Abstract
Nurr1 is an orphan nuclear receptor that is essential for the differentiation and maintenance of dopaminergic neurons in the brain, and it is a therapeutic target for Parkinson’s disease (PD). During the screening for Nurr1 activators from natural sources using cell-based assay systems, a methanol extract of the combined stems and roots of Daphne genkwa was found to activate the transcriptional function of Nurr1 at a concentration of 3 μg/mL. The active components were isolated and identified as genkwanine N (1) and yuanhuacin (2). Both compounds 1 and 2 significantly enhanced the function of Nurr1 at 0.3 μM. Nurr1-specific siRNA abolished the activity of 1 and 2, strongly suggesting that transcriptional activation by 1 and 2 occurred through the modulation of Nurr1 function. Additionally, treatment with 1 and 2 inhibited 6-hydroxydopamine (6-OHDA)-induced neuronal cell death and lipopolysaccharide (LPS)-induced neuroinflammation. Moreover, in a 6-OHDA-lesioned rat model of PD, intraperitoneal administration of 2 (0.5 mg/kg/day) for 2 weeks significantly improved behavioral deficits and reduced tyrosine hydroxylase (TH)-positive dopaminergic neuron death induced by 6-OHDA injection and had a beneficial effect on the inflammatory response in the brain. Accordingly, compounds 1 and 2, the first reported Nurr1 activators of natural origin, are potential lead compounds for the treatment of PD.
Graphical abstract
Parkinson’s disease (PD) is a neurodegenerative disorder characterized by the progressive loss of dopaminergic (DA) neurons in the substantia nigra pars compacta (SNc), and is the most prevalent movement disorder, affecting 1–2% of the global population over the age of 65.1–3 Despite extensive research attempting to treat or delay the progress of PD, currently available pharmacological treatments largely address the symptoms, and almost all show severe side effects, such as dyskinesia.
Nurr1, an orphan nuclear receptor, is well-known as a key regulator of DA neuronal differentiation,4–6 and is also essential for the maintenance of DA neurons.7 Post-mortem PD studies have shown diminished expression of Nurr1 in the brain.8,9 Recently, other roles of Nurr1 have received attention. In microglia and astrocytes, Nurr1 inhibits the expression of proinflammatory genes, together with repressor element-1 silencing transcription (REST) corepressor (CoREST), and the repression of inflammation by Nurr1 activation decreases PD symptoms and DA neuronal loss in both in vivo and in vitro PD models.10 Thus, activation of Nurr1 could improve the pathogenesis of PD. Indeed, very recently, small-molecule Nurr1 agonists, such as amodiaquine, have been reported to improve behavioral deficits in animal models of PD.11
Daphne genkwa Siebold et Zucc. (Thymelaeaceae) is widely distributed in Korea and mainland China and is a well-known traditional oriental medicine. The flower buds of this plant (“Genkwa Flos”) are mainly used for diuretic, antitussive, expectorant, and antitumor purposes.12 Chemical investigation of D. genkwa has resulted in the isolation of flavonoids, coumarins, amides, and daphnane-type diterpenes.13–15 Daphnane-type diterpenes have been reported to show various biological activities, including antineoplastic, contraceptive, and neurotrophic activities.16–18 Neuroprotective and antiinflammatory effects of daphnane-type diterpenes, however, have not yet been reported.
During the course of screening for Nurr1 activators from Korean medicinal plants, the methanol extract of the stem and root portion of D. genkwa was found to have potent Nurr1-activating activity, as well as neuroprotective effects, in an animal model of PD. The present study isolated and identified the Nurr1-activating components in D. genkwa and investigated their neuroprotective activity both in vitro and in an in vivo animal model.
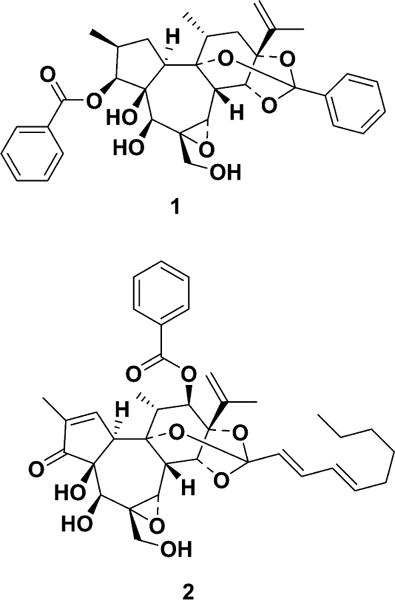
