Significance
Synaptotagmins-1 and -7, act as Ca2+ sensors for the fast and slow components of neurotransmitter release, respectively, through the two C2 domains that form their cytoplasmic region. Surprisingly, Ca2+-binding to the synaptotagmin-7 C2A domain is more critical for slow release than Ca2+-binding to the C2B domain, whereas the opposite was found for the synaptotagmin-1 C2 domains and fast release. This paper suggests an explanation for this apparent contradiction, showing that the C2A domain dominates binding of synaptotagmin-7 to membranes, whereas such binding is dominated by the C2B domain in the case of synaptotagmin-1. Thus, membrane affinity may be a key determinant of the relative functional importance of synaptotagmin C2 domains.
Keywords: synaptotagmin-7, synaptotagmin-1, membrane binding, X-ray crystallography, neurotransmitter release
Abstract
Synaptotagmins (Syts) act as Ca2+ sensors in neurotransmitter release by virtue of Ca2+-binding to their two C2 domains, but their mechanisms of action remain unclear. Puzzlingly, Ca2+-binding to the C2B domain appears to dominate Syt1 function in synchronous release, whereas Ca2+-binding to the C2A domain mediates Syt7 function in asynchronous release. Here we show that crystal structures of the Syt7 C2A domain and C2AB region, and analyses of intrinsic Ca2+-binding to the Syt7 C2 domains using isothermal titration calorimetry, did not reveal major differences that could explain functional differentiation between Syt7 and Syt1. However, using liposome titrations under Ca2+ saturating conditions, we show that the Syt7 C2A domain has a very high membrane affinity and dominates phospholipid binding to Syt7 in the presence or absence of l-α-phosphatidylinositol 4,5-diphosphate (PIP2). For Syt1, the two Ca2+-saturated C2 domains have similar affinities for membranes lacking PIP2, but the C2B domain dominates binding to PIP2-containing membranes. Mutagenesis revealed that the dramatic differences in membrane affinity between the Syt1 and Syt7 C2A domains arise in part from apparently conservative residue substitutions, showing how striking biochemical and functional differences can result from the cumulative effects of subtle residue substitutions. Viewed together, our results suggest that membrane affinity may be a key determinant of the functions of Syt C2 domains in neurotransmitter release.
Neurons communicate with each other via the release of neurotransmitters by synaptic vesicle exocytosis. Neurotransmitter release is triggered by entry of Ca2+ into a presynaptic terminal, exhibiting a fast synchronous component that occurs within less than 1 ms after Ca2+ influx and a slower, asynchronous component (1). The sophisticated machinery that controls release includes core proteins, such as Munc18-1, Munc13s, and the SNAP receptors (SNAREs) syntaxin-1, SNAP-25, and synaptobrevin, which form a tight SNARE complex that brings the vesicle and plasma membranes together and is key for membrane fusion (2–4). In addition, the exquisite regulation of synaptic exocytosis depends on specialized factors, including the Ca2+ sensors from the synaptotagmin (Syt) family (5). Syt1 and the closely related Syt2 and Syt9 act as the Ca2+ sensors for the major, synchronous components of neurotransmitter release (6–8), whereas Syt7 mediates at least in part the asynchronous component (9). While major advances have been made in characterizing how all these proteins function, and reconstitution experiments have suggested that basic steps of synaptic vesicle fusion can be recapitulated with eight central proteins (10), fundamental questions remain about the overall mechanism of Ca2+-dependent membrane fusion and in particular about the specific actions of Syts.
Syts contain tandem C2 domains (referred to as C2A and C2B domains) that span most of their cytoplasmic region and adopt characteristic β-sandwich structures (11–13). The Syt1 C2A and C2B domains bind three and two Ca2+ ions, respectively, through loops at the top of the these β-sandwiches (13–15), which does not cause substantial conformational changes (12) but drastically changes the electrostatic potential of the Ca2+-binding region (13, 15, 16). These top loops also mediate Ca2+-dependent binding to negatively charged phospholipid membranes (17, 18). The key functional importance of this activity was demonstrated by the finding that mutations that increase or decrease the apparent Ca2+ affinity of Syt1 in phospholipid binding lead to parallel changes in the Ca2+ sensitivity of release (6, 19). The Syt1 C2B domain can bind simultaneously to two membranes in a Ca2+-dependent manner via the Ca2+-binding loops at the top and two arginines (R398 and R399) at the bottom of the β-sandwich, which may cooperate with SNARE complex formation in bringing the membranes together and inducing membrane fusion upon Ca2+ influx (20–22). In support of this model, NMR studies of Syt1–SNARE complex interactions in solution revealed a dynamic binding mode that allows simultaneous interactions with the SNAREs and two closely apposed membranes (23). However, SNARE binding in this structure is mediated by a polybasic region of the C2B domain that also binds to l-α-phosphatidylinositol 4,5-diphosphate (PIP2) (24, 25), and multiple Syt1–SNARE complex binding modes likely exist (23). Indeed, a crystal structure revealed three different Syt1–SNARE complex binding modes, all distinct from that observed in solution (26). Hence, it is still unclear how the functions of Syt1 and the SNARE complex are coupled.
The structural and biochemical properties uncovered by the extensive studies of the Syt1 C2 domains are paradigmatic in general terms for C2 domains of other Ca2+-dependent Syts, but differences in these properties can range from subtle to dramatic (5). Such differences can be difficult to predict from sequence alignments alone (e.g., ref. 27) and can result in fine or drastic functional differences that may also be difficult to rationalize from sequence analyses. Particularly intriguing in this respect are the differences between Syt1 and Syt7, as both isoforms function as Ca2+ sensors for exocytosis in chromaffin cells (28) and presynaptic terminals (6, 9), but Syt7 cannot functionally replace Syt1 to trigger fast synchronous neurotransmitter release (8). In fact, replacing the C2B domain in Syt1 with the Syt7 C2B domain prevents rescue of synchronous release in Syt1 KO neurons even after extensive mutagenesis to replace residues of the Syt7 C2B domain back with those present in Syt1 (29). The crystal structure of the Syt7 C2B domain did not reveal substantial differences from that of the Syt1 C2B domain that could clarify the basis for these results (29). Moreover, mutating the Ca2+-binding sites of the Syt1 C2B domain impairs synchronous release much more drastically than analogous mutations in the Ca2+-binding sites of the Syt1 C2A domain (30–33), whereas asynchronous release is more strongly impaired by mutations in the Ca2+-binding sites of the Syt7 C2A domain than mutations in the Syt7 C2B domain Ca2+-binding sites (9). In principle, these results suggest that the relative functional importance of the two C2 domains is switched in Syt1 and Syt7, but this conclusion represents a simplification because substitutions in the C2B domain Ca2+-binding sites render Syt1 dominant-negative (30, 34). Strikingly, expressing such mutant Syt1 in WT cells blocks both synchronous and asynchronous Ca2+-triggered release, suggesting that C2B-domain mutant Syt1 locks synaptic vesicles into a Ca2+-unresponsive state. Overall, these findings show that understanding the key determinants for functional differentiation between Syt1 and Syt7 remains as a major challenge in this field.
In the study presented herein, we have investigated the structural and biochemical basis for the differences in the relative functional importance of Ca2+-binding to the Syt7 C2 domains. We have determined crystal structures of the Syt7 C2A domain and of a fragment spanning the two Syt7 C2 domains (C2AB), and found no structural features that could readily explain their distinct functional properties. Isothermal titration calorimetry (ITC) data reveal that the intrinsic Ca2+ affinities of the Syt7 C2 domains are similar to those of the Syt1 C2 domains. However, in contrast to Syt1, we find that under Ca2+ saturating conditions the Syt7 C2A domain binds to phospholipids with much higher affinity than the Syt7 C2B domain, and that disruption of the C2A domain Ca2+-binding sites impairs binding of the Syt7 C2AB fragment to phospholipids more strongly than disruption of the C2B domain Ca2+-binding sites. Moreover, the Ca2+-saturated Syt7 C2A domain clusters liposomes and hence has the ability to bring two membranes together, whereas the Syt7 C2B domain does not, again in contrast to results obtained with Syt1. These results suggest that the dominant function of Ca2+ binding to the Syt7 C2A domain versus the C2B domain in asynchronous release arises because of its preponderant role in membrane binding and perhaps in bringing the vesicle and plasma membranes together. Furthermore, mutagenesis shows that cooperative effects of apparently conservative amino acid substitutions underlie at least in part the much higher phospholipid binding affinity of the Syt7 C2A domain compared with the Syt1 C2A domain, illustrating how the cumulative effects of subtle mutations can have profound effects in protein biochemistry and function.
Results
Crystal Structure of the Ca2+-Bound Syt7 C2A Domain.
The crystal structure of the Ca2+-bound Syt7 C2B domain has been determined and found to be very similar to that of the Syt1 C2B domain, although three Ca2+-binding sites were observed in the Syt7 C2B domain instead of two (29). A solution structure of the Ca2+-free Syt7 C2A domain determined by NMR spectroscopy was deposited in the Protein Data Bank as part of a structural genomics initiative (PDB ID code 2D8K). To investigate whether Ca2+ induces conformational changes in the Syt7 C2A domain, we performed crystallization screens in the presence of Ca2+ and were able to obtain crystals that diffracted to 1.7 Å. The structure of the Ca2+-bound Syt7 C2A domain was determined by molecular replacement using the Ca2+-free solution structure as the search model. Data collection and refinement statistics are described in Table S1.
