Significance
Regulation of virulence factor production is critical for bacterial pathogenesis. The human pathogen group A Streptococcus (GAS) produces a potent secreted protease, streptococcal pyrogenic exotoxin B (SpeB), that is crucial for pathogenesis. Although it is known that GAS produces SpeB at high population density, the molecular mechanism whereby GAS coordinates temporal SpeB production is unknown. Here, we identify a GAS-encoded short leaderless intercellular peptide signal [SpeB-inducing peptide (SIP)], and define the mechanism by which SIP induces population-wide SpeB production and contributes to GAS virulence. Furthermore, discovery of SIP provides a framework for the identification of SIP-like leaderless peptide signals in other microorganisms. Thus, our data reveal a paradigm of bacterial signaling and identify previously unknown molecules that may serve as therapeutic targets.
Keywords: Streptococcus pyogenes, SpeB, virulence regulation, quorum sensing, leaderless peptide
Abstract
Successful pathogens use complex signaling mechanisms to monitor their environment and reprogram global gene expression during specific stages of infection. Group A Streptococcus (GAS) is a major human pathogen that causes significant disease burden worldwide. A secreted cysteine protease known as streptococcal pyrogenic exotoxin B (SpeB) is a key virulence factor that is produced abundantly during infection and is critical for GAS pathogenesis. Although identified nearly a century ago, the molecular basis for growth phase control of speB gene expression remains unknown. We have discovered that GAS uses a previously unknown peptide-mediated intercellular signaling system to control SpeB production, alter global gene expression, and enhance virulence. GAS produces an eight-amino acid leaderless peptide [SpeB-inducing peptide (SIP)] during high cell density and uses the secreted peptide for cell-to-cell signaling to induce population-wide speB expression. The SIP signaling pathway includes peptide secretion, reimportation into the cytosol, and interaction with the intracellular global gene regulator Regulator of Protease B (RopB), resulting in SIP-dependent modulation of DNA binding and regulatory activity of RopB. Notably, SIP signaling causes differential expression of ∼14% of GAS core genes. Several genes that encode toxins and other virulence genes that enhance pathogen dissemination and infection are significantly up-regulated. Using three mouse infection models, we show that the SIP signaling pathway is active during infection and contributes significantly to GAS pathogenesis at multiple host anatomic sites. Together, our results delineate the molecular mechanisms involved in a previously undescribed virulence regulatory pathway of an important human pathogen and suggest new therapeutic strategies.
Spatiotemporal regulation of virulence factor production is a fundamental trait required to be a successful pathogen. Bacterial pathogens sense their environment at infection sites and respond by modulating expression of genes involved in pathogen–host interactions (1–6). In addition to host-derived signals, bacteria monitor their population density using secreted small molecules and modulate gene expression during high cell density by a process called quorum sensing (2, 7, 8). Quorum sensing includes production and secretion of signaling molecules, signal detection, and altered gene regulation (7–9). Typically, gram-positive bacteria use either linear or modified oligopeptides as quorum-sensing molecules to monitor their population density (7–9). Quorum sensing controls several bacterial properties, including virulence (7, 8). However, a direct link between quorum signaling and bacterial virulence is limited to the agr signaling pathway in Staphylococcus aureus and other related gram-positive pathogens (10).
Group A Streptococcus (GAS) is an exclusive human pathogen that causes a spectrum of diseases, including mild pharyngitis (“strep throat”) and life-threatening necrotizing fasciitis (“flesh-eating disease”) (11, 12). GAS produces many bacterial surface-bound or secreted virulence factors, including superantigens, cytolytic toxins, and proteases (11, 12). Streptococcal pyrogenic exotoxin B (SpeB) is a potent secreted cysteine protease that functions as a major GAS virulence factor (13–20). SpeB is produced abundantly in infected humans and during experimental animal infection (13–19). The protease degrades various host proteins to contribute to tissue damage and cleaves bacterial cell-surface proteins to promote disease dissemination (14–19, 21, 22). Consistent with its documented significance in infection, interference strategies targeting SpeB or its proteolytic activity confer protection against GAS infection (23–26).
SpeB production in vitro is greatly up-regulated late in growth at high bacterial cell density (18, 27). Since the discovery of SpeB nearly a century ago, regulation of its biogenesis has been the focus of extensive investigation (18, 21, 22, 27–38). Several regulatory circuits converge on transcriptional and posttranscriptional control of SpeB production (18, 21, 22, 27–36). However, the exact molecular mechanism underlying cell density-dependent up-regulation of SpeB has remained elusive. The Regulator of Protease B (RopB), a global gene regulator, directly controls speB expression during the stationary growth phase (18, 22, 27, 35). Although RopB is indispensable for growth phase-dependent transcription of speB, RopB alone is not sufficient to activate speB expression; additional unknown cell density-specific regulatory factors are required (27, 35). Consistent with this, recent findings suggest that transcriptional regulation of speB is controlled by a RopB-dependent quorum-sensing signaling pathway (18, 35). Several lines of evidence support this model, which includes cell density-dependent regulation of speB, structural homology of RopB with the Rgg-Rap-NprR-PlcR-PrgX (RRNPP) family of quorum-sensing transcription regulators, and antagonistic effects of cell density-specific secreted proteinaceous factors on speB expression (18, 35, 39). However, due to the lack of precise delineation of all necessary genetic and biochemical regulatory signals, quorum-sensing regulation of speB expression remains only a formal possibility.
We here report that GAS uses a short peptide-mediated intercellular communication mechanism to modulate virulence gene expression in a cell density-dependent fashion. The short peptide lacks a secretion signal sequence. Our results support a model in which the leaderless peptide is produced by GAS at high cell density, secreted extracellularly, imported into the cytosol, and subsequently interacts with RopB. Peptide binding to RopB promotes high-affinity RopB–DNA interactions and RopB polymerization, resulting in activation of RopB-dependent speB expression. Importantly, we show that this peptide signaling pathway is active during infection and contributes significantly to GAS virulence in multiple mouse models of infection.
Results
The ropB-speB Intergenic Region Encodes a Factor That Up-Regulates speB Expression.
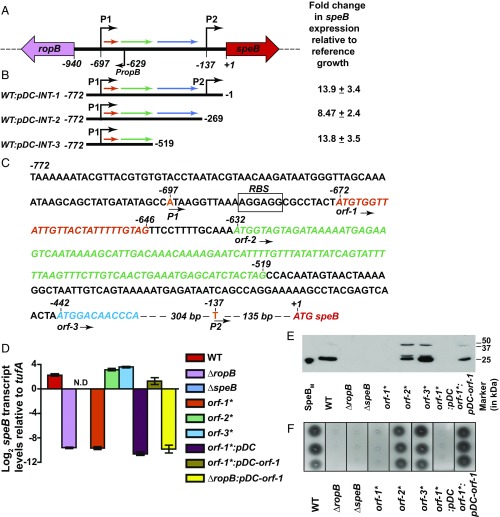
The structural homologs of RopB have their cognate regulatory peptides encoded in their immediate genomic vicinity (SI Appendix, Fig. S1A). Thus, we analyzed the 940-bp ropB-speB intergenic region for ability to alter speB expression (Fig. 1A). To identify the region that may encode an activation factor, we constructed three trans-complementation plasmids that contain different fragments of the intergenic region (Fig. 1 B and C). Typically, gene expression is higher from a multicopy plasmid than the chromosome (SI Appendix, Fig. S1B). Thus, we hypothesized that higher expression levels of a gene encoding the putative activation factor will decouple growth phase-dependent speB expression. To test this hypothesis, we introduced each of the three plasmids into wild-type (WT) GAS and characterized the resulting strains for early onset of speB expression by qRT-PCR. Relative to the WT strain, each of the three intergenic fragments caused an 8- to 14-fold increase in speB transcript level in the late exponential phase of growth. These results suggest that the activation factor for speB expression is likely encoded within the shortest of the three fragments, that is, the 257-bp fragment located between −775 and −519 bp upstream of the SpeB translation start site (Fig. 1 A and B).
Fig. 1.
The ropB-speB intergenic region has the genetic element encoding the activation peptide signal for RopB-dependent speB expression. (A) Organization of the ropB and speB gene region in GAS. The ropB and speB genes are divergently transcribed. The angled arrows above the line indicate two transcription start sites for speB, designated P1 and P2. The angled arrow below the line indicates the transcription start site for ropB (PropB). The intergenic region with three predicted ORFs, designated orf-1 (orange), orf-2 (green), and orf-3 (blue), is shown as horizontal arrows. (B) Characterization of the sequences in the ropB-speB intergenic region for their role in speB expression. High-copy-number plasmids containing different fragments of the intergenic region were introduced individually into WT GAS, and the resulting strains were characterized for premature induction of speB expression. Cells were grown to the late exponential growth phase (A600 ∼ 1.0), and speB transcript levels were assessed by qRT-PCR. Numbers at either end of the constructs indicate the nucleotide positions relative to the first nucleotide of the speB start codon. Fold changes in speB transcript levels in the trans-complemented strains relative to reference GAS growth are shown. WT GAS (WT: empty vector) grown to late exponential growth was used as the reference. (C) Nucleotide sequence characteristics of the ropB-speB intergenic region. The numbers above the nucleotides indicate positions relative to the first nucleotide of the speB start codon. Nucleotides corresponding to transcription start sites P1 and P2 are highlighted in orange. Nucleotide sequences of orf-1, orf-2, orf-3, and speB are italicized and colored in red. An inferred ribosomal-binding site (RBS) located upstream of orf-1 is boxed and labeled. (D) Analysis of speB transcript levels in the indicated strains as determined by qRT-PCR. N.D., not detected. (E) Western immunoblot analysis of secreted SpeB in filtered growth media from indicated strains. Growth media samples were probed with anti-SpeB polyclonal rabbit antibody and chemiluminescence. The masses of molecular weight markers in kilodaltons (kDa) are shown. The mature form of purified recombinant SpeB (SpeBM; 25 kDa) was used as a marker. (F) Milk plate clearing assay to assess SpeB protease activity in indicated strains. Protease activity was determined by the presence of a clear zone around the bacterial growth.
