Summary
Homeostasis of the gut microbiota critically influences host health and aging. Developing genetically engineered probiotics holds great promise as a new therapeutic paradigm to promote healthy aging. Here, through screening 3,983 Escherichia coli mutants, we discovered that 29 bacterial genes, when deleted, increase longevity in the host Caenorhabditis elegans. A dozen of these bacterial mutants also protect the host from age-related progression of tumor growth and amyloid-beta accumulation. Mechanistically, we discovered that five bacterial mutants promote longevity through increased secretion of the polysaccharide colanic acid (CA), which regulates mitochondrial dynamics and unfolded protein response (UPRmt) in the host. Purified CA polymers are sufficient to promote longevity via ATFS-1, the host UPRmt-responsive transcription factor. Furthermore, the mitochondrial changes and longevity effects induced by CA are conserved across different species. Together, our results identified molecular targets for developing pro-longevity microbes, and a bacterial metabolite acting on host mitochondria to promote longevity.
Keywords: Longevity, probiotics, colanic acid, mitochondrial dynamics, mitochondrial unfolded protein response, microbiota-host
Graphical abstract
Introduction
In large part because of progress in curing infectious and contagious diseases, modern society is progressively aging and is made up of an ever-growing proportion of elderly individuals. Improving healthy aging and preventing aging-associated chronic disabilities have become a priority of current biomedical research. Although our understanding of aging has been deepened by discoveries of several conserved molecular mechanisms (Kenyon, 2010), few longevity-promoting compounds have been discovered to date (Cabreiro et al., 2013; Harrison et al., 2009; Harrison et al., 2014; Howitz et al., 2003).
A growing body of evidence suggests that gut microbiota -diverse microorganisms inhabiting the digestive track- are tightly linked to the aging process of their host (Cho and Blaser, 2012; Heintz and Mair, 2014). This community of microbial species, among which bacteria are predominant and most extensively studied, not only generates metabolites essential for various host functions (Lee and Hase, 2014), but also mediates effects of exogenous chemicals on the animals they reside in (Cabreiro et al., 2013). Changes in the bacterial composition of the microbiota have been observed in elderly people, and diet-driven microbiota alterations have been shown to improve their health (Claesson et al., 2012; Yatsunenko et al., 2012). However, most of our knowledge on microbiota is limited to taxonomic and metagenomic profiling, whereas functions of individual microbial genes and their molecular mechanisms in modulating host aging remain elusive. This is due in part to the high complexity of mammalian gut microbiota and to technical challenges of isolating specific pro-longevity microbial variations.
The nematode Caenorhabditis elegans with its short and easily monitored lifespan as well as defined microbiota, is a powerful model for disentangling the interactions between microbes and host aging (Heintz and Mair, 2014). Under standard laboratory conditions, C. elegans are reared on a single bacterial strain of Escherichia coli. Starting from early adulthood of the nematodes, bacterial cells colonize the intestinal lumen and form their entire gut microbiota (Clark and Hodgkin, 2014). Using this simple model system, studies have demonstrated the crucial role that the microbiota play in regulating host longevity (Garigan et al., 2002; Portal-Celhay et al., 2012). Some bacterial variants have been serendipitously identified as determinants of host lifespan (Larsen and Clarke, 2002; Virk et al., 2012), and small molecules secreted from bacteria, such as certain non-coding RNAs and nitric oxide have been linked to host longevity in the context of specific bacterial backgrounds (Gusarov et al., 2013; Liu et al., 2012). However, a systematic analysis on the longevity-related effects of bacterial genetic factors has not been performed.
In this study, we designed a high-throughput screening platform to identify microbial pro-longevity factors from the entire collection of E. coli non-essential gene deletion mutants (Baba et al., 2006). We discovered a series of microbial genetic factors that regulate host health and longevity, and demonstrated their interactions with several known aging-regulatory pathways of the host. We also discovered a probiotic mechanism distinct from these known pathways, which functions through induction of a secreted polysaccharide that promotes host longevity by tuning mitochondrial homeostasis. Our data thus reveal the crucial link between microbiota genetic composition and host longevity, provide a roadmap for developing genetically engineered pro-longevity probiotics, and uncover a chemical dialogue between bacteria and mitochondria.
Results
Genome-wide analysis of microbial factors regulating host longevity
To systematically identify microbial factors that promote host longevity, we conducted a genome-wide screen of the E. coli single-gene knockout library for lifespan extension in C. elegans. We used the parental strain of this library, E. coli K-12 BW25113, as an experimental control. C. elegans grown on this control bacteria show similar development, reproduction, and lifespan as those on standard laboratory E. coli strains (Figure S1A-C). Because of the crucial role of the reproductive system in regulating C. elegans longevity, we applied a high-throughput screening strategy different from the conventional method that interrupts reproduction. We selected the sqt-3(e2117) collagen mutant of C. elegans that reproduces normally but is embryonically lethal at 25 °C (Wang et al., 2014). Using this mutant, we performed the primary screen at the restrictive temperature to avoid labor-intensive animal transfer without interrupting normal reproduction (Figure S1D and E).
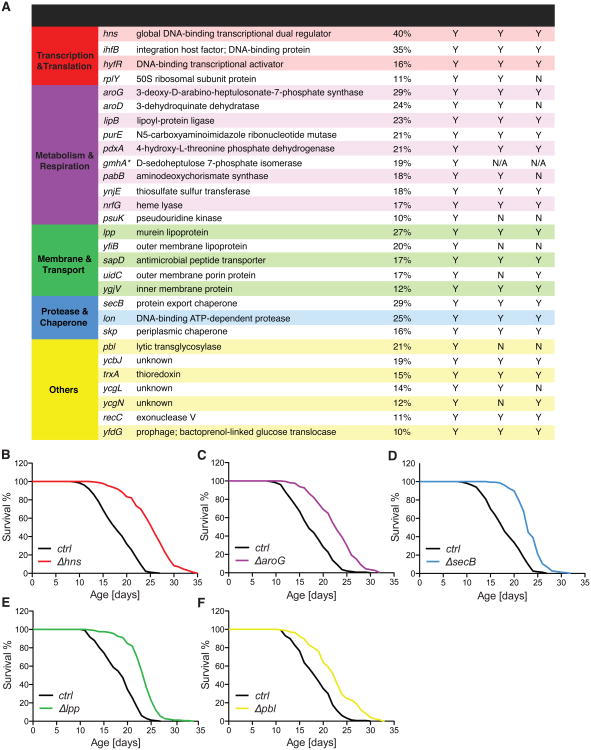
Out of the 3,983 E. coli mutants, 68 prolonged lifespan in the sqt-3 background (Table S1). We next validated the longevity effects of these 68 E. coli mutants in wild-type C. elegans at 20 °C, and identified 35 bacterial mutants that significantly prolonged host lifespan by more than 10% (p<0.001, Table S2). To determine whether the longevity effects of these bacterial mutants are indeed conferred by the deletion of their annotated genes, we reintroduced these mutations into the parental strain BW25113, and confirmed the lifespan-extending effects of 29 bacterial mutants (Figure 1A, Table S3). These longevity-promoting bacterial mutants appear functionally heterogeneous, and deletions of genes in different functional groups can lead to comparable levels of lifespan extension in C. elegans (Figure 1 B-F). Moreover, 21 of these bacterial mutants preserved their longevity effects when only provided during adulthood of C. elegans (Figure 1A, Table S3). We also introduced these mutations into a wild-type E. coli strain, MG1655, and showed that 23 of them significantly prolong C. elegans lifespan (Figure 1A, Table S3). Together, these results show that alterations in bacterial genetic composition can modulate the aging process of the host, and these effects are not restricted to specific microbial backgrounds or host developmental stages.
Figure 1. Bacterial mutants promote host longevity.
(A) A genome-wide screen of the E. coli BW25113 single-gene knockout library identified 29 mutants that prolong C. elegans lifespan by more than 10% (p<0.001, log-rank test). 21 of these mutants significantly prolong lifespan when provided only to C. elegans adults (p<0.05, log-rank test), and 23 of them also promote host longevity in the wild-type E. coli MG1655 strain background (p<0.05, log-rank test). These longevity-promoting bacterial mutants are classified into different functional categories, delineated by different colors. An asterisk marks the gmhA gene, when deleted, resulting in resistance to P1 phage.
(B to F) Lifespan extensions by different functional categories of bacterial mutants are represented by Δhns (B), ΔaroG (C), ΔsecB (D), Δlpp (E), and Δpbl (F), respectively.
See Figure S1 for information on the control bacteria and the design of the screen, Figure S2 for information of viability and colonization capacities of the mutants, Figure S7 for the enrichment of genes involved in chorismate metabolism, and Table S1 to S3 for lifespan data including animal numbers, mean and median lifespans, and statistics.
In order to rule out the possibility that increased lifespan is caused by differences in bacterial amount, we ensured the same numbers of bacteria supplied to the worms based on both light-scattering measurements and cell counting by microscopy. We also measured viability of the identified longevity-promoting bacterial mutants, showing that the numbers of live bacterial cells initially provided to the worms are similar (Figure S2A). When comparing host colonization capacities among these bacteria, we observed a decrease in 5 mutants and an increase in 2 mutants (Figure S2B and C). Therefore, there is no direct association between the viability or the host colonization of the bacterial mutants and their lifespan-extending effects.
Microbial factors ameliorate age-related pathologies of tumor progression and amyloid-b accumulation
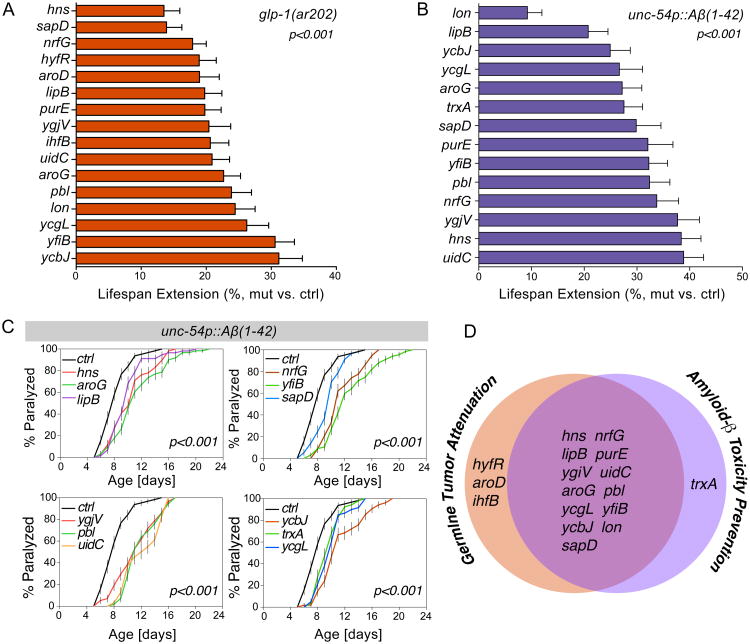
Advanced age is a major risk factor for various chronic diseases, such as cancer and neurodegeneration (Niccoli and Partridge, 2012). Next, we tested whether bacterial mutants identified in our screen could increase the survival of host animals under age-associated pathological conditions. In the C. elegans glp-1(ar202) mutant, excessive proliferation of germ cells results in germline tumors that migrate into somatic tissues and causes early death (Pepper et al., 2003). We found that 16 bacterial mutants increase the survival rate of the glp-1(ar202) mutant (Figure 2A, Table S4).
Figure 2. Bacterial mutants attenuate age-related pathologies in the host.