A high-throughput cell-based assay systems was used, described in a previous report,11 to screen a plant extract library, and it was discovered that a methanol extract of a combined stem and root sample of D. genkwa activated Nurr1. This methanol extract enhanced the luciferase reporter activity by approximately 3.5-fold, with the highest activity being at 3 μg/mL (Figure S1, Supporting Information). To determine whether the methanol extract could improve motor behavior deficits in an animal model of PD, adjusting step tests were performed using vehicle-treated and methanol extract-treated 6-OHDA-lesioned rats. Adjusting steps in the contralateral hind limb were significantly improved following administration of D. genkwa (Figure S2, Supporting Information). The methanol extract was successively partitioned into hexane, ethyl acetate (EtOAc), and butanol (BuOH). The hexane and EtOAc extracts activated the luciferase reporter activity by 5-fold and 3.8-fold at 1 μg/mL and 10 μg/mL, respectively, while the BuOH extract and aqueous layer showed no activity (Figure S1, Supporting Information). Two active components, 1 and 2, were isolated from the hexane extract using bioassay-guided fractionation, and they were identified as genkwanine N19 and yuanhuacin,20–22 respectively, by spectroscopic data interpretation (Tables S1 and S2, Supporting Information). To determine the effects of 1 and 2 on Nurr1, a cell-based luciferase reporter assay was used. Both compounds activated Nurr1 in a concentration-dependent manner (Figure 1A,B), by 3- and 3.5-fold, respectively, at 0.3 μM. No cellular toxicity was observed up to 30 μM (data not shown). A cell-based luciferase reporter assay in the absence or presence of Nurr1 specific siRNA was performed to investigate whether the induction of reporter gene expression by 1 or 2 was abolished by Nurr1 knockdown. As shown in Figure 1C, Nurr1 knockdown by the addition of siRNA reduced 1- and 2-induced luciferase reporter gene activation by up to 83%. The knockdown of Nurr1 expression by the siRNA treatment was confirmed by Western blotting analysis (Figure 1D). This data indicated that transcriptional activation by 1 and 2 was indeed mediated through the modulation of Nurr1 function. To examine the possibility that activation of Nurr1 by 1 and 2 could inhibit cell death induced by 6-OHDA, a hydroxylated analogue of dopamine and a selective catecholaminergic neurotoxin used to generate lesions in the nigrostriatal dopaminergic neurons of animals,23 the SH-SY5Y dopaminergic cell line was treated with 6-OHDA in the presence or absence of 1 and 2. A MTT assay showed that 1 and 2 could significantly inhibit 6-OHDA-induced cell death at 6.25 μM (Figure 2A). At higher concentrations, cellular toxicity was seen with both 1 and 2 (data not shown). In general, 6-OHDA induced a high level of reactive oxygen species (ROS) stress, resulting in caspase-dependent apoptosis. It was examined whether cleaved caspase-3, the active form, was diminished by 1 and 2. Co-treatment of 1 or 2 with 6-OHDA prevented the activation of caspase-3 in 6-OHDA-induced SH-SY5Y cell death (Figure 2B).
Figure 1.
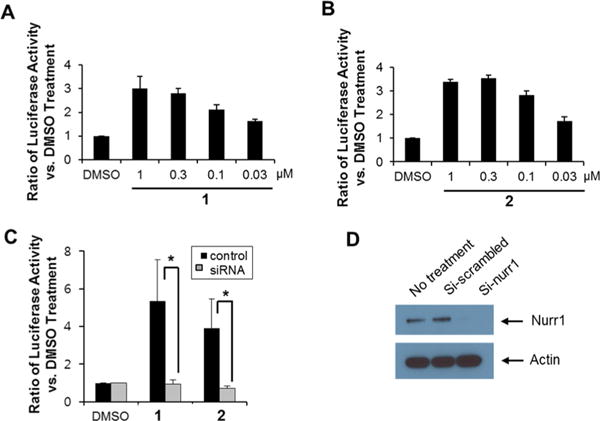
Activation of Nurr1 with compounds 1 and 2. (A) Luciferase assay to measure Nurr1 activity with 1, (B) with 2. Experiments were performed with human neuroblastoma SK-N-BE(2)C cells transfected with pTH2600LUC, pCMV Nurr1-myc, pZeoSV-Nurr1(LBD), and p8xGAL-Luc for the luciferase assay. (C) Luciferase assay to measure Nurr1 activity with 1 and 2 at 0.3 μM in Nurr1-knockdown cells. Human neuroblastoma SK-N-BE(2)C cells were transfected with pTH2600LUC, pCMV Nurr1-myc, pZeoSV-Nurr1(LBD), p8xGAL-Luc, and Nurr1-siRNA for the luciferase assay. (D) Western blotting analysis of Nurr1 in Nurr1-siRNA- treated SK-N-BE(2)C cells.
Figure 2.
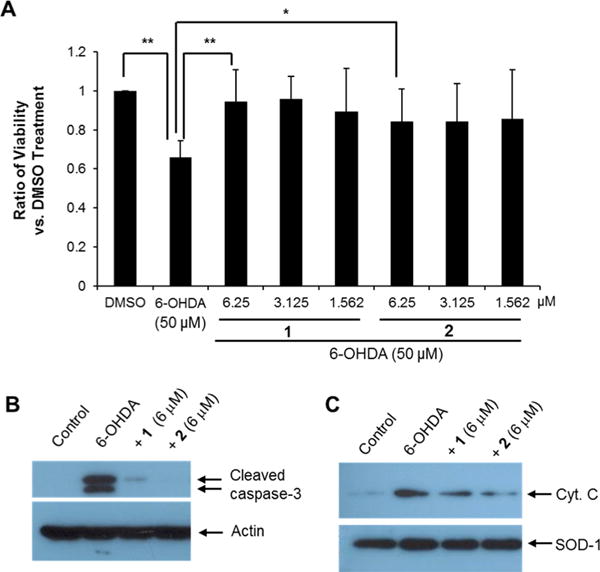
Inhibition of 6-OHDA-induced cell death by treatment on SH-SY5Y cells with compound 1 or 2. (A) Cellular viability assay with 6-OHDA in the presence or absence of 1 and 2 in SH-SY5Y cells (ANOVA and Tukey–Kramer method, n = 4; * p < 0.05, ** p < 0.01). (B and C) Western blotting analysis of cleaved caspase-3 and the release of mitochondrial cytochrome c into the cytosolic fractions during 6-OHDA-induced cell death with or without 1 and 2 at 6 μM. Actin and SOD-1 showed relatively equal protein loading in each lane.
