Abstract
Microbes and their hosts share the same environment, and microbial metabolic molecules (metabolites) exert crucial effects on host physiology1. Environmental factors not only shape the composition of the host’s resident microbes, but also modulate their metabolism2. However, the exact molecular relationship among the environment, microbial metabolites and host metabolism remains largely unknown. Here, we discovered that environmental methionine tunes bacterial methyl metabolism to regulate host mitochondrial dynamics and lipid metabolism in Caenorhabditis elegans through an endocrine crosstalk involving NR5A nuclear receptor and hedgehog signaling. We discovered that methionine deficiency in bacterial medium decreases the production of bacterial metabolites that are essential for phosphatidylcholine synthesis in C. elegans. Reductions of diundecanoyl and dilauroyl phosphatidylcholines attenuate NR5A nuclear receptor/NHR-25-mediated transcriptional suppression on the Hedgehog-like protein/GRL-21. The induction of GRL-21 consequently inhibits the Patched receptor/PTR-24 cell non-autonomously, resulting in mitochondrial fragmentation and lipid accumulation. Together, our work reveals an environment-microbe-host metabolic axis regulating host mitochondrial dynamics and lipid metabolism, and discovers NR5A-Hedgehog intercellular signaling that controls these metabolic responses with critical consequences for host health and survival.
Microbes live in close association with eukaryotic organisms across plant and animal kingdoms3. Responding to environmental fluctuations, microbes quickly alter their transcriptomes, proteomes and biochemical profiles4–6, which modify microbe-derived metabolites and consequently contribute to physiological homeostasis and disease susceptibility of the host7,8. In order to characterize the molecular relationship between environmental inputs, microbe-derived metabolites and host signaling pathways in regulating host lipid metabolism, we utilized the Escherichia coli-Caenorhabditis elegans system.
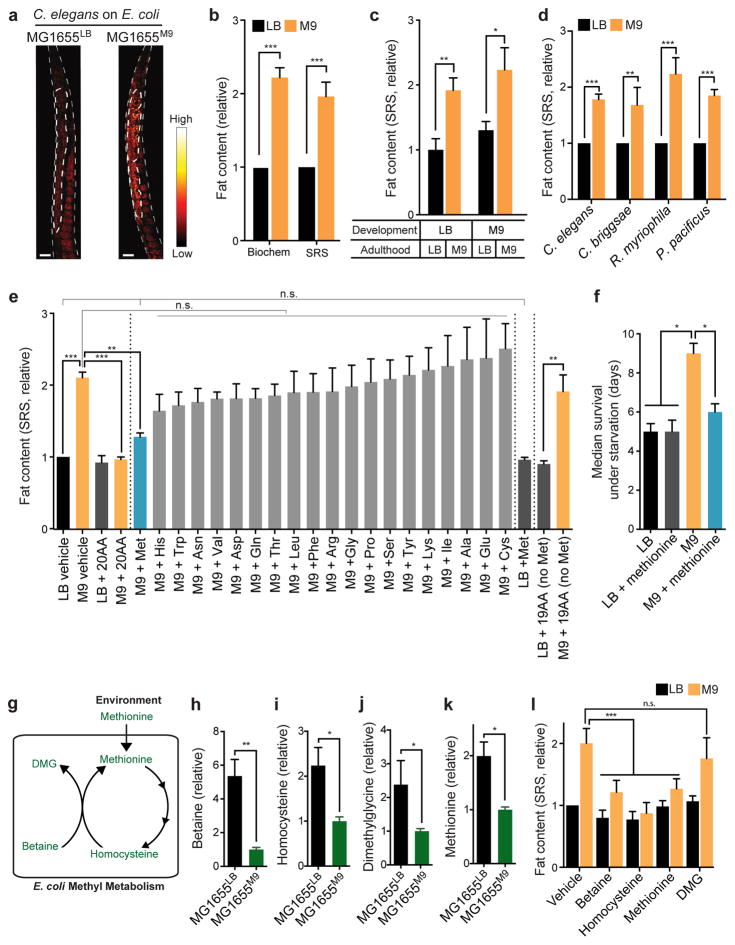
C. elegans consume and harbor microbes in their gut, and display conserved microbe-host interactions as in human9–11. To study whether same bacteria exposed to different environments exert distinct effects on host lipid metabolism, we cultured wild type (WT) E. coli strain MG1655 in either lysogeny broth (LB) medium or minimal salts (M9) medium. Comparable amounts of LB- and M9-cultured MG1655 (hereafter called MG1655LB and MG1655M9, respectively) were provided to WT C. elegans. We found that C. elegans grown on MG1655M9 have 2-fold higher fat content levels than those on MG1655LB (Fig. 1a,b), which were quantified using a label-free lipid imaging technique, stimulated Raman scattering (SRS) microscopy12, and further confirmed biochemically (Fig. 1b). These changes in fat content levels are independent of developmental exposure (Fig. 1c), and occur rapidly within 24 hours of bacterial switch (Supplementary Fig. 1a). The animals on MG1655LB and MG1655M9 have similar pharyngeal pumping, defecation and lipid absorption rates (Supplementary Fig. 1b–d), indicating indistinguishable food and lipid uptake. Their motilities, lifespan and brood sizes are also similar (Supplementary Fig. 1e–g), suggesting LB- and M9-cultured bacteria provide comparable levels of support to maintain C. elegans physical activities, somatic maintenance and reproduction. Interestingly, nematode species that are evolutionarily distant from C. elegans, including Pristionchus pacificus, Rhabditis myriophila and Caenorhabditis briggsae, all increase fat content levels when grown on MG1655M9, as compared to those on MG1655LB (Fig. 1d), suggesting a well-conserved environment-bacteria-host metabolic responsive mechanism across different host species.
Figure 1. Bacterial methyl metabolism links host lipid accumulation with environmental methionine availability.
(a, b) Wild type (WT) C. elegans raised on M9-cultured E. coli (MG1655M9) show increased fat content levels compared with those on LB-cultured E. coli (MG1655LB). Lipid storage was visualized using stimulated Raman scattering (SRS) microscopy (a, scale bar=30μm). Quantifications based on SRS microscopy and biochemical analyses show similar results (b). ***p<0.001, Student’s t-test; n=3 biologically independent experiments.
(c) Adult C. elegans sufficiently increase or decrease fat content levels 48 hours after switching to MG1655M9 or MG1655LB, respectively. **p<0.01, *p<0.05, Student’s t-test; n=3 biologically independent experiments.
(d) Different nematode species, C. elegans, Caenorhabditis briggsae, Rhabditis myriophila and Pristionchus pacificus show higher fat content levels on MG1655M9 than on MG1655LB. ***p<0.001, **p<0.01, Student’s t-test; n=3 biologically independent experiments.
(e) When cultured in M9 medium supplemented with 20 amino acids (AAs), MG1655M9 fail to induce lipid accumulation in C. elegans; while AA supplementation does not alter the effect of MG1655LB. When supplemented individually, only methionine, but not the other 19 AAs, sufficiently suppresses MG1655M9-conferred lipid accumulation in C. elegans. Without methionine, the other 19 AAs combined cannot exert any suppressing effects. ***p<0.001, **p<0.01, n.s. p>0.05, one-way ANOVA; n=3 biologically independent experiments.
(f) C. elegans raised on MG1655M9 show increased resistance to starvation, compared with those on MG1655LB. Supplementation of methionine to M9 medium suppresses the starvation resistance conferred by MG1655M9. *p<0.05, Log-rank test; n=120 biologically independent animals. The experiments were independently replicated in laboratory 3 times with similar results. Error bars represent standard error.
(g–k) Metabolomic analyses show the levels of bacterial metabolites in methyl metabolism, including betaine (h), homocysteine (i), dimethylglycine (j) and methionine (k), are decreased in MG1655M9, compared with MG1655LB. **p<0.01, *p<0.05, Student’s t-test; n=5 biologically independent experiments. Their relationships in E. coli methyl cycle are shown in (g).
(l) MG1655M9-conferred lipid accumulations in C. elegans are suppressed by direct supplementations of betaine, homocysteine and methionine but not dimethylglycine. ***p<0.001, n.s. p>0.05, two-way ANOVA; n=3 biologically independent experiments.
Error bars represent mean ± standard error of the mean (SEM) unless indicated.
Next, we found that MG1655M9 are not different from MG1655LB in the levels of caloric content, triacylglycerides or proteins (Supplementary Fig. 2a–c), but show a marginally increased level of carbohydrate content (Supplementary Fig. 2d). However, increasing carbohydrate content in MG1655LB by supplementing extra glucose (MG1655LB+Glucose) is not sufficient to increase fat storage in C. elegans (Supplementary Fig. 2e). Therefore, the altered fat storage is less likely due to global nutritional differences between bacteria, but may be actively regulated by functional metabolites derived from bacteria.