(A) 16 bacterial mutants significantly increase the survival of the C. elegans glp-1(ar202) mutants, which exhibit early death due to age-related progression of germline tumors (p<0.001, Log-rank test).
(B) Ectopic expression of human amyloid-β (Aβ) in the C. elegans CL2006 transgenic strain dvIs2[unc-54p∷Aβ(1-42)] leads to a shortened lifespan. 14 bacterial mutants significantly increase the survival of this strain (p<0.001, Log-rank test).
Survival analyses were conducted three times independently. The average percentage of lifespan extension by each bacterial mutant vs. the parental control is shown, and error bars represent standard error of the mean (SEM).
(C) 12 of the bacterial mutants that increase the survival of the CL2006 strain also significantly delay the onset of paralysis caused by Aβ proteotoxicity (p<0.001, Log-rank test). Error bars represent SEM.
(D) The sets of the bacterial mutants that protect C. elegans against the lethality of germline tumor and Aβ accumulation are largely overlapped.
See Table S4 and S5 for survival data of three independent trails including animal numbers, mean lifespans and statistics.
In addition, C. elegans transgenic strains expressing toxic human Amyloid-β (Aβ) exhibit early decline in physical activities and shortened lifespan (Link, 1995). Among the 29 lifespan-extending bacterial mutants, we identified 14 that significantly prolong the lifespan of the Aβ transgenic strains (Figure 2B, Table S5), and 12 of them substantially delay age-associated paralysis (Figure 2C). Altogether, we discovered 13 bacterial mutants that protect C. elegans from lethality of both germline tumors and Aβ accumulation (Figure 2D), suggesting that they may improve the overall fitness of the host.
Interaction of beneficial microbial factors with host longevity mechanisms
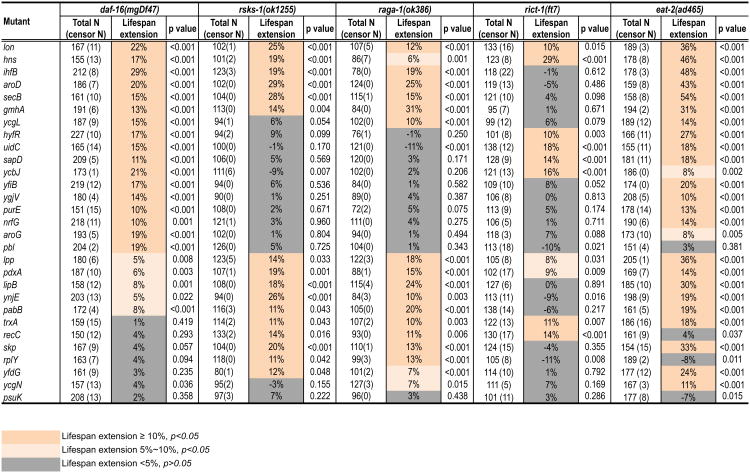
To gain insight into the molecular mechanisms mediating the beneficial effects of these bacterial mutants, we examined whether they act through several known longevity mechanisms in the host, including the insulin/IGF-1 and the TOR signaling pathways as well as caloric restriction (CR). The FoxO transcription factor DAF-16 is a necessary downstream mediator of insulin/IGF-1 and JNK longevity signaling pathways (Lin et al., 1997; Ogg et al., 1997; Oh et al., 2005; Wang et al., 2005). RSKS-1, the ribosomal protein S6 kinase and RAGA-1, the Ras-related GTPase are crucial regulators of the TOR complex 1 longevity mechanism (Pan et al., 2007; Robida-Stubbs et al., 2012; Stanfel et al., 2009). RICT-1, a subunit of the TOR complex 2, is also involved in regulating the lifespan of C. elegans (Mizunuma et al., 2014; Soukas et al., 2009). We investigated the lifespan of the daf-16(mgDf47), rsks-1(ok1255), raga-1(ok386), and rict-1(ft7) C. elegans mutants grown on each bacterial mutant to determine whether these host factors contribute to the longevity effects of the bacterial mutants. Out of the 29 bacterial mutants, there are 7, 13, 11 and 19 mutants unable to significantly prolong the lifespan of the C. elegans daf-16, rsks-1, raga-1, and rict-1 mutants, respectively (p>0.05, Figure 3), suggesting that they act through the host insulin/IGF-1 and/or TOR signaling pathway to regulate longevity.
Figure 3. Genetic interaction analyses with host longevity mechanisms.
Bacterial mutants were examined for their effects on the lifespans of the daf-16(mgDf47), rsks-1(ok1255), raga-1(ok386), rict-1(ft7), and eat-2(ad465) C. elegans mutants. Grey colors mark the candidates showing dependence on these known host longevity regulatory pathways, and apricot colors mark the ones acting independently. Total N, the number of total animals; censor N, the number of censored animals; Lifespan extension, the percentage of lifespan extension in different C. elegans mutants on each bacterial mutant compared to those on parental control bacteria; p-values, bacterial mutant vs. parental control by Log-rank test.
See Figure S3 for effects of bacterial mutants on C. elegans feeding and defecation behaviors.
CR is an extensively studied intervention that promotes longevity in a variety of species (Fontana et al., 2010). We further examined whether CR plays a role in mediating the longevity effects of these bacterial mutants. We first assessed the effects of the bacterial mutants on feeding and defecation behaviors of wild-type C. elegans. When providing similar amounts of bacteria (Figure S2A), we found that only 2 bacterial mutants lowered the pharyngeal pumping rate (Figure S3A), and another 2 slowed down defecation (Figure S3B), suggesting minimal involvement of low food intake in most bacteria-induced longevity effects.
In addition, CR can be genetically mimicked using C. elegans eat-2 mutants that have defective pharyngeal contraction during feeding (Lakowski and Hekimi, 1998). We examined how the bacterial mutants affect the host lifespan under this genetically mimicked CR condition. We first confirmed that none of the bacterial mutants alter the pharyngeal contraction rate of the eat-2 mutant (Figure S3C). Next, we found that only 4 out of the 29 bacterial mutants do not significantly prolong the lifespan of the eat-2(ad465) mutant (p>0.05, Figure 3), suggesting that CR does not contribute to the longevity effects of the other 25 bacterial mutants.
Bacterial colanic acid overproduction as a longevity-promoting mechanism
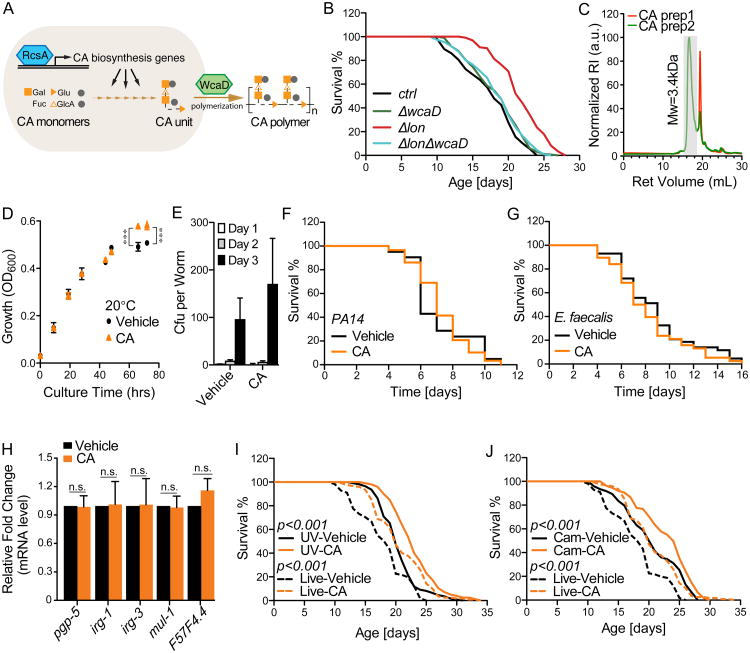
The genetic interaction analyses between longevity-promoting bacterial mutations and host mutations lead to the discovery of 2 bacterial mutants, Δhns and Δlon, which act independently of DAF-16, TOR, or CR (Figure 3). Interestingly, both lon and hns genes regulate biosynthesis of colanic acid (CA), a polysaccharide secreted by many enterobacterial species including E. coli K-12 (Figure 4A) (Grant et al., 1969). RcsA, the central activator of the CA biosynthesis gene cluster, is degraded by the ATP-dependent protease Lon (Gottesman et al., 1985; Torres-Cabassa and Gottesman, 1987), and is also repressed by the global transcriptional regulator H-NS (Sledjeski and Gottesman, 1995) (Figure 4A). We thus hypothesized that CA overproduction underlies the longevity promoting effects of these 2 mutants. To test this hypothesis, we first measured the CA levels in bacterial culture medium, and found that CA secretion is elevated in both Δlon and Δhns, compared to the parental control (Figure 4B). Furthermore, we discovered that the ΔrcsA deletion fully suppresses the CA overproduction in these mutants (Figure 4B). The ΔrcsA deletion completely abrogates the lifespan-extending effect of the Δlon mutant, suggesting a full requirement of CA overproduction (Figure 4C, Table S6). For the Δhns mutant, the ΔrcsA deletion exerts a partial suppression, indicating that both CA-dependent and -independent mechanisms contribute to its longevity effect (Figure 4D, Table S6). On the other hand, ΔaroD, a mutant that does not induce CA secretion acts independently of RcsA to promote longevity (Figure 4E, Table S6) and has an additive lifespan-extending effect with Δlon (Figure 4F, Table S6). Interestingly, there are other 3 CA overproducing bacterial mutants that extend C. elegans lifespan in an RcsA dependent manner (Figure S4, Table S6). Together, these results suggest CA overproduction as a probiotic mechanism linking microbial metabolism and host longevity.
Figure 4. Colanic acid-overproducing bacterial mutants promote host longevity.
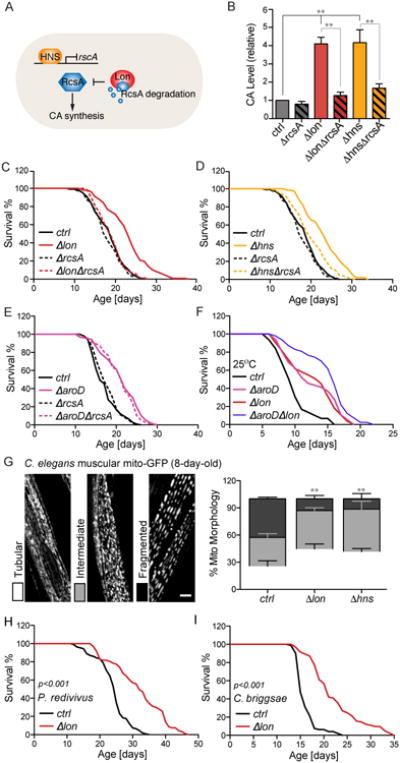
(A) Schematic depiction of biosynthesis of colanic acid (CA) in bacteria. RcsA, a positive activator required for CA biosynthesis, can be transcriptionally repressed by H-NS and degraded by the Lon protease.
(B) Both Δlon and Δhns show increased CA secretion in the culture medium, which are completely suppressed by the deletion of rcsA (**p<0.01, one-way ANOVA followed by Bonferroni's multiple comparison test). Error bars represent SEM.
(C and D) The ΔrcsA deletion suppresses the lifespan extension conferred by Δlon (C) and Δhns (D) (p<0.001, double mutants vs. corresponding single mutants, Log-rank test). Experiments were conducted in parallel with same parental controls and ΔrcsA mutants.
