Significance
Diverse species from all over the bacterial tree of life produce antibiotics to limit the growth of competitors and thereby enhance their resource availability. Here we examined the pairwise inhibition between bacterial species from natural settings. We find that bacteria mainly inhibit the growth of metabolically similar and evolutionary related species, in line with Darwin’s age old competition-relatedness hypothesis. We further find that inhibiting the growth of other species is associated with a generalist lifestyle, suggesting a trade-off between specialists efficiently growing on few resources and generalists who are able to use many resources but have to inhibit the specialists to obtain them.
Keywords: antagonism, competition, competition-relatedness hypothesis, antibiotics, bacteria
Abstract
In the Origin of Species, Charles R. Darwin [Darwin C (1859) On the Origin of Species] proposed that the struggle for existence must be most intense among closely related species by means of their functional similarity. It has been hypothesized that this similarity, which results in resource competition, is the driver of the evolution of antagonism among bacteria. Consequently, antagonism should mostly be prevalent among phylogenetically and metabolically similar species. We tested the hypothesis by screening for antagonism among all possible pairwise interactions between 67 bacterial species from 8 different environments: 2,211 pairs of species and 4,422 interactions. We found a clear association between antagonism and phylogenetic distance, antagonism being most likely among closely related species. We determined two metabolic distances between our strains: one by scoring their growth on various natural carbon sources and the other by creating metabolic networks of predicted genomes. For both metabolic distances, we found that the probability of antagonism increased the more metabolically similar the strains were. Moreover, our results were not compounded by whether the antagonism was between sympatric or allopatric strains. Intriguingly, for each interaction the antagonizing strain was more likely to have a wider metabolic niche than the antagonized strain: that is, larger metabolic networks and growth on more carbon sources. This indicates an association between an antagonistic and a generalist strategy.
More than 150 y ago, Charles R. Darwin (1) speculated that since congeneric species are generally more similar than species from distinct genera, they are likely to compete more fiercely. This competition-relatedness hypothesis has a long history in ecology, but explicit tests of this fundamental hypothesis are missing for the most prevalent and diverse group of all organisms, bacteria. Empirical support for this hypothesis in other organisms is inconsistent, with some finding evidence supporting this hypothesis (2–5), but others find none (6–8).
In bacteria fierce competition is exemplified by antagonism, for example, with the production of antibiotics to directly inhibit the growth of other species (9). Several experimental studies have examined congeneric antagonism in bacteria (i.e., antagonism between species from the same genera) and they have revealed that antagonism is often reciprocal and related to prior coexistence (10), and that antagonism is mostly prevalent between communities (allopatric) rather than within communities (sympatric) (11, 12). It is therefore clear that congeneric antagonism abounds, but it is unknown whether there is more antagonism between closely related bacteria than between more distantly related ones.
Case and Gilpin (13) hypothesized that resource competition drives the evolution of interference competition (i.e., antagonism), and that there is a trade-off between interference competition and resource exploitation efficiency. Accordingly, antagonism should be most prevalent among metabolically similar species, but we would further hypothesize that antagonists are generalists; it is a trade-off between specializing in efficiently exploiting few resources or growing on many resources and antagonizing the specialists. To test this, we used 67 different natural bacterial strains isolated from 8 different environments, 7 of them soil. The strains represent a diverse range of phyla (Fig. 1 and Table S1), and the strains from each of the seven soil environments were sampled from a small area (0.5–2 cm2), and are thus more likely to have influenced each other than strains from distinct environments. The criterion for inclusion in the study was visible growth on a rich medium (tryptic soy agar, TSA) at 24 °C in 1 d, so all strains were aerobic, heterotrophic, fast growing, and potential competitors due to this overlapping niche. For all possible pairwise interactions (4,422; i.e., 2,211 pairs) we screened whether one bacterial strain produced a diffusible substance that inhibited the growth of the other strain. Although the expression of an inhibitory phenotype can change depending on the growth conditions, having the ability to directly inhibit another strain is a strong sign of antagonism (11).
Fig. 1.
