Abstract
Mutations in the PIGN gene involved in the glycosylphoshatidylinositol (GPI) anchor biosynthesis pathway cause Multiple Congenital Anomalies–Hypotonia–Seizures syndrome 1 (MCAHS1). The syndrome manifests developmental delay, hypotonia, and epilepsy, combined with multiple congenital anomalies. We report on the identification of a homozygous novel c.755A>T (p.D252V) deleterious mutation in a patient with Israeli–Arab origin with MCAHS1. The mutated PIGN caused a significant decrease of the overall GPI-anchored proteins and CD24 expression. Our results, strongly support previously published data, that partial depletion of GPI-anchored proteins is sufficient to cause severe phenotypic expression.
Keywords: MCAHS1, PIGN, Exome sequencing, glycosylphoshatidylinositol (GPI)
INTRODUCTION
MCAHS1 (OMIM 614080) is an autosomal recessive disorder characterized by developmental delay, hypotonia and epilepsy, combined with multiple congenital anomalies owing to mutations in the PIGN gene [Maydan et al., 2011; Ohba et al., 2014]. PIGN is one of more than 20 genes involved in the GPI anchor biosynthesis pathway, of which PIGN controls the addition of phosphoethanolamine to the first mannose in GPI [Freeze et al., 2012; Hong et al., 1999]. Mutations in PIGN, and seven additional genes involved in GPI biosynthesis, have been identified in individuals presenting with varied neurological abnormalities [Maydan et al., 2011; Freeze et al., 2012; Hansen et al., 2013; Krawitz et al., 2013; Kvarnung et al., 2013; Brady et al., 2014; Ohba et al., 2014]. We here report on a girl with MCAHS1 who was born to consanguineous parents and harbors a homozygous novel c.755A>T PIGN mutation. Our family is the second consanguineous Israeli–Arab family, and the fourth family, reported to date with MCAHS1 resulting from a mutation in the PIGN gene [Maydan et al., 2011; Ohba et al., 2014; Brady et al., 2014].
MATERIALS AND METHODS
Ethical Review
This research project was approved by the ethical committee of Emek Medical Center, following the declaration of Helsinki.
Exome Sequencing
Exonic sequences were enriched in the DNA sample of patient IV4 using Nextera Rapid Capture Expanded Exome Kit (Illumina, San Diego, CA). Sequencing was performed on the HiSeq2500 (Illumina, San Diego) with 100 bp paired-end reads. Read alignment to reference genome hg19 (GRCh37) was performed using the Burrows–Wheeler Aligner [Li, 2013] and variant calling was performed using Samtools [Li et al., 2009]. Annotation of variants was performed using ANNOVAR [Wang et al., 2010] in combination with in-house scripts.
Sanger Sequencing
Sequence analysis of exon nine of PIGN, using genomic DNA from the patient, two healthy sisters, one healthy brother and their parents was performed by amplification of a 229 bp fragment containing the putative mutation identified through exome sequencing. The sense 5’-AAGCATTTCAGAAGTTACTG-3’ and the antisense 5’-AAGACATCTAATCCTCTCAA-3’ primers were used under the following PCR conditions for DNA amplification: denaturation at 94°C for 5 min; 35 subsequent amplification cycles performed at 94°C for 30 sec, at 55°C for 45 sec and at 72°C for 30 sec; and at 72°C for 5 min. The sequencing reaction was performed using the Bigdye terminator kit and analyzed by the ABI PRISM 3130xl Genetic Analyzer (Applied Biosystems, Warrington, UK), according to the manufacturer’s instructions.
Flow Cytometry (FC) Analysis
We examined the effect of the c.755A>T mutation in PIGN on the surface expression of GPI-anchored proteins by staining granulocyte cells with fluorescently-labeled inactive toxin aerolysin (FLAER–ALEXA) (CEDARLANE, Burlington, NC), mouse antidecay accelerating factor CD16,CD18, CD24, and CD45 antibodies (BD Biosciences, Franklin Lakes, NJ). The antibodies against the following were FITC, PE, APC, and PerCP fluorescently labeled, and used in comprehensive four-color multiparameter Flow Cytometric Analysis on a BD FACSCalibur (BD Biosciences).
