Significance
Identifying the drivers of tumorigenesis provides insight into mechanisms of transformation and can suggest novel therapeutic targets. IDH1 mutations in gliomas are one such promising target. Drivers of tumor initiation may be distinct from those at tumor recurrence, however. Here, we demonstrate that in a subset of initially IDH1 mutant gliomas IDH1 is deleted or amplified at recurrence, yielding a higher grade tumor with a reprogrammed epigenome. We also report systematic selection for cells with IDH1 CNA in vitro and in vivo. Thus, while IDH1 mutation likely initiates gliomagenesis, neither mutant IDH1 nor the oncometabolite 2HG that it produces are required at recurrence. These findings have important implications for emerging therapeutic strategies targeting mutant IDH1.
Keywords: IDH1, DNA methylation, 2HG, glioma, copy number
Abstract
IDH1 mutation is the earliest genetic alteration in low-grade gliomas (LGGs), but its role in tumor recurrence is unclear. Mutant IDH1 drives overproduction of the oncometabolite d-2-hydroxyglutarate (2HG) and a CpG island (CGI) hypermethylation phenotype (G-CIMP). To investigate the role of mutant IDH1 at recurrence, we performed a longitudinal analysis of 50 IDH1 mutant LGGs. We discovered six cases with copy number alterations (CNAs) at the IDH1 locus at recurrence. Deletion or amplification of IDH1 was followed by clonal expansion and recurrence at a higher grade. Successful cultures derived from IDH1 mutant, but not IDH1 wild type, gliomas systematically deleted IDH1 in vitro and in vivo, further suggestive of selection against the heterozygous mutant state as tumors progress. Tumors and cultures with IDH1 CNA had decreased 2HG, maintenance of G-CIMP, and DNA methylation reprogramming outside CGI. Thus, while IDH1 mutation initiates gliomagenesis, in some patients mutant IDH1 and 2HG are not required for later clonal expansions.
Heterozygous mutations in IDH1 or IDH2 (collectively, IDH) are the earliest and most common genetic alteration in low-grade gliomas (LGGs) and are frequently found in other cancers as well, including acute myelogenous leukemia (AML) (1–4). Mutant IDH produces high intracellular concentrations of the oncometabolite d-2-hydroxyglutarate (2HG), which can activate some α-ketoglutarate (αKG)-dependent enzymes and competitively inhibit others, including TET2 and histone demethylases (5–7). 2HG-mediated inhibition of these epigenetic modifiers in IDH mutant tumors is associated with increases in CpG island (CGI) DNA methylation (in gliomas, the glioma CpG island methylator phenotype, or G-CIMP), changes in histone methylation patterns, and changes in chromosome topology (7–9). Among all adult gliomas, IDH mutant tumors are associated with the longest patient survival (10, 11). IDH mutations and high levels of 2HG are associated with a differentiation block, which can be released by treatment with mutant IDH inhibitors; however, the effects of IDH inhibition may depend on the disease, cell type, or tumor model (12–17).
The impact of IDH mutations on glioma progression remains difficult to assess due to the difficulty of modeling these tumors. While cell lines overexpressing the mutant enzyme have provided key insights, they do not accurately represent the heterozygous mutant state of the disease. Considerable efforts have been invested to establish brain tumor initiating cell (BTIC) lines from IDH mutant gliomas and more closely recapitulate the biology of these tumors. However, patient-derived cells with endogenous expression of mutant IDH are largely refractory in culture, although xenografts from IDH mutant gliomas with tertiary mutations have been established (18, 19).
In paired LGG samples, IDH mutations were shown to be retained through tumor recurrence, in contrast to TP53, ATRX, BRAF, and SMARCA4 mutations that are occasionally “lost” (2, 3, 20). However, case reports suggest that the mutant or wild-type IDH1 allele can be deleted during tumor recurrence and may be selected against in vitro (21–25), raising the critical question of the role of IDH mutations beyond tumor initiation. Here we used longitudinally collected tumor samples to discover and characterize the dynamic molecular, cellular, metabolic, and clinical features of initially IDH1 mutant LGG with and without subsequent copy number alterations (CNAs) affecting IDH1. In addition, we developed cell culture and xenograft models that spontaneously and systematically lost a copy of IDH1, and compared their metabolic and epigenome changes with those seen in patient samples.
Results
Tumors with Altered Copy Number at IDH1.
We performed exome sequencing on a cohort of 50 paired initial LGGs and their patient-matched recurrences (3, 26, 27) (SI Appendix, Table S1 and Dataset S1). All of the initial tumors had heterozygous mutations in IDH1 or IDH2; however, in two recurrences (in patients 14 and 169), the IDH1 mutant allele frequency (MAF) dropped below the 10% minimum detection threshold (Fig. 1A and SI Appendix, Table S2). Other mutations were retained (Fig. 1B and SI Appendix, Dataset S2), suggesting that recurrence in these cases involved deletion of the mutant IDH1 allele.