(E) The ΔrcsA deletion does not affect the lifespan extension conferred by ΔaroD (p>0.05, ΔaroDΔrcsA vs. ΔaroD, Log-rank test).
(F) Δlon further enhances the lifespan extension conferred by ΔaroD (p<0.05, ΔaroDΔlon vs. ΔaroD, Log-rank test).
(G) Both Δlon and Δhns mutants significantly lower the levels of mitochondrial fragmentation in the transgenic C. elegans expressing mito-GFP in body wall muscles (raxIs[myo-3p∷mitoGFP]), visualized at day-8 adulthood. Different mitochondrial morphologies were classified as tubular, intermediate and fragmented (representative images are shown, scale bar = 10μm), and quantified double-blindly. Error bars represent SEM; **p<0.01 bacterial mutant vs. parental control by Student's t-test.
(H and I) The Dlon bacterial mutant significantly prolongs the lifespan of P. redivivus (H) and C. briggsae (I) (p<0.001, Log-rank test).
See Figure S4 for information on additional CA-overproducing mutants, and Table S6 for lifespan data including animal numbers, mean and median lifespans, and statistics.
In addition to prolonging lifespan, these CA-overproducing bacterial mutants improve C. elegans tissue maintenance during aging. Mitochondrial dysfunction in body wall muscles is a hallmark of somatic aging, which is associated with increased mitochondrial fragmentation (Regmi et al., 2014). We found that the CA-overproducing bacterial mutants attenuate this age-associated muscular mitochondrial fragmentation (Figure 4G, Figure S4E). We further showed that the lifespan-extending effect of the Δlon mutant is evolutionarily conserved in nematode species that have diverged from C. elegans more than 100 million years ago, including Panagrellus redivivus (Figure 4H, Table S6) and C. briggsae (Figure 4I, Table S6).
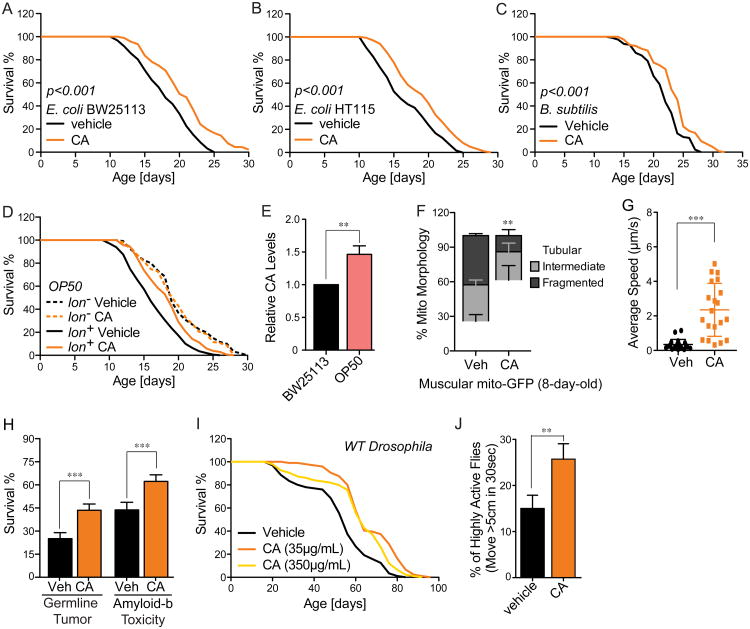
Purified CA sufficiently promotes host longevity
In order to directly evaluate the efficacy of CA in promoting host longevity, we supplemented purified CA to wild-type C. elegans. Strikingly, CA supplementation sufficiently prolonged the lifespan of C. elegans grown on E. coli K-12 BW25113 (Figure 5A, Table S7). CA also extended C. elegans lifespan when supplemented together with a diverse range of bacteria, including another E. coli K-12 strain, HT115 (Figure 5B, Table S7), and gram-positive Bacillus subtilis (Figure 5C, Table S7). In contrast, when supplemented with a commonly used laboratory bacterial strain, E. coli OP50, CA lost its lifespan-extending effect (Figure 5D, Table S7). Interestingly, as a B-type E. coli, OP50 is naturally lon deficient, and produces more CA than the K-12 strain (Figure 5E). When we restored lon in OP50, worms exhibited reduced lifespan that was restored upon CA supplementation (Figure 5D, Table S7). These observations on E. coli OP50 suggest that microbial lon deficiency and dietary CA supplementation are both effective to promote host longevity, and likely act through the same mechanism.
Figure 5. Purified colanic acid promotes host longevity across species.
(A to C) CA significantly prolongs the lifespan of wild-type C. elegans when supplemented with gram-negative E. coli BW25113 (A) and HT115 (B), and with gram-positive B. subtilis (C) (p<0.001, CA vs. vehicle control (ddH2O), Log-rank test).
(D) CA supplementation does not prolong the lifespan of C. elegans grown on lon-deficient E. coli B OP50 (p>0.5, Log-rank test), but resumes its effects when lon expression is restored in OP50 (p<0.001, Log-rank test).
(E) E. coli OP50 produces 50% more CA than E. coli BW25113 (**p<0.01, Student's t-test).
(F) Muscular cells of 8-day-old C. elegans with CA supplementation display significantly increased levels of tubular mitochondrial morphology, compared to those on parental control bacteria (**p<0.01, Student's t-test).
(G) CA supplementation increases locomotion velocity of aged C. elegans, measured at day 15 of adulthood (***p<0.0001, non-parametric t-test).
(H) CA supplementation increases the survival of the glp-1(ar202) tumorous mutants and the transgenic strains expressing human Aβ (***p<0.001, Log-rank test).
(I) CA supplementation during adulthood at different concentrations significantly prolongs the lifespan of wild-type Drosophila OreR males (p<0.05, CA vs. vehicle control, Log-rank test).
(J) 15 days old Drosophila adults supplemented with CA have increased mobility, compared to the vehicle controls (**p<0.01, non-parametric t-test).
Error bars represent SEM. See Table S7 for detailed lifespan data including animal numbers, mean and median lifespans and statistics.
CA supplementation also delayed the onset of muscular mitochondrial fragmentation (Figure 5F), and consequently improved locomotion activities of aged C. elegans (Figure 5G), and increased the survival of C. elegans carrying germline tumor or toxic human Aβ (Figure 5H). Intriguingly, we found that adult-only dietary supplementation of CA substantially increases the lifespan of wild-type Drosophila melanogaster (Figure 5I) and improves their spontaneous activities (Figure 5J), supporting a well-conserved longevity effect of CA across evolutionarily distant hosts.
Direct impact of CA polymers on the host but not bacteria
The CA polymer contains a repeating unit of glucose, galactose, fucose, and glucuronic acid, which are further decorated by pyruvate and acetate (Sutherland, 1969) (Figure 6A). After being synthesized in the cytoplasm, CA repeating units are transported to periplasm and linked together by the CA polymerase, WcaD (Stevenson et al., 1996). When we supplemented C. elegans with the six monomers of CA, either separately or in combination, no lifespan extension was detected (Figure S5A). Moreover, neither the ΔlonΔwcaD mutant nor extract from this double mutant prolonged C. elegans lifespan (Figure 6B, Figure S5B, Tables S6 and S7). Therefore, the monomers or the single repeating units of CA are not sufficient to induce host longevity. Gel permeation chromatography analysis determined that the molecular weight of purified CA is approximately 3.4 kDa (Figure 6C), suggesting that CA polymers with 3 or more repeating units are required for their longevity effects. Moreover, supplementation of two other extracellular polysaccharides, hyaluronic acid and alginic acid did not extend C. elegans lifespan (Figure S5C), supporting the specificity of CA as a longevity-promoting polysaccharide.
Figure 6. Polymer forms of CA act directly on the host to promote longevity.
(A) Schematic depiction of CA polymerization in bacteria. RcsA activates the CA biosynthesis gene cluster. The CA unit contains 6 monomers, glucose (Glc), galactose (Gal), fucose (Fuc), glucuronic acid (GlcA), pyruvate (Pyr), and acetate (Ac), and is polymerized by WcaD in the periplasm.
(B) Inhibition of CA polymerization by the bacterial ΔwcaD deletion fully suppresses the lifespan extension of C. elegans induced by the Δlon bacterial mutant (p>0.05, ΔwcaDΔlon vs. ΔwcaD, Log-rank test).
(C) Gel permeation chromatography analysis determines the molecular weight of purified CA as ∼3.4kDa. Peaks after 19mL of retention volume are derived from the solvent.
(D) CA does not reduce bacterial growth. The parental control E. coli BW25113 supplemented with water or CA were grown on worm standard plates at 20°C, and collected at designated time points to measure OD600 (*** p<0.001 CA vs. vehicle, Student's t-test). Error bars represent SEM.
(E) The colonization of bacteria in the C. elegans gut is not affected by CA supplementation (p>0.5, Student's t-test). Error bars represent SEM.
(F and G) CA supplementation does not affect the survival of C. elegans infected by Pseudomonas aeruginosa PA14 (F) or Enterococcus faecalis (G) (p>0.5, Log-rank test).
(H) Expressions of five pathogenic response genes are not affected by CA supplementation (n.s. p>0.5, Student's t-test). Error bars represent standard deviation.
(I and J) When supplemented to bacteria killed by UV (I) or inactivated by chloramphenicol (J), CA is still sufficient to prolong lifespan (p<0.001, CA vs. vehicle control, Log-rank test). Experiments were conducted in parallel with same live-bacteria controls (vehicle and CA treated).
See Figure S5 for lifespan data on CA monomers/unit, other polysaccharides, and CA with endocytosis mutants, and Table S6 and Table S7 for lifespan data including animal numbers, mean and median lifespans, and statistics.
Next, to test whether CA increases the survival of C. elegans by reducing bacterial growth or pathogenicity, we monitored the growth of bacteria treated with CA. Interestingly, instead of inhibiting growth, CA had a slight growth-stimulating effect after the stationary phase (Figure 6D). Furthermore, with CA supplementation, the capacity of bacteria colonized in the C. elegans gut was not reduced (Figure 6E). In addition, CA neither attenuated the lethality caused by pathogenic bacteria, Pseudomonas aeruginosa and Enterococcus faecalis (Figure 6F and G), nor affected the expression of several well-known pathogenic responsive genes (Figure 6H). Therefore, the longevity-promoting effect of CA does not result from inhibiting bacterial growth or pathogenicity.
To determine whether live bacteria mediate the beneficial effect of CA on the host, we killed bacteria with ultraviolet radiation or inactivated them with chloramphenicol before CA supplementation. We found that CA remains its lifespan-extending capacity when supplemented with dead bacteria, and the magnitude of lifespan extension is unaffected (Figure 6I and J, Table S7). Furthermore, we discovered that host endocytotic genes, rab-5 and rab-7 (Grant et al., 2001) are required for CA to prolong lifespan (Figure S5D-F, Table S7). Together, these results show that CA alone is sufficient to exert its beneficial effects, and requires endocytosis-mediated uptake into host intestinal cells.