RESULTS
Family History
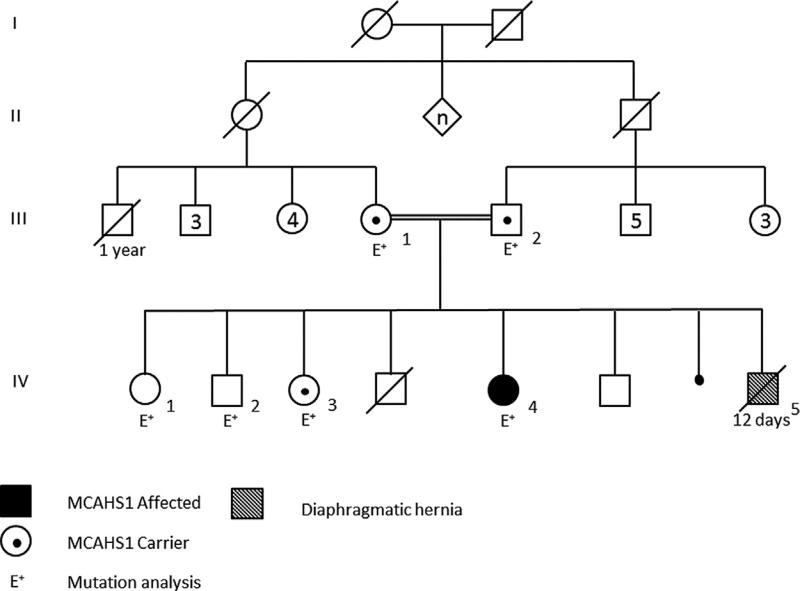
The parents of our proband are first cousins of Israeli–Arab origin. They have two sons and two daughters who are healthy. They had two prior spontaneous abortions and reported another male baby, who died at age of 12 days, following the diagnosis of diaphragmatic hernia (no further details are available). The mother had a brother who died during his 1st year of life but no further details are available. Otherwise, the family history is unremarkable (Fig. 1).
FIG. 1.
Pedigree of the Israeli-Arab family presenting MCAHS1.
Clinical Description of the Proband
The pregnancy was normal, with no known teratogenic exposure; the mother was 32 years old. The proband (IV4 in Fig. 1) was born at term with normal birth weight of 3300 g. Weakness of muscles was noticed at age four months. At age nine months metabolic work-up that included complete blood count, serum routine chemistry, glucose, lactate, ammonia, biotinidase, creatine kinase, acylcarnitines, amino acids, very long chain fatty acids, and isoelectric focusing of transferrins and urinary organic acids profile were all normal. Cerebrospinal fluid analysis for cells, glucose, protein, lactate, and amino acids were normal. Enzymatic assays in white blood cells of enzyme activities for various lysosomal diseases including GM1 and GM2 deficiency, Krabbe, and MLD were negative. Clinical examination showed hypotonia and dysmorphic features “reminiscent of Down syndrome” (Fig. 2A,B). At age six months the 1st seizure was noticed; at age 10 months the diagnoses of convulsions, developmental delay, and hypotonia were recorded. EEG analysis revealed epileptiformic bursts. Brain MRI at nine months revealed widening of the sub-arachnoidal space in the frontal and temporal lobes, the lateral ventricles widths being enlarged to 12mm. These findings were interpreted as brain atrophy. Echocardiography and ophthalmological examination were normal. Peripheral blood analysis of karyotype was 46,XX and normal, while FISH analyses specific to chromosomal regions 21q22.3 and 9q34 were also normal. Cytogenetic array CGH yielded no known pathogenic copy number variant (CNV).