One route to activate caspase-3 is the intrinsic pathway via mitochondria. To verify inhibition of the intrinsic apoptotic pathway by treatment with 1 or 2, cytochrome c release from the mitochondria to the cytosol was examined. In immunoblotting analyses, cytochrome c appeared in the cytosolic fraction of 6-OHDA-treated cells, and the intensity of the band decreased in the presence of 1 and 2 (Figure 2C), indicating that activation of Nurr1 by treatment with 1 and 2 prevented 6-OHDA-induced mitochondrial dysfunction. This result is similar to previous reports that Nurr1 is responsible for the regulation of mitochondrial survival and death via interaction with the p53 tumor suppressor protein.24 Sustained inflammation leads to disease progression in disorders including Alzheimer’s disease, PD, amyotrophic lateral sclerosis, and multiple sclerosis.25 During pathogen invasion or tissue damage, microglia promote inflammation, and sustained and uncontrolled inflammatory responses may result in the production of neurotoxic factors in the brain.26 Nurr1 is related to the neurodegeneration of dopaminergic neurons and acts to suppress inflammatory responses in microglia and astrocytes.26
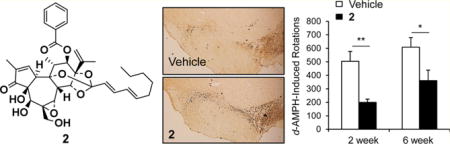
Because Nurr1 plays an important role in inflammation in microglia and astrocytes,10,11 the effects of 1 and 2 on the expression of proinflammatory cytokine genes were analyzed in a BV-2 microglial cell line. When the cells were treated with inflammation-inducing lipopolysaccharide (LPS; 10 ng/mL) for 8 h, the expression of proinflammatory genes including interleukin-1β (IL-1β) and interleukin-6 (IL-6) were dramatically induced, as expected (Figure 3A). It was also found that treatment of 1 and 2 prominently reduced the expression of the proinflammatory genes, as seen by qRT-PCR and Western blotting assays (Figure 3B). To determine the effects of 2 on PD, a rat model with a unilateral 6-OHDA lesion was used. The unilateral dopaminergic cell death caused by 6-OHDA in rodents results in asymmetric rotational behavior and progressive retrograde neuronal degeneration in the SNc.27,28 Injection of 6-OHDA into the dorsal striatum caused dopaminergic neuronal degeneration in the SNc of both vehicle- and 2-treated rats. Two and 6 weeks after surgery, d-amphetamine-induced rotation tests were performed on both groups (Figure 4A). d-Amphetamine-induced ipsilateral rotations were significantly decreased on day 14 and 42 following 6-OHDA lesion induction in the 2-treated group when compared with the vehicle-treated group (Figure 4B).
Figure 3.
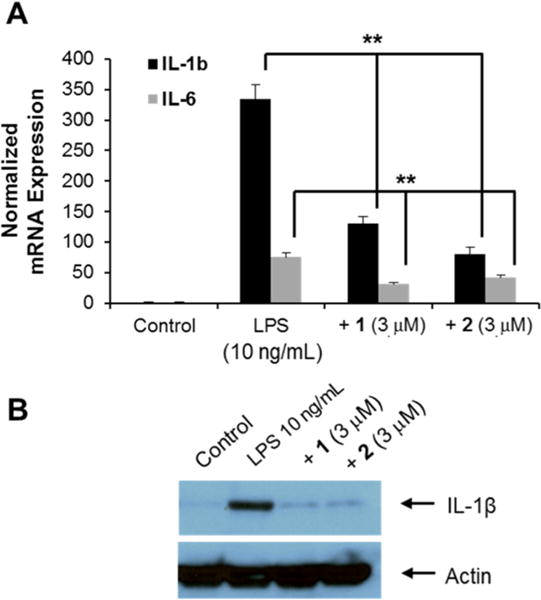
Anti-inflammatory effects of 1 and 2 in a microglia cell line. (A) Analysis of IL-1β and IL-6 mRNA expression in LPS-treated BV-2 cells with or without compounds 1 and 2 at 3 μM (ANOVA and Tukey–Kramer method, n = 4; * p < 0.05, ** p < 0.01). (B) Western blotting analysis of IL-1β in LPS-induced inflammatory cells with or without 1 and 2 at 3 μM.
Figure 4.
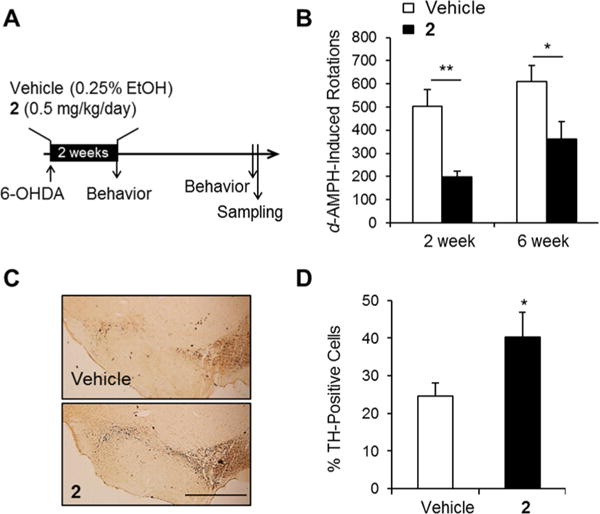
Effects of 2 on the behavioral asymmetry and dopaminergic cell death in a 6-OHDA-lesioned rat model of PD. (A) Experimental design. 6-OHDA-lesioned rats were administered vehicle or 2 for 2 weeks. Rotational behavior was tested at 2 and 6 weeks following 6-OHDA lesion indution. (B) d-Amphetamine-induced ipsilateral rotations in the two groups (Student’s t-test, *p < 0.05 and **p < 0.01). (C) Photomicrograph showing TH-immunoreactive cells in the SNc. Scale bar, 1 mm. (D) Percentage of SNc TH-positive cells in the lesioned side compared with those in the unlesioned side of vehicle-treated and rats treated with 2 (n = 10–14, Student’s t-test, *p < 0.05).