Table S1.
Data collection and refinement statistics
| Data collection | ||
| Crystal | Syt7 C2A | Syt7 C2AB |
| Space group | P6(5) | P2(1) |
| Cell constants, Å, ° | 55.30, 89.81 | 60.64, 75.06, 70.98, 90.00, 114.51, 90.00 |
| Wavelength, Å | 0.97915 | 0.97918 |
| Resolution range, Å | 27.65–1.70 (1.73–1.70) | 44.45–2.25 (2.33–2.25) |
| Unique reflections | 17,195 (866) | 25,080 (774) |
| Multiplicity | 6.6 (4.1) | 3.7 (2.4) |
| Data completeness, % | 99.7 (99.5) | 91.6 (56.3) |
| Rmerge,* % | 5.4 (98.5) | 7.9 (76.6) |
| Rpim,† % | 2.2 (52.8) | 4.6 (54.1) |
| CC1/2 (last resolution shell) | 0.542 | 0.698 |
| I/σ(I) | 34.6 (1.42) | 15.4 (1.28) |
| Wilson B-value, Å2 | 44.6 | |
| Wilson B-value, Å2, sharpened‡ | 11.4 | 33.3 |
| Mean |L|§ | 0.483 (0.500/0.375) | 0.537 (0.500/0.375) |
| Mean L2 | 0.314 (0.333/0.200) | 0.375 (0.333/0.200) |
| Refinement statistics | ||
| Resolution range, Å | 27.65–1.69 (1.75–1.69) | 44.45–2.25 (2.33–2.25) |
| No. of reflections Rwork/Rfree | 14,830/1,638 (897/96) | 20,026/2,011 (737/67) |
| Data completeness, % | 95.7 (51.0) | 80.2 (26.0) |
| Atoms (non-H Protein/Ca2+/solvent) | 1,169/3/188 | 4,341/8/103 |
| Rwork, % | 14.4 (22.9) | 21.4 (30.2) |
| Rfree, % | 18.1 (34.3) | 25.6 (31.3) |
| Rmsd bond length, Å | 0.017 | 0.014 |
| Rmsd bond angle, ° | 0.675 | 0.489 |
| Mean B-value, Å2 (protein/Ca2+/solvent) | 16.2/18.9/29.9 | Chain A, 50.1/chain B, 51.8/50.5/52.2 |
| Ramachandran plot, % (favored/additional/disallowed)¶ | 97.8/2.2/0 | 94.0/6.0/0 |
| Clashscore/overall score¶ | 0.00/0.53 | 0.68/1.12 |
| Maximum-likelihood coordinate error | 0.16 | 0.29 |
| Missing residues | −19 to −9 | −19 to −1, 134, 260–266 |
Data for the outermost shell are given in parentheses.
Rmerge = 100 ΣhΣi|Ih,i −〈Ih〉|/ΣhΣi〈Ih,i〉, where the outer sum (h) is over the unique reflections and the inner sum (i) is over the set of independent observations of each unique reflection.
Rpim = 100 ΣhΣi [1/(nh − 1)]1/2|Ih,i −〈Ih〉|/ΣhΣi〈Ih,i〉, where nh is the number of observations of reflections h.
B-factor sharpening was performed in the autocorrection mode of HKL-3000 (57).
L values are calculated using PHENIX (60) for acentric reflections in the resolution range 10–3.5 Å. Values in parentheses are the expected values for untwinned and perfectly twinned data, respectively.
As defined by the validation suite MolProbity (64).
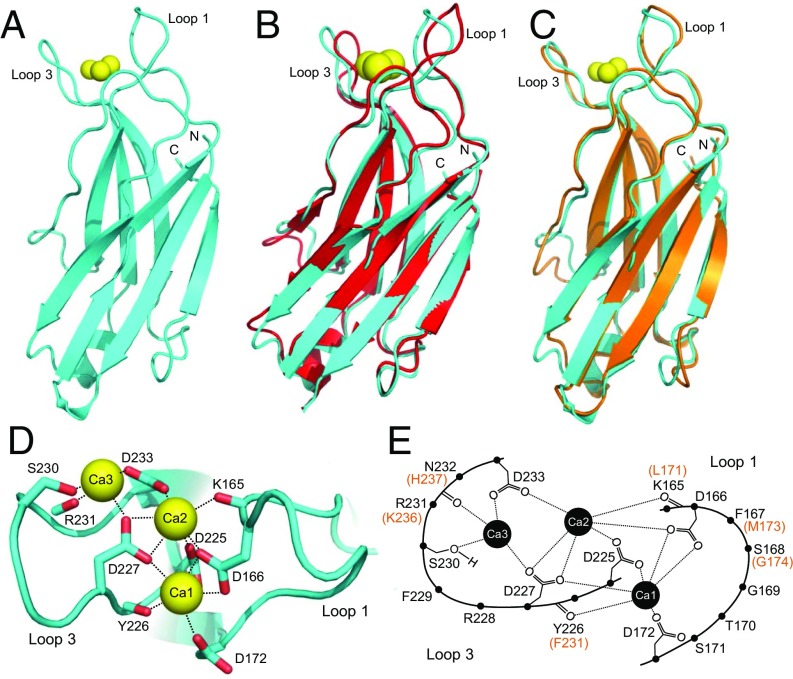
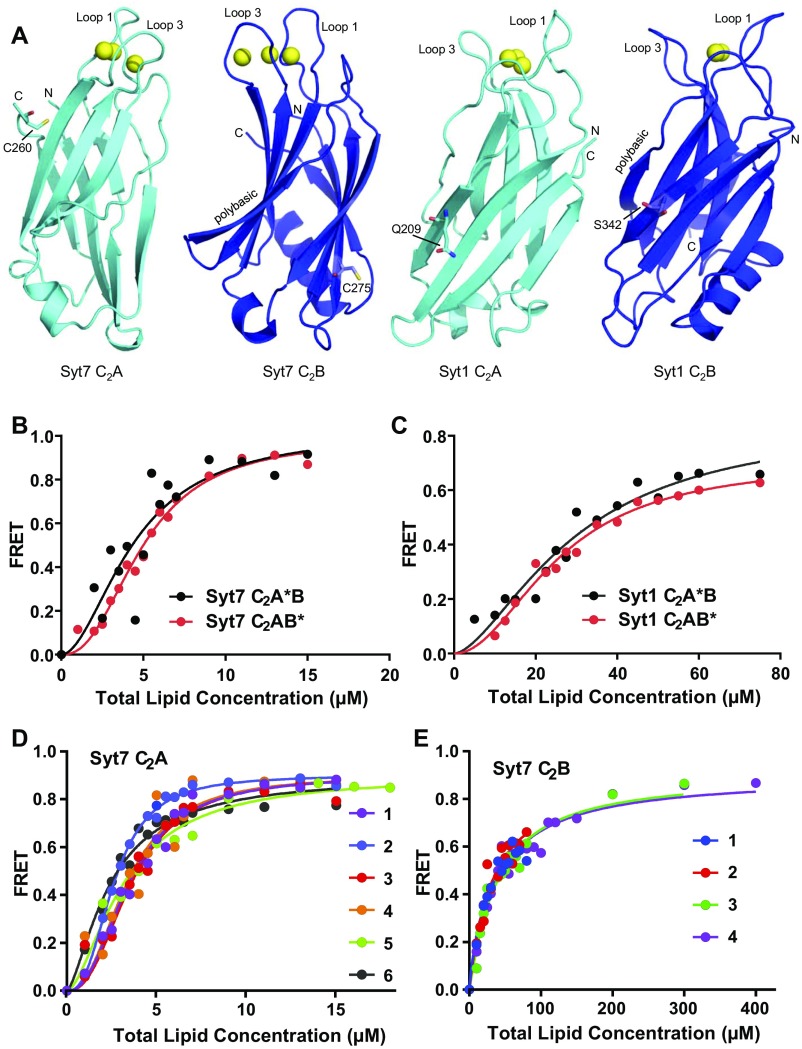
Fig. 1A shows the ribbon diagram of the Ca2+-bound Syt7 C2A domain, which has a typical β-sandwich architecture composed of two four-stranded β-sheets characteristic of C2 domains (35). The structure is very similar to the Ca2+-free solution structure (Fig. 1B), with a 1.13 Å rmsd for 127 equivalent Cα carbons. Differences observed in some of the loops can readily arise from the known flexibility of these loops in C2 domains (12, 35) and potential adaptations induced by crystal contacts. Hence, Ca2+ binding does not appear to induce substantial structural changes in the Syt7 C2A domain, as observed for the Syt1 C2A domain (12). Indeed, the structures of the Ca2+-bound C2A domains of Syt1 and Syt7 are also very similar (Fig. 1C), with a 0.92 Å rmsd for 113 equivalent Cα carbons. Note that the perturbations induced by Ca2+ in the 1H-15N heteronuclear single quantum coherence (HSQC) spectra of the Syt7 C2A domain (36) are also comparable to those observed for the Syt1 C2A domain (15). Correspondingly, the Syt7 C2A domain binds three Ca2+ ions via the same ligands (Fig. 1 D and E) observed in the three Ca2+-binding sites of the Syt1 C2A domain (12, 15). Overall, these results show that there are no overt differences in the structure and Ca2+-binding mode of the C2A domains from Syt1 and Syt7 that might explain the predominant role of Ca2+ binding to the Syt7 C2A domain in asynchronous neurotransmitter release.