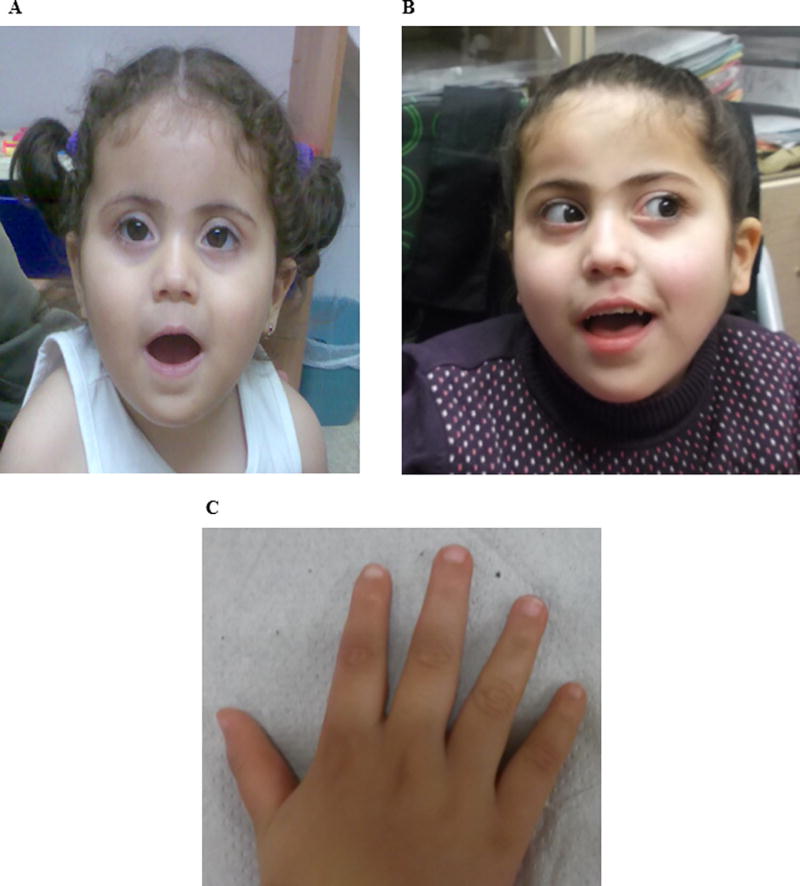
FIG. 2.
Photographs of: A. the patient face at age of 1 year and 9 months. B. the patient face at age of 6 years and 4 months. C. the patient hand at age of 5 years and 2 months. [Color figure can be seen in the online version of this article, available at http://wileyonlinelibrary.com/journal/ajmga]
Neurological assessment at age 13 months indicated DQ of 40, developmental delay and convulsions. At age 13 months she has started to roll over, but there was no crawling, sitting or standing. Her weight and height were normal for her age. Her head circumference was 44.5 cm (10–25 centile). Clinical examination revealed some unusual findings, including, brachycephaly, flat face, up-slanting palpebral fissures, synophrys, squint, large cheeks, small nose and mouth, relatively small ears (4 cm, 2nd centile), hyperfolded and coarse helices, short neck, and dimples of elbows (Fig. 2A). There were no transverse creases of palms and the 5th fingers appeared almost normal. There was general mild hypoplasia of distal parts of all fingers. Brain MRI at age 23 months was interpreted as progressive white matter disease. At age five years there was only partial response to combined anti-convulsive therapy. She was not ambulant, there was no speech, and she needed assistance with daily life activities including eating. Ophthalmological examination showed intermediate esotropia, nystagmus with vertical component, blepharitis, and normal fundi. Gastro-esophageal reflux was diagnosed. Clinical examination documented brachycephaly, some hypopigmented macules over the leg, open mouth, and drooling. There was good control of the head but hypotonia of the upper body with postural kyposis while sitting. There was reduced strength of upper body, but in the lower body there was proximal weakness and increased tone distally. Tendon reflexes were mildly increased, with bilateral clonus. Her growth parameters were normal, with penciled eyebrows and epicanthal folds. The palpebral fissures were up-slanted, the eyes were deep set with nystagmus. We also noted a small nose and somewhat small auricles (5th centiles). Palms and fingers length were normal, but the fingers appeared tapering in shape with hypoplastic fingernails; the thumb appeared unusually sharp (Fig. 2C). There were prominent blood vessels over the skin, and there was indentation of the middle part of the chest.
