Significance
This paper describes a mechanism of regulation of Sirt1 activity by Sirt7. Deacetylases Sirt1 and Sirt7 belong to the mammalian family of seven sirtuins, which play important regulatory roles in several biological processes such as metabolism and aging. We discovered that Sirt1 is able to augment its own catalytical activity by autodeacetylation. Sirt7 binds to Sirt1 and inhibits its activity. The biological importance of this regulation was revealed in the differentiation and maintenance of white adipose tissue (WAT). Sirt7 knockout mice contain a significantly diminished amount of WAT due to the increased Sirt1 activity and Sirt1 inhibition restores WAT in Sirt7 knockout mice.
Keywords: sirtuin, acetylation, adipogenesis
Abstract
Sirtuins (Sirt1–Sirt7) are NAD+-dependent protein deacetylases/ADP ribosyltransferases, which play decisive roles in chromatin silencing, cell cycle regulation, cellular differentiation, and metabolism. Different sirtuins control similar cellular processes, suggesting a coordinated mode of action but information about potential cross-regulatory interactions within the sirtuin family is still limited. Here, we demonstrate that Sirt1 requires autodeacetylation to efficiently deacetylate targets such as p53, H3K9, and H4K16. Sirt7 restricts Sirt1 activity by preventing Sirt1 autodeacetylation causing enhanced Sirt1 activity in Sirt7−/− mice. Increased Sirt1 activity in Sirt7−/− mice blocks PPARγ and adipocyte differentiation, thereby diminishing accumulation of white fat. Thus, reduction of Sirt1 activity restores adipogenesis in Sirt7−/− adipocytes in vitro and in vivo. We disclosed a principle controlling Sirt1 activity and uncovered an unexpected complexity in the crosstalk between two different sirtuins. We propose that antagonistic interactions between Sirt1 and Sirt7 are pivotal in controlling the signaling network required for maintenance of adipose tissue.
The seven sirtuins in mammals (Sirt1–Sirt7) are involved in the regulation of essential cellular processes. Sirtuins rapidly adjust the activity of chromatin, transcription factors, metabolic enzymes, and structural proteins to cellular needs by deacetylating a broad range of targets. The ability to sense metabolic alterations and various stressors enable sirtuins to adapt cellular homeostasis to varying conditions. It seems likely that this feature of sirtuins is crucial to prevent age-dependent pathologies and promote a healthy lifespan (1, 2).
Sirt1 is the most widely studied mammalian sirtuin showing the highest homology to the founding member of the sirtuin family, the yeast silence information regulator, Sir2. Sirt1 deacetylates histones H3K9, H3K56, H4K16, and H1K26 as well as many nonhistone targets thereby contributing to the maintenance of metabolic homeostasis and genomic integrity (3, 4). Sirt1 was also identified as a critical component of lifespan extension in response to calorie restriction in several model organisms, although its exact contribution is still under debate (5). The functions of Sirt7 have attracted less attention compared with Sirt1. It is known that Sirt7 specifically deacetylates H3K18 and a few other targets, which are primarily involved in activation of rDNA transcription (6, 7).
Sirtuins are generally assumed to allow organisms to cope with different stressors and prevent aging but their specific functions differ considerably. Although the full complexity of the regulatory potency of individual sirtuins still needs to be uncovered, similar effects have been described for several sirtuins. For example, Sirt1 and Sirt2 can both inhibit adipogenesis. Sirt1 inhibits adipogenesis in 3T3-L1–derived adipocytes by binding and repressing PPARγ via association with the PPARγ corepressor nuclear receptor corepressor 1 (NCoR1) (8, 9). Similarly, Sirt2 does inhibit adipogenesis and accumulation of lipids in 3T3-L1 adipocytes by deacetylation of FOXO1, which represses PPARγ (10). The role of Sirt6 in the regulation of adipogenic differentiation is less clear although it is known that Sirt6 knockout (KO) mice suffer from reduced adipose tissue stores, while Sirt6 overexpressing mice are protected against high-fat diet-induced obesity (11, 12). Interestingly, Sirt1 and Sirt6 both mediate effects of rosiglitazone on hepatic lipid accumulation (13). Sirtuins also synergistically cooperate to improve the mitochondrial metabolism at different regulatory levels: Sirt1 activates mitochondrial gene expression, while Sirt3, Sirt4, and Sirt5 directly modify various mitochondrial enzymes (14, 15). In contrast to synergistic functions, antagonistic effects of sirtuins have been reported as well. For example, Sirt1 inhibits whereas Sirt7 activates rDNA transcription (16).
Despite the obvious need to coordinate the actions of different sirtuins, no concepts have been developed to explain the crosstalk between individual sirtuins. The mechanistic basis of potential regulatory interactions between sirtuins has remained enigmatic so far preventing a comprehensive understanding of the integration of different sirtuin-driven regulatory pathways. Here, we describe that Sirt1 activates itself by deacetylation of the K230 residue. This self-activation of Sirt1 is mitigated by Sirt7, which binds to Sirt1 and inhibits autodeacetylation. Furthermore, we show that the inhibition of Sirt1 by Sirt7 serves an important physiological function during adipogenesis. We found that Sirt7 is instrumental for efficient adipocyte differentiation in vivo and in vitro by restraining the activity of Sirt1, thereby allowing efficient activation of PPARγ.
Results
Sirt7 Is an Interaction Partner of Sirt1.
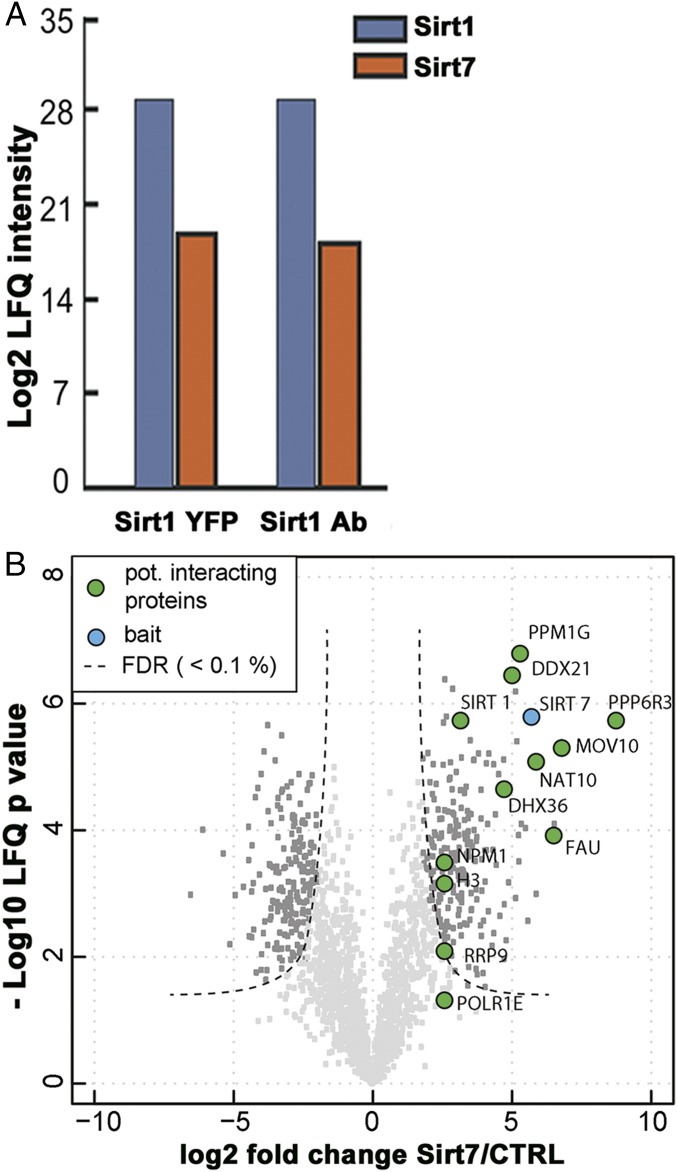
Sirt1 is regulated by NAD+ as well as by microRNAs, protein–protein interactions, and posttranslational modifications (17–19), but the full complexity of Sirt1 regulation in vivo still needs to be uncovered. To identify unique proteins controlling Sirt1 activity we searched for interaction partners by coimmunoprecipitation and label-free quantitative mass spectrometry. Untagged and YFP-tagged Sirt1 constructs were transfected into HEK293T cells and cellular proteins forming complexes with Sirt1 were precipitated using anti-Sirt1 or anti-YFP antibody, respectively. Among known and unknown Sirt1 interaction partners we detected several Sirt7 peptides, indicating that Sirt1 interacts with Sirt7 (Fig. 1A and SI Appendix, Fig. S1 and Table S1). Complementary results were obtained when Sirt7-YFP was used as bait (Fig. 1B and SI Appendix, Fig. S2). The volcano plot presented in Fig. 1B depicts the different Sirt7 interactors including Sirt1, some of which have been described before (16, 20, 21). A full list of Sirt7 interacting proteins identified in this screen is included in SI Appendix, Dataset S1. GST-pull down assays using bacterially produced GST-Sirt7 protein and extracts of cells expressing Sirt1-CFP suggested that Sirt1 and Sirt7 interact directly (SI Appendix, Fig. S3A). To prove that Sirt1 and Sirt7 interact with each other at physiological concentrations, we coimmunoprecipitated endogenous Sirt1 and Sirt7 from U2OS cells using antibodies directed against human Sirt1 and Sirt7 (Fig. 2A). Notably, coimmunoprecipitations were performed in the presence of the nuclease benzonase used to degrade all forms of DNA and RNA, thus excluding that Sirt1/Sirt7 interact via chromatin association. Moreover, we found that Sirt1 and Sirt7 are partially colocalized within the same regions of the nucleus (SI Appendix, Fig. S3B), which is a prerequisite for any potential interaction.