The number of tyrosine hydroxylase (TH)-immunoreactive neuronal cells on the ipsilateral side (6-OHDA-lesioned side, Figure 4C) was analyzed as a percentage of those on the intact contralateral side (Figure 4D). The percentages of TH-immunoreactive cells in the ipsilateral SNc regions of the vehicle- and 2-treated group were 24.62 ± 3.48% and 40.28 ± 6.49%, respectively (Figure 4D). Loss of TH-positive cells on the ipsilateral side of the SNc was significantly decreased in the 2-treated group compared with the vehicle-treated group (p < 0.05), indicating that 2 had a neuroprotective effect in the 6-OHDA-lesioned rat model of PD. In the present study, 2 effectively activated Nurr1 in neuroblastoma cells (Figure 1), and this activation of Nurr1 reduced LPS-induced inflammation (Figure 3). The number of reactive microglia was analyzed using immunohistochemistry in the 6-OHDA-lesioned striatum of rats. The number of Iba-1-positive cells, a marker of activated microglia, was significantly decreased in the 6-OHDA-lesioned striatum of 2-injected rats compared with vehicle-injected rat striata (Figure 5B, p < 0.05), suggesting that 2 showed an antiinflammatory response in vivo as well as in vitro.
Figure 5.
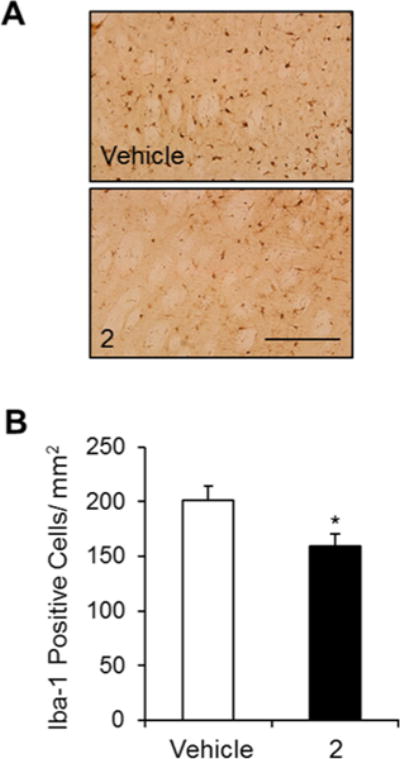
Effects of 2 on inflammation in a 6-OHDA-lesioned rat model of PD. (A) Photomicrograph showing Iba-1-immunoreactive cells in the dorsal striatum. Scale bar, 200 μm. (B) Number of Iba-1-positive cells in the lesioned striatum of vehicle-treated and 2-treated rats (n = 10–14, Student’s t-test, *p < 0.05).
Although daphnane-type diterpenes are the important compounds for the biological activities of D. genkwa, their in vivo toxicity has not been studied yet. Recently, total diterpenoids extract, composed of seven daphnane-type diterpenes (yuanhuacine, yuanhuadine, yuanhuafine, yuanhuapine, genkwanine F, genkwanine N, and Wikstroemia factor M1) from Genkwa Flos was reported to show no lethal effect at 0.1 g/kg but toxicity above 0.25 g/kg after oral administration to rats. Interestingly, the pharmacokinetic study suggested that yuanhuafine and yuanhuapine, among the seven diterpenes, were the key toxic components of D. genkwa in vivo.29
In summary, the present study shows that a methanol extract of the combined stems and roots portion of D. genkwa potently activated the transcriptional function of Nurr1, as well as improved behavioral deficits in a 6-OHDA-lesioned rat model of PD. The active principles were identified as 1 and 2, which enhanced the function of Nurr1 at 0.3 μM and ameliorated 6-OHDA-induced neuronal death and lipopolysaccharide-induced neuroinflammation in vitro. Importantly, the intraperitoneal administration of 2 (0.5 mg/kg/day) for 2 weeks significantly improved behavioral deficits in the animal model of PD and reduced both tyrosine hydroxylase-positive dopaminergic neuron death and microglial inflammation in the 6-OHDA-lesioned striatum. Compounds 1 and 2, the first Nurr1 activators of natural origin, can therefore be suggested as promising novel lead compound candidates for the chemotherapeutic treatment of PD.
EXPERIMENTAL SECTION
General Experimental Procedures
For cell culture, fetal bovine serum (FBS) was purchased from Hyclone (Piscataway, NY, U.S.A.). Horse serum, penicillin, streptomycin, and MTT were purchased from Invitrogen (Carlsbad, CA, U.S.A.). Dulbecco’s modified Eagle’s medium (DMEM), L-glutamine, and LPS were purchased from Sigma-Aldrich Co. (St. Louis, MO, U.S.A.). For the animal study, 6-hydroxydopamine (6-OHDA) was purchased from Sigma-Aldrich Co. and diluted in saline containing 0.02% ascorbic acid. Desipramine (25 mg/kg) and 3,4-dihydroxyphenyl-L-alanine (L-DOPA, 20 mg/kg) were purchased from Sigma-Aldrich Co. and dissolved in saline. d-Amphetamine (d-AMPH) was purchased from USP (Rockville, MD, U.S.A.) and dissolved in saline. Compound 2 was dissolved in saline containing 0.25% ethanol.
Plant Material
A sample of the stems and roots of Daphne genkwa was purchased from Hantaek Botanical Garden, Yongin, Gyeonggi Province, South Korea. The sample was identified by staff at the Plant Extract Bank, Korea Research Institute of Bioscience and Biotechnology, Daejeon, Korea. A voucher specimen (DG-2010-8) was deposited in the Superbacteria Research Center, Korea Research Institute of Bioscience and Biotechnology, Daejeon, Korea. Plants were immediately transferred to the laboratory, washed in tap water and finally rinsed with distilled water. The stem and root portions were dried for 1 week in the shade and then cut.