CA acts on host mitochondria to promote longevity
To further investigate the molecular mechanism underlying the longevity effect of CA, we examined the lifespan of various C. elegans mutants raised on the CA-overproducing Δlon bacterial mutant. Similar to the daf-16, rict-1, rsks-1, raga-1 and eat-2 mutants (Figure 3), the hypoxia-inducible factor mutant, hif-1(ia4) (Leiser and Kaeberlein, 2010), the NRF2 mutant, skn-1(zu135) (Tullet et al., 2008), and the mitophagy mutant, pink-1(ok3538) (Palikaras et al., 2015) all exhibit significant lifespan extension induced by Δlon (p<0.001, Figure 7A, Table S6), suggesting their negligible roles in regulating CA-induced longevity. In contrast, the Δlon bacterial mutant was unable to further prolong the lifespan of the nuo-6(qm200) or isp-1(qm150) C. elegans mutants (Figure 7A, Table S6), neither did the CA supplementation (Figure 7B and C, Table S7). Interestingly, both of these C. elegans mutants are deficient in the mitochondrial electron transport chain (ETC) (Feng et al., 2001; Ishii et al., 1998; Yang and Hekimi, 2010), suggesting host mitochondria as essential cellular targets of CA.
Figure 7. CA acts on host mitochondria to promote longevity.
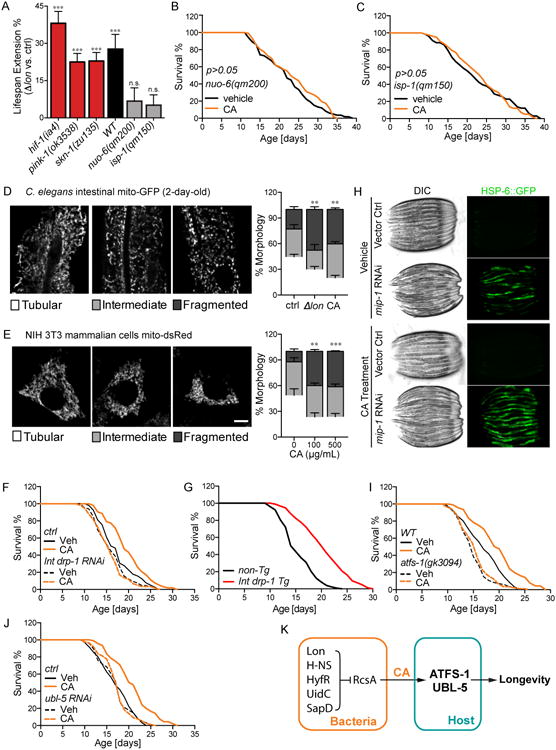
(A) The Δlon bacterial mutant extends the lifespan of C. elegans hif-1(ia4), skn-1(zu135), and pink-1(ok3538) mutants, but is not able to do so in the mutants of C. elegans ETC components, nuo-6(qm200) or isp-1(qm15) (***p<0.001, n.s. p>0.05, Δlon vs. parental control, Log-rank test). Error bars represent SEM.
(B and C) CA supplementation cannot further increase the lifespan extension conferred by the mitochondrial ETC mutations, nuo-6(qm200) (B) and isp-1(qm150) (C) (p>0.05, CA vs. vehicle control, Log-rank test).
(D) Both the Δlon mutant and CA supplementation increase mitochondrial fragmentation in C. elegans intestinal cells (**p<0.01, Student's t-test). The transgenic strain expressing mito-GFP in the intestine (raxIs[ges-1p∷mitoGFP]) was analyzed at day-2 adulthood. As shown in representative images, mitochondrial morphologies are categorized as either tubular, intermediate, or fragmented. Scale bar = 10μm. Error bars represent SEM.
(E) CA increases mitochondrial fragmentation in mammalian cells (**p<0.01, ***p<0.001, Student's t-test). Mitochondrial morphologies were analyzed in the NIH/3T3 mammalian cells stably expressing dsRed2-mito, categorized as either tubular, intermediate, or fragmented. Representative images are shown, with scale bar = 10μm. Error bars represent SEM.
(F) Intestinal-specific knockdown of drp-1 by RNAi fully suppresses the lifespan extension conferred by CA (p>0.05, CA vs. vehicle control (drp-1 RNAi), Log-rank test).
(G) Intestinal-specific overexpression of drp-1 is sufficient to increase the lifespan of C. elegans (p<0.001 compared to non-transgenic siblings by Log-rank test).
(H) The level of mitochondrial chaperone HSP-6∷GFP was used to measure UPRmt. Inactivation of mitochondrial intermediate peptidase, mip-1 (Y67H2A.7) by RNAi induces the HSP-6∷GFP level, which is further elevated by CA supplementation.
(I) In the loss-of-function mutant of atfs-1(gk3094), the lifespan extension conferred by CA supplementation is fully suppressed (p>0.05, CA vs. vehicle control (atfs-1), Log-rank test).
(J) RNAi knockdown of ubl-5 completely abrogates the lifespan-extending effect of CA (p>0.05, CA vs. vehicle control (ubl-5 RNAi), Log-rank test).
(K) A diagram of CA-mediated bacteria-host communication in regulating host longevity.
See Figure S6 for additional information of CA effects on mitochondria, lipid storage and proteasome, and Table S6 and Table S7 for lifespan data including animal numbers, mean and median lifespans, and statistics.
Mitochondria are highly dynamic organelles, and their architecture is shaped by a balance between fusion and fission (Chan, 2006). Although the Δlon bacterial mutant did not alter mitochondrial DNA levels, ATP production, or lipid storage (Figure S6A-C), we observed that bacterial lon deficiency and CA supplementation significantly increase mitochondrial fragmentation in the intestine of C. elegans (Figure 7D). Remarkably, mammalian cells treated with CA also display a more fragmented mitochondrial network (Figure 7E), suggesting a highly conserved function of CA in regulating mitochondrial dynamics. Recent studies suggest an association between increased proteasome activity and mitochondrial fragmentation in regulating longevity (Yao, et al., 2015). However, we found that CA supplementation has no effect on proteasome activity (Figure S6D-F). Next, we examined whether changes of mitochondrial dynamics in the C. elegans intestine conferred by CA supplementation are important for its longevity effects. In C. elegans, drp-1 encodes the homolog of yeast Dnm1p and mammalian DRP1 that are required for mitochondrial fission (Breckenridge et al., 2008), and fzo-1 encodes mitofusin required for mitochondrial fusion (Rolland et al., 2009). We found that intestine-specific knockdown of drp-1 by RNAi completely abrogates CA-induced lifespan extension (Figure 7F), while intestine-specific knockdown of fzo-1 does not show such an effect (Figure S6G), suggesting that induction of mitochondrial fission, but not fusion, in the host gut is responsible for CA-induced longevity. Furthermore, mild induction of intestinal mitochondrial fragmentation by tissue-specific overexpression of drp-1 sufficiently increased C. elegans lifespan (Figure 7G).
In addition to its effects on mitochondrial dynamics, we found that CA also modulates mitochondrial unfolded protein responses (UPRmt) under stress conditions. UPRmt has previously been linked to mitochondrial homeostasis and longevity, which can be denoted by induced expression of C. elegans mitochondrial chaperone HSP-6 (Pellegrino et al., 2013). When protein folding in mitochondria is comprised by inactivation of the mitochondrial intermediate peptidase gene, mip-1(Y67H2A.7), CA supplementation significantly exaggerated the induction of HSP-6 (Figure 7H), suggesting enhanced UPRmt by CA under mitochondrial stress conditions. However, in absence of mitochondrial stress, we found no noticeable induction of UPRmt by CA (Fig. 7H), suggesting that CA does not directly disrupt mitochondrial protein-folding homeostasis.
ATFS-1 is a key transcription factor that regulates UPRmt and forms a complex with UBL-5 and DVE-1 to induce transcription of mitochondrial chaperones (Andreux et al., 2013). We found that the loss-of-function atfs-1(gk3094) mutant completely suppresses the lifespan-extending effect of CA (Figure 7I, Table S7). Similarly, RNAi knockdown of ubl-5 also fully abrogated CA-induced longevity (Figure 7J, Table S7). These results reveal that the effect of bacterial CA on host longevity requires the UPRmt signaling pathway (Figure 7K).
Discussion
Our studies reveal the significance of microbial genetic variations in modulating host longevity. The microbiota not only comprise diverse microbial species but also exhibit profound genetic heterogeneity within each species. Genetic drift frequently occurs in microbes, and microbial gene expression is dynamically influenced by environmental stimuli in the host gut. In a model organism C. elegans, we systematically identified bacterial genes whose deletion leads to lifespan extension, and revealed the potential of a dozen of these bacterial mutants in attenuating lethality due to age-associated progression of germline tumor and accumulation of pathological Aβ. Interestingly, these identified bacterial genes are highly heterogeneous in their functions spanning many aspects of bacterial morphology and physiology (Figure 1), which implies a number of discrete mechanisms that await individual characterization.
Using genetic and biochemical analyses, we discovered overproducing CA polysaccharide as a longevity-promoting mechanism. In parallel, functional gene ontology analysis on our screening hits reveals the enrichment of genes involved in chorismate metabolism (Figure S7). This molecule is a biochemical precursor for many aromatic metabolites (Bentley, 1990). Synthetic deficiencies in two of the chorismate derivatives, ubiquinone and folate, underlie the prolonged host lifespan by the E. coli mutants previously identified (Saiki et al., 2008; Virk et al., 2012). A life-prolonging drug metformin exerts its beneficial effects by interfering with bacterial methionine biosynthesis, which is also coupled with chorismate metabolism (Cabreiro et al., 2013). Thus manipulating chorismate biosynthesis in intestinal microbes may be a promising strategy to promote host longevity. Our results also demonstrate that microbial CA overproduction and chorismate deficiency function in parallel pathways to promote host longevity and exert additive effects when combined.
The majority of our identified bacterial genes sustain their abilities in influencing host longevity when deleted in distinct bacterial backgrounds, and do not require presence during C. elegans development to exert their longevity-promoting effects. Therefore, our studies mark their probable merits in taking effects across diverse gut inhabitants and for late-life intervention. In addition, these bacterial mutants interact with one or more longevity regulatory mechanisms in the host, including insulin/IGF-1 signaling, TOR signaling, caloric restriction, and mitochondrial hormesis, suggesting that altering bacterial genetic composition could be an effective way to activate major host longevity signaling and promote healthy aging. On the other hand, mechanistic dissection of each longevity-promoting bacterial mutant may highlight specific molecules that can directly act on the host, such as CA. Effective in several host-microbe environments and pursuant further tests in mammalian systems, CA may provide an exciting approach for improving human health and longevity.
Mitochondria, which have been linked to aging processes, are not only the metabolic center of the cell, but also key signaling organelles (Riera and Dillin, 2015). Mitochondrial hormesis in the nervous system has been previously shown to generate systemic signals and exert beneficial effects cell non-autonomously in distal tissues (Durieux et al., 2011). Our studies show that CA enhances UPRmt and triggers mitochondrial fragmentation in the intestine, and prolongs host lifespan via interacting with mitochondrial ETC complexes. These different impacts of CA on mitochondria can be integrated by the action of ATFS-1, as supported by previous studies showing that reduction in mitochondrial ETC components causes a strong activation of UPRmt (Durieux et al., 2011; Houtkooper et al., 2013), that the activation of ATFS-1 by UPRmt suppresses the expression of ETC genes in both the nuclear and mitochondrial genomes (Nargund et al., 2015), and that ATFS-1 induces the expression of the mitochondrial fission protein DRP-1 (Nargund et al., 2015). Our studies demonstrate ATFS-1 as a key mediator of the CA longevity effect. Therefore, the intestine is another key tissue for mitochondrial signals to regulate the rate of aging systemically, and CA can trigger this mitochondrial signaling.