We systematically compared metabolite profiles between MG1655LB and MG1655M9. Depletion of sucrose, fructose and related metabolites in MG1655M9 (Supplementary Table 1) suggests increased fermentation; however, C. elegans grown on fermenting MG1655LB+Glucose did not show increased fat content levels (Supplementary Fig. 2e,f), ruling out the effect of bacterial fermentation on host fat storage. We also detected alterations in the levels of different amino acids (Supplementary Table 1), suggesting that MG1655M9 adjust their amino acid metabolism in response to environmental amino acid deprivation. Moreover, we found that restoration of amino acids to M9 medium by adding either peptone or casamino acids sufficiently suppresses the increased fat storage in C. elegans on MG1655M9 (Supplementary Fig. 1h). Next, we supplemented 20 amino acids individually to M9 medium and examined their effects on C. elegans fat content levels. We found that methionine, but not the other 19 amino acids, specifically suppresses the increased fat storage conferred by MG1655M9, without affecting the fat content level on MG1655LB (Fig. 1e). Without methionine, the other 19 amino acids combined fail to exert any suppressing effects (Fig. 1e). Together, these results show that methionine deprivation is the key environmental input that triggers bacterial metabolic alterations and consequently changes host metabolism.
Interestingly, we found that C. elegans on MG1655M9 are more resistant to starvation than those on MG1655LB (Fig. 1f), and this survival advantage is fully abrogated when methionine is restored in the environment (Fig. 1f). These results indicate that environmental methionine deprivation reprograms bacterial metabolism and induces “anticipatory” metabolic adaption in the host to ensure better survival through starvation. The improved survival outcome might be determined simply by increased fat content levels, and/or more complicatedly by secondary metabolic alterations associated fat storage changes.
Further functional classification of metabolite profiles revealed that MG1655M9 have reduced levels of all the components in the bacterial methyl cycle, including betaine, homocysteine, dimethylglycine (DMG), and methionine (Fig. 1g–k). We supplemented these metabolites with MG1655M9 to C. elegans, and found that betaine, homocysteine and methionine, but not DMG, are sufficient to suppress the MG1655M9-conferred lipid accumulation (Fig. 1l). Moreover, the supplementation of betaine or homocysteine requires live bacteria to exert their effects on C. elegans (Supplementary Fig. 1i). Together, these results suggest that specific metabolites derived from the bacterial methyl cycle actively regulate fat storage in the host C. elegans.
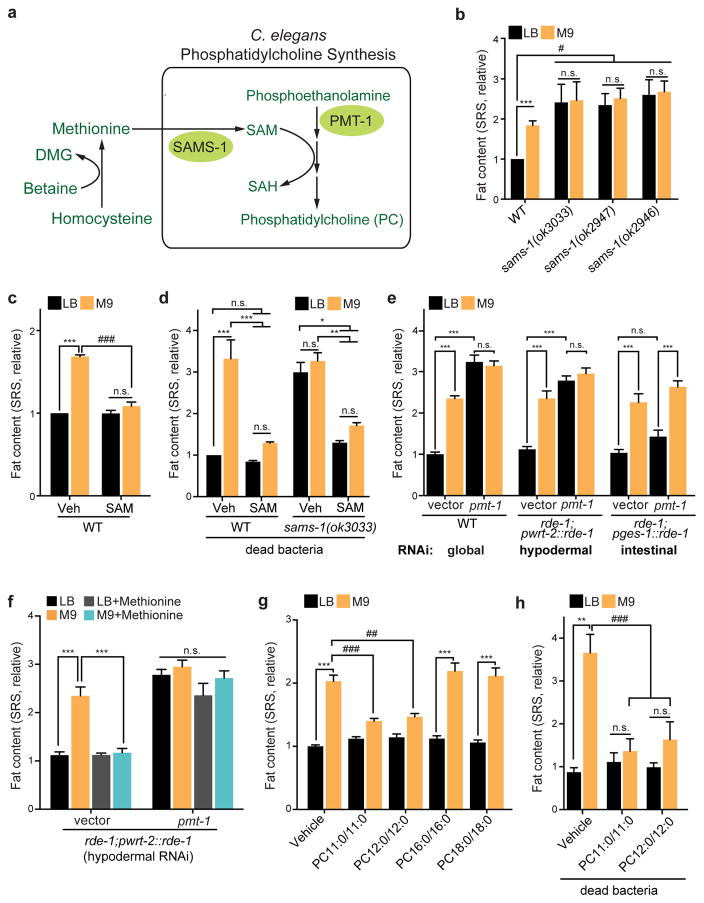
Methyl metabolism is tightly linked to the synthesis of the universal methyl donor S-adenosylmethionine (SAM) from methionine, a reaction catalyzed by S-adenosylmethionine synthetase SAMS-1 (Fig. 2a)13. We found that the C. elegans sams-1(ok3033), sams-1(ok2946), and sams-1(ok2947) mutants have 2~2.5-fold higher fat content levels than WT worms on MG1655LB, and do not further increase their fat content levels when on MG1655M9 (Fig. 2b). In addition, direct supplementation of SAM or methionine to WT C. elegans fully suppresses the MG1655M9-conferred lipid accumulation, without affecting C. elegans on MG1655LB, and these effects can bypass the requirement of live bacteria (Fig. 2c,d and Supplementary Fig. 1i). As the product of SAMS-1, SAM supplementation directly to C. elegans also rescues the increased fat storage conferred by the sams-1 mutation (Fig 2d). Therefore, bacterial methyl cycle deficiency leads to a reduction of SAM synthesis and consequently promotes lipid accumulation in the host.
Figure 2. Specific phosphatidylcholines mediate host lipid metabolic responses.
(a) A diagram of phosphatidylcholine (PC) biosynthesis via transmethylation. S-adenosyl methionine (SAM), synthesized by S-adenosyl methionine synthetase (SAMS-1) from methionine, acts as a methyl donor for the phosphoethanolamine N-methyltransferases like PMT-1 to synthesize phosphocholine from phosphoethanolamine. Phosphocholine is the precursor for PC synthesis.
(b) C. elegans sams-1(ok3033), sams-1(ok2947) and sams-1(ok2946) mutants show increased fat content levels compared to WT when on MG1655LB; and the fat content levels of the mutants on MG1655LB and MG1655M9 are not significantly different. ***p<0.001, n.s. p>0.05, Student’s t-test. #p<0.05, two-way ANOVA; n=3 biologically independent experiments.
(c) Direct supplementation of SAM (1mg/mL) to C. elegans suppresses MG1655M9-conferred lipid accumulation. ***p<0.001, n.s. p>0.05, Student’s t test. ###p<0.001, two-way ANOVA; n=3 biologically independent experiments.
(d) With bacteria killed by carbenicillin (60μg/ml), direct supplementation of SAM suppresses lipid accumulations in WT C. elegans conferred by MG1655M9 and in the sams-1(ok3033) mutant. ***p<0.001, **p<0.01, *p<0.05, n.s. p>0.05, two-way ANOVA; n=3 biologically independent experiments.
(e) Global RNAi knockdown of pmt-1 increases fat content to similar levels in C. elegans on MG1655LB and MG1655M9. Hypodermal but not intestinal RNAi knockdown of pmt-1 sufficiently increases fat content levels. ***p<0.001, n.s. p>0.05, One-way ANOVA; n=3 biologically independent experiments.
(f) Upon hypodermal-specific RNAi knockdown of pmt-1, supplementation of methionine to M9 medium fails to suppress MG1655M9-conferred lipid accumulation. ***p<0.001, n.s. p>0.05, one-way ANOVA; n=3 biologically independent experiments.
(g) Direct supplementation of PC11:0/11:0 or PC12:0/12:0, but not PC16:0/16:0 or PC18:0/18:0, to C. elegans suppresses MG1655M9-conferred lipid accumulation. ***p<0.001, Student’s t-test. ##p<0.01, ###p<0.001, two-way ANOVA; n=3 biologically independent experiments.
(h) With bacteria killed by carbenicillin (60μg/ml), direct supplementation of PC11:0/11:0 or PC12:0/12:0 suppresses MG1655M9-conferred lipid accumulations in C. elegans. **p<0.01, n.s. p>0.05, Student’s t test. ###p<0.001, two-way ANOVA; n=3 biologically independent experiments.
Error bars represent mean ± SEM.
SAM is required for three transmethylation steps during phosphatidylcholine (PC) biosynthesis, catalyzed by phosphoethanolamine methyltransferases (PMTs; Fig. 2a)14,15. When hindering transmethylation by RNAi knockdown of pmt-1, we observed a 3-fold fat content increase in C. elegans on MG1655LB (Fig. 2e). pmt-1 is known to be specifically expressed in hypodermis16. We found that hypodermis-specific RNAi knockdown of pmt-1 sufficiently mimics the effects of the global RNAi knockdown; however, RNAi knockdown of pmt-1 in the intestine has no such effects (Fig. 2e). Furthermore, upon pmt-1 inactivation in hypodermis, methionine supplementation could no longer suppress the MG1655M9-confered lipid accumulation (Fig. 2f), suggesting that responding to bacterial methyl cycle deficiency, reduction of hypodermal PC biosynthesis in C. elegans leads to increased fat storage. In supporting this idea, direct supplementation of diundecanoyl (PC11:0/11:0) or dilauroyl PC (PC12:0/12:0) suppresses the MG1655M9-conferred lipid accumulation (Fig. 2g), which does not require live bacteria (Fig. 2h). In contrast, neither dipalmitoyl (PC16:0/16:0) nor distearoyl PC (PC18:0/18:0) shows such an effect (Fig. 2g). The requirement of specific PC molecules in this metabolic regulation hints the involvement of specific receptor(s) for PC signaling transduction.