Fig. 1.
Screening for Sirt1 binding partners reveals an interaction between Sirt1 and Sirt7. (A) Label-free quantification (LFQ) of Sirt1 and Sirt7 peptide intensities identified by mass spectrometry after overexpression of untagged Sirt1 and Sirt1-YFP in HEK293T cells and immunoprecipitation using anti-Sirt1 and anti-YFP antibody. (B) Volcano plot showing proteins detected in three independent experiments after expression of Flag-Sirt7 and precipitation using Flag beads followed by elution with the Flag peptide.
Fig. 2.
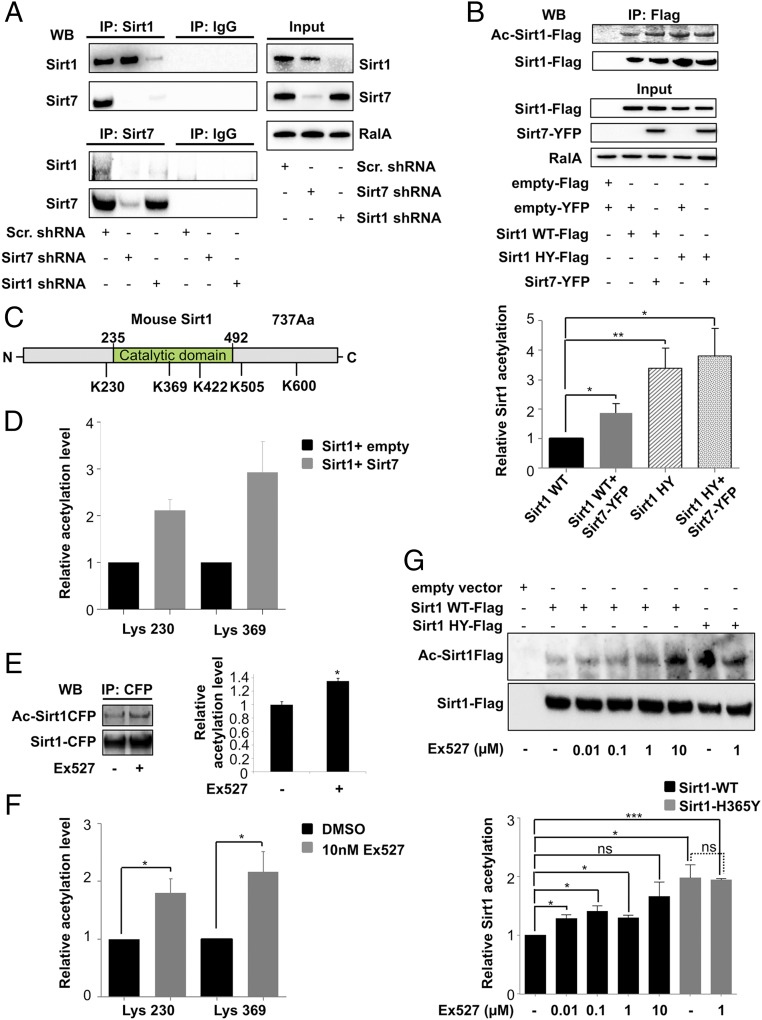
Sirt7 prevents autodeacetylation of Sirt1. (A) Coimmunoprecipitation of endogenous Sirt1 and Sirt7 from U2OS cells in presence and absence of Sirt1 or Sirt7 shRNAs or scramble controls. All experiments were performed in presence of benzonase (10 units/μL); (n = 3). (B) Acetylation of Sirt1 and the Sirt1 catalytic inactive mutant, Sirt1 H355Y (Sirt1 HY) after coexpression with Sirt7 in HEK293T cells. Sirt1-Flag and Sirt1 HY-Flag constructs were precipitated using Flag beads and analyzed by Western blot with anti-Flag or anti-AcK antibodies (n = 3); mean values ± SD; *P < 0.05; **P < 0.01. (C) Localization of acetylation sites in Sirt1 protein as determined by mass spectrometry. (D) Extent of Sirt1 acetylation on lysine residues 230 and 369 with and without Sirt7-YFP coexpression measured by mass spectrometry (n = 3); mean values ± SD. Sirt1-Flag was transfected into HEK293T cells and purified using Flag resin. (E) Increased acetylation of Sirt1 after inhibition of Sirt1 deacetylase activity by 1 μM Ex527 in cells transfected with Sirt1-CFP. Relative intensities of acetylated Sirt1 protein were quantified and are shown in histogram on the Right. The relative acetylation level of Sirt1-CFP without Ex527 inhibitor was set to 1; (n = 3) mean values ± SD; *P < 0.05. (F) Relative acetylation levels measured by mass spectrometry at lysine residues 230 and 369 of Sirt1 with and without addition of 10 nM Ex527 (n = 3); mean values ± SD; *P < 0.05. (G) Acetylation levels of Sirt1 WT-Flag and Sirt1 HY mutant-Flag transfected into the HEK293T cells. Cells were treated with different concentrations of the Sirt1 inhibitor Ex527 as indicated. Cell lysates were purified using Flag beads, followed by Western blot analysis using anti-Flag and anti-AcK antibodies. Quantifications of Sirt1 acetylation levels calculated from three individual experiments are shown in the histogram below; mean values ± SD; *P < 0.05; ***P < 0.001. Data were analyzed using Student’s t test (two-tailed paired t test). N = independent biological replicates.
To determine which domain within the Sirt7 protein is responsible for binding to Sirt1 we analyzed several Sirt7 mutants, revealing that both the N- and C-terminal domains of Sirt7 are required for binding to Sirt1. The N-terminal domain (amino acids 1–210) of Sirt7 appears to play the most important role for the interaction since its deletion almost completely abolished binding (SI Appendix, Fig. S3C). In contrast, mutation of the amino acid residue critical for the enzymatic activity of Sirt7 (H188Y) (22) reduced binding to Sirt1 only by 40% (SI Appendix, Fig. S3C).
Sirt7 Inhibits Autodeacetylation of Sirt1.
To investigate whether acetylation of Sirt1 is reduced after binding to Sirt7, both proteins were coexpressed in HEK293T cells and the extent of Sirt1 acetylation with and without Sirt7 coexpression was analyzed by Western blot analysis (Fig. 2B). Initially, we expected that Sirt7 might deacetylate Sirt1. However, much to our surprise, addition of Sirt7 caused a major increase but not a decrease of Sirt1 acetylation, while Sirt1 displayed only a limited degree of acetylation in the absence of Sirt7 (Fig. 2B and SI Appendix, Fig. S4A). Interestingly, the Sirt1 catalytic mutant, Sirt1HY, showed much higher acetylation level than wild-type Sirt1 and the acetylation level of Sirt1HY was not further influenced by the concomitant overexpression of Sirt7 (Fig. 2B). The catalytically inactive Sirt7-H188Y mutant was less efficient in increasing Sirt1 acetylation compared with WT Sirt7 (SI Appendix, Fig. S4A), which corresponded to the weaker binding of Sirt7-H188Y to Sirt1 (SI Appendix, Fig. S3C). Notably, another nuclear sirtuin, Sirt6, showed no effects on Sirt1 acetylation (SI Appendix, Fig. S4B). The unexpected finding that Sirt7 increased acetylation of Sirt1 but not of its catalytic inactive mutant (Fig. 2B), prompted us to investigate whether Sirt1 owns an autodeacetylase activity, which is inhibited by Sirt7. Quantitative liquid chromatography-tandem mass spectrometry analysis (LC-MS/MS) identified five lysine residues within Sirt1 (K230, K369, K422, K505, and K600), which showed a higher degree of acetylation in the presence of Sirt7 (Fig. 2C). However, statistical significance was only reached for lysines K230 and K369, while acetylation of the remaining three lysines did not increase in all experiments in the presence of Sirt7 (Fig. 2D). To study whether Sirt1 undergoes autodeacetylation we devised an in vitro deacetylation assay based on acetylated, isotope-labeled Sirt1-derived peptides and recombinant Sirt1 protein (SI Appendix, Fig. S4C). Quantification of deacetylation events by mass spectrometry revealed that Sirt1 efficiently deacetylated the K230 residue (SI Appendix, Fig. S4C). Furthermore, we inhibited the catalytic activity of Sirt1 by addition of the Sirt1-specific inhibitor Ex527. We observed a clear increase in acetylation of Sirt1 already at nanomolar concentration of Ex527, which specifically inhibits Sirt1 but does not inhibit other sirtuins (23), supporting our conclusion that Sirt1 deacetylates itself in the absence of Sirt7 (Fig. 2 E and F). Next, we wanted to know whether the catalytically inactive Sirt1 mutant is able or unable to deacetylate itself. Western blot analysis revealed a significant increase of acetylation in the Sirt1 mutant compared with enzymatically active wild-type Sirt1, which was not enhanced any further by the Sirt1 inhibitor Ex527 (Fig. 2 E–G). Importantly, sirtuin and HDAC inhibitors did not increase acetylation of the Sirt1 HY catalytic negative mutant, strongly supporting the decisive role of Sirt1 autodeacetylation (SI Appendix, Fig. S5A). Similarly, we did not observe major changes in the acetylation level of Sirt1 after inhibition of protein acetyltransferases (KATs), which essentially rules out a scenario in which Sirt7 increases Sirt1 acetylation by facilitating interaction of KATs with Sirt1 (SI Appendix, Fig. S5B). In contrast, Sirt7 strongly increased binding of Sirt1 to the methyltransferase Set7/9, which is a known inhibitor of Sirt1 (24) (SI Appendix, Fig. S5C). Interestingly, this binding is abolished in the dominant hyperactive Sirt1 K230R mutant (SI Appendix, Fig. S5D). Sirt7-dependent association of Sirt1 with Set7/9 correlated with enhanced formation of Sirt1 oligomers (SI Appendix, Fig. S5E), which raises the intriguing possibility that Sirt1 autodeacetylation is prevented by Sirt7-mediated oligomerization of Sirt1 and subsequent recruitment of inhibitors such as Set7/9. Taken together, our data indicate that Sirt7 directly interacts with Sirt1 to prevent its autodeacetylation, although the exact details of how this is accomplished need to be elucidated in future experiments.