Fig. 1.
Crystal structure of the Ca2+-bound Syt7 C2A domain. (A) Ribbon diagram of the crystal structure of the Ca2+-bound Syt7 C2A domain. The three bound Ca2+ ions are shown as yellow spheres. The two loops involved in Ca2+ binding are indicated (loop 1 and loop 3), and the N and C termini are labeled N and C, respectively. (B) Superposition of the crystal structure of the Ca2+-bound Syt7 C2A domain (cyan) with the solution structure of Ca2+-free Syt7 C2A domain (red) (PDB ID code 2D8K). (C) Superposition of the crystal structure of the Ca2+-bound Syt7 C2A domain (cyan) with the solution structure of Ca2+-bound Syt1 C2A domain (orange) (PDB ID code 1BYN). (D) Diagram illustrating the Ca2+-binding sites of the Syt7 C2A domain. The three bound Ca2+ ions are labeled Ca1, Ca2, and Ca3. The Ca2+ ligands are shown as stick models and labeled. (E) Diagram of the Ca2+-binding sites of Syt7 C2A domain summarizing all of the residues in loops 1 and 3 (black letters). The residues that are different in the Syt1 C2A domain are indicated in parenthesis in red letters.
Crystal Structure of the Syt7 C2AB Fragment.
A crystal structure of a Syt1 C2AB fragment in the absence of Ca2+ revealed extensive contacts between the two C2 domains that distort the Ca2+-binding region of the C2A domain and hence would be expected to be disrupted upon Ca2+ binding to this domain, which could provide a mechanism for Ca2+-regulation of neurotransmitter release (37). Although such contacts did not appear to occur in solution (20), we performed extensive crystallization screens with the Syt7 C2AB fragment to examine the possibility of intramolecular interactions between the two Syt7 C2 domains. Unfortunately, we were not able to obtain crystals for Ca2+-free Syt7 C2AB, but we did obtain crystals in the presence of Ca2+ and we determined its structure by molecular replacement using the crystal structures of the Syt7 C2A and C2B domains (Fig. 1A and ref. 29) as search models (Fig. 2A). Data collection and refinement statistics are summarized in Table S1.
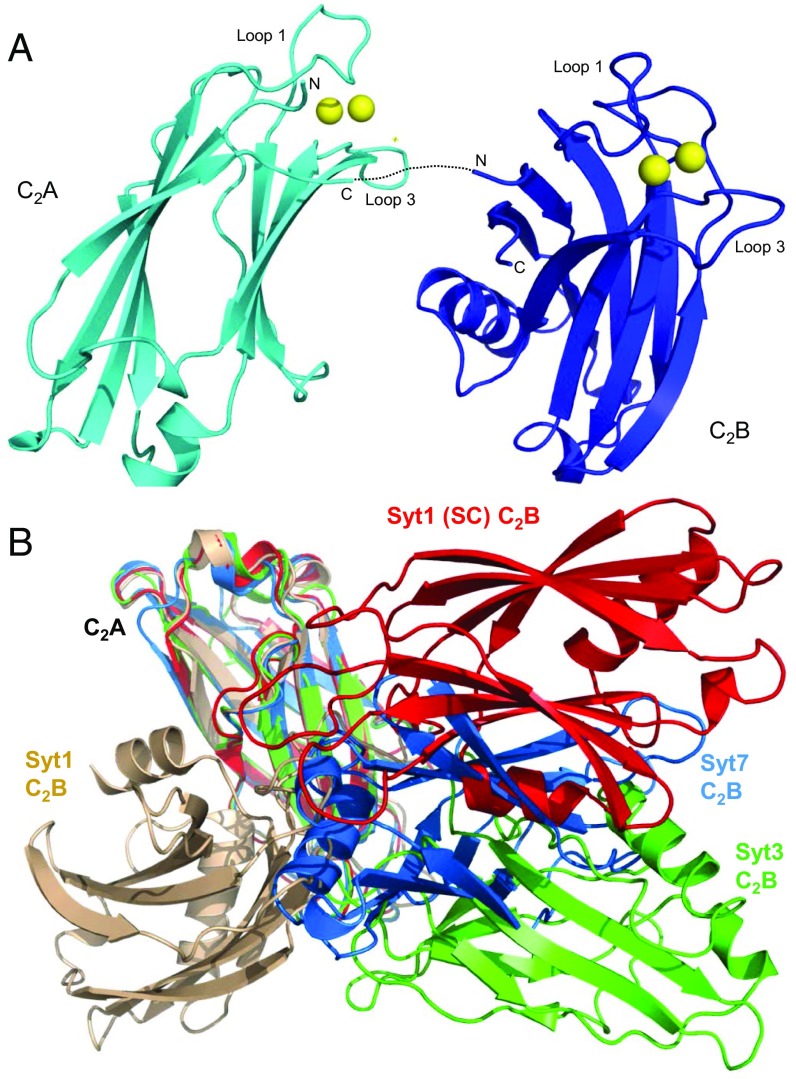
Fig. 2.
Crystal structure of the Ca2+-bound Syt7 C2AB fragment. (A) Ribbon diagram of the crystal structure of the Ca2+-bound Syt7 C2AB fragment, with the C2A domain in cyan and the C2B domain in blue. The bound Ca2+ ions are shown as yellow spheres. The loops involved in Ca2+ binding are indicated (loop 1 and loop 3), and the N and C termini of both domains are labeled N and C, respectively. The sequence linking the two C2 domains, for which there is insufficient electronic density to build the structure, is illustrated by a dotted line. (B) Superposition of the crystal structure of the Ca2+-bound Syt7 C2AB (cyan and blue) with the crystal structures of Ca2+-free Syt1 C2AB (wheat; PDB ID code 2R83), Syt1 C2AB bound to the SNARE complex (red; PDB ID code 5CCG) and Ca2+-free Syt3 C2AB (green; PDB ID code 1DQV). Only the C2A domain of each structure was used for the superposition to illustrate the diversity of relative orientations between the two C2 domains in these structures. The position of all of the C2A domains is indicated by C2A and those of the different C2B domains are also labeled; Syt1 (SC) C2B domain refers to Syt1 C2AB bound to the SNARE complex.
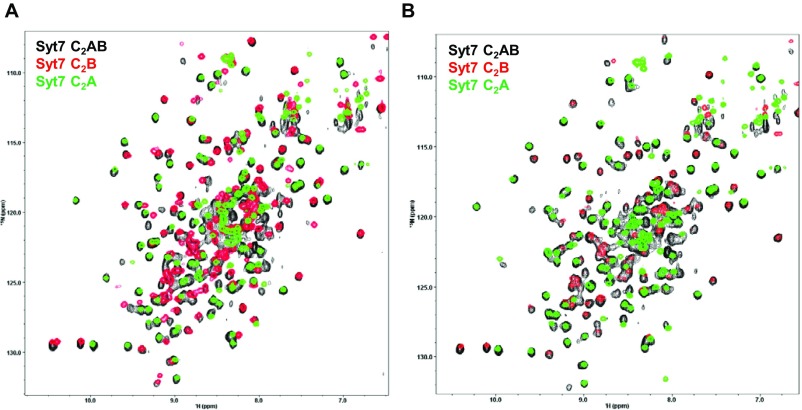
The structures of the two C2 domains in the Syt7 C2AB structure are very similar to those observed in the crystal structures of the isolated C2A (described above) and C2B (29) domains. Comparison of the isolated C2A domain with this domain in the two structures of C2AB in the asymmetric unit of its crystals revealed rmsds of 0.49 and 0.58 Å for 125 equivalent Cα carbons, whereas for the C2B domain the rmsds were 0.45 and 0.41 Å for 138 and 137 equivalent Cα carbons, respectively. No density could be modeled for the linker sequence between the two C2 domains in the structure of the Syt7 C2AB fragment, and the observed contacts between the domains were typical of crystal packing interfaces, suggesting that the relative orientation between the two C2 domains is a result of crystal packing. Comparison of the crystal structure of Syt7 C2AB with those of isolated Syt1 C2AB (37), one of the C2AB molecules in the crystal structure of Syt1 C2AB bound to the SNARE complex (26) and the crystal structure of Syt3 C2AB (38), shows that the relative orientation of the two C2 domains is very different in each structure (Fig. 2B), supporting the conclusion drawn from NMR studies of Syt1 C2AB that this relative orientation is flexible, and hence adaptable (20). We also analyzed the Syt7 C2AB fragment by NMR spectroscopy and found that its 1H-15N HSQC spectra in the presence and absence of Ca2+ are very similar to the superposition of 1H-15N HSQC acquired separately for the isolated C2A and C2B domains (Fig. S1). There are a few cross-peaks that do not overlap, but they can be attributed to residues from the linker or adjacent sequences, and the overall coincidence of cross-peaks is inconsistent with extensive contacts between the two C2 domains, thus supporting the conclusion that their relative orientation is flexible.
Fig. S1.
There are no stable interactions between the C2A and C2B domains of Syt7 in solution. Superpositions of 1H-15N HSQC spectra of the Syt7 C2AB fragment (black contours), the Syt7 C2A domain (green contours) or C2B domain (red contours) in the presence of 1 mM EDTA (A) or 2 mM Ca2+ (B).