Analysis of the 940-bp intergenic region identified three hypothetical ORFs (orf), termed orf-1, orf-2, and orf-3 (Fig. 1A). Given that the intergenic region contains cis-acting regulatory elements required for speB expression (27), genetic alterations within the promoter in the form of nucleotide deletions or insertions might disrupt the regulatory elements and spacing between the regulatory elements in the promoter. Thus, to preserve the overall architecture of the promoter, we constructed isogenic mutant strains that replace the start codon (ATG) of each orf with a stop codon (TAG) in the chromosome. None of the resulting three mutant strains grew differently than the WT strain (SI Appendix, Fig. S1C). Translational disruption of orf-1 (i.e., orf-1* mutant) abolished speB expression, SpeB protein levels, and SpeB protease activity (Fig. 1 D–F and SI Appendix, Fig. S2). The transcript level of speB made by the orf-1* mutant strain was comparable to that of the ∆ropB mutant, suggesting that orf-1 is a crucial factor in speB transcriptional regulation (Fig. 1 D–F). In contrast, the orf-2* and orf-3* mutant strains had WT-like speB transcript levels, suggesting that the putative polypeptides encoded by orf-2 and orf-3 are dispensable for speB expression (Fig. 1 D–F). Provision of orf-1 alone in trans (pDC–orf-1) was sufficient to rescue the defective phenotype of the orf-1* mutant. Phenotype restoration in the orf-1* mutant by the trans-complementation plasmid was reversed by translational disruption of orf-1 in trans (pDC–orf-1*) (Fig. 1 D and F and SI Appendix, Figs. S2 and S3). To exclude the possibility that defective speB expression in the orf-1* mutant is due to altered ropB expression, we measured the transcript levels of ropB in the WT, orf-1* mutant, and trans-complemented strains. The transcript levels of ropB during different phases of growth in the orf-1* mutant were comparable to those of WT and trans-complemented strains, suggesting that the nonsense substitution in the start codon of orf-1 does not affect the expression of divergently transcribed ropB (SI Appendix, Fig. S4). Together, these results suggest that the observed phenotype of the orf-1* mutant strain is caused by genetic inactivation of orf-1, not by promoter alterations (Fig. 1 D–F and SI Appendix, Figs. S3 and S4). Importantly, trans-complementation plasmid pDC–orf-1 failed to restore speB expression in the ∆ropB mutant strain (Fig. 1D), indicating that the regulatory activity of the orf-1 gene product requires RopB. Collectively, these data indicate that orf-1 encodes the activation factor for RopB-dependent speB expression. For the purpose of clarity, due to its ability to induce speB expression, we will refer to the putative eight-amino acid polypeptide corresponding to the orf-1 gene product as SpeB-inducing peptide (SIP).
A GAS-Encoded Short Peptide Signal Activates RopB-Dependent speB Expression.
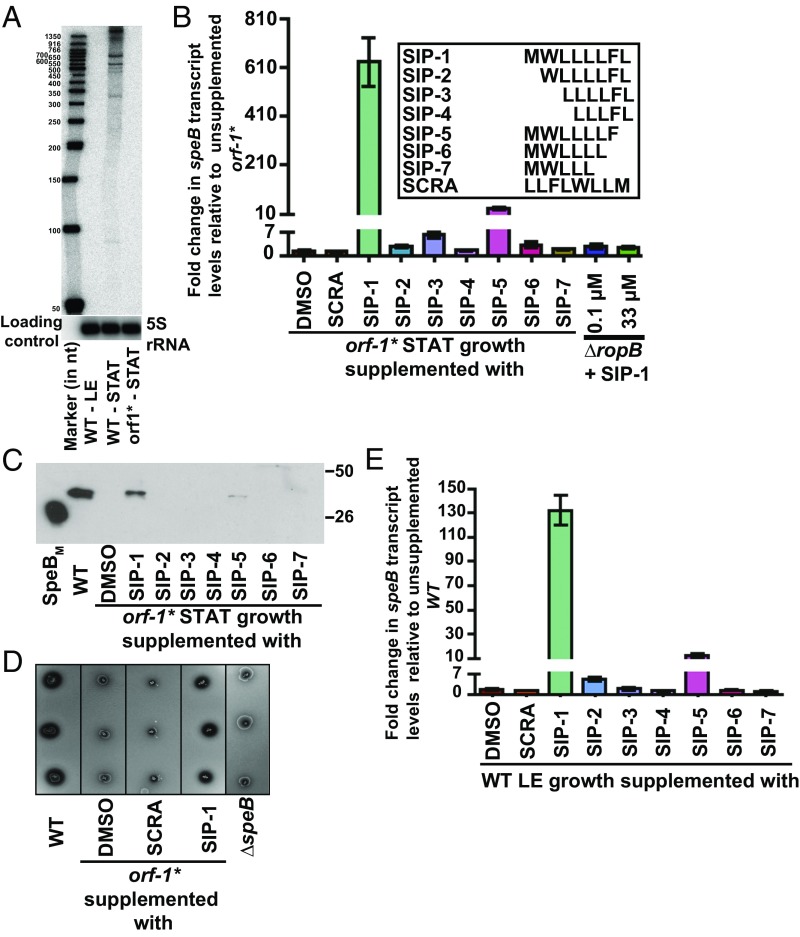
To determine if orf-1 is transcribed, we performed Northern blot analysis using a probe complementary to orf-1. Consistent with the polycistronic nature of speB transcripts (34, 40), several bands ranging in size between 300 and 1,500 bases corresponding to the orf-1 transcript were detected (Fig. 2A). Importantly, the orf-1 transcript was detected only in WT GAS grown to the stationary phase, indicating that orf-1 is expressed at high cell density (Fig. 2A). To investigate if the predicted short peptide encoded by orf-1 acts as an intercellular signal and activates speB expression, we conducted two synthetic peptide addition experiments. First, synthetic peptides containing different fragments of SIP were tested for their ability to restore speB expression to an orf-1* mutant (Fig. 2B). When the orf-1* mutant strain was grown to stationary phase and supplemented with synthetic peptides of varying length, only the full-length SIP (SIP-1) caused robust induction of speB expression (>630 fold) and restored WT-like speB transcript levels, secreted SpeB levels, and SpeB protease activity (Fig. 2 B–D). Synthetic peptide SIP-5 containing the N-terminal seven amino acids had weaker activity (>30 fold), whereas all other tested synthetic peptides failed to activate speB transcription (Fig. 2 B–D). Second, we also tested the ability of SIP to cause early induction of speB expression by WT GAS. Consistent with our observations derived from the orf-1* mutant strain, peptide SIP-1 decoupled growth phase dependency of speB expression and induced early onset of speB expression by the WT strain (>130 fold) in the late exponential growth phase (Fig. 2E).
Fig. 2.
Synthetic peptides containing the amino acid sequences of SIP activate speB expression. (A) The orf1 gene, encoding SIP, is expressed during the stationary phase of GAS growth. Total RNA extracted from WT GAS grown to either the late exponential (LE; A600 ∼ 1.0) or stationary (STAT) phase of growth and orf-1* mutant grown to the STAT phase of growth were analyzed by Northern blot. (B, Inset) Amino acid sequences of the synthetic peptides (SIP-1–SIP-7) used in the experiment. (B) SCRA peptide of an identical length and amino acid composition as SIP-1 but differing in the order of sequence was used as a negative control. The orf-1* mutant strain was grown in chemically defined medium (CDM) to the early STAT phase (A600 ∼ 1.7) and supplemented with either 100 nM indicated synthetic peptide or the carrier for the synthetic peptides (DMSO). After 60 min of incubation, transcript levels of speB were assessed by qRT-PCR. The orf-1* mutant strain supplemented with DMSO was used as a reference, and fold changes in speB transcript levels relative to the reference are shown. (C) Western immunoblot analysis of secreted SpeB in filtered growth media from the indicated samples. Cell growth and synthetic peptide supplementation were performed as described in A. Growth media collected were probed with anti-SpeB polyclonal rabbit antibody and detected by chemiluminescence. The masses of molecular weight markers in kilodaltons (kDa) are marked. (D) Milk plate clearing assay to assess the ability of various SIPs to induce SpeB protease activity in the orf-1* mutant. (E) Addition of SIP-1 and SIP-5 decouples the growth phase dependency of speB expression in WT GAS. The WT GAS was grown in CDM to the mid-exponential growth phase (A600 ∼ 0.6), and cells were incubated with 100 nm of each synthetic peptide for 60 min. Transcript levels of speB were assessed by qRT-PCR, and the fold change in speB expression relative to DMSO-supplemented growth is shown.
