Significance
The optix gene is well known for its genetic association with wing pattern variation in butterflies; however, its actual function has never been directly confirmed. Using CRISPR genome editing in multiple butterfly species, we show that this gene plays a fundamental and deeply conserved role in the butterfly family Nymphalidae, where it acts as an activator of wing color. We were also surprised to discover that optix simultaneously controls blue iridescence in some species as well, providing an example of how a single gene can act as a switch to coordinate between structural and pigmentary coloration.
Keywords: optix, CRISPR, iridescence, ommochrome, butterfly
Abstract
The optix gene has been implicated in butterfly wing pattern adaptation by genetic association, mapping, and expression studies. The actual developmental function of this gene has remained unclear, however. Here we used CRISPR/Cas9 genome editing to show that optix plays a fundamental role in nymphalid butterfly wing pattern development, where it is required for determination of all chromatic coloration. optix knockouts in four species show complete replacement of color pigments with melanins, with corresponding changes in pigment-related gene expression, resulting in black and gray butterflies. We also show that optix simultaneously acts as a switch gene for blue structural iridescence in some butterflies, demonstrating simple regulatory coordination of structural and pigmentary coloration. Remarkably, these optix knockouts phenocopy the recurring “black and blue” wing pattern archetype that has arisen on many independent occasions in butterflies. Here we demonstrate a simple genetic basis for structural coloration, and show that optix plays a deeply conserved role in butterfly wing pattern development.
Butterfly wing patterns provide an important model system for studying the interplay among ecological, developmental, and genetic factors in the evolution of complex morphological traits. Dozens of genes have been implicated in wing pattern development thanks to a combination of comparative expression and, more recently, knockout studies (1–4). Interestingly, however, mapping and association work has highlighted only a small subset of these genes that seem to play a causative role in wing pattern adaptation in nature: optix, WntA, cortex, and doublesex (5–10). These genes are particularly compelling for two reasons. First, they have all been genetically associated with local adaptation in multiple populations and/or species, and are thus characterized as “adaptive hotspot” genes that repeatedly drive morphological evolution across different lineages (11, 12). Second, based on detailed crossing and expression studies, we infer that these genes behave as complex trait regulators, with different alleles associated with different spatial expression domains that determine highly varied and complex color patterns, not simply the presence or absence of individual features. Although there is strong interest in these genes for these reasons, their specific developmental roles and the depth of conservation of their color patterning functions remain unclear.
Here we present a comparative functional analysis of the optix gene in butterflies. This gene is linked to adaptive geographic variation of red ommochrome color patterns in the genus Heliconius, although its actual function remained unconfirmed before the present study (5, 13). optix is also interesting because it is expressed in association with nonpigmentation wing traits in various species, including morphologically derived wing conjugation scales, suggesting that it may have multiple regulatory roles in both wing scale coloration and structure (5, 14). In the present work, we used Cas9-mediated targeted deletion of optix to test its color patterning function in four species of nymphalid butterflies. Not only did we confirm deeply conserved roles for optix in coordinating pigmentation and scale morphology in all species surveyed, but we were surprised to find that this gene simultaneously regulates blue structural iridescence in some butterflies. Importantly, this coordinated regulation of pigmentation and iridescence strongly phenocopies wing patterns seen in other distantly related species, leading us to hypothesize that optix may have played a role in wing pattern evolution in many different butterfly lineages.
Results
optix Simultaneously Represses Melanins and Promotes Ommochromes.
optix was first identified as a wing pattern gene candidate in Heliconius butterflies, in which mapping, association, and in situ expression data suggest a role in the determination of red color patterns (5, 14, 15). Subsequent mRNA-seq work also showed up-regulation of optix in red color patterns of the painted lady butterfly Vanessa cardui, raising the possibility of a more widespread role for this gene in red color pattern specification (16). To functionally confirm the role of optix in color patterning, we used a Cas9-mediated long-deletion mosaic knockout approach (4, 16, 17) in four nymphalid species: Heliconius erato, Agraulis vanillae, V. cardui, and Junonia coenia (Dataset S1, Tables S1 and S2).
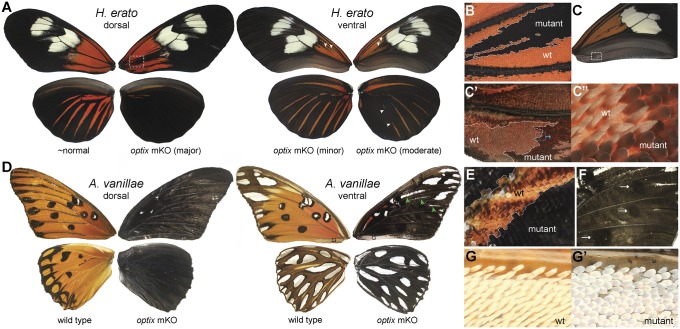
optix knockout in H. erato produced results predicted by previous genetic and in situ hybridization studies. Mosaics revealed loss of the red color patterns previously shown to be presaged by pupal optix expression, including the color field at the base of the forewing (the so-called “dennis” element) and the hindwing rays (Fig. 1 A and B and Dataset S1, Tables S1 and S2). Not only was red pigmentation lost in knockout clones, but red pigments were replaced by black pigments. These results show that optix is required for red color pattern specification in H. erato, and acts as a coordinating “or” switch between ommochrome (orange and red) and melanin (black and gray) pigment types.
Fig. 1.
optix determines wing scale color identity and morphology in H. erato and A. vanillae. (A) optix mosaic knockouts in H. erato result in conversion of red ommochrome color patterns to black melanin. The comparisons shown are left-right asymmetrical knockout effects from single individual injected butterflies. (B) Detail of mutant clone highlighted in the mutant in A showing red replaced by black in a proximal red “dennis” pattern of the dorsal forewing. (C–C′′) optix knockout mosaics showing transformation of pointed wing conjugation scales to normal wing scales. Each panel in the series shows successive detail. (D) optix replaces orange and brown ommochromes in A. vanillae with melanins, resulting in a black and silver butterfly. Arrows highlight presumptive clone boundaries discussed in the text. (E) Detail of a knockout clone boundary highlighting the switch between red and black pigmentation in the ventral forewing from D. (F) Ventral view of black spots in optix knockout mutant showing a phenotype similar to WT. (G and G′) Wing conjugation scales in WT (G) and optix knockout mutant (G′) demonstrating a role for optix in determining A. vanillae scale morphology.
