Significance
Our study assesses the long-held hypothesis that evolution of new gene functions underlies the diversification of animal forms. To do this, we systematically compared the patterning roles of a single gene across seven butterfly species. Under a null hypothesis of gene stasis, each knockout experiment should yield directly comparable phenotypes. We instead observed a varied repertoire of lineage-specific effects in different wing regions, demonstrating that the repeated modification of a key instructive signal was instrumental in the complex evolution of wing color patterns. These comparative data confirm the heuristic potential of CRISPR mutagenesis in nontraditional model organisms and illustrate the principle that biodiversity can emerge from the tinkering of homologous genetic factors.
Keywords: Wnt signaling, pattern formation, evolutionary tinkering, gene co-option, CRISPR mutagenesis
Abstract
Butterfly wing patterns provide a rich comparative framework to study how morphological complexity develops and evolves. Here we used CRISPR/Cas9 somatic mutagenesis to test a patterning role for WntA, a signaling ligand gene previously identified as a hotspot of shape-tuning alleles involved in wing mimicry. We show that WntA loss-of-function causes multiple modifications of pattern elements in seven nymphalid butterfly species. In three butterflies with a conserved wing-pattern arrangement, WntA is necessary for the induction of stripe-like patterns known as symmetry systems and acquired a novel eyespot activator role specific to Vanessa forewings. In two Heliconius species, WntA specifies the boundaries between melanic fields and the light-color patterns that they contour. In the passionvine butterfly Agraulis, WntA removal shows opposite effects on adjacent pattern elements, revealing a dual role across the wing field. Finally, WntA acquired a divergent role in the patterning of interveinous patterns in the monarch, a basal nymphalid butterfly that lacks stripe-like symmetry systems. These results identify WntA as an instructive signal for the prepatterning of a biological system of exuberant diversity and illustrate how shifts in the deployment and effects of a single developmental gene underlie morphological change.
The multitude of patterns found in developing organisms is achieved by a small number of conserved signaling pathways, which raises an important question. How does biodiversity arise from the sharing of constituents across a single tree of life? One explanation for this apparent paradox is that conserved regulatory genes evolve new “tricks” or roles during development (1). Assessing this phenomenon requires comparing the function of candidate genes across a dense phylogenetic sampling of divergent phenotypes. Here, the patterns on butterfly wings provide an ideal test case. The development of scale-covered wings, their structural and pigment complexity, and an elaborate patterning system are key features of the Lepidoptera (moths and butterflies), which form about 10% of all species known to humankind (2). Wing patterns across the group are fantastically diverse and are often shaped by natural and sexual selection (3). Studies in fruit flies, butterflies, and moths have implicated secreted Wnt-signaling ligands as color pattern inducers (4–8). In butterfly wings, two lines of evidence suggest a prominent patterning role for the Wnt ligand gene WntA in particular. First, WntA was repeatedly mapped as a locus driving pattern-shape adaptations involved in mimicry, and a total of 18 WntA causative alleles have been identified across a wide phylogenetic spectrum (9–13). Second, WntA expression marks developing wing domains that prefigure the position and shape of pattern elements of various color compositions (10, 14).
The nymphalid groundplan provides a conceptual framework to understand pattern variation in butterflies (3). Under this framework, patterns are organized into parallel subdivisions of autonomous color pattern complexes known as “symmetry systems,” which are arranged across the dorsal and ventral surfaces of both the fore- and hindwing (14–19) (Fig. 1 A–C). This arrangement is thought to represent a putative archetype of a butterfly wing pattern, and diversity is created by modifying elements within and among these symmetry systems (3). WntA is typically expressed in three of the four symmetry systems (14): the small proximal pattern called Basalis (B), the large median pattern called the Central Symmetry System (CSS), and the Marginal Band System (MBS), which features laminar stripes bordering the wing. Here we used CRISPR/Cas9 mutagenesis to impair WntA function and assess its patterning roles in Nymphalidae, the largest butterfly family that radiated around 90 Mya (20). We characterize the developmental function of WntA in species representative of the nymphalid groundplan and then show that WntA has acquired divergent patterning roles in several lineages.