To determine if the observed speB induction is specific for the primary amino acid sequence of SIP, we conducted analogous experiments with a scrambled (SCRA) peptide of identical length and amino acid composition of SIP (Fig. 2B). Importantly, SCRA peptide did not activate speB expression in either the orf-1* mutant or WT strain. These results are consistent with the interpretation that induction is specific for SIP (Fig. 2 B–E). As expected, addition of SIP-1 to the isogenic ∆ropB mutant strain did not induce speB transcript production (Fig. 2B), even when SIP was added at 300-fold excess (Fig. 2B). Together, these data indicate that amino acid sequences corresponding to SIP-1 are the high cell density-specific activation peptide signal for RopB-dependent speB expression.
Molecular Mechanism of GAS Intercellular Signaling.
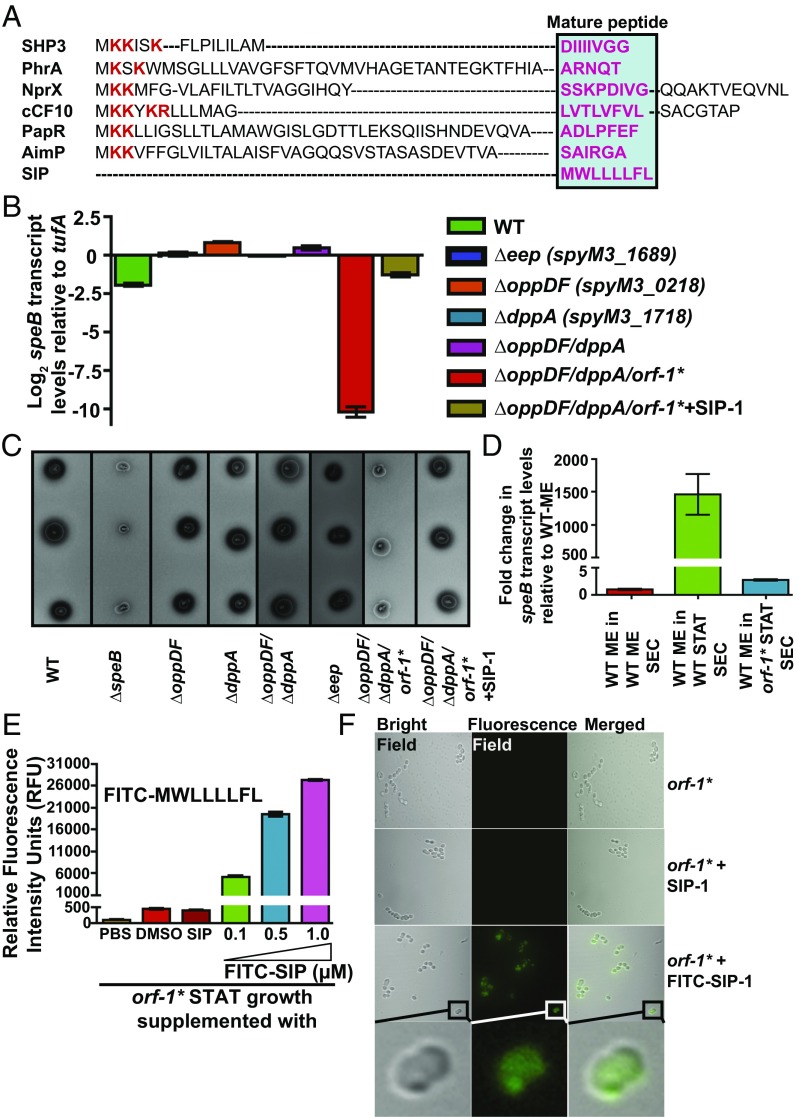
Most of the characterized peptide signals in gram-positive bacteria are generated by proteolytic processing of a precursor propeptide form into mature active peptide by membrane-bound enhanced expression of pheromone (Eep) protease (41–44). However, SIP is unique in that it is made as a mature leaderless peptide (Fig. 3A). The extracellular mature peptides are subsequently reinternalized into the bacterial cytosol by oligopeptide permeases (Opp) (41, 45, 46). Consistent with this process, inactivation of opp or dipeptide permease (dpp) in GAS caused down-regulation of speB expression (47, 48). To test the hypothesis that SIP biosynthesis involves similar molecular mechanisms, we constructed isogenic single-mutant strains of eep (∆eep), opp (∆oppDF), or dpp (∆dppA) and the double mutant, ∆oppDF/∆dppA. When tested for speB expression, all four mutant strains had WT-like speB transcript levels and protease activity (Fig. 3 B and C), indicating that the eep, opp, and dpp genes are not involved in SIP signaling.
Fig. 3.
Eep protease, Opp, and Dpp do not participate in SIP biosynthesis. (A) Alignment of amino acid sequences of characterized propeptides specific for each founding member of RRNPP family regulators. The propeptides of small hydrophobic peptide 3 (SHP3) from Streptococcus pyogenes, phosphatase regulator A (PhrA) from Bacillus subtilis, peptide signal for neutral protease regulator (NprX) from B. cereus, peptide controlling conjugative transfer of plasmids (cCF10) from E. faecalis, peptide activating PlcR (PapR) from B. cereus, arbitrium communication peptide (AimP) from phage Phi3T, and SIP from S. pyogenes are shown. The positively charged residues characteristic of bacterial peptide signals are shown in red, and the amino acid sequence corresponding to each mature peptide is boxed and highlighted in pink. Transcript levels of the speB (B) and SpeB protease activity of SpeB (C) were assessed in the indicated strains by qRT-PCR and milk plate clearing assay, respectively. (D) Genetic inactivation of sip results in loss of regulatory activity in the secreted component of GAS growth. A qRT-PCR analysis of speB transcript level in WT GAS grown in cell‐free culture supernatants obtained from the indicated strains is shown. Secretome preparation and secretome swap assay were performed as described in SI Appendix, Supplemental Materials and Methods. Triplicate biological replicates were grown on two different occasions and analyzed in duplicate. The data were graphed as the mean ± SD. ME, mid-exponential phase of growth; ME SEC, total secretome prepared from mid‐exponential growth phase; orf-1* STAT SEC, total secretome prepared from the stationary growth phase of the orf-1* mutant; WT STAT SEC, total secretome prepared from the stationary growth phase of WT GAS. (E, Inset) Amino acid sequence of the synthetic peptide SIP-1 with fluorescein modification at its amino terminus (FITC–SIP-1) used in the experiment. (E) The orf-1* mutant strain was grown in chemically defined medium (CDM) to the early stationary phase (STAT, A600 ∼ 1.7) and supplemented with either the indicated synthetic peptide or the carrier for the synthetic peptides (DMSO). Unmodified SIP-1 was added at a final concentration of 1 μM, whereas varying concentrations of FITC–SIP-1 were used. After 60 min of incubation at 37 °C, cells were washed three times with sterile PBS, suspended in PBS, and lysed. Fluorescence measurements were obtained with clarified cell lysates using excitation and emission wavelengths of 480 nm and 520 nm, respectively. The unsupplemented orf-1* mutant strain was used as a reference, and changes in relative fluorescence units (RFU) relative to the reference are shown. (F) Confocal microscopy images of the orf-1* mutant strain either unsupplemented or supplemented with the indicated synthetic peptide. Synthetic peptide addition to the orf-1* mutant strain was performed as described in E. For each sample, bright-field, fluorescence-field, and merged images are shown. (Bottom) Magnified view of the FITC–SIP-1–supplemented growth. [Scale bars: 63.4 μm × 63.4 μm (y axis × x axis) at 100× magnification.]
To address the lack of influence of Opp and Dpp permeases on SIP signaling, we considered two possibilities: (i) SIP is not secreted, and thus does not require active import by peptide permeases, or (ii) SIP is secreted and internalized by a yet-to-be-identified mechanism. The results from secretome swap assays and synthetic peptide addition experiments suggest that SIP is secreted and internalized into the cytosol (35) (Fig. 2). To test the hypothesis that the regulatory factor in the secretome is encoded by the sip gene, we performed secretome swap assays using conditioned medium obtained from the WT or orf-1* mutant strain. Consistent with our hypothesis, compared with the WT secretome, inactivation of SIP in the orf-1* mutant resulted in the loss of regulatory activity in the secretome derived from this strain, indicating that SIP is the high cell density-specific activation factor in the secretome that induces speB expression (Fig. 3D). We next investigated if the secreted SIP is reinternalized into the bacterial cytoplasm using synthetic SIP peptide containing fluorescein modification at its amino terminus (FITC–SIP-1) (Fig. 3E). Peptide addition experiments with FITC–SIP-1 demonstrated that the modified FITC–SIP-1 is functional and retained the ability to induce speB expression in the orf-1* mutant strain similar to unmodified SIP-1 (SI Appendix, Fig. S5). To determine if FITC–SIP-1 is reimported into the bacterial cytoplasm, we incubated the orf-1* mutant with FITC–SIP-1 and assessed the cytosolic presence of FITC–SIP-1 by measuring the relative fluorescence in the clarified cell lysates. An FITC–SIP-1 concentration-dependent increase in fluorescence was observed in the cell lysates from FITC–SIP-1–treated samples compared with SIP-1–treated or untreated GAS (Fig. 3E). Next, we investigated the cytosolic presence of FITC–SIP-1 in the orf-1* mutant strain by confocal microscopy. The fluorescence signal was present in the cytoplasm of GAS cells incubated with FITC–SIP-1, whereas no fluorescent signal was detected in the unsupplemented or unmodified SIP-1–supplemented GAS growth (Fig. 3F). These results are consistent with the interpretation that exogenously added FITC–SIP-1 is transported into the bacterial cytoplasm. To test the presence of additional import mechanisms, we constructed a triple-mutant strain (∆oppDF/∆dppA/orf-1*) in which the orf-1* mutation was introduced into the isogenic double-mutant strain, ∆oppDF/∆dppA. Expression of speB in this triple-mutant strain is dependent entirely on exogenously provided synthetic SIP peptide. Thus, we measured speB transcript levels in the triple-mutant strain grown in the presence or absence of exogenously added SIP. As observed in the orf-1* mutant strain, addition of SIP restored WT levels of speB expression and SpeB protease activity in the triple mutant (Fig. 3 B and C). Together, these data indicate that secreted SIP is reimported into the bacterial cytosol, and that GAS uses specialized export and import mechanisms for SIP signaling that are distinct from other characterized bacterial peptide signaling pathways.