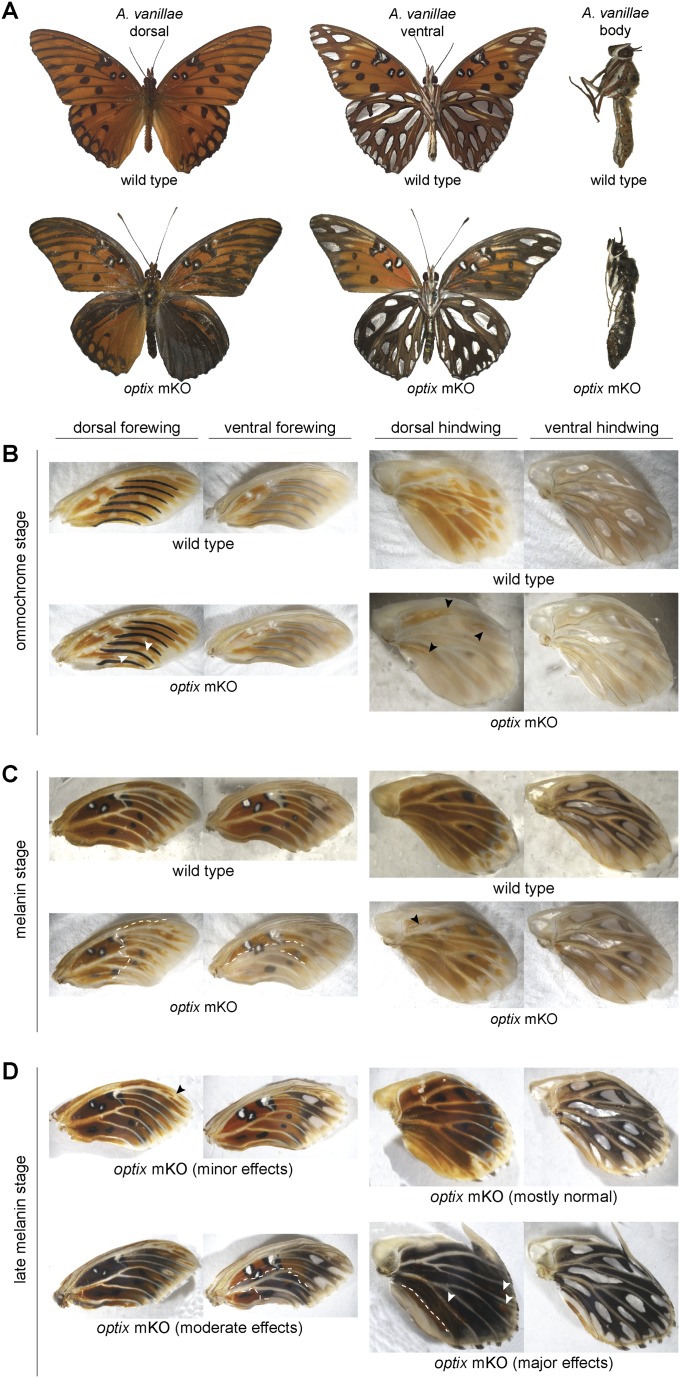
To test whether optix has a role in color patterning in a more basal heliconiine butterfly, we generated knockouts in the gulf fritillary A. vanillae (Fig. 1 D–F, Fig. S1, and Dataset S1, Tables S1 and S2). Previous in situ hybridization work in A. vanillae suggested that optix is not expressed in association with ommochrome patterns during early pupal development, leading to the hypothesis that the gene might not play a major color patterning role in this species (14). Thus, we were surprised to find that optix knockout resulted in a complete transformation of ommochrome scales to black melanin scales, producing a very unusual and dramatic phenotype of a completely black and silver butterfly (Fig. 1D). We also observed a handful of orange or brown scales that changed to silver patches (Fig. 1D, ventral forewing, green arrows), although we cannot confidently conclude that these are cell-autonomous knockout effects since it has been shown that silver scales can be induced through long-range signaling (18, 19), in this case potentially from neighboring knockout clones. The wild-type (WT) black spots and marginal bands in the ventral forewing were unaffected in knockouts and remained a darker color relative to the neighboring mutant melanic scales (Fig. 1F). optix knockout also resulted in melanic hyperpigmentation in adult bodies (Fig. S1A). Thus, our results in A. vanillae are consistent with those in H. erato in supporting a role for optix as a switch-like regulator that toggles between ommochrome and melanin patterns.
Fig. S1.
A. vanillae WT and optix knockout phenotypes in developing wings and body. Illustrative optix mosaic knockout phenotypes are shown at different stages of development: (A) adult, (B) ommochrome stage (when ommochrome pigments first appear), (C) melanin stage (when black melanin pigments first began to appear), and (D) late melanin stage (when black melanin pigments are prevalent across the wing). Arrowheads and dashed lines highlight illustrative mutant clone boundaries.
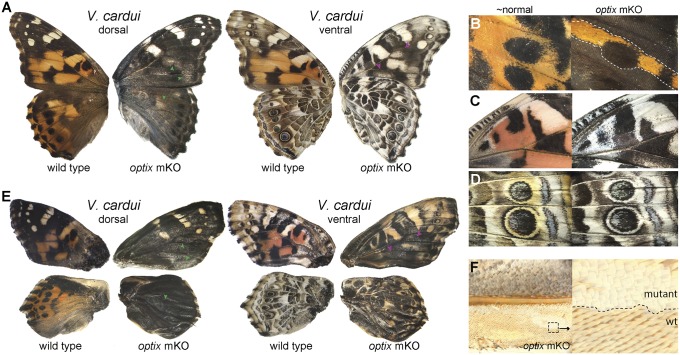
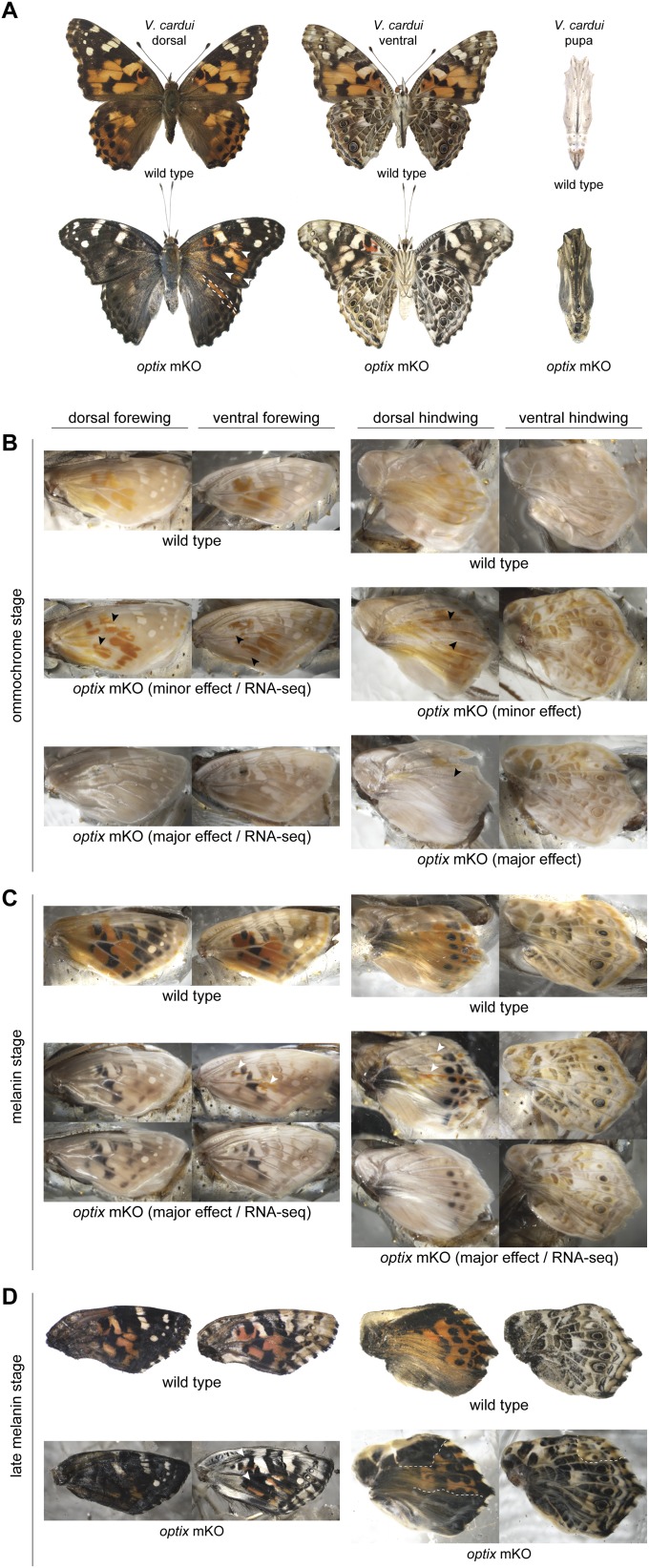
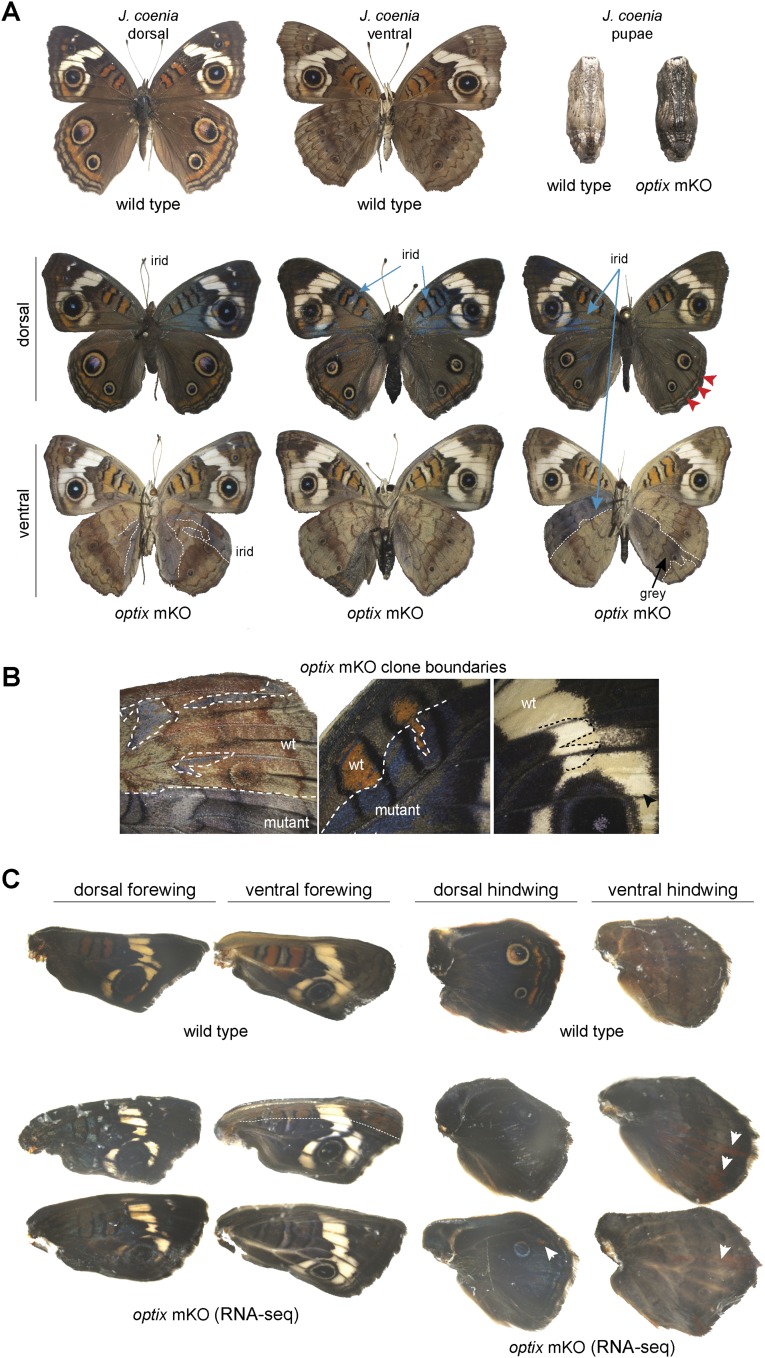
We next aimed to test whether optix regulates wing patterning in more distantly related lineages by performing knockouts in the nymphalines V. cardui (Fig. 2, Fig. S2, and Dataset S1, Tables S1 and S2) and J. coenia (Fig. 3, Fig. S3, and Dataset S1, Tables S1 and S2), which diverged from heliconiines by ∼75–80 mya (20, 21). Our results were consistent with those from H. erato and A. vanillae, where optix knockouts in both species showed mutant clones with complete