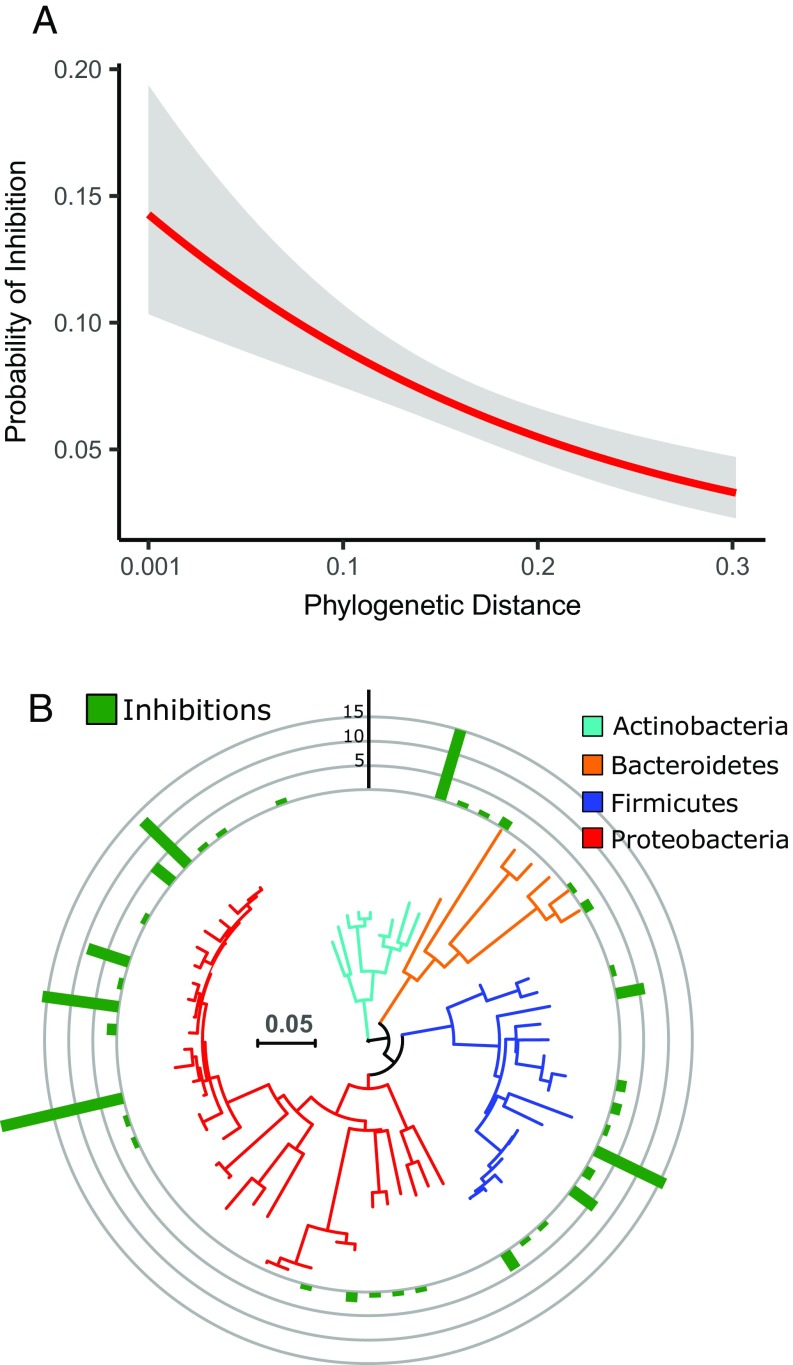
(A) Bacteria mainly antagonize closely related genotypes; the probability of inhibition increases with decreasing 16S rRNA gene phylogenetic distance. The red line is a logistic regression and the gray shaded area denotes the 95% confidence interval. (P < 0.001, n = 2,211). (B) Few strains inhibit many others, but these superkillers are randomly distributed across the phylogeny.
Table S1.
Overview of the 67 isolates used in the study
| Isolate | GenBank accession no. | Environment | Source | Closest relative in EzBioCloud database | Identity, % |
| 1 | LT547828 | Soil 1 | (32) | Flavobacterium oncorhynchi | 98.5 |
| 2 | JQ890536 | Soil 1 | (33) | Pseudomonas abietaniphila | 98.7 |
| 3 | LT547829 | Soil 1 | (32) | Pseudomonas brassicacearum | 99.8 |
| 4 | LT547830 | Soil 1 | (32) | Pseudomonas jessenii | 99.1 |
| 5 | LT547831 | Soil 1 | (32) | Microbacterium oxydans | 99.7 |
| 6 | LT547832 | Soil 1 | (32) | Chryseobacterium ginsenosidimutans | 98.4 |
| 7 | LT547833 | Soil 1 | (32) | Chryseobacterium jejuense | 99.5 |
| 8 | JQ890540 | Soil 1 | (33) | Paenibacillus xylanexedens | 99.5 |
| 9 | LT547834 | Soil 1 | (32) | Sphingobacteriumc anadense | 97.4 |
| 10 | JQ890537 | Soil 1 | (33) | Pseudomonas geniculata | 99.2 |
| 11 | JQ890541 | Soil 1 | (33) | Arthrobacter nitroguajacolicus | 99.0 |
| 12 | JQ890542 | Soil 1 | (33) | Ochrobactrum rhizospharae | 100 |
| 13 | JQ890538 | Soil 1 | (33) | Stenotrophomonas rhizophila | 99.8 |
| 14 | JQ890539 | Soil 1 | (33) | Microbacterium oxydans | 100 |
| 15 | LT547835 | Soil 1 | (32) | Ensifer morelense | 99.9 |
| 16 | LT547836 | Soil 1 | (32) | Janthinobacterium lividum | 99.7 |
| 17 | LK391509 | Freshwater streambed 1 | (16) | Paenibacillus amylolyticus | 99.4 |
| 18 | LK391510 | Freshwater streambed 1 | (16) | Pseudomonas migulae | 99.9 |
| 19 | LK391511 | Freshwater streambed 1 | (16) | Bacillus simplex | 99.2 |
| 20 | LK391512 | Freshwater streambed 1 | (16) | Pseudomonas fragi | 99.9 |
| 21 | LK391513 | Freshwater streambed 1 | (16) | Bacillus aryabhattai | 100 |
| 22 | LK391516 | Freshwater streambed 1 | (16) | Bacillus mycoides | 100 |
| 23 | LK391514 | Freshwater streambed 1 | (16) | Arthrobacter antarticus | 99.5 |
| 24 | LK391515 | Freshwater streambed 1 | (16) | Erwinia billingiae | 99.6 |
| 25 | LT547844 | Freshwater streambed 1 | Present study | Janthinobacterium lividum | 99.8 |
| 26 | LT547845 | Freshwater streambed 2 | Present study | Shewanella putrefaciens | 99.2 |
| 27 | LT547846 | Freshwater streambed 2 | Present study | Bacillus marisflavi | 99.9 |
| 28 | LT547847 | Freshwater streambed 2 | Present study | Bacillus mycoides | 99.8 |
| 29 | LT547848 | Freshwater streambed 2 | Present study | Bacillus mycoides | 99.9 |
| 30 | LT547849 | Freshwater streambed 2 | Present study | Brevibacterium frigoritolerans | 99.9 |
| 31 | LT547850 | Freshwater streambed 2 | Present study | Pseudomonas jessenii | 99.5 |
| 32 | LT547851 | Freshwater streambed 2 | Present study | Pseudomonas veronii | 99.9 |
| 33 | LT547852 | Freshwater streambed 2 | Present study | Bacillus isronensis | 99.8 |
| 34 | KJ831327 | Soil 2 | Present study | Pseudomonas oryzihabitans | 99.9 |