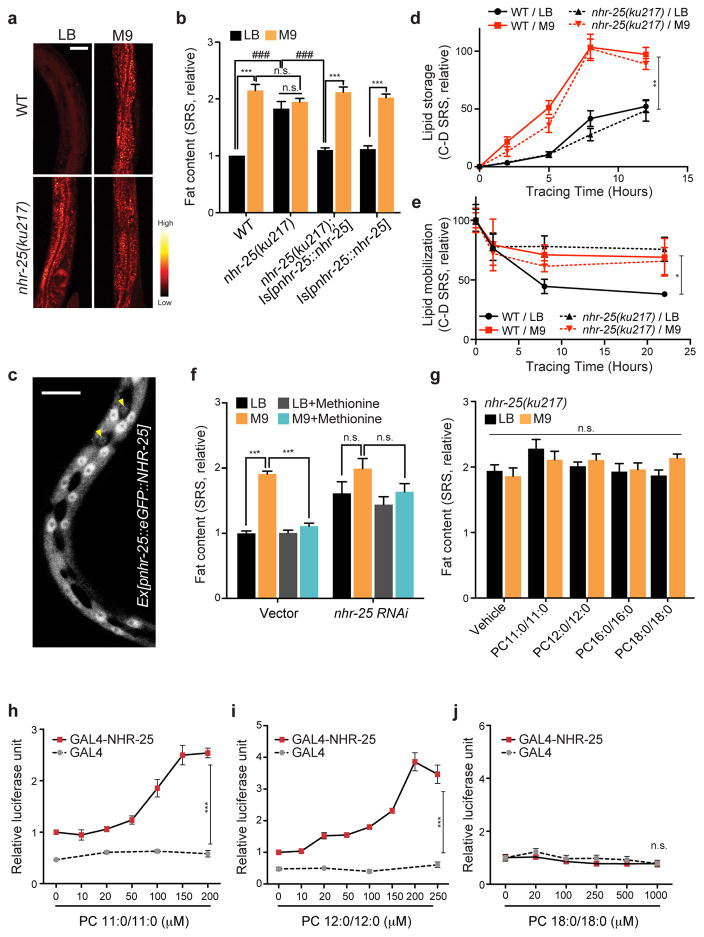
From screening a series of mutants of nuclear receptors, cell surface receptors and transcription factors (Supplementary Table 2a), we identified that the nhr-25(ku217) mutant shows a 2-fold fat storage increase when grown on MG1655LB, which is not further enhanced when grown on MG1655M9 (Fig. 3a,b). nhr-25 encodes an orphan nuclear hormone receptor that belongs to the NR5A family, orthologous to mammalian LRH-1 (liver receptor homologue 1) and SF-1 (steroidogenic factor 1)17–19. Although nhr-25 plays a crucial role in C. elegans development18, the observed fat storage changes in the nhr-25(ku217) mutant are not due to developmental deficiency because this temperature-sensitive mutant was kept under permissive temperature throughout development and shifted to non-permissive temperature only at the adult stage. Next, we generated a transgenic strain expressing GFP-fused NHR-25, which restored the difference of fat content levels in the nhr-25 mutant between MG1655LB and MG1655M9 (Fig. 3b). The GFP fusion revealed the localization of the NHR-25 protein in the nuclei of hypodermal cells (Fig. 3c), which is consistent with previous studies17,19.
Figure 3. PCs act on NHR-25 to regulate host lipid metabolic responses.
(a) SRS microscopic images show that the C. elegans nhr-25(ku217) mutant has higher levels of fat content than WT on MG1655LB, but does not have further increased fat content levels on MG1655M9. Scale bar=50μm. The experiments were independently replicated in laboratory 3 times with similar results.
(b) Restoring nhr-25 expression completely suppresses the increased lipid storage in the nhr-25(ku217) mutant on MG1655LB, without affecting the fat content levels of the nhr-25(ku217) mutant on MG1655M9 or WT on either MG1655LB or MG1655M9. ***p<0.001, n.s. p>0.05, Student’s t-test. ###p<0.001, two-way ANOVA; n=3 biologically independent experiments.
(c) GFP-fused NHR-25 proteins are enriched in the nuclei of hypodermal cells. Yellow arrowheads highlight hypodermal seam cells. Scale bar=10μm. The experiments were independently replicated in laboratory 3 times with similar results.
(d) The rates of lipid storage in WT and nhr-25(ku217) mutant C. elegans on MG1655M9 are faster than those in C. elegans on MG1655LB. **p<0.01, Student’s t-test; n=3 biologically independent experiments.
(e) In WT C. elegans, the rate of lipid mobilization is faster on MG1655LB than on MG1655M9; however, this increased lipid mobilization is abrogated in the nhr-25(ku217) mutant on MG1655LB. *p<0.05, Student’s t-test; n=3 biologically independent experiments.
(f) RNAi knockdown of nhr-25 increases fat content levels in C. elegans raised on MG1655LB, and blocks the effect of methionine in suppressing MG1655M9-conferred lipid accumulation. ***p<0.001, n.s. p>0.05, one-way ANOVA; n=3 biologically independent experiments.
(g) The effects of PC11:0/11:0 and PC12:0/12:0 on suppressing lipid accumulation are fully abrogated in the nhr-25(ku217) mutant. n.s. p>0.05, two-way ANOVA; n=3 biologically independent experiments.
(h–j) In HeLa cells transfected with GAL4-DBD/NHR-25-LBD fusion constructs and UAS-luciferase reporters, administration of PC11:0/11:0 (h) or PC12:0/12:0 (i), but not PC18:0/18:0 (j), activates NHR-25-LBD to induce luciferase expression. ***p<0.001, n.s. p>0.05, Student’s t-test; n=6 biologically independent wells (h, i); n=5 biologically independent wells (j). The experiments were independently replicated in laboratory 3 times with similar results.
Error bars represent mean ± SEM.
Fat content levels are tightly balanced between the processes of lipid storage (synthesis and incorporation) and lipid mobilization (lipolysis and oxidation). We applied a chemical imaging method coupling deuterium-tracing and SRS microscopy20 to monitor these two processes in vivo. We found that compared to those on MG1655LB, C. elegans on MG1655M9 display an increased rate of lipid storage (Fig. 3d) and a decreased rate of lipid mobilization (Fig. 3e). Interestingly, we found that the nhr-25 mutant on MG1655LB show a low rate of lipid mobilization similar to WT worms on MG1655M9 (Fig. 3e), but does not affect the rate of lipid storage (Fig. 3d). Together, these results suggest that NHR-25 functions specifically in the hypodermis and regulates intestinal lipid mobilization cell non-autonomously.
When examining whether NHR-25 transduces specific PC signals to regulate fat metabolic responses, we found that methionine, PC11:0/11:0 or PC12:0/12:0 supplementation fails to suppress the MG1655M9-confered lipid accumulation in the nhr-25 mutant (Fig. 3f,g). Next, to confirm direct interactions between the PCs and NHR-25, we conducted transactivation assays by transfecting Hela cells with constructs of GAL4 DNA-binding domain (DBD)/NHR-25 ligand-binding domain (LBD) fusion proteins and the UAS-luciferase reporter. We found that PC11:0/11:0 and PC12:0/12:0, but not PC18:0/18:0, induce luciferase levels in dose-dependent manners (Fig. 3h–j), suggesting a direct binding between NHR-25 LBD and PC11:0/11:0 or PC12:0/12:0. Considering functional specificities of both PC biosynthesis and NHR-25 in hypodermal cells (Fig. 2e and 3c), we conclude that reductions in the levels of hypodermal PCs decrease NHR-25 activation cell-autonomously, and NHR-25 subsequently regulates lipid metabolic responses, primarily in the intestine, via a cell non-autonomous mechanism.