Intrinsic Calcium Binding to the Syt7 C2A and C2B Domains.
In the crystal structure of the Syt7 C2AB fragment we observed only two Ca2+ ions bound to the C2A domain and two Ca2+ ions bound to the C2B domain (Fig. 2A), corresponding in both cases to sites Ca1 and Ca2 as defined for the C2A domain in Fig. 1E. These findings contrast with the observation of three bound Ca2+ ions in the structure of the isolated Syt7 C2A domain (Fig. 1D) and four bound Ca2+ ions in the crystal structure of the isolated C2B domain (29). These differences are not surprising, as the third Ca2+-binding site observed in the isolated Syt7 C2A domain (Ca3) is likely of lower affinity because it is formed by only four protein ligands (Fig. 1D) and, in the final refined structure, Ca3 displayed a higher B-factor (11.8 Å2) than Ca1 (7.8 Å2) or Ca2 (7.2 Å2). The Ca3 site may not be occupied in the crystals of the Syt7 C2AB fragment because of the high sulfate concentration present in the crystallization conditions. For the isolated Syt7 C2B domain, one of the two additional Ca2+ ions was bound to site Ca3, which is also formed by four protein ligands, and the other was bound to an additional site formed by only three protein ligands on the outside of the cup shape formed by the Ca2+-binding loops, near a lattice contact (29). In the final refined Syt7 C2B structure, Ca3 also displayed a higher B-factor (16.2 Å2) than Ca1 (8.8 Å2) or Ca2 (10.1 Å2). Although the crystals of the isolated Syt7 C2B domain also contained high sulfate concentration, the lower pH used to crystalize the Syt7 C2AB fragment (6.0 instead of 6.5) may have decreased the already weak Ca2+ affinity of these two sites and, hence, no binding is observed.
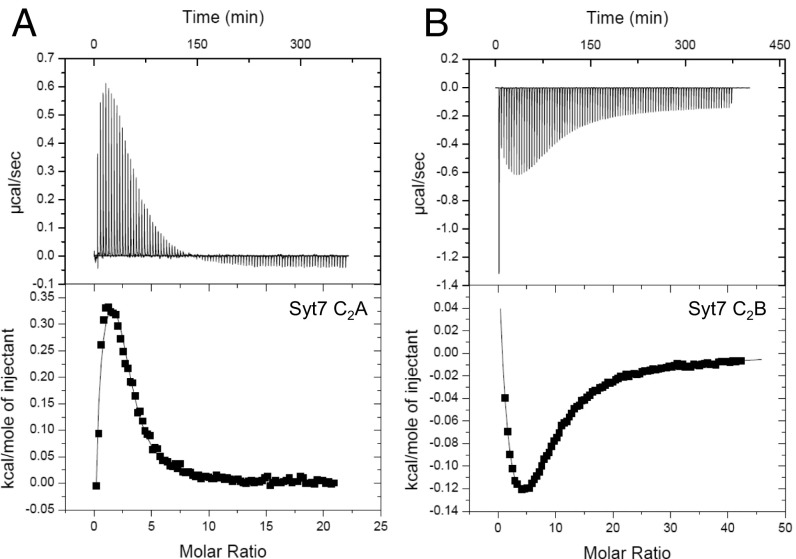
To analyze the intrinsic affinities of the Ca2+-binding sites of the Syt7 C2 domains in solution, we used ITC (Fig. 3). For this purpose, we used the isolated Syt7 C2A and C2B domains because the expected existence of multiple Ca2+ binding sites would hinder fitting the data obtained with the C2AB fragment, and because the two C2 domains are not expected to influence each other in Ca2+ binding given their flexible relative orientation. We found that fitting the ITC data obtained for the Syt7 C2A domain (Fig. 3A) required a three-sequential-binding-site model, consistent with the crystal structure of the isolated domain (Fig. 1A). The KDs measured for the three Ca2+ binding sites were 90 ± 15 μM, 180 ± 42 μM, and 11 ± 5 mM. These KDs should be examined with caution due to the natural difficulty in obtaining accurate KDs from such multiple binding-site data, but it is reassuring that the values are comparable to those measured previously by NMR spectroscopy for the Syt7 C2A domain (∼150 μM, 200–300 μM and >2 mM) (36). They are also similar to KDs obtained for the Syt1 C2A domain by NMR spectroscopy (54 μM, 530 μM and >2 mM) (6) and ITC (119 μM, 465 μM and 1.7 mM) (39).
Fig. 3.
ITC analysis of intrinsic Ca2+ binding to the Syt7 C2 domains. Illustrative ITC data obtained by titration of Ca2+ versus the isolated Syt7 C2A (A) or C2B (B) domain. The curves represent the fits of the data to a three- (A) or four- (B) sequential-binding-site model.
Fitting the ITC data obtained for the Syt7 C2B domain required a four-sequential-binding-site model, as attempts to fit the data assuming only three binding sites yielded a systematic error at the end of the titration, which reflects binding to a very low-affinity Ca2+-binding site. Hence, these data are consistent with the crystal structure of the isolated Syt7 C2B domain (29). Unfortunately, it was difficult to obtain reliable KDs from these data because of the multiplicity of binding sites and because the values obtained depended strongly on the position chosen for the zero baseline (note the drift observed even at molar ratio = 40) (Fig. 3B). Nevertheless, the ranges of KD values for the four sites (75–900 μM, 0.5–1.2 mM, 3–5 mM, and 5–27 mM) obtained in different fits give an idea of the intrinsic affinities of the four sites. The KDs of the two higher affinity sites are comparable to those of the two Ca2+-binding sites of the Syt1 C2B domain, for which NMR data yielded estimated KDs of 300–400 and 500–600 μM (13) and ITC experiments yielded an estimated overall KD of 200 μM for the two sites (39). It is currently unclear whether all four Ca2+-binding sites of the Syt7 C2B domain, or perhaps only three or two, are physiologically relevant but, regardless of these possibilities, these results indicate that the intrinsic Ca2+-binding properties of the C2 domains from Syt7 are similar to those of the Syt1 C2 domains and do not provide an explanation for the differences in the relative functional importance of the Syt7 C2 domains.
The Ca2+-Bound Syt7 C2A Domain Binds to Membranes with Higher Affinity than the Ca2+-Bound C2B Domain.
The apparent Ca2+ affinities of Syt C2 domains in Ca2+-dependent phospholipid binding assays are much stronger (apparent KDs < 10 μM) than their intrinsic Ca2+ affinities because the phospholipids contribute to coordinate the bound Ca2+ ions (6, 17, 40). Notably, the apparent Ca2+-affinity of the Syt7 C2A domain in Ca2+-dependent phospholipid binding is considerably higher than that of the Syt1 C2A domain (40). This higher apparent Ca2+ affinity can be associated to slower dissociation rates from the membranes (41), which has been also observed in experiments performed with Syt1 and Syt7 C2AB fragments (42). Since the intrinsic Ca2+ affinities of the Syt1 and Syt7 C2 domains are similar, their differences in Ca2+-dependent phospholipid binding can be ascribed to stronger interactions with the membranes in the Ca2+-bound state. Thus, we decided to investigate whether differences in membrane affinity might underlie the relative contributions of the C2A and C2B domains to Syt7 function. For this purpose, we used a similar approach to that described previously for the Syt1 C2 domains, which used FRET to detect phospholipid binding and titrations with liposomes under Ca2+ saturating conditions to compare relative membrane affinities (39).
In our FRET assays, we used liposomes containing Rhodamine-PE as a FRET acceptor and a BODIPY-FL donor probe was attached to the native cysteines of the Syt7 C2A and C2B domain (C260 and C275, respectively), which are located in regions that are not expected to participate in phospholipid binding based on extensive analyses of Syt1 (e.g., refs. 17, 18, 43, and 44) (Fig. S2A). For experiments with the Syt7 C2AB fragment, the BODIPY-FL probe was attached to either C260 or C275 (indicated by an asterisk; i.e., C2A*B or C2AB*) and the other cysteine was mutated to alanine. The position of the probe did not substantially alter the phospholipid binding curves (Fig. S2B). Most of the data shown were obtained with the probe on C260 of the C2A domain, except for experiments where the Ca2+-binding sites of the C2A domain were mutated, in which case the probe was attached to C275 of the C2B domain. In initial experiments and those described below for Syt1 fragments, we performed titrations with increasing amounts of liposomes onto 100 nM protein under Ca2+ saturating conditions. However, because of the very high liposome affinities observed for fragments including the Syt7 C2A domain, we lowered the protein concentration to 20 nM to allow better discrimination of relative liposome affinities. Even at these low protein concentrations, excellent consistency was observed in separate experiments performed under the same conditions with different liposome preparations (e.g., Fig. S2 D and E). All of the data could be fit well to a Hill function (Figs. 4–7), with Hill coefficients ranging from 1 to 3 (Table 1) that in principle could indicate some cooperativity. However, it is important to note that it is difficult to interpret Hill coefficients in these experiments because at low concentrations of liposomes (high protein-to-lipid ratios) their surface may be covered by bound proteins, which is expected to occlude some of the available binding sites because of molecular crowding and at the same time can favor liposome clustering. On the opposite end, at high liposome concentrations (low protein-to-lipid ratios) molecular crowding effects are not expected and liposome clustering is less favorable (20). Hence, we did not attempt to rationalize the Hill coefficients and only used the KDs derived from fitting the data (Table 1) as apparent values that report on the relative affinities of the Ca2+-saturated proteins for the liposomes.