Exome Analysis
Following alignment and variant calling, we removed variants with a Phred quality score less than 30 or coverage less than 10 to exclude low quality variant calls. Because the disorder was assumed to be recessive, we further filtered all variants to only homozygous and compound heterozygous variants with an allele frequency less than 1% in 1000 Genomes (www.1000Genomes.org) and Exome Sequencing Project (http://evs.gs.washington.edu/EVS/) databases. For single nucleotide variants (SNVs), we retained splice site, stop-gain, stop-loss, and non-synonymous variants deemed most likely to affect protein function (i.e., phyloP or GERP++ phylogenic conservation score greater than four and polyPhen structural prediction of Damaging variants “D”). After filtering, 6 SNVs were retained across four genes (NBPF10, TEP1, CDC27, and PIGN). Only one gene, PIGN, is known to be associated with morbidities in OMIM database. The PIGN variant is a homozygous mutation located at Chr18:59814254A>T, c.755A>T, p. D252V. We also examined small insertion deletion variants (indels), which were filtered by SIFT Indel [Hu and Ng, 2013] for those predicted as damaging with confidence score greater than 0.8. There were 24 such indels, but none of these had an apparent connection to the phenotype.
Sanger Sequence Analysis
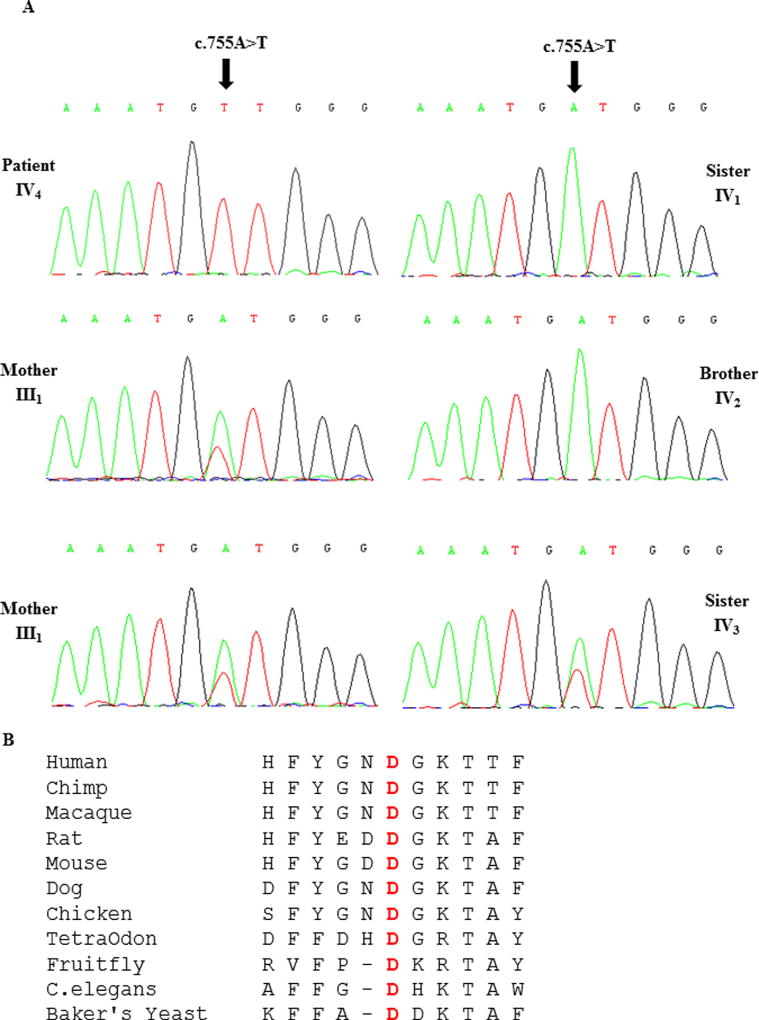
The mutation was validated by Sanger sequencing which confirmed homozygosity for the c.755A>T in the proband IV4. The mother III1, father III2, and one sister IV3 were heterozygous for the same variant. One brother IV1 and one sister IV2 were homozygous for the wild-type allele (Fig. 3).
FIG. 3.