Catalytic Activity of Sirt1 Depends on Sirt1 Autodeacetylation at K230.
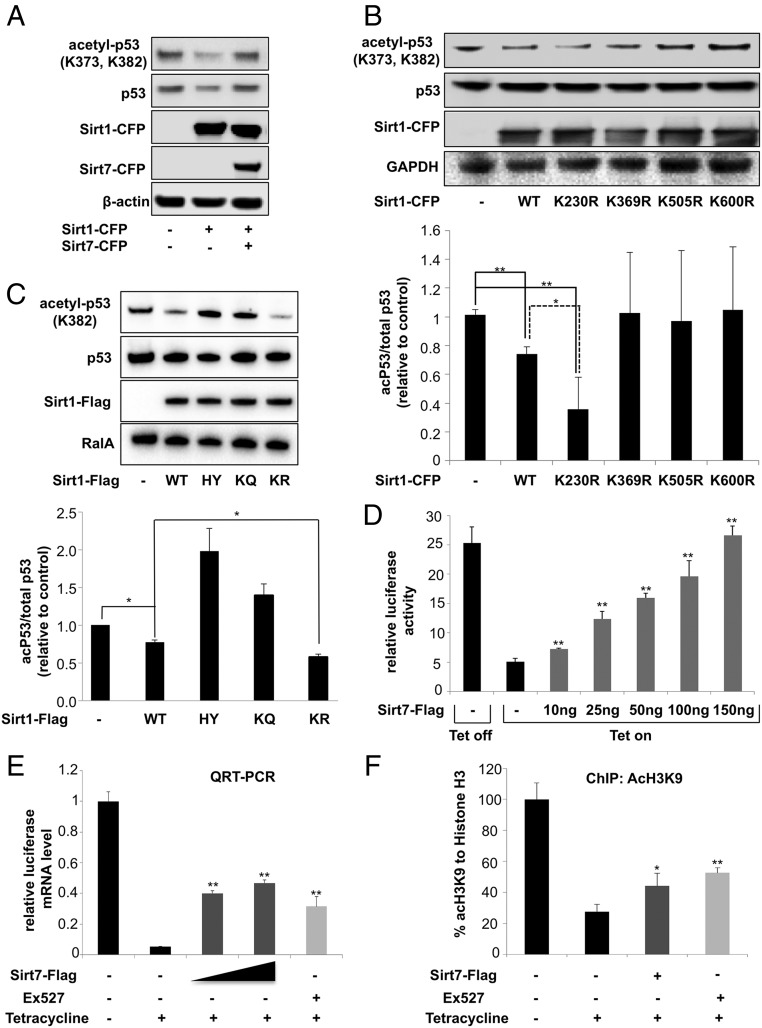
To investigate the consequences of Sirt7-dependent regulation of Sirt1 activity, we analyzed the acetylation status of p53, an established target of Sirt1 (25). Importantly, acetylation of p53 was increased in cells expressing both Sirt1 and Sirt7 compared with decreased acetylation of p53 in cells expressing only Sirt1 (Fig. 3A). Sirt6 had no effects on Sirt1-dependent p53 deacetylation, suggesting a unique role for Sirt7 in the regulation of Sirt1 activity (SI Appendix, Fig. S4B). Next, we assessed the impact of Sirt1 acetylation on Sirt1 enzymatic activity by replacing the acetylated lysine residues at positions 230, 369, 505, and 600 with arginine to mimic deacetylation. Interestingly, expression of Sirt1 K230R but none of the other lysine mutants decreased p53 acetylation levels, suggesting that deacetylation of K230, which is also the subject of Sirt1 autodeacetylation, is necessary to acquire the full catalytic activity of Sirt1 (Fig. 3B). To further prove the importance of Sirt1 K230 deacetylation for its catalytical activity, we generated the acetylation mimicking mutant, Sirt1 K230Q, in addition to the deacetylation mutant Sirt1 K230R. Indeed, the Sirt1 K230Q, similarly to the inactive Sirt1HY mutant, was not able to deacetylate p53, while WT and Sirt1 K230R both efficiently deacetylated p53 (Fig. 3 B and C). Finally, we used an in vitro tetracycline-inducible Sirt1/GAL4 expression system to investigate the activity of Sirt1 in the presence of Sirt7 (26). Induction of expression of Sirt1 by tetracycline led to inhibition of luciferase reporter activity and luciferase transcript level (Fig. 3 D and E). The decreased luciferase activity corresponded to lower H3K9 acetylation at the TK promoter regulating luciferase expression (Fig. 3F). Strikingly, addition of the Sirt1 inhibitor Ex527 or overexpression of Sirt7 relieved Sirt1-mediated inhibition of reporter gene expression in a concentration-dependent manner (Fig. 3 D–F), corroborating the inhibition of Sirt1 deacetylase activity by Sirt7. Interestingly, autodeacetylation of Sirt1 was sufficient to achieve maximal catalytic activity, since addition of sirtuin activators did not further increase the activity of Sirt1 K230R, the deacetylation mimicking mutant (SI Appendix, Fig. S5F). In contrast to Sirt1, we did not detect autodeacetylation of Sirt7, although Sirt7 is clearly an acetylated protein (SI Appendix, Fig. S5G).
Fig. 3.
Sirt7 inhibits the catalytic activity of Sirt1. (A) Western blot analysis of p53 acetylation after expression of Sirt1-CFP with and without Sirt7-CFP. Lysates of transfected cells were analyzed by Western blot with antibodies as indicated (n = 3). (B) Western blot analysis of p53 acetylation after expression of Sirt1 mutants. (Lower) Quantification of p53 acetylation. The relative acetylation level of p53 was set to 1 (n = 3); mean values ± SD; *P < 0.05; **P < 0.01. (C) Western blot analysis of p53 acetylation after expression of Sirt1, Sirt1 catalytic inactive mutant H355Y (HY), Sirt1 acetylation mimicking mutant, Sirt1 K230Q-Flag (KQ), or the deacetylation mimicking Sirt1 mutant (KR). Antibodies used for WB analysis are indicated. (D–F) Activity of Sirt1/Gal4 reporter constructs after induction by tetracycline without and with Sirt7 overexpression (D–F) and after addition of the Sirt1 inhibitor Ex527 (E and F). Sirt1 activity was determined by measuring inhibition of the relative luciferase activity (D), decrease of luciferase mRNA level (E), and diminished H3K9 acetylation at the TK promoter (F) (n = 3); mean values ± SD; *P < 0.05; **P < 0.01.
Adipocytes Require Sirt7 to Generate Physiological Amounts of White Adipose Tissue.
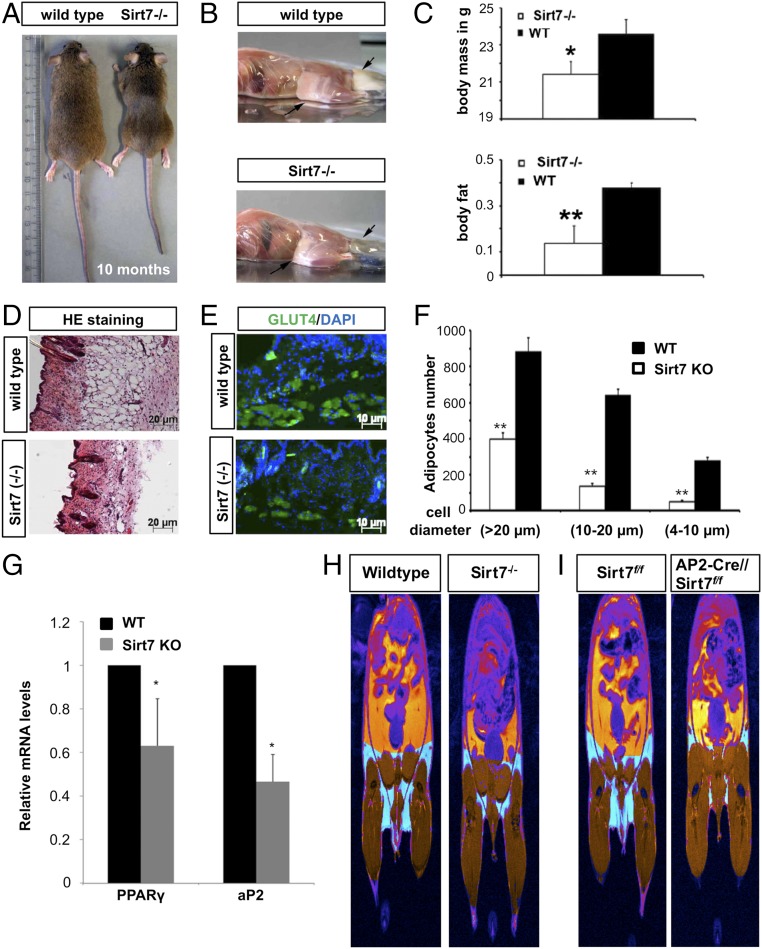
To investigate whether the regulation of Sirt1 by Sirt7 is physiologically relevant, we turned to an in vivo system. Since activation of Sirt1 inhibits adipogenesis (9) we reasoned that the lack of Sirt7 would decrease Sirt1 acetylation, increase its activity, and thereby block adipogenesis, which might result in diminished formation of white adipose tissue (WAT) in Sirt7 KO mice, although impaired adipogenesis does not necessarily translate into reduced fat mass. Nevertheless, analysis of Sirt7 mutant mice (22, 27–29) revealed a massive reduction of visceral and s.c. WAT (n = 5) (Fig. 4 A–C), which left only a thin layer of residual adipocytes (Fig. 4 D–F). In line with these observations we detected lower mRNA expression levels of PPARγ and adipocyte-specific protein AP2 in the remaining Sirt7−/− WAT (Fig. 4G). Sirt7 KO mice also show changes in energy metabolism as reported previously, which might contribute to the observed loss of WAT (27). To analyze whether Sirt7 serves a cell-autonomous function in adipocytes to promote adipogenesis, we generated adipocyte-specific Sirt7 KO using a floxed Sirt7 allele and AP2-Cre–expressing mice. Magnetic resonance imaging (MRI) analysis revealed a strong reduction of fat depots in adipocyte-specific Sirt7 KO mice similar to germ line Sirt7 KO animals (Fig. 4 H and I), suggesting that increased activity of Sirt1 within Sirt7-mutant adipogenic cells prevents proper formation of WAT.