Fig. 1.
Deletion or amplification of mutant IDH1 during malignant progression. (A) Bar plot showing IDH1 MAF in each exome. The RNA-seq MAF is shown with a black triangle for samples with data. (B) Phylogenetic trees of patients 14 and 169. Branch lengths are proportional to the number of mutations detected. (C) LOH (Left) and TCN (Right) plots of chr2 for patient 169 initial A and recurrence 1 A. LOH is plotted as decrease in heterozygosity from normal (0, no change; 1, complete loss of heterozygosity). (D) Representative images for two distinct regions of patient 169 recurrence 1 (Left, heterozygous IDH1 mutation; Right, deletion of mutant allele) and patient 21 recurrence 1 (Left, heterozygous IDH1 mutation; Right, chr2 gain and amplification of IDH1). (Top) FISH for the centromere of chr2 (green) and IDH1 (magenta). (Middle) IHC with an antibody specific to R132H mutant IDH1. (Scale bar: 50 μm.) (Bottom) Ki-67 staining, with the percentage of positive cells labeled. (Scale bar: 50 μm.) amp, amplification; del, single copy deletion; mut, IDH1 mutant; N/A, not available; rec, recurrence.
To test for the suspected CNA, we used loss of heterozygosity (LOH) analysis (28). For both recurrent tumors, the apparent LOH suggested an allelic imbalance, which was confirmed as single copy loss in total copy number (TCN) analysis (Fig. 1C and SI Appendix, Fig. S1A). We next searched all tumors for LOH at IDH1 or IDH2 and identified four additional cases (SI Appendix, Fig. S1 and Table S2). To conclusively determine the copy number at IDH1, we performed fluorescence in situ hybridization (FISH) at the IDH1 locus (Fig. 1D and SI Appendix, Table S3). (The exome data for one case was acquired from an outside institution and was not analyzed further.) In patient 27, IDH1 CNAs were present in both the initial and recurrent tumor, while the remaining cases presented with IDH1 CNAs at recurrence. From these analyses, we identified the genetic basis of potentially functional changes in IDH1 TCN during malignant progression: deletion of the mutant allele (patients 14 and 169), subclonal gain of the wild-type allele (patient 68), subclonal gain of the mutant allele (patient 17), and gain with additional subclonal amplification of the mutant allele (patient 21) (SI Appendix, Table S2).
To investigate how well these single samples represented the tumor overall, we analyzed new exome sequencing of spatially distinct tumor tissue from patients 14, 68, and 169, along with previously published data from patient 17 (3) (SI Appendix, Table S2 and Dataset S1). These intratumoral analyses suggest that IDH1 CNAs were present throughout the recurrent tumors, yet were undetectable in the initial tumor (Fig. 1A and SI Appendix, Fig. S1). Thus, the CNAs were followed by substantial clonal expansion.
Copy Number Changes at IDH1 Impact mRNA and Protein Expression.
We next examined the impact of these genetic alterations on IDH1 mRNA and protein levels. In each case with transcriptome sequencing, the MAF from mRNA was very similar to the MAF in DNA (Fig. 1A and SI Appendix, Table S2), suggesting that all alleles were equally expressed. We then performed immunohistochemistry (IHC) with an IDH1 R132H-specific antibody on all available formalin-fixed, paraffin-embedded tumor blocks (SI Appendix, Table S3). We found that the ploidy and chromosome 2 (chr2) CNAs manifested at the protein level, ranging from absence of mutant protein following deletion of the mutant allele to dramatically increased staining intensity following amplification of the mutant allele (Fig. 1D and SI Appendix, Fig. S2).
Decreased 2HG and Increased Proliferation Following Amplification or Deletion of Mutant IDH1.
We next addressed the functional consequences of IDH1 CNAs. IDH1 mutations are strictly heterozygous, maintaining a specific ratio of wild-type to mutant enzymes for maximal production of 2HG (22, 29–32). IDH1 CNAs may shift this balance and lead to a paradoxical decrease in the production of 2HG. We used NMR analysis of snap-frozen tumor tissue to calculate the level of 2HG and then exome-sequenced that tissue. In patient 14, 2HG was undetectable following loss of the mutant allele (SI Appendix, Fig. S3A and Table S2). Patient 21 had low levels of 2HG despite malignant progression to high-grade glioblastoma (GBM), which typically has elevated 2HG levels (33) (SI Appendix, Fig. S3 B and C and Table S2). This is consistent with biochemical evidence indicating that a relative abundance of mutant IDH1 decreases 2HG production (22, 31, 32). In contrast, in patient 17 2HG levels fell in a normal range for IDH mutant GBMs (34) (SI Appendix, Fig. S3 C and D), potentially due to the subclonal nature of the CNA. Interestingly, the initial tumor from patient 169 showed little detectable 2HG (SI Appendix, Fig. S3E), despite subsequent exome sequencing of the tissue showing intact chr2, although with decreased IDH1 MAF (Fig. 1A and SI Appendix, Fig. S1F and Table S2). This discrepancy may be due to a subclonal deletion, as IHC for mutant IDH1 was negative in several regions of the initial tumor. For all other tumors, exome-derived IDH1 MAFs on post-NMR tissue were consistent with other tissue samples from the same surgical resections (SI Appendix, Fig. S1 and Table S3). These data further support in vivo clonal expansion of the cell that sustained IDH1 CNA and had impaired 2HG production.