A. DNA sequence electropherograms of the c.755A>T mutation identified in exon 9 of PIGN in the patient, her brother, her two sisters, and her parents. B. Alignment of different PIGN amino acids sequences with human PIGN. The conserved aspartic acid at position 252 is shaded in gray. [Color figure can be seen in the online version of this article, available at http://wileyonlinelibrary.com/journal/ajmga]
FC Results
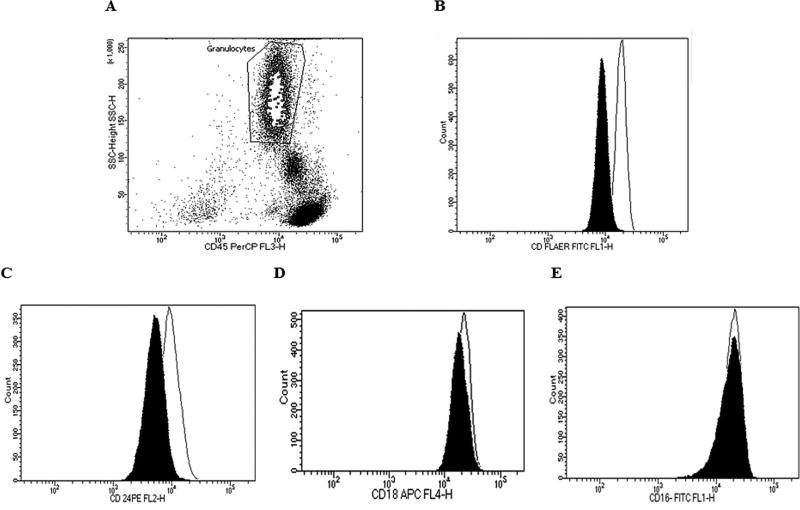
In order to examine the effect of the c.755A>T mutation on the function of PIGN, the surface expression of GPI-anchored proteins on granulocytes were analyzed by flow cytometry. Granulocytes were gated after staining with mouse anti-CD45 PerCp to allow us to perform the FC analysis only on blood granulocytes (Fig. 4A). The overall expression of GPI-anchored proteins was significantly decreased to 53% of normal levels as revealed by FLAER expression on patient granulocytes (Fig. 4B). CD 24 and CD18 expression on granulocytes was decreased to 44% and 14% respectively (Fig. 4C, D). No abnormal expression of CD16 on granulocytes was observed (Fig. 4E).
FIG. 4.
A. Gated granulocyte cells stained with mouse anti CD45. B. Surface expression of overall GPI-anchored proteins as revealed by FLAER expression on blood granulocytes. C–E. Expression of CD24, CD18 and CD16, respectively, on blood granulocytes. Dark shadow represents our patient, solid line represents normal controls.
DISCUSSION
The involvement of PIGN in MCAHS1 has been previously reported in two families. The first family, of Israeli-Arab origin, included seven affected individuals with a missense homozygous c.2126G>A (p.R709Q) mutation [Maydan et al., 2011]. The second family, of Japanese origin, included two patients with compound heterozygosity for c.808T>C (p.S270P) and c.963G>A variants (led to aberrant splicing, in which two mutant transcripts with premature stop codons p.E308Gfs*2 and p. A322Vfs*24 were generated [Ohba et al., 2014]. A third family of North African origin with a splicing homozygous mutation c.1574+1G>A in PIGN was described by Brady et al., with an intrauterine phenotype associated with diaphragmatic hernia [Brady et al., 2014].
Here, we describe another family, the second of Israeli–Arab origin, with a PIGN mutation. The novel mutation c.755A>T that was detected in our proband was predicted to be “probably damaging” with a score of 1 ((polyPhen-2), “deleterious” with a score of 0 (SIFT) and disease causing with a P-value 1 (Mutation Taster). Contrary to the results published by Ohba (2014, the overall expression of GPI-anchored proteins on our patient blood granulocytes was significantly affected by the mutant PIGN as compared to control samples, as revealed by the significant decrease in FLAER expression. Only CD24, but not CD16 and CD18, expression was drastically decreased on granulocytes from patients as compared to controls. Similar results for CD24, but not CD16, were reported previously [Ohba et al., 2014]. These data support the conclusion that the novel mutation detected in our patient causes major damage to the GPI-anchored protein PIGN, thus leading to MCAHS1 in our patient.
The girl we described has marked phenotypic overlap with the previously reported affected individuals, including developmental delay, hypotonia, epilepsy, and nystagmus [Maydan et al., 2011; Ohba et al., 2014]. However, our proband did not present with congenital anomalies of the cardiac, urinary or gastrointestinal systems (excluding gastro-esophageal reflux) as in other patients. These phenotypic differences could arise from allele specific effects, involvement of genetic modifiers or be developmental chance effects. The dysmorphic phenotype can be compared with only two of the families previously described, and overlap is present, in particular with respect to the unusual auricles and tapering fingers that were described in at least one individual [Maydan et al., 2011].