Fig. 4.
Loss of Sirt7 restricts formation of s.c. and visceral white adipose tissue. (A) Macroscopic appearance of 10-month-old Sirt7 KO mice compared with wild-type littermates. (B) Representative images of skinned WT and Sirt7−/− mice and distribution of s.c. and visceral WAT (arrows). (C) Body weight and fraction of body fat were determined in WT (n = 4) and Sirt7 KO (n = 5) mice and are presented in histograms; mean values ± SD; *P < 0.05; **P < 0.01. (D) H&E stained cross-sections of the body wall of Sirt7 KO and control mice. (E) Cross-sections of the body wall of Sirt7−/− and control animals stained with an adipocyte-specific anti-Glut4 antibody (green) and DAPI. (F) Number of adipocytes in wild-type and Sirt7 KO mice. Average adipocyte numbers from Sirt7−/− and control animals (n = 3 for each group) were analyzed in three groups: small (diameter 4–10 μm), middle (diameter 10–20 μm), and large (diameter >20 μm); mean values ± SD; **P < 0.01. (G) Quantitative RT-PCR analysis of PPARγ and AP2 expression in WAT of Sirt7 KO and control mice (n = 3 for each group). Relative gene expression levels were normalized to β-actin. Expression in wild type was set to 1; mean values ± SD; *P < 0.05. (H and I) Representative MR images visualizing the fat (yellow) and muscle (brown) tissues in wild-type controls (n = 8) and in Sirt7 constitutive (n = 7) (H) and adipocyte specific (Sirt7f/f and AP2-Cre//Sirt7f/f, n = 8 each) (I) KO mice. N = independent biological replicates. Data were analyzed using Student’s t test (two-tailed paired t test).
Sirt7 Enables Adipogenic Differentiation by Inhibiting the Enzymatic Activity of Sirt1.
To further analyze the interplay between Sirt1 and Sirt7 in the regulation of adipogenesis we used mouse embryonic fibroblasts (MEFs), which can be converted into adipocytes in vitro. As expected, Sirt7−/− MEFs showed impaired adipogenic differentiation compared with wild-type MEFs (Fig. 5A), which was reflected by decreased expression of key adipogenic factors in Sirt7−/− MEFs (Fig. 5 B–F and SI Appendix, Fig. S6 A–D). To exclude that secondary developmental defects prevented differentiation of Sirt7−/− MEFs into adipocytes, we suppressed Sirt7 expression using a knockdown shRNA approach. Sirt7 knockdown 3T3L1 cells showed a pronounced arrest of adipogenic differentiation (Fig. 5G and SI Appendix, Fig. S6E). Furthermore, we detected a much stronger interaction between Sirt1 and Sirt7 in differentiating adipocytes in contrast to negligible binding of both sirtuins in growing preadipocytes (Fig. 5H). Importantly, the impaired adipocyte differentiation in Sirt7-deficient cells was not caused by unspecific cellular defects since overexpression of PPARγ efficiently rescued adipogenesis (SI Appendix, Fig. S7A). Likewise, we did not observe any overt changes in the proliferation capacity of Sirt7-deficient preadipocytes (SI Appendix, Fig. S7 B and C).
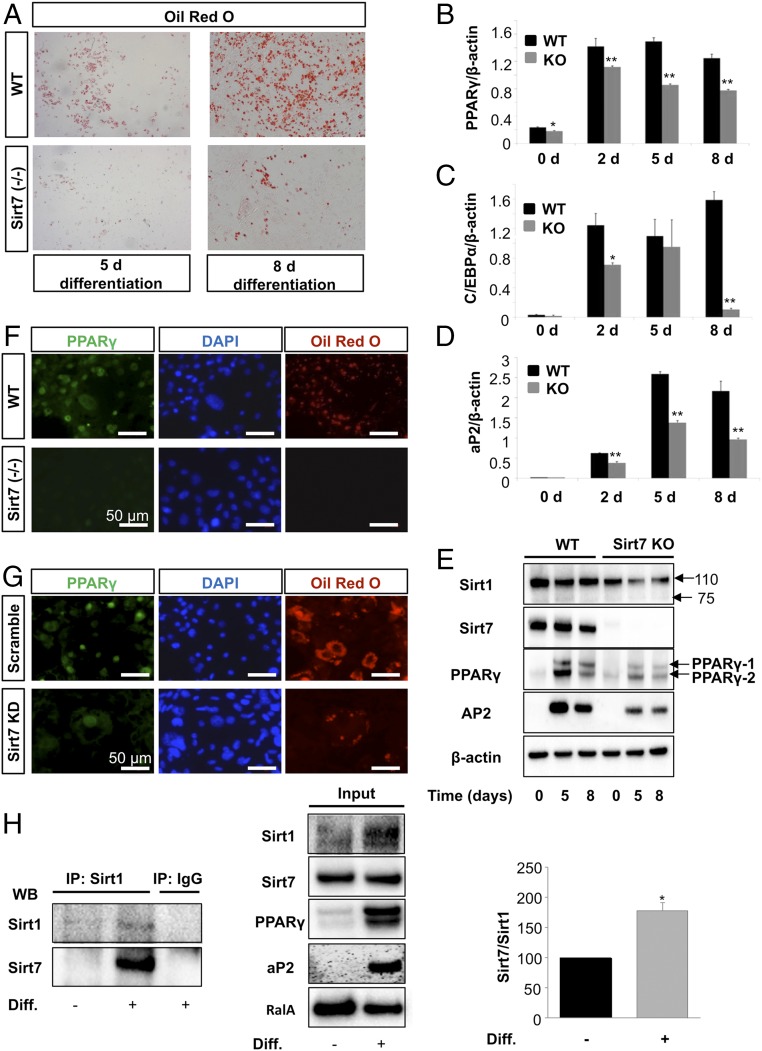
Fig. 5.
Sirt7 acts upstream of PPARγ to stimulate adipogenesis. (A) Oil-Red-O staining of primary MEFs isolated from Sirt7−/− and wild-type mice subjected to adipocyte differentiation (n = 4). (B–D) Analysis of PPARγ, C/EBPα, and AP2 expression in cultured wild-type and KO MEFs by quantitative RT-PCR (n = 3 for each group). Relative gene expression levels were normalized to β-actin; mean values ± SD; *P < 0.05; **P < 0.01. (E) Western blot analysis of Sirt1, Sirt7, PPARγ, aP2, and β-actin expression in wild-type and Sirt7 KO MEFs at different time points after induction of adipogenesis (n = 4). (F) Immunofluorescence analysis of the expression of PPARγ (green) in adipocytes derived from Sirt7−/− and Sirt7+/+ immortalized MEFs (n = 3). (G) Effects of shRNA-mediated knockdown of Sirt7 (Sirt7KD) on adipocyte differentiation. Oil-Red-O staining and PPARγ immunofluorescence of 3T3L1 cells infected with scramble control and Sirt7 shRNA lentivirus (Sirt7KD) and incubated for 8 d in differentiation medium (n = 3). (H) Analysis of endogenous Sirt1/Sirt7 interaction in growing 3T3L1 preadipocytes and adipocytes differentiated for 5 d. Sirt1 immunoprecipitates were analyzed on Western blot with indicated antibodies. Inputs are shown in the Middle and quantification on the Right (n = 3). *P < 0.05.
To investigate whether the activity of Sirt1 decreases during adipogenesis depending on the presence of Sirt7, we determined the ability of native Sirt1 isolated from preadipocytes and differentiated adipocytes to deacetylate H4K16ac histones in vitro using a previously developed deacetylation assay (26) (Fig. 6A). Sirt1 showed high deacetylation activity when isolated from dividing preadipocytes. In contrast, Sirt1 activity was strongly reduced when Sirt1 was isolated from differentiated adipocytes. Addition of Sirt7 inhibited H4K16ac deacetylation by Sirt1 purified from preadipocytes but had no additional effect on the already very low deacetylase activity of Sirt1 isolated from differentiated adipocytes (Fig. 6A). Significantly, addition of recombinant Sirt7 strongly inhibited Sirt1-mediated deacetylation of H4K16 (Fig. 6A). We concluded that the presence of Sirt7 in differentiated adipocytes is sufficient to inhibit Sirt1 preventing further effects of exogenously added Sirt7. To further validate that Sirt1 activity depends on the absence or inactivity of Sirt7, we isolated Sirt1 from WT and Sirt7 KO MEFs. We observed stronger deacetylation of two different Sirt1 substrates, H4K16 and H3K9, when Sirt1 isolated from Sirt7 KO cells was added to in vitro deacetylation assays compared with Sirt1 isolated from WT cells (Fig. 6 B and C).