Mutant IDH1 and 2HG reduce BTIC proliferation in vitro (25, 31, 35). Indeed, recurrent tumors with IDH1 CNAs underwent malignant progression to a higher grade (SI Appendix, Table S2). Therefore, we asked whether the observed IDH1 CNAs and decrease in 2HG were associated with more proliferative tumor clones. We counted Ki-67 positivity, a marker of actively cycling cells, in tumors with regionally heterogeneous IDH1 R132H IHC staining. In all three cases, the region with abnormal staining had a higher Ki-67 index than the region with typical staining (P = 0.008, paired t test) (Fig. 1D and SI Appendix, Table S4). Thus, clonal IDH1 CNA and reduction of 2HG are associated with a more proliferative state and outgrowth of a more aggressive tumor.
Systematic IDH1 Deletion in Vitro.
A retrospective analysis showed successful establishment of BTICs from 8% (4 of 49) of IDH mutant gliomas, compared to 63% (62 of 99) from IDH wild-type gliomas (P = 1 × 10−5, Fisher’s exact test) (SI Appendix, Table S5), illustrating the refractory nature of IDH mutant cells to in vitro culture. However, four BTIC lines were established from grade III or IV IDH mutant tumors, three of which harbored tertiary mutations that likely contributed to successful establishment (19, 25, 36, 37) (SI Appendix, Tables S6 and S7 and Dataset S3). However, these BTIC lines systematically lost either the mutant or wild-type IDH1 allele (SI Appendix, Fig. S4A). Here we confirmed previous findings (23, 25), using a higher resolution copy number array, that each BTIC line derived from IDH1 mutant glioma has chr2 deletions encompassing IDH1 (Fig. 2A, and SI Appendix, Fig. S4B and Table S6) despite globally similar copy number profiles (SI Appendix, Fig. S4C). In contrast, chr2 alterations were rare in a panel of 38 IDH wild-type tumors and matched BTIC lines (Fig. 2 B and C and SI Appendix, Fig. S5). Thus, selection of cells with chr2 deletions was exclusive to IDH mutant gliomas, suggesting that this deletion confers a selective growth advantage for IDH mutant gliomas in vitro (25).
Fig. 2.
Hemizygous deletion of the IDH1 locus in BTIC lines and xenografts derived from IDH mutant gliomas. (A) Averaged copy number along chr2 for four IDH1 mutant tumors (blue) and their derived BTICs (red). (B) Circos plot representing the frequency of CNA among 38 IDH wild-type tumor samples (outer track) and their matching derived BTIC lines (inner track). The bar height represents the frequency of deletions (green) and amplifications (red). (C) Copy number at IDH1 from the same 38 IDH wild-type gliomas (blue) and BTIC lines derived from them (red). (D) Magnetic resonance imaging of the patient tumor from which BT257 was derived. (E) Microscopic image of BT257 primary BTICs. (Scale bar: 500 μm.) (F) Sanger sequencing traces showing a decrease in mutant allele frequency at xenograft passage 6. (G) H&E staining of xenografted brain. (Scale bar: 2 mm.) (H) Serial orthotopic xenografting in SCID mice was used to propagate BT257. (Scale bar: mouse brain, 2 mm; neurospheres, 500 μm.) (I) Chr2 copy number in BT257 tumor tissue (blue) and adjacent normal brain (black). (J) Chr2 copy number in BT257 primary BTICs (blue) and xenograft passages 1 (orange) and 6 (red).
We next asked whether IDH1 CNAs arose in vitro or if cells with the deletion preexisted in the tumor. A low-frequency mosaic deletion in BT92 tumor tissue suggested the latter scenario (SI Appendix, Fig. S4B). CNA analysis of a separate piece of BT92 tumor tissue identified a subclonal chr2 deletion matching the deletion observed in the derived BTIC line (SI Appendix, Fig. S6). Together with the CNA identified in our cohort of paired primary gliomas, this suggests that cells with IDH1 deletions exist in the tumor and are selected for in vitro.
IDH1 Deletion in Vivo.