The discovery of CA's modes of action reveals an intriguing crosstalk between mitochondria and symbiotic bacteria in animal hosts. Mitochondria evolved from and share many metabolic pathways with bacteria (Andersson et al., 1998). Conceptually, the microbiota may influence the host similarly as mitochondria do, by chemically communicating with their ancient intra-cellular relatives. CA is a case in point, serving as a messenger for this type of dialogue. Synthesized in bulk upon environmental stress (Chen et al., 2004), bacterial CA induces mitochondrial fragmentation and UPRmt, which consequently generates systemic signals to improve host fitness. In this sense, animals can perceive stress-associated information from their gut microbiota, and get physiologically ready for upcoming unfavorable conditions, likely a result of coevolution. Empirically, future investigations on bacteria-mitochondria communications might open up new avenues for treating human mitochondrial diseases and aging.
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the corresponding author, Meng C. Wang (wmeng@bcm.edu).
Experimental Model and Subject Details
Animals
The nematodes C. elegans, C. briggsae, and P. redivivus were mostly obtained from the Caenorhabditis Genome Center. Some nematode strains were gifts from other labs or generated in the lab. The nematodes were maintained and experimentally examined at 20°C on standard nematode growth medium (NGM) agar plates seeded with pre-cultured bacterial strains unless otherwise noted. Hermaphrodites of C. elegans and C. briggsae were age-synchronized by isolating eggs as previously described (Sulston and Brenner, 1974). Female P. redivivus were synchronized by collecting newly produced larvae within a time window of 2 hours. The Drosophila melanogaster OreR strain was reared and assayed at 25°C with a 12h:12h light-dark cycle. During the experiments, the flies were allowed to develop and mate for two days after eclosion, and then were sexually segregated. In all cases, individual animals were assigned randomly into different experimental groups.
Microbe strains
For primary screens, E. coli Keio mutants were grown overnight at 37°C in Luria-Broth (LB) medium added with 10 μg/mL kanamycin. 150μL of the bacteria culture was seeded to each well of 12-well NGM plates. For longitudinal assays, E. coli were cultivated in LB medium at 37°C for 9 hours. Centrifuged bacteria pellets were washed using M9 buffer for three times, and re-suspended in M9 buffer to a concentration whose 20-fold dilution exhibits an OD600 between 0.15 and 0.16. A microscopic counting chamber was used to confirm the variations of bacterial cell numbers less than 10%. 350μL of the bacteria suspension was seeded on each 6cm NGM plate.
For lifespan assays with CA supplementation, E. coli, B. subtilis, and P. aeruginosa were cultured in LB medium overnight while E. faecalis was grown in Brain-heart infusion medium (BHI) for 5 hours. The bacteria culture was then mixed with half volume of either CA solution or ddH2O as control before seeded onto plates. NGM plates were used for E. coli and B. subtilis, slow-killing (SK) plates were used for P. aeruginosa, and BHI plates with gentamicin were used for E. faecalis. The NGM plates were incubated at 20°C for 2 days while both the SK and BHI plates were incubated at 37°C overnight.
For RNAi knockdown of selected worm genes, bacteria from the Ahringer collection were grown at 37°C for 8 hours in LB medium with 50 μg/mL carbenicillin and directly seeded onto NGM plates. 5 mM IPTG was used to induce dsRNA expression overnight at room temperature. All prepared plates were stored at 4°C till subsequent usage.
Cell lines
Mouse NIH/3T3 cells stably expressing dsRed2-fused mitochondrial targeting sequence (Mito-dsRed2) was a gift from the M. A. Frohman laboratory. The cells were cultured in Dulbecco's modified Eagle's Medium (Hylcone) supplemented with 10% Calf Serum (Hyclone) and maintained in 37°C with 5% CO2.
Method Details
Genomic screen
The sqt-3(e2117) temperature-sensitive mutant of C. elegans was utilized in the primary stage of the screening. These worms were kept at 15°C during embryonic and larval development, and assayed at 25°C as adults. The average time when their survival rate on E. coli BW25113 decreases to 10% was experimentally determined, which is on day-13 of adulthood. We used this time point to examine the survival of all the screening candidates. Positive hits with an increased survival rate were tested longitudinally first in the sqt-3 mutant background, and then in wild-type worms (N2) at 20°C. Introduction of each candidate bacterial mutation into the parental strain as well as another E. coli strain MG1655 was performed, to confirm their lifespan-extending effects. The experimental design of the screen and the working flow are illustrated in Figure S1.
Bacterial growth and colonization analyses
For growth assay in liquid medium, bacteria of a single colony were inoculated in LB and grown at 37°C. OD600 was determined using LB medium as the blank control. For a plate assay, 30μL of LB cultured bacteria at OD600=0.4 were seeded on each NGM plate, and were allowed to grow at 20°C. The bacteria were thoroughly washed off the plates with 1mL M9 buffer and OD600 was measured, with M9 buffer as the blank control.
For a viability assay, bacteria from seeded NGM plates (stored at 4°C) were washed off the plates with adequate M9 buffer till OD600 reaches below 0.1. The numbers of total bacterial cells were determined using a counting chamber (Hausser Scientific). The numbers of total live bacteria were determined based on colony forming unit on LB plates as described below. The percentage of living bacteria was then calculated.
To measure colonizing capacity of the bacteria in C. elegans, the nematodes were washed off the NGM plates at the selected age, and extensively rinsed with M9 buffer. The animals were then put on empty NGM plates with 100 μg/mL ampicillin for 1 hour. Using sterile techniques, 5 worms were individually picked into M9 buffer and homogenized. Part or all of the mixture was then plated onto LB plates. After incubated at 37°C overnight, the number of bacterial colonies were determined.
P1 phage transduction
Wild-type bacteriophage P1 was cultured overnight at 37°C together with the donor bacteria in 0.5×LB agar to lysate the cells, and purification was done using 5% (v/v) chloroform. In case of taking a Keio mutant as the recipient cell, its kanamycin-resistant cassette was first removed using electro-transformation of plasmid pCP20 that contains the FLP recombinase. The phage lysate was incubated with the recipient cells at 32°C in LB medium containing 6mM sodium citrate before plated onto LB-citrate plates. The resulting mutants due to homologous recombination were obtained after sequential streaking on LB-citrate plates to remove phages, using kanamycin as a selection marker.
Restoration of lon functionality
For lifespan assays on OP50 expressing lon, the full-length lon gene was cloned into the plasmid pCA24N, which was used to transform E. coli OP50. The empty plasmid vector was used as the control. The successful insertion was confirmed by PCR. The resulting bacteria were cultured in LB medium at 37°C till the OD600 reached 0.6, and then seeded onto NGM plates with 1mM IPTG for induction. The plates were then incubated at 37°C for 24 hours before used for lifespan experiments.
Quantification of CA
Adapted methods were used to quantitatively compare CA production among bacterial strains (Dische and Shettles, 1948). For most E. coli strains, the bacteria was grown in M9 minimal medium at 37°C for 8 hours and at 20°C for 2 days. After centrifugation at 10,000 rpm for 20 minutes, the supernatant was considered as the assay sample. For the strains that cannot be grown in the M9 medium, including OP50 and a few auxotrophic Keio mutants, an alternative method was used to measure their secreted CA. In brief, the bacteria were cultivated in LB medium at 20°C overnight. After centrifugation to remove bacterial cells, the liquid medium was further centrifuged at 15,000 rpm for 30 minutes at 4°C. Equal volume of ethanol was then mixed with the supernatant, and put in 4°C overnight. Centrifugation was performed again to collect the pellet, which was then rinsed with cold 70% ethanol for 3 times and air dried. The pellet from every 5 mL culture was dissolved in 1 mL ddH2O as the assay sample. The assay samples were then quantified colormetrically. Briefly, 30 μL sample was mixed with 1 mL sulfuric/water solution (6:1 v/v) and boiled for 20 minutes. After cooling, the absorbance difference at 396nm and 430nm, ΔOD396-430, was measured using a spectrophotometer (BioTek). Cysteine hydrochloride was added to a final concentration of 3.5mg/mL, and ΔOD396-430 was measured again. The difference between these two values is proportional to the fucose concentration, and therefore the CA contents.
Purification and administration of CA
Adapted methods were used to purify CA polysaccharide from bacterial culture (Dische and Shettles, 1948). In brief, the Δlon mutant was grown in M9 minimal medium at 37°C overnight and at 20°C for 2 days. After heated in boiling water for 5 minutes, cells were removed by centrifugation and the supernatant was lyophilized. Every 5 mg of the resulting solid was dissolved in 1 mL water and centrifuged at 15,000 rpm for 20 minutes at 4°C. The supernatant was mixed with 1% (v/v) glacial acetic acid, boiled for 2 hours, and centrifuged at 15,000 rpm for 30 minutes at 4°C to precipitate lipopolysaccharides. The supernatant was then mixed with equal volume of chloroform/methanol (2:1 v/v) to eliminate proteins. 10K daltons dialysis cassettes (Thermo Scientific) were used to dialyze the aqueous phase against water at 4°C for 2 days. Solid CA was obtained after lyophilization, and sterilized under ultraviolet light for 2 hours. Purified CA was quantified as described above, after dissolved in water in a concentration of 1 mg/mL, with fucose solutions up to 100 μg/mL as standards.
For dietary supplementation of CA to worms, purified CA was dissolved in ddH2O, and mixed with bacteria before seeding. Bacteria mixed with the same volume of ddH2O were used as the vehicle control. For supplementation of CA to flies, highly concentrated CA solution was thoroughly mixed with fly cornmeal at 70°C. Same volume of ddH2O were added as the vehicle control.
Lifespan and paralysis assays
Nematodes were age-synchronized as described above. Adults were kept separated from their offsprings by manually transferring to new rearing settings as necessary. Survival of the nematodes was examined on a daily basis. Death was scored by not responding to gentle mechanical prodding. Post-reproductive animals that are lost accidentally, dead of internal hatching or extruded organs were censored. The physical paralysis time of the Aβ transgenic C. elegans was determined by observing their movement every day and analyzed in a similar manner. For lifespan analysis of the flies, animals were reared on the standard food till sexual segregation, and then were supplemented with CA, and kept transferred to new food every 4 days. Death was scored when a fly fails to move. Kaplan-Meier curves were analyzed using a Log-rank test.
Developmental time and brood size analyses
To determine the developmental time of C. elegans, 10 synchronized L1 worms were allowed to develop at 20°C. The animals were examined every 30 minutes to note their transition to L4, and the lapsed time from L1 to L4 was recorded for each individual. To assess the brood size, 10 synchronized L4 hermaphrodites were grown at 20°C, and kept transferred to fresh NGM plates every day. The number of progeny was counted until reproductive cessation, and the total number of progeny per individual was calculated. The experiments were performed for three times independently, and analyzed using a Student's t-test.
Pharyngeal pumping, defecation cycle, and locomotion activity measurement
C. elegans on day-1 adulthood were monitored using an SMZ1500 stereo microscope (Nikon). The number of pharyngeal contractions in a single minute interval was counted. The time between the same phase of two consecutive defecations were also determined. Each randomly selected animal was measured for 3 times. The mean values of 10 animals were compared with the control using a Student's t-test. A C11440 camera (Hamamatsu) connected to the microscope was used to record the spontaneous movement of the worms for one minute. Individual worms were tracked using NIS Elements AR imaging software (Nikon) and the average moving speed was calculated. Speeds of 20 animals were compared to the control using a Student's t-test.