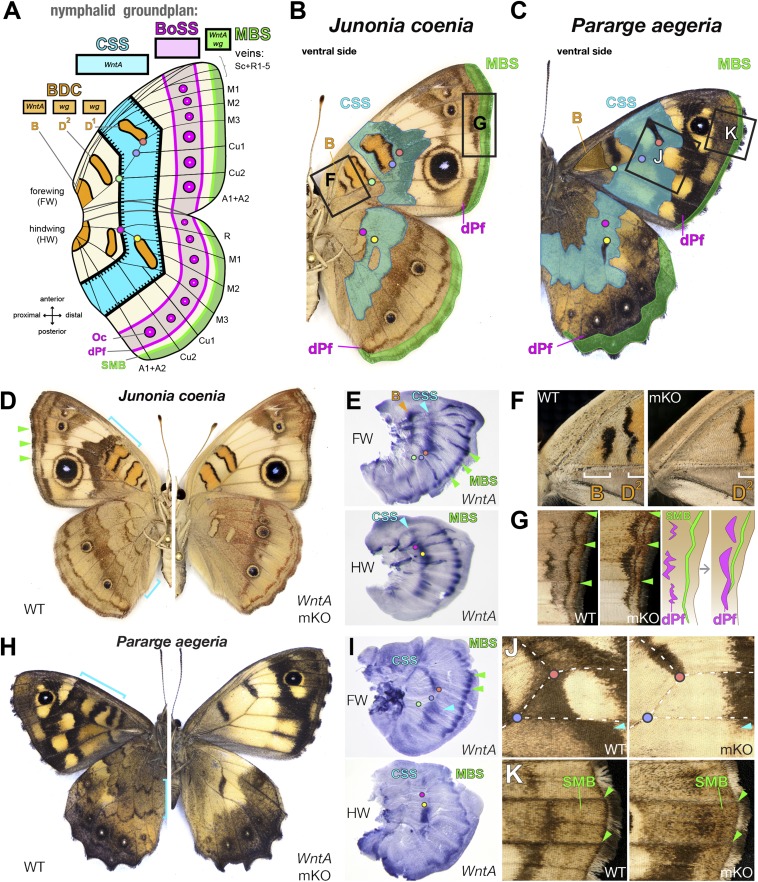
Fig. 1.
WntA loss-of-function effects in groundplan-like nymphalids. (A–C) The nymphalid groundplan consists of consecutive symmetry systems organized along the antero-posterior axis. Color code indicates groundplan elements in subsequent panels. Orange: Baso-Discal Complex (BDC) patterns; blue: CSS; fuchsia: Bordel Ocelli Symmetry System (BoSS), including dPf; green: MBS, including Sub-Marginal Band (SMB). Dots show wing topological landmarks corresponding to vein crossings. (D–G) WntA mKO in J. coenia results in the loss of WntA+ patterns. (D) Whole-wing phenotypes. (E) In situ hybridization of WntA in WT fifth instar imaginal disks. (F) Blow-up of proximal forewing area showing the loss of B upon WntA mKO. (G) Blow-up of proximal forewing area showing the distalization of dPf and SMB elements. (H–K) Replication of the J. coenia results in P. aegeria. (H) Whole-wing phenotypes. (I) In situ hybridization of WntA in WT fifth instar imaginal disks. (J) Loss of the forewing CSS. (K) Distalization of dark-brown dPF and SMB; arrowheads point at corresponding WntA expression domains in I.
Results and Discussion
We injected Cas9/sgRNA duplexes into 1–6 h butterfly embryos at a syncytial stage (n = 5,794 eggs). As only a fraction of the dividing nuclei are edited, the resulting mosaicism can bypass the deleterious effects of developmental mutations and yields G0 escapers that survive until the adult stage for phenotypic analysis (21–23). We performed CRISPR injections in seven nymphalid species to induce frameshift mutations in WntA-coding exons. About 10% of hatchlings (240 of 2,293 survivors) yielded adult butterflies with mosaic knockout (mKO) pattern defects on their wings (SI Appendix, Figs. S1–S9 and Tables S1 and S2).
WntA Induces Central Symmetry Systems.
First we used CRISPR to test the effects of WntA loss-of-function on the wing patterns of the Common Buckeye Junonia coenia (tribe: Junoniini). WntA mKOs resulted in a complete loss of the CSS, consistent with WntA expression that prefigures its shape and position in the wing imaginal disks (Fig. 1 D and E and SI Appendix, Fig. S1). The WntA-positive forewing B element was lost while the wg-positive D1-D2 elements (8, 24) were unaffected (Fig. 1F). The B-D1-D2 patterns have a similar color composition, indicating that WntA and wg play interchangeable roles in their induction. In contrast, the double loss of the distinct B and CSS patterns also illustrates the regional specificity of WntA-signaling color outputs across the wing surface. In the marginal section of the wing (Fig. 1G), WntA mKOs resulted in a contraction of the MBS and in a shift of chevron patterns known as the distal parafocal elements (dPF) (17, 19). WntA may impact these distal elements by participating in complex patterning dynamics in the marginal section of the wing (25).
Variations on the WntA Groundplan Theme.