Fig. S2.
Placing the fluorescent probe on the C2A or C2B domain does not alter phospholipid binding to the Syt1 or Syt7 C2AB fragment. (A) Ribbon diagrams of the structures of the Syt7 and Syt1 C2A or C2B domains, as indicated. The structure of the Syt7 C2A domain is described here. Those of the Syt7 C2B domain, the Syt1 C2A domain, and the Syt1 C2B domain correspond to PDB ID codes 3N5A, 1BYN, and 1K5W, respectively. The bound Ca2+ ions are shown as yellow spheres. The two loops involved in Ca2+ binding are indicated (loop 1 and loop 3), and the N and C termini are labeled N and C, respectively. The native cysteine side chains of the Syt7 C2A and C2B domains, as well as the side chains that were mutated to cysteine in the Syt1 C2A and C2B domains to attach a fluorescent probe, are shown as stick models. (B) Titrations of 20 nM Syt7 C2A*B and C2AB* fragments (where the * indicates the domain where the fluorescent probe was attached) with liposomes in the presence of 1 mM Ca2+. Binding was monitored from the FRET developed between a donor BODIPY-FL probe attached to the proteins and a rhodamine acceptor present in the liposomes. Each point represents the average of at least three measurements performed with different liposome preparations. (C) Analogous liposome titrations of 100 nM Syt1 C2A*B and C2AB* fragments. All of the data were fit to a Hill function. (D and E) Superpositions of the data obtained in analogous titrations of 20 nM Syt7 C2A domain (D) or Syt7 C2B domain (E), illustrating the reproducibility of the results. Each symbol represents one dataset obtained with a different liposome preparation. The apparent KDs derived for the six datasets shown in D are 1.49, 2.35, 2.62, 2.42, 2.37, and 3.17 μM, and those derived for the four datasets shown in E are 34.1, 31.4, 38.1, and 36.4 μM. The Syt7 C2B domain data provide an example with small variability among different titrations while the Syt7 C2A domain data illustrate an example with the largest variability, which is reflected in a larger SD in relative terms (Table 1). Even in this case, the data exhibit excellent consistency that allows accurate comparison with the binding curves obtained for other proteins.
Fig. 4.
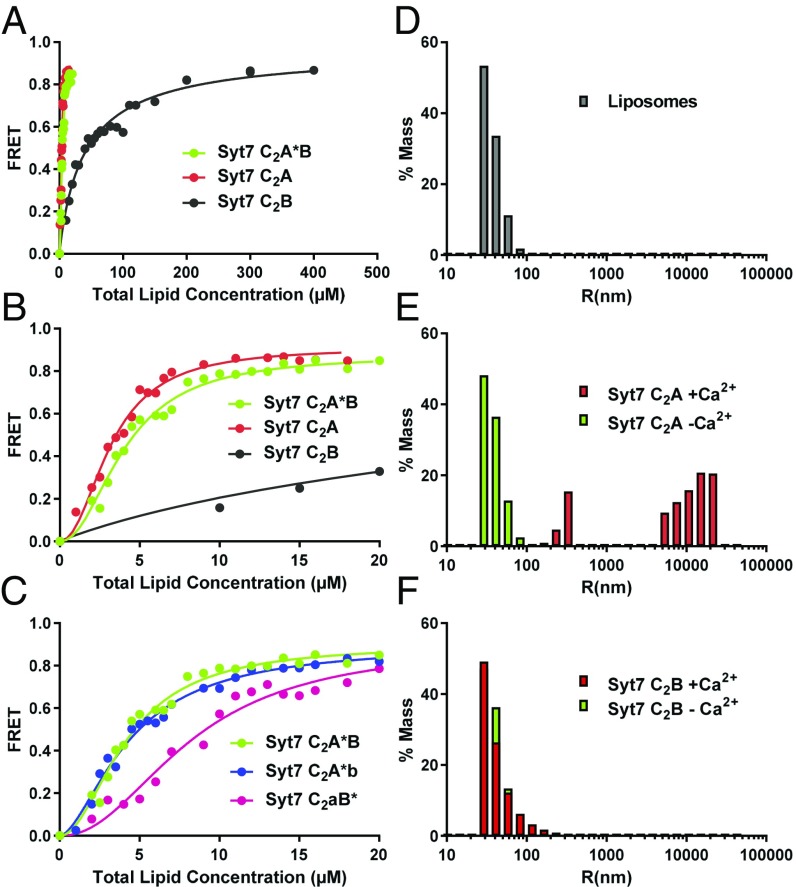
The Syt7 C2A domain dominates binding of Syt7 to membranes and clusters liposomes. (A) Titrations of 20 nM Syt7 C2A domain, C2B domain and C2A*B fragment (the * in C2A*B denotes that the fluorescent probe is attached to C260 of the C2A domain) with liposomes lacking PIP2 in the presence of 1 mM Ca2+. Binding was monitored from the FRET developed between a donor BODIPY-FL probe attached to the proteins and a rhodamine acceptor present in the liposomes. Each point represents the average of at least three measurements performed with different liposome preparations. (B) The same titrations shown in A but changing the x axis to better show the points of the titrations obtained at low liposome concentrations. (C) Liposome titrations of 20 nM WT Syt7 C2AB fragment and mutant fragments where the Ca2+-binding sites of the C2A domain (C2aB*) or C2B domain (C2A*b) were disrupted with D225A,D227A,D233A or D357A,D359A mutations. All of the data in A–C were fit to a Hill function. (D–F) Distribution of particle size measured by DLS on samples containing liposomes alone (D) or liposomes in the presence of the Syt7 C2A domain (E) or Syt7 C2B domain (F). In E and F, the diagrams show superpositions of data obtained in the absence (green bars) or presence (red bars) of Ca2+.
Fig. 7.
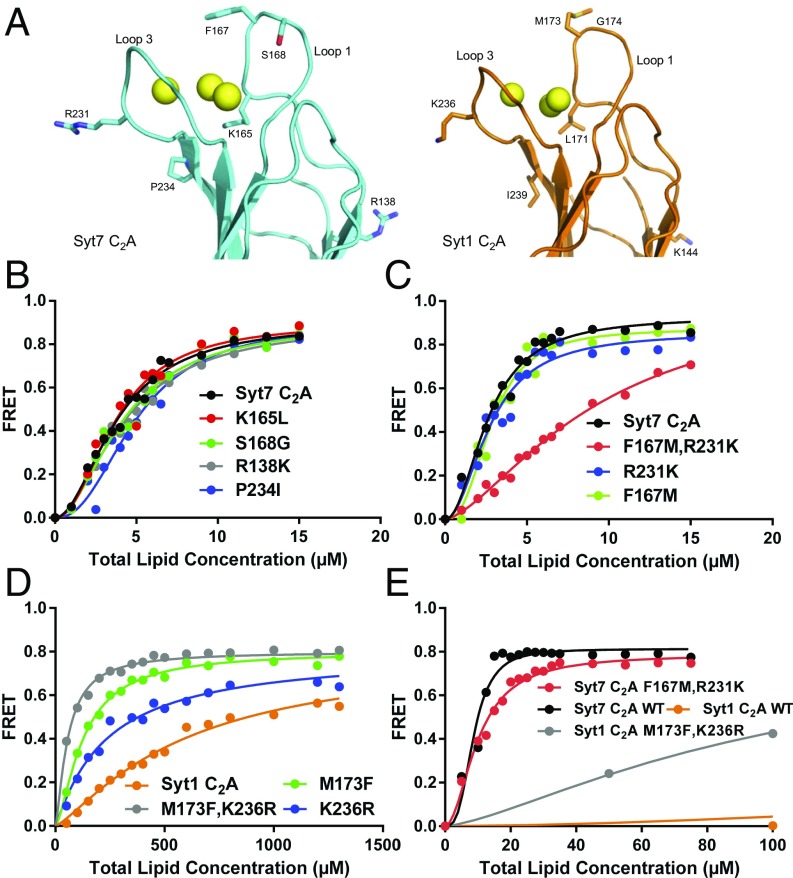
Subtle residue substitutions underlie in part the stronger membrane binding affinity of the Syt7 C2A domain compared with the Syt1 C2A domain. (A) Ribbon diagrams of the Syt7 C2A domain (cyan) and Syt1 C2A domain (orange) showing the Cα and side chain atoms of residues that are different in their Ca2+-binding regions and were mutated to analyze the basis for their different membrane affinities (oxygens are in red, nitrogens in blue, sulfur atoms in yellow, and carbon atoms in cyan for Syt7 and in orange for Syt1). (B and C) Titrations of 20 nM WT and mutant Syt7 C2A domains with liposomes in the presence of 1 mM Ca2+. Binding was monitored from the FRET developed between a donor BODIPY-FL probe attached to the proteins and a rhodamine acceptor present in the liposomes. Each point represents the average of at least three measurements performed with different liposome preparations. (D) Analogous liposome titrations of 100 nM WT and mutant Syt1 C2A domains. (E) Analogous liposome titrations of 100 nM WT and mutant Syt1 and Syt7 C2A domains. Note that the data obtained for WT and M173F,K236R Syt1 C2A domains (orange and gray circles, respectively) are the same as those shown in D, but the scale of the x axis is different to allow comparison with the data obtained for the WT and mutant Syt7 C2A domains. All of the data were fit to a Hill function.
