Abstract
The Zika outbreak, spread by the Aedes aegypti mosquito, highlights the need to create high-quality assemblies of large genomes in a rapid and cost-effective way. Here we combine Hi-C data with existing draft assemblies to generate chromosome-length scaffolds. We validate this method by assembling a human genome, de novo, from short reads alone (67× coverage). We then combine our method with draft sequences to create genome assemblies of the mosquito disease vectors Ae. aegypti and Culex quinquefasciatus, each consisting of three scaffolds corresponding to the three chromosomes in each species. These assemblies indicate that almost all genomic rearrangements among these species occur within, rather than between, chromosome arms. The genome assembly procedure we describe is fast, inexpensive, and accurate, and can be applied to many species.
Generating a high-quality genome sequence is a critical foundation for the analysis of any organism. Yet it remains a challenge, especially for genomes containing substantial repetitive sequences, such as Aedes aegypti, the principal vector of the Zika virus. Recently, an international consortium was organized to better understand Zika’s principal vector by improving the quality of the Ae. aegypti genome (1).
Currently, most genomes are assembled from a deep collection of short DNA sequence reads. These data are combined with linking information, which makes it possible to estimate the distance between individual sequences; such linking information is typically obtained by sequencing paired ends from a DNA clone library with a characteristic insert size. On the basis of sequence overlap, the reads are assembled into contiguous sequences (contigs); by means of the linking information, the contigs are ordered and oriented with respect to one another into larger scaffolds (2). Within scaffolds, adjacent contigs are often separated by a gap, which corresponds to a region that is hard to assemble from the available sequence reads (for example, because of repetitive sequences or low coverage) but that can nevertheless be spanned by using the linking information to determine the contigs at either end of the gap. Long links, from large-insert clones such as Fosmids, have been especially valuable (3). Such clone libraries provide physical coverage (defined as the average number of clones spanning a point in the genome), often in the range of 1000-fold. With this strategy, it has been possible to produce mammalian genome assemblies with scaffolds ranging from 1 to 15 Mb (2, 3). However, it has generally not been feasible to achieve scaffolds that span entire chromosomes, because some repetitive regions are too large and difficult to be spanned by the available clone libraries.
Hi-C is a sequencing-based approach for determining how a genome is folded by measuring the frequency of contact between pairs of loci (4, 5). Contact frequency depends strongly on the one-dimensional (1D) distance, in base pairs, between a pair of loci. For instance, loci separated by 10 kb in the human genome form contacts eight times more often than those at a distance of 100 kb. In absolute terms, a typical distribution of Hi-C contacts from a given locus is 15% to loci within 10 kb; 15% to loci 10 kb to 100 kb away; 18% to loci 100 kb to 1 Mb away; 13% to loci 1 to 10 Mb away; 16% to loci 10 to 100 Mb away; 2% to loci on the same chromosome but more than 100 Mb away; and 21% to loci on a different chromosome.
Hi-C data can provide links across a variety of length scales, spanning even whole chromosomes. However, unlike paired-end reads from clone libraries, any given Hi-C contact spans an unknown length and may connect loci on different chromosomes. This challenge may be mitigated, in part, by the physical coverage achieved by Hi-C data sets. For the maps reported in (4, 5), summing the span of each individual contact reveals that 1× of sequence coverage of the target genome translates, on average, into 23,000× of physical coverage. This suggests that a statistical approach analyzing the pattern of Hi-C contacts as a whole could generate extremely long scaffolds.
Computational experiments with simulated input data have suggested that Hi-C should be able to produce chromosome-length scaffolds (6–8). Indeed, Hi-C has been used to improve draft genome assemblies (7, 9) and to create chromosome-length scaffolds for large genomes (10). In this process, Hi-C data are used to assign draft scaffolds to chromosomes and then to order and orient the draft scaffolds within each chromosome. Unfortunately, the resulting predictions contain large errors, including chromosome-scale inversions and misjoins that fuse chromosomes (10). Such mis-assemblies may be caused by errors in the original draft assembly (10). One approach to avoiding such errors might be additional types of information, such as longer reads or optical mapping data [see, e.g., (11, 12)].