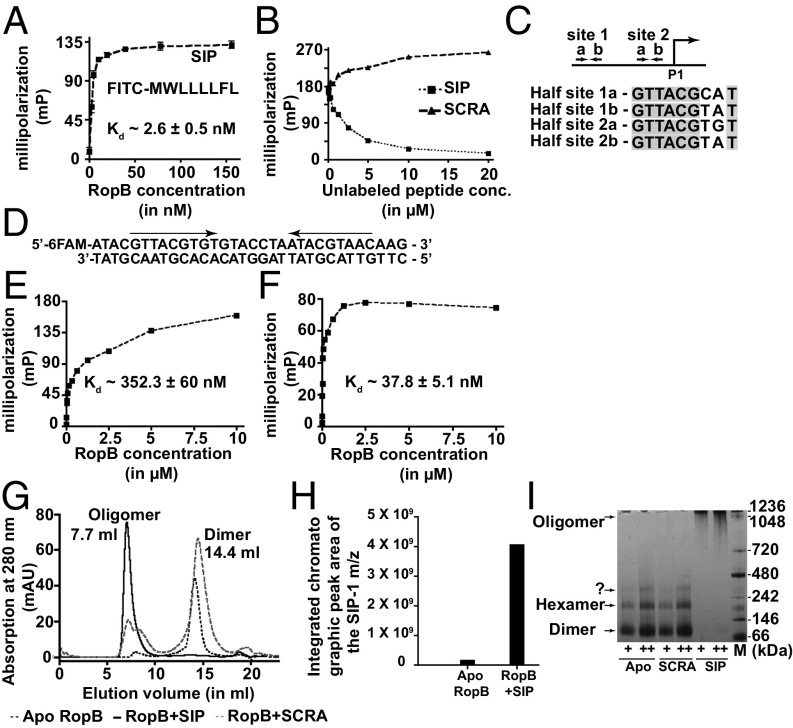
To obtain information about the molecular mechanism of gene regulation by SIP, we used a fluorescence polarization (FP) assay to test the hypothesis that SIP directly interacts with RopB. RopB bound directly to SIP with a Kd of ∼2.6 nM, indicating a very high-affinity interaction between the two partners (Fig. 4A). This strong interaction was disrupted only by the addition of unlabeled SIP, indicating that the RopB–SIP interaction is sequence specific (Fig. 4B). To understand the downstream mechanistic consequences of RopB–SIP interaction, we first assessed the effect of SIP binding on RopB–DNA interactions using an oligoduplex containing the putative RopB-binding site located upstream of the P1 promoter (27) (Fig. 4 C and D). Although apo-RopB had the ability to mediate sequence-specific DNA interactions (Kd of ∼352 nM) (Fig. 4E), SIP binding caused a ninefold increase in the affinity of the SIP-bound form of RopB for DNA (Kd of ∼38 nM) (Fig. 4F). These results suggest that SIP binding promotes high-affinity RopB–DNA interactions.
Fig. 4.
SIP directly interacts with RopB and controls gene regulation by inducing allosteric changes in RopB. (A) Analysis of the binding between purified RopB and fluoresceinated SIP by an FP assay. (B) Ability of SIP or SCRA peptide to compete with the FITC-labeled SIP–RopB complex for binding. A preformed RopB (350 nM)-labeled SIP (10 nM) complex was titrated with the indicated unlabeled peptides. (C) Schematics of the location of RopB-binding sites within the P1 promoter. The transcription start site of the P1 promoter is shown as bent arrows, whereas the two RopB-binding sites with the inverted repeats are marked as arrows. Alignment of the nine-base-long nucleotide sequences of the RopB-binding half-sites from site 1 and site 2 are aligned from a 5′→3′ direction, and the identical bases among the half-sites are shaded in gray. (D) Nucleotide sequence of the RopB-binding site used in the binding studies is shown. The pseudoinverted repeat within the RopB-binding site is marked by arrows. Analysis of the binding between the FITC-labeled oligoduplex containing the putative RopB-binding site and apo-RopB (E) or SIP-bound RopB (F) by FP assay is shown. (G) Size exclusion chromatography analysis of purified RopB with or without the presence of either synthetic SIP or SCRA peptide. Molecular masses were calculated based on the calibration curve using molecular weight standards. (H) Mass spectrometry analyses of the apo- or SIP-bound RopB complex purified as described in E for the presence of SIP. (I) Increasing concentrations of the apo- or peptide-bound form of RopB (+, 1.5 μg: ++, 3 μg) were analyzed by Blu-native PAGE. The oligomeric forms of RopB, assessed based on the molecular weight marker [M; in kilodaltons (kDa)], are labeled.
To further study RopB–DNA interactions, we analyzed the DNA-binding properties of RopB by electrophoretic mobility shift assay (EMSA). RopB bound to promoter sequences containing RopB-binding sites that are located upstream of the P1 promoter, whereas it failed to bind sequences upstream of the P2 promoter that lack the RopB-binding site, indicating that these interactions are sequence specific (SI Appendix, Fig. S6). However, no SIP-induced differences in RopB–DNA interactions were observed, as both apo- and SIP-bound RopB had similar DNA-binding properties (SI Appendix, Fig. S6). Given that apo-RopB binds DNA with relatively high affinity (Fig. 3E), it is likely that the sensitivity of EMSA is not sufficient to distinguish the differences in DNA-binding affinities of apo- and SIP-bound RopB. We also observed that RopB at higher concentrations formed multiple supershifted RopB–DNA complexes, suggesting that RopB multimerizes on the promoter sequences (SI Appendix, Fig. S6 B and C). Finally, to rule out the possibility that the defective speB expression in the orf-1* mutant is not due to impaired RopB binding to speB promoter sequences, we carried out EMSA studies using speB promoter sequences from the WT or orf-1* mutant strain. We observed no significant differences in RopB interactions with the WT and orf-1* promoters (SI Appendix, Fig. S7). These data add further support to our conclusion that translation disruption of orf-1 is the underlying cause for the defective speB expression in the orf-1* mutant strain.
Purified apo-RopB typically eluted as a homodimer during size exclusion chromatography (Fig. 4G). However, we observed that the addition of SIP caused a shift from RopB dimer to a higher order oligomer much larger than a dodecamer (Fig. 4G). Mass spectrometry analysis of the oligomer fractionated by size exclusion chromatography confirmed that the oligomeric RopB contains bound SIP, indicating that SIP binding induces RopB oligomerization (Fig. 4 G and H). These results were further confirmed by Blu-native PAGE analysis of RopB in the presence or absence of SIP. Although apo-RopB had the tendency to oligomerize, the higher order oligomeric form of RopB was stabilized only by SIP binding (Fig. 4I and SI Appendix, Fig. S8). To probe the structural components of RopB involved in SIP-dependent oligomerization, we conducted similar experiments using the C-terminal domain (residues 56–280) of RopB (RopB-CTD) (35). Interestingly, SIP failed to induce RopB-CTD oligomerization (SI Appendix, Fig. S9), suggesting that an intact DNA-binding domain (residues 1–55) is required for SIP-dependent RopB oligomerization. To investigate if RopB binds DNA in both dimeric and oligomeric forms, we conducted EMSA with purified RopB dimer coincubated with SIP and DNA, or preformed RopB-SIP oligomer fractionated by size exclusion chromatography (Fig. 4G). Consistent with previous results, RopB dimer formed higher order RopB-SIP–DNA complex, whereas preformed RopB-SIP oligomer failed to bind DNA (SI Appendix, Fig. S10). These results indicate that the preformed RopB-SIP oligomer is not compatible for DNA binding and that SIP-induced high-affinity RopB–DNA interactions precede RopB multimerization. Together, the data suggest that SIP directly interacts with RopB and modulates gene regulation by inducing a two-pronged allostery in RopB, namely, high-affinity RopB–DNA interactions and RopB polymerization.
Global Gene Regulatory Influence of SIP Signaling Pathway.