We further set out to determine whether the selection for IDH1 CNA would be recapitulated in vivo. Primary cells isolated from a secondary GBM (BT257; Fig. 2D) formed spheres within the first week in culture (38) (Fig. 2E). Sequencing identified a rare IDH1 R132S mutation, which was maintained in vitro (Fig. 2F). Although these cells remained viable in culture for three months, they could not be successfully expanded. Thus, all remaining viable cells were used for orthotopic xenografts. Within a year, all animals developed large 2HG-producing IDH1 mutant tumors consistent with GBM (Fig. 2G and SI Appendix, Fig. S7 A–D). We maintained BT257 through serial xenografting, with survival ranging from three to six months (Fig. 2H and SI Appendix, Fig. S7D). While heterozygosity was maintained in the first xenograft, we observed a decrease in the height of the mutant peak in later xenografts (Fig. 2F). The BT257 tumor had no detectable alteration on chr2, with the exception of a focal amplification of MYCN (Fig. 2I), yet we observed mosaic loss of chr2 being enriched from the primary BTICs to X6-BTICs, correlating with the loss of the IDH1 mutant allele (Fig. 2J and SI Appendix, Fig. S7E). IDH1 R132H and R132S produce different amounts of 2HG, which could result in subtle differences in selective pressure for IDH1 CNA events (35).
IDH1 Copy Number-Altered Tumors Retain G-CIMP.
Given the widespread DNA methylation changes associated with IDH mutations, we investigated the DNA methylation dynamics of these tumors. All tumors retained G-CIMP based on an eight-site definition (8) and unsupervised hierarchical clustering with IDH mutant and wild-type gliomas from TCGA (11, 39) (SI Appendix, Fig. S8). Similarly, among 50 genes with G-CIMP–associated decreased expression (8, 25), none significantly changed expression following IDH1 CNA (SI Appendix, SI Materials and Methods). These analyses, based on a subset of CpG sites, suggest a static methylome during recurrence despite IDH1 CNA.
Recent analysis of G-CIMP–positive gliomas suggested that subclassification into G-CIMP high and G-CIMP low can better stratify by outcome (40). We wondered whether IDH1 CNA may drive a shift from G-CIMP high to G-CIMP low with its associated worse outcomes. We applied a random forest model built on TCGA data to classify each sample from the five patients with IDH1 CNAs and found that all samples from the three recurrences with clonal CNA were classified as G-CIMP low, while all other samples were G-CIMP high (Fig. 3A and SI Appendix, Table S8). This raises the possibility that G-CIMP–low tumors have altered methylomes due to IDH1 CNA.
Fig. 3.
Loss of DNA methylation following deletion or amplification of mutant IDH1. (A) G-CIMP–high and G-CIMP–low scores for the initial-recurrent pairs with IDH1 CNA. Samples are color-coded based on genetic status and shape reflects the G-CIMP–high/G-CIMP–low classification. “Pre-change” indicates the initial tumor from patients with IDH1 CNA recurrent tumors. (B) PCA built on TCGA GBM and LGG initial tumors and applied to our cohort of 22 initial-recurrent tumor pairs. (C) Boxplot of mean β values separated into normal brain (n = 53), IDH mutant LGG and patient-matched recurrences (n = 55) and the IDH1 CNA patients (n = 28). (D and E) Mean change in methylation from initial to recurrence for each patient in the cohort, using all CpG sites (D) or divided into those within CGIs, in CGI shores or shelves, or unassociated with a CGI (E). The number of CpG sites in each subset is provided in parentheses. amp, amplification; del, single copy deletion; mut, IDH1 mutant; rec, recurrence.
IDH1 Copy Number-Altered Tumors Exhibit Methylation Reprogramming Outside of CGI.
We used principal component analysis (PCA) to provide a global view of DNA methylation patterns. Principal components (PCs) 1 and 2 defined using TCGA gliomas separated tumors by IDH mutation status; when we plotted the IDH1 CNA cases on the same PCs, several of the recurrences appeared more like the IDH wild-type TCGA tumors, while their initial IDH1 mutant counterparts resembled IDH mutant TCGA tumors (Fig. 3B). This dramatic shift within individual patients suggests that despite retention of G-CIMP, these malignantly progressed tumors have dynamically altered DNA methylation. Indeed, density plots, average methylation per tumor, and average change in methylation from the initial tumor to the IDH1 copy number-altered recurrence indicate a decrease in methylation of much larger magnitude than the decrease characteristic of malignant progression to GBM (26) (Fig. 3 C and D and SI Appendix, Fig. S9A).
Because the persistence of G-CIMP indicated retained hypermethylation within CGIs, we asked whether the hypomethylation occurred primarily outside of CGIs. Indeed, non-CGI CpG sites clearly drove the hypomethylation (Fig. 3E). While the pattern of hypomethylation was very strong in cases with clonal CNA (patients 14, 21, and 169), it was moderate or not detected in cases with subclonal CNA. Since the genomic DNA for methylation analysis was derived from a larger tumor area than the FISH analysis, subclonality might have muted the signal of any aberrant hypomethylation.