| 35 | KJ831339 | Soil 2 | Present study | Mycobacterium immunogenum | 99.3 |
| 36 | KJ831340 | Soil 2 | Present study | Rhodococcus globerulus | 99.7 |
| 37 | KJ831344 | Soil 2 | Present study | Paenibacillus macquariensis | 99.9 |
| 38 | LT547837 | Soil 2 | Present study | Collimonas pratensis | 100 |
| 39 | LT547838 | Soil 2 | Present study | Collimonas areanae | 100 |
| 40 | LT547839 | Soil 2 | Present study | Nevskia soli | 99.8 |
| 41 | KJ831347 | Soil 2 | Present study | Bradyrhizobium erythrophlei | 99.4 |
| 42 | KJ831348 | Soil 2 | Present study | Micrococcus yunnanensis | 100 |
| 43 | LK391522 | Soil 3 | (16) | Pseudomonas granadensis | 99.7 |
| 44 | LK391523 | Soil 3 | (16) | Bacillus mycoides | 99.9 |
| 45 | LK391524 | Soil 3 | (16) | Viridibacillus arenosi | 100 |
| 46 | LK391525 | Soil 3 | (16) | Bacillus simplex | 99.9 |
| 47 | LK391526 | Soil 3 | (16) | Rahnella victoriana | 99.9 |
| 48 | LK391527 | Soil 3 | (16) | Pseudomonas helmanticensis | 99.6 |
| 49 | LK391528 | Soil 3 | (16) | Pseudomonas jessenii | 99.2 |
| 50 | LK391529 | Soil 3 | (16) | Janthinobacterium lividum | 99.8 |
| 51 | DQ328319 | Marine | (34) | Microbacterium phyllosphaerae | 99.4 |
| 52 | DQ328322 | Marine | (34) | Prolinoborus fasciculus | 99.9 |
| 53 | LT547840 | Marine | Present study | Bacillus anthracis | 100 |
| 54 | LT547841 | Marine | Present study | Micrococcus yunnanensis | 99.9 |
| 55 | LT547842 | Marine | Present study | Bacillus anthracis | 100 |
| 56 | LT547843 | Marine | Present study | Prolinoborus fasciculus | 99.9 |
| 57 | DQ981456 | Soil 4 | (34) | Bacillus nealsonii | 99.2 |
| 58 | DQ981457 | Soil 4 | (34) | Pseudomonas helmanticensis | 99.1 |
| 59 | DQ981458 | Soil 4 | (34) | Pseudomonas helmanticensis | 99.5 |
| 60 | DQ981460 | Soil 4 | (34) | Pseudomonas mohnii | 99.6 |
| 61 | DQ981461 | Soil 4 | (34) | Flavobacterium pectinovorum | 97.7 |
| 62 | DQ981462 | Soil 4 | (34) | Pseudomonas mohnii | 99.6 |
| 63 | DQ981463 | Soil 4 | (34) | Dyadobacter psychrophilus | 99.0 |
| 64 | DQ981464 | Soil 4 | (34) | Chryseobacterium shandongense | 99.9 |
| 65 | LT547853 | Rhizosphere | Present study | Pseudomonas congelans | 99.7 |
| 66 | LT547854 | Rhizosphere | Present study | Pseudomonas granadensis | 100 |
| 67 | LT547855 | Rhizosphere | Present study | Pseudomonas congelans | 99.7 |
Results
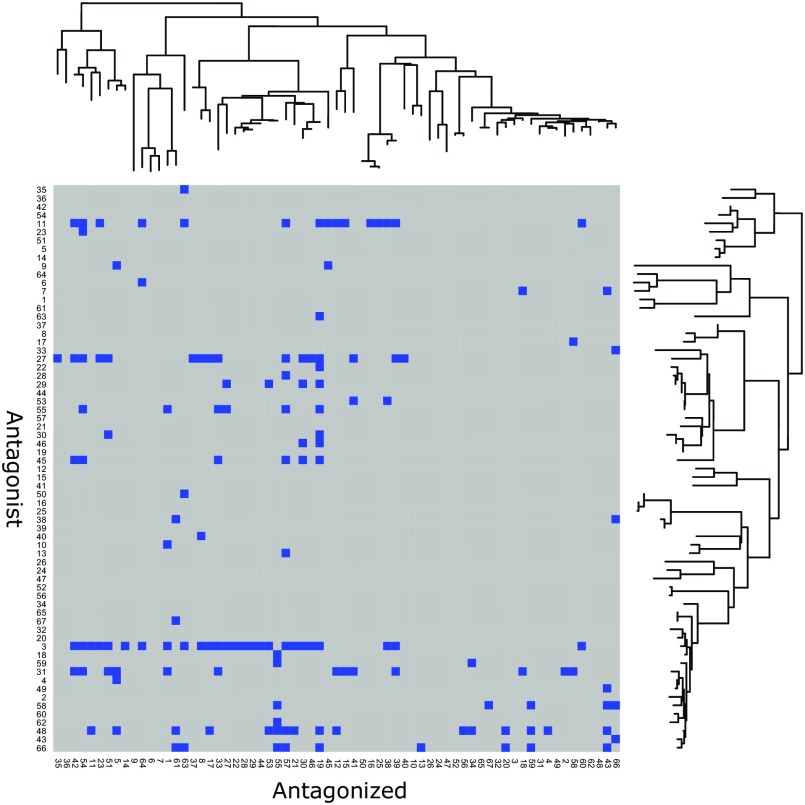
Of all of the possible interactions, 148 of 4,422 (3.4%) were inhibitory, resulting in 147 of 2,211 (6.6%) competing pairs, two interactions per pair, whether A inhibits B and whether B inhibits A (Fig. S1). No strains were found to inhibit themselves, and these self–self controls were not included in the analysis. Among the 2,211 competing bacterial pairs, we found that antagonism was enriched between closely related strains (P < 0.001, pseudo-R2 = 0.024, logistic regression used throughout unless otherwise noted) (Fig. 1A). We note that most of the variation in the data was explained by between-strain variation in antagonizing and being antagonized; a mixed-effect logistic regression, with antagonists and antagonized as random intercepts and phylogenetic distance as fixed effect, had a good fit (P < 0.001, pseudo-R2 = 0.572).