To test the hypothesis that SIP controls a global quorum-sensing regulon, we constructed an isogenic ∆orf-1 mutant strain with an in-frame deletion of the entire SIP coding region. Deletion of orf-1 abolished speB expression, and addition of synthetic SIP peptide restored WT-like speB transcription and SpeB protease activity in the ∆orf-1 mutant, indicating the nonpolar nature of the ∆orf-1 mutant (SI Appendix, Fig. S11). We next used RNA-sequencing (RNA-seq) analysis to compare the global transcript profiles of the WT and ∆orf-1 mutant strains grown to stationary phase. Compared with the WT strain, 271 genes (14% of the GAS core chromosome, P < 0.05, and twofold differences) were differentially regulated in the ∆orf-1 mutant strain, of which 228 genes were up-regulated and 43 genes were down-regulated (SI Appendix, Tables S1 and S2). As expected, the level of speB transcript was drastically down-regulated in the ∆orf-1 mutant strain (>2,000-fold reduction) relative to the WT strain (SI Appendix, Fig. S12). Additional genes that were significantly down-regulated in the ∆orf-1 mutant include genes that encode known virulence factors, such as SagP, NdoS, Sdn, and HlyIII (SI Appendix, Table S1). Conversely, the up-regulated genes in the ∆orf-1 mutant strain belong to the following categories: (i) de novo protein synthesis (rps, rpl, and rpm operons that encode ribosomal subunits), (ii) DNA synthesis (pyr, pur, guaAB, nrd, carAB, and rexAB operons), (iii) cell wall synthesis and cell division (murB, murN, pgdA, smc, and divIVA), (iv) group A carbohydrate antigen synthesis (rgpBCDEFG operon), (v) transporters involved in nutrient acquisition (dpp, opp, and pot operons), (vi) proteases (pcp, pepB, pepN, pepO, pepXP, cppA, and clpX), and (vii) cell surface protein (grab) (SI Appendix, Table S2).
To further understand the effect of SIP on global gene regulation, we also performed RNA-seq analysis of WT and orf-1* mutant strains grown to stationary phase. Compared with the WT strain, 72 genes (4% of the GAS core chromosome, P < 0.05, and twofold differences) were differentially expressed in the orf-1* mutant strain, of which 31 genes were up-regulated and 41 genes were down-regulated (SI Appendix, Tables S3 and S4). As observed in the ∆orf-1 mutant strain, the level of speB, spi, and orf-3 (SpyM3_1743) transcripts was drastically down-regulated in the orf-1* mutant strain (>1,300-fold reduction) relative to the WT strain (SI Appendix, Fig. S12). However, the SIP regulon identified in the orf-1* mutant strain is relatively smaller compared with that of the ∆orf-1 mutant strain. To understand the differences in the global gene regulatory influence of SIP between orf-1* and ∆orf-1 mutant strains, we compared the RNA-seq reads from the orf-1* and ∆orf-1 mutant strains within the ropB-speB gene loci (SI Appendix, Fig. S12). Interestingly, the level of ropB transcript was significantly up-regulated in the ∆orf-1 mutant strain relative to the WT strain (greater than threefold increase) (SI Appendix, Fig. S12 and Table S2), whereas no significant difference in the ropB transcript level was observed in the orf-1* mutant strain. Thus, it is possible that transcription activation of speB, spi, and orf-3 (SpyM3_1743) by RopB is SIP-dependent; however, RopB has additional SIP-independent regulatory roles that may contribute to the larger regulon observed in the ∆orf-1 mutant strain. Together, the global transcriptome data demonstrate that the SIP signaling pathway directly or indirectly up-regulates the expression of several secreted virulence factors, including SpeB.
RopB-SIP Signaling Pathway Is Required for WT GAS Virulence.
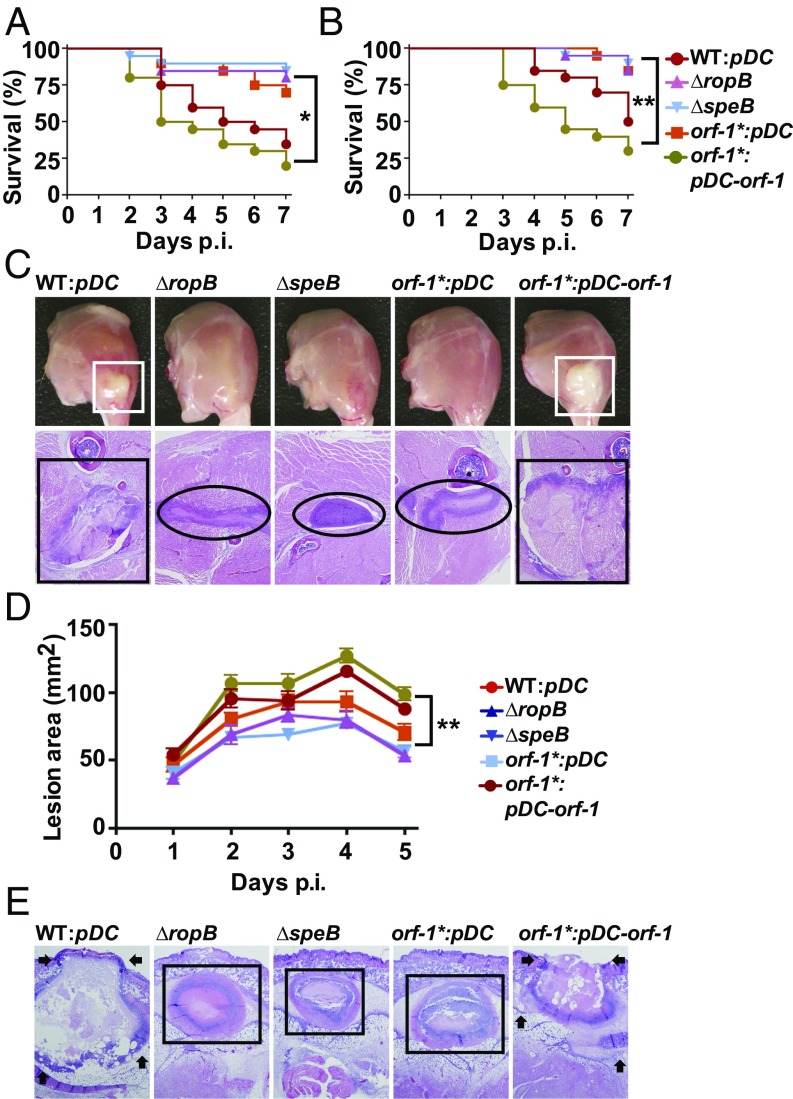
To test the hypothesis that the RopB-SIP signaling pathway participates in GAS pathogenesis, we compared the virulence of the WT and orf-1* mutant strains in a mouse model of bacteremia (17, 49, 50). Consistent with its key role in SpeB production, the virulence of the orf-1* mutant strain was significantly attenuated relative to the WT strain and comparable to that of the ∆ropB and ∆speB mutant strains that lack SpeB production (Fig. 5A). Next, we tested if SIP signaling is also critical for GAS virulence in invasive disease using a mouse model of necrotizing myositis (17, 49, 50). The orf-1* mutant strain was significantly less virulent than the WT and trans-complemented strains (Fig. 5B). Inasmuch as SpeB contributes to host tissue damage and disease dissemination (14–18, 22, 51), we also investigated lesion character by visual and microscopic examination. Consistent with the virulence phenotype, the orf-1*, ∆ropB, and ∆speB mutant strains caused smaller muscle lesions with less severe tissue destruction relative to WT and trans-complemented strains (Fig. 5C). Finally, we tested if the RopB-SIP signaling pathway contributes to GAS virulence in a mouse model of skin and soft tissue infection (17, 49, 50). Compared with WT and trans-complemented strains, the orf-1*, ∆ropB, and ∆speB mutant strains caused significantly smaller and more confined lesions (Fig. 5D) with less tissue damage and ulceration (Fig. 5E). Together, these virulence data demonstrate that the RopB-SIP signaling pathway significantly contributes to GAS pathogenesis at multiple anatomic sites.
Fig. 5.
SIP-mediated regulation of virulence genes is critical for GAS pathogenesis in mouse models of infection. (A) Twenty outbred CD-1 mice were inoculated i.p. with each indicated strain. Kaplan–Meier survival curves with P values derived by the log-rank test are shown. p.i., postinfection. (B) Twenty outbred CD-1 mice per strain were injected i.m. with each indicated strain. Kaplan–Meier survival curves with P values derived by the log rank-test are shown. (C) Gross (Top) and microscopic (Bottom) analyses of hind-limb lesions from mice infected with each indicated strain. (Top) Larger lesions with extensive tissue damage in SpeB-expressing strains are boxed (white boxes). (Bottom) Areas of disseminated lesions in the infected tissues are boxed (black box), whereas confined, less destructive lesions are circled. (D) Fifteen immunocompetent hairless mice were infected s.c. with each indicated strain, and the lesion area produced by each strain was determined. The lesion area was measured and graphed (mean ± SEM). The P value was derived by two-way ANOVA. (E) Histopathologic analysis of lesions from mice infected s.c. with each indicated strain. Areas of disseminated lesions and ulcerations on the skin surfaces caused by SpeB-producing strains are marked by arrows, whereas confined, less destructive lesions caused by SpeB-deficient strains are boxed. [Scale bars: C and E, 2.2 mm × 1.7 mm (y axis × x axis) at 4× magnification.]
RopB-SIP Signaling Pathway Is Active During Mouse Infection.