Gel permeation chromatography analysis
Gel permeation chromatography was performed using an Agilent 1260 Infinity chromatography system equipped with PLaquagel-OH Mixed (300 × 7.5 mm) and PL1120-6830 (300 × 7.5 mm) column set, 0.1 M NaNO3 + 0.05% NaN3 aqueous solution as eluent at 1.50 mL/min flow rate. Refractive index, right angle light scattering (RALS) and Low Angle Light Scattering (LALS) signals were acquired using Viscotek TDA 302 Triple Detection Array system. Both chromatography and the signal acquisition were performed at 40°C. Purified CA was dissolved in eluent at a concentration of 10.0 mg/mL and 100 μL of this solution were used for analysis. Signals were recorded and processed using a Malvern OmniSEC software. Assuming a typical dn/dc value of 0.147 for polysaccharides, the molecular weight of the purified CA was calculated.
Quantitative PCR analyses
For gene expression analysis, cDNA was synthesized using a Power SYBR Green Cells-to-Ct Kit (ThermoFisher Scientific) from lysate of 30 age-synchronized young adult worms. For mitochondrial content analysis, total DNA was released from worms lysed with proteinase K, followed by heat inactivation (Williams et al., 1992). Quantitative PCR was performed using a Kapa SYBR fast PCR kit (Kapa Biosystems) in an Eppendorf Realplex 4 PCR machine (Eppendorf). Values were normalized to those of rpl-32 and ant-1.3 as the internal control in gene expression and mitochondrial content analyses, respectively. All data shown represent three biologically independent samples. Statistical analysis was performed using a Student's t-test.
ATP quantification
∼3,000 age-synchronized worms were washed off the NGM plates as young adults, and extensively rinsed with M9 buffer to eliminate ingested bacteria. The worms were then sonicated in 500μL 6M guanidine hydrochloride in extraction buffer (100mM Tris, 4mM EDTA, pH 7.8), flash frozen in liquid nitrogen, and heated in boiling water for 2 minutes. After cooling and centrifuged at 13,000 rpm for 5 minutes, the supernatant was diluted 1000 times with the extraction buffer. An ATP determination kit (Invitrogen) was used to quantify ATP in the samples, based requirement of luciferase for ATP in emitting light. With up to 1 μM ATP solutions provided by the manufacturer as the standards, the ATP concentration was determined using a luminometer (Berthold). The ATP contents were then normalized to the total protein concentration, which was determined by a protein assay (Thermo Scientific). ATP was quantified in four independent samples, and compared between groups using a Student's t-test.
Quantification of peptidase activities of the proteasome
∼500 age-synchronized worms were washed off the NGM plates as young adults using M9 buffer. After rinsed with 1 mL washing buffer (50mM Tris-HCl, pH7.5, 250mM sucrose, 5mM MgCl2) for 3 times and 500 μL lysis buffer (50mM Tris-HCl, pH7.5, 250mM sucrose, 5mM MgCl2, 0.5mM EDTA, 2mM ATP, 1mM DTT) for 1 additional time, the worms were homogenized in 100 μL ice-cold lysis buffer. After centrifuged at 8,000 rpm for 15 minutes at 4°C, the protein content in the supernatant was determined using a protein assay (Thermo Scientific). The fluorogenic proteasomal substrates (Bachem) were diluted in assay buffer (50mM Tris-HCl, pH7.5, 40mM KCl, 5mM MgCl2, 0.5mM ATP, 1mM DTT, 0.05mg/mL BSA): Suc-LLVY-AMC and Ac-RLR-AMC in 100 μM, while Ac-nLPnLD-AMC in 300 μM. 200 μL substrate was added into each well of a 96-well microtitre plate, together with lysate containing 50 μg total protein. After incubated at room temperature for 5 minutes, the fluorescence (excitation/emission: 380/460nm) was monitored using a fluorometer every 5 minutes for 1 hour. The slope representing relative proteasome activity was determined in three independent trials, and compared between groups using Student's t-test.
Mitochondrial morphology assays and stimulated Raman scattering microscopy
Adult C. elegans at desired ages were individually picked into 0.1% sodium azide dropped on glass coverslips. The coverslips were then mounted onto pads of 2% agorose on microscope slides. Mouse NIH/3T3 cells stably expressing dsRed2-fused mitochondrial targeting sequence (dsRed2-Mito) were cultured in Dulbecco's modified Eagle's Medium (Hylcone) supplemented with 10% Calf Serum (Hyclone) and maintained in 37°C with 5% CO2. Cells grown on glass coverslips were treated overnight with 0, 100, and 500 μg/mL of CA, respectively, and fixed with 4% paraformaldehyde in cytoskeleton stabilizing buffer (10mM PIPES, 150mM NaCl, 5mM EGTA, 5mM glucose, pH 6.8) for 10 minutes. Coverslips were washed with cytoskeleton stabilizing buffer for three times and mounted onto microscope slides using 4% n-propyl gallate. The slides were visualized under a confocal fluorescence microscope (Olympus) for mitochondrial morphology, or imaged under a Stimulated Raman Scattering (SRS) microscope (Ramachandran et al., 2015; Wang et al., 2011) for determining fat storage levels.
Quantification and Statistical Analysis
All the data were obtained from three or four biologically independent replicates. The Log-rank test (Kaplan-Meier survival method) was used to analyze lifespan and paralysis data. The non-parametric t-test was used to analyze locomotion velocity. One-way ANOVA followed by Bonferroni's multiple comparison test was used to analyze CA levels in bacterial medium. For all the other quantifications, data were analyzed using a Student's t-test. In all cases, p values <0.05 was considered significant.
Supplementary Material
Figure S1. Characterization of parental E. coli strain and experimental design of genetic screens. Related to Figure 1
(A) Average times are 50.4 ± 1.2, 53.1 ± 0.9 and 51.2 ± 0.9 hours for wild type (WT) L1 larvae developing into L4 larvae when grown on E. coli strains BW25113, OP50 and HT115, respectively. p>0.05, Student's t-test.
(B)Total numbers of progeny from WT adult worms grown on E. coli strains BW25113 (307 ± 11), OP50 (292 ± 8) and HT115 (295 ± 15) are not significantly different. p>0.05, Student's t-test.
(C)Lifespans of WT worms grown on E. coli strains BW25113, OP50, and HT115 are not significantly different (p>0.05, Log-rank test). Mean lifespan of BW25113, 18.63 ± 0.31 days, n= 167; OP50, 19.32 ± 0.44 days, n=121; HT115, 17.29 ± 0.96 days, n=102.
(D)Schematic diagram of the procedures for the primary screen using the sqt-3(e2117)ts collagen mutants, which are embryonic lethal at 25°C non-permissive temperature. Various light colors represent different bacterial mutants from the single-gene knockout E. coli library.
(E) The working flow of primary screens and subsequent retests. The entire process includes: (1) three rounds of primary screening; (2) further longitudinal confirmation; (3) validation of longevity in WT worms at 20°C with controlled bacteria amount; (4) validation of annotated bacterial mutations after reintroduction; (5) testing sufficiency upon adult-only supplementation; and (6) universality testing in another bacterial strain background. Boxes enclose the total numbers of mutants scored positive along the procedures. Utilized C. elegans strains, bacterial strains, and the criteria for positive hits are listed at each step.
Error bars represent standard error of the mean (SEM).
Figure S2. Viability and host colonization capacity of the longevity-promoting bacterial mutants. Related to Figure 1
(A) Except ΔrecC, the bacterial mutants that prolong C. elegans lifespan do not exhibit decreased survival when provided to the host (p>0.05, Student's t-test).
(B and C) Longitudinal studies of colony forming in the WT C. elegans gut show limited alterations of colonization capacities by the pro-longevity bacterial mutations. Different degrees of shading delineate age of the C. elegans. * p<0.05 compared to the control strain BW25113, Student's t-test.
Error bars represent SEM.
Figure S3. Effects of longevity-promoting bacterial mutants on C. elegans feeding and defecation behaviors. Related to Figure 3
(A) Pharyngeal pumping rates were measured in day-1-old WT worms grown on the bacterial mutants that prolong lifespan.
(B) Defecation interval length was examined in day-1-old WT worms grown on the bacterial mutants that prolong lifespan.
(C) Pharyngeal pumping rates were measured in day-1-old eat-2(ad465) mutants grown on the bacterial mutants that prolong lifespan.
*p<0.05 compared to the control, Student's t-test; error bars represent SEM.
Figure S4. RcsA dependent CA production and beneficial effects conferred by three bacterial mutants. Related to Figure 4
(A) CA production is elevated in three bacterial mutants: ΔhyfR, ΔuidC and ΔsapD (**p<0.01, *p<0.05, one-way ANOVA compared to the parental control followed by Bonferroni's multiple comparison test).
(B-D) The deletion mutation of ΔrcsA abrogates host longevity conferred by ΔhyfR (B), ΔsapD (C), and ΔuidC (D) (p>0.05, ΔrcsA vs. the double mutants, Log-rank test). Experiments were conducted in parallel with same parental controls and ΔrcsA mutants.
(E) The CA-overproducing mutants of ΔhyfR, ΔuidC and ΔsapD all significantly lower the levels of age-associated muscular mitochondrial fragmentation in the transgenic C. elegans line (raxIs[myo-3p∷mitoGFP]) (**p<0.01 compared to the parental control, Student's t-test).
Error bars represent SEM.
Figure S5. Lifespan effects of CA monomers and other polysaccharides, and the requirement of host endocytosis for CA-induced longevity. Related to Figure 6
(A) Supplementation with CA monomers does not extend C. elegans lifespan (p>0.1, Log-rank test). Glc, glucose; Gal, galactose; Fuc, fucose; GlcA, glucuronic acid; Pyr, sodium pyruvate; Ac, acetic acid; Mix, the combination of all these monomers.
(B) Extracts were purified from the Δlon and the ΔlonΔwcaD bacterial mutants in parallel. Supplementation of Δlon extracts extends the lifespan of C. elegans by 15% (p<0.001, Log-rank test), while extracts from the ΔlonΔwcaD double mutant do not significantly affect C. elegans lifespan (p>0.5, Log-rank test).
(C) Supplementation with other polysaccharides—hyaluronic acid (HA) and alginic acid (AA)—does not extend the lifespan of C. elegans (p>0.1, Log-rank test).
(D-F) CA supplementation prolongs lifespan of WT C. elegans (D), but is failed to do so with inactivation of either rab-5 (E) or rab-7 (F) that blocks early endosome to late endosome or late endosome to lysosome fusion, respectively. p>0.5, CA vs. vehicle control, Log-rank test.
Figure S6. Effects of CA overproduction on C. elegans mitochondria and proteasome. Related to Figure 7
(A) Mitochondrial DNA amounts assessed using two mitochondrial genome-encoded genes, nduo-1 and ctb-1 show no difference between C. elegans on the parental control and Δlon mutant bacteria. Error bars represent standard deviation (s.d.), p>0.05, Student's t-test.
(B) C. elegans on the parental control and Δlon mutant bacteria have similar levels of ATP. Error bars represent s.d., p>0.05, Student's t-test.
(C) Total fat levels stored in the intestine were measured by stimulated Raman scattering (SRS) microscopy, and are similar between C. elegans on the parental control and Δlon mutant bacteria. Error bars represent s.d., p>0.05, Student's t-test.
(D-F) CA supplementation does not affect C. elegans proteasome functions assayed by chymotrypsin (D), trypsin (E) and caspase-like 3 (F) activities.
(G) Intestinal-specific knockdown of fzo-1 by RNAi does not affect the lifespan-extending effect of CA. p<0.001, CA vs. vehicle control (fzo-1 RNAi), Log-rank test.
Figure S7. A group of longevity-promoting E. coli mutants are involved in the biosynthesis pathway of chorismate and its derivatives. Related to Figure 1
Highlights.