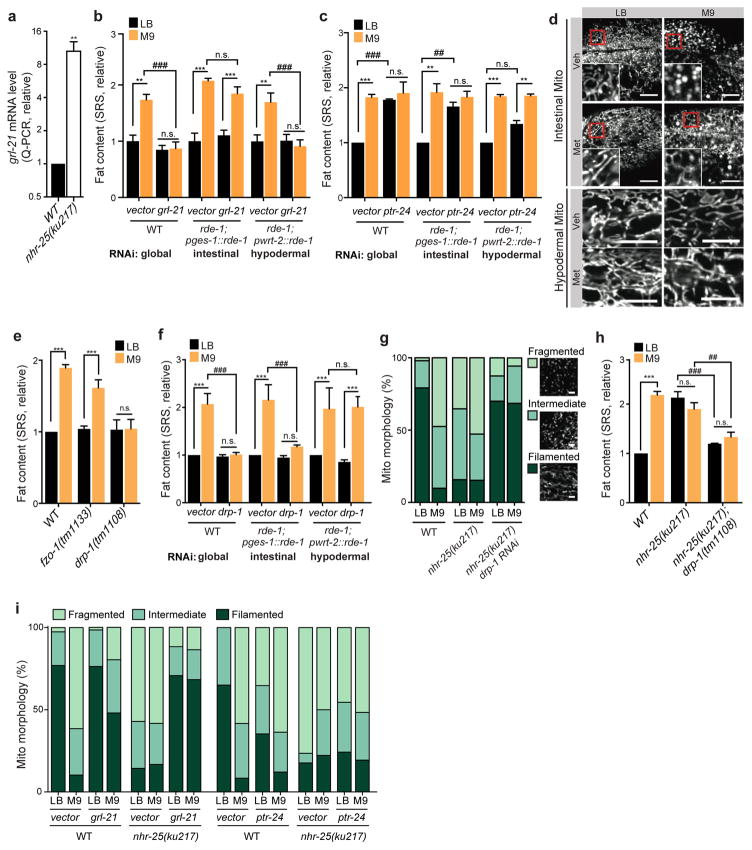
We next investigated how NHR-25 regulates lipid metabolic changes cell non-autonomously. From comparing RNA-seq profiles of C. elegans on different bacteria and ChIP-seq profiles of NHR-25, we identified 188 candidate genes that are not only differentially regulated between C. elegans exposed to MG1655LB and MG1655M9, but also contain NHR-25 binding sites in promoter regions or gene bodies (Supplementary Fig. 3, Supplementary Table 3a). Among them, 11 genes encode secreted proteins and express in the hypodermis, and were further examined for their expression levels in the nhr-25 mutant vs. WT (Supplementary Table 3b) and their effects on fat storage. We discovered that grl-21, encoding a Hedgehog-like protein, is induced 10-fold transcriptionally in the nhr-25 mutant (Fig. 4a, Supplementary Table 3b), and either global or hypodermal but not intestinal RNAi knockdown of grl-21 completely abrogates the MG1655M9-conferred lipid accumulation (Fig. 4b). In search of Patched receptor(s) that is antagonized by the Hedgehog-like protein, we screened 15 Patched homologs in C. elegans (Supplementary Table 2b,c). We discovered that RNAi knockdown of the Patched gene ptr-24 increases lipid accumulation in C. elegans on MG1655LB (Fig. 4c and Supplementary Table 2b), and intestine-specific RNAi knockdown of ptr-24 sufficiently mimics this effect (Fig. 4c). Together, these results suggest that NHR-25 controls grl-21 transcription in the hypodermis, and GRL-21 inhibits the intestinal PTR-24 receptor to regulate fat storage cell non-autonomously.
Figure 4. Endocrine crosstalk of NHR-25 and Hedgehog signaling regulates host lipid metabolic responses by tuning mitochondrial dynamics.
(a) The nhr-25(ku217) mutant increases the expression of a hedgehog-like gene, grl-21. N=4 biologically independent experiments.
(b) Global or hypodermis-specific, but not intestine-specific RNAi knockdown of grl-21 suppresses MG1655M9-conferred lipid accumulation. N=3 biologically independent experiments.
(c) Global or intestine-specific RNAi knockdown of a patched-related receptor, ptr-24 increases fat content in C. elegans raised on MG1655LB to the level as on MG1655M9. RNAi knockdown of ptr-24 in the hypodermis marginally but insignificantly increase fat content levels in C. elegans raised on MG1655LB. N=3 biologically independent experiments.
(d) C. elegans raised on MG1655M9 exhibit fragmented mitochondria in the intestine, in contrast to their filamentous mitochondrial network on MG1655LB. Methionine supplementation to M9 medium suppresses MG1655M9-conferred mitochondrial fragmentation. Hypodermal mitochondrial morphology is not affected by bacterial conditions. Mitochondrial morphologies in the intestine and hypodermis were visualized by confocal microscopy in raxIs51[Pges-1::mitoGFP] and raxIs49[Pcol-12::mitoGFP] adults, respectively. Scale bar=10μm.
(e) The drp-1(tm1108) mutation, which inhibits mitochondrial fission, suppresses the MG1655M9-conferred lipid accumulation. In contrast, the fzo-1(tm1133) mutation, which inhibits mitochondrial fusion, does not affect fat content levels in C. elegans on either MG1655LB or MG1655M9. N=3 biologically independent experiments.
(f) Global or intestine-specific, but not hypodermis-specific RNAi knockdown of drp-1 suppresses MG1655M9-conferred lipid accumulation. N=3 biologically independent experiments.
(g) Compared to WT, the nhr-25(ku217) mutant has increased mitochondrial fragmentation when raised on MG1655LB (p<0.001, Chi-squared test), but has the WT level of fragmentation when on MG1655M9 (p>0.05, Chi-squared test). RNAi knockdown of drp-1 abrogates nhr-25-conferred mitochondrial fragmentation. Scale bar=3μm. N=70 biologically independent cells.
(h) The drp-1(tm1108) mutation suppresses the nhr-25(ku217)-conferred lipid accumulation. N=3 biologically independent experiments.
(i) RNAi knockdown of grl-21 suppresses mitochondrial fragmentation conferred by MG1655M9 or by the nhr-25(ku217) mutation (p<0.001, Chi-squared test). RNAi knockdown of ptr-24 induces mitochondrial fragmentation in the WT C. elegans raised on MG1655LB (p<0.001, Chi-squared test), but does not further increase mitochondrial fragmentation conferred by MG1655M9 or by the nhr-25(ku217) mutation. N=60 biologically independent cells.
***p<0.001, **p<0.01, Student’s t-test; ###p<0.001, ##p<0.01, two-way ANOVA; error bars represent mean ± SEM.
Lipid metabolism is tightly linked to mitochondrial dynamics, which is governed by the balance between organellar fusion and fission. Although there are no significant differences in the levels of mitochondrial DNA content between C. elegans on MG1655M9 and MG1655LB (p>0.05, Supplementary Fig. 1j), we found that C. elegans on MG1655M9 show excessive mitochondrial fragmentation in the intestine, in contrast to C. elegans on MG1655LB that have mitochondria composed of a tubular and filamentous network (Fig. 4d). Interestingly, methionine supplementation compromises the MG1655M9-conferred mitochondrial fragmentation in the intestine (Fig. 4d). On the other hand, mitochondrial architecture in the hypodermis shows no obvious differences irrespective of bacterial conditions (Fig. 4d). Furthermore, we found that the mutation of drp-1, encoding the C. elegans homolog of yeast Dnm1p and mammalian DRP1 required for mitochondrial fission21, completely suppresses the MG1655M9-conferred lipid accumulation (Fig. 4e). However, the mutant defective in fzo-1, encoding the C. elegans mitofusin required for mitochondrial fusion22 (Supplementary Fig. 1k), still increases fat content levels when exposed to MG1655M9 (Fig. 4e). These findings suggest that mitochondrial fragmentation is necessary for the MG1655M9-confered fat content increase, but under the MG1655LBcondition, it is not sufficient to induce lipid accumulation. More importantly, consistent with the tissue-specific alterations in mitochondrial architecture (Fig. 4d), we demonstrated that intestine-specific RNAi knockdown of drp-1 suppresses the MG1655M9-conferred lipid accumulation as the global RNAi knockdown does; however, hypodermis-specific drp-1 RNAi knockdown has no such an effect (Fig. 4f). Therefore, the induction of mitochondrial fission in the intestine is specifically responsible for the increased lipid accumulation conferred by bacterial metabolic inputs.
Strikingly, host NHR-25/GRL-21/PTR-24 signaling regulates mitochondrial architecture in response to bacterial metabolic inputs. The nhr-25 mutant exhibits highly fragmented mitochondrial architecture in the intestine regardless of bacterial conditions (Fig. 4g), and inactivation of drp-1 suppresses mitochondrial fragmentation (Fig. 4g) and increased lipid accumulation in the nhr-25 mutant (Fig. 4h). Thus NHR-25 regulates fat storage through tuning mitochondrial fission-fusion dynamics in the intestine. Moreover, RNAi knockdown of grl-21 reduces the levels of intestinal mitochondrial fragmentation conferred by MG1655M9 and by the nhr-25 mutation (Fig. 4i); and conversely RNAi knockdown of ptr-24 promotes mitochondrial fragmentation in C. elegans grown on MG1655LB, but cannot further enhance the effect conferred by MG1655M9 or by the nhr-25 mutation (Fig. 4i).
In summary, our results reveal the molecular mechanism by which bacterial metabolic activities link environmental variations with host fat metabolism, and elucidate the crucial roles of NR5A family nuclear receptor and Hedgehog signaling in regulating mitochondrial dynamics and fat metabolism (Fig 5). The endocrine crosstalk coordinating phospholipid (hypodermis) and neutral lipid (intestine) metabolism highlights the complexity of metabolic adaptation responses at the whole organism level.