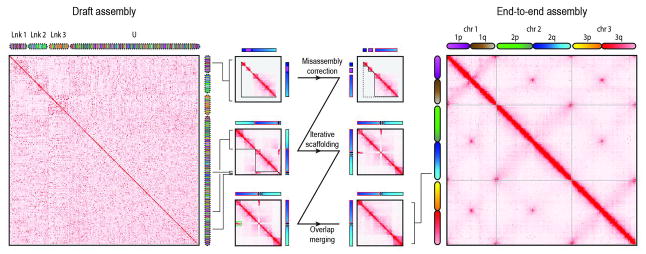
We therefore sought to develop a robust procedure for using Hi-C linking information to generate accurate genome assemblies with chromosome-length scaffolds. A key aspect of the approach is to first use Hi-C data to identify and correct errors in the scaffolds of the initial assembly. Briefly, we correct misjoins by identifying positions where a scaffold’s long-range contact pattern changes abruptly, which is unlikely for a correctly assembled scaffold. Next, we use a novel algorithm to anchor, order, and orient the resulting sequences, employing the contact frequency between a pair of sequences as an indicator of their proximity in the 1D genome. Finally, we merge contigs and scaffolds that correspond to overlapping regions of the genome by identifying pairs of scaffolds that exhibit both strong sequence homology as well as strong similarity in long-range contact pattern (Fig. 1 and fig. S1).
Fig. 1. Starting with a draft assembly, we used Hi-C data to correct mis-joins, scaffold, and merge overlaps, thereby generating an assembly of the Ae. aegypti mosquito genome with chromosome-length scaffolds.
Here we show contact matrices generated by aligning a Hi-C data set to both the AaegL2 assembly (18) that we used as input (left) and the final AaegL4 assembly generated by our algorithm (right). Pixel intensity in the contact matrix indicates how often a pair of loci colocate in the nucleus. The loci corresponding to each row and column are illustrated using chromograms. The chromograms on the left depict the three linkage groups [Lnk1, Lnk2, Lnk3, or unassigned (U)] reported in AaegL2; the chromograms on the right depict the three chromosome-length scaffolds in AaegL4 (chr1, chr2, and chr3). To create the chromogram, we assigned each AaegL4 arm a linear color gradient, thereby specifying a color for each AaegL4 locus. The same colors are then used for the corresponding loci in AaegL2 (left) and in the illustration of our procedure (center, though with increased contrast). Chromogram discontinuities indicate differences with AaegL4. In the center, we illustrate our assembly algorithm using an input scaffold from Lnk1 of AaegL2 (“supercontig 1.12,” see bracket). First, the scaffold is examined for misjoins and split such that the resulting segments each exhibit a continuous Hi-C signal (center, top row). Next, the segments are used as input for iterative scaffolding. Ultimately, only one of the segments is assigned to chromosome 1 of AaegL4. The rest of supercontig 1.12 is assigned to 2q, in the vicinity of several scaffolds that were not anchored in AaegL2 (center, middle row). Finally, segments exhibiting a similar 3D signal are examined for evidence of overlapping sequence (green rectangle) and merged (center, bottom row). The final contact map is consistent with the Rabl configuration, i.e., the spatial clustering of centromeres and telomeres.
We validated our approach by creating a de novo assembly of a human genome (the GM12878 cell line), comprising 23 chromosome-length scaffolds, using only short Illumina reads (67× coverage). We created a draft assembly from 250–base pair (bp) paired-end reads [60× coverage, generated by Illumina sequencing with a polymerase chain reaction (PCR)–free protocol, downloaded from Sequence Read Archive (SRX297987); assembled with DISCOVAR de novo (13)]. This assembly, termed Hs1, comprises 2.82 Gb of sequence (contig N50 length: 103 kb) partitioned among 73,770 scaffolds (scaffold N50: 126 kb, Table 1).
Table 1.
3D de novo assembly statistics for the Hs2-HiC, AaegL4, and CpipJ3 assemblies.