loss of presumptive ommochrome pigments and replacement by melanins (Figs. 2 A–E and 3 A–C). One interesting exception to this finding was in V. cardui, where the complete ommochrome-to-melanin switch consistently occurred in dorsal wings (Fig. 2 A and B), but much of the ventral wing area showed only a loss of ommochrome and little obvious hypermelanization (Fig. 2 A, C, and D). Importantly, however, we recovered late-stage pupal wings from V. cardui that had died before emergence that displayed hypermelanization of ventral wing surfaces (Fig. 3E). We speculate that this variable strength of ventral wing pattern melanization among individuals may reflect a dosage effect, with the stronger phenotypes representing biallelic optix deletion clones. We have no direct evidence for this, however, given the challenges in rigorously characterizing specific alleles from individual mutant clones (16). We also recovered hypermelanic optix knockout pupae in both V. cardui (Fig. S2) and J. coenia (Fig. S3). Taken together, our knockout data from four nymphalids clearly demonstrate that optix plays a conserved role in coordinating the color identities of butterfly wing scales, where it operates as an “or” function between ommochrome and melanin identities, but also may be modulated to serve as an “and” function in some contexts, as demonstrated by phenotypes seen in the ventral wings of V. cardui.
Fig. 2.
optix determines wing scale color identity and morphology in V. cardui. (A) optix knockout mutant showing loss of ommochrome pigments. (B–D) Left-right asymmetrical comparisons from individual optix mutant butterflies, showing melanization of red patterns (B), loss of color pigmentation without widespread hypermelanization in the ventral forewing (C) and hindwing (D). (E) Severe defects in late-stage pupal wings displaying hypermelanization in red regions of dorsal and ventral wing surfaces (green and purple arrowheads) compared with mosaic adult mutants in A. (F) optix knockout showing conversion of pointed wing conjugation scales to normal scales.
Fig. S2.
V. cardui WT and optix knockout phenotypes in developing wings and pupae. Illustrative optix mosaic knockout phenotypes are shown at different stages of development: (A) adult and pupa, (B) ommochrome stage (when ommochrome pigments first appear), (C) melanin stage (when black melanin pigments first began to appear), and (D) late melanin stage (when black melanin pigments are prevalent across the wing). Arrowheads and dashed lines highlight illustrative mutant clone boundaries. The actual wings used for the RNA-seq experiments shown in Fig. 5 are labeled “RNA-seq”.