Fig. S1.
Heatmap of all interactions ordered by the phylogenetic tree. Antagonistic interactions are shown in blue.
Half of the strains inhibited at least one other. A few “superkillers” were responsible for a disproportionally large number of the inhibitions, but being a superkiller appears not to be phylogenetically clustered (Fig. 1B).
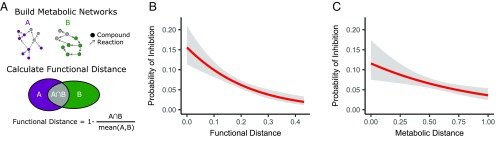
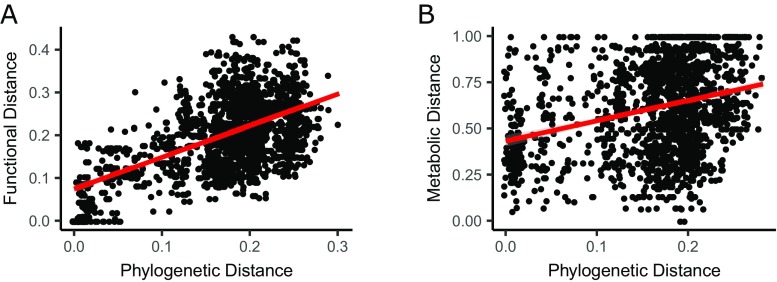
To assess whether the association between antagonism and phylogeny is driven by the metabolic niche overlap between the strains, we created metabolic networks from genomes predicted with PICRUSt (14). We calculated a “functional similarity” between the strains, defined as the number of shared metabolites (nodes in the networks) divided by the mean number of metabolites of both networks. A “functional distance” was then defined as one minus the functional similarity (Fig. 2A). We found a clear association between the functional distance and antagonism (P < 0.001, pseudo-R2 = 0.032) (Fig. 2B). These results were validated by growing the strains on 31 different carbon sources (BiOLOG EcoPlate) and calculating a metabolic distance among all pairs (one minus overlap/total). As seven strains did not grow on any of the carbon sources, we excluded them from this analysis. Among the 60 strains able to grow on the BiOLOG EcoPlate, there was a significant negative correlation between antagonism and metabolic distances (P = 0.0023, pseudo-R2 = 0.015) (Fig. 2C): The greater the overlap in growth on the 31 EcoPlate substances, the higher a probability of antagonism. Furthermore, removing the five superkillers that inhibited more than 10 other strains did not alter the results for either phylogenetic distance (P < 0.001), functional distance (P = 0.003), or metabolic distance (P = 0.003). As both the functional distances and metabolic distances correlate with phylogenetic distances (Fig. S2), we repeated the analyses after removing the effect of phylogeny on the functional distances and metabolic distances; instead of the original functional and metabolic distances, we used the residuals from a linear regression on the phylogenetic distances, (i.e., the variation unexplained by phylogenetic distances). Antagonism still correlated with both functional distances (P = 0.0026) and metabolic distances (P = 0.046).
Fig. 2.
(A) A functional distance was calculated between all strain pairs by overlapping the compounds (nodes) in their metabolic networks. (B) The probability of inhibition increases with decreasing functional distance. The red line is a logistic regression and the gray shaded area denotes the 95% confidence intervals (P < 0.001, n = 2,211). (C) The probability of inhibition increases with decreasing metabolic distance as calculated from their growth patterns on BiOLOG Ecoplates. The red line is a logistic regression and the gray shaded area denotes the 95% confidence intervals (P = 0.0023, n = 1,770).
Fig. S2.
(A) Correlation between functional distance and phylogenetic distance. The red line is a linear regression. (B) Correlation between metabolic distance and phylogenetic distance. The red line is a linear regression.