We did not attempt to further assemble tiny scaffolds contained in each draft assembly. The other scaffolds in each draft were assembled by using Hi-C to create huge, chromosome-length scaffolds and additional small scaffolds.
| Hs2-HiC | AaegL4 | CpipJ3 | |
|---|---|---|---|
| Draft scaffolds | |||
| Base pairs | 2,819,306,710 | 1,310,076,332 | 539,974,961 |
| Number of contigs | 80,223 | 36,204 | 48,672 |
| Contig N50 | 102,922 | 82,618 | 28,546 |
| Number of scaffolds | 73,770 | 4,756 | 3,172 |
| Scaffold N50 | 125,775 | 1,547,048 | 486,756 |
| Chromosome-length scaffolds | |||
| Base pairs | 2,654,127,695 | 1,157,961,392 | 492,400,177 |
| Number of contigs | 36,616 | 25,585 | 41,051 |
| Contig N50 | 108,937 | 93,132 | 30,599 |
| Number of scaffolds | 23 | 3 | 3 |
| Scaffold N50* | 141,244,516 | 404,248,146 | 190,989,159 |
| Small scaffolds | |||
| Base pairs | 13,416,754 | 82,464,476 | 31,168,201 |
| Number of contigs | 850 | 9,416 | 5,609 |
| Contig N50 | 27,968 | 14,202 | 10,570 |
| Number of scaffolds | 811 | 3,981 | 1,224 |
| Scaffold N50 | 30,467 | 65,348 | 45,079 |
| Tiny scaffolds | |||
| Base pairs | 151,762,261 | 14,122,292 | 112,343 |
| Number of contigs | 43,259 | 2,223 | 61 |
| Contig N50 | 6,129 | 6,574 | 2,110 |
| Number of scaffolds | 43,231 | 2,222 | 25 |
| Scaffold N50 | 6,144 | 6,577 | 9,403 |
The scaffold N50 for the output assemblies is not a particularly meaningful assembly statistic: It is determined almost entirely by the chromosome-length scaffolds, which reflect the length distribution of the chromosomes rather than the quality of the genome assembly. The particular value shown is the length of chromosome X (Hs2-HiC) and chromosome 3 (for AaegL4 and CpipJ3).
We then used in situ Hi-C data (6.7× sequence coverage) to improve Hs1. We set aside the tiny scaffolds (43,231 scaffolds shorter than 15 kb, whose N50 length is 6.1 kb). Together, these contain 5.4% of sequenced bases in Hs1. Because of their small size, they have relatively few Hi-C contacts and are more difficult to analyze. We then used Hi-C data to split, anchor, order, and orient the remaining 30,539 scaffolds.
The resulting assembly (Hs2-HiC) consisted of 23 huge scaffolds (lengths from 28.8 to 225.2 Mb) containing 99.5% of the total sequence, together with an additional 811 small scaffolds (N50 length of 30 kb; maximum length of 231 kb) making up the remaining 0.5% of the genome (Table 1 and tables S1 to S6). Crucially, the assembly was generated entirely de novo.
We assessed the quality of Hs2-HiC by comparing it to the human genome reference, hg38 (fig. S9). The 23 scaffolds correspond to the 23 human chromosomes, spanning 99% of the length and containing 91% of the sequence in the chromosome-length scaffolds (table S1). These scaffolds are comparable in length to those reported by the International Human Genome Sequencing Consortium (14) and longer than those reported by (15).
Of the 29,344 scaffolds that were incorporated into chromosome-length scaffolds in Hs2-HiC and that could be uniquely placed in hg38, 99.70% (comprising 99.88% of the sequenced bases) were assigned to the correct chromosome. For randomly selected pairs of scaffolds assigned to the same chromosome-length scaffold in Hs2-HiC, the order in Hs2-HiC agreed with the order in hg38 in 99% of cases. The agreement was 96% for pairs of scaffolds that were adjacent in Hs2-HiC, reflecting the fact that the Hi-C data provide less information to resolve the fine-structure order of short scaffolds. However, the agreement was 99% for scaffolds of at least 120 kb in length. Similarly, the orientation was correct for 93% of scaffolds, with errors mostly resulting from short scaffolds.
Taken together, the chromosome-length, small, and tiny scaffolds accounted for 97.3% of the chromosome-length scaffolds of hg38; the remainder was mostly due to repetitive sequences that could not be adequately assembled from short reads. Our method was further validated by obtaining similar results with a draft assembly generated with Pacific Biosciences long reads, which contained longer contigs (16).