If the epigenetic changes in tumors with clonal CNA are mechanistically related to the decrease in 2HG as a consequence of IDH1 CNA, then there should be a common set of altered CpG sites. We calculated the change in methylation between initial and recurrent tumors at each assayed CpG site and found that patients 14, 21, and 169 grouped together in unsupervised hierarchical clustering and PCA (Fig. 4A and SI Appendix, Fig. S9B). Similarly, the three highest pairwise correlations were among patients 14, 21, and 169 (SI Appendix, Fig. S9C). Taken together, these analyses demonstrate that similar CpG sites are altered following IDH1 CNA, suggesting a common effector.
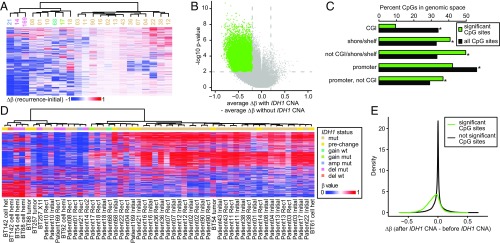
Fig. 4.
Shared pattern of DNA methylation changes in patient tumors and model systems with IDH1 CNA. (A) Unsupervised hierarchical clustering of the change in methylation from initial to recurrence in each patient using 1,482 CpG sites (top 0.5% most variable ranked by SD). Patient numbers are provided under the dendrogram. (B) Volcano plot of the difference between the changes in methylation from initial to recurrent gliomas with or without clonal IDH1 CNA. Colored points denote significantly differentially methylated CpG sites. P values are Benjamini–Hochberg corrected. (C) Percentage of significant CpG sites (green) and all CpG sites (black) in the genomic spaces listed. All genomic spaces show significant differences (P < 0.0001, permutation test). (D) Unsupervised hierarchical clustering of initial-recurrent pairs and BTICs and xenografts using the 1,479 most variable probes (top 0.5% most variable ranked by SD). (E) Density plots of the change in methylation of the significant (green line, 29,018 CpG sites; median change, −0.07) and not significant (black line, 268,324 CpG sites; median change, −0.0001) CpG sites defined in B in the BTICs and xenografts with CNA. amp, amplification; del, single copy deletion; mut, IDH1 mutant.
To identify the CpGs that lose methylation following IDH1 CNA, we compared the change in methylation between initial and recurrent tumors with and without IDH1 CNA and identified 29,018 CpG sites with significant hypomethylation specific to recurrence with IDH1 CNA (Fig. 4B). These CpG sites were underenriched in CGI and overenriched in shores/shelves and non-CGI genomic spaces (P < 0.0001, permutation test) (Fig. 4C). While globally promoter CpG sites were underenriched, there was a clear enrichment once CpG sites in CGI were excluded (P < 0.0001, permutation test) (Fig. 4C). We performed gene ontology enrichment analysis on genes with significant CpG sites in their promoters while excluding CpG sites in CGI and found modest enrichments (Dataset S4).
Methylation Reprogramming Following IDH1 CNA in Vitro and in Vivo.
To further confirm that the identified DNA methylation changes are related to IDH1 CNA, we profiled the BTICs and xenografts, along with a second sample from each patient lacking the CNA: tumor tissue, an earlier passage of BT142 before CNA, and a primary culture derived from an earlier surgery that expanded but did not establish (BT61, matching recurrent sample BT92). All samples were G-CIMP–positive (SI Appendix, Fig. S8). Unsupervised hierarchical clustering with the initial-recurrent tumor pairs showed that samples with CNA, whether cell line, xenograft, or primary tumor tissue, clustered together (Fig. 4D).
We then examined the 29,018 CpG sites that were associated with CNA in the initial-recurrent tumor pairs. If the methylation changes identified in the initial-recurrent tumor pairs are a signature of IDH1 CNA, then those same CpG sites should also lose methylation in these models. We calculated the change in methylation at each CpG site for each patient (SI Appendix, Table S6). Examination of the methylation changes in the 29,018 CpG sites revealed decreased methylation in those CpG sites, with a median methylation change of −0.07, compared with −0.0001 in the background CpG sites (Fig. 4E). Thus, a common pattern of methylation change is evident across in vitro BTICs, an in vivo xenograft, and paired initial-recurrent patient tumors, suggesting that IDH1 CNA truly leads to reprogramming of the DNA methylome.
Discussion
IDH mutations are typically retained upon recurrence, leading to the initial hypothesis that these mutations play a role in tumor recurrence. Our paired initial-recurrent gliomas show that glioma cells can delete or amplify the mutant or wild-type allele, altering the balanced ratio of wild-type and mutant IDH1 and leading to disrupted or abolished production of 2HG. Cells with IDH1 CNAs then clonally expand to occupy a substantial portion of the recurrent tumor, suggesting a growth advantage. This is confirmed by the in vitro and in vivo models described here, in which deletion of either the mutant or wild-type IDH1 allele occurs systematically in IDH mutant-derived BTICs, but not in IDH wild-type–derived BTICs. This selection for CNA in vitro is reminiscent of tumor-suppressor genes for which the frequency of homozygous deletion or biallelic loss in primary cultures is elevated relative to tumor tissue (41, 42). In the case of IDH1 CNA, however, the unifying feature is loss of the heterozygous mutant state. In almost all of these cases, patients received temozolomide (TMZ) before the recurrence with IDH1 CNA (SI Appendix, Table S1), raising the possibility that TMZ treatment may contribute to outgrowth of the CNA cells, perhaps by preferentially killing the tumor cells that retain heterozygous IDH1 mutations (43).