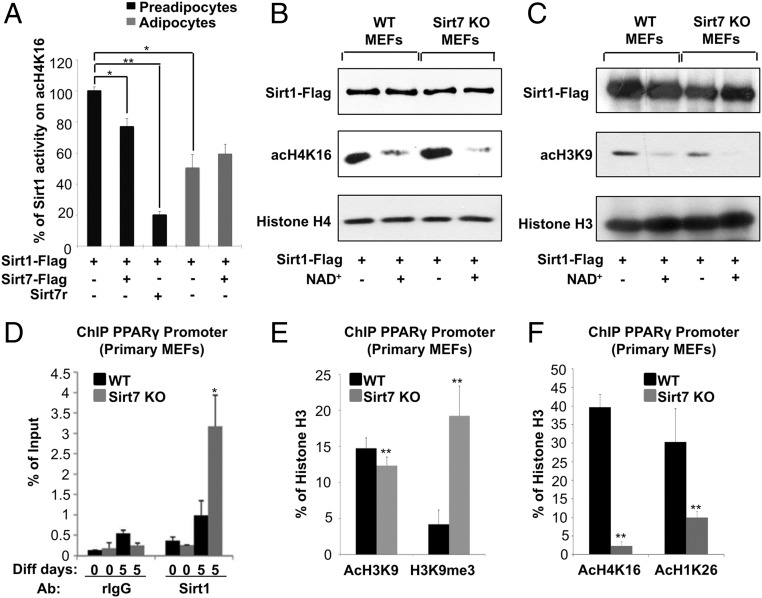
Fig. 6.
The activity of Sirt1 declines during adipocyte differentiation concomitant with increased Sirt1/Sirt7 interaction. (A) In vitro deacetylation of H4K16 by Sirt1. Sirt1 and Sirt7 proteins were immunoprecipitated from Sirt1Flag or Sirt1/Sirt7Flag overexpressing preadipocytes and differentiated adipocytes. Additionally, a bacterially produced, recombinant Sirt7 protein was used. n = 3; mean values ± SD; *P < 0.02; **P < 0.001. (B and C) Western blot analysis showing in vitro deacetylation of H4K16 (B) and H3K9 (C) by Sirt1. Sirt1 protein was immunoprecipitated from Sirt1-Flag expressing WT or Sirt7 KO MEFs. Hyperacetylated core histones were purified from HeLa cells, incubated with precipitated sirtuins before analysis of histone acetylation by Western blot (n = 3). (D) ChIP analysis of Sirt1 bound to the PPARγ promoter in differentiating (5 d) Sirt7 KO cells (n = 6); mean values ± SD; *P < 0.05. (E and F) ChIP analysis of histone H3K9 acetylation and methylation (E) and histone H4K16 and H1K26 acetylation (F) at the PPARγ promoter in wild-type and Sirt7−/− cells (n = 6); mean values ± SD; **P < 0.01. N = independent biological replicates. Data were analyzed using Student’s t test (two-tailed paired t test).
Increased Sirt1 Activity Is Responsible for the Impairment of Adipogenesis in Sirt7 KO.
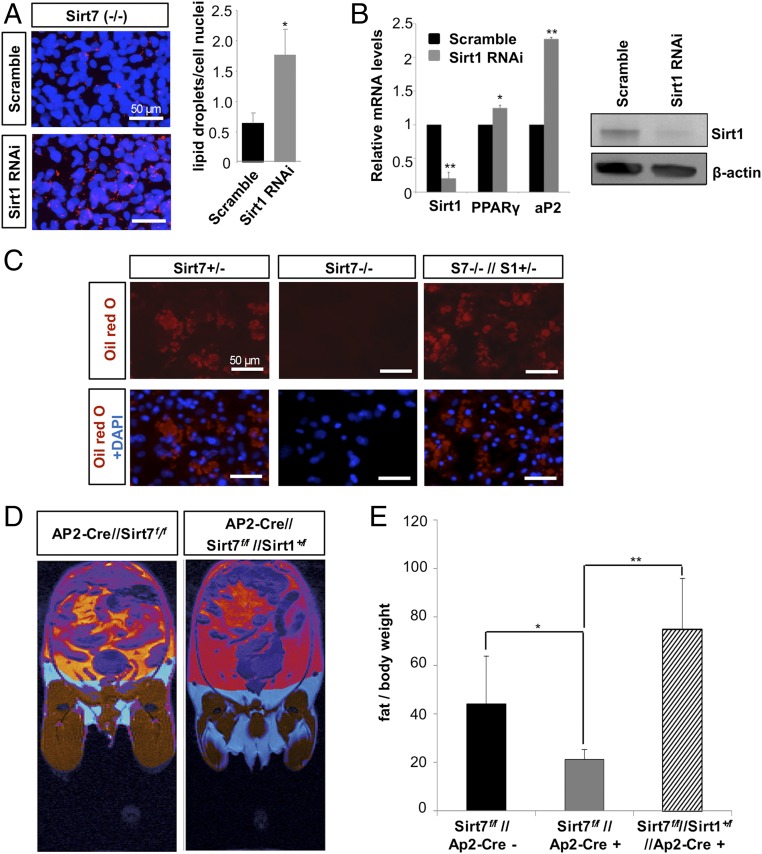
So far our data indicated that Sirt7 restricts the activity of Sirt1 during adipogenesis. Since the PPARγ promoter is a well-known target of Sirt1 in adipocytes and assumed to mediate the inhibitory effect of Sirt1 on adipogenesis, we studied the chromatin configuration at the PPARγ promoter in Sirt7−/− cells. Lack of Sirt7 in KO MEFs caused more than threefold increase of Sirt1 occupancy at the PPARγ promoter after induction of differentiation (Fig. 6D). In addition, we found lower H3K9 acetylation and an increased methylation of the PPARγ promoter, which provides an explanation for the massive reduction of WAT in Sirt7−/− mice and the failure of Sirt7−/− MEFs to undergo adipogenic differentiation (Fig. 6E). Since deacetylation of H3K9 at the PPARγ promoter was only moderately enhanced in the absence of Sirt7, we analyzed the acetylation level of two other histone targets of Sirt1 at the PPARγ promoter. We found that inactivation of Sirt7 caused a strong reduction of H4K16 and H1K26 acetylation, indicating high Sirt1 activity and efficient inhibition of PPARγ transcription (Fig. 6F). To validate that increased Sirt1 activity is responsible for impaired adipogenesis of Sirt7-deficient cells, we inhibited Sirt1 activity in Sirt7−/− MEFs by retrovirus-mediated shRNA knockdown. Knockdown of Sirt1 increased adipogenic differentiation as demonstrated by an increased Oil-Red-O staining and a higher expression of key adipogenic markers PPARγ and AP2 (Fig. 7 A and B, respectively), indicating a partial rescue of the Sirt7 mutant phenotype. Moreover, removal of one Sirt1 allele restored the highly compromised adipogenic potential of primary Sirt7−/− preadipocytes (Fig. 7C). Most importantly, adipocyte-specific inactivation of one Sirt1 allele restored WAT depots in conditional AP2-Cre Sirt7 KO mice in vivo with a high statistical significance, while heterozygous Sirt1 mice showed normal WAT content when the Sirt7 gene was left intact (Fig. 7 D and E). In summary, our data indicate that increased activity of Sirt1 suppresses PPARγ promoter activity, thereby accounting for impaired adipogenesis and diminished WAT depots in Sirt7 KO mice.
Fig. 7.
Sirt7-dependent inhibition of Sirt1 promotes differentiation of adipocytes. (A) Effect of retroviral shRNA-mediated knockdown of Sirt1 on adipogenic differentiation in Sirt7 KO cells. Representative images of Oil-Red-O stained cells (n = 4); *P < 0.05. Quantification of lipid droplets is shown on the Right; mean values ± SD. (B) Quantitative RT-PCR analysis of shRNA-treated cells (n = 3); mean values ± SD; *P < 0.05; **P < 0.01. Efficient knockdown of Sirt1 was controlled by Western blot analysis (Right). (C) Oil-Red-O staining of primary preadipocytes isolated from Sirt7+/−, Sirt7−/−, and Sirt7−/−//Sirt1+/− mice cultured in adipocyte differentiation medium for 8 d (n = 3). (D) MR images of fat tissue (red) in Sirt7 conditional KO (AP2-Cre//Sirt7f/f) (n = 6) and in Sirt7−/− FKO containing only one Sirt1 allele in adipose cells (AP2-Cre//Sirt7f/f//Sirt1+/f) (n = 5). (E) Quantification of MRI measurements; mean values ± SD; *P < 0.05; **P < 0.01. N = independent biological replicates. Data were analyzed using Student’s t test (two-tailed paired t test).