Systematic analysis of longevity-promoting microbial genetic variations.
Colanic acid as a pro-longevity natural compound effective in different species.
Bacterial metabolites regulate host mitochondrial dynamics and UPRmit.
Acknowledgments
We thank H. Doebbler and A. Dervisefendic for laboratory support; C. Kenyon, C. Haynes, and H. Dierick for providing animal strains; J. J. Wang and D. A. Garsin for bacteria; M. A. Frohman for NIH/3T3 cells expressing dsRed2-mito; M. K. Estes for providing a luminometer; H. Dierick, J. Halliday, A. Kuspa, J. Petrosino, S. Rosenberg, and H. Zoghbi for critical reading the manuscript; and the members of the Wang laboratory for insightful discussions. Some worm strains were provided by CGC, which is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). Supported by NIH grants R01AG045183 (M. C. W.), R01AT009050 (M. C. W.), DP1DK113644 (M. C. W.), R01HL119478 (G. D), R01GM088653 (C. H.), R01GM115622 (J.W.), R01CA207701 (J.W.), and HHMI Faculty Scholar Award (M.C.W.).
Footnotes
Author Contributions: B. H., C. H., and M. C. W. conceived the project. B. H., P. S., C. J. L., I. A. A. N., J. H., L. W. R. T., J. N. S., A. S., G. D., J. W., C. H., and M. C. W. conducted the experiments. B. H., C. H. and M. C. W. wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Andersson SG, Zomorodipour A, Andersson JO, Sicheritz-Ponten T, Alsmark UC, Podowski RM, Naslund AK, Eriksson AS, Winkler HH, Kurland CG. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature. 1998;396:133–140. doi: 10.1038/24094. [DOI] [PubMed] [Google Scholar]
- Andreux PA, Houtkooper RH, Auwerx J. Pharmacological approaches to restore mitochondrial function. Nat Rev Drug Discov. 2013;12:465–483. doi: 10.1038/nrd4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Molecular systems biology. 2006;2:2006 0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentley R. The shikimate pathway--a metabolic tree with many branches. Critical reviews in biochemistry and molecular biology. 1990;25:307–384. doi: 10.3109/10409239009090615. [DOI] [PubMed] [Google Scholar]
- Breckenridge DG, Kang BH, Kokel D, Mitani S, Staehelin LA, Xue D. Caenorhabditis elegans drp-1 and fis-2 regulate distinct cell-death execution pathways downstream of ced-3 and independent of ced-9. Molecular cell. 2008;31:586–597. doi: 10.1016/j.molcel.2008.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabreiro F, Au C, Leung KY, Vergara-Irigaray N, Cocheme HM, Noori T, Weinkove D, Schuster E, Greene ND, Gems D. Metformin retards aging in C. elegans by altering microbial folate and methionine metabolism. Cell. 2013;153:228–239. doi: 10.1016/j.cell.2013.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan DC. Mitochondria: dynamic organelles in disease, aging, and development. Cell. 2006;125:1241–1252. doi: 10.1016/j.cell.2006.06.010. [DOI] [PubMed] [Google Scholar]
- Chen J, Lee SM, Mao Y. Protective effect of exopolysaccharide colanic acid of Escherichia coli O157:H7 to osmotic and oxidative stress. International journal of food microbiology. 2004;93:281–286. doi: 10.1016/j.ijfoodmicro.2003.12.004. [DOI] [PubMed] [Google Scholar]
- Cho I, Blaser MJ. The human microbiome: at the interface of health and disease. Nature reviews Genetics. 2012;13:260–270. doi: 10.1038/nrg3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claesson MJ, Jeffery IB, Conde S, Power SE, O'Connor EM, Cusack S, Harris HM, Coakley M, Lakshminarayanan B, O'Sullivan O, et al. Gut microbiota composition correlates with diet and health in the elderly. Nature. 2012;488:178–184. doi: 10.1038/nature11319. [DOI] [PubMed] [Google Scholar]
- Clark LC, Hodgkin J. Commensals, probiotics and pathogens in the Caenorhabditis elegans model. Cellular microbiology. 2014;16:27–38. doi: 10.1111/cmi.12234. [DOI] [PubMed] [Google Scholar]
- Dische Z, Shettles LB. A specific color reaction of methylpentoses and a spectrophotometric micromethod for their determination. The Journal of biological chemistry. 1948;175:595–603. [PubMed] [Google Scholar]
- Durieux J, Wolff S, Dillin A. The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell. 2011;144:79–91. doi: 10.1016/j.cell.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Bussiere F, Hekimi S. Mitochondrial electron transport is a key determinant of life span in Caenorhabditis elegans. Developmental cell. 2001;1:633–644. doi: 10.1016/s1534-5807(01)00071-5. [DOI] [PubMed] [Google Scholar]
- Fontana L, Partridge L, Longo VD. Extending healthy life span--from yeast to humans. Science. 2010;328:321–326. doi: 10.1126/science.1172539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garigan D, Hsu AL, Fraser AG, Kamath RS, Ahringer J, Kenyon C. Genetic analysis of tissue aging in Caenorhabditis elegans: a role for heat-shock factor and bacterial proliferation. Genetics. 2002;161:1101–1112. doi: 10.1093/genetics/161.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman S, Trisler P, Torres-Cabassa A. Regulation of capsular polysaccharide synthesis in Escherichia coli K-12: characterization of three regulatory genes. Journal of bacteriology. 1985;162:1111–1119. doi: 10.1128/jb.162.3.1111-1119.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant B, Zhang Y, Paupard MC, Lin SX, Hall DH, Hirsh D. Evidence that RME-1, a conserved C. elegans EH-domain protein, functions in endocytic recycling. Nature cell biology. 2001;3:573–579. doi: 10.1038/35078549. [DOI] [PubMed] [Google Scholar]
- Grant WD, Sutherland IW, Wilkinson JF. Exopolysaccharide colanic acid and its occurrence in the Enterobacteriaceae. Journal of bacteriology. 1969;100:1187–1193. doi: 10.1128/jb.100.3.1187-1193.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusarov I, Gautier L, Smolentseva O, Shamovsky I, Eremina S, Mironov A, Nudler E. Bacterial nitric oxide extends the lifespan of C. elegans. Cell. 2013;152:818–830. doi: 10.1016/j.cell.2012.12.043. [DOI] [PubMed] [Google Scholar]
- Harrison DE, Strong R, Allison DB, Ames BN, Astle CM, Atamna H, Fernandez E, Flurkey K, Javors MA, Nadon NL, et al. Acarbose, 17-alpha-estradiol, and nordihydroguaiaretic acid extend mouse lifespan preferentially in males. Aging cell. 2014;13:273–282. doi: 10.1111/acel.12170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, Nadon NL, Wilkinson JE, Frenkel K, Carter CS, et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–395. doi: 10.1038/nature08221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintz C, Mair W. You are what you host: microbiome modulation of the aging process. Cell. 2014;156:408–411. doi: 10.1016/j.cell.2014.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houtkooper RH, Mouchiroud L, Ryu D, Moullan N, Katsyuba E, Knott G, Williams RW, Auwerx J. Mitonuclear protein imbalance as a conserved longevity mechanism. Nature. 2013;497:451–457. doi: 10.1038/nature12188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howitz KT, Bitterman KJ, Cohen HY, Lamming DW, Lavu S, Wood JG, Zipkin RE, Chung P, Kisielewski A, Zhang LL, et al. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003;425:191–196. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- Ishii N, Fujii M, Hartman PS, Tsuda M, Yasuda K, Senoo-Matsuda N, Yanase S, Ayusawa D, Suzuki K. A mutation in succinate dehydrogenase cytochrome b causes oxidative stress and ageing in nematodes. Nature. 1998;394:694–697. doi: 10.1038/29331. [DOI] [PubMed] [Google Scholar]
- Kenyon CJ. The genetics of ageing. Nature. 2010;464:504–512. doi: 10.1038/nature08980. [DOI] [PubMed] [Google Scholar]
- Lakowski B, Hekimi S. The genetics of caloric restriction in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1998;95:13091–13096. doi: 10.1073/pnas.95.22.13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen PL, Clarke CF. Extension of life-span in Caenorhabditis elegans by a diet lacking coenzyme Q. Science. 2002;295:120–123. doi: 10.1126/science.1064653. [DOI] [PubMed] [Google Scholar]
- Lee WJ, Hase K. Gut microbiota-generated metabolites in animal health and disease. Nature chemical biology. 2014;10:416–424. doi: 10.1038/nchembio.1535. [DOI] [PubMed] [Google Scholar]
- Leiser SF, Kaeberlein M. The hypoxia-inducible factor HIF-1 functions as both a positive and negative modulator of aging. Biological chemistry. 2010;391:1131–1137. doi: 10.1515/BC.2010.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin K, Dorman JB, Rodan A, Kenyon C. daf-16: An HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science. 1997;278:1319–1322. doi: 10.1126/science.278.5341.1319. [DOI] [PubMed] [Google Scholar]
- Link CD. Expression of human beta-amyloid peptide in transgenic Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1995;92:9368–9372. doi: 10.1073/pnas.92.20.9368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Wang X, Wang HD, Wu J, Ren J, Meng L, Wu Q, Dong H, Wu J, Kao TY, et al. Escherichia coli noncoding RNAs can affect gene expression and physiology of Caenorhabditis elegans. Nature communications. 2012;3:1073. doi: 10.1038/ncomms2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizunuma M, Neumann-Haefelin E, Moroz N, Li Y, Blackwell TK. mTORC2-SGK-1 acts in two environmentally responsive pathways with opposing effects on longevity. Aging cell. 2014;13:869–878. doi: 10.1111/acel.12248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nargund AM, Fiorese CJ, Pellegrino MW, Deng P, Haynes CM. Mitochondrial and nuclear accumulation of the transcription factor ATFS-1 promotes OXPHOS recovery during the UPR(mt) Molecular cell. 2015;58:123–133. doi: 10.1016/j.molcel.2015.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niccoli T, Partridge L. Ageing as a risk factor for disease. Current biology : CB. 2012;22:R741–752. doi: 10.1016/j.cub.2012.07.024. [DOI] [PubMed] [Google Scholar]
- Ogg S, Paradis S, Gottlieb S, Patterson GI, Lee L, Tissenbaum HA, Ruvkun G. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 1997;389:994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- Oh SW, Mukhopadhyay A, Svrzikapa N, Jiang F, Davis RJ, Tissenbaum HA. JNK regulates lifespan in Caenorhabditis elegans by modulating nuclear translocation of forkhead transcription factor/DAF-16. Proc Natl Acad Sci U S A. 2005;102:4494–4499. doi: 10.1073/pnas.0500749102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palikaras K, Lionaki E, Tavernarakis N. Coupling mitogenesis and mitophagy for longevity. Autophagy. 2015;11:1428–1430. doi: 10.1080/15548627.2015.1061172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrino MW, Nargund AM, Haynes CM. Signaling the mitochondrial unfolded protein response. Biochimica et biophysica acta. 2013;1833:410–416. doi: 10.1016/j.bbamcr.2012.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepper AS, Lo TW, Killian DJ, Hall DH, Hubbard EJ. The establishment of Caenorhabditis elegans germline pattern is controlled by overlapping proximal and distal somatic gonad signals. Developmental biology. 2003;259:336–350. doi: 10.1016/s0012-1606(03)00203-3. [DOI] [PubMed] [Google Scholar]