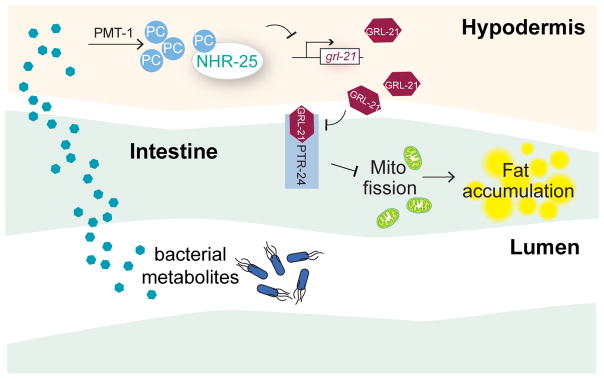
Figure 5. Summary model.
Environmental methionine regulates bacterial methyl cycle, which generates substrates for SAM synthesis in the host. SAM acts as the methyl donor for PC biosynthesis in the hypodermis. Specific PC molecules activate NHR-25, which suppresses the expression of a hedgehog-related gene, grl-21. Reduction of GRL-21 cell non-autonomously liberates the Patched receptor PTR-24 from inhibiting mitochondrial fission. In contrast, under the methionine deficient environment, low NHR-25 activity leads to highly expressed GRL-21, which antagonistically represses PTR-24 to promote severe mitochondria fragmentation and excessive lipid accumulation.
Interestingly, although methionine and its associated 1-carbon metabolism have been linked to SREBP-mediated lipogenesis23, SREBP plays a negligible role in the metabolic adaptation responses to different bacterial environments (Supplementary Table 2a). Instead, our findings demonstrate a close link between mitochondrial architecture and bacterial inputs during host metabolic adaptation, and methionine serves as a crucial cue mediating this connection. Bearing in mind that mitochondria and bacteria are evolutionary relatives and share some of the similar mechanisms for chemical communication24,25, it would be interesting to further elucidate the roles of mitochondria in bacteria-host interactions for improving host fitness.
METHODS
Nematode strains and generation of transgenic lines
Caenorhabditis elegans wild type (N2), Caenorhabditis briggsae wild isolate (PB826), Pristionchus pacificus wild isolate (PS312), Rhabditis myriophila wild isolate (DF5020), SJ4100 (zcIs13[hsp-6::GFP] V), MH1955 (nhr-25(ku217) X), RB2240 (sams-1(ok3033) X), VC2234 (sams-1(ok2947) X), VC2428(sams-1(ok2946) X), CU6372 (drp-1(tm1108) IV), CU5991 (fzo-1(tm1133) II), CE833 (sbp-1(ep176) III), BC165(nhr-80(tm1011) III), STE68 (nhr-49(nr2041) I), AA86 (daf-12(rh61; rh231) X), LT121 (dbl-1(wk70) V), CB1372 (daf-7(e1372) III), DR63 (daf-4(m63) III), DR40 (daf-1(m40) IV), GR1321 (tph-1(mg280) II), FX1106 (nhr-64(tm1106) I), STE69 (nhr-66(ok940) IV), STE71 (nhr-13(gk796) V), AE501 (nhr-8(ok186) IV), VC1320 (nhr-123(gk577) V), RB2085 (nhr-74(ok2751) I), RB1661 (nhr-85(ok2051) I), VC1120 (nhr-17(gk509) X), RB1201 (nhr-181(ok1250) V), FX977 (nhr-83(tm977) V), FX1804 (nhr-226(tm1804) V), FX1375 (nhr-42(tm1375) V), VC3219 (ptr-7(e1377) X), and VC3219 (ptr-23(ok3663) I) were obtained from the Caenorhabditis Genetics Center (Minneapolis, MN). The strain nhr-139(tm3370), ptr-3(tm7942), ptr-16(tm7951), and ptr-23(tm3762) was obtained from National BioResource Project (Japan). The strain daf-16(mgDf47) was a gift from Dr. Gary Ruvkun. The strains JM45 (rde-1(ne219); Is[Pges-1::RDE-1::unc54 3′UTR; Pmyo2::RFP3]) and JM43 (rde-1(ne219); Is[Pwrt-2::RDE-1::unc54 3′UTR; Pmyo2::RFP3]) were gifts from Dr. Justine Mello.
The transgenic strains were generated by gonadal microinjection of DNA mixtures at the young adult stage. The integrations of extrachromosomal arrays were induced by gamma irradiation exposures (4500 rads, 5.9 minutes) at the L4 stage, and the integrated progeny were backcrossed to N2 at least 5 times. We generated and used the following transgenic strains: raxEx78[Pnhr-25::eGFP::nhr-25; Pmyo-2::mcherry], raxIs32[Pnhr-25::eGFP::nhr-25; Pmyo-2::mcherry], raxIs51[Pges-1::mitoGFP], raxIs49[Pcol-12::mitoGFP].
The strains were incubated in 20°C for both maintenance and experiments; except for experiments containing the temperature-sensitive allele nhr-25(ku217) were shifted to 25°C after development.
Bacteria strains
E. coli wild type (MG1655) was a gift from Dr. Jade J. Wang. The RNAi competent E. coli OP50 strain [rnc14::DTn10 laczgA::T7pol camFRT] was generated as described in literature38.
Culture media, plates and supplementations
MG1655LB refers to the bacteria that were inoculated into 2ml of LB medium for overnight culture (37°C, 220 rpm), and then transferred into 150ml of LB medium for additional 3 hours (37°C, 220 rpm) to reach exponential growth phase. After centrifugation (4,000rpm, 10min), bacterial pellet was weighted, re-suspended in fresh LB medium at 15mg/ml, and plated onto nematode growth medium (NGM) standard plates. The LB medium was made with 2.5% of Luria broth base (Invitrogen #12795-084) in H2O and autoclaved. The NGM plate contains 2% agar, 0.23% bactopeptone, 48.28mM NaCl, 1mM MgSO4, 25mM KH2PO4, 5μg/ml cholesterol, 0.001% Nystatin, and 1mM CaCl2. MG1655M9 refers to the bacteria that were inoculated into 4 tubes, each containing 2ml of M9 minimal medium for overnight culture (37°C, 220 rpm), and then pooled altogether into 150ml of M9 minimal medium for additional 3 hours (37°C, 220 rpm) to reach exponential growth phase. Similarly, bacterial culture was concentrated to 15mg/ml in fresh M9 minimal medium for plating onto M9 minimal plates. The M9 minimal medium contains 0.2% glucose, 18.69mM NH4Cl, 8.62mM NaCl, 1mM MgSO4, 22.06mM KH2PO4, 47.76mM Na2HPO4.7H2O, 0.1mM CaCl2, 10μM FeCl2, 40μg/ml Thaimine.HCl in H2O and 0.22μm filtered. The M9 plate contains 2% agar, 0.4% glucose, 18.69mM NH4Cl, 8.62mM NaCl, 1mM MgSO4, 22.06mM KH2PO4, 47.76mM Na2HPO4.7H2O, 5μg/ml cholesterol, 0.001%Nystatin, and 1mM CaCl2. The MG1655 bacterial cultures were stored in 4°C, and used within 2 weeks.
Supplementations of peptone, casamino acids (CAA), individual amino acids and glucose were added into both bacterial M9 minimal medium and M9 plates (Fig. 1e, 1f, S1h, S2e and S2f). Supplementation of peptone (BD #211677) was prepared by adding peptone solution (0.4%) into both M9 medium and M9 plates. Supplementation of CAA (BD #223050) was prepared by adding CAA solution (0.4%) and tryptophan (40μg/ml) into both M9 medium and M9 plates. Supplementation of 19 or 20 amino acids was prepared by adding 1.5mM of each selected amino acids into both media and plates. Supplementation of single amino acid was prepared by adding 3mM of the amino acid into both media and plates, and replenished 1.5mM on plate everyday. Supplementation of glucose (MP Biomedicals #905594) was prepared by adding glucose into LB medium (0.2%) and NGM plates (0.4%). The percentages/concentrations in parenthesis indicate the final concentrations.
Supplementations of betaine, homocysteine, methionine, DMG and SAM were plated on the top of bacterial lawns on NGM or M9 plates, and fed to L4 larvae for 48 hours followed by measuring C. elegans fat content. DMG (Now Foods # 0472) and SAM (Now Foods # 0141) were extracted from capsules or well-grinded tablets with H2O, followed by filtrations 0.22μm filters. Concentrations of betaine (Sigma #14300, 200mM), homocystein (Sigma #H4628, 10mM), methionine (Sigma #64319, 3mM), DMG (2mg/ml), and SAM (2mM) were calculated based on agar volume.
PCs were mixed with bacteria at concentration of 1mM and seeded onto the plates. PC supplemented bacteria were freshly fed to L4 larvae for 24 hours, followed by C. elegans fat content measurement. PCs used in this study were 1,2-diundecanoyl-sn-glycero-3-phosphocholine (PC11:0/11:0, Avanti #850330P), 1,2-dilauroyl-sn-glycero-3-phosphocholine (PC12:0/12:0, Avanti #850335P), 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (PC16:0/16:0, Avanti #850355P), and 1,2-distearoyl-sn-glycero-3-phosphocholine (PC18:0/18:0, Avanti #850365P).
