Vibriobactin, a siderophore from Vibrio cholerae. J Biol Chem 259:383–385. [PubMed] [Google Scholar]
- 18.Ehlert G, Taraz K, Budzikiewicz H. 1994. Serratiochelin, a new catecholate siderophore from Serratia marcescens. Z Naturforsch C 49:11–17. [Google Scholar]
- 19.Seyedsayamdost MR, Cleto S, Carr G, Vlamakis H, João Vieira M, Kolter R, Clardy J. 2012. Mixing and matching siderophore clusters: structure and biosynthesis of serratiochelins from Serratia sp. V4. J Am Chem Soc 134:13550–13553. doi: 10.1021/ja304941d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fischbach MA, Walsh CT, Clardy J. 2008. The evolution of gene collectives: how natural selection drives chemical innovation. Proc Natl Acad Sci U S A 105:4601–4608. doi: 10.1073/pnas.0709132105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Budzikiewicz H. 2010. Microbial siderophores. Fortschr Chem Org Naturst 92:1–75. doi: 10.1007/978-3-211-99661-4_1. [DOI] [PubMed] [Google Scholar]
- 22.Luke RK, Gibson F. 1971. Location of three genes concerned with the conversion of 2,3-dihydroxybenzoate into enterochelin in Escherichia coli K-12. J Bacteriol 107:557–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gehring AM, Bradley KA, Walsh CT. 1997. Enterobactin biosynthesis in Escherichia coli: isochorismate lyase (EntB) is a bifunctional enzyme that is phosphopantetheinylated by EntD and then acylated by EntE using ATP and 2,3-dihydroxybenzoate. Biochemistry 36:8495–8503. doi: 10.1021/bi970453p. [DOI] [PubMed] [Google Scholar]
- 24.Wyckoff EE, Stoebner JA, Reed KE, Payne SM. 1997. Cloning of a Vibrio cholerae vibriobactin gene cluster: identification of genes required for early steps in siderophore biosynthesis. J Bacteriol 179:7055–7062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wyckoff EE, Smith SL, Payne SM. 2001. VibD and VibH are required for late steps in vibriobactin biosynthesis in Vibrio cholerae. J Bacteriol 183:1830–1834. doi: 10.1128/JB.183.5.1830-1834.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keating TA, Marshall CG, Walsh CT. 2000. Vibriobactin biosynthesis in Vibrio cholerae: VibH is an amide synthase homologous to nonribosomal peptide synthetase condensation domains. Biochemistry 39:15513–15521. doi: 10.1021/bi001651a. [DOI] [PubMed] [Google Scholar]
- 27.Keating TA, Marshall CG, Walsh CT. 2000. Reconstitution and characterization of the Vibrio cholerae vibriobactin synthetase from VibB, VibE, VibF, and VibH. Biochemistry 39:15522–15530. doi: 10.1021/bi0016523. [DOI] [PubMed] [Google Scholar]
- 28.Marshall CG, Burkart MD, Keating TA, Walsh CT. 2001. Heterocycle formation in vibriobactin biosynthesis: alternative substrate utilization and identification of a condensed intermediate. Biochemistry 40:10655–10663. doi: 10.1021/bi010937s. [DOI] [PubMed] [Google Scholar]
- 29.De Voss JJ, Rutter K, Schroeder BG, Barry CE III. 1999. Iron acquisition and metabolism by mycobacteria. J Bacteriol 181:4443–4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gobin J, Wong DK, Gibson BW, Horwitz MA. 1999. Characterization of exochelins of the Mycobacterium bovis type strain and BCG substrains. Infect Immun 67:2035–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Snow GA. 1970. Mycobactins: iron-chelating growth factors from mycobacteria. Bacteriol Rev 34:99–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ciche TA, Blackburn M, Carney JR, Ensign JC. 2003. Photobactin: a catechol siderophore produced by Photorhabdus luminescens, an entomopathogen mutually associated with Heterorhabditis bacteriophora NC1 nematodes. Appl Environ Microbiol 69:4706–4713. doi: 10.1128/AEM.69.8.4706-4713.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Page WJ, Tigerstrom MV. 1988. Aminochelin, a catecholamine siderophore produced by Azotobacter vinelandii. Microbiology 134:453–460. doi: 10.1099/00221287-134-2-453. [DOI] [Google Scholar]
- 34.Cornish AS, Page WJ. 2000. Role of molybdate and other transition metals in the accumulation of protochelin by Azotobacter vinelandii. Appl Environ Microbiol 66:1580–1586. doi: 10.1128/AEM.66.4.1580-1586.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Felnagle EA, Jackson EE, Chan YA, Podevels AM, Berti AD, McMahon MD, Thomas MG. 2008. Nonribosomal peptide synthetases involved in the production of medically relevant natural products. Mol Pharm 5:191–211. doi: 10.1021/mp700137g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Demain AL. 1999. Pharmaceutically active secondary metabolites of microorganisms. Appl Microbiol Biotechnol 52:455–463. doi: 10.1007/s002530051546. [DOI] [PubMed] [Google Scholar]