Discussion
Our results highlight the importance of cross-regulatory circuits between individual members of the sirtuin family for organismal homeostasis (Fig. 8) and emphasize the role of posttranslational acetylation in the regulation of Sirt1 activity. Autocatalytic modifications of proteins are common and have been primarily described for kinases (30), acetyltransferases (31), and also for phosphatases (32) but not for deacetylases. An involvement of acetylation/deacetylation in the regulation of enzymatic activity of this class of proteins has mostly remained enigmatic, although acetylation of several protein deacetylases has been reported previously (33). So far, the only example for a regulatory acetylation of a protein deacetylase is the inhibition of HDAC6 tubulin deacetylase activity by p300-mediated acetylation but the reversibility of this modification was not studied (34). The finding that Sirt1 deacetylation is instrumental for regulation of Sirt1 activity indicates that the principle of autocatalytic modification does also apply to deacetylases. Although we cannot completely rule out the possibility that Sirt1 is also deacetylated by another deacetylase or that Sirt7 inhibits the activity of an acetyltransferase, autodeacetylation of Sirt1 creates attractive regulatory advantages: Autocatalytic activation of Sirt1 might provide a versatile tool to amplify signal responses, which is otherwise often accomplished by a cascade of different molecules as, for example, in phosphokinase signaling. Autocatalytic activation alleviates the need for other molecules but provides means for the input of additional regulators and for nonlinear responses. This might be particularly important for sirtuins, whose activities depend on NAD+/NADH ratios (35). Although it is relatively difficult to determine the “active” concentration of NAD+ and NADH in different cellular compartments and to distinguish free from protein-bound NAD+, several studies indicate that sirtuins react to rather minor changes in NAD+/NADH ratios (36). Autocatalytic activation might amplify such moderate alterations of NAD+ or NADH concentrations in the submillimolar range (35) and therefore allow efficient Sirt1 signaling, which would be difficult to achieve by a solely linear response. An important component in such a regulatory circuit is the acetyltransferase that targets and inactivates Sirt1. Since inhibition of Sirt1 autodeacetylase activity by expression of Sirt7 increased Sirt1 acetylation, it seems likely that the tentative acetyltransferase is permanently active, at least in some cell types. Currently, the acetyltransferase that targets and inactivates Sirt1 is unknown, which at present prevents the study of its regulatory function. Obviously, acetylation and inactivation of Sirt1 are essential to assure efficient adipocyte differentiation under physiological conditions, but it is unclear whether, in addition to the critical role of Sirt7 down-regulation for removal of the inhibitory acetylation moiety in Sirt1, a dynamic regulation of the activity of the unknown Sirt1 acetyltransferase contributes to this process.
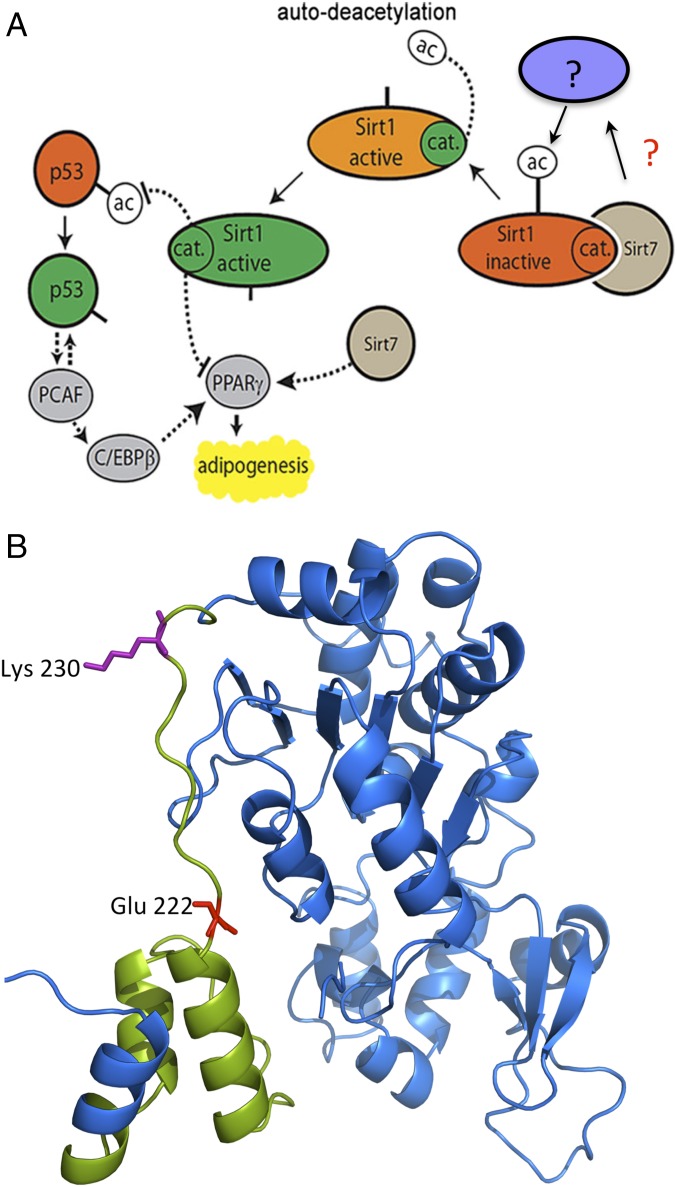
Fig. 8.
Potential mechanism of Sirt1 inhibition. (A) Schematic representation of the crosstalk between Sirt1 and Sirt7 controlling adipocyte differentiation. Sirt7 restricts autodeacetylation of Sirt1 either directly or through yet uncharacterized factors (represented by question marks) thereby promoting PPARγ expression and adipogenesis. Sirt7 might also control targets such as p53 and PPARγ independently of Sirt1. Sirt1 and Sirt7 regulate p53, which stimulates adipogenesis by activation of PPARγ via PCAF and C/EBPβ (42). Distortion of the Sirt1–Sirt7 balance results in lipodystrophy and disturbed metabolic homeostasis. (B) Sirt1 crystal structure and position of lysine 230 (magenta) and glutamic acid 222 (red). The structure of the Sirt1 regulatory domain (green) was described by Hubbard et al. (39).
At present it is also unclear how autodeacetylation increases activity of Sirt1 but recent findings suggest that two regulatory motifs A and B within the N-terminal part of Sirt1 boost its catalytic activity via intra- or intermolecular interactions (37). Since our findings indicate that autodeacetylation of Sirt1 at K230, which is located immediately downstream of the regulatory motif B, is instrumental for Sirt1 activity, it is tempting to speculate that acetylation of lysine 230 prevents such interactions. Sirt7 might inhibit autodeacetylation of Sirt1 at K230 by altering the monomeric state of Sirt1. Intriguingly, Sirt1 shows lower activity as an oligomer and only acquires full activity when present as a monomer (38), which makes it likely that heterodimer formation of Sirt1 and Sirt7 will reduce the activity of Sirt1. Notably, lysine 230 (magenta) of Sirt1 is located within the conserved domain (highlighted in green) (Fig. 8B), which is assumed to play a critical role for the regulation of Sirt1 activity by resveratrol and other sirtuin-activating compounds (STACs) (39). The same domain also contains a glutamic acid (red) at position 222 required for Sirt1 activation (39), which might indicate that deacetylation of K230 is necessary to activate Sirt1 by STACs. The inability of Sirt1 activators to further stimulate enzymatic activity of the Sirt1 deacetylation mimicking hyperactive mutant, Sirt1 K230R clearly supports such a mechanism. We hypothesize that Sirt7 inhibits Sirt1 autodeacetylation and activity by promoting Sirt1 oligomerization, which might facilitate binding of the Sirt1 inhibitor Set7/9. In conclusion, our study revealed that Sirt7 efficiently inhibits Sirt1 activity at different levels. Further investigations are required to disentangle the full complexity of regulatory network that fine tunes the activity of different sirtuins.
Although current evidence points to antagonistic functions of Sirt1 and Sirt7 as also exemplified by opposing effects of Sirt1 and Sirt7 for regulation of rDNA transcription (40), other findings suggest synergistic roles. Malik et al. found that Sirt1 and Sirt7 collaborate to synergistically repress cadherin expression for regulation of metastatic phenotypes in epithelial and mesenchymal tumors (41). Depending on the presence or absence of different cofactors that facilitate or disable interactions between sirtuins or modify (e.g., acetylate) different components of the sirtuin signaling machinery, Sirt7 might antagonize Sirt1 in one setting but synergize with Sirt1 on other occasions. In fact, preliminary observations suggest that Sirt1 and Sirt7 act together in distinct complexes, which might differentially affect their functions and substrate specificity.
Here, we provide evidence for the physiological relevance of Sirt7-mediated Sirt1 inhibition by demonstrating impaired adipogenesis after inactivation of Sirt7 in vitro and in vivo. Loss of Sirt7 led to increased Sirt1 activity and recruitment to the PPARγ promoter, resulting in strong attenuation of the PPARγ expression, which seems to be the primary cause of the lipodystrophic phenotype in Sirt7 KO mice. Reduced PPARγ expression might also contribute to the impaired WAT accumulation under a high-fat diet in Sirt7 KO mice, although further experiments are required to fully elucidate the underlying molecular mechanism (27). We also found that deacetylation of another Sirt1 target, p53, is repressed by Sirt7. The resulting attenuation of the function of p53 in Sirt7-deficient mice might also contribute to the development of lipodystrophy in Sirt7 mutants (Fig. 8). p53 plays a well-documented but rather complex role in adipogenesis. Several reports suggested that p53 inhibits adipogenesis, while others indicate a role of p53 in maintaining adipogenic depots in vivo (42, 43). Most significantly, p53 hypomorphic mice develop lipodystrophy during aging, which strongly resembles the phenotype of Sirt7-deficient mice (44). Further studies are required to explore the full physiological significance of p53 function and its dependence on sirtuins in the adipose tissue. In this context, it will be also interesting to inactivate Sirt7 in adult adipocytes to analyze a potential reduction of WAT mass and disclose effects of Sirt7 on the maintenance rather than on the formation of adipocytes.