Next, we applied our approach to Ae. aegypti, which was previously assembled from Sanger reads (8× coverage) (17). This assembly, AaegL2, contains 1.3 Gb of sequence (contig N50: 83 kb) partitioned among 4756 scaffolds (scaffold N50: 1.5 Mb).
To improve AaegL2, we generated in situ Hi-C data (40× sequence coverage). After setting aside 2222 scaffolds shorter than 10 kb (spanning 1% of the bases in the initial assembly), we used Hi-C data to split, anchor, order, orient, and merge the remaining 2534 scaffolds. Notably, our pipeline identified apparent misjoins in 1422 of these input scaffolds (56%).
The resulting assembly, AaegL4, contained three huge scaffolds (307, 472, and 404 Mb in length) comprising 93.6% of the input sequence, together with an additional 3981 small scaffolds (N50 of 65 kb, maximum of 474 kb) comprising the remainder. The three huge scaffolds correspond to chromosomes 1, 2, and 3 of the Ae. aegypti genome (18) (Table 1 and tables S2 to S7).
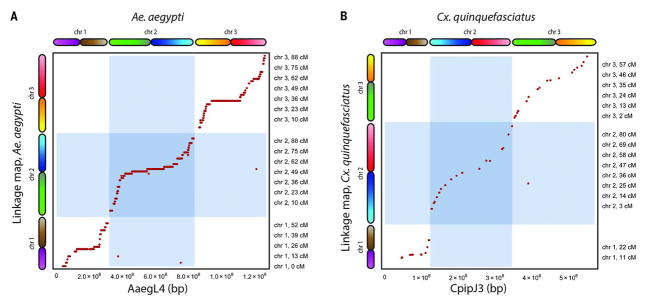
We compared our assembly to a genetic map of Ae. aegypti (19). Of the 2006 markers in the genetic map, 1826 markers could be unambiguously mapped in AaegL4. Notably, our assembly agreed with the genetic map for 1822 of these 1826 markers (Fig. 2). All exceptions were due to misjoins in AaegL2 that had not been detected in AaegL4. We also observed close correspondence with a physical map of the Ae. aegypti genome (fig. S12).
Fig. 2. Comparison of AaegL4 and CpipJ3 with genetic maps.
(A) We compared AaegL4 with a genetic map of Ae. aegypti (19). Our assembly agreed with the genetic map on 1822 out of 1826 markers. The exceptions are due to misjoins in AaegL2 that were not corrected in AaegL4. (B) Similarly, CpipJ3 is in agreement with a genetic map of Cx. quinquefasciatus (21).
Next, we used our approach to create a genome assembly of the mosquito Culex quinquefasciatus, which, like Ae. aegypti, is a disease vector—in this case for West Nile virus, St. Louis encephalitis, and lymphatic filariasis. We generated in situ Hi-C data (~100× sequence coverage) and used them to improve the previous assembly, CpipJ2 (20), obtaining a new assembly, CpipJ3, with three chromosome-length scaffolds that together contain 94% of the sequence in the initial assembly (Table 1 and tables S2 to S7). We validated CpipJ3 by comparing it to existing genetic and physical maps of the Cx. quinquefasciatus genome (20, 21) (Fig. 2 and figs. S13 and S14).
The mosquito Hi-C data and the draft assemblies were generated from different strains. Ideally, the same strain would be used in both cases.
The creation of chromosome-length scaffolds for Ae. aegypti and Cx. quinquefasciatus allowed us to use our Hi-C data to create a Hi-C heatmap showing proximity relationships between chromosomal loci throughout both genomes (22, 23) (Fig. 1 and fig. S15). Notably, the distal ends of the three chromosomes show spatial clustering in both species. Both species also exhibited a second spatial cluster, comprising three loci: one locus from each chromosome, positioned roughly in the middle. This clustering is consistent with the spatial clustering of centromeres, which is known to be present in many organisms. Taken together, the 3D maps are consistent with a spatial arrangement known as the Rabl configuration (24). Our findings also suggest the position of each chromosome’s centromere and thereby partition each mosquito chromosome into two arms.