- 37.Walsh C. 2003. Antibiotics: actions, origins, resistance. ASM Press, Washington, DC. [Google Scholar]
- 38.Gao X, Wang P, Tang Y. 2010. Engineered polyketide biosynthesis and biocatalysis in Escherichia coli. Appl Microbiol Biotechnol 88:1233–1242. doi: 10.1007/s00253-010-2860-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Khosla C, Herschlag D, Cane DE, Walsh CT. 2014. Assembly line polyketide synthases: mechanistic insights and unsolved problems. Biochemistry 53:2875–2883. doi: 10.1021/bi500290t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Winn M, Fyans JK, Zhuo Y, Micklefield J. 2016. Recent advances in engineering nonribosomal peptide assembly lines. Nat Prod Rep 33:317–347. doi: 10.1039/c5np00099h. [DOI] [PubMed] [Google Scholar]
- 41.Rantala A, Fewer DP, Hisbergues M, Rouhiainen L, Vaitomaa J, Bo T, Sivonen K. 2003. Phylogenetic evidence for the early evolution of microcystin synthesis. Proc Natl Acad Sci U S A 101:568–573. doi: 10.1073/pnas.0304489101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baltz RH. 2010. Genomics and the ancient origins of the daptomycin biosynthetic gene cluster. J Antibiot 63:506–511. doi: 10.1038/ja.2010.82. [DOI] [PubMed] [Google Scholar]
- 43.Dudnik A, Bigler L, Dudler R. 2013. Heterologous expression of a Photorhabdus luminescens syrbactin-like gene cluster results in production of the potent proteasome inhibitor glidobactin A. Microbiol Res 168:73–76. doi: 10.1016/j.micres.2012.09.006. [DOI] [PubMed] [Google Scholar]
- 44.Traxler MF, Watrous JD, Alexandrov T, Dorrestein PC, Kolter R. 2013. Interspecies interactions stimulate diversification of the Streptomyces coelicolor secreted metabolome. mBio 4:e00459-13. doi: 10.1128/mBio.00459-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nguyen DD, Wu CH, Moree WJ, Lamsa A, Medema MH, Zhao X, Gavilan RG, Aparicio M, Atencio L, Jackson C, Ballesteros J, Sanchez J, Watrous JD, Phelan VV, van de Wiel C, Kersten RD, Mehnaz S, De Mot R, Shank EA, Charusanti P, Nagarajan H, Duggan BM, Moore BS, Bandeira N, Palsson BØ, Pogliano K, Gutiérrez M, Dorrestein PC. 2013. MS/MS networking guided analysis of molecule and gene cluster families. Proc Natl Acad Sci U S A 110:E2611–E2620. doi: 10.1073/pnas.1303471110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sullivan JT, Jeffery EF, Shannon JD, Ramakrishnan G. 2006. Characterization of the siderophore of Francisella tularensis and role of fslA in siderophore production. J Bacteriol 188:3785–3795. doi: 10.1128/JB.00027-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang YL, Xu Y, Straight P, Dorrestein PC. 2009. Translating metabolic exchange with imaging mass spectrometry. Nat Chem Biol 5:885–887. doi: 10.1038/nchembio.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ahmed E, Holmström SJM. 2014. Siderophores in environmental research: roles and applications. Microb Biotechnol 7:196–208. doi: 10.1111/1751-7915.12117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marshall CG, Hillson NJ, Walsh CT. 2002. Catalytic mapping of the vibriobactin biosynthetic enzyme VibF. Biochemistry 41:244–250. doi: 10.1021/bi011852u. [DOI] [PubMed] [Google Scholar]
- 50.Roche ED, Walsh CT. 2003. Dissection of the EntF condensation domain boundary and active site residues in nonribosomal peptide synthesis. Biochemistry 42:1334–1344. doi: 10.1021/bi026867m. [DOI] [PubMed] [Google Scholar]