Table 1.
Apparent liposome affinities of Ca2+-saturated Syt7 and Syt1 C2 domain fragments
| Protein fragment | KD (µM) | SD | h | SD |
| Syt7 C2A | 2.40 | 0.54 | 2.18 | 0.41 |
| Syt7 C2B | 34.97 | 2.92 | 1.02 | 0.09 |
| Syt7 C2A*B | 4.12 | 0.80 | 3.1 | 0.52 |
| Syt7 C2A*b | 4.29 | 0.18 | 1.74 | 0.26 |
| Syt7 C2aB* | 8.06 | 0.39 | 2.03 | 0.26 |
| Syt7 C2A F167M,R231K | 8.68 | 0.44 | 1.61 | 0.17 |
| Syt1 C2A | 551.1 | 134.26 | 1.34 | 0.159 |
| Syt1 C2B | 637.37 | 146.89 | 1.78 | 0.38 |
| Syt1 C2A*B | 24.05 | 1.77 | 2.15 | 0.46 |
| Syt1 C2A*b | 245.5 | 4.49 | 1.95 | 0.24 |
| Syt1 C2aB* | 267.23 | 38.96 | 2.09 | 0.25 |
| Syt1 C2A M173F,K236R | 95.48 | 25.9 | 1.51 | 0.3 |
The listed apparent KDs and Hill coefficients (h), as well as their SDs, were obtained from fitting the FRET data obtained in liposome titrations of the indicated Ca2+-saturated Syt7 and Syt1 C2 domain fragments. At least three titrations were performed for each fragment. KDs and h values were derived for each titration, and the averages as well as SDs were calculated. The protein concentrations were 20 nM for all Syt7 fragments and 100 nM for all Syt1 fragments.
We first used liposomes containing a lipid composition that resembles that of physiological membranes (33) but did not include PIP2 (Fig. 4). Importantly, liposome titrations revealed that the Syt7 C2A domain binds with much higher affinity than the Syt7 C2B domain and a similar affinity to that observed for Syt7 C2AB (Fig. 4 A and B and Table 1). In fact, the curves observed for Syt7 C2AB were consistently shifted to the right (i.e., larger KD) compared with those obtained for the Syt7 C2A domain (Fig. 4B), but this observation does not necessarily imply a higher affinity of the C2A domain versus C2AB. Considering that a C2 domain can bind multiple phospholipids and has a much larger area than that of a phospholipid headgroup, that the liposomes contained 17% of negatively charged phospholipids, that only half of the lipids are in the outer leaflets, and that molecular crowding limits the availability of more than 70% of the liposome surface (20), the amounts of phospholipids required to observed maximum FRET for the Syt7 C2A domain and C2AB fragment suggest that we are observing saturation binding curves. Hence, the right shift of the curves observed for the Syt7 C2AB fragment compared with the C2A domain most likely reflects the fact that C2AB is larger than C2A, thus requiring more lipids for quantitative binding. We also performed titrations with Syt7 C2AB fragments containing mutations that disrupt the Ca2+-binding sites of the C2A domain (denoted C2aB* because C2A cannot bind Ca2+ and the fluorescent probe is on C2B) or the C2B domain (C2A*b). Interestingly, impairing Ca2+-binding to the C2A domain markedly decreased the membrane affinity of Syt7 C2AB, whereas disrupting Ca2+ binding to the C2B domain had little effect (Fig. 4C and Table 1). These results show that the C2A domain dominates binding of Syt7 to membranes and reveal a clear correlation with the finding that disrupting Ca2+ binding to the C2A domain has a much stronger effect on the function of Syt7 in asynchronous release than abrogating Ca2+ binding to the C2B domain (9).
The Ca2+-Bound Syt7 C2A Domain Has Higher Membrane-Bridging Activity than the Ca2+-Bound C2B Domain.
The finding that the Syt1 C2B domain can bridge two membranes, bringing them into close proximity, suggested that this property could be crucial for Syt1 function and could explain the critical functional importance of Ca2+-binding to the Syt1 C2B domain (20, 22). Thus, we also tested the membrane bridging activity of the Syt7 C2 domains. For this purpose, we analyzed the formation of liposome clusters due to membrane bridging using dynamic light scattering (DLS). Isolated liposomes had radii below 100 nm (Fig. 4D) that did not change substantially upon addition of the Syt7 C2A domain or C2B domain in the absence of Ca2+ (Fig. 4 E and F). However, in the presence of Ca2+, the Syt7 C2A domain caused a dramatic increase in particle size reflecting robust vesicle clustering, whereas the C2B domain had practically no activity (Fig. 4 E and F). These results again correlate with the electrophysiological data and suggest that the preponderant role of Ca2+ binding to the Syt7 C2A domain in triggering asynchronous release may arise not only from its the higher membrane affinity, compared with the C2B domain, but also from its stronger ability to bridge two membranes.
The Ca2+-Bound Syt1 C2B Domain Binds to PIP2-Containing Membranes with Higher Affinity than the Ca2+-Bound Syt1 C2A Domain.
Phospholipid binding to the Syt1 C2 domains has been extensively investigated (1, 45, 46), but most studies analyzed the Ca2+ dependence of lipid binding, and analysis of phospholipid binding under Ca2+ saturating conditions was mostly focused on Syt1 C2AB fragments (39). To compare the properties of the Syt1 C2 domains using the same methodology used for our experiments with the Syt7 C2 domains and examine whether the asymmetry in the functional importance of Ca2+-binding to the Syt1 C2 domains may also be reflected in their membrane affinities, we performed liposome titrations of the Ca2+-saturated Syt1 C2A domain, C2B domain, and C2AB fragment. Because the Syt1 C2 domains only contain one cysteine side chain (C277) and this side chain is buried, we mutated this cysteine to alanine and introduced single cysteine mutations in regions of the C2A and C2B domain that have not been implicated in lipid binding (Fig. S2A). As in the case of Syt7 C2AB, placing the fluorescent probe on the C2A or C2B domain did not substantially affect the phospholipid curves observed for the Syt1 C2AB fragment (Fig. S2C).
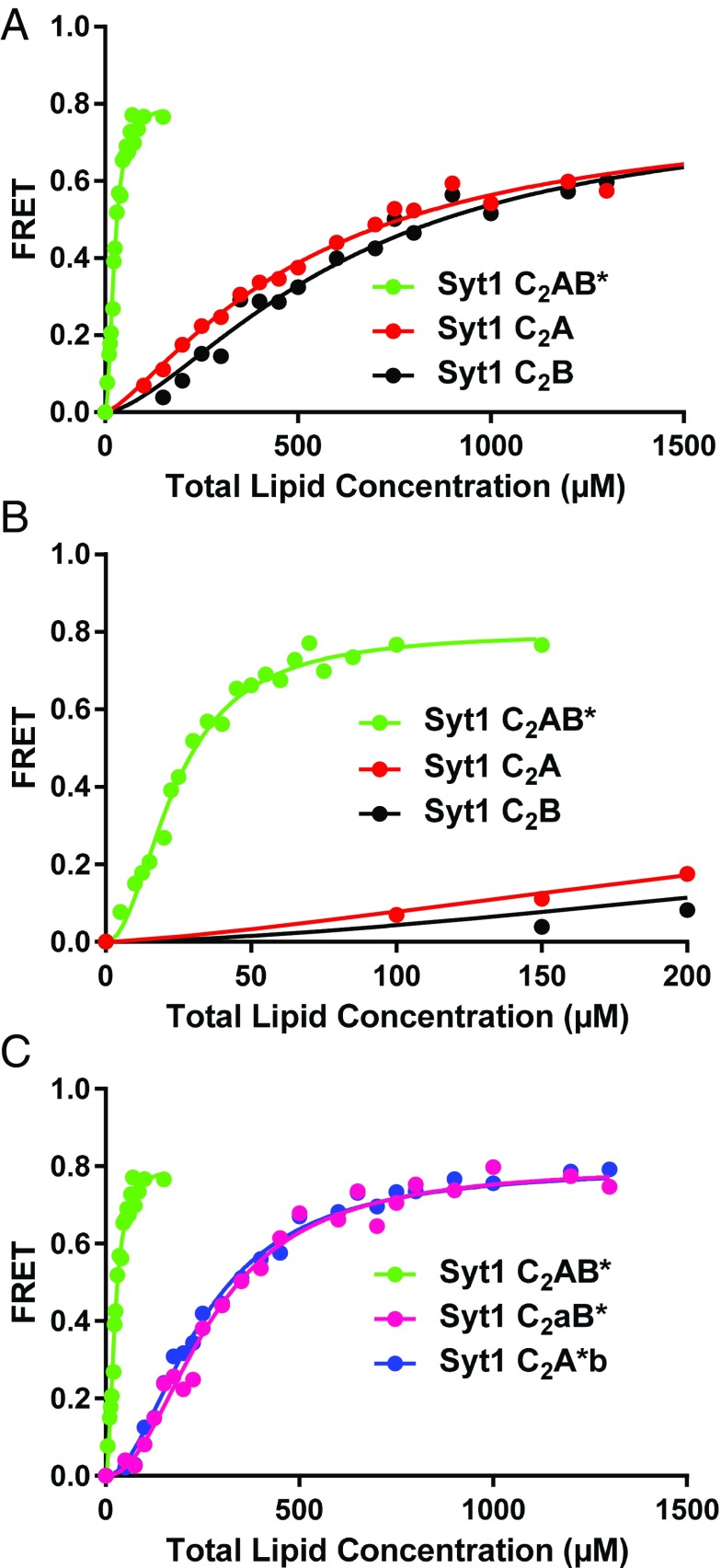
Our data showed that the Ca2+-saturated Syt1 C2A and C2B domains bind to the liposomes with similar affinities, which were much weaker than that observed for the Syt7 C2A domain and were far from leading to saturation binding curves (Fig. 5A). Syt1 C2AB bound much more avidly than the individual Syt1 C2 domains (Fig. 5 A and B and Table 1), showing that they cooperate strongly in membrane binding. Mutation of the Ca2+-binding sites of the C2A or C2B domain considerably impaired phospholipid binding to the Syt1 C2AB fragment, but to similar extents (Fig. 5C) that do not correlate with the stronger disruption of Syt1 function caused by mutation of the C2B domain Ca2+-binding sites.