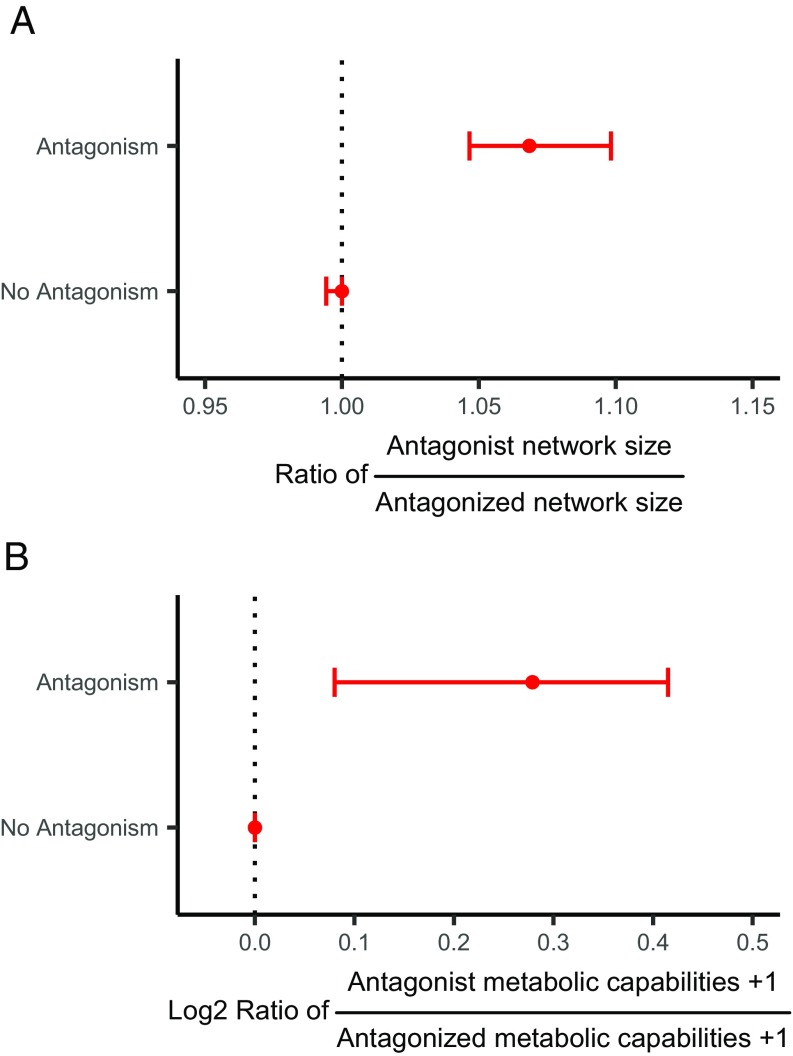
We further investigated the trade-off between being an antagonist and a generalist or specialist. For each antagonistic interaction, we asked whether the antagonist had a wider metabolic niche space (metabolic network size or growth on BiOLOG EcoPlates) or not, and compared this to interactions with no observed antagonism. For both the metabolic networks and the BiOLOG EcoPlates, we found that the antagonists are most likely to have larger networks and grow on more carbon sources, respectively [Fisher’s exact test, odds ratio (OR) = 2.65, P < 0.001 and OR = 1.53, P = 0.012, respectively] (Fig. 3).
Fig. 3.
(A) The ratios between metabolic network sizes of the antagonists compared with the antagonized are larger than 1 when antagonism was observed (P = 0), but not different from 1 for the remaining interactions (P = 0.99). (B) The log2 ratios between the number of carbon sources antagonists grow on compared with how many the antagonized grow on are larger than 0 when antagonism was observed (P = 0.03), but not different from 0 for the remaining interactions (P = 1). P values are one-tailed tests of whether medians are larger than 1 (A) or 0 (B) estimated from 1,000 bootstrap realizations. Points are medians and error bars are 90% bootstrapped confidence limits of the medians (equivalent to 5% one-tailed test of the median). (n = 148 and n = 4,274 for “antagonism” and “no antagonism,” respectively).
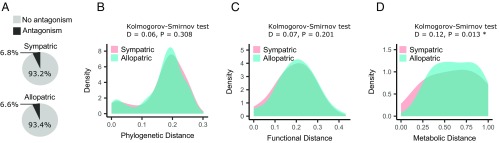
We then asked if the results were influenced by whether the interacting strains were sympatric or allopatric; there was no difference in the amount of inhibitory interactions between sympatric or allopatric strains (OR = 0.97, P = 0.90, Fisher’s exact test) (Fig. 4A). Furthermore, we found no difference between the distributions of the phylogenetic and functional distances, whether they were sympatric or allopatric strains (Fig. 4 B and C), but for metabolic distances there was a significant enrichment of metabolically similar sympatric strains (Fig. 4D).
Fig. 4.
(A) The proportion of inhibitory interactions is no different whether the strains were sympatric or allopatric (OR = 0.97, P = 0.90, Fisher’s exact test). (B and C) There is no difference in the distribution of phylogenetic or functional distances between sympatric and allopatric strains, but there is an overrepresentation of sympatric strains that are (D) metabolically similar.
Discussion
In microbes antagonism by production of antimicrobial compounds appears to be a widespread phenomenon (see for example, Fig. 1B). The syntheses of these secondary metabolites likely confer a cost for the producer that must be outweighed by the benefits, and this warrants an exploration of the evolutionary drivers. We would expect that a bacterial strain would antagonize other strains that use the same resources as it does. We further hypothesized that if the production of an antibacterial compound confers a cost to the producer, we would find a trade-off between antagonism and resource specialization. Two opposing strategies thus emerge: a specialist, which can efficiently use few resources, and a generalist, which inefficiently uses many resources but inhibits the specialists to obtain those resources.