Measurement of lipid content levels in C. elegans by SRS microscopy
C. elegans (day-1-old hermaphrodite adults unless indicated, sample size larger than 10 for each of three biological replicates) were anesthetized in 1% sodium azide in M9 buffer (3g KH2PO4, 6g Na2HPO4, 5g NaCl, 1ml 1M MgSO4, H2O to 1 liter, sterilized by autoclaving), and mounted on 2% agarose pads sandwiched between glass microscopic slides and coverslips. Images were taken using the SRS system as described in literature26. In brief, when using Stokes beam at 1064nm and pump beam at 817nm, the energy difference between the Stokes and pump photons resonates with the vibrational frequency of CH2 bonds (2845cm-1). Due to the fact that the CH2 chemical bonds are highly enriched in lipid carbon chains, we quantitatively image total fat content levels by detecting the emitted Raman signals of CH2 bonds.
In lipid storage and lipid mobilization rates measurement, deuterium-labeled fat contents were traced by examining the Raman signals of CD2 bonds at 2100 cm-1, which were generated by using Stokes beam at 1064nm and pump beam at 870 nm. In lipid storage tracing experiment, C. elegans were transferred to deuterium diet at L3 stage, and imaged under SRS at desired time points until saturation. In lipid mobilization tracing experiment, C. elegans were grew on deuterium diet for two generations, and transferred to regular diet at L3 stage, which were then followed under SRS at time points of interest. Deuterium diet was prepared by mixing 2% oleic acid-d34 (Sigma #683582) with MG1655 bacteria, which were cultured from either LB or M9 medium made with deuterium water (D2O, Sigma #151882).
Measurement of lipid content levels in C. elegans by biochemical assays
Around 5,000 C. elegans (day-1-old adults) were washed off each plate, and divided into two parts for both lipid and protein analyses. For lipid extraction, the C. elegans samples were sonicated in organic solvent made of chloroform : isopropanol : NP-40 (7:11:0.1). The supernatants of the lysates underwent vacuum-drying step to evaporate the organic solvents, and then were re-dissolved in PBS for TAG assay (Infinity™ Triglycerides Liquid Stable Reagent, Thermo Scientific, TR-22421). The values of TAG levels were normalized to the values of protein levels. For protein extraction, the C. elegans samples were sonicated in PBS. The supernatants of the lysates were subjected to Bradford method (Bio-Rad Protein Assay Dye Reagent Concentrate, 500-0006) to determine protein concentrations.
Physiological measurement in C. elegans
Food intake and defecation rates
Videos recording anterior regions of at least 10 individual C. elegans (day-1-old hermaphrodite adults) under stereoscope were played in slow motion to calculate the average number of pharyngeal contractions per second, as an indication of food intake rate. Videos recording posterior regions of at least 10 individual C. elegans (day-1-old adults) were used to count the time interval between anal muscle contractions to determine defecation frequencies.
Lipid absorption
C1-BODIPY-C12 solution (Life Technologies, D-3823) was added on top of UV-killed bacteria lawn on NGM or M9 plates at a final concentration of 10μM, and kept in dark while being dried in laminar flow hood. C. elegans (day-1-old hermaphrodite adults) were transferred onto the C1-BODIPY-C12-containing plates. After desired times, at least 10 BODIPY-fed C. elegans were imaged under Axioplan2 fluorescence compound microscope (Zeiss) with Axiocam MRc5 camera (Zeiss), with “auto exposure” function in AxioVision imaging software. The average fluorescent signals from the first 3 pairs of intestinal cells were measured by using software ImageJ and were normalized to exposure times.
Locomotion activities
C. elegans (day-1-old hermaphrodite adults) on freshly seeded bacterial lawns were video recorded using an SMZ1500 stereo microscope (Nikon) connected to a C11440 camera (Hamamatsu). At least 10 individual C. elegans were tracked by NIS Elements AR imaging software (Nikon). The moving speed of each of C. elegans was calculated by dividing the distance traveled by the time length.
Brood size measurement
More than 15 synchronized L4 hermaphrodite C. elegans larvae were transferred to new, individual plates every day until reproduction cessation. The numbers of hatched progeny were counted across the whole reproductive span to calculate the total brood size for each of individual C. elegans. The experiments were performed at 20°C.
Lifespan
Survival, death and censor of more than 100 synchronized hermaphrodite C. elegans were recorded every day. Meanwhile, C. elegans were transferred to new plates every day until reproductive cessation, and then transferred to new plates every four days to keep providing comparable environmental condition over time until population dies off. The experiments were performed at 20°C.
Macronutrient and caloric measurements in E. coli
E. coli cultures were seeded onto NGM or M9 plates for at least five biological replicates. After being incubated at 20°C for 3 days, the E. coli lawns were scraped and collected in distilled deionized water and washed 3 times. The E. coli pellets were pooled and vacuum-dried. Dried weights were measured for normalization.
Protein
Protein extraction method was modified from the previous study27. E. coli samples were re-suspended in TENG-300 buffer (50mM Tris-HCl pH 7.5, 300mM NaCl, 1mM EDTA pH 8.0, 0.5% NP-40, 10% glycerol), PBS and glass beads (0.1mm) in a ratio of 2:3:1 by volume. E. coli were broken with a bead beater, followed by ultrasonic sonication. The whole lysates were centrifuged at 16,000g, and the supernatants were collected and subjected to Bradford method (Bio-Rad Protein Assay Dye Reagent Concentrate, 500-0006) to determine protein concentrations.
TAG
Lipids were extracted using organic solvent consisting of chloroform : isopropanol : NP-40 (7:11:0.1), followed by bead beating (0.1mm bead) and sonication. After centrifuging the whole lysate at 16,000 g, the supernatants were collected for vacuum-drying process to evaporate the organic solvents. The final pellets of lipid extracts were dissolved in PBS for performing TAG assay (Infinity™ Triglycerides Liquid Stable Reagent, Thermo Scientific, TR-22421).
Carbohydrate
Carbohydrate levels were determined by Anthrone method modified from the previous study28. Boil E. coli samples in 2.5M HCl for 3 hours to hydrolyze saccharides, and then filter the whole lysate through 0.2 μm poles. Mix filtered lysate with 75% H2SO4 and anthrone solution, and incubate in 90°C for 17 minutes. After cooling, measure the absorbance at 620nm.
Calorimetry
Bacterial calorimetry was performed by using an oxygen bomb calorimeter (Parr Model 1541) at Mississippi State Chemical Laboratory (Miss State, MS). In brief, the calorific values of freeze-dried bacterial samples (>500mg) were determined by burning in a high-pressure oxygen atmosphere within an oxygen bomb. The released heat of combustion resulted in temperature rise within the absorbing medium. The calorific values were calculated by multiplying the temperature changes by its heat capacity, followed by adjustment using correction factor for heat loss.
Metabolomics analysis
E. coli metabolite profiling was performed at Metabolon, Inc (Durham, NC) with 5 samples each of MG1655LB and MG1655M9. E. coli lawns were incubated at 20°C for 3 days, and then scraped and collected in distilled deionized water with 3 times of washing. Each sample contained around 0.1mg of pooled bacterial pellet. The samples were flash frozen in liquid nitrogen, and the profiling platforms have been described in the previous study29. 261 named compounds were identified in this study. Statistical analysis was performed using Welch’s two-sample t-test to identify compounds that differed significantly between MG1655LB and MG1655M9.
Transactivation assay in mammalian cells
The GAL4/UAS system driven luciferase expression in Hela cells was utilized to measure the transactivation activities of phosphatidylcholines on NHR-25. Hela cells were cultured in DMEM (Sigma #D5796) with 10% newborn calf serum (Sigma #N4637) and 1% penicillin-streptomycin (Sigma #P0781). When Hela cells reached 50–70% confluency, lipotransfection of plasmids for expressing GAL4::NHR-25 (200ng/well), UAS::Luciferase (200ng/well) and β-galactosidase (100ng/well) was performed together with FuGENE® 6 transfection reagent (Promega #E2692) (2μl/well) in 24-well plates (1ml/well). The GAL4::NHR-25 fusion protein consists of NHR-25 linker region (aa 110-351) and NHR-25 LBD (aa 352-572), fused to the C-terminus of GAL4. GAL4 expression alone without NHR-25 fragment was used as a negative control. β-galactosidase expressions were quantified as internal controls for transfection efficiencies. 20 hours after transfection, Hela cells were treated with phosphatidylcholines (Avanti, Inc) or vehicle solvent in medium containing 10% charcoal stripped fetal bovine serum (Gibco #S-1206-500) for 6 hours before conducting cell lysis and luciferase assay. All data shown represent samples from at least four biological replicates.