Although derived from a relatively small number of cases, the molecular, cellular, and metabolic data presented here, along with genetics-focused case reports, suggest that mutant IDH1 is not strictly required for recurrence (21–24). Model systems expressing mutant IDH1 also suggest an abbreviated period during which inhibition of mutant IDH1 affects cell proliferation (44). Recurrent tumors with IDH1 CNA have some resemblance to the G-CIMP–low subtype (40). Taken together, these data suggest that loss or inhibition of mutant IDH may be associated with a more aggressive phenotype.
Small-molecule inhibition of mutant IDH1 in glioma cell lines results in few DNA methylation changes (15, 17), yet our samples demonstrated massive hypomethylation and decreased 2HG in cells with clonal IDH1 CNA. This may reflect the differences of in vitro drug therapy relative to tumor evolution in patients or differences among IDH1 inhibitors (45). A central question is whether mutant IDH is a reasonable target for therapy. Early clinical trial results suggest some efficacy in patients with IDH mutant AML (4), but not yet in IDH mutant gliomas. IDH mutant AML and gliomas have several key differences. While IDH mutations in gliomas are prognostic of better outcomes, studies in AML report conflicting impacts on survival (46). AML has a higher frequency of IDH2 mutations and a more even distribution of different IDH1 mutations. This, combined with the different subcellular localization of IDH1 and IDH2, affects the concentration of αKG, the NADP/NADPH ratio, and production of 2HG, and may account for different prognostic effects (32, 46). However, IDH mutations in AML, as in gliomas, are early events that drive an altered epigenome and block cellular differentiation (7). The efficacy of IDH inhibitors in AML may be related to successful reversal of the differentiation block, a key feature of other AML therapies (16, 47).
It is unlikely that an inhibitor of mutant IDH1 would have an impact on tumor cell populations with IDH1 deletion, since the target enzyme will not be present. Thus, we propose that longitudinal monitoring of the IDH1 locus during treatment with IDH1 inhibitors be incorporated into clinical trials to better understand the role that such deletions may play in drug response. In addition, mutations in IDH are associated with a wide range of epigenetic, gene expression, and metabolic changes with potential as therapeutic targets, possibly independent of whether the tumor retains or loses mutant IDH1 (17, 48, 49).
Materials and Methods
Sample use was approved by the University of California San Francisco’s Committee on Human Research, and this research was approved by the University of California, San Francisco’s Institutional Review Board. Additional samples were collected following protocols approved by the University of Calgary’s Human Research Ethics Board. All patients provided informed written consent. Animal studies were performed following institutional ethical guidelines and protocols approved by the University of Calgary’s Animal Care Committee. BTIC lines were processed as described previously (38). Genomic DNA was bisulfite-converted and processed on Infinium HumanMethylation450 bead arrays according to the manufacturer’s protocol as described previously (26).
New exome sequencing, RNA sequencing, and DNA methylation array data (accession no. EGAS00001001854) and whole genome sequencing (accession no. EGAS00001002618) have been deposited in the European Genome-Phenome Archive database. EGA accession numbers for previously published data are provided in SI Appendix, Dataset S1. Code is available on GitHub (https://github.com/UCSF-Costello-Lab/IDH1_CNA).
Full details of all the protocols used in this study are provided in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank the staff of the University of California, San Francisco’s Brain Tumor Tissue Bank for timely and significant contributions of key samples; J. Song for advice on statistical analyses; the Calgary Brain Tumor and Tissue Bank for sample contribution; Yaron Butterfield for genomic analysis of BT54 and BT88 cell lines; and Rozina Hassam, Orsolya Cseh, and Jiqing Zhang for technical assistance. This project was generously supported by a gift from the Dabbiere family. Additional support was provided by National Institutes of Health Grants R01 CA169316 (to J.F.C.), R01 CA172845 (to S.M.R.), P01 CA118816-06 (to J.F.C.), P50 CA097257 (to A. Perry, S.M.C., A.M.M., J.J.P., and J.F.C.), and 5T32CA151022-07 (to L.E.J.); National Cancer Institute Grant 5P30CA82103 (to H.B. and A.B.O.); the Terry Fox Research Institute (S.J.M.J., M.A.M., J.G.C., S.W., and H.A.L.); the Stem Cell Network (S.W. and H.A.L); the Alberta Cancer Foundation (C.C.); Henry Ford Health System (H.N.); and Sao Paulo Research Foundation and Coordination of Improvement of Higher Education Personnel Grants 2014/08321-3, 2015/07925-5, and 2016/15485-8 (to C.F.S. and H.N.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: New exome sequencing, RNA sequencing, and DNA methylation array data (accession no. EGAS00001001854) and whole genome sequencing (accession no. EGAS00001002618) have been deposited in the European Genome-Phenome Archive database. Code is available on GitHub (https://github.com/UCSF-Costello-Lab/IDH1_CNA).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1708914114/-/DCSupplemental.