Fig. 3.
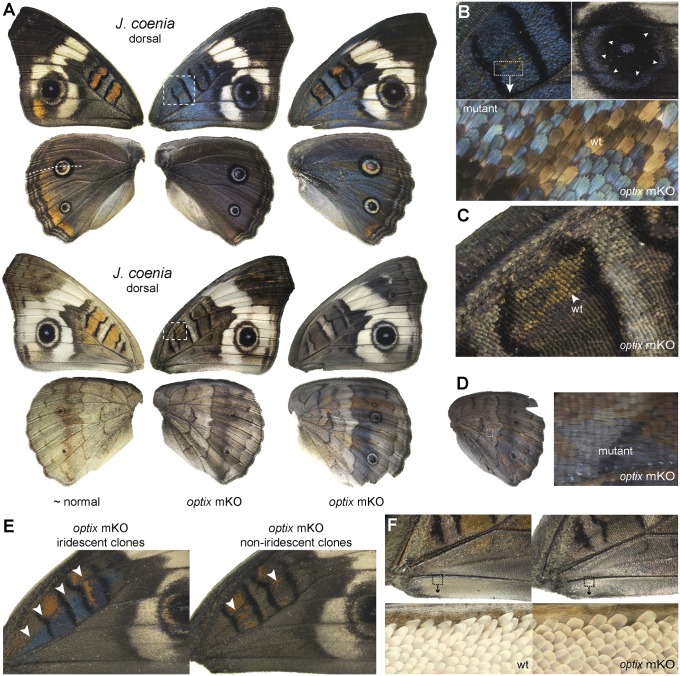
optix coordinates pigment color and structural iridescence in J. coenia. (A) optix knockout results in loss of red ommochrome pigments, melanization, and gain of structural iridescence. The left-right comparisons shown are bilaterally asymmetric mosaic phenotypes from single individuals. Wings to the right are additional examples of mosaic mutants with clearly identifiable knockout clones that highlight iridescence in both dorsal and ventral wing surfaces. (B–D) Details of mosaic defects in the asymmetric mutant shown in A, including strong induced iridescence in dorsal discal spot (DII) and eyespot ring (B), replacement of orange ommochrome with presumptive melanin in ventral discal spot (DII) red patterns (C), and detail of knockout-induced iridescence on the ventral hindwing (D). (E) Mosaic knockout clones showing asymmetrical variation in promoted melanin and iridescence induction in dorsal forewings. (F) Wing conjugation scales in WT and optix knockout mutant.
Fig. S3.
J. coenia WT and optix knockout phenotypes in developing wings and pupae. Illustrative optix mosaic knockout phenotypes are shown at different stages of development: (A) whole adults and pupa, (B) details of clone boundaries on adult wings showing iridescent and noniridescent color shifts, and (C) late melanin stage wings (when black melanin pigments are prevalent across the wing). Arrowheads and dashed lines highlight illustrative mutant clones and boundaries. The actual wings used for RNA-seq experiments in Fig. 5 are labeled “RNA-seq.”
optix Function Is Required for Determination of Derived Scale Structures.
Along with its expression in color patterns, in situ optix expression also precisely predicts the location of patches of derived, pointed scales thought to play a role in conjugating forewings and hindwings during flight (5, 14). To determine whether optix plays a role in determining the unusual morphology of these scales, we examined optix knockouts for changes in wing scale structure. Indeed, we found that in all four species, optix knockout resulted in transformation of wing conjugation scales to normal wing scales (Figs. 1 C and G, 2F, and 3F). Furthermore, in H. erato, A. vanillae, and V. cardui, where wing conjugation scales display color pigmentation, we observed both structural and pigmentation changes in the same scales, suggesting that optix can coregulate both scale morphology and pigmentation simultaneously. One final observation of note relates to the optix-expressing pheroscales that occur along the veins of male A. vanillae (14). These scales did not show any grossly apparent transformation in optix knockouts (data not shown), even though the scales occurred within obvious knockout clones. Therefore, whether optix plays a functional role in the development of pheroscales, as was predicted previously (14), remains an open question. In sum, our observations that optix knockout results in transformation of wing conjugation scales to normal wing scales indicate that optix plays a deeply conserved role in switching between discrete, complex scale morphologies in butterfly wings.
optix Regulates Iridescence in J. coenia.
The most surprising results from the present study came from our work in J. coenia, where knockout of optix induced strong blue iridescence in wing scales (Fig. 3 A–E and Fig. S3). This induction of structural color occurred in addition to the loss of ommochrome pigmentation described above. Broad, strong iridescence occurred in knockouts across dorsal wing surfaces, including in scales that are normally buff or orange in WT butterflies, such as the bright-orange discal spots and eyespot rings (Fig. 3 A and B) and the marginal bands of the dorsal hindwing (Fig. 3A). Iridescence induction was less pronounced on ventral wing surfaces, although it clearly occurred (Fig. 3 A and D). Iridescence was least apparent in areas of the wing that are normally heavily melanized in WT individuals, such as the black borders of the discal spots, distal tip of the forewing, and black rings around the eyespots (Fig. 3 A and B). Although most of our J. coenia mutants showed a strong ommochrome-to-iridescence transition, we also recovered some mosaic individuals with clones showing a partial transformation in which ommochromes were replaced by presumptive melanins, but lacked iridescence (Fig. 3E). These individuals often also showed additional mutant clones with iridescence, thus ruling out a background transregulatory effect. We speculate that these clones may represent lower dosage effects due to clones being monoallelic for deletions; however, we have not confirmed this hypothesis. In sum, our knockout experiments in J. coenia show that optix is a repressor of structural iridescence in this species, and that this regulatory function of optix occurs in addition to its other functions in pigment regulation.
J. coenia optix Mutants Phenocopy Distantly Related Species.
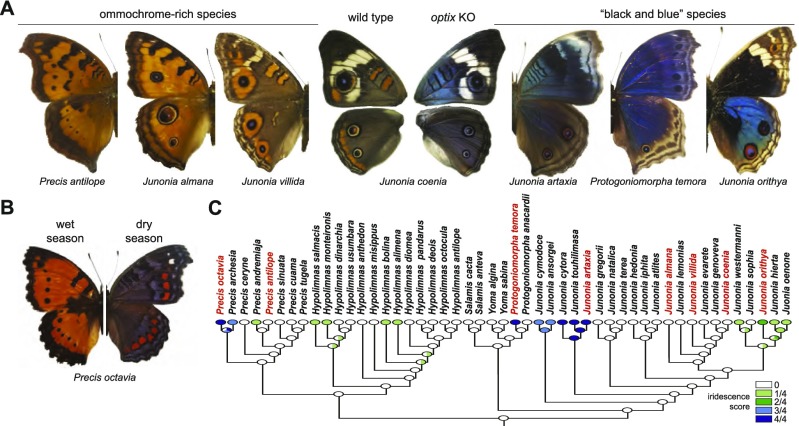
One striking aspect of the J. coenia optix knockout phenotype is the degree to which it phenocopies the archetypal “black and blue” wing patterns that seem to have continually recurred in many distantly related species across all butterfly families. Even simply focusing on the nymphalid tribe Junoniini, which includes J. coenia, phylogenetic analysis suggests multiple origins of predominantly black and blue wing patterns as both fixed phenotypes and plastic seasonal variants (Fig. 4). Notable examples of fixed black and blue phenotypes are seen in such species as Junonia artaxia, which are almost indistinguishable from J. coenia optix knockout phenotypes on casual observation (Fig. 4A). Along with closely phenocopying other species, optix knockouts are also strikingly reminiscent of seasonal phenotypes in such butterflies as Precis octavia (Fig. 4B), in which the wet season form is predominantly red-orange and the dry season form is an archetypal black and blue phenotype with highly reduced red patterns. These seasonal color pattern differences might be explained by local changes in optix expression, although further work is needed to test this hypothesis. In sum, optix knockout phenotypes show many striking parallels with natural interspecific variation, leading us to speculate that differences in optix expression may be responsible for much of the wing pattern diversity seen in nymphalid butterflies.