Fig. 5.
The Syt1 C2 domains have similar membrane affinities and cooperate in membrane binding. (A) Titrations of 100 nM Syt1 C2A domain, C2B domain and C2AB* fragment (the * in C2AB* denotes that the fluorescent probe is attached to the C2B domain) with liposomes lacking PIP2 in the presence of 1 mM Ca2+. Binding was monitored from the FRET developed between a donor BODIPY-FL probe attached to the proteins and a rhodamine acceptor present in the liposomes. Each point represents the average of at least three measurements performed with different liposome preparations. (B) The same titrations shown in A but changing the x axis to better show the points of the titrations obtained at low liposome concentrations. (C) Liposome titrations of 100 nM WT Syt1 C2AB* fragment and mutant fragments where the Ca2+-binding sites of the C2A domain (C2aB*) or C2B domain (C2A*b) were disrupted with D178A,D230A,D232A or D309A,D363A,D365A mutations. All of the data were fit to a Hill function.
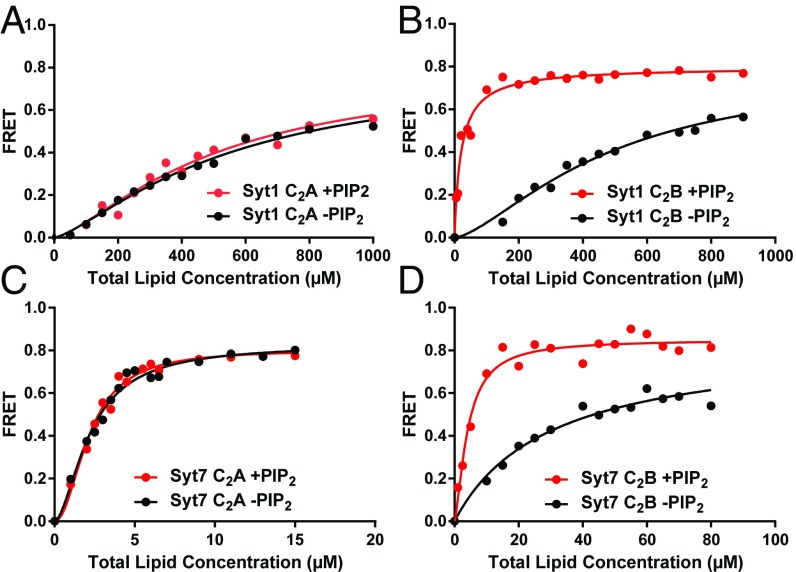
Because PIP2 is known to increase the membrane affinity of the C2B domain but not the C2A domain (47), we performed additional experiments with liposomes that contained the same lipid composition except for the inclusion of 1% PIP2. Our data confirmed that, indeed, the presence of PIP2 in the liposomes did not alter the affinity for the Syt1 C2A domain but dramatically increased the affinity for the Syt1 C2B domain (Fig. 6 A and B). Hence, in the presence of PIP2, the stronger membrane affinity of the Ca2+-saturated Syt1 C2B domain compared with the C2A domain does correlate with the functional effects of disrupting Ca2+ binding. These results led us to also test the effects of including PIP2 on the membrane affinity of the Syt7 C2 domains. PIP2 clearly increased the membrane affinity of the Syt7 C2B domain but did not appear to alter liposome binding to the Syt7 C2A domain (Fig. 6 C and D). However, the latter result needs to be interpreted with caution because the titrations with Syt7 C2A domain most likely reflect saturation binding (see above), which is expected to prevent observation of any increase in membrane affinity due to inclusion of PIP2. Note also that, even in the presence of PIP2, the Ca2+-saturated Syt7 C2A domain still binds membranes with higher affinity than the Syt7 C2B domain (Fig. S3).
Fig. 6.
PIP2 enhances the membrane affinity of the Syt1 and Syt7 C2B domains. Titrations of 100 nM Syt1 C2A domain (A) or C2B domain (B), or 20 nM Syt7 C2A domain (C) and C2B domain (D) with liposomes lacking (black circles) or containing (red circles) PIP2 in the presence of 1 mM Ca2+. Binding was monitored from the FRET developed between a donor BODIPY-FL probe attached to the proteins and a rhodamine acceptor present in the liposomes. Each point represents the average of at least three measurements performed with different liposome preparations. All of the data were fit to a Hill function.
Fig. S3.
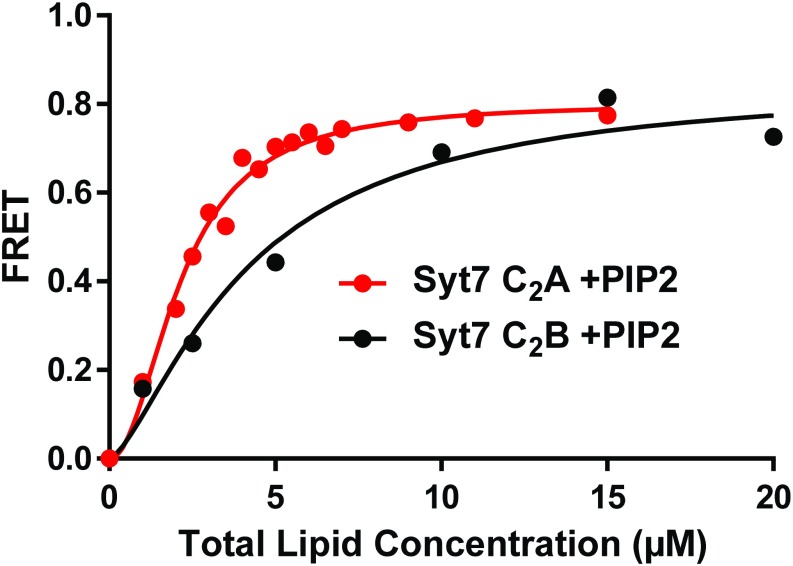
The Syt7 C2A domain binds to PIP2-containing liposomes with higher affinity than the Syt7 C2B domain. Titrations of 20 nM Syt7 C2A domain (red circles) or C2B domain (blue circles) with liposomes containing PIP2 in the presence of 1 mM Ca2+. Binding was monitored from the FRET developed between a donor BODIPY-FL probe attached to the proteins and a rhodamine acceptor present in the liposomes. Each point represents the average of at least three measurements. All of the data were fit to a Hill function. The data are the same shown in Fig. 6 C and D with liposomes containing PIP2.
Subtle Residue Substitutions Underlie in Part the Differential Membrane Affinities of the Syt1 and Syt7 C2A Domains.
Overall, the data presented above and other previous results suggest that phospholipid binding affinity is a key factor that determines the functional importance of Syt C2 domains. When comparing the properties of Syt1 and Syt7, it is particularly striking that the Syt7 C2A domain has such a higher membrane-binding affinity than the Syt1 C2A domain (Figs. 4 and 5), with an apparent KD at least 200 times smaller (Table 1). This finding is not surprising given previous results showing a higher apparent Ca2+ affinity (40) and a slower membrane dissociation rate (41) for the Syt7 C2A domain compared with the Syt1 C2A domain, but it seems difficult to rationalize considering the similarities of the 3D structures, the intrinsic Ca2+ affinities, and the sequences of these domains. Given the key functional importance of membrane binding, elucidating the sequence determinants of membrane affinity is critical to understand the functional differences between Syts, and the C2A domains of Syt1 and Syt7 provide an ideal benchmark to address this question.
We hypothesized that differences in the membrane affinities of the two domains arise because of cumulative small or moderate effects caused by residues that are distinct in the Ca2+-binding regions of the Syt1 and Syt7 C2A domains. To test this hypothesis, we prepared six single-site mutants of the Syt7 C2A domain where single residues were replaced by the (distinct) residue present in the homologous position of the Syt1 C2A domain (Fig. 7A; see also Fig. 1E). Titrations with liposomes under Ca2+ saturating conditions did not reveal substantial differences between the membrane affinities of any of the single mutants and the WT Syt7 C2A domain (Fig. 7 B and C). We reasoned that perhaps some of these single site mutations decreased the membrane affinity of the Syt7 C2A domain but, since the WT affinity is so high, a modest decrease in affinity may not be observable because the experiments still yield saturation binding curves. To try to cause a stronger decrease in membrane affinity that may be observable, we combined two of the single mutations (F167M and R231K) into a double mutant. Note that these mutations would normally be considered conservative, but they might decrease membrane affinity to some extent because arginines can form hydrogen bonds more efficiently with phosphate groups than lysines, and phenylalanines have a larger hydrophobic surface than methionines. Indeed, the double F167M,R231K mutation markedly impaired liposome binding to the Syt7 C2A domain (Fig. 7C and Table 1).