Starvation survivability
40~60 age-synchronized C. elegans (day-10-old hermaphrodite adults) were randomly selected from each population that has been maintained in the environments of interest for more than two generations. After washed, individual C. elegans were transfer to each well of 96-well plates pre-filled with M9 buffer for starvation challenges. The survivals and deaths were recorded daily. The experiments were performed at 20°C. The p-values for log-rank tests were achieved by using Kaplan-Meier survival analysis.
RNAi treatments
The RNAi vectors minipreped from the Ahringer or Vidal library were transformed into the genetic modified competent OP50 [rnc14::DTn10 laczgA::T7pol camFRT], which lacks RNAIII RNase activity but gains IPTG-inducible T7 RNA polymerase30. The RNAi OP50 colonies were selected by carbenicillin (50μg/ml), tetracycline (50μg/ml) and chloramphenicol (17μg/ml) resistance, and verified by Sanger sequencing.
RNAi OP50 bacteria were cultured for 16~20 hours in LB with 25μg/ml carbenicillin, 5μg/ml tetracycline and 17μg/ml chloramphenicol; and the three-fold concentrated bacteria cultures were seeded onto RNAi agar plates that contain 1mM IPTG and 50μg/ml carbenicillin. The plates were incubated at room temperature for one day to induce dsRNA expressions. Approximately 100 synchronized L1 larvae were placed; when reaching adulthood, 20~30 hermaphrodite adults were transferred to new RNAi bacterial plates to lay progeny. The reason for two generations of RNAi treatment was to enhance RNAi efficiency and phenotypic consistency. Synchronized L4 hermaphrodite larvae from the second generation were subjected to MG1655 bacteria under environments of interest for 48 hours, followed by phenotypic examinations.
Determination of the mtDNA copy number
50 age-synchronized C. elegans (day-2-old hermaphrodite adults) were collected into lysis buffer (50 mM KCl, 10 mM Tris pH 8.3, 2.5 mM MgCl2, 0.45% NP-40, 0.45% Tween-20, 0.01% Gelatin, 100μg/ml proteinase K), followed by sonication (Branson Sonifier 450, output control = 5, cycle = 10). The DNA content was determined using quantitative real-time polymerase chain reaction (qPCR) analysis of mtDNA genes ctb-1 and nduo-1, normalized to nucleus gDNA gene ant-1.3 as an internal control, from three biological replicates. The protocol was modified from the previous study31.
Primers
ant-1.3 FWD CGACACTGCCAAGATGGTAT
ant-1.3 REV CGGTGACGACTTGAGCAATA
ctb-1 FWD ATTGCCGTGAGCTATTCTAGTT
ctb-1 REV ACCGTGGCAATATAACCTAGATG
nduo-1 FWD AGCGTCATTTATTGGGAAGAAGAC
nduo-1 REV AAGCTTGTGCTAATCCCATAAATGT
Quantitative RT-PCR
Total RNAs extracted from at least 1,000 young C. elegans adults with Trizol (Life Technologies #15596) followed by column purification (Qiagen # 74106) were subjected to cDNA synthesis with the use of qScript cDNA SuperMix (Quanta # 95048). The qPCR reactions were performed with Kapa SYBR fast PCR kit (Kapa Biosystems # KK4602) in an Eppendorf Realplex 4 PCR machine. All data shown represent four biologically independent samples, and were normalized to rpl-32 as an internal control.
Primers
grl-21 FWD AGACCTCAACAATGCTGCTG
grl-21 REV GGTCCGTAGGATCCAGCATA
grd-13 FWD TGTTCTCGCTGCCTATGATG
grd-13 REV ATCCAAATGCCTTTTGAACG
nlp-29 FWD CGCACAATGGGGATATGG
nlp-29 REV CTTTCCCCATCCTCCATACA
nlp-31 FWD AAAAAGACAAGTAAAGTGGCGAATA
nlp-31 REV CTAGAAGGACGACGAGAACAAGAAT
lpr-3 FWD GACAACCATCTGCTGCTCAA
lpr-3 REV TGGAAGAAGTGGGATTCCTG
lpr-4 FWD CATCAACGTCATGACCAAGG
lpr-4 REV ATACCCGACAGCGAGATGAC
lpr-6 FWD CATTATCGGAGCTGGACCAT
lpr-6 REV CCATGAGGGAAAGTCGGTTA
cnc-3 FWD TATGGATACGGCCCAATGAT
cnc-3 REV GGCGGTACATTCCATATCCA
col-12/col-13 FWD AGACGCATATCACCGTAGCC
col-12/col-13 REV CTCCAGATGATCCACCGAAT
mlt-10 FWD TGATGGATTCGCATTTTTGA
mlt-10 REV CGAGCAACTCGGAACTTTTC
Confocal microscopy
Transgenic strains raxEx78[Pnhr-25::eGFP::nhr-25; Pmyo-2::mcherry], raxIs51[Pges-1::mitoGFP]), raxIs49[Pcol-12::mitoGFP], and nhr-25(ku217); raxIs51[Pges-1::mitoGFP], were anesthetized in 1% sodium azide in M9 buffer with sample size larger than 20 for each of three biological replicates, and mounted on 2% agarose pads sandwiched between glass microscopic slides and coverslips. Confocal images were taken using an IX81 microscope (Olympus) connected to an AxioCam ICc3 camera (Zeiss).
Mitochondria morphology measurement
Transgenic strains raxIs51[Pges-1::mitoGFP], nhr-25(ku217); raxIs51[Pges-1::mitoGFP], and raxIs49[Pcol-12::mitoGFP] were imaged in hermaphrodites at the day-3-old adult stage, and presented with single-layer images in anterior regions for consistency. For morphological categorization, if the lengths of majority of mitochondrial filaments in a cell were longer than 4 μm, it would be considered “filamented”; if the lengths of majority of mitochondrial filaments in a cell were shorter than 2 μm, it would be considered “fragmented”; the rest belonged to the category “intermediate”. By using cell as a unit, the accumulated counts of at least 60 cells from each genotype and environmental condition were summed up across biological replicates, and calculated into percentages for bar representations. The chi-squared test for trend was conducted for statistic analysis.
Fermentation test
Fermentation test was performed with phenol red as an acidification indicator32. Phenol red (Sigma # P3532) was added into the media of interest at concentration of 0.001%. After being inoculated into culture tubes with snap caps (BD Falcon #352059), bacteria were cultured (37°C, 220 rpm) for 9 hours, followed by color examinations. If colors become yellow (pH < 6), it indicates bacteria undergo fermentative reactions; if colors become red (pH > 8), it indicates bacteria undergo oxidative respiration.
RNA extraction and sequencing analysis
Total RNAs were extracted from at least 5,000 young C. elegans adults with Trizol (Life Technologies #15596) followed by column purification (Qiagen # 74106). cDNA library and sequencing were carried out by Genomic and RNA Profiling Core at Baylor College of Medicine. Sequence reads were aligned to the C. elegans reference genome (ce10) from UCSC by methods Burrows-Wheeler Aligner (BWA) and Botie2/TopHat. By using Cuffdiff, the aligned sequences were mapped to 19,861 of annotated genes, including 1632 differentially expressed genes with change ≥ 50%, q-value < 0.05.
Statistics and reproducibility
In our experimental design, the sample sizes were determined carefully based on experience, although no statistical method was used to predetermine sample sizes. Animal populations were randomized by pipetting in liquid before seeded onto the plates, and picked blindly for phenotypic examinations. From biologically independent experiments/animals/cells/wells, the quantified values that passed statistical assumption test and showed comparable variances were subjected to appropriate statistical analyses as indicated. The n numbers used to derive statistical analyses are noted in the legends. The numbers of times that experiments were replicated in laboratory are also noted in the legends, except for Fig. 4d, 4g, and 4i, in which the experiments were independently replicated in the laboratory 3 times with similar results.
Data availability
RNA–seq data that support the findings of this study have been deposited in the NCBI Sequence Read Archive (SRA, http://www.ncbi.nlm.nih.gov/sra/) under the BioProject ID PRJNA378539. The accession codes for each of the biological samples are SAMN06551858, SAMN06551859, SAMN06551860, SAMN06551861, SAMN06551862 and SAMN06551863. NHR ChIP-seq data that support the findings of this study are from the modENCODE project under the NCBI BioProject ID PRJNA13758. The extracted datasets generated for this manuscript are provided in Supplementary Table 3a and 3b. Source data for Fig. 1h–k have been provided as Supplementary Table 1. All other data supporting the findings of this study are available from the corresponding author on request.