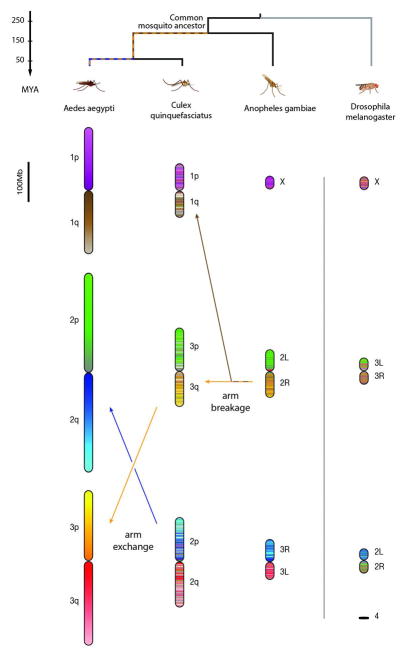
The assemblies of the Ae. aegypti and Cx. quinquefasciatus genomes allowed us to study genome evolution. We began by examining a whole-genome alignment between the published Anopheles gambiae genome, which is 278 Mb long, and the Ae. aegypti genome, which is ~1.2 Gb long. This analysis identified 1389 large blocks of conserved synteny (fig. S16). Similar results were observed for Cx. quinquefasciatus. Despite extensive rearrangements, we observed correspondence of sequence content among chromosome arms in An. gambiae, Cx. quinquefasciatus, and Ae. aegypti. Specifically, for the vast majority of DNA sequences on a particular chromosome arm in one of the three species, the homologous sequences were all found on a single chromosome arm in the other two species. The only exception is the observation that a single arm in An. gambiae (2R) corresponds to two arms in both Ae. aegypti (1q and 3p) and Cx. quinquefasciatus (1q and 3q). This is consistent with the breakage of this arm in the lineage leading to the shared ancestor of Ae. aegypti and Cx. quinquefasciatus (Fig. 3 and tables S8 to S11). These observations are consistent with cytogenetic analyses (18–20) (figs. S17 and S18).
Fig. 3. The content of chromosome arms is strongly conserved across mosquitoes.
Here each 100-kb locus in Ae. aegypti is assigned a color. For the other species, each 100-kb locus is assigned a combination of the colors of the corresponding DNA sequences in Ae. aegypti, weighted by length. MYA, million years ago.
Taken together, these results suggest that—with the exception of the breakage event noted above—each chromosome arm in the Aedes, Culex, and Anopheles species descends from a single arm present in their common ancestor about 150 to 200 million years ago. The preference for within-arm rearrangement in mosquitoes is stronger than has been observed in mammals (25).
Notably, the left arm of chromosome 2 in Drosophila melanogaster has a clear counterpart in all three mosquito species. Thus, all four arms derive from a single chromosome arm present in their dipteran ancestor a quarter of a billion years ago (Fig. 3 and fig. S19).
Overall, our results show that incorporating Hi-C data into genome assembly provides a rapid, inexpensive methodology for generating highly accurate de novo assemblies with chromosome-length scaffolds. At present, the total sequencing costs for a 3D de novo assembly are below $10,000 for mammalian genomes and less for smaller genomes (table S12).
It is important to bear in mind that these assemblies still contain errors. For example, although the Hi-C data provide extensive links covering large distances, the current approach is not perfect for local ordering of small adjacent contigs. This might be circumvented by more sophisticated analysis of Hi-C data. Additional data (such as long or paired-end reads) could also improve the results. The ability to rapidly and reliably generate genome assemblies with chromosome-length scaffolds should accelerate genomic analysis of many organisms.