- 51.Rausch C, Weber T, Kohlbacher O, Wohlleben W, Huson DH. 2005. Specificity prediction of adenylation domains in nonribosomal peptide synthetases (NRPS) using transductive support vector machines (TSVMs). Nucleic Acids Res 33:5799–5808. doi: 10.1093/nar/gki885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pettis GS, McIntosh MA. 1987. Molecular characterization of the Escherichia coli enterobactin cistron entF and coupled expression of entF and the fes gene. J Bacteriol 169:4154–4162. doi: 10.1128/jb.169.9.4154-4162.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhou Z, Lai JR, Walsh CT. 2007. Directed evolution of aryl carrier proteins in the enterobactin synthetase. Proc Natl Acad Sci U S A 104:11621–11626. doi: 10.1073/pnas.0705122104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 56.Altschul SF, Wootton JC, Gertz EM, Agarwala R, Morgulis A, Schäffer AA, Yu YK. 2005. Protein database searches using compositionally adjusted substitution matrices. FEBS J 272:5101–5109. doi: 10.1111/j.1742-4658.2005.04945.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mercado G, Tello M, Marín M, Monasterio O, Lagos R. 2008. The production in vivo of microcin E492 with antibacterial activity depends on salmochelin and EntF. J Bacteriol 190:5464–5471. doi: 10.1128/JB.00351-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Charlier D, Glansdorff N. 9 September 2004, posting date Biosynthesis of arginine and polyamines. EcoSal Plus 2013 doi: 10.1128/ecosalplus.3.6.1.10. [DOI] [PubMed] [Google Scholar]
- 60.Abraham KA. 1968. Studies on DNA-dependent RNA polymerase from Escherichia coli. Eur J Biochem 5:143–146. doi: 10.1111/j.1432-1033.1968.tb00348.x. [DOI] [PubMed] [Google Scholar]
- 61.Frydman L, Rossomando PC, Frydman V, Fernandez CO, Frydman B, Samejima K. 1992. Interactions between natural polyamines and tRNA: an 15N NMR analysis. Proc Natl Acad Sci U S A 89:9186–9190. doi: 10.1073/pnas.89.19.9186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Huang SC, Panagiotidis CA, Canellakis ES. 1990. Transcriptional effects of polyamines on ribosomal proteins and on polyamine-synthesizing enzymes in Escherichia coli. Proc Natl Acad Sci U S A 87:3464–3468. doi: 10.1073/pnas.87.9.3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sakamoto A, Terui Y, Yoshida T, Yamamoto T, Suzuki H, Yamamoto K, Ishihama A, Igarashi K, Kashiwagi K. 2015. Three members of polyamine modulon under oxidative stress conditions: two transcription factors (SoxR and EmrR) and a glutathione synthetic enzyme (GshA). PLoS One 10:e0124883. doi: 10.1371/journal.pone.0124883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tabor CW, Tabor H. 1985. Polyamines in microorganisms. Microbiol Rev 49:81–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Campilongo R, Di Martino ML, Marcocci L, Pietrangeli P, Leuzzi A, Grossi M, Casalino M, Nicoletti M, Micheli G, Colonna B, Prosseda G. 2014. Molecular and functional profiling of the polyamine content in enteroinvasive E. coli: looking into the gap between commensal E. coli and harmful Shigella. PLoS One 9:e106589. doi: 10.1371/journal.pone.0106589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Schwarz FP, Wasik SP. 1976. Fluorescence measurements of benzene, naphthalene, anthracene, pyrene, fluoranthene, and benzo(e)pyrene in water. Anal Chem 48:524–528. doi: 10.1021/ac60367a046. [DOI] [PubMed] [Google Scholar]
- 67.Stark J. 1907. Ultra-violet fluorescence of benzene. Nature 75:295. doi: 10.1038/075295b0. [DOI] [Google Scholar]
- 68.Chung Chun Lam CKS, Jickells TD, Richardson DJ, Russell DA. 2006. Fluorescence-based siderophore biosensor for the determination of bioavailable iron in oceanic waters. Anal Chem 78:5040–5045. doi: 10.1021/ac060223t. [DOI] [PubMed] [Google Scholar]
- 69.Wang G. 2008. Structures of human host defense cathelicidin LL-37 and its smallest antimicrobial peptide KR-12 in lipid micelles. J Biol Chem 283:32637–32643. doi: 10.1074/jbc.M805533200. [DOI] [PubMed] [Google Scholar]
- 70.Lim JM, Hong MJ, Kim S, Oh DB, Kang HA, Kwon O. 2008. Iron chelator-inducible expression system for Escherichia coli. J Microbiol Biotechnol 18:1357–1363. [PubMed] [Google Scholar]
- 71.Cleto S, Van Der Auwera G, Almeida C, Vieira MJ, Vlamakis H, Kolter R. 2014. Genome sequence of Serratia plymuthica V4. Genome Announc 2:e00340-14. doi: 10.1128/genomeA.00340-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Weiss DS, Chen JC, Ghigo JM, Boyd D, Beckwith J. 1999. Localization of FtsI (PBP3) to the septal ring requires its membrane anchor, the Z ring, FtsA, FtsQ, and FtsL. J Bacteriol 181:508–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The supplemental materials and methods, as well as supplemental data, can be accessed at figshare (https://figshare.com/s/43bb422cac98e3dfeebd).