- Portal-Celhay C, Bradley ER, Blaser MJ. Control of intestinal bacterial proliferation in regulation of lifespan in Caenorhabditis elegans. BMC microbiology. 2012;12:49. doi: 10.1186/1471-2180-12-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran PV, Mutlu AS, Wang MC. Label-free biomedical imaging of lipids by stimulated Raman scattering microscopy. In: Ausubel Frederick M, et al., editors. Current protocols in molecular biology. Vol. 109. 2015. pp. 30 33 31–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regmi SG, Rolland SG, Conradt B. Age-dependent changes in mitochondrial morphology and volume are not predictors of lifespan. Aging. 2014;6:118–130. doi: 10.18632/aging.100639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riera CE, Dillin A. Tipping the metabolic scales towards increased longevity in mammals. Nature cell biology. 2015;17:196–203. doi: 10.1038/ncb3107. [DOI] [PubMed] [Google Scholar]
- Robida-Stubbs S, Glover-Cutter K, Lamming DW, Mizunuma M, Narasimhan SD, Neumann-Haefelin E, Sabatini DM, Blackwell TK. TOR signaling and rapamycin influence longevity by regulating SKN-1/Nrf and DAF-16/FoxO. Cell metabolism. 2012;15:713–724. doi: 10.1016/j.cmet.2012.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolland SG, Lu Y, David CN, Conradt B. The BCL-2-like protein CED-9 of C. elegans promotes FZO-1/Mfn1,2- and EAT-3/Opa1-dependent mitochondrial fusion. The Journal of cell biology. 2009;186:525–540. doi: 10.1083/jcb.200905070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki R, Lunceford AL, Bixler T, Dang P, Lee W, Furukawa S, Larsen PL, Clarke CF. Altered bacterial metabolism, not coenzyme Q content, is responsible for the lifespan extension in Caenorhabditis elegans fed an Escherichia coli diet lacking coenzyme Q. Aging cell. 2008;7:291–304. doi: 10.1111/j.1474-9726.2008.00378.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sledjeski D, Gottesman S. A small RNA acts as an antisilencer of the H-NS-silenced rcsA gene of Escherichia coli. Proc Natl Acad Sci U S A. 1995;92:2003–2007. doi: 10.1073/pnas.92.6.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soukas AA, Kane EA, Carr CE, Melo JA, Ruvkun G. Rictor/TORC2 regulates fat metabolism, feeding, growth, and life span in Caenorhabditis elegans. Genes & development. 2009;23:496–511. doi: 10.1101/gad.1775409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson G, Andrianopoulos K, Hobbs M, Reeves PR. Organization of the Escherichia coli K-12 gene cluster responsible for production of the extracellular polysaccharide colanic acid. Journal of bacteriology. 1996;178:4885–4893. doi: 10.1128/jb.178.16.4885-4893.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulston JE, Brenner S. The DNA of Caenorhabditis elegans. Genetics. 1974;77:95–104. doi: 10.1093/genetics/77.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutherland IW. Structural studies on colanic acid, the common exopolysaccharide found in the enterobacteriaceae, by partial acid hydrolysis. Oligosaccharides from colanic acid. The Biochemical journal. 1969;115:935–945. doi: 10.1042/bj1150935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres-Cabassa AS, Gottesman S. Capsule synthesis in Escherichia coli K-12 is regulated by proteolysis. Journal of bacteriology. 1987;169:981–989. doi: 10.1128/jb.169.3.981-989.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tullet JM, Hertweck M, An JH, Baker J, Hwang JY, Liu S, Oliveira RP, Baumeister R, Blackwell TK. Direct inhibition of the longevity-promoting factor SKN-1 by insulin-like signaling in C. elegans. Cell. 2008;132:1025–1038. doi: 10.1016/j.cell.2008.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virk B, Correia G, Dixon DP, Feyst I, Jia J, Oberleitner N, Briggs Z, Hodge E, Edwards R, Ward J, et al. Excessive folate synthesis limits lifespan in the C. elegans: E. coli aging model. BMC biology. 2012;10:67. doi: 10.1186/1741-7007-10-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang MC, Bohmann D, Jasper H. JNK extends life span and limits growth by antagonizing cellular and organism-wide responses to insulin signaling. Cell. 2005;121:115–125. doi: 10.1016/j.cell.2005.02.030. [DOI] [PubMed] [Google Scholar]
- Wang MC, Min W, Freudiger CW, Ruvkun G, Xie XS. RNAi screening for fat regulatory genes with SRS microscopy. Nature methods. 2011;8:135–138. doi: 10.1038/nmeth.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang MC, Oakley HD, Carr CE, Sowa JN, Ruvkun G. Gene pathways that delay Caenorhabditis elegans reproductive senescence. PLoS Genet. 2014;10:e1004752. doi: 10.1371/journal.pgen.1004752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams BD, Schrank B, Huynh C, Shownkeen R, Waterston RH. A genetic mapping system in Caenorhabditis elegans based on polymorphic sequence-tagged sites. Genetics. 1992;131:609–624. doi: 10.1093/genetics/131.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Hekimi S. Two modes of mitochondrial dysfunction lead independently to lifespan extension in Caenorhabditis elegans. Aging cell. 2010;9:433–447. doi: 10.1111/j.1474-9726.2010.00571.x. [DOI] [PubMed] [Google Scholar]
- Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, Magris M, Hidalgo G, Baldassano RN, Anokhin AP, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Characterization of parental E. coli strain and experimental design of genetic screens. Related to Figure 1
(A) Average times are 50.4 ± 1.2, 53.1 ± 0.9 and 51.2 ± 0.9 hours for wild type (WT) L1 larvae developing into L4 larvae when grown on E. coli strains BW25113, OP50 and HT115, respectively. p>0.05, Student's t-test.
(B)Total numbers of progeny from WT adult worms grown on E. coli strains BW25113 (307 ± 11), OP50 (292 ± 8) and HT115 (295 ± 15) are not significantly different. p>0.05, Student's t-test.
(C)Lifespans of WT worms grown on E. coli strains BW25113, OP50, and HT115 are not significantly different (p>0.05, Log-rank test). Mean lifespan of BW25113, 18.63 ± 0.31 days, n= 167; OP50, 19.32 ± 0.44 days, n=121; HT115, 17.29 ± 0.96 days, n=102.
(D)Schematic diagram of the procedures for the primary screen using the sqt-3(e2117)ts collagen mutants, which are embryonic lethal at 25°C non-permissive temperature. Various light colors represent different bacterial mutants from the single-gene knockout E. coli library.
(E) The working flow of primary screens and subsequent retests. The entire process includes: (1) three rounds of primary screening; (2) further longitudinal confirmation; (3) validation of longevity in WT worms at 20°C with controlled bacteria amount; (4) validation of annotated bacterial mutations after reintroduction; (5) testing sufficiency upon adult-only supplementation; and (6) universality testing in another bacterial strain background. Boxes enclose the total numbers of mutants scored positive along the procedures. Utilized C. elegans strains, bacterial strains, and the criteria for positive hits are listed at each step.
Error bars represent standard error of the mean (SEM).
Figure S2. Viability and host colonization capacity of the longevity-promoting bacterial mutants. Related to Figure 1
(A) Except ΔrecC, the bacterial mutants that prolong C. elegans lifespan do not exhibit decreased survival when provided to the host (p>0.05, Student's t-test).
(B and C) Longitudinal studies of colony forming in the WT C. elegans gut show limited alterations of colonization capacities by the pro-longevity bacterial mutations. Different degrees of shading delineate age of the C. elegans. * p<0.05 compared to the control strain BW25113, Student's t-test.
Error bars represent SEM.
Figure S3. Effects of longevity-promoting bacterial mutants on C. elegans feeding and defecation behaviors. Related to Figure 3
(A) Pharyngeal pumping rates were measured in day-1-old WT worms grown on the bacterial mutants that prolong lifespan.
(B) Defecation interval length was examined in day-1-old WT worms grown on the bacterial mutants that prolong lifespan.
(C) Pharyngeal pumping rates were measured in day-1-old eat-2(ad465) mutants grown on the bacterial mutants that prolong lifespan.
*p<0.05 compared to the control, Student's t-test; error bars represent SEM.
Figure S4. RcsA dependent CA production and beneficial effects conferred by three bacterial mutants. Related to Figure 4
(A) CA production is elevated in three bacterial mutants: ΔhyfR, ΔuidC and ΔsapD (**p<0.01, *p<0.05, one-way ANOVA compared to the parental control followed by Bonferroni's multiple comparison test).
(B-D) The deletion mutation of ΔrcsA abrogates host longevity conferred by ΔhyfR (B), ΔsapD (C), and ΔuidC (D) (p>0.05, ΔrcsA vs. the double mutants, Log-rank test). Experiments were conducted in parallel with same parental controls and ΔrcsA mutants.
(E) The CA-overproducing mutants of ΔhyfR, ΔuidC and ΔsapD all significantly lower the levels of age-associated muscular mitochondrial fragmentation in the transgenic C. elegans line (raxIs[myo-3p∷mitoGFP]) (**p<0.01 compared to the parental control, Student's t-test).
Error bars represent SEM.
Figure S5. Lifespan effects of CA monomers and other polysaccharides, and the requirement of host endocytosis for CA-induced longevity. Related to Figure 6
(A) Supplementation with CA monomers does not extend C. elegans lifespan (p>0.1, Log-rank test). Glc, glucose; Gal, galactose; Fuc, fucose; GlcA, glucuronic acid; Pyr, sodium pyruvate; Ac, acetic acid; Mix, the combination of all these monomers.
(B) Extracts were purified from the Δlon and the ΔlonΔwcaD bacterial mutants in parallel. Supplementation of Δlon extracts extends the lifespan of C. elegans by 15% (p<0.001, Log-rank test), while extracts from the ΔlonΔwcaD double mutant do not significantly affect C. elegans lifespan (p>0.5, Log-rank test).
(C) Supplementation with other polysaccharides—hyaluronic acid (HA) and alginic acid (AA)—does not extend the lifespan of C. elegans (p>0.1, Log-rank test).
(D-F) CA supplementation prolongs lifespan of WT C. elegans (D), but is failed to do so with inactivation of either rab-5 (E) or rab-7 (F) that blocks early endosome to late endosome or late endosome to lysosome fusion, respectively. p>0.5, CA vs. vehicle control, Log-rank test.
Figure S6. Effects of CA overproduction on C. elegans mitochondria and proteasome. Related to Figure 7
(A) Mitochondrial DNA amounts assessed using two mitochondrial genome-encoded genes, nduo-1 and ctb-1 show no difference between C. elegans on the parental control and Δlon mutant bacteria. Error bars represent standard deviation (s.d.), p>0.05, Student's t-test.
(B) C. elegans on the parental control and Δlon mutant bacteria have similar levels of ATP. Error bars represent s.d., p>0.05, Student's t-test.
(C) Total fat levels stored in the intestine were measured by stimulated Raman scattering (SRS) microscopy, and are similar between C. elegans on the parental control and Δlon mutant bacteria. Error bars represent s.d., p>0.05, Student's t-test.
(D-F) CA supplementation does not affect C. elegans proteasome functions assayed by chymotrypsin (D), trypsin (E) and caspase-like 3 (F) activities.
(G) Intestinal-specific knockdown of fzo-1 by RNAi does not affect the lifespan-extending effect of CA. p<0.001, CA vs. vehicle control (fzo-1 RNAi), Log-rank test.
Figure S7. A group of longevity-promoting E. coli mutants are involved in the biosynthesis pathway of chorismate and its derivatives. Related to Figure 1