Multiple sclerosis and microbiome
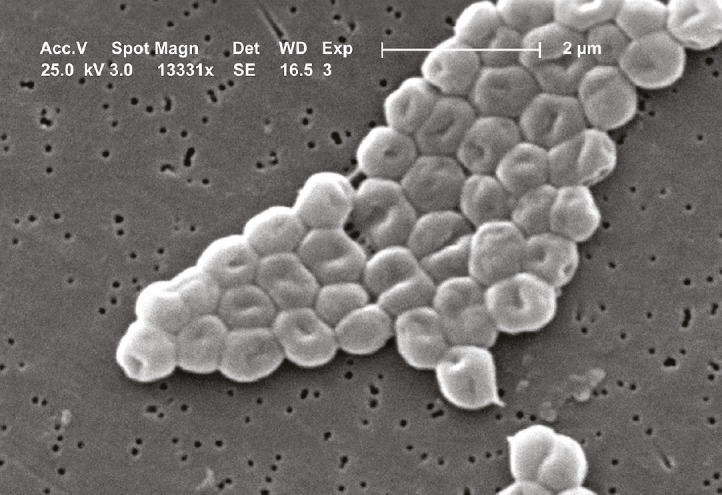
Acinetobacter. Image courtesy of CDC/Janice Carr.
Previous work has shown that mice engineered to develop experimental autoimmune encephalitis (EAE), a model for multiple sclerosis (MS), remain disease-free when raised in a germ-free environment, suggesting that commensal microbiota mediate brain autoimmunity. To understand the role of human gut microbiota in MS, Egle Cekanaviciute et al. (pp. 10713–10718) analyzed the microbiomes of 71 MS patients and 71 healthy controls, ages 19–71, and found significant differences in the relative abundances of certain bacterial genera. Acinetobacter and Akkermansia were significantly more abundant in MS patients than in controls, whereas Parabacteroides was less abundant in patients than in controls. Exposing human peripheral blood mononuclear cells to extracts from Acinetobacter and Akkermansia bacteria increased differentiation of proinflammatory T helper cells. Acinetobacter extracts also inhibited regulatory T cell (Treg) differentiation, whereas Parabacteroides extracts promoted Treg differentiation. Similar differences in T cell differentiation were observed in germ-free mice colonized with individual bacterial genera. According to the authors, the results could pave the way for microbiome-based therapies for autoimmune diseases. In a related article, Kerstin Berer et al. (pp. 10719–10724) examined 34 human identical twin pairs, ages 21–63, in which only one twin had MS. When microbiota derived from the twins were transplanted into EAE-susceptible mice, the mice receiving transplants from MS patients had a greater incidence of EAE than those receiving transplants from healthy donors. The results suggest that components of the human microbiome can contribute to MS, according to the authors. — B.D.
Improved dating of Neanderthal remains

Neanderthal bones found at Vindija Cave, Croatia. Photograph by Ian Cartwright (University of Oxford, Oxford).
Neanderthal remains from Vindija Cave in northern Croatia have been previously dated at approximately 32,000 years old, making them the most recent known Neanderthal remains and implying considerable temporal overlap between Neanderthals and modern humans in central Europe. Thibaut Deviese et al. (pp. 10606–10611) dated four Neanderthal bone samples from Vindija, one of which was previously unidentified, by extracting the amino acid hydroxyproline (HYP) from bone collagen. Because HYP occurs almost exclusively in collagen, dating purified HYP removed modern contaminants, including conservation materials, from the specimens. The authors obtained dates older than 40,000 years for all four sets of Neanderthal remains, far older than previously obtained dates. Dating of animal bones from the same layer as the Neanderthal bones yielded a wide range of dates. The finding suggests that postdepositional mixing of material has occurred, and therefore Upper Paleolithic tools found alongside the Neanderthal bones may not necessarily date from the same period. The Neanderthals at Vindija Cave likely did not overlap with modern humans, and were not part of a late-surviving, refugial population as previously thought, according to the authors. — B.D.
Genetic control of butterfly wing color

Mosaic deletion of optix replaces pigmentation with blue structural iridescence in J. coenia wing scales. Image courtesy of Roxanne Conowitch (Cornell University, Ithaca, New York).
Gene association, mapping, and expression studies have helped link the optix gene to pattern variation in butterfly wings. However, the gene’s actual function remains unclear. Linlin Zhang et al. (pp. 10707–10712) used CRISPR/Cas9 genome editing to delete the optix gene in nymphalid butterflies from four species: the red postman (Heliconius erato), the Gulf fritillary (Agraulis vanillae), the painted lady (Vanessa cardui), and the common buckeye (Junonia coenia). In all four species, black pigment replaced red and orange pigment when optix was deleted, with corresponding changes in expression of genes involved in pigment production. In the common buckeye, deleting optix also induced structural changes in wing scales that resulted in blue iridescence. The resulting wing patterns resembled the archetypal “black and blue” wing patterns found in multiple butterfly species and that appear to have evolved independently of each other. According to the authors, optix might function as a switch that toggles wing scales between different color states through coordinated changes in pigmentation and structure, and whose function is conserved across diverse butterfly genera. — B.D.
Decidualization and preeclampsia

Normally decidualized endometrial stromal cells (Left) and stromal cells with decidualization resistance (Right).
Before and during pregnancy, cells lining the uterus undergo a process of differentiation and transformation called decidualization. Tamara Garrido-Gomez et al. (pp. E8468–E8477) explored the role of decidualization in preeclampsia, a condition that afflicts 8 million mother–infant pairs worldwide every year. The authors removed uterine cells from 26 women who had been pregnant within the previous 5 years. Cells from women with normal pregnancies could be induced to decidualize in vitro, unlike cells from women who had experienced severe preeclampsia (sPE). Decidual tissue and cell samples were also collected from nine women immediately after delivery. Samples from women who did not have sPE showed signs of decidualization, unlike those from women with sPE. Conditioned medium from nonsPE cells promoted robust invasion by cultured placental cytotrophoblasts, unlike medium from sPE cells. The authors found that cytotrophoblast invasion required the presence of the decidualization biomarker proteins prolactin and insulin-like growth factor binding protein 1. The results suggest that sPE is associated with a decidualization defect that prevents cytotrophoblast invasion during embryo implantation. According to the authors, this defect might be detectable prior to conception using an endometrial genetic signature, potentially enabling therapeutic interventions. — B.D.
Evolving gene function spurs butterfly wing diversity

Comparison of a wild-type (Left) vs. WntA mutant (Right) color pattern along the wing margin of Vanessa butterflies.
Conserved signaling pathways suggest the shared evolutionary histories of species. However, small fragments of shared, essential genetic information can also lead to divergent morphologies, including changes in size, shape, color, and structure. To test the hypothesis that the evolution of new gene function drives the complexity of animal forms, Anyi Mazo-Vargas et al. (pp. 10701–10706) tested whether WntA, a gene implicated in butterfly wing mimicry, plays a broad cross-species role in butterfly wing patterning. Using CRISPR/Cas9 somatic mutagenesis, the authors conducted knockout experiments in seven nymphalid butterfly species and found that WntA underlies key, lineage-specific effects spanning multiple pattern elements. In three species, the authors report, this gene is required both for stripe-like patterns known as symmetry systems, which distinguish one species from another, and for a forewing eyespot that characterizes the Vanessa genus. Furthermore, WntA delineates color boundaries for the pigment melanin in two species of Heliconius and explains interveinous patterns in monarch butterflies, a subfamily that lacks symmetry systems. The findings demonstrate how repeated evolutionary tinkering in a single gene can lead to elaborate morphological diversity, according to the authors. — T.J.