Fig. 4.
History of iridescence in Junonia and related butterfly genera. (A) J. coenia optix knockouts phenocopy other junoniine species of the black and blue pattern archetype. (B) The iridescent dry season form of P. octavia is largely consistent with the optix knockout effects seen in J. coenia. (C) Parsimony reconstruction of iridescence in Junoniini suggests multiple origins as a fixed phenotype. Species highlighted in red are shown in A. Prevalence of iridescence across dorsal wing surfaces is color coded where the light green → dark-blue continuum represents the approximate proportion of the wing surface that is iridescent (Materials and Methods).
Global Expression Profiling of Butterfly Wings in Response to optix Knockout.
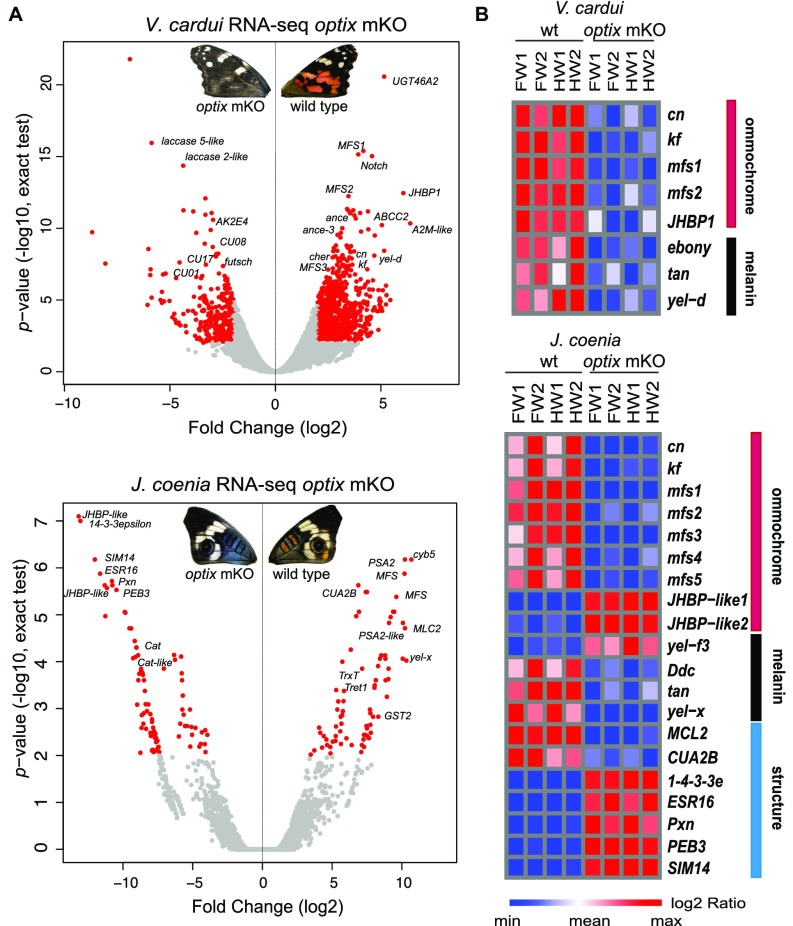
To investigate how wing patterns are controlled by optix, we used RNA-seq to compare transcript abundance in WT and optix knockout wings of V. cardui and J. coenia. We sampled forewings and hindwings separately at a late stage of pupal development when ommochrome and melanin pigments are visible, in two biological replicates of both WT and strong knockout phenotypes (Figs. S2 and S3 and Dataset S1, Table S3). We first examined the expression of optix itself and confirmed a significant depletion of optix transcripts in all knockout wings (Dataset S1, Table S4). A closer analysis of optix transcript reads failed to reveal any partial transcripts showing lesions at the Cas9 cut site, suggesting that mutant transcripts resulting from edited alleles do not persist in wing tissue. We then aimed to identify all highly differentially expressed genes (DEGs) in comparisons between WT and optix knockout wings using cutoff values of a fold change of >4 and a false discovery rate (FDR) of <0.001. In V. cardui, 97 unigenes were up-regulated and 243 were down-regulated in optix knockout wings compared with WT wings. We noted that Gene Ontology (GO) terms related to “structural constituent of cuticle” were significantly enriched in optix knockouts, while “organ morphogenesis” and “transport” were down-regulated (Dataset S1, Table S5). In J. coenia, only 31 unigenes were significantly up-regulated and 37 were down-regulated in optix knockouts. As in V. cardui, optix knockout down-regulated transcripts related primarily to cellular transport. Meanwhile, transcripts related to “muscle thin filament assembly” were enriched in optix knockouts (Dataset S1, Table S5).
To identify pigmentation genes potentially regulated by the optix network, we sorted for transcripts that show differential expression in optix knockout vs. WT wings and are orthologs or paralogs of putative pigmentation genes expressed during pigment maturation and/or spatially associated with red and black color regions in V. cardui (16). Using these criteria, we identified 12 genes associated with ommochrome pigmentation and 3 genes associated with melanin pigmentation in V. cardui (Fig. 5 and Dataset S1, Tables S4 and S6). We found that Drosophila ommochrome pathway genes cinnabar and kynurenine formamidase (kf) showed significant down-regulation in optix mutants. Four unigenes coding for ommochrome-associated transporters were also down-regulated, including three major facilitator superfamily (mfs) transporters and one ATP-binding cassette transporter C family member. Another strongly down-regulated transcript was juvenile hormone binding protein (JHBP1), a gene of unknown function that showed one of the strongest signals of red color association in a previous study (16). Knockouts also showed strong down-regulation of several melanin pathway genes, including tan, ebony, and yellow-d. Of note, all three of these genes are involved in the synthesis of N-β-alanyl dopamine sclerotin, which produces yellowish-tan hues.
Fig. 5.
RNA-seq analysis reveals differential gene expression in response to optix knockout. (A) Volcano plots of individual gene expression levels with log-twofold change (x-axis) against P value (FDR, y-axis, exact test) in V. cardui and J. coenia. DEGs (log-twofold ≥2, P < 0.01) are in red. (B) Expression levels of candidate pigmentation and scale structure genes across replicate WT and optix knockout V. cardui and J. ocenia pupal wings.