References
- 1.Suzuki H, et al. Mutational landscape and clonal architecture in grade II and III gliomas. Nat Genet. 2015;47:458–468. doi: 10.1038/ng.3273. [DOI] [PubMed] [Google Scholar]
- 2.Watanabe T, Nobusawa S, Kleihues P, Ohgaki H. IDH1 mutations are early events in the development of astrocytomas and oligodendrogliomas. Am J Pathol. 2009;174:1149–1153. doi: 10.2353/ajpath.2009.080958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnson BE, et al. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science. 2014;343:189–193. doi: 10.1126/science.1239947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dang L, Yen K, Attar EC. IDH mutations in cancer and progress toward development of targeted therapeutics. Ann Oncol. 2016;27:599–608. doi: 10.1093/annonc/mdw013. [DOI] [PubMed] [Google Scholar]
- 5.Dang L, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Losman JA, Kaelin WG., Jr What a difference a hydroxyl makes: Mutant IDH, (R)-2-hydroxyglutarate, and cancer. Genes Dev. 2013;27:836–852. doi: 10.1101/gad.217406.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Figueroa ME, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Noushmehr H, et al. Cancer Genome Atlas Research Network Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flavahan WA, et al. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature. 2016;529:110–114. doi: 10.1038/nature16490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eckel-Passow JE, et al. Glioma groups based on 1p/19q, IDH, and TERT promoter mutations in tumors. N Engl J Med. 2015;372:2499–2508. doi: 10.1056/NEJMoa1407279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brat DJ, et al. Cancer Genome Atlas Research Network Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med. 2015;372:2481–2498. doi: 10.1056/NEJMoa1402121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu C, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483:474–478. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sasaki M, et al. IDH1(R132H) mutation increases murine haematopoietic progenitors and alters epigenetics. Nature. 2012;488:656–659. doi: 10.1038/nature11323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Losman JA, et al. (R)-2-hydroxyglutarate is sufficient to promote leukemogenesis and its effects are reversible. Science. 2013;339:1621–1625. doi: 10.1126/science.1231677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rohle D, et al. An inhibitor of mutant IDH1 delays growth and promotes differentiation of glioma cells. Science. 2013;340:626–630. doi: 10.1126/science.1236062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang F, et al. Targeted inhibition of mutant IDH2 in leukemia cells induces cellular differentiation. Science. 2013;340:622–626. doi: 10.1126/science.1234769. [DOI] [PubMed] [Google Scholar]
- 17.Tateishi K, et al. Extreme vulnerability of IDH1 mutant cancers to NAD+ depletion. Cancer Cell. 2015;28:773–784. doi: 10.1016/j.ccell.2015.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Piaskowski S, et al. Glioma cells showing IDH1 mutation cannot be propagated in standard cell culture conditions. Br J Cancer. 2011;104:968–970. doi: 10.1038/bjc.2011.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wakimoto H, et al. Targetable signaling pathway mutations are associated with malignant phenotype in IDH-mutant gliomas. Clin Cancer Res. 2014;20:2898–2909. doi: 10.1158/1078-0432.CCR-13-3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yan H, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pusch S, et al. Glioma IDH1 mutation patterns off the beaten track. Neuropathol Appl Neurobiol. 2011;37:428–430. doi: 10.1111/j.1365-2990.2010.01127.x. [DOI] [PubMed] [Google Scholar]
- 22.Jin G, et al. Disruption of wild-type IDH1 suppresses D-2-hydroxyglutarate production in IDH1-mutated gliomas. Cancer Res. 2013;73:496–501. doi: 10.1158/0008-5472.CAN-12-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luchman HA, Chesnelong C, Cairncross JG, Weiss S. Spontaneous loss of heterozygosity leading to homozygous R132H in a patient-derived IDH1 mutant cell line. Neuro Oncol. 2013;15:979–980. doi: 10.1093/neuonc/not064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Favero F, et al. Glioblastoma adaptation traced through decline of an IDH1 clonal driver and macro-evolution of a double-minute chromosome. Ann Oncol. 2015;26:880–887. doi: 10.1093/annonc/mdv127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chesnelong C, et al. Lactate dehydrogenase A silencing in IDH mutant gliomas. Neuro Oncol. 2014;16:686–695. doi: 10.1093/neuonc/not243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mazor T, et al. DNA methylation and somatic mutations converge on the cell cycle and define similar evolutionary histories in brain tumors. Cancer Cell. 2015;28:307–317. doi: 10.1016/j.ccell.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Thuijl HF, et al. Evolution of DNA repair defects during malignant progression of low-grade gliomas after temozolomide treatment. Acta Neuropathol. 2015;129:597–607. doi: 10.1007/s00401-015-1403-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cavenee WK, et al. Expression of recessive alleles by chromosomal mechanisms in retinoblastoma. Nature. 1983;305:779–784. doi: 10.1038/305779a0. [DOI] [PubMed] [Google Scholar]