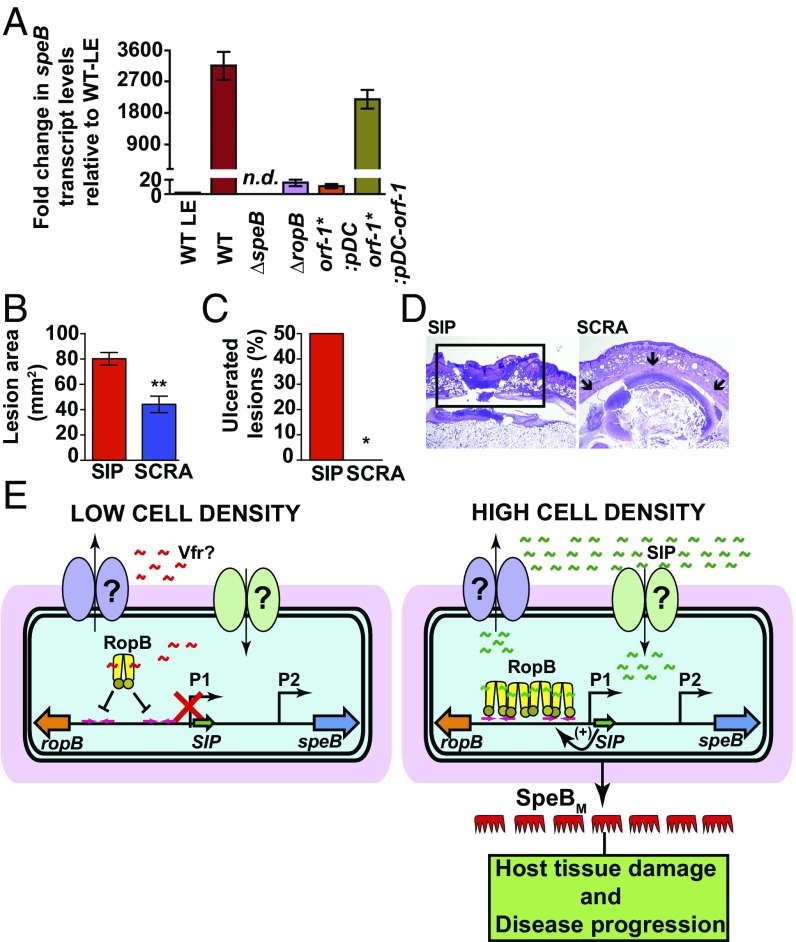
We next investigated if the RopB-SIP–mediated quorum-sensing pathway controls speB expression in vivo during the course of infection. Mice were inoculated s.c. with each of the indicated strains, and speB transcript levels in the infected lesions were measured by qRT-PCR. Compared with the WT strain grown to the late exponential phase in laboratory medium, the WT strain isolated from infected lesions had a 3,000-fold higher level of speB transcript (Fig. 6A). Consistent with the in vitro observations, lesions from mice infected with ∆ropB and orf-1* mutants had drastically decreased speB expression in vivo and speB transcript levels were comparable to those of the WT strain in the late exponential growth phase (Fig. 6A). Importantly, trans-complementation of the orf-1* mutant strain with pDC–orf-1 fully restored WT-like speB transcript levels, indicating that SIP-mediated speB regulation occurs in vivo (Fig. 6A). Finally, we assessed the ability of synthetic peptide SIP to activate speB expression in vivo and promote bacterial virulence using the s.c. mouse model of infection. Coinjection with SIP restored a WT-like virulence phenotype to the orf-1* mutant strain, resulting in larger ulcerated lesions relative to orf-1* mutant coinjected with the SCRA peptide (Fig. 6 B–D). These data indicate that synthetic peptides containing the SIP amino acid sequences have biological activity in vivo, and are sufficient to restore GAS pathogenesis in the orf-1* mutant strain. Collectively, our data demonstrate that SIP signaling is active during host infection and, moreover, show that SIP-mediated up-regulation of virulence gene expression contributes significantly to GAS virulence.
Fig. 6.
SIP signaling controls speB expression during infection. (A) Analysis of the speB transcript level in the s.c. lesions from mice infected with the indicated strains. Samples were collected 24 h postinfection (P.I.) from the lesions of four mice per strain and analyzed in triplicate. Data were graphed as mean SD, with P values derived from a two-sample t test. Ten immunocompetent hairless mice per group were infected s.c. with the orf-1* mutant coinjected with either 10 μg of synthetic SIP or SCRA peptide. LE, late exponential GAS growth in laboratory medium. The lesion area (B) and ulceration (C) caused by each peptide at 24 h P.I. were determined. The lesion area was measured and graphed (mean ± SEM). The P value was derived by the Mann–Whitney test. *P < 0.05; **P < 0.001. (D) Histopathologic analysis of lesions from mice coinjected s.c. with SIP or SCRA peptide. SIP-induced ulcerated lesions that extend beyond the field of view are boxed. Coinjection with SCRA peptide caused small abscesses that are confined to the inoculation site (indicated by arrows). [Scale bars: 3.3 mm × 4.4 mm (y axis × x axis) at 2× magnification.] (E) Proposed model for the mechanism of intercellular communication and GAS virulence regulation. (Left) At low cell density, the secretion signal sequence of Vfr binds to RopB and negatively influences RopB-dependent transcription activation from the P1 promoter, possibly by disrupting RopB–DNA interactions. (Right) At high cell density, SIP is produced, secreted, and reimported into the cytosol. The high-affinity RopB–DNA interactions and RopB polymerization aided by SIP binding lead to up-regulation of sip expression, which results in robust induction of SIP production by a positive-feedback mechanism. In addition to up-regulation of virulence genes, the SIP signaling circuit down-regulates the expression of categories of genes involved in GAS growth and host cell attachment. Finally, the SIP-dependent up-regulation of speB leads to abundant secretion of mature SpeB (SpeBM), which facilitates host tissue damage and disease dissemination by cleavage of various host and GAS proteins.
Discussion
In the aggregate, we here show that GAS uses complex intercellular communication machinery to monitor its population density and determine whether to initiate a virulent lifestyle that involves host tissue damage and disease dissemination. The results presented herein show that an intercellular peptide signal, SIP, and the intracellular global gene regulator RopB form a signal/receptor pair that contributes significantly to GAS pathogenesis by modulating virulence gene expression. Importantly, the nucleotide sequence encoding the inferred eight amino acids of SIP is absolutely conserved, and the promoter sequences upstream of the SIP-coding region are highly conserved among 11 different GAS serotypes (SI Appendix, Fig. S13). Thus, the SIP signaling pathway likely represents a virulence regulatory mechanism conserved among many GAS M-protein serotypes.
All previously characterized bacterial peptide signals are produced as propeptides that have a cleavable amino-terminal secretion signal sequence and protease cleavage sites, resulting in a mature peptide (8) (Fig. 3A). The secretion signal sequence targets the propeptide to the secretion machinery at the cell membrane, where the propeptide undergoes proteolytic cleavage. The released mature peptide is exported, and subsequently reimported into the cytosol by Opp permeases (39). In contrast, SIP lacks the distinctive sequence motifs of bacterial peptide signals. GAS produces SIP in a mature form that is devoid of a secretion signal sequence and protease cleavage site (Fig. 3A). Nevertheless, SIP is secreted and reimported by an Opp permease-independent import mechanism. These differences indicate that SIP biosynthesis is an important exception to the established paradigm of bacterial peptide signaling. Thus, SIP belongs to a previously undescribed new class of leaderless bacterial peptide signals. Such lack of conformity likely hobbled previous extensive efforts to identify SIP (18, 21, 22, 27, 30–33, 35, 36). Given that RRNPP family proteins are widespread among low guanine + cytosine gram-positive bacteria and bacteriophages (42, 52), and the cognate peptide signals for the vast majority of these regulators have not yet been identified, we speculate that leaderless peptides analogous to SIP will participate in other microbial signaling pathways. Thus, our delineation of SIP signaling may accelerate discovery of similar peptide signals in other microorganisms.
Typically, the internalized cognate peptides bind to the respective regulators and trigger regulator-specific conformational changes that modulate gene regulation (39). The characterized activation peptide-induced allostery among RRNPP family regulators includes disruption of tetramerization in PrgX from Enterococcus faecalis, induction of tetramerization in NprR from Bacillus cereus, and unmasking of the N-terminal DNA-binding domain that aids promoter binding by PlcR from B. cereus (39, 53–55). Our results indicate that SIP controls RopB regulatory activity by facilitating high-affinity RopB–DNA interactions and RopB polymerization (Fig. 4). The sequences upstream of the P1 promoter have two putative RopB-binding sites, site 1 and site 2, and these two sites are separated by a 121-bp-long spacer region. The two sites are highly similar (Fig. 4C), and RopB binds to each site with comparable affinity. Thus, it is plausible that SIP promotes RopB interactions with the two high-affinity operator sequences. Using these interactions as nucleation events, RopB polymerizes on the spacer between the two sites, resulting in RopB-dependent transcription activation from the P1 promoter. However, additional investigations will be required to test the proposed model and elucidate the mechanism by which RopB polymerization contributes to its regulatory activity.
Based on earlier work (18) and results presented here, we propose the following model (Fig. 6E). During low cell density, the inhibition peptide signal derived from the secretion signal sequence of Vfr interacts with RopB and negatively influences speB expression, possibly by inhibiting RopB–DNA interactions (Fig. 6E). Conversely, during high cell density, expression of vfr is down-regulated, which results in low-level SIP production. The initial SIP production acts as a positive feedback loop, resulting in robust induction of SIP. The SIP-bound RopB binds to operator sequences, polymerizes on the promoter, and mediates transcription activation of target genes.
To summarize, the data we present here provide detailed molecular and mechanistic understanding of a key virulence regulatory pathway of an abundant human pathogen responsible for greater than 700 million human infections annually worldwide (56, 57). Moreover, the work identified previously unknown molecular targets that may be exploited to develop new therapeutics.
Materials and Methods
Bacterial Strains, Plasmids, and Experimental Procedures.