Extraction and Isolation
To isolate the Nurr1-activating constituents of D. genkwa, dried material (2.75 kg) from the stem and root portion was extracted twice with 6 L of methanol. After the methanol solution was concentrated in vacuo, the dried residue (172.7 g) was suspended in water and successively partitioned three times using 600 mL each of hexane, EtOAc, and BuOH. Subsequently, the biological activity was found mainly in the hexane layer. After the hexane layer was concentrated in vacuo, the resultant dried residue (20 g) was subjected to silica gel (Merck art no. 7734.9025) column chromatography, followed by stepwise elution with n-hexane/EtOAc (10:1, 5:1, 2:1, 1:1, 1:2, 1:5, and 1:10). Each fraction was dried in vacuo, dissolved in DMSO, and subsequently bioassayed. The fractions eluted with n-hexane-EtOAc (1:2) showed Nurr1-activating activity. The active fractions (536 mg) were further purified by preparative ODS TLC with acetonitrile–water (85:15) to give fractions B-1 (20.8 mg) and B-2 (72.8 mg) with Rf values of 0.44 and 0.33, respectively. Fractions B-1 and B-2 were purified finally by preparative HPLC (column 10 × 250 mm, S-4 μm, YMC C18), with acetonitrile–water (80:20) at a flow rate of 3 mL/min to give 1 (9.2 mg) and 2 (36.4 mg), with retention times of 14.5 and 30.8 min, respectively. The purities of 1 and 2 were determined to be over 99% at 230 nm by analytical HPLC (column 4.6 × 250 mm, S-4 μm, YMC C18), with acetonitrile–water (75:25) containing 0.025% TFA at a flow rate of 1.3 mL/min with retention times of 6.1 and 13.9 min, respectively.
Genkwanine N (1)
White powder; [α]25D −27.5 (c 0.1, CHCl3); 1H, 13C NMR, and MS data in accordance with those of genkwanine N.19
Yuanhuacin (2)
White powder; [α]25D + 62.5 (c 0.1, CHCl3) [lit.22 [α]D + 61.7 (CHCl3)]; [α]25D, 1H, 13C NMR, and MS data in accordance with those of yuanhuacin.20–22
Cell Culture and Extract Screening
SK-N-BE(2)C human neuroblastoma cells were maintained in DMEM supplemented with 10% FBS. SH-SY5Y cells were grown in DMEM supplemented with 10% horse serum and 5% FBS. All culture media contained 100 units/mL penicillin and 100 μg/mL streptomycin. For transfection, cells were plated onto 24-wells plates at 1.5 × 105 cells/well in DMEM without antibiotics 1 day prior to transfection. Transfections were carried out using Lipofectamine Plus (Invitrogen) according to the manufacturer’s protocol. The total amount of DNA was 0.5 μg/well, with 0.1 μg pRSV-ß-gal as an internal control. Six hours after transfection, extracts diluted at 0.5% concentration in DMEM media with 1% charcoal-stripped fetal calf serum were added and incubated overnight. Cells from each well were lysed with 100 μL lysis buffer, which contained 25 mM Tris-phosphate (pH 7.8), 2 mM DTT, 2 mM CDTA (1,2-diaminocyclohexane-N,N,N′,N′-tetraacetic acid), 10% glycerol, and 1% Triton X-100. An equal volume of firefly luciferase substrate was added, and luciferase activity was measured using a luminometer plate reader, and normalized for β-galactosidase activity.
Plasmid Constructs
The pTH2600LUC reporter construct contained the 2.6 kb upstream sequences of the rat tyrosine hydroxylase gene in front of a firefly luciferase gene in the pGL3-basic vector. A Nurr1-expression plasmid, pSV40Nurr1, was kindly provided by Dr. Orla M. Conneely of the Baylor College of Medicine (Houston, TX, U.S.A.).30 Full-length mouse Nurr1 cDNA was amplified by RT-PCR from pSV40Nurr1 using forward 5′-GCT TTC AAG CTT ATG CCT TGT GTT CAG GCG CAG TAT GG –3′ and reverse 5′- GTC AAT CTC GAG GAA AGG TAA GGT GTC CAG GAA AAG-3′ oligonucleotides. The amplified PCR fragment was cut with HindIII and XhoI and subsequently cloned into the plasmid pCDNA3.1/myc-His (Invitrogen), resulting in the expression of a C-terminal myc-tagged fusion protein, pCMV Nurr1-myc. The Nurr1 LBD fragment from amino acid position 328 to 598 was PCR-amplified and subcloned into pZeoSV (Invitrogen) containing the GAL4 DNA binding domain. The reporter plasmid, p8xGAL-Luc, was constructed by cloning synthetic oligonucleotides to generate eight tandem repeat GAL4 binding sites (5′-CTC GGA GGA CAG TAC TCC G-3′)31 upstream of luciferase in the pGV-B2 (Toyo Ink) plasmid.
Cell Culture and Drug Treatments
SH-SY5Y cells were initially plated at a density of 2 × 106 and 2 × 105 cells in 50 μg/mL poly-D-lysine-coated culture dishes and 24-well plates, respectively, and maintained at 37 °C in a humidified atmosphere of 5% CO2/95% air in DMEM supplemented with 10% horse serum and 5% fetal bovine serum for 2 days. Cells were subsequently switched to serum-free N-2-defined medium containing 6-OHDA, with or without 1 or 2. Cell viability was determined using the conventional MTT reduction assay. Following incubation for the indicated times, cells were treated with the MTT solution (final concentration, 1 mg/mL, Sigma) for 1 h. Formazan grains that formed in intact cells were solubilized with MTT solubilization solution containing 10% Triton X-100 and 0.1 N HCl in anhydrous isopropanol for 24 h, and the absorbance at 570 nm was then measured using a microplate reader (Molecular Devices, Menlo Park, CA, U.S.A.).