Here we screened 2,211 pairs of bacterial strains for antagonism and found that closely related bacteria do indeed have a higher probability of antagonism than more distantly related bacteria (Fig. 1A). However, this finding only supports the hypothesis if traits of resource use are phylogenetically conserved. We used dissimilarity of the 16S rRNA gene as a proxy for phylogenetic distance since it is widely used in microbial marker gene studies, but it is a coarse proxy and might not be strongly associated with resource use. Therefore, we screened for the metabolic capabilities of our strains with the BiOLOG EcoPlate system. The EcoPlates contain 31 carbon sources specifically chosen to distinguish between soil communities (15). Since most of our strains originate from soil (61 of 67), this system was chosen to evaluate the metabolic capabilities of the strains. As expected, metabolic distance had a significant correlation with antagonism (Fig. 2C). To further strengthen this finding, we created metabolic networks of each strain based on their predicted genomes. We find that strains with a high overlap between their networks were more likely to antagonize each other, thus further corroborating the hypothesis. Additionally, the correlations were still present even after transforming the metabolic and functional distances to remove the effect of phylogeny.
Intriguingly, most of the observed antagonism was mainly driven by few strains (Fig. 1B), which suggests that these few strains have a particular antagonistic life-strategy. We therefore explored whether the antagonizing strains were also generalists, as we had hypothesized. For each antagonistic interaction we would therefore expect the antagonist to have a wider metabolic niche than the antagonized. This approach is more robust than simply correlating the width of the metabolic niche with the number of inhibitions for each strain, as we can ensure that variations in metabolic niche size across the phylogeny does not influence the results; antagonists are only expected to have wider metabolic niches than their competitors (which are phylogenetically related) (Fig. 1A), and not necessarily wide metabolic niches in general. Our hypothesis was supported by both the metabolic network analysis and the BiOLOG EcoPlate screening, where the metabolic networks size of the antagonists were ∼5–10% larger than that of the antagonized, and the antagonists grew on ∼7–30% more carbon sources than the antagonized (Fig. 3). Importantly, no such difference was found for the remaining interactions where no antagonism was observed (Fig. 3). This strongly suggests that antagonists are also generalists.
We did not find any difference in the probability of sympatric vs. allopatric antagonism (Fig. 4A). As several studies on congeneric competition have found an effect of prior coexistence (10–12), it is possible that coexistence mainly affects interactions between closely related bacteria. This is also concordant with results from Madsen et al. (16); these authors studied cocultures of bacterial strains and found the strongest effect of prior coexistence among closely related strains. Vetsigian et al. (10) and Cordero et al. (11) examined congeneric competition in Streptomyces and Vibrio, respectively, and both found that closely related strains were competing less than more distantly related strains. Distantly related strains were, in these studies, still from the same genera, and the results are therefore not necessarily in disagreement with ours. It is therefore possible that on the large taxonomical scale studied here (strains from four phyla), metabolic similarity appears to be the main driver of antagonism, while the effect of prior coexistence is stronger for congeneric interactions. We could also expect that for very closely related coexisting strains, other evolutionary drivers, such as inclusive fitness, could play a role in the evolution of antagonism.
Overall, only a few of the interactions were identified as competitive (6.6%), but this does not indicate that most interactions are noncompetitive in these environments. First, we only looked for antagonism, so competition by outgrowth and other such exploitative behaviors were not detected. Second, we only identified inhibition if mediated by an inhibitor that was diffusible in agar. Third, we only identified inhibition that was produced by the strain growing by itself, in a specifically chosen medium, temperature, and temporal span (TSB, 24 °C, 2 d).
Despite the clear association between phylogenetic relatedness and antagonism, the processes that lead to interspecific competition are likely multifaceted. Notably, both phylogeny and our measures of resource use explained only a small fraction of the variation in antagonism. Furthermore, bacteria coexist with many other types of bacteria, so it is therefore crucial to move from studying pairwise interactions to exploring higher order interactions (17). Including more strains in the experiments change results substantially (18–20), and this is certainly the path toward elucidating the role of biotic interactions in microbial communities. The competition-relatedness hypothesis is a fundamental piece of the puzzle of community assembly, but relating these findings with studies on natural communities must be a next step to reveal the processes that govern coexistence of both bacterial species and species in general.
Materials and Methods
Bacterial Strains.
The strains used in this study all originate from the Molecular Microbial Ecology group collection (Table S1). All strains, except “Marine,” originating from the same environment were all isolated from an area between 0.5 and 2 cm2 at the same time. “Soil 1” isolates were isolated from a baby maize leaf incubated in soil. Marine strains were isolated from the algae Ulva lactuca, as described in Rao et al. (21). “Soil 2” strains were isolated from a forest topsoil, as described in Lladó et al. (22), but on GYM Streptomyces medium. “Soil 3” strains were isolated from small stones (diameter: 5 mm). Other strains from the “Present study” were isolated from stones (diameter: 5 mm) sampled from a streambed (“Freshwater streambed”) or from roots of Aegopodium podagraria (Ground elder, identified by J.R., “Rhizosphere”), all sampled in February 2013 at Brobæk Mose, Denmark. Pebbles and roots were gently washed in phosphate-buffered saline (PBS), and then samples were vortexed in PBS to release the attached bacteria, which were subsequently plated on TSA and incubated at 24 °C. Colonies with different morphologies were restreaked at least three times and afterward cryopreserved in 20% glycerol at −80 °C. 16S rRNA genes were amplified with 27F (5′-AGAGTTTGATCMTGGCTCAG-3′) and 1492R (5′-TACGGYTACCTTGTTACGACTT-3′) primers, and subsequently Sanger-sequenced with 27F, 518F (5′-CCAGCAGCCGCGGTAATACG-3′), and 1492R primers. Sequences were assembled with CLC Main Workbench (CLC bio) and curated manually. Taxonomical affiliations of the strains were determined through the EzBioCloud database (23).
Screening of Inhibition.