The bacterial strains and plasmids used in this study are listed in SI Appendix, Table S5. Probes and primers used in this study are listed in SI Appendix, Table S6. The composition of the chemically defined medium (58) is provided in SI Appendix, Table S7. Details of the isogenic mutant strain construction, trans-complementation plasmids, and plasmids for overexpression are included in SI Appendix, Supplemental Materials and Methods. Details of protein overexpression and purification are provided in SI Appendix, Supplemental Materials and Methods. Preparation of synthetic peptides used for SIP addition experiments, FP assays, EMSA, and animal infection studies is described in SI Appendix, Supplemental Materials and Methods. Details of speB transcript level analysis by qRT-PCR, secreted SpeB protein levels by Western immunoblotting, and SpeB protease activity by milk plate clearing assay are provided in SI Appendix, Supplemental Materials and Methods. Secretome swap assays were performed as described previously (8), and the details are given in SI Appendix, Supplemental Materials and Methods. Details of confocal microscopy studies and fluorescence measurements to monitor the uptake of fluorescein-labeled SIP are included in SI Appendix, Supplemental Materials and Methods.
Northern Blot and RNA-Seq Analysis.
Northern blot analysis was performed as previously described (59). Membranes were hybridized in ULTRAhyb Ultrasensitive Hybridization Buffer (Thermo Fisher) at 42 °C with 32P-end-labeled DNA oligonucleotides. Signals were visualized with a Typhoon phosphorimager (GE Healthcare), and band intensities were quantified using GelQuant software (BiochemLabSolutions). RNA-seq experiments were performed as described previously (60). Experimental details are given in SI Appendix, Supplemental Materials and Methods.
RopB–DNA Interaction Studies.
Interactions between RopB and SIP and dsDNA were studied by FP assay and EMSA. Details of binding isotherm measurement conditions and protocols are given in SI Appendix, Supplemental Materials and Methods.
Analysis of RopB Oligomerization State.
Size exclusion chromatography and blue-native polyacrylamide gel electrophoresis were used to determine the oligomerization state of recombinant RopB in the presence and absence of synthetic peptides. Experiment details are provided in SI Appendix, Supplemental Materials and Methods. Details of sample preparation and mass spectrometry analysis of the different oligomeric forms of RopB fractionated by size exclusion chromatography are described in SI Appendix, Supplemental Materials and Methods.
Animal Virulence Studies.
Mouse experiments were performed according to protocols approved by the Houston Methodist Research Institute Institutional Animal Care and Use Committee. The studies were carried out in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals (61). Virulence of the isogenic mutant GAS strains was assessed using three mouse models of infection, namely, i.p., i.m., and s.c. inoculation (17, 18, 62) (approved nos. AUP-0716-0038, AUP-0615-0041, and AUP-0416-0019). Details of mouse infection studies and data analyses are given in SI Appendix, Supplemental Materials and Methods.
Supplementary Material
Acknowledgments
We thank Frank DeLeo for critical reading of the manuscript, and an anonymous reviewer for scholarly suggestions to improve the manuscript. This work was supported by NIH Grant 1R01AI109096-01A1 (to M.K.), by the Deutsche Forschungsgemeinschaft (Exc114-2) (to K.P.), and by funds from the Fondren Foundation (to J.M.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1705972114/-/DCSupplemental.
References
- 1.Parker CT, Sperandio V. Cell-to-cell signalling during pathogenesis. Cell Microbiol. 2009;11:363–369. doi: 10.1111/j.1462-5822.2008.01272.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hughes DT, Sperandio V. Inter-kingdom signalling: Communication between bacteria and their hosts. Nat Rev Microbiol. 2008;6:111–120. doi: 10.1038/nrmicro1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sanson M, et al. Phosphorylation events in the multiple gene regulator of group A Streptococcus significantly influence global gene expression and virulence. Infect Immun. 2015;83:2382–2395. doi: 10.1128/IAI.03023-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dubbs JM, Mongkolsuk S. Peroxide-sensing transcriptional regulators in bacteria. J Bacteriol. 2012;194:5495–5503. doi: 10.1128/JB.00304-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hood MI, Skaar EP. Nutritional immunity: Transition metals at the pathogen-host interface. Nat Rev Microbiol. 2012;10:525–537. doi: 10.1038/nrmicro2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Somerville GA, Proctor RA. At the crossroads of bacterial metabolism and virulence factor synthesis in staphylococci. Microbiol Mol Biol Rev. 2009;73:233–248. doi: 10.1128/MMBR.00005-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rutherford ST, Bassler BL. Bacterial quorum sensing: Its role in virulence and possibilities for its control. Cold Spring Harb Perspect Med. 2012;2:a012427. doi: 10.1101/cshperspect.a012427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dunny GM, Leonard BA. Cell-cell communication in gram-positive bacteria. Annu Rev Microbiol. 1997;51:527–564. doi: 10.1146/annurev.micro.51.1.527. [DOI] [PubMed] [Google Scholar]
- 9.Papenfort K, Bassler BL. Quorum sensing signal-response systems in gram-negative bacteria. Nat Rev Microbiol. 2016;14:576–588. doi: 10.1038/nrmicro.2016.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thoendel M, Kavanaugh JS, Flack CE, Horswill AR. Peptide signaling in the staphylococci. Chem Rev. 2011;111:117–151. doi: 10.1021/cr100370n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Olsen RJ, Shelburne SA, Musser JM. Molecular mechanisms underlying group A streptococcal pathogenesis. Cell Microbiol. 2009;11:1–12. doi: 10.1111/j.1462-5822.2008.01225.x. [DOI] [PubMed] [Google Scholar]
- 12.Cunningham MW. Pathogenesis of group A streptococcal infections. Clin Microbiol Rev. 2000;13:470–511. doi: 10.1128/cmr.13.3.470-511.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carroll RK, Musser JM. From transcription to activation: How group A Streptococcus, the flesh-eating pathogen, regulates SpeB cysteine protease production. Mol Microbiol. 2011;81:588–601. doi: 10.1111/j.1365-2958.2011.07709.x. [DOI] [PubMed] [Google Scholar]
- 14.Lukomski S, et al. Inactivation of Streptococcus pyogenes extracellular cysteine protease significantly decreases mouse lethality of serotype M3 and M49 strains. J Clin Invest. 1997;99:2574–2580. doi: 10.1172/JCI119445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lukomski S, et al. Extracellular cysteine protease produced by Streptococcus pyogenes participates in the pathogenesis of invasive skin infection and dissemination in mice. Infect Immun. 1999;67:1779–1788. doi: 10.1128/iai.67.4.1779-1788.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lukomski S, et al. Genetic inactivation of an extracellular cysteine protease (SpeB) expressed by Streptococcus pyogenes decreases resistance to phagocytosis and dissemination to organs. Infect Immun. 1998;66:771–776. doi: 10.1128/iai.66.2.771-776.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Olsen RJ, et al. Decreased necrotizing fasciitis capacity caused by a single nucleotide mutation that alters a multiple gene virulence axis. Proc Natl Acad Sci USA. 2010;107:888–893. doi: 10.1073/pnas.0911811107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shelburne SA, 3rd, et al. An amino-terminal signal peptide of Vfr protein negatively influences RopB-dependent SpeB expression and attenuates virulence in Streptococcus pyogenes. Mol Microbiol. 2011;82:1481–1495. doi: 10.1111/j.1365-2958.2011.07902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Svensson MD, et al. Role for a secreted cysteine proteinase in the establishment of host tissue tropism by group A streptococci. Mol Microbiol. 2000;38:242–253. doi: 10.1046/j.1365-2958.2000.02144.x. [DOI] [PubMed] [Google Scholar]
- 20.Olsen RJ, et al. The majority of 9,729 group A Streptococcus strains causing disease secrete SpeB cysteine protease: Pathogenesis implications. Infect Immun. 2015;83:4750–4758. doi: 10.1128/IAI.00989-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loughman JA, Caparon M. Regulation of SpeB in Streptococcus pyogenes by pH and NaCl: A model for in vivo gene expression. J Bacteriol. 2006;188:399–408. doi: 10.1128/JB.188.2.399-408.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lyon WR, Gibson CM, Caparon MG. A role for trigger factor and an rgg-like regulator in the transcription, secretion and processing of the cysteine proteinase of Streptococcus pyogenes. EMBO J. 1998;17:6263–6275. doi: 10.1093/emboj/17.21.6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ulrich RG. Vaccine based on a ubiquitous cysteinyl protease and streptococcal pyrogenic exotoxin A protects against Streptococcus pyogenes sepsis and toxic shock. J Immune Based Ther Vaccines. 2008;6:8. doi: 10.1186/1476-8518-6-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang AY, González-Páez GE, Wolan DW. Identification and co-complex structure of a new S. pyogenes SpeB small molecule inhibitor. Biochemistry. 2015;54:4365–4373. doi: 10.1021/acs.biochem.5b00607. [DOI] [PubMed] [Google Scholar]