In contrast, using exome sequencing, a homozygous splicing mutation c.1574+1G>A in the PIGN gene was identified in a fetus of consanguineous parents of North African descent, with multiple congenital anomalies including bilateral congenital diaphragmatic hernia (CDH) [Brady et al., 2014]. These authors suggested that the increased severity of the phenotypic features represented by CDH in the tested fetus is due to the homozygous splicing mutation predicting a truncated protein, in comparison to reports of non-synonymous and splicing mutations which likely produce hypomorphic alleles. The family we describe had a male baby, who reportedly died at age of 12 days, following the diagnosis of diaphragmatic hernia. No DNA sample was available and thus mutation analysis could not be performed. Since the mutation detected in our patient is a nonsynonymous mutation, and speculating that the affected baby IV5 (Fig. 1) in our family was homozygous for the same mutation, other hypotheses of environmental and genetic modification need to be considered.
To date, mutations in eight genes (GIPA, GIPL, GIPM, GIPN, GIPO, GIPT, GIPV, and PGAP2) involved in the GPI biosynthesis pathway have been identified in humans. All the affected individuals involved share clinical features including seizures, cardiac defects, skeletal defects, and dysmorphic features [Almeida et al., 2006; Krawitz et al., 2010; Horn et al., 2011; Maydan et al., 2011; Johnston et al., 2012; Ng et al., 2012; Hansen et al., 2013; Krawitz et al., 2013; Brady et al., 2014; Ohba et al., 2014]. This suggests that some tissues are more sensitive than others to the loss of PIGN activity during embryonic development. This study strengthens the association between PIGN mutation and the intellectual disability–hypotonia–seizures syndrome, and expands the mutational spectrum found in this gene.
Acknowledgments
The authors thank the family for their kind cooperation. We also thank Prof. Chemke for his kind assistance. This Research was supported by a collaboration grant between the Technion, Israel Institute of Technology and Johns Hopkins University.
Footnotes
Conflict of interest: none.
References
- Almeida AM, Murakami Y, Layton DM, Hillmen P, Sellick GS, Maeda Y, Richards S, Patterson S, Kotsianidis I, Mollica L, Crawford DH, Baker A, Ferguson M, Roberts I, Houlston R, Kinoshita T, Karadimitris A. Hypomorphic promoter mutation in PIGM causes inherited glycosylphosphatidylinositol deficiency. Nat Med. 2006;12:846–851. doi: 10.1038/nm1410. [DOI] [PubMed] [Google Scholar]
- Brady PD, Moerman P, De Catte L, Deprest J, Devriendt K, Vermeesch JR. Exome sequencing identifies a recessive PIGN splice site mutation as a cause of syndromic congenital diaphragmatic hernia. Eur J Med Genet. 2014;57:487–493. doi: 10.1016/j.ejmg.2014.05.001. [DOI] [PubMed] [Google Scholar]
- Freeze HH, Eklund EA, Ng BG, Patterson MC. Neurology of inherited glycosylation disorders. Lancet Neurol. 2012;11:453–466. doi: 10.1016/S1474-4422(12)70040-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen L, Tawamie H, Murakami Y, Mang Y, ur Rehman S, Buchert R, Schaffer S, Muhammad S, Bak M, Nothen MM, Bennett EP, Maeda Y, Aigner M, Reis A, Kinoshita T, Tommerup N, Baig SM, Abou Jamra R. Hypomorphic mutations in PGAP2, encoding a GPI-anchor remodeling protein, cause autosomal-recessive intellectual disability. Am J Hum Genet. 2013;92:575–583. doi: 10.1016/j.ajhg.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Y, Maeda Y, Watanabe R, Ohishi K, Mishkind M, Riezman H, Kinoshita T. Pig-n, amammalian homologue of yeastMcd4p, is involved in transferring phosphoethanolamine to the first mannose of the glycosylphosphatidylinositol. J Biol Chem. 1999;274:35099–35106. doi: 10.1074/jbc.274.49.35099. [DOI] [PubMed] [Google Scholar]