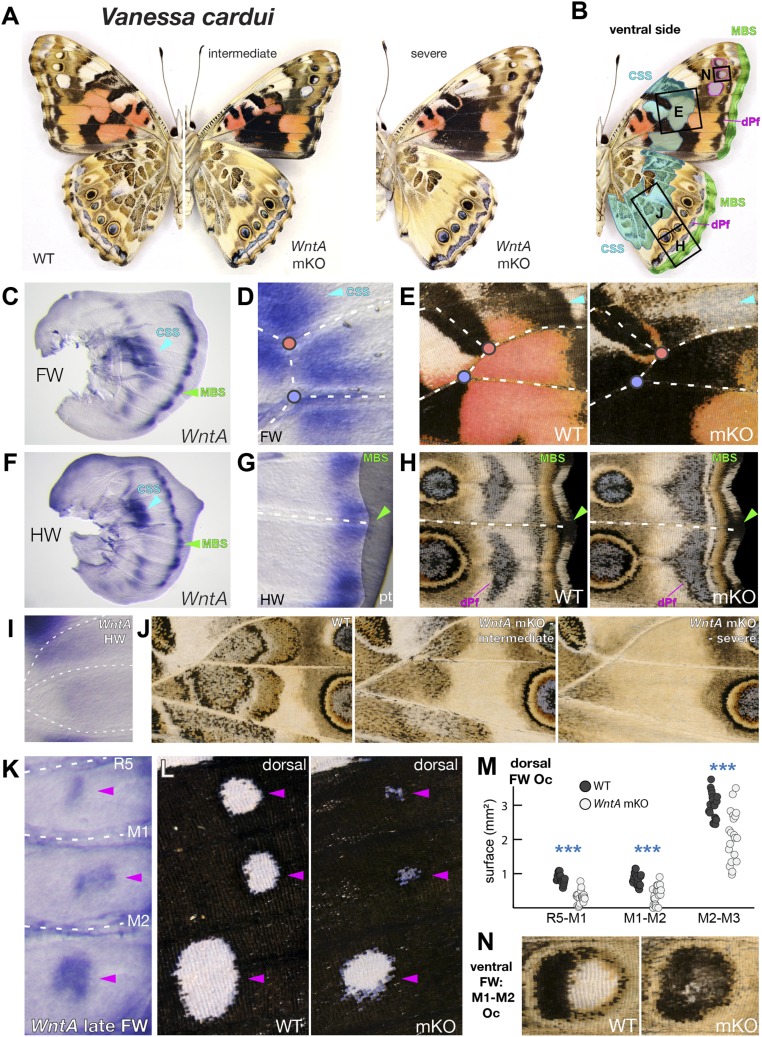
Next we asked if the instructive roles of WntA were phylogenetically conserved, using two other nymphalid butterflies with a groundplan organization, the Specked Wood Pararge aegeria (tribe: Satyrini) (15) and the Painted Lady Vanessa cardui (tribe: Nymphalini) (18). WntA mKOs yielded consistent effects by eliminating the CSS and distalizing the parafocal elements in these two species (Figs. 1 H–K and 2 A–H and SI Appendix, Fig. S10). Of note, in the V. cardui hindwing, the complex wave-like patterns of the CSS were lost upon severe WntA mKO and reduced in more intermediate forms (Fig. 2 I and J). These two species also highlighted other aspects of WntA phenotypic effects. In P. aegeria hindwings, the mKO-mediated disruption of the marginal system resulted in an apparent expansion of the eyespot outer rings (SI Appendix, Fig. S10D). V. cardui WntA mKOs resulted in the reduction of each dorsal forewing eyespot (P values < 10−4; Fig. 2 K–M) and generated color composition defects in the ventral forewing eyespots (Fig. 2N and SI Appendix, Fig. S3). Only V. cardui forewings are known to express WntA in their eyespots (14). We thus infer that WntA was co-opted in the eyespot gene regulatory network of the V. cardui lineage to elaborate upon the patterning of this complex feature (26). Overall, comparisons in three species show that multifaceted modulations of WntA function have shaped variations on the basic nymphalid groundplan theme.
Fig. 2.
Conserved and novel aspects of WntA function in Painted Lady butterflies. (A and B) WntA mKOs in V. cardui result in defects or loss of the CSS, highlighted in cyan in B. (C and D) WntA forewing expression is associated with the CSS (magnified in D) and the MBS in WT fifth instar wing disks. (E) Blow-up of a CSS section showing pattern disruption upon WntA mKO. (F and G) WntA hindwing expression in the CSS and the MBS (magnified in G; pt, peripheral tissue) in WT fifth instar wing disks. (H) WntA mKO results in distal shifts of dPf elements. (I) Blow-up of WntA expression in the hindwing CSS. (J) Magnification of intermediate and severe levels of CSS reduction observed upon WntA mKO. (K) WntA expression as observed in the presumptive forewing eyespots in late fifth instar wing disks of V. cardui. (L and M) Reduction of dorsal forewing eyespots following WntA mKO. (N) Color change in ventral mKO forewing eyespots.