Our DEG results in J. coenia overlap with those of V. cardui in many areas, but also include several different transcripts (Fig. 5 and Dataset S1, Tables S4 and S7). In J. coenia, we identified nine ommochrome-associated genes, including cinnabar and kf, and five mfs transporter transcripts that showed down-regulation in optix knockouts. Importantly, the mfs2 transcript appears to be orthologous between V. cardui and J. coenia, suggesting that this may be a conserved ommochrome synthesis gene in butterflies. The melanin-associated genes were somewhat different between the two species; instead of yellow-d, two other yellow gene family members, yellow-x and yellow-f3, were differentially expressed in optix knockout wings. Surprisingly, two transcripts belonging to the JHBP superfamily showed dramatic up-regulation in optix knockout mutants. Of note, these two JHBP transcripts are not orthologs of the ommochrome-associated genes in V. cardui and showed relatively low expression levels [fragments per kilobase of transcript per million mapped fragments (FPKM) <3] during wing development. In sum, these transcriptomic comparisons show that optix directly or indirectly regulates a sizable suite of downstream genes during butterfly wing development, and both positively and negatively regulates distinct batteries of pigmentation genes, consistent with its role in switching between ommochrome and melanin pigment patterns. Furthermore, our data suggest that there may be differences in the downstream targets of the optix network between species, suggesting that downstream interactions in the pathway have diverged.
While a fair amount is known about the genetic basis of insect pigmentation, virtually nothing is known about the types of genes that may control structural coloration. Thus, we wanted to use our experimental system to identify potential candidate genes that may underlie butterfly iridescence. This task is more challenging than identifying pigmentation genes, because there are few precedents for how to informatically highlight DEGs that may have a role in determining scale microstructure. Recent work identified actin filaments as playing a role in determining iridescence-producing scale cuticular structures in butterflies (22); thus, we scanned our DEG set for genes likely involved in filament or cuticle generation and regulation (Dataset S1, Table S7). In iridescent optix knockouts, we found significant down-regulation of two F-actin filament organization-related genes, myosin light chain 2 and thioredoxin, and a cuticle-related gene, larval cuticle protein A2B. We also noted strong up-regulation in knockouts of 1–4-3–3 epsilon, which has been shown to be involved in Ras/MAP kinase pathway and Drosophila eye development (23). Dataset S1, Table S7 highlights other candidates as well. A number of these genes are interesting candidates for effectors of iridescence; however, they should be considered preliminary candidates until further functional work confirms their roles. Nonetheless, we now have an experimental system in which we can modulate iridescence by knocking out a single gene, making uncovering the gene regulatory networks underlying structural coloration a more tractable problem.
Discussion
Here we present functional evidence that the widely studied adaptive color pattern locus optix plays a fundamental and deeply conserved role in regulating both pigmentary and structural coloration in butterfly wings. Even with numerous studies examining the expression and population genetic dynamics of optix, this gene’s actual function has remained a matter of speculation until now. Using Cas9-mediated mosaic knockouts, we have shown that this gene acts as a switch-like master regulator of butterfly color. In every species that we investigated, optix knockouts produced a striking phenotype in which all color pigmentation was lost and replaced with black and/or gray melanins. Furthermore, in buckeye butterflies, another distinct class of knockout phenotype occurred with clones showing intense blue iridescence, demonstrating that optix also can act as a repressor of structural coloration in some species. All of our findings are consistent with a model in which optix operates as an “or” switch to toggle between discrete pigmentary and structural color fates for scale cells. We were surprised to find that a complex and multifaceted trait like color identity, which is interwoven with so many different genetic, biochemical, and morphological features, has such a simple and discrete regulatory underpinning.
We see two conceptual consequences emerging from our findings. First, optix demonstrates how a single master regulator can be selectively redeployed to radically alter a range of independent traits. Compared with other wing patterning genes, which are largely implicated in the determination of specific pattern elements [e.g., spalt for eyespots (4), WntA for stripes (18, 19)], optix function is not limited to specific color pattern elements. In the various species that we examined, optix could determine the color identity of any number of pattern elements, the entire wing, or even the pupa or body. Therefore, optix behaves like a paintbrush that can be applied anywhere on a butterfly to modulate color, and the “hands” guiding optix can be any number of upstream patterning agents. In one species, these agents might allow optix to disperse orange across large portions of the wing, while in another they may decorate eyespots with fine chromatic filigree. optix exerts its control over phenotypes by toggling between discrete states by activating alternative downstream gene effector modules, as illustrated by our RNA-seq work. Thus, this gene represents a striking case of a regulator that can be deployed across numerous morphological features to effect discrete phenotypic shifts through switch-like coordination of multiple gene regulatory networks.
Second, optix provides an example of how a gene underlying adaptive microevolution also has a deeply conserved regulatory role in morphological development. In this respect, it is interesting that optix knockouts phenocopy the discrete red ↔ black ↔ iridescent evolutionary color state changes commonly observed in many wing pattern elements, including the whole-wing black and blue pattern archetype that has arisen on many occasions in butterflies. These phenocopies lead us to hypothesize that evolutionary changes in optix expression may have played a repeated role in nymphalid wing pattern evolution, although additional functional work is needed to rigorously assess this hypothesis.
Because optix plays a repeated role in wing pattern adaption in different species (24, 25), it is presented as an example of an adaptive hotspot similar to other genes, such as shavenbaby (26) and Pitx1 (27). There is ongoing discussion about what characteristics lead genes like these to have a disproportionate, recurring role in morphological evolution (12). One model is that some of these loci behave as “input-output genes” that have a modular regulatory architecture that predisposes them to be able to fine tune the relationships between many transregulatory “inputs” and downstream gene regulatory “outputs” (28). What we know about optix is consistent with this model. It resides in a large (∼200 kb) gene desert suggested to contain numerous regulatory elements on chromatin immunoprecipitation analysis (29), and different color pattern elements show associations with different intervals in this region, suggesting that a modular “input” architecture controls wing pattern variation (15, 30). Our present study provides a functional demonstration of the phenotypic “output” of optix expression. We speculate that the simple switch-like function of this gene to determine color identity may favor its recurring role in wing pattern evolution, especially when coupled with a modular regulatory architecture that facilitates fine tuning of spatial expression.
The ability of a single gene to independently determine the placement of color anywhere on a butterfly also leads us to consider the origin of butterfly color itself. Each major family of butterflies produces color by deploying a different chemical class of pigments: papiliochromes in papilionids, pterins in pierids, and ommochromes in nymphalids (31). Given that each of these pigment types requires a very different set of transporters and enzymes, we speculate that wing color may have had multiple independent origins in butterflies. We propose that optix likely played a causative role in the origin of wing color in the Nymphalidae, the most speciose and morphologically diverse butterfly family. This gene’s simple and deeply conserved regulatory function of replacing melanin with ommochrome pigmentation leads to a simple model where co-option of optix into a color-patterning role could have been the key event sufficient for the deployment of ommochromes in wing scales. Based on in situ gene expression comparisons, it has been proposed that the ancestral role for optix in lepidopteran wing development is to specify wing conjugation scales (14); therefore, the regulatory ability of optix to be expressed in wing tissues predates nymphalid color patterns and can be dated back to moths. It is now an open question of how optix came to regulate pigmentation—whether the regulatory associations with genes identified in the comparative transcription work were forged de novo in the wings or were carried with optix from some ancestral role elsewhere in the insect, perhaps the ommochrome-bearing eyes (32). Whatever the case, with optix we now have a case study of a switch-like regulator gene that can be deployed anywhere in an organism to toggle between multiple discrete color states, and that has also played a role in color pattern evolution in multiple species (24, 25). The stage is now set for asking a deeper set of questions about how an adaptive hotspot gene can gain novel functions over time, and what kind historical and mechanistic phenomena might drive it to play a repeated role in morphological evolution.