- Horn D, Krawitz P, Mannhardt A, Korenke GC, Meinecke P. Hyperphosphatasia-mental retardation syndrome due to PIGV mutations: Expanded clinical spectrum. Am J Med Genet. 2011;A155:1917–1922. doi: 10.1002/ajmg.a.34102. [DOI] [PubMed] [Google Scholar]
- Hu J, Ng PC. SIFT Indel: Predictions for the Functional Effects of Amino Acid Insertions/Deletions in Proteins. PLoS ONE. 2013;8:e77940. doi: 10.1371/journal.pone.0077940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston JJ, Gropman AL, Sapp JC, Teer JK, Martin JM, Liu CF, Yuan X, Ye Z, Cheng L, Brodsky RA, Biesecker LG. The phenotype of a germline mutation in PIGA: The gene somatically mutated in paroxysmal nocturnal hemoglobinuria. Am J Hum Genet. 2012;90:295–300. doi: 10.1016/j.ajhg.2011.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawitz PM, Murakami Y, Riess A, Hietala M, Kruger U, Zhu N, Kinoshita T, Mundlos S, Hecht J, Robinson PN, Horn D. PGAP2 mutations, affecting the GPI-anchor-synthesis pathway, cause hyperphosphatasia with mental retardation syndrome. Am J Hum Genet. 2013;92:584–589. doi: 10.1016/j.ajhg.2013.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krawitz PM, Schweiger MR, Rödelsperger C, Marcelis C, Kölsch U, Meisel C, Stephani F, Kinoshita T, Murakami Y, Bauer S, Isau M, Fischer A, Dahl A, Kerick M, Hecht J, Köhler S, Jäger M, Grünhagen J, de Condor BJ, Doelken S, Brunner HG, Meinecke P, Passarge E, Thompson MD, Cole DE, Horn D, Roscioli T, Mundlos S, Robinson PN. Identity-by-descent filtering of exome sequence data identifies PIGV mutations in hyperphosphatasia mental retardation syndrome. Nat Genet. 2010;42:827–829. doi: 10.1038/ng.653. [DOI] [PubMed] [Google Scholar]
- Kvarnung M, Nilsson D, Lindstrand A, Korenke GC, Chiang SC, Blennow E, Bergmann M, Stodberg T, Makitie O, Anderlid BM, Bryceson YT, Nordenskjold M, Nordgren A. A novel intellectual disability syndrome caused by GPI anchor deficiency due to homozygous mutations in PIGT. J Med Genet. 2013;50:521–528. doi: 10.1136/jmedgenet-2013-101654. [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 1000 Genome Project Data Processing Subgroup. 2009. The Sequence alignment/map (SAM) format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv preprint arXiv:1303. 2013:3997. [Google Scholar]
- Maydan G, Noyman I, Har-Zahav A, Neriah ZB, Pasmanik-Chor M, Yeheskel A, Albin-Kaplanski A, Maya I, Magal N, Birk E, Simon AJ, Halevy A, Rechavi G, Shohat M, Straussberg R, Basel-Vanagaite L. Multiple congenital anomalies-hypotonia-seizures syndrome is caused by a mutation in PIGN. J Med Genet. 2011;48:383–389. doi: 10.1136/jmg.2010.087114. [DOI] [PubMed] [Google Scholar]
- Ng BG, Hackmann K, Jones MA, Eroshkin AM, He P, Wiliams R, Bhide S, Cantagrel V, Gleeson JG, Paller AS, Schnur RE, Tinschert S, Zunich J, Hegde MR, Freeze HH. Mutations in the glycosylphosphatidylinositol gene PIGL cause CHIME syndrome. Am J Hum Genet. 2012;90:685–688. doi: 10.1016/j.ajhg.2012.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohba C, Okamoto N, Murakami Y, Suzuki Y, Tsurusaki Y, Nakashima M, Miyake N, Tanaka F, Kinoshita T, Matsumoto N, Saitsu H. PIGN mutations cause congenital anomalies, developmental delay, hypotonia, epilepsy, and progressive cerebellar atrophy. Neurogenetics. 2014;15:85–92. doi: 10.1007/s10048-013-0384-7. [DOI] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164–e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]