WntA Induces Pattern Boundaries in Heliconius.
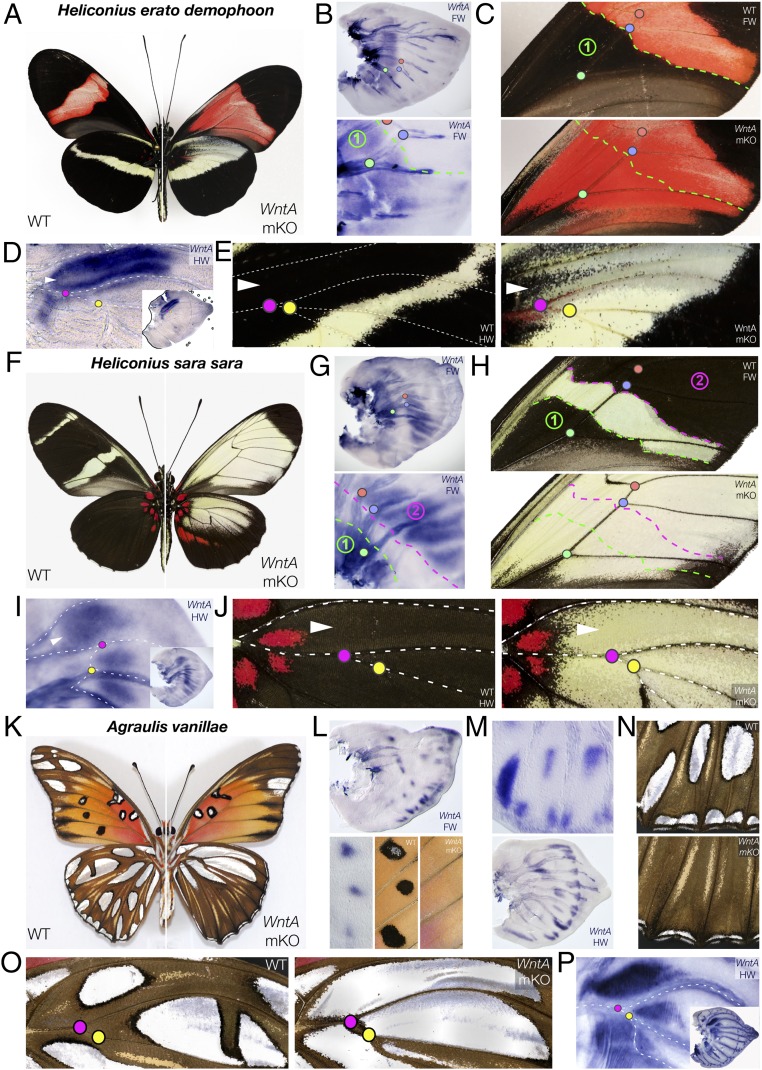
We next focused on species that departed more markedly from the nymphalid groundplan configuration, starting with the hyperdiverse Heliconius clade (tribe: Heliconiini). We performed CRISPR mKOs in Central American morphs of two species, Heliconius erato demophoon and Heliconius sara sara. WntA removal resulted in an expansion of light-color patterns in both cases (Fig. 3 A and F). In H. erato demophoon, WntA expression marked melanic patches that contour forewing red and hindwing yellow stripes (Fig. 3 B and D). Predictably, its loss-of-function resulted in the loss of the corresponding boundaries with black contours being replaced by expansions of red or yellow (Figs. 3 C and E). H. sara forewing disks showed a proximal and central WntA expression domains that each correspond to melanic fields that frame the signature yellow stripes of this butterfly (Fig. 3G). Both melanic intervals were lost following WntA mKOs (Fig. 3H), yielding an almost uniformly yellow forewing surface. Hindwings showed a similar effect with WntA deficiency resulting in melanic-to-yellow switches in the antero-proximal half of the wing (Fig. 3J). Interestingly, this treatment also revealed a cryptic stripe of red patches. A similar phenotype is observed in subspecies of H. sara, as well as in its sister species Heliconius leucadia (SI Appendix, Fig. S11), suggesting that modulations of Wnt signaling could underlie these cases of natural variation. Overall, these data support previous predictions that groundplan elements such as the CSS can be homologized to what form the apparent contours of Heliconius patterns (27–29). WntA is best thought as a prepatterning factor that determines boundaries between color fields, a view that is compatible with the replacement effects of mKOs, where WntA-deficient cells acquire the color fate of the adjacent territory. This property may explain why cis-regulatory tinkering of WntA expression seems to underlie the repeated modification of color pattern shapes across this explosive radiation (9–12), as it allows the coordinated modulation of color fate on either side of a moving boundary.
Fig. 3.