Materials and Methods
CRISPR/Cas9 Genome Editing.
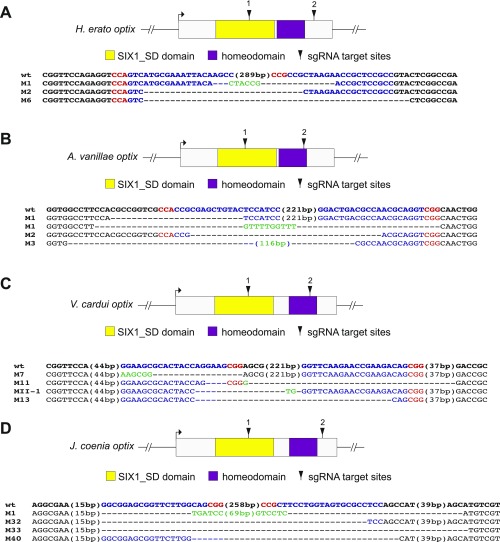
We opted to generate long deletions using dual sgRNAs following the protocol of Zhang and Reed (4, 16, 17, 33). sgRNA target sequences were identified by searching for GGN18NGG or N20NGG patterns targeting the optix exon and then tested for uniqueness by BLAST against the genome or transcriptome reference (Dataset S1, Table S1). Target regions were amplified by genotyping primers flanking the target regions, gel-purified, subcloned into a TOPO TA vector (Invitrogen), and sequenced (Fig. S4 and Dataset S1, Table S1).
Fig. S4.
sgRNA design and sequence genotyping of optix knockout mutants in H. erato (A), A. vanillae (B), V. cardui (C), and J. coenia (D). Locations of sgRNAs are shown relative to the predicted functional domain single-exon optix coding region. Sequences of optix alleles from the knockout mutants shown in the main figures confirm lesions at target sites. Purple indicates sgRNA targets; red, PAM sequences; green, novel sequences not observed in WT alleles.
Phylogenetic Analysis.
The latest available phylogeny of Junoniini (34) was used to estimate the gains and losses in butterfly wing iridescence. Ancestral states were mapped using maximum parsimony in Mesquite (35). Specimens from the Cornell University Insect Collection and specimen photos from Encyclopedia of Life were used to score iridescence levels in Junoniini butterflies as a character. We first divided butterfly wings into four regions based on a nymphalid grand plan model: basal, central, border symmetry system, and discal spot (also called DI and DII patterns). We further classified the iridescence trait into five distinct levels—0, 1/4, 1/2, 3/4, and 1—depending on the extent of iridescence occurrence in those four regions (36). For example, J. orithya was counted as 1/2 because iridescence is well represented in two of the four defined domains (i.e., central and border symmetry regions).
Pupal Wing Isolation and mRNA Extraction.
V. cardui and J. coenia forewings and hindwings were rapidly dissected and stored in RNAlater (Life Technologies) at −80 °C. Wings from melanin stages were then selected for RNA sequencing. RNA isolation was performed using the Ambion Purelink RNA Mini Kit (Life Technology). Two biological replicates were sampled from both forewing and hindwing in both WT and optix knockout mutants, resulting in eight samples for each species. Asymmetrical major and minor mosaic effect forewings from a V. cardui optix knockout individual were further collected as two samples to take advantage of asymmetrical mosaic information. In summary, 10 RNA-seq samples were collected in V. cardui, and 8 RNA-seq samples were collected in J. coenia. Library construction and sequencing were conducted as described previously (16).
Analysis of Transcript Expression Data.
The V. cardui transcriptome assembly (16) was downloaded from www.butterflygenome.org and served as a reference. To build a reference for J. coenia RNA-seq analysis, sequencing reads from this study and National Center for Biotechnology Information’s Gene Expression Omnibus (GEO) database (accession no. GSE54819) were merged. Assembly was built using Trinity (37) after in silico normalization. The TransDecoder-predicted geneset was then searched against Swissprot, Pfam, and GO databases for gene, domain, and GO annotation, respectively. Sequencing data were subjected to quality control by removing PCR primers, adapters, and low-quality reads. Clean reads were further aligned with reference genes with Bowtie2 (38). Gene expression levels were calculated using FPKM. Differential gene expression was calculated for comparisons between WT and optix knockout mutants based on edge R using cutoffs of a fold change of ≥2 and an FDR of ≤0.001. Only DEGs shared between forewing and hindwing in WT vs. mutant comparisons were kept. In V. cardui, DEGs were further filtered with their expression levels of fold change ≥1.2 between large and small asymmetrical mosaicism in the same individual. V. cardui and J. coenia wing RNA-seq raw sequencing data, full transcriptome assembly, and expression profiles are available in the GEO database (accession no. GSE98678).
Supplementary Material
Acknowledgments
We thank Jacob Berv for assistance with phylogenetic analysis, Jason Dombroskie for assistance with collection work and imaging, Benjamin Brack for assistance with raising butterflies, and Arnaud Martin and members of the R.D.R. laboratory for discussion and input. This work was supported by National Science Foundation Grants IOS-1354318 and IOS-1557443 (to R.D.R.), and DGE-1650441 (to A.M.-V.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE98678).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1709058114/-/DCSupplemental.