Supplementary Material
Acknowledgments
This work was supported by a Center for Theoretical Biological Physics postdoctoral fellowship to O.D., an NIH New Innovator Award (1DP2OD008540-01), an NSF Physics Frontier Center Award (PHY-1427654, Center for Theoretical Biological Physics), the National Human Genome Research Institute (NHGRI) Center for Excellence for Genomic Sciences (HG006193), the Welch Foundation (Q-1866), an NVIDIA Research Center Award, an IBM University Challenge Award, a Google Research Award, a Cancer Prevention Research Institute of Texas Scholar Award (R1304), a McNair Medical Institute Scholar Award, an NIH 4D Nucleome Grant (U01HL130010), an NIH Encyclopedia of DNA Elements Mapping Center Award (UM1HG009375), and the President’s Early Career Award in Science and Engineering to E.L.A. We thank C. Nusbaum, L. Vosshall, B. Matthews, and R. Andino for their comments on this manuscript; J. Robinson and D. Turner for work on the Web interface; D. Neafsey, D. Jaffe, I. MacCallum, and members of the Aiden lab for helpful discussions; and S. Knemeyer for assistance with figures. All data sets reported in this paper are available at the Gene Expression Omnibus (GEO), series accession number GSE95797. The Hi-C maps for the final assemblies can be viewed at aidenlab.org/juicebox; the code is available at github.com/theaidenlab/3D-DNA, and additional resources are available at aidenlab.org/3D-DNA. O.D., S.S.B., A.D.O., S.K.N., N.C.D., E.S.L., A.P.A., and E.L.A. are inventors on U.S. provisional patent application 62/347,605, filed 8 June 2016, by the Baylor College of Medicine and the Broad Institute, relating to the assembly methods in this manuscript.
Footnotes
REFERENCES AND NOTES
- 1.Harmon A. Team of Rival Scientists Comes Together to Fight Zika. New York Times. 2016 Mar 30; www.nytimes.com/2016/03/31/us/mapping-a-genetic-strategy-to-fight-the-zika-virus.html?_r=0.
- 2.Gnerre S, et al. Proc Natl Acad Sci USA. 2011;108:1513–1518. doi: 10.1073/pnas.1017351108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Williams LJS, et al. Genome Res. 2012;22:2241–2249. doi: 10.1101/gr.138925.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lieberman-Aiden E, et al. Science. 2009;326:289–293. [Google Scholar]
- 5.Rao SSP, et al. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaplan N, Dekker J. Nat Biotechnol. 2013;31:1143–1147. doi: 10.1038/nbt.2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marie-Nelly H, et al. Nat Commun. 2014;5:5695. doi: 10.1038/ncomms6695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peichel CL, Sullivan ST, Liachko I, White MA. 2016 doi: 10.1093/jhered/esx058. http://biorxiv.org/content/early/2016/08/09/068528. [DOI] [PMC free article] [PubMed]
- 9.Putnam NH, et al. Genome Res. 2016;26:342–350. doi: 10.1101/gr.193474.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burton JN, et al. Nat Biotechnol. 2013;31:1119–1125. doi: 10.1038/nbt.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bickhart DM, et al. 2016 http://biorxiv.org/content/early/2016/07/18/064352.
- 12.Session AM, et al. Nature. 2016;538:336–343. doi: 10.1038/nature19840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Love RR, Weisenfeld NI, Jaffe DB, Besansky NJ, Neafsey DE. BMC Genomics. 2016;17:187. doi: 10.1186/s12864-016-2531-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lander ES, et al. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 15.Venter JC, et al. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 16.Pendleton M, et al. Nat Methods. 2015;12:780–786. doi: 10.1038/nmeth.3454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jaffe DB, et al. Genome Res. 2003;13:91–96. doi: 10.1101/gr.828403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nene V, et al. Science. 2007;316:1718–1723. doi: 10.1126/science.1138878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Juneja P, et al. PLOS Negl Trop Dis. 2014;8:e2652. doi: 10.1371/journal.pntd.0002652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arensburger P, et al. Science. 2010;330:86–88. doi: 10.1126/science.1191864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hickner PV, Mori A, Chadee DD, Severson DW. J Hered. 2013;104:649–655. doi: 10.1093/jhered/est040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Durand NC, et al. Cell Syst. 2016;3:95–98. doi: 10.1016/j.cels.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Durand NC, et al. Cell Syst. 2016;3:99–101. doi: 10.1016/j.cels.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hübner MR, Spector DL. Annu Rev Biophys. 2010;39:471–489. doi: 10.1146/annurev.biophys.093008.131348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ferguson-Smith MA, Trifonov V. Nat Rev Genet. 2007;8:950–962. doi: 10.1038/nrg2199. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.