BV-2 cells, a microglial-like mouse cell line, were thawed and passaged until logarithmic growth was achieved. Cell cultures were maintained in six-well culture plates (Corning Costar, Corning, NY, U.S.A.) at a density of 1.0 × 105 cells/well in DMEM supplemented with 10% FBS heat-inactivated at 56 °C for 30 min, 2 mM L-glutamine, and 1X Pen-Strep (Invitrogen). To begin the experiment, BV-2 cells were washed with DMEM and treated with LPS in the presence or absence of 1 and 2. The plates were incubated in a tissue culture incubator at 37 °C in an atmosphere of 5% CO2/95% air for the listed periods of time. Cells from each well were withdrawn for RNA extraction.
Western Blotting
Cells were washed twice with phosphate-buffered saline (PBS) and harvested in a solution containing 1% Triton X-100, 20 mM Tris (pH 7.6), 150 mM sodium chloride, and 1 mM phenylmethylsulfonyl fluoride (PMSF). The cell suspensions were sonicated and boiled in an equal volume of SDS sample buffer. Samples were subjected to SDS-polyacrylamide gel electrophoresis (PAGE) and transferred to polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, MA, U.S.A.). After blocking, the membranes were incubated with primary antibodies diluted in PBS containing 0.1% BSA. The following primary antibodies were used: rabbit anticleaved caspase-3 [Cell Signaling (Danver, MA, U.S.A.); 1:1000], anticytochrome c [Pharmingen (San Diego, CA, U.S.A.)], anti-interleukin 1 β [Santa Cruz (Santa Cruz, CA, U.S.A.)], anti-nurr1 (Sigma), anti-SOD-1 (Santa Cruz), and mouse antiactin (Sigma 1:5000). The membranes were incubated with a 1:3000 dilution of horseradish peroxidase-conjugated antimouse or antirabbit immunoglobulin G (IgG) antibody (Amersham, Piscataway, NY, U.S.A.). Detection was achieved using an enhanced-chemiluminescent substrate (Amersham).
Real-Time PCR Analysis
Real-time PCR analysis was performed using the ABI-Prism7700 sequence detection system (Applied Biosystems, Foster City, CA, U.S.A.) as described previously.32,33 Briefly, it was performed in 0.5 mL tubes (Applied Biosystems) on cDNA equivalent to 50 ng DNase-digested RNA in a volume of 1/25, containing 2/25 of SyBRGreen universal master mix, and forward and reverse primers following the manufacturer’s protocol. All primers for mouse cytokines and GAPDH were obtained from Invitrogen. The mRNA expression of cytokines was normalized to the level of GAPDH mRNA. Data were processed by the ABI Sequence Detection System 1.6 software and analyzed by ANOVA.
siRNA Knockdown
To knockdown Nurr1 in SK-N-BE(2)C cells, MISSION siRNA targeting human NR4A2 (Sigma) and N-TER/siRNA nanoparticles (Sigma) were used according to the manufacturer’s protocol. Following transfection of scrambled siRNA or siRNA for Nurr1, a luciferase assay was conducted.
Animals
Sprague-Dawley rats, weighing 140–160 g, were obtained from Koatech (Pyeongtaek, Gyeonggi-do, Korea). All rats were housed in regular polycarbonate plastic cages in a temperature- (21–22 °C) and humidity- (50–60%) controlled environment with a 12 h light/dark cycle (lights on at 7 a.m.), and they were maintained on an ad libitum diet of lab chow (Purina Inc., Hwasung-shi, Korea) and water. The animal room was maintained in a specific-pathogen-free condition. All animal experiments were approved by the Institutional Animal Use and Care Committee of the Korea Research Institute of Bioscience and Biotechnology (KRIBB-AEC-11019 and KRIBB-AEC-12025) and were performed in accordance with the Guide for the Care and Use of Laboratory Animals, published by the U.S. National Institutes of Health.
6-Hydroxydopamine Lesion Induction
Rats were anesthetized with Zoletil 50 (100 mg/kg, ip, Virbac, Carros, France) and mounted in a stereotactic frame (Stoelting Europe, Dublin, Ireland) equipped with a rat adaptor. Rats were pretreated with desipramine (25 mg/kg, intraperitoneal) 30 min prior to the surgery in order to prevent noradrenergic neuron damage. Rats received unilateral injections of 6-OHDA in 8 μL (2.5 μg/μL, at the injection speed of 1 μL/min) into the left side of the striatum at the following coordinates according to the mouse brain atlas of Paxinos and Franklin (2001): anteroposterior (AP), 0.7 mm; lateral (L), −2.4 mm; and dorsoventral (DV), −5.5 mm. Rats were left on a warming plate until they awoke from the anesthesia.
The 6-OHDA-lesioned rats were randomly assigned to two groups: vehicle (0.25% ethanol)-treated rats (n = 14) and 2-treated rats (n = 10). Vehicle (0.25% ethanol) or 2 (0.5 mg/kg/day) was administered intraperitoneally for 2 weeks.
d-Amphetamine-Induced Rotation Test
d-AMPH (5 mg/kg)-induced rotation was measured 2 and 6 weeks after the 6-OHDA injections. The number of ipsilateral turns was recorded, 30 min following amphetamine administration, for 30 min.