Variegated WntA loss-of-function phenotypes in passionvine butterflies. (A–E) Effects of WntA mKO in H. e. demophoon. (A) Whole wings. (B) Detection of WT proximal WntA expression by larval forewing in situ hybridization (zone 1). (C) Loss of proximal pattern boundary in WntA-positive zone 1. (D) Antero-proximal expression of WntA in WT late larval hindwings. (E) Loss of antero-proximal pattern boundary in mKO hindwings. (F–J) Effects of WntA mKO in H. sara. (F) Whole wings. (G) Detection of proximal (zone 1) and median (zone 2) WntA in larval forewings. (H) Loss of proximal (green line) and median (fuchsia line) pattern boundaries resulting in loss of melanic identity in zones 1 and 2. (I) Antero-proximal expression of WntA in larval hindwings. (J) Widespread antero-proximal color identity shift in mKO hindwings. (K–P) Effects of WntA mKO in A. vanillae. (K) Whole wings. (L) Silver-spot–related expression of WntA in larval forewings and loss in mKO forewings. (Bottom) M3-A1 spot triad. (M and N) Silver spot-related expression of WntA in larval hindwings (M) and loss/reduction in mKO hindwings (N). (Top) M3-A1 spot complex. (O) Silver-spot pattern expansion in proximal mKO hindwings. (P) Secondary expression of WntA in the proximal region of late larval hindwings. Colored dots: wing topological landmarks (vein crossings).
Antagonistic Roles of WntA in Adjacent Patterns.
Compared with Heliconius, the closely related Gulf Fritillary butterfly (Agraulis vanillae) has modified the nymphalid groundplan differently to produce its distinctive wing pattern (27). Rather than continuous stripes, A. vanillae shows dispersed silver spots of identical color composition, each consisting of a core of highly reflective “mirror” scales (30) and an outline of black scales. A subset of silver spots express WntA or wg (14), and accordingly, all of the WntA+ patterns contracted or disappeared in WntA mKOs (Fig. 3 K–N and SI Appendix, Fig. S6). Among the wg+ elements (forewing D1 and D2), only D1 coexpressed WntA and was specifically reduced in WntA mKOs (SI Appendix, Fig. S12), suggesting that silver spots respond to overall Wnt dosage. WntA mKOs also resulted in a drastic expansion of WntA-free (WntA−) patterns (Fig. 3O). Importantly, butterflies treated with exogenous heparin, a ligand-binding molecule with Wnt gain-of-function effects (9, 14, 31, 32), showed the opposite outcome: expanded WntA+ and reduced WntA− patterns (14). These reverse effects of CRISPR loss-of-function vs. heparin gain-of-function suggest that WntA activates and represses two distinct sets of patterns, and the repressed domain in fact shows a secondary wave of WntA expression in late larval instar wing disks (Fig. 3P). This observation leads us to propose that the dual effect of WntA may be due to a biphasic deployment, with a first wave of WntA pattern-activating expression followed by an inhibitory event in the Wnt-repressed territory. Testing this working model will require the identification and expression profiling of WntA-signaling targets in A. vanillae.
Repurposing of WntA in a Reduced Groundplan.
Finally, we used the lack of visible CSS in monarchs (Danaus plexippus; tribe: Danaini) as an example of extreme divergence from the nymphalid groundplan. WntA lacked a CSS median stripe expression as expected and was instead detected around the presumptive veins, indicative of a potential role in the induction of vein-dependent patterns (33). WntA mKO adults showed drastic expansions of the white interveinous patterns (Fig. 4), which are usually visible as thin outlines of the veins in WT ventral wings. In addition, white dot elements that ornate the marginal region expanded and fused following WntA mKO. Other WntA mKO monarchs showed a small dorsal patch of ectopic interveinous scales in the crossvein region, demonstrating maximal WntA expression in hindwings (SI Appendix, Fig. S13). Consistent with a Wnt loss-of-function, this mild phenotype was reproduced by injection of dextran sulfate, a drug treatment that emulates Wnt signal inhibition in other butterflies (14, 32) (SI Appendix, Fig. S14). Overall, expression and functional data suggest that WntA was again repurposed, in this case as a repressor of interveinous white scales in the monarch lineage.
Fig. 4.
WntA loss-of-function in monarch butterflies induces interveinous white scales. (A and B) In situ detection of WntA in Danaus plexippus last larval instar forewing (A) and hindwing disks (B). (C) Whole-wing phenotypes following WntA mKO. All effects consist of expansion of white scale patterns. (D) Close-up view of larval hindwing WntA interveinous expression in the periphery of tracheal vein precursors. (E and F) Expansion of interveinous white scale fate in the hindwing region corresponding to D. Colored dots: wing topological landmarks (vein crossings).
Lessons from Somatic CRISPR Phenotypes.