We are proposing a model in which Sirt7 adjusts the activity of Sirt1. Aberrant or pathologically sustained activation of Sirt1 will repress its principally beneficial effects such as fat mobilization and restriction of adipogenesis, thereby causing metabolic dysfunctions. It remains to be explored, whether Sirt1/Sirt7 interaction plays a similar regulatory role in other fat depots that are regulated by Sirt1, especially the regulation of brown and beige fat (45). Continuous repression of key regulatory molecules downstream of the sirtuin signaling network, such as PPARγ and p53, will initiate exaggerated metabolic responses (42) disrupting tissues homeostasis and facilitating disease. The prospect of pharmacological regulation of Sirt1 activity to manipulate metabolic conditions and aging has attracted a lot attention. Our model raises some caveats but provides opportunities to change the activity of sirtuin signaling network. The unexpected level of complexity in sirtuin regulation indicates that consequences of the disturbance of the delicate, finely tuned cross-regulatory network between individual members of the sirtuin family has to be taken into account before attempting pharmacological interventions. Future experiments will reveal whether regulatory circuits exist not only between Sirt1 and Sirt7 but also between other members of the sirtuin family. Therefore, development of specific inhibitors for individual sirtuins seems mandatory, since global activators or inhibitors might provoke mixed responses resulting in adverse effects depending on the composition and activity of different sirtuin complexes and the presence of respective cofactors. Manipulation of Sirt7, on the other hand, becomes more attractive by our findings: specific activators of Sirt7 will most likely reduce the activity of Sirt1 on distinct targets, while small molecules interfering with the function of Sirt7 might induce activation of Sirt1.
Materials and Methods
Animals and Generation of Sirt1 and Sirt7 Conditional KO Mouse Strains.
C57BL/6 Sirt7−/− mice have been described previously (22). Wild-type C57BL/6 mice were obtained from Harlan-Winkelmann. For generation of conditional Sirt1 KO mice, a targeting Sirt1 vector containing a ≈19-kb DNA sequence spanning the 5′-UTR and the region from exon1 to intron4 and two loxP sites flanking exon4 including the neo cassette was used for homologous recombination in ES cells. Mice carrying Sirt1 floxed exon4 were generated by standard procedures and bred with the CMV-Cre strain. Mice heterozygous for the deleted Sirt1 allele were crossed with Sirt7 KO mice to generate Sirt7(−/−)//Sirt1(+/−) and Sirt7(+/−)//Sirt1(+/−) animals. To specifically inactivate Sirt7 in WAT, mice carrying Sirt7 floxed exons 6–9 were bred with AP2-Cre recombinase transgenic mice (46). All animal experiments were performed in accordance with the Guide for the Care and Use of Laboratory Animals (47) published by the National Institutes of Health and were approved by the local authorities (RP Darmstadt).
Plasmids and Cloning.
All plasmids and the cloning details are described in SI Appendix, SI Material and Methods.
Cell Culture, Viral Infection, and Transfection.
HEK293, U2OS, and 3T3-L1 cells (American Type Culture Collection) were cultured in Dulbecco’s modified Eagle’s medium with 10% FCS and antibiotics. Primary preadipocytes were isolated from s.c. adipose tissue of 7wk-old Sirt7(+/−), Sirt7(−/−), and Sirt7(−/−)//Sirt1(+/−) mice. MEFs were isolated from wild-type, Sirt7(−/−), and Sirt7(+/−) E13.5 embryos. The protocols are described in SI Appendix, SI Material and Methods.
To generate retroviral particles, Phoenix cells were transfected with either pQsupR-Scramble or pQsupR-mSirt1shRNA using the CaPO4 precipitation method. Forty-eight hours after transfection, medium containing retroviruses was collected, filtered, and transferred to target cells in the presence of polybrene (4 μg/mL). Infected cells were selected with 2.5 μg/mL puromycin for 7 d.
RNA and Protein Preparation and Analysis.
Total RNA was extracted from cells or tissues using TRIzol reagent (Invitrogen) and analyzed by quantitative real-time PCR in an iCycle iQ Multi Color Real-Time PCR machine (Bio-Rad) using Absolute QPCR SYBR Green Mix (Abgene). Primer sequences are given in SI Appendix, Table S2. Proteins from cells and mouse tissues were extracted in 100 mM Tris⋅HCl, pH 8.0, 10% SDS, 10 mM EDTA, and protease inhibitor mixture and homogenized by sonication on ice. Lysates were incubated at 99 °C for 2 min, then cleared by centrifugation (20,817 × g, 5 min at room temperature). Antibodies used for Western blot analyses are listed in SI Appendix, Table S3.
Pull-Down and Coimmunoprecipitation Assays.
For immunoprecipitation assays, samples were prepared using standard protocols followed by incubation with different primary antibodies (SI Appendix, Table S3) overnight at 4 °C. Complexes were precipitated by addition of 25 μL protein G Sepharose 4 Fast Flow beads (GE Healthcare Life Sciences), separated by SDS/PAGE and analyzed by Western blot. For isolation of Flag-tagged proteins an ANTI-FLAGRM2 Affinity Gel (Sigma-Aldrich) was used and the proteins were eluted by incubation with FlagR peptide (Sigma-Aldrich) in RIPA buffer. Identical amounts of GST or GST–Sirt7, GST–Sirt6, GST–GFAT fusion proteins were bound to glutathione-Sepharose (Pharmacia) and incubated with cell lysates from HEK293 cells expressing Sirt1–CFP fusion protein. Beads were washed with binding buffer and bound proteins were eluted and resolved on denaturing SDS-polyacrylamide gel electrophoresis for Western blot analysis.
Sirt 1 Deacetylase Assay and Mass Spectrometric Analysis.
The following Sirt1 peptides were used for the Sirt1 deacetylation assay: RKKRK(ac)DINTIEDAVK(8); RKKRK(ac)DINTIEDAVK(0); and DINTIEDAVK(0). Peptides were synthesized and labeled using the SILAC technique (48). Further information is provided in SI Appendix, SI Material and Methods.
In Vitro Deacetylation Assay.
Stable cell lines were generated by infection of 3T3-L1 cells with pMSCV-Flag Sirt1 or pMSCV-Flag Sirt7 retroviruses followed by puromycin selection. Purified proteins were incubated for 30 min at 37 °C with HeLa hyperacetylated core histones in HDAC buffer (10 mM Tris⋅HCl pH 8.0, 150 mM NaCl, 10% glycerol) in presence or absence of 5 mM NAD+. The reactions were stopped with Laemmli buffer and separated by SDS/PAGE. The acetylation status of H4K16ac was monitored by Western blot analysis using the anti-H4K16ac antibody (07-329, Millipore).
Luciferase Assay in Stable Gal4–Sirt1 Inducible Cell Line.
ChIP2 cells (293TREX containing pcDNA4/TetOn-Gal4-Sirt1 and pGal4-Tk-Luc-Neo; ref. 26) were cultured in DMEM supplied with 10% FCS, 5 μg/mL blasticidin (Invitrogen), 500 μg/mL G418 (Life Technologies), and 100 μg/mL Zeocin (Invitrogen). Cells were transiently transfected with different amounts of pCMVTag2a-hSirt7-Flag using Lipofectamine 2000 (Invitrogen). To control for transfection efficiency and to normalize the luciferase activity, 1 ng pGL3 control plasmid (Renilla luciferase under TK promoter, Promega) was mixed with 150 ng of experimental DNA and cotransfected. Twenty-four hours after transfection cells were induced with 1 μg/mL tetracycline (Merck) for an additional 24 h. Luciferase activity was determined using a Dual Luciferase Reporter System (Promega). Firefly luciferase activity was normalized for Renilla luciferase activity. Each transfection and luciferase assay was repeated three times.
Chromatin Immunoprecipitation.
The protocol is described in SI Appendix, SI Material and Methods.
Immunofluorescence, Morphological Analysis, and Magnetic Resonance Imaging.
Tissue samples were characterized by hematoxylin and eosin staining (H&E) (Chroma/Waldeck) according to standard protocols. Samples for fluorescence microscopy were processed as previously described (49). All MRI measurements were performed on a 7.0T superconducting magnet (Pharmascan, 70/16, 16 cm; Bruker Biospin) equipped with an actively shielded imaging gradient field of 760 mT/m using established methods (50).
Supplementary Material
Acknowledgments
We thank M. Euler and S. Jeratsch for technical assistance and Y. Zhou and X. Yuan for discussions. This work was supported by the Deutsche Forschungsgemeinschaft (Excellence Cluster Cardiopulmonary System and Grant SFB TR81), the LOEWE Center for Cell and Gene Therapy, the German Center for Cardiovascular Research, the German Center for Lung Research, and the Foundation Leducq 13 CVD 01. Work in the laboratory of A.V. was supported by Grant SAF2011-25619 from the Spanish Ministry of Economy and Competitiveness.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1706945114/-/DCSupplemental.