Immunohistochemistry
Immunohistochemistry was conducted as described previously.34 Rats were sacrificed 43 days after 6-OHDA-induced lesion formation, followed by intracardiac perfusion with 4% paraformaldehyde in PBS. Brains were removed, fixed overnight, and cut into 40-μm coronal sections using a vibratome (Vibratome VT1000A, Leica Microsystems GmbH, Wetzlar, Germany). Free-floating sections were incubated in PBS containing 3% H2O2 (v/v), rinsed 3 times in PBS, and blocked with 5% horse serum (HS) for 1 h at room temperature. Sections were incubated overnight at 4 °C with anti-TH (5% HS, Pel-Freez, catalog no. P40101-0) or anti-Iba-1 (ionized calcium-binding adapter molecule 1, 5% HS, catalog no. 019-19741, Wako) antibodies. After washing the sections, they were incubated with biotinylated secondary antirabbit IgG (Vector Laboratories, Inc., Burlingame, CA, U.S.A.), followed by the avidin–biotinylated peroxidase complex (ABC kit, Vector Laboratories, Inc.) and 3,3′-diaminobenzidine (Sigma-Aldrich). Sections containing the striatum (2.8 mm × 2.08 mm) at AP + 1.0 to +0.6 mm and the SNc at AP −4.8 to −5.5 mm from the bregma were selected. Immunoreactive cells were counted in the left and right SNc (TH) or striatum (Iba-1) of three sections per animal under a microscope, following the procedure described previously.35 To avoid the double counting of neurons with unusual shapes, TH-stained cells were counted only when their nuclei were visualized in a focal plane. Qualitative evaluation of immunoreactive cells was performed in which the investigator was blind to both genotype and treatment, following the procedure introduced by Kim et al.36 Analysis values obtained on the ipsilateral side (6-OHDA-lesioned side) are expressed as a percentage of those on the intact contralateral side.
Statistical Methods
Two-sample comparisons were carried out using Student’s t-test, ANOVA, and Tukey–Kramer comparisons. All data are presented as means ± S.E.M., and statistical differences were accepted at the 5% level unless otherwise indicated.
Supplementary Material
Acknowledgments
This work was supported by the Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, Forestry, and Fisheries (IPET) through the Agri-Bio industry Technology Development Program, funded by the Ministry of Agriculture, Food, and Rural Affairs (MAFRA) (114145-3). Support was also obtained from the Cooperative Research Program for Agriculture Science and Technology Development (PJ008022), Rural Development Administration and from the KRIBB Research Initiative Program, Republic of Korea.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jnatprod.6b00110.
1H- and 13C NMR data of genkwanine N (1) and yuanhuacin (2); neuroprotective activity of the methanol extract of D. genkwa in an in vivo animal model (PDF)
Notes
The authors declare no competing financial interest.
References
- 1.Dauer W, Przedborski S. Neuron. 2003;39:889–909. doi: 10.1016/s0896-6273(03)00568-3. [DOI] [PubMed] [Google Scholar]
- 2.Meissner WG, Frasier M, Gasser T, Goetz CG, Lozano A, Piccini P, Obeso JA, Rascol O, Schapira A, Voon V, Weiner DM, Tison F, Bezard E. Nat Rev Drug Discovery. 2011;10:377–393. doi: 10.1038/nrd3430. [DOI] [PubMed] [Google Scholar]
- 3.Obeso JA, Rodriguez-Oroz MC, Goetz CG, Marin C, Kordower JH, Rodriguez M, Hirsch EC, Farrer M, Schapira AH, Halliday G. Nat Med. 2010;16:653–661. doi: 10.1038/nm.2165. [DOI] [PubMed] [Google Scholar]
- 4.Saucedo-Cardenas O, Quintana-Hau JD, Le WD, Smidt MP, Cox JJ, De Mayo F, Burbach JP, Conneely OM. Proc Natl Acad Sci U S A. 1998;95:4013–4018. doi: 10.1073/pnas.95.7.4013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zetterstrom RH, Solomin L, Jansson L, Hoffer BJ, Olson L, Perlmann T. Science. 1997;276:248–250. doi: 10.1126/science.276.5310.248. [DOI] [PubMed] [Google Scholar]
- 6.Chung S, Leung A, Han BS, Chang MY, Moon JI, Kim CH, Hong S, Pruszak J, Isacson O, Kim KS. Cell Stem Cell. 2009;5:646–658. doi: 10.1016/j.stem.2009.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kadkhodaei B, Ito T, Joodmardi E, Mattsson B, Rouillard C, Carta M, Muramatsu S, Sumi-Ichinose C, Nomura T, Metzger D, Chambon P, Lindqvist E, Larsson NG, Olson L, Bjorklund A, Ichinose H, Perlmann T. J Neurosci. 2009;29:15923–15932. doi: 10.1523/JNEUROSCI.3910-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chu Y, Kompoliti K, Cochran EJ, Mufson EJ, Kordower JH. J Comp Neurol. 2002;450:203–214. doi: 10.1002/cne.10261. [DOI] [PubMed] [Google Scholar]