Somatic mutagenesis yielded loss-of-function data in the G0 adults of seven butterfly species, an achievement that would have been unrealistic in the pre-CRISPR era. Experimental replication using various single-guide RNA (sgRNA) targets ruled out a contribution of off-target lesions, and genotyping experiments revealed a predominance of frameshift, presumably null WntA alleles (SI Appendix, Figs. S8 and S9). Variations in clone size, allelic dosage, and the possible occurrence of hypomorphic mutations could underlie complex cases of mosaicism, explaining the range of observed effects (Fig. 2J and SI Appendix, Figs. S1–S7). Inferring the allelic composition of wing mutant clones from their genotyping is complicated by the movement of insect wing epithelial cells following adult emergence (34), as well as by the presence of cell contaminants that are unlikely to underlie the pattern phenotype (e.g., tracheal cells, neurons, hemocytes). We attempted the generation of germline mutations in V. cardui to bypass the experimental limitations of somatic heterogeneity. Following the injection of a single sgRNA targeting the WntA stop codon, we obtained an adult female bearing a modification of the forewing CSS (SI Appendix, Fig. S15). Six G1 offsprings displayed the same phenotype and were all heterozygous for a 16-bp indel mutation, resulting in a C-terminal Cys-Asn-Stop → Gly-Ser-Arg-Stop editing of the predicted WntA protein. This allele was passed to a second generation but was subsequently lost due to an episode of high mortality in our stock. Nonetheless, this preliminary result illustrates the potential of CRISPR to induce a variety of loss-of-function alleles, which could be propagated via the germline for tackling future developmental questions where mosaicism is a concern.
Conclusions.
The Nymphalidae family comprises about 6,000 butterfly species, most of which can be identified by their wing patterns. We used this system as a proxy of morphological evolution and found that a single signal articulates its underlying complexity, as shown by the variety of WntA mKO phenotypes obtained across different wing regions and species. Our data highlight three major results. First, WntA is associated with multiple pattern elements within the same individual, including within the same wing surface, e.g., both the adjacent Basalis and CSS patterns require WntA in J. coenia forewings, despite distinct color compositions, whereas CSS stripes often differ between wing surfaces (dorsal vs. ventral, forewing vs. hindwing). Wnt signaling may combine with selector genes that mark distinct wing domains to mediate these regional-specific outputs within a single individual (24, 35). Second, spatial shifts in WntA expression cause pattern-shape evolution, exemplified by the multitude of species-specific manifestations of the CSS. Cis-regulatory variants of WntA (9–12), or alternatively, modulations of the trans-regulatory landscape that controls WntA expression, may have fashioned these macroevolutionary shifts. Finally, WntA evolves new patterning functions. It was co-opted into forewing eyespot formation in the V. cardui lineage, evolved a localized pattern-inhibiting role in A. vanillae, and was repurposed for the patterning of vein-contouring markings in monarchs. In summary, WntA instructs the formation of multiple wing-pattern elements in the nymphalid radiation, demonstrating the importance of prepatterning processes in the unfolding of complex anatomy. The versatility of this signaling factor illustrates how the repeated tinkering of a developmental gene can foster boisterous evolutionary change.
Experimental Procedures
Butterflies.
Insect stock origins, rearing conditions, and oviposition host plants are described in SI Appendix, Table S3.
In Situ Hybridizations.
WntA cDNA sequences, cloned or amplified with T7 overhang primers, were used as a template to synthesize digoxigenin-labeled RNA probe as described previously (14, 36). Primers for amplification of template DNA are shown in SI Appendix, Table S4. In situ hybridization of imaginal discs from fifth instar larvae were performed as described (14).
Egg Injections.
Butterfly eggs laid on host plant leaves were collected after 1–6 h (SI Appendix, Tables S2 and S3). J. coenia and V. cardui eggs were then washed for 20–100 s in 5% benzalkonium chloride (Sigma-Aldrich), rinsed in water, and dried in a desiccation chamber or by air ventilation for softening the chorion. To soften and separate egg mass in H. sara, clumps were treated with a 1:20 dilution of Milton sterilizing fluid (Procter and Gamble) for 4 min, rinsed with water, and dried. Eggs were arranged on a double-sided adhesive tape or glued to a glass slide, usually with the micropyle facing up. CRISPR mixtures containing preassembled sgRNAs and recombinant Cas9 protein (PNA Bio) were injected, using pulled quartz or borosilicate needles. The concentration of sgRNAs and Cas9 varied between butterfly species and experiments (SI Appendix, Table S2).
Genotyping.
DNA was extracted from wing muscles or single legs using the Phire animal tissue direct PCR kit (Thermo Fisher Scientific), and amplified using oligonucleotides flanking the sgRNAs target region (SI Appendix, Table S2). PCR amplicons were gel-purified, subcloned into the pGEM-T Easy Vector System (Promega), and sequenced on an ABI 3730 sequencer.