- 25.Kapur V, et al. Vaccination with streptococcal extracellular cysteine protease (interleukin-1 β convertase) protects mice against challenge with heterologous group A streptococci. Microb Pathog. 1994;16:443–450. doi: 10.1006/mpat.1994.1044. [DOI] [PubMed] [Google Scholar]
- 26.Björck L, et al. Bacterial growth blocked by a synthetic peptide based on the structure of a human proteinase inhibitor. Nature. 1989;337:385–386. doi: 10.1038/337385a0. [DOI] [PubMed] [Google Scholar]
- 27.Neely MN, Lyon WR, Runft DL, Caparon M. Role of RopB in growth phase expression of the SpeB cysteine protease of Streptococcus pyogenes. J Bacteriol. 2003;185:5166–5174. doi: 10.1128/JB.185.17.5166-5174.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Graham MR, et al. Virulence control in group A Streptococcus by a two-component gene regulatory system: Global expression profiling and in vivo infection modeling. Proc Natl Acad Sci USA. 2002;99:13855–13860. doi: 10.1073/pnas.202353699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller AA, Engleberg NC, DiRita VJ. Repression of virulence genes by phosphorylation-dependent oligomerization of CsrR at target promoters in S. pyogenes. Mol Microbiol. 2001;40:976–990. doi: 10.1046/j.1365-2958.2001.02441.x. [DOI] [PubMed] [Google Scholar]
- 30.Kietzman CC, Caparon MG. CcpA and LacD.1 affect temporal regulation of Streptococcus pyogenes virulence genes. Infect Immun. 2010;78:241–252. doi: 10.1128/IAI.00746-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shelburne SA, et al. A combination of independent transcriptional regulators shapes bacterial virulence gene expression during infection. PLoS Pathog. 2010;6:e1000817. doi: 10.1371/journal.ppat.1000817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Podbielski A, Woischnik M, Pohl B, Schmidt KH. What is the size of the group A streptococcal vir regulon? The Mga regulator affects expression of secreted and surface virulence factors. Med Microbiol Immunol (Berl) 1996;185:171–181. doi: 10.1007/s004300050028. [DOI] [PubMed] [Google Scholar]
- 33.Ribardo DA, McIver KS. Defining the Mga regulon: Comparative transcriptome analysis reveals both direct and indirect regulation by Mga in the group A Streptococcus. Mol Microbiol. 2006;62:491–508. doi: 10.1111/j.1365-2958.2006.05381.x. [DOI] [PubMed] [Google Scholar]
- 34.Ma Y, et al. Identification and characterization of bicistronic speB and prsA gene expression in the group A Streptococcus. J Bacteriol. 2006;188:7626–7634. doi: 10.1128/JB.01059-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Makthal N, et al. Structural and functional analysis of RopB: A major virulence regulator in Streptococcus pyogenes. Mol Microbiol. 2016;99:1119–1133. doi: 10.1111/mmi.13294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Podbielski A, Woischnik M, Kreikemeyer B, Bettenbrock K, Buttaro BAL. Cysteine protease SpeB expression in group A streptococci is influenced by the nutritional environment but SpeB does not contribute to obtaining essential nutrients. Med Microbiol Immunol (Berl) 1999;188:99–109. doi: 10.1007/s004300050111. [DOI] [PubMed] [Google Scholar]
- 37.Elliott SD. A proteolytic enzyme produced by group A streptococci with special reference to its effect on the type-specific M antigen. J Exp Med. 1945;81:573–592. doi: 10.1084/jem.81.6.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tongs MS. Ectoenzymes of streptococci. JAMA. 1919;73:1277–1279. [Google Scholar]
- 39.Do H, Kumaraswami M. Structural mechanisms of peptide recognition and allosteric modulation of gene regulation by the RRNPP family of quorum-sensing regulators. J Mol Biol. 2016;428:2793–2804. doi: 10.1016/j.jmb.2016.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen Z, Itzek A, Malke H, Ferretti JJ, Kreth J. Dynamics of speB mRNA transcripts in Streptococcus pyogenes. J Bacteriol. 2012;194:1417–1426. doi: 10.1128/JB.06612-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chang JC, LaSarre B, Jimenez JC, Aggarwal C, Federle MJ. Two group A streptococcal peptide pheromones act through opposing Rgg regulators to control biofilm development. PLoS Pathog. 2011;7:e1002190. doi: 10.1371/journal.ppat.1002190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fleuchot B, et al. Rgg proteins associated with internalized small hydrophobic peptides: A new quorum-sensing mechanism in streptococci. Mol Microbiol. 2011;80:1102–1119. doi: 10.1111/j.1365-2958.2011.07633.x. [DOI] [PubMed] [Google Scholar]
- 43.An FY, Clewell DB. Identification of the cAD1 sex pheromone precursor in Enterococcus faecalis. J Bacteriol. 2002;184:1880–1887. doi: 10.1128/JB.184.7.1880-1887.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chandler JR, Dunny GM. Characterization of the sequence specificity determinants required for processing and control of sex pheromone by the intramembrane protease Eep and the plasmid-encoded protein PrgY. J Bacteriol. 2008;190:1172–1183. doi: 10.1128/JB.01327-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gominet M, Slamti L, Gilois N, Rose M, Lereclus D. Oligopeptide permease is required for expression of the Bacillus thuringiensis plcR regulon and for virulence. Mol Microbiol. 2001;40:963–975. doi: 10.1046/j.1365-2958.2001.02440.x. [DOI] [PubMed] [Google Scholar]
- 46.Leonard BA, Podbielski A, Hedberg PJ, Dunny GM. Enterococcus faecalis pheromone binding protein, PrgZ, recruits a chromosomal oligopeptide permease system to import sex pheromone cCF10 for induction of conjugation. Proc Natl Acad Sci USA. 1996;93:260–264. doi: 10.1073/pnas.93.1.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Podbielski A, et al. Molecular characterization of group A streptococcal (GAS) oligopeptide permease (opp) and its effect on cysteine protease production. Mol Microbiol. 1996;21:1087–1099. doi: 10.1046/j.1365-2958.1996.661421.x. [DOI] [PubMed] [Google Scholar]
- 48.Podbielski A, Leonard BA. The group A streptococcal dipeptide permease (Dpp) is involved in the uptake of essential amino acids and affects the expression of cysteine protease. Mol Microbiol. 1998;28:1323–1334. doi: 10.1046/j.1365-2958.1998.00898.x. [DOI] [PubMed] [Google Scholar]
- 49.Olsen RJ, Musser JM. Molecular pathogenesis of necrotizing fasciitis. Annu Rev Pathol. 2010;5:1–31. doi: 10.1146/annurev-pathol-121808-102135. [DOI] [PubMed] [Google Scholar]
- 50.Watson ME, Jr, Neely MN, Caparon MG. Animal models of Streptococcus pyogenes infection. In: Ferretti J, Stevens D, Fishcetti V, editors. Streptococcus pyogenes: Basic Biology to Clinical Manifestations. University of Oklahoma Health Sciences Center; Oklahoma City: 2016. pp. 629–660. [Google Scholar]
- 51.Kapur V, et al. A conserved Streptococcus pyogenes extracellular cysteine protease cleaves human fibronectin and degrades vitronectin. Microb Pathog. 1993;15:327–346. doi: 10.1006/mpat.1993.1083. [DOI] [PubMed] [Google Scholar]
- 52.Erez Z, et al. Communication between viruses guides lysis-lysogeny decisions. Nature. 2017;541:488–493. doi: 10.1038/nature21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grenha R, et al. Structural basis for the activation mechanism of the PlcR virulence regulator by the quorum-sensing signal peptide PapR. Proc Natl Acad Sci USA. 2013;110:1047–1052. doi: 10.1073/pnas.1213770110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zouhir S, et al. Peptide-binding dependent conformational changes regulate the transcriptional activity of the quorum-sensor NprR. Nucleic Acids Res. 2013;41:7920–7933. doi: 10.1093/nar/gkt546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shi K, et al. Structure of peptide sex pheromone receptor PrgX and PrgX/pheromone complexes and regulation of conjugation in Enterococcus faecalis. Proc Natl Acad Sci USA. 2005;102:18596–18601. doi: 10.1073/pnas.0506163102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sanyahumbi AS, Colquhoun S, Wyber R, Carapetis JR. Global disease burden of group A Streptococcus. In: Ferretti J, Stevens D, Fishcetti V, editors. Streptococcus pyogenes: Basic Biology to Clinical Manifestations. University of Oklahoma Health Sciences Center; Oklahoma City: 2016. pp. 661–704. [PubMed] [Google Scholar]
- 57.Ralph AP, Carapetis JR. Group A streptococcal diseases and their global burden. In: Chhatwal SG, editor. Host-Pathogen Interactions in Streptococcal Diseases. Springer; Heidelberg: 2012. pp. 1–27. [Google Scholar]
- 58.van de Rijn I, Kessler RE. Growth characteristics of group A streptococci in a new chemically defined medium. Infect Immun. 1980;27:444–448. doi: 10.1128/iai.27.2.444-448.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fröhlich KS, Haneke K, Papenfort K, Vogel J. The target spectrum of SdsR small RNA in Salmonella. Nucleic Acids Res. 2016;44:10406–10422. doi: 10.1093/nar/gkw632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sanson M, et al. Adhesin competence repressor (AdcR) from Streptococcus pyogenes controls adaptive responses to zinc limitation and contributes to virulence. Nucleic Acids Res. 2015;43:418–432. doi: 10.1093/nar/gku1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.National Research Council . Guide for the Care and Use of Laboratory Animals. 8th Ed National Academies Press; Washington, DC: 2011. [Google Scholar]
- 62.VanderWal AR, et al. Iron efflux by PmtA is critical for oxidative stress resistance and contributes significantly to group A Streptococcus virulence. Infect Immun. 2017;85:e00091–17. doi: 10.1128/IAI.00091-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.