Supplementary Material
Acknowledgments
We thank J. Mello (Harvard Medical School, USA) for providing strains JM45 and JM43, J. Marmrosh (Caltech, USA) for providing MH1955 strain and discussion, G. Ruvkun (Harvard Medical School, USA) for providing daf-16(mgDf47), J.J. Wang (U of Wisconsin-Madison, USA) for providing MG1655 bacteria and discussion, J. Sowa and I. Neve for providing OP50 RNAi bacteria, K.H.L. Mak for providing strain raxIs49, W. Dang and H. Liu for biochemical support, Y. Yu and A.S. Mutlu for the SRS support, A. Dervisefendic, H.D. Oakley and P. Svay for maintenance support, H. Liang and L. Han for RNA-seq bioinformatic support, C. Herman, D. Moore, A. Yu, A. Folick and S. Choi for discussion, and H. Dierick, C. Herman and C.-L.F. Li for critical reading of the manuscript. We appreciate the NBRP (Japan) and the CGC (USA) for providing mutant strains. The CGC is funded by NIH Office of Research Infrastructure Programs (P40 OD010440). This work was supported by grants from HHMI (M.C.W.) and National Institute of Health (R01AG045183, R01AT009050, DP1DK113644, M.C.W.), and in part by a training fellowship from the Burroughs Wellcome Fund – The Houston Laboratory and Population Science Training Program in Gene-Environment Interaction of the University of Texas Health Science Center at Houston (BWF Grant 1008200, C.-C.J.L.).
Footnotes
Note: Supplementary Information is available in the online version of the paper.
AUTHOR CONTRIBUTION
C.-C.J.L. and M.C.W. wrote the manuscript. C.-C.J.L. and M.C.W. conceived and designed the study. C.-C.J.L. conducted the experiments.
COMPETING FINANCIAL INTEREST
The authors declare no competing financial interests.
References
- 1.Lee WJ, Hase K. Gut microbiota-generated metabolites in animal health and disease. Nat Chem Biol. 2014;10:416–424. doi: 10.1038/nchembio.1535. [DOI] [PubMed] [Google Scholar]
- 2.Muegge BD, et al. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science. 2011;332:970–4. doi: 10.1126/science.1198719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hacquard S, et al. Microbiota and host nutrition across plant and animal kingdoms. Cell Host and Microbe. 2015;17:603–616. doi: 10.1016/j.chom.2015.04.009. [DOI] [PubMed] [Google Scholar]
- 4.Cases I, De Lorenzo V, Ouzounis CA. Transcription regulation and environmental adaptation in bacteria. Trends in Microbiology. 2003;11:248–253. doi: 10.1016/s0966-842x(03)00103-3. [DOI] [PubMed] [Google Scholar]
- 5.Guo MS, Gross CA. Stress-induced remodeling of the bacterial proteome. Current Biology. 2014;24 doi: 10.1016/j.cub.2014.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.David LA, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–63. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krishnan S, Alden N, Lee K. Pathways and functions of gut microbiota metabolism impacting host physiology. Current Opinion in Biotechnology. 2015;36:137–145. doi: 10.1016/j.copbio.2015.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown JM, Hazen SL. The gut microbial endocrine organ: bacterially derived signals driving cardiometabolic diseases. Annu Rev Med. 2015;66:343–359. doi: 10.1146/annurev-med-060513-093205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cabreiro F, Gems D. Worms need microbes too: Microbiota, health and aging in caenorhabditis elegans. EMBO Mol Med. 2013;5:1300–1310. doi: 10.1002/emmm.201100972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heintz C, Mair W. You are what you host: Microbiome modulation of the aging process. Cell. 2014;156:408–411. doi: 10.1016/j.cell.2014.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dirksen P, et al. The native microbiome of the nematode Caenorhabditis elegans: gateway to a new host-microbiome model. BMC Biol. 2016;14:1–16. doi: 10.1186/s12915-016-0258-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang MC, Min W, Freudiger CW, Ruvkun G, Xie XS. RNAi screening for fat regulatory genes with SRS microscopy. Nat Methods. 2011;8:135–138. doi: 10.1038/nmeth.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chiang PK, et al. S-Adenosylmethionine and methylation. FASEB J. 1996;10:471–80. [PubMed] [Google Scholar]
- 14.Bobenchik AM, Augagneur Y, Hao B, Hoch JC, Ben Mamoun C. Phosphoethanolamine methyltransferases in phosphocholine biosynthesis: Functions and potential for antiparasite therapy. FEMS Microbiol Rev. 2011;35:609–619. doi: 10.1111/j.1574-6976.2011.00267.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pessi G, Kociubinski G, Mamoun C Ben. A pathway for phosphatidylcholine biosynthesis in Plasmodium falciparum involving phosphoethanolamine methylation. Proc Natl Acad Sci U S A. 2004;101:6206–6211. doi: 10.1073/pnas.0307742101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Y, Na K, Lee HJ, Lee EY, Paik YK. Contribution of sams-1 and pmt-1 to lipid homoeostasis in adult Caenorhabditis elegans. J Biochem. 2011;149:529–538. doi: 10.1093/jb/mvr025. [DOI] [PubMed] [Google Scholar]
- 17.Gissendanner CR, Sluder aE. nhr-25, the Caenorhabditis elegans ortholog of ftz-f1, is required for epidermal and somatic gonad development. Dev Biol. 2000;221:259–72. doi: 10.1006/dbio.2000.9679. [DOI] [PubMed] [Google Scholar]
- 18.Chen Z, Eastburn DJ, Han M. The Caenorhabditis elegans nuclear receptor gene nhr-25 regulates epidermal cell development. Mol Cell Biol. 2004;24:7345–58. doi: 10.1128/MCB.24.17.7345-7358.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ward JD, et al. Sumoylated NHR-25/NR5A Regulates Cell Fate duringC. elegans Vulval Development. PLoS Genet. 2013;9 doi: 10.1371/journal.pgen.1003992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fu D, et al. In vivo metabolic fingerprinting of neutral lipids with hyperspectral stimulated raman scattering microscopy. J Am Chem Soc. 2014;136:8820–8828. doi: 10.1021/ja504199s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Breckenridge DG, et al. Caenorhabditis elegans drp-1 and fis-2 Regulate Distinct Cell-Death Execution Pathways Downstream of ced-3 and Independent of ced-9. Mol Cell. 2008;31:586–597. doi: 10.1016/j.molcel.2008.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rolland SG, Lu Y, David CN, Conradt B. The BCL-2-like protein CED-9 of C. elegans promotes FZO-1/Mfn1,2- and EAT-3/Opa1-dependent mitochondrial fusion. J Cell Biol. 2009;186:525–540. doi: 10.1083/jcb.200905070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walker AK, et al. A conserved SREBP-1/phosphatidylcholine feedback circuit regulates lipogenesis in metazoans. Cell. 2011;147:840–852. doi: 10.1016/j.cell.2011.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rudel T, Kepp O, Kozjak-Pavlovic V. Interactions between bacterial pathogens and mitochondrial cell death pathways. Nat Rev Microbiol. 2010;8:693–705. doi: 10.1038/nrmicro2421. [DOI] [PubMed] [Google Scholar]
- 25.Govindan JA, Jayamani E, Zhang X, Mylonakis E, Ruvkun G. Dialogue between E. coli free radical pathways and the mitochondria of C. elegans. Proc Natl Acad Sci U S A. 2015;201517448 doi: 10.1073/pnas.1517448112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ramachandran PV, Mutlu AS, Wang MC. Label-free biomedical imaging of lipids by stimulated Raman scattering microscopy. Curr Protoc Mol Biol. 2015;109:30 33 31–30 33 17. doi: 10.1002/0471142727.mb3003s109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Edwards CR, Dang W, Berger SL. Histone H4 lysine 20 of Saccharomyces cerevisiae is monomethylated and functions in subtelomeric silencing. Biochemistry. 2011;50:10473–10483. doi: 10.1021/bi201120q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brooks JR, Griffin VK, Kattan MW. A modified method for total carbohydrate analysis of glucose syrups, maltodextrins, and other starch hydrolysis products. Cereal Chem. 1986;63:465–467. [Google Scholar]
- 29.Evans AM, DeHaven CD, Barrett T, Mitchell M, Milgram E. Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Anal Chem. 2009;81:6656–6667. doi: 10.1021/ac901536h. [DOI] [PubMed] [Google Scholar]
- 30.Neve I, Sowa J, Wang MC. Modified E. coli B strain OP50 facilitates RNA interference induction in C. elegans. The Worm Breeder’s Gazette. 2015 [Google Scholar]
- 31.Bratic I, et al. Mitochondrial DNA level, but not active replicase, is essential for Caenorhabditis elegans development. Nucleic Acids Res. 2009;37:1817–1828. doi: 10.1093/nar/gkp018gkp018. [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lemos ML, Toranzo AE, Barja JL. Modified medium for the oxidation-fermentation test in the identification of marine bacteria. Appl Environ Microbiol. 1985;49:1541–1543. doi: 10.1128/aem.49.6.1541-1543.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA–seq data that support the findings of this study have been deposited in the NCBI Sequence Read Archive (SRA, http://www.ncbi.nlm.nih.gov/sra/) under the BioProject ID PRJNA378539. The accession codes for each of the biological samples are SAMN06551858, SAMN06551859, SAMN06551860, SAMN06551861, SAMN06551862 and SAMN06551863. NHR ChIP-seq data that support the findings of this study are from the modENCODE project under the NCBI BioProject ID PRJNA13758. The extracted datasets generated for this manuscript are provided in Supplementary Table 3a and 3b. Source data for Fig. 1h–k have been provided as Supplementary Table 1. All other data supporting the findings of this study are available from the corresponding author on request.