Supplementary Material
Acknowledgments
We thank Saad Arif, Nora Braak, Chris Day, Melanie Gibbs, Jonah Heller, José Hermina-Perez, Colin Morrison, Oscar Paneso, Manu Sanjeev, David Tian, Camille Tulure, Matthew Verosloff, and Hans Van Dyck for assistance with rearing, injecting, dissecting, and imaging butterflies; Lawrence Gilbert for sharing photographs and insights on Heliconius wing patterning; and Karin van der Burg, Bernardo Clavijo, David Jaffe, James Lewis, and James Mallet for providing access to genomic data. This work was supported by grants from the NSF (Grants DGE-1650441, IOS-1354318, IOS-1557443, and IOS-1452648); the NIH (Grant GM108626); the Leverhulme Trust (Grant RPG-2014-167); the Pew Charitable Trust; a Nigel Groome PhD studentship (Oxford Brookes University); and the Smithsonian Institution.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1708149114/-/DCSupplemental.
References
- 1.Carroll SB. Evo-devo and an expanding evolutionary synthesis: A genetic theory of morphological evolution. Cell. 2008;134:25–36. doi: 10.1016/j.cell.2008.06.030. [DOI] [PubMed] [Google Scholar]
- 2.Kristensen NP, Scoble MJ, Karsholt O. Lepidoptera phylogeny and systematics: The state of inventorying moth and butterfly diversity. Zootaxa. 2007;1668:699–747. [Google Scholar]
- 3.Nijhout HF. The Development and Evolution of Butterfly Wing Patterns. Smithsonian Institution; Washington, DC: 1991. [Google Scholar]
- 4.Werner T, Koshikawa S, Williams TM, Carroll SB. Generation of a novel wing colour pattern by the Wingless morphogen. Nature. 2010;464:1143–1148. doi: 10.1038/nature08896. [DOI] [PubMed] [Google Scholar]
- 5.Yamaguchi J, et al. Periodic Wnt1 expression in response to ecdysteroid generates twin-spot markings on caterpillars. Nat Commun. 2013;4:1857. doi: 10.1038/ncomms2778. [DOI] [PubMed] [Google Scholar]
- 6.Koshikawa S, et al. Gain of cis-regulatory activities underlies novel domains of wingless gene expression in Drosophila. Proc Natl Acad Sci USA. 2015;112:7524–7529. doi: 10.1073/pnas.1509022112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Özsu N, Chan QY, Chen B, Gupta MD, Monteiro A. Wingless is a positive regulator of eyespot color patterns in Bicyclus anynana butterflies. Dev Biol. 2017 doi: 10.1016/j.ydbio.2017.06.030. [DOI] [PubMed] [Google Scholar]
- 8.Martin A, Reed RD. Wingless and aristaless2 define a developmental ground plan for moth and butterfly wing pattern evolution. Mol Biol Evol. 2010;27:2864–2878. doi: 10.1093/molbev/msq173. [DOI] [PubMed] [Google Scholar]
- 9.Martin A, et al. Diversification of complex butterfly wing patterns by repeated regulatory evolution of a Wnt ligand. Proc Natl Acad Sci USA. 2012;109:12632–12637. doi: 10.1073/pnas.1204800109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gallant JR, et al. Ancient homology underlies adaptive mimetic diversity across butterflies. Nat Commun. 2014;5:4817. doi: 10.1038/ncomms5817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huber B, et al. Conservatism and novelty in the genetic architecture of adaptation in Heliconius butterflies. Heredity (Edinb) 2015;114:515–524. doi: 10.1038/hdy.2015.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Belleghem SM, et al. Complex modular architecture around a simple toolkit of wing pattern genes. Nat Ecol Evol. 2017;1:0052. doi: 10.1038/s41559-016-0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martin A, Courtier-Orgogozo V. Morphological evolution repeatedly caused by mutations in signaling ligand genes. In: Sekimura T, Nijhout F, editors. Diversity and Evolution of Butterfly Wing Patterns: An Integrative Approach. Springer Singapore; Singapore: 2017. [Google Scholar]
- 14.Martin A, Reed RD. Wnt signaling underlies evolution and development of the butterfly wing pattern symmetry systems. Dev Biol. 2014;395:367–378. doi: 10.1016/j.ydbio.2014.08.031. [DOI] [PubMed] [Google Scholar]
- 15.Schwanwitsch B. Evolution of the wing-pattern in Palaearctic Satyridae: Pararge and five other genera. Acta Zool. 1935;16:143–281. [Google Scholar]
- 16.Schwanwitsch B. Color-pattern in Lepidoptera. Entomol Obozr. 1956;35:530–546. [Google Scholar]