References
- 1.Haigis MC, Sinclair DA. Mammalian sirtuins: Biological insights and disease relevance. Annu Rev Pathol. 2010;5:253–295. doi: 10.1146/annurev.pathol.4.110807.092250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Imai S, Guarente L. Ten years of NAD-dependent SIR2 family deacetylases: Implications for metabolic diseases. Trends Pharmacol Sci. 2010;31:212–220. doi: 10.1016/j.tips.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bosch-Presegué L, Vaquero A. Sirtuin-dependent epigenetic regulation in the maintenance of genome integrity. FEBS J. 2015;282:1745–1767. doi: 10.1111/febs.13053. [DOI] [PubMed] [Google Scholar]
- 4.Chang HC, Guarente L. SIRT1 and other sirtuins in metabolism. Trends Endocrinol Metab. 2014;25:138–145. doi: 10.1016/j.tem.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giblin W, Skinner ME, Lombard DB. Sirtuins: Guardians of mammalian healthspan. Trends Genet. 2014;30:271–286. doi: 10.1016/j.tig.2014.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kiran S, Anwar T, Kiran M, Ramakrishna G. Sirtuin 7 in cell proliferation, stress and disease: Rise of the seventh sirtuin! Cell Signal. 2015;27:673–682. doi: 10.1016/j.cellsig.2014.11.026. [DOI] [PubMed] [Google Scholar]
- 7.Barber MF, et al. SIRT7 links H3K18 deacetylation to maintenance of oncogenic transformation. Nature. 2012;487:114–118. doi: 10.1038/nature11043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang S, Wang W, Miner J, Fromm M. Cross regulation of sirtuin 1, AMPK, and PPARγ in conjugated linoleic acid treated adipocytes. PLoS One. 2012;7:e48874. doi: 10.1371/journal.pone.0048874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Picard F, et al. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature. 2004;429:771–776. doi: 10.1038/nature02583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang F, Tong Q. SIRT2 suppresses adipocyte differentiation by deacetylating FOXO1 and enhancing FOXO1’s repressive interaction with PPARgamma. Mol Biol Cell. 2009;20:801–808. doi: 10.1091/mbc.E08-06-0647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kanfi Y, et al. SIRT6 protects against pathological damage caused by diet-induced obesity. Aging Cell. 2010;9:162–173. doi: 10.1111/j.1474-9726.2009.00544.x. [DOI] [PubMed] [Google Scholar]
- 12.Mostoslavsky R, et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124:315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 13.Yang SJ, Choi JM, Chang E, Park SW, Park CY. Sirt1 and Sirt6 mediate beneficial effects of rosiglitazone on hepatic lipid accumulation. PLoS One. 2014;9:e105456. doi: 10.1371/journal.pone.0105456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gomes AP, et al. Declining NAD(+) induces a pseudohypoxic state disrupting nuclear-mitochondrial communication during aging. Cell. 2013;155:1624–1638. doi: 10.1016/j.cell.2013.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Parihar P, Solanki I, Mansuri ML, Parihar MS. Mitochondrial sirtuins: Emerging roles in metabolic regulations, energy homeostasis and diseases. Exp Gerontol. 2015;61:130–141. doi: 10.1016/j.exger.2014.12.004. [DOI] [PubMed] [Google Scholar]
- 16.Zhou Y, et al. Reversible acetylation of the chromatin remodelling complex NoRC is required for non-coding RNA-dependent silencing. Nat Cell Biol. 2009;11:1010–1016. doi: 10.1038/ncb1914. [DOI] [PubMed] [Google Scholar]
- 17.Cantó C, Auwerx J. Targeting sirtuin 1 to improve metabolism: All you need is NAD(+)? Pharmacol Rev. 2012;64:166–187. doi: 10.1124/pr.110.003905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flick F, Lüscher B. Regulation of sirtuin function by posttranslational modifications. Front Pharmacol. 2012;3:29. doi: 10.3389/fphar.2012.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Revollo JR, Li X. The ways and means that fine tune Sirt1 activity. Trends Biochem Sci. 2013;38:160–167. doi: 10.1016/j.tibs.2012.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsai YC, Greco TM, Cristea IM. Sirtuin 7 plays a role in ribosome biogenesis and protein synthesis. Mol Cell Proteomics. 2014;13:73–83. doi: 10.1074/mcp.M113.031377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee N, et al. Comparative interactomes of Sirt6 and Sirt7: Implication of functional links to aging. Proteomics. 2014;14:1610–1622. doi: 10.1002/pmic.201400001. [DOI] [PubMed] [Google Scholar]
- 22.Vakhrusheva O, et al. Sirt7 increases stress resistance of cardiomyocytes and prevents apoptosis and inflammatory cardiomyopathy in mice. Circ Res. 2008;102:703–710. doi: 10.1161/CIRCRESAHA.107.164558. [DOI] [PubMed] [Google Scholar]
- 23.Napper AD, et al. Discovery of indoles as potent and selective inhibitors of the deacetylase SIRT1. J Med Chem. 2005;48:8045–8054. doi: 10.1021/jm050522v. [DOI] [PubMed] [Google Scholar]
- 24.Liu X, et al. Methyltransferase Set7/9 regulates p53 activity by interacting with Sirtuin 1 (SIRT1) Proc Natl Acad Sci USA. 2011;108:1925–1930. doi: 10.1073/pnas.1019619108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vaziri H, et al. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- 26.Vaquero A, et al. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol Cell. 2004;16:93–105. doi: 10.1016/j.molcel.2004.08.031. [DOI] [PubMed] [Google Scholar]
- 27.Yoshizawa T, et al. SIRT7 controls hepatic lipid metabolism by regulating the ubiquitin-proteasome pathway. Cell Metab. 2014;19:712–721. doi: 10.1016/j.cmet.2014.03.006. [DOI] [PubMed] [Google Scholar]
- 28.Shin J, et al. SIRT7 represses Myc activity to suppress ER stress and prevent fatty liver disease. Cell Rep. 2013;5:654–665. doi: 10.1016/j.celrep.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cioffi M, et al. MiR-93 controls adiposity via inhibition of Sirt7 and Tbx3. Cell Rep. 2015;12:1594–1605. doi: 10.1016/j.celrep.2015.08.006. [DOI] [PubMed] [Google Scholar]
- 30.Liu S, et al. ATR autophosphorylation as a molecular switch for checkpoint activation. Mol Cell. 2011;43:192–202. doi: 10.1016/j.molcel.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Santos-Rosa H, Valls E, Kouzarides T, Martínez-Balbás M. Mechanisms of P/CAF auto-acetylation. Nucleic Acids Res. 2003;31:4285–4292. doi: 10.1093/nar/gkg655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pazy Y, et al. Matching biochemical reaction kinetics to the timescales of life: Structural determinants that influence the autodephosphorylation rate of response regulator proteins. J Mol Biol. 2009;392:1205–1220. doi: 10.1016/j.jmb.2009.07.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Dyke MW. Lysine deacetylase (KDAC) regulatory pathways: An alternative approach to selective modulation. ChemMedChem. 2014;9:511–522. doi: 10.1002/cmdc.201300444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu W, et al. HDAC6 regulates epidermal growth factor receptor (EGFR) endocytic trafficking and degradation in renal epithelial cells. PLoS One. 2012;7:e49418. doi: 10.1371/journal.pone.0049418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin SJ, Ford E, Haigis M, Liszt G, Guarente L. Calorie restriction extends yeast life span by lowering the level of NADH. Genes Dev. 2004;18:12–16. doi: 10.1101/gad.1164804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koch-Nolte F, Fischer S, Haag F, Ziegler M. Compartmentation of NAD+-dependent signalling. FEBS Lett. 2011;585:1651–1656. doi: 10.1016/j.febslet.2011.03.045. [DOI] [PubMed] [Google Scholar]
- 37.Ghisays F, et al. The N-terminal domain of SIRT1 is a positive regulator of endogenous SIRT1-dependent deacetylation and transcriptional outputs. Cell Rep. 2015;10:1665–1673. doi: 10.1016/j.celrep.2015.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guo X, et al. The NAD(+)-dependent protein deacetylase activity of SIRT1 is regulated by its oligomeric status. Sci Rep. 2012;2:640. doi: 10.1038/srep00640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hubbard BP, et al. Evidence for a common mechanism of SIRT1 regulation by allosteric activators. Science. 2013;339:1216–1219. doi: 10.1126/science.1231097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Grummt I. The nucleolus—Guardian of cellular homeostasis and genome integrity. Chromosoma. 2013;122:487–497. doi: 10.1007/s00412-013-0430-0. [DOI] [PubMed] [Google Scholar]
- 41.Malik S, et al. SIRT7 inactivation reverses metastatic phenotypes in epithelial and mesenchymal tumors. Sci Rep. 2015;5:9841. doi: 10.1038/srep09841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bazuine M, Stenkula KG, Cam M, Arroyo M, Cushman SW. Guardian of corpulence: A hypothesis on p53 signaling in the fat cell. Clin Lipidol. 2009;4:231–243. doi: 10.2217/clp.09.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hallenborg P, Feddersen S, Madsen L, Kristiansen K. The tumor suppressors pRB and p53 as regulators of adipocyte differentiation and function. Expert Opin Ther Targets. 2009;13:235–246. doi: 10.1517/14712590802680141. [DOI] [PubMed] [Google Scholar]
- 44.Tyner SD, et al. p53 mutant mice that display early ageing-associated phenotypes. Nature. 2002;415:45–53. doi: 10.1038/415045a. [DOI] [PubMed] [Google Scholar]
- 45.Qiang L, et al. Brown remodeling of white adipose tissue by SirT1-dependent deacetylation of Pparg. Cell. 2012;150:620–632. doi: 10.1016/j.cell.2012.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.He W, et al. Adipose-specific peroxisome proliferator-activated receptor gamma knockout causes insulin resistance in fat and liver but not in muscle. Proc Natl Acad Sci USA. 2003;100:15712–15717. doi: 10.1073/pnas.2536828100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.National Research Council 2011. Guide for the Care and Use of Laboratory Animals (National Academies Press, Washington, DC), 8th Ed.
- 48.Krüger M, et al. SILAC mouse for quantitative proteomics uncovers kindlin-3 as an essential factor for red blood cell function. Cell. 2008;134:353–364. doi: 10.1016/j.cell.2008.05.033. [DOI] [PubMed] [Google Scholar]
- 49.Lörchner H, et al. Myocardial healing requires Reg3β-dependent accumulation of macrophages in the ischemic heart. Nat Med. 2015;21:353–362. doi: 10.1038/nm.3816. [DOI] [PubMed] [Google Scholar]
- 50.Zhang T, et al. Prmt5 is a regulator of muscle stem cell expansion in adult mice. Nat Commun. 2015;6:7140. doi: 10.1038/ncomms8140. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.