- 29.Jin G, et al. 2-hydroxyglutarate production, but not dominant negative function, is conferred by glioma-derived NADP-dependent isocitrate dehydrogenase mutations. PLoS One. 2011;6:e16812. doi: 10.1371/journal.pone.0016812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pietrak B, et al. A tale of two subunits: How the neomorphic R132H IDH1 mutation enhances production of αHG. Biochemistry. 2011;50:4804–4812. doi: 10.1021/bi200499m. [DOI] [PubMed] [Google Scholar]
- 31.Bralten LB, et al. IDH1 R132H decreases proliferation of glioma cell lines in vitro and in vivo. Ann Neurol. 2011;69:455–463. doi: 10.1002/ana.22390. [DOI] [PubMed] [Google Scholar]
- 32.Ward PS, et al. The potential for isocitrate dehydrogenase mutations to produce 2-hydroxyglutarate depends on allele specificity and subcellular compartmentalization. J Biol Chem. 2013;288:3804–3815. doi: 10.1074/jbc.M112.435495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Elkhaled A, et al. Magnetic resonance of 2-hydroxyglutarate in IDH1-mutated low-grade gliomas. Sci Transl Med. 2012;4:116ra5. doi: 10.1126/scitranslmed.3002796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jalbert LE, et al. Metabolic profiling of IDH mutation and malignant progression in infiltrating glioma. Sci Rep. 2017;7:44792. doi: 10.1038/srep44792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pusch S, et al. D-2-hydroxyglutarate producing neo-enzymatic activity inversely correlates with frequency of the type of isocitrate dehydrogenase 1 mutations found in glioma. Acta Neuropathol Commun. 2014;2:19. doi: 10.1186/2051-5960-2-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kelly JJ, et al. Oligodendroglioma cell lines containing t(1;19)(q10;p10) Neuro Oncol. 2010;12:745–755. doi: 10.1093/neuonc/noq031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luchman HA, et al. An in vivo patient-derived model of endogenous IDH1-mutant glioma. Neuro Oncol. 2012;14:184–191. doi: 10.1093/neuonc/nor207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kelly JJP, et al. Proliferation of human glioblastoma stem cells occurs independently of exogenous mitogens. Stem Cells. 2009;27:1722–1733. doi: 10.1002/stem.98. [DOI] [PubMed] [Google Scholar]
- 39.Brennan CW, et al. TCGA Research Network The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ceccarelli M, et al. TCGA Research Network Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma. Cell. 2016;164:550–563. doi: 10.1016/j.cell.2015.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hartmann C, Kluwe L, Lücke M, Westphal M. The rate of homozygous CDKN2A/p16 deletions in glioma cell lines and in primary tumors. Int J Oncol. 1999;15:975–982. doi: 10.3892/ijo.15.5.975. [DOI] [PubMed] [Google Scholar]
- 42.Ishii N, et al. Frequent co-alterations of TP53, p16/CDKN2A, p14ARF, PTEN tumor suppressor genes in human glioma cell lines. Brain Pathol. 1999;9:469–479. doi: 10.1111/j.1750-3639.1999.tb00536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.SongTao Q, et al. IDH mutations predict longer survival and response to temozolomide in secondary glioblastoma. Cancer Sci. 2012;103:269–273. doi: 10.1111/j.1349-7006.2011.02134.x. [DOI] [PubMed] [Google Scholar]
- 44.Johannessen TA, et al. Rapid conversion of mutant IDH1 from driver to passenger in a model of human gliomagenesis. Mol Cancer Res. 2016;14:976–983. doi: 10.1158/1541-7786.MCR-16-0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Okoye-Okafor UC, et al. New IDH1 mutant inhibitors for treatment of acute myeloid leukemia. Nat Chem Biol. 2015;11:878–886. doi: 10.1038/nchembio.1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Medeiros BC, et al. Isocitrate dehydrogenase mutations in myeloid malignancies. Leukemia. 2017;31:272–281. doi: 10.1038/leu.2016.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nowak D, Stewart D, Koeffler HP. Differentiation therapy of leukemia: 3 decades of development. Blood. 2009;113:3655–3665. doi: 10.1182/blood-2009-01-198911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Inoue S, et al. Mutant IDH1 down-regulates ATM and alters DNA repair and sensitivity to DNA damage independent of TET2. Cancer Cell. 2016;30:337–348. doi: 10.1016/j.ccell.2016.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sulkowski PL, et al. 2-hydroxyglutarate produced by neomorphic IDH mutations suppresses homologous recombination and induces PARP inhibitor sensitivity. Sci Transl Med. 2017;9:eaal2463. doi: 10.1126/scitranslmed.aal2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.