- 9.Chu Y, Le W, Kompoliti K, Jankovic J, Mufson EJ, Kordower JH. J Comp Neurol. 2006;494:495–514. doi: 10.1002/cne.20828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saijo K, Winner B, Carson CT, Collier JG, Boyer L, Rosenfeld MG, Gage FH, Glass CK. Cell. 2009;137:47–59. doi: 10.1016/j.cell.2009.01.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim CH, Han BS, Moon J, Kim DJ, Shin J, Rajan S, Nguyen QT, Sohn M, Kim WG, Han M, Jeong I, Kim KS, Lee EH, Tu Y, Naffin-Olivos JL, Park CH, Ringe D, Yoon HS, Petsko GA, Kim KS. Proc Natl Acad Sci U S A. 2015;112:8756–8761. doi: 10.1073/pnas.1509742112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiangsu New Medical College. The Encyclopedia of Traditional Chinese Medicine. 2nd. Shanghai Science and Technology; Shanghai: 1985. p. 2573. [Google Scholar]
- 13.Hall IH, Kasai R, Wu RY, Tagahara K, Lee KH. J Pharm Sci. 1982;71:1263–1267. doi: 10.1002/jps.2600711120. [DOI] [PubMed] [Google Scholar]
- 14.Zhan Z-J, Fan C-Q, Ding J, Yue J-M. Bioorg Med Chem. 2005;13:645–655. doi: 10.1016/j.bmc.2004.10.054. [DOI] [PubMed] [Google Scholar]
- 15.Akhtar K, Khan SB, Ali I. Magn Reson Chem. 2006;44:1063–1066. doi: 10.1002/mrc.1896. [DOI] [PubMed] [Google Scholar]
- 16.Zhang S, Li X, Zhang F, Yang P, Gao X, Song Q. Bioorg Med Chem. 2006;14:3888–3895. doi: 10.1016/j.bmc.2006.01.055. [DOI] [PubMed] [Google Scholar]
- 17.He W, Cik M, Appendino G, Van Puyvelde L, Leysen JE, De Kimpe N. Mini-Rev Med Chem. 2002;2:185–200. doi: 10.2174/1389557024605492. [DOI] [PubMed] [Google Scholar]
- 18.Hong J-Y, Chung H-J, Lee H-J, Park H-J, Lee S-K. J Nat Prod. 2011;74:2102–2108. doi: 10.1021/np2003512. [DOI] [PubMed] [Google Scholar]
- 19.Li L-Z, Gao P-Y, Peng Y, Wang L-H, Yang J-Y, Wu C-F, Zhang Y, Song S-J. Helv Chim Acta. 2010;93:1172–1179. [Google Scholar]
- 20.Zhou BN, Xue L, Lin LZ, Lin LJ, Johnson ME, Cordell GA. Magn Reson Chem. 1993;31:194–199. [Google Scholar]
- 21.Borris RP, Cordell GA. J Nat Prod. 1984;47:270–278. doi: 10.1021/np50032a006. [DOI] [PubMed] [Google Scholar]
- 22.Ying B-P, Wang C-S, Chou P-N, Pan P-C, Liu J-S. Huaxue Xuebao. 1978;35:103–108. [Google Scholar]
- 23.Ungerstedt U. Eur J Pharmacol. 1968;5:107–110. doi: 10.1016/0014-2999(68)90164-7. [DOI] [PubMed] [Google Scholar]
- 24.Zhang T, Wang P, Ren H, Fan J, Wang G. Mol Cancer Res. 2009;7:1408–1415. doi: 10.1158/1541-7786.MCR-08-0533. [DOI] [PubMed] [Google Scholar]
- 25.Fakhoury M. Neurodegener Dis. 2015;15:63–69. doi: 10.1159/000369933. [DOI] [PubMed] [Google Scholar]
- 26.Glass CK, Saijo K, Winner B, Marchetto MC, Gage FH. Cell. 2010;140:918–934. doi: 10.1016/j.cell.2010.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sauer H, Oertel WH. Neuroscience. 1994;59:401–415. doi: 10.1016/0306-4522(94)90605-x. [DOI] [PubMed] [Google Scholar]
- 28.Przedbroski S, Leviver M, Jiang H, Ferreira M, Jackson-Lewis V, Donaldson D, Togasaki DM. Neuroscience. 1995;67:631–647. doi: 10.1016/0306-4522(95)00066-r. [DOI] [PubMed] [Google Scholar]
- 29.Chen YY, Guo JM, Qian YF, Guo S, Ma CH, Duan JA. Phytomedicine. 2013;21:82–89. doi: 10.1016/j.phymed.2013.06.012. [DOI] [PubMed] [Google Scholar]
- 30.Murphy EP, Dobson AD, Keller C, Conneely OM. Gene Expr. 1996;5:169–179. [PMC free article] [PubMed] [Google Scholar]
- 31.Webster N, Jin JR, Green S, Hollis M, Chambon P. Cell. 1988;52:169–178. doi: 10.1016/0092-8674(88)90505-3. [DOI] [PubMed] [Google Scholar]
- 32.Hering R, Petrovic S, Mietz EM, Holzmann C, Berg D, Bauer P, Woitalla D, Muller T, Berger K, Kruger R, Riess O. Neurology. 2004;62:1231–1232. doi: 10.1212/01.wnl.0000118285.18383.90. [DOI] [PubMed] [Google Scholar]
- 33.Kim SY, Han YM, Oh M, Kim WK, Oh KJ, Lee SC, Bae KH, Han BS. Stem Cells Dev. 2015;24:686–700. doi: 10.1089/scd.2014.0434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park HY, Kang YM, Kang Y, Park TS, Ryu YK, Hwang JH, Kim YH, Chung BH, Nam KH, Kim MR, Lee CH, Han PL, Kim KS. J Neurosci. 2014;34:11744–11753. doi: 10.1523/JNEUROSCI.0864-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Granado N, O’Shea E, Bove J, Vila M, Colado MI, Moratalla R. J Neurochem. 2008;107:1102–1112. doi: 10.1111/j.1471-4159.2008.05705.x. [DOI] [PubMed] [Google Scholar]
- 36.Kim KS, Han PL. J Neurosci Res. 2009;87:2983–2993. doi: 10.1002/jnr.22119. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