- 17.Otaki JM. Color pattern analysis of nymphalid butterfly wings: Revision of the nymphalid groundplan. Zoolog Sci. 2012;29:568–576. doi: 10.2108/zsj.29.568. [DOI] [PubMed] [Google Scholar]
- 18.Abbasi R, Marcus JM. Color pattern evolution in Vanessa butterflies (Nymphalidae: Nymphalini): Non-eyespot characters. Evol Dev. 2015;17:63–81. doi: 10.1111/ede.12109. [DOI] [PubMed] [Google Scholar]
- 19.Taira W, Kinjo S, Otaki JM. The marginal band system in nymphalid butterfly wings. Zoolog Sci. 2015;32:38–46. doi: 10.2108/zs140058. [DOI] [PubMed] [Google Scholar]
- 20.Wahlberg N, et al. Nymphalid butterflies diversify following near demise at the Cretaceous/Tertiary boundary. Proc Biol Sci. 2009;276:4295–4302. doi: 10.1098/rspb.2009.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perry M, et al. Molecular logic behind the three-way stochastic choices that expand butterfly colour vision. Nature. 2016;535:280–284. doi: 10.1038/nature18616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang L, Reed RD. Genome editing in butterflies reveals that spalt promotes and Distal-less represses eyespot colour patterns. Nat Commun. 2016;7:11769. doi: 10.1038/ncomms11769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang L, et al. Genetic basis of melanin pigmentation in butterfly wings. Genetics. 2017;205:1537–1550. doi: 10.1534/genetics.116.196451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carroll SB, et al. Pattern formation and eyespot determination in butterfly wings. Science. 1994;265:109–114. doi: 10.1126/science.7912449. [DOI] [PubMed] [Google Scholar]
- 25.Nijhout HF. A comprehensive model for colour pattern formation in butterflies. Proc R Soc Lond B Biol Sci. 1990;239:81–113. [Google Scholar]
- 26.Monteiro A, Gupta MD. Identifying coopted networks and causative mutations in the origin of novel complex traits. Curr Top Dev Biol. 2016;119:205–226. doi: 10.1016/bs.ctdb.2016.03.003. [DOI] [PubMed] [Google Scholar]
- 27.Nijhout HF, Wray GA. Homologies in the color patterns of the genus Heliconius (Lepidoptera, Nymphalidae) Biol J Linn Soc Lond. 1988;33:345–365. [Google Scholar]
- 28.Nijhout HF, Wray GA, Gilbert LE. An analysis of the phenotypic effects of certain color pattern genes in Heliconius (Lepidoptera, Nymphalidae) Biol J Linn Soc Lond. 1990;40:357–372. [Google Scholar]
- 29.Gilbert LE. Adaptive novelty through introgression in Heliconius wing patterns: Evidence for shared genetic “tool box” from synthetic hybrid zones and a theory of diversification. In: Boggs CL, Watt WB, Ehrlich PR, editors. Ecology and Evolution Taking Flight: Butterflies as Model Systems. Univ. of Chicago Press; Chicago: 2003. [Google Scholar]
- 30.Dinwiddie A, et al. Dynamics of F-actin prefigure the structure of butterfly wing scales. Dev Biol. 2014;392:404–418. doi: 10.1016/j.ydbio.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 31.Binari RC, et al. Genetic evidence that heparin-like glycosaminoglycans are involved in wingless signaling. Development. 1997;124:2623–2632. doi: 10.1242/dev.124.13.2623. [DOI] [PubMed] [Google Scholar]
- 32.Serfas MS, Carroll SB. Pharmacologic approaches to butterfly wing patterning: Sulfated polysaccharides mimic or antagonize cold shock and alter the interpretation of gradients of positional information. Dev Biol. 2005;287:416–424. doi: 10.1016/j.ydbio.2005.09.014. [DOI] [PubMed] [Google Scholar]
- 33.Nijhout HF. Molecular and physiological basis of colour pattern formation. Adv Insect Physiol. 2010;38:219–265. [Google Scholar]
- 34.Tögel M, Pass G, Paululat A. The Drosophila wing hearts originate from pericardial cells and are essential for wing maturation. Dev Biol. 2008;318:29–37. doi: 10.1016/j.ydbio.2008.02.043. [DOI] [PubMed] [Google Scholar]
- 35.Rebeiz M, Patel NH, Hinman VF. Unraveling the tangled skein: The evolution of transcriptional regulatory networks in development. Annu Rev Genomics Hum Genet. 2015;16:103–131. doi: 10.1146/annurev-genom-091212-153423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carter J-M, Gibbs M, Breuker CJ. Divergent RNA localisation patterns of maternal genes regulating embryonic patterning in the butterfly Pararge aegeria. PLoS One. 2015;10:e0144471. doi: 10.1371/journal.pone.0144471. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.