References
- 1.Oliver JC, Tong X-L, Gall LF, Piel WH, Monteiro A. A single origin for nymphalid butterfly eyespots followed by widespread loss of associated gene expression. PLoS Genet. 2012;8:e1002893. doi: 10.1371/journal.pgen.1002893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hines HM, et al. Transcriptome analysis reveals novel patterning and pigmentation genes underlying Heliconius butterfly wing pattern variation. BMC Genomics. 2012;13:288. doi: 10.1186/1471-2164-13-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Connahs H, Rhen T, Simmons RB. Transcriptome analysis of the painted lady butterfly, Vanessa cardui during wing color pattern development. BMC Genomics. 2016;17:270. doi: 10.1186/s12864-016-2586-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang L, Reed RD. Genome editing in butterflies reveals that spalt promotes and Distal-less represses eyespot colour patterns. Nat Commun. 2016;7:11769. doi: 10.1038/ncomms11769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reed RD, et al. optix drives the repeated convergent evolution of butterfly wing pattern mimicry. Science. 2011;333:1137–1141. doi: 10.1126/science.1208227. [DOI] [PubMed] [Google Scholar]
- 6.Martin A, et al. Diversification of complex butterfly wing patterns by repeated regulatory evolution of a Wnt ligand. Proc Natl Acad Sci USA. 2012;109:12632–12637. doi: 10.1073/pnas.1204800109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nadeau NJ, et al. The gene cortex controls mimicry and crypsis in butterflies and moths. Nature. 2016;534:106–110. doi: 10.1038/nature17961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van’t Hof AE, et al. The industrial melanism mutation in British peppered moths is a transposable element. Nature. 2016;534:102–105. doi: 10.1038/nature17951. [DOI] [PubMed] [Google Scholar]
- 9.Kunte K, et al. doublesex is a mimicry supergene. Nature. 2014;507:229–232. doi: 10.1038/nature13112. [DOI] [PubMed] [Google Scholar]
- 10.Nishikawa H, et al. A genetic mechanism for female-limited Batesian mimicry in Papilio butterfly. Nat Genet. 2015;47:405–409. doi: 10.1038/ng.3241. [DOI] [PubMed] [Google Scholar]
- 11.Papa R, Martin A, Reed RD. Genomic hotspots of adaptation in butterfly wing pattern evolution. Curr Opin Genet Dev. 2008;18:559–564. doi: 10.1016/j.gde.2008.11.007. [DOI] [PubMed] [Google Scholar]
- 12.Martin A, Orgogozo V. The loci of repeated evolution: A catalog of genetic hotspots of phenotypic variation. Evolution. 2013;67:1235–1250. doi: 10.1111/evo.12081. [DOI] [PubMed] [Google Scholar]
- 13.Reed RD, McMillan WO, Nagy LM. Gene expression underlying adaptive variation in Heliconius wing patterns: Nonmodular regulation of overlapping cinnabar and vermilion prepatterns. Proc Biol Sci. 2008;275:37–45. doi: 10.1098/rspb.2007.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martin A, et al. Multiple recent co-options of Optix associated with novel traits in adaptive butterfly wing radiations. Evodevo. 2014;5:7. doi: 10.1186/2041-9139-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van Belleghem SM, et al. Complex modular architecture around a simple toolkit of wing pattern genes. Nat Ecol Evol. 2017;1:0052. doi: 10.1038/s41559-016-0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang L, et al. Genetic basis of melanin pigmentation in butterfly wings. Genetics. 2017;205:1537–1550. doi: 10.1534/genetics.116.196451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang L, Reed R. A practical guide to CRISPR/Cas9 genome editing in Lepidoptera. In: Sekimura T, Nijhout HF, editors. Diversity and Evolution of Butterfly Wing Patterns: An Integrative Approach. Springer; Singapore: 2017. [Google Scholar]
- 18.Martin A, Reed RD. Wnt signaling underlies evolution and development of the butterfly wing pattern symmetry systems. Dev Biol. 2014;395:367–378. doi: 10.1016/j.ydbio.2014.08.031. [DOI] [PubMed] [Google Scholar]
- 19.Mazo-Vargas A, et al. Macroevolutionary shifts of WntA function potentiate butterfly wing-pattern diversity. Proc Natl Acad Sci USA. 2017;114:10701–10706. doi: 10.1073/pnas.1708149114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wahlberg N, et al. Nymphalid butterflies diversify following near demise at the Cretaceous/Tertiary boundary. Proc Biol Sci. 2009;276:4295–4302. doi: 10.1098/rspb.2009.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peña C, Espeland M. Diversity dynamics in Nymphalidae butterflies: Effect of phylogenetic uncertainty on diversification rate shift estimates. PLoS One. 2015;10:e0120928. doi: 10.1371/journal.pone.0120928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dinwiddie A, et al. Dynamics of F-actin prefigure the structure of butterfly wing scales. Dev Biol. 2014;392:404–418. doi: 10.1016/j.ydbio.2014.06.005. [DOI] [PubMed] [Google Scholar]
- 23.Chang HC, Rubin GM. 14-3-3 epsilon positively regulates Ras-mediated signaling in Drosophila. Genes Dev. 1997;11:1132–1139. doi: 10.1101/gad.11.9.1132. [DOI] [PubMed] [Google Scholar]
- 24.Kronforst MR, Papa R. The functional basis of wing patterning in Heliconius butterflies: The molecules behind mimicry. Genetics. 2015;200:1–19. doi: 10.1534/genetics.114.172387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jiggins CD, Wallbank RW, Hanly JJ. Waiting in the wings: What can we learn about gene co-option from the diversification of butterfly wing patterns? Philos Trans R Soc Lond B Biol Sci. 2017;372:20150485. doi: 10.1098/rstb.2015.0485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sucena E, Delon I, Jones I, Payre F, Stern DL. Regulatory evolution of shavenbaby/ovo underlies multiple cases of morphological parallelism. Nature. 2003;424:935–938. doi: 10.1038/nature01768. [DOI] [PubMed] [Google Scholar]
- 27.Chan YF, et al. Adaptive evolution of pelvic reduction in sticklebacks by recurrent deletion of a Pitx1 enhancer. Science. 2010;327:302–305. doi: 10.1126/science.1182213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stern DL, Orgogozo V. Is genetic evolution predictable? Science. 2009;323:746–751. doi: 10.1126/science.1158997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lewis JJ, van der Burg KR, Mazo-Vargas A, Reed RD. ChIP-Seq-annotated Heliconius erato genome highlights patterns of cis-regulatory evolution in Lepidoptera. Cell Reports. 2016;16:2855–2863. doi: 10.1016/j.celrep.2016.08.042. [DOI] [PubMed] [Google Scholar]
- 30.Wallbank RW, et al. Evolutionary novelty in a butterfly wing pattern through enhancer shuffling. PLoS Biol. 2016;14:e1002353. doi: 10.1371/journal.pbio.1002353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nijhout HF. 1991. The Development and Evolution of Butterfly Wing Patterns. Smithsonian Series in Comparative Evolutionary Biology (Smithsonian Institution Scholarly Press, Washington, DC)
- 32.Monteiro A. Gene regulatory networks reused to build novel traits: Co-option of an eye-related gene regulatory network in eye-like organs and red wing patches on insect wings is suggested by optix expression. BioEssays. 2012;34:181–186. doi: 10.1002/bies.201100160. [DOI] [PubMed] [Google Scholar]
- 33.Bassett AR, Tibbit C, Ponting CP, Liu J-L. Highly efficient targeted mutagenesis of Drosophila with the CRISPR/Cas9 system. Cell Reports. 2013;4:220–228. doi: 10.1016/j.celrep.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kodandaramaiah U. Eyespot evolution: Phylogenetic insights from Junonia and related butterfly genera (Nymphalidae: Junoniini) Evol Dev. 2009;11:489–497. doi: 10.1111/j.1525-142X.2009.00357.x. [DOI] [PubMed] [Google Scholar]
- 35.Maddison WP, Maddison DR. 2017 Mesquite: A modular system for evolutionary analysis. Version 3.2. Available at mesquiteproject.org. Accessed May 2017.
- 36.Nijhout H, Wray G. Homologies in the colour patterns of the genus Charaxes (Lepidoptera: Nymphalidae) Biol J Linn Soc Lond. 1986;28:387–410. [Google Scholar]
- 37.Haas BJ, et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 2013;8:1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.