A screening of all possible pairwise interactions was done once using the Burkholder plate assay (24, 25) to test for inhibition between the selected strains. Broth cultures of the 67 strains growing in TSB at 24 °C for 2 d were each spread onto three square TSA plates (10 × 10 cm) to create bacterial lawns (aliquots of 200 µL). Next, 24 wells (diameter: 0.5 cm) were made in each plate, and aliquots (70 µL) of each broth culture were transferred into separate wells, resulting in a total of 4,489 interactions (67 × 67), with 67 of those being self-inhibition controls. Plates were placed at 24 °C and inhibition was scored as presence/absence (1/0) of clearing zones after 3 d of incubation.
Screening of Metabolic Capabilities.
Metabolic capabilities were screened with the BiOLOG EcoPlate system (Biolog Inc.). Overnight cultures (TSB, 24 °C) were washed twice in PBS (1 mL, 5000 × g, 5 min) and the EcoPlates were then inoculated with 150 µL washed culture and incubated at 24 °C. Absorbance was measured after 24 h at 590 nm.
Metabolic Networks and Functional Distance.
First, the 16S rRNA gene sequences were blasted against the Greengenes v13.5 16S rRNA gene database (26) with prfectBLAST (27). Second, the best hit that matched a 16S rRNA gene from an OTU in the precalculated Kyoto Encyclopedia of Genes and Genomes (KEGG) Ortholog table provided by PICRUSt (14) was selected. Third, the metabolic networks were generated from the list of KEGG Orthologs with RevEcoR (28) (threshold = 0, is giant = FALSE). A functional similarity was then calculated for each pair as the overlap between nodes (metabolites) in the two networks divided by the mean number of nodes in both networks. The functional distance was then defined as 1 subtracted by the functional similarity.
Data Analysis.
For each pair of strains, we had two values for inhibition: x = 1 if A inhibited B and y = 1 if B inhibited A, both 0 if no inhibition was observed. We denoted pairs to be antagonistic if x, y, or both were 1. If both x and y were 1 it would mean reciprocal inhibition, which was only observed between one pair. Phylogenetic distances were calculated from 16S rRNA gene sequences with the Ribosomal Database Project aligner and distance calculator (29). A phylogenetic tree was made in Genious 9.0.2 (Biomatters) with neighbor-joining and the Tamura-Nei model, and visualized with iTOL (30). Additional data analysis was done in R 3.1.0 (31). Calculating the metabolic distances between the strains were done based on growth in each of the different BiOLOG substrates. Growth was scored as positive if the absorbance, subtracted the blank control, was above the 97.5% percentile of the blank values (equivalent to 5% level for two tailed distributions). Jaccard distances were then calculated for each pair of strains. Of the 67 strains, 7 were not able to grow on any of the 31 substrates of the BiOLOG EcoPlate. These seven strains were therefore excluded in analyses involving the metabolic distances. Associations between antagonism and the phylogenetic, metabolic, and functional distances were evaluated with logistic regression (glm, binomial family).
To infer whether the correlation between antagonism and the functional and metabolic distances was not confounded by the correlation between the phylogenetic distances and the functional and metabolic distances, we first made a linear regression for both functional distance and metabolic distance with phylogenetic distance as the independent variable. The residuals (i.e., the variation not explained by phylogenetic distance) were then used as independent variables in the logistic regressions.
Data Availability.
All sequences have been deposited in GenBank (Table S1 for accession numbers). Experimental data and R scripts are available upon request.
Acknowledgments
We thank Salvador Lladó for sharing his bacterial strains (Soil 2), and Karin Vestberg and Anette Hørdum Løth for technical assistance. The study was funded by grants from The Danish Council for Independent Research, Grants DFF-1335-00071 and DFF-1323-00235 (SIMICOM) (to S.J.S. and M.B.); and grants from the Villum Foundation, Young Investigator Programme (to M.B.). J.R. was supported by a Novo scholarship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper has been deposited in the GenBank database (accession nos. LT547837–LT547855, KJ831327, KJ831339, KJ831340, KJ831344, KJ831347, and KJ831348).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1706016114/-/DCSupplemental.
References
- 1.Darwin C. On the Origin of Species. John Murray; London: 1859. [Google Scholar]
- 2.Cavender-Bares J, Ackerly DD, Baum DA, Bazzaz FA. Phylogenetic overdispersion in Floridian oak communities. Am Nat. 2004;163:823–843. doi: 10.1086/386375. [DOI] [PubMed] [Google Scholar]
- 3.Stubbs WJ, Wilson JB. Evidence for limiting similarity in a sand dune community. J Ecol. 2004;92:557–567. [Google Scholar]
- 4.Violle C, Nemergut DR, Pu Z, Jiang L. Phylogenetic limiting similarity and competitive exclusion. Ecol Lett. 2011;14:782–787. doi: 10.1111/j.1461-0248.2011.01644.x. [DOI] [PubMed] [Google Scholar]
- 5.Kunstler G, et al. Plant functional traits have globally consistent effects on competition. Nature. 2016;529:204–207. doi: 10.1038/nature16476. [DOI] [PubMed] [Google Scholar]
- 6.Cahill JF, Kembel SW, Lamb EG, Keddy PA. Does phylogenetic relatedness influence the strength of competition among vascular plants? Perspect Plant Ecol Evol Syst. 2008;10:41–50. [Google Scholar]
- 7.Venail PA, et al. The influence of phylogenetic relatedness on species interactions among freshwater green algae in a mesocosm experiment. J Ecol. 2014;102:1288–1299. [Google Scholar]
- 8.Alexandrou MA, et al. Evolutionary relatedness does not predict competition and co-occurrence in natural or experimental communities of green algae. Proc Biol Sci. 2015;282:20141745. doi: 10.1098/rspb.2014.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hibbing ME, Fuqua C, Parsek MR, Peterson SB. Bacterial competition: Surviving and thriving in the microbial jungle. Nat Rev Microbiol. 2010;8:15–25. doi: 10.1038/nrmicro2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vetsigian K, Jajoo R, Kishony R. Structure and evolution of Streptomyces interaction networks in soil and in silico. PLoS Biol. 2011;9:e1001184. doi: 10.1371/journal.pbio.1001184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cordero OX, et al. Ecological populations of bacteria act as socially cohesive units of antibiotic production and resistance. Science. 2012;337:1228–1231. doi: 10.1126/science.1219385. [DOI] [PubMed] [Google Scholar]
- 12.Pérez-Gutiérrez RA, et al. Antagonism influences assembly of a Bacillus guild in a local community and is depicted as a food-chain network. ISME J. 2013;7:487–497. doi: 10.1038/ismej.2012.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Case TJ, Gilpin ME. Interference competition and niche theory. Proc Natl Acad Sci USA. 1974;71:3073–3077. doi: 10.1073/pnas.71.8.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Langille MGI, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Insam H. A new set of substrates proposed for community characterization in environmental samples. In: Insam H, Rangger A, editors. Microbial Communities. Functional Versus Structural Approaches. Springer; Heidelberg: 1997. pp. 260–261. [Google Scholar]
- 16.Madsen JS, et al. Coexistence facilitates interspecific biofilm formation in complex microbial communities. Environ Microbiol. 2016;18:2565–2574. doi: 10.1111/1462-2920.13335. [DOI] [PubMed] [Google Scholar]
- 17.Rivett DW, et al. Resource-dependent attenuation of species interactions during bacterial succession. ISME J. 2016;10:2259–2268. doi: 10.1038/ismej.2016.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ren D, Madsen JS, Sørensen SJ, Burmølle M. High prevalence of biofilm synergy among bacterial soil isolates in cocultures indicates bacterial interspecific cooperation. ISME J. 2015;9:81–89. doi: 10.1038/ismej.2014.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abrudan MI, et al. Socially mediated induction and suppression of antibiosis during bacterial coexistence. Proc Natl Acad Sci USA. 2015;112:11054–11059. doi: 10.1073/pnas.1504076112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kelsic ED, Zhao J, Vetsigian K, Kishony R. Counteraction of antibiotic production and degradation stabilizes microbial communities. Nature. 2015;521:516–519. doi: 10.1038/nature14485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rao D, Webb JS, Kjelleberg S. Competitive interactions in mixed-species biofilms containing the marine bacterium Pseudoalteromonas tunicata. Appl Environ Microbiol. 2005;71:1729–1736. doi: 10.1128/AEM.71.4.1729-1736.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lladó S, Žifčáková L, Větrovský T, Eichlerová I, Baldrian P. Functional screening of abundant bacteria from acidic forest soil indicates the metabolic potential of Acidobacteria subdivision 1 for polysaccharide decomposition. Biol Fertil Soils. 2016;52:251–260. [Google Scholar]
- 23.Kim O-S, et al. Introducing EzTaxon-e: A prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol. 2012;62:716–721. doi: 10.1099/ijs.0.038075-0. [DOI] [PubMed] [Google Scholar]
- 24.Burkholder PR, Pfister RM, Leitz FH. Production of a pyrrole antibiotic by a marine bacterium. Appl Microbiol. 1966;14:649–653. doi: 10.1128/am.14.4.649-653.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Long RA, Azam F. Antagonistic interactions among marine pelagic bacteria. Appl Environ Microbiol. 2001;67:4975–4983. doi: 10.1128/AEM.67.11.4975-4983.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DeSantis TZ, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Santiago-Sotelo P, Ramirez-Prado JH. prfectBLAST: A platform-independent portable front end for the command terminal BLAST+ stand-alone suite. Biotechniques. 2012;53:299–300. doi: 10.2144/000113953. [DOI] [PubMed] [Google Scholar]
- 28.Cao Y, Wang Y, Zheng X, Li F, Bo X. RevEcoR: An R package for the reverse ecology analysis of microbiomes. BMC Bioinformatics. 2016;17:294. doi: 10.1186/s12859-016-1088-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cole JR, et al. The Ribosomal Database Project: Improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2009;37:D141–D145. doi: 10.1093/nar/gkn879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Letunic I, Bork P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.R Core Team 2014. R: A Language and Environment for Statistical Computiing v3.1.0 (R Foundation for Statistical Computing, Vienna)
- 32.de la Cruz-Perera CI, et al. The ability of soil bacteria to receive the conjugative IncP1 plasmid, pKJK10, is different in a mixed community compared to single strains. FEMS Microbiol Lett. 2013;338:95–100. doi: 10.1111/1574-6968.12036. [DOI] [PubMed] [Google Scholar]
- 33.Ren D, et al. High-throughput screening of multispecies biofilm formation and quantitative PCR-based assessment of individual species proportions, useful for exploring interspecific bacterial interactions. Microb Ecol. 2014;68:146–154. doi: 10.1007/s00248-013-0315-z. [DOI] [PubMed] [Google Scholar]
- 34.Burmølle M, et al. Enhanced biofilm formation and increased resistance to antimicrobial agents and bacterial invasion are caused by synergistic interactions in multispecies biofilms. Appl Environ Microbiol. 2006;72:3916–3923. doi: 10.1128/AEM.03022-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All sequences have been deposited in GenBank (Table S1 for accession numbers). Experimental data and R scripts are available upon request.