If the differences in membrane affinities between the Syt1 and Syt7 C2A domains arise in part because of the nature of these two side chains, rather than differences in 3D structure or intrinsic Ca2+ affinities, we could expect that making the reverse mutations in the Syt1 C2A domain would increase its membrane affinity. Indeed, single M173F and K236R mutations both increased the membrane affinity of the Syt1 C2A domain, and a double M173F,K236R mutation revealed an additional increase in membrane affinity that reflects the cumulative effects of the two mutations (Fig. 7D). Titrations of Syt1 and Syt7 fragments were generally performed at 100 nM and 20 nM protein concentrations, respectively, to better dissect the differences in membrane affinities between the fragments of each isoform. To be able to directly compare the membrane affinities of the Syt1 and Syt7 C2A domains in this set of experiments, we repeated the titrations of the WT and F167M,R231K mutant Syt7 C2A domain at 100 nM protein concentration. Comparison of the results with those obtained for the WT and M173F,K236R Syt1 C2A domains (Fig. 7E) showed that the Syt7 double mutant still has a higher affinity than the Syt1 double mutant, but it is clear that the double mutations helped to close the large gap existing between the membrane affinities of the WT Syt1 and Syt7 C2A domains. Thus, the ratio between the apparent KDs was reduced from more than 200 to 11 (Table 1). These results show that apparently conservative residue substitutions in the membrane-binding regions of C2 domains can have significant effects on membrane affinity that can accumulate to yield dramatic biochemical and functional differences.
Discussion
Much has been learned about the neurotransmitter release machinery and, among its components, Syts have been studied extensively because of their roles as Ca2+ sensors. Although Syt function in release is broadly understood and plausible models for the mechanism of action of Syts in fusion have emerged, the structural determinants that shape the functional differences between fast and slow Syts remain unclear. Particularly intriguing was the observation that disrupting Ca2+ binding to the Syt7 C2A domain impaired its function in asynchronous release much more strongly than disruption of the C2B domain Ca2+ binding sites, in contrast to the opposite effects caused by mutations in the Ca2+-binding sites of the Syt1 C2 domains on synchronous release (9). The results presented here strongly suggest that the predominant role of the Syt7 C2A domain in asynchronous release, compared with the C2B domain, arises because of its stronger contribution to membrane binding, and perhaps to bridging the vesicle and plasma membranes. More generally, our data suggest that the contribution to membrane affinity is a major determinant of the relative functional importance of the C2 domains of Syts. Furthermore, our results illustrate how small or moderate effects from what appear to be subtle residue substitutions can accumulate to yield dramatic functional differences in Syt C2 domains.
Ca2+-dependent phospholipid binding was clearly shown to be critical for Syt1 function based on the correlations that were established between the effects of mutations that increase or decrease the apparent Ca2+ affinity of Syt1 in the presence of phospholipids and their functional effects on neurotransmitter release (6, 19). This activity is also believed to underlie at least in part the functions of other Ca2+-dependent Syts (5). Hence, our finding that Ca2+-dependent phospholipid binding to Syt7 is dominated by the C2A domain (Fig. 4) provides a natural explanation for the stronger disruption of asynchronous release caused by mutations in the Ca2+-binding sites of the Syt7 C2A domain compared with those induced by analogous mutations in the Syt7 C2B domain (9). Moreover, although the Syt1 C2A and C2B domains bind with similar affinities to membranes lacking PIP2 (Fig. 5), the Syt1 C2B domain clearly dominates membrane binding in the presence of PIP2 (Fig. 6), and mutations in the Ca2+-binding sites of the C2B domain impair membrane binding to Syt1 C2AB more strongly than mutations in the C2A domain Ca2+ binding sites, in correlation with the functional effects of these mutations (33, 39, 47). A remaining unresolved key issue, however, is the precise role of Ca2+-binding to the Syt1 C2B-domain. Since the C2B-domain mutant Syt1 abolishes even asynchronous release in a dominant-negative fashion, although it does not normally mediate such release, such mutant Syt1 likely locks the prefusion complex at the active zone in a Ca2+-unresponsive state, possibly by preventing the Ca2+-dependent unlocking of a complexin-induced prefusion intermediate (48). Although the lack of rescue of Ca2+-dependent release by C2B-domain mutant Syt1 can thus not be used to argue that Ca2+-dependent phospholipid binding by the Syt1 C2B-domain is of central importance in release, it seems likely that the “unlocking” of the prefusion complex is actually mediated by Ca2+-dependent phospholipid binding by the Syt1 C2B-domain. In this sense, Ca2+-dependent phospholipid binding would constitute an intrinsic aspect of Syt1 function even if it did not underlie the differential effects of mutations in the Ca2+-binding sites of the Syt1 C2 domains, which are opposite to those observed for Syt7.
The intrinsic Ca2+ affinities of the C2A and C2B domains are similar for both Syt1 and Syt7. What appears to be critical for their relative functional importance is the membrane affinity of the Ca2+-saturated C2 domain, which is indirectly reflected in the apparent Ca2+ affinities observed in Ca2+-dependent phospholipid binding experiments. Thus, the very high affinity of the Ca2+-saturated Syt7 C2A domain for membranes, dramatically stronger than that of the Syt1 C2A domain (Figs. 4, 5, and 7), was already reflected in its higher apparent Ca2+ affinity (40). Our data show that this dramatic difference in membrane affinity arises in part from cooperative effects resulting from apparently subtle substitutions, including a methionine-to-phenylalanine substitution that is expected to increase hydrophobic interactions with the acyl chains of the membranes and a lysine-to-arginine substitution that is expected to enhance hydrogen bonding to phospholipid headgroups (Fig. 7). It is likely that the overall basic nature of the Syt7 C2A domain, compared with the acidic nature of the Syt1 C2A domain (see below), also contributes to the stronger affinity for negatively charged membranes. Regardless of this possibility, the effects of the F167M,R231K mutation in the Syt7 C2A domain and the reverse M173F,K236R mutation in the Syt1 C2A domain on membrane binding provide an emphatic illustration of how such subtle mutations can have profound consequences for the biochemical properties of C2 domains and proteins in general.
Another biochemical property that is intrinsically related to phospholipid binding and could underlie a dominant role for the Syt1 C2B domain is its ability to bring two membranes into close proximity in a Ca2+-dependent manner (20, 22), which arises because of binding of its Ca2+-binding loops at the top of the β-sandwich to one membrane and interactions of R398 and R399 at the bottom of the sandwich with another membrane, and may also be aided by its basic nature (#basic − #acidic residues = 5). However, there was a priori no clear reason why this property might have switched to the C2A domain for Syt7. Thus, although the Syt7 C2A domain (#basic − #acidic residues = 4) is more basic than the Syt1 C2A domain (#basic − #acidic residues = −2), the Syt7 C2B domain is even more basic (#basic − #acidic residues = 10) and also contains arginines at the bottom of the β-sandwich. Nevertheless, our DLS measurements (Fig. 4 D–F) show that the Syt7 C2A domain does have a much stronger activity in bridging two membranes than the Syt7 C2B domain. The sequence determinants for this unexpected activity of the Syt7 C2A domain are currently unclear, but the switch in this property, which is characteristic of the C2B domain in Syt1 and of the C2A domain in Syt7, provides a correlation with the critical importance of Ca2+ binding to the Syt1 C2B domain for synchronous release and of Ca2+ binding to the Syt7 C2A domain for asynchronous release. Because Ca2+-dependent bridging of the two membranes could be crucial to trigger Ca2+-dependent fusion, in cooperation with the SNAREs, it is tempting to speculate that this activity is key to determine the relative functional importance of Ca2+ binding to the Syt1 and Syt7 C2 domains. However, it is worth noting that there are other mechanisms by which Ca2+-dependent membrane binding could help to cause membrane fusion, for example by creating tension in the membranes (1), perturbing the packing of the bilayers, or inducing membrane curvature (20, 49).
The notion that membrane affinity or the ability to bridge two membranes might determine the relative contributions of the C2 domains to Syt function does not imply that other interactions of Syts, such as those with PIP2 or the SNARE complex, are not important. However, it is worth noting that the primary regions that have been implicated in these interactions [the polybasic region and one face of the β-sandwich of the Syt1 C2B domain (23–26)] do not include the Ca2+-binding regions, even though Ca2+ enhances these interactions. At the high local concentrations of these reagents existing in the primed state of a synaptic vesicle, PIP2 or SNARE complex binding to the C2B domain (of either Syt1 or Syt7) could occur in the absence of Ca2+ and could help setting up Ca2+-dependent membrane fusion, but the key action of Ca2+ binding to Syts that triggers neurotransmitter release is most likely the stimulation of interactions of the Syt Ca2+-binding loops with one or both membranes. Binding of Syt1 to the SNARE complex could facilitate this action, as SNARE complex and membrane binding to Syt1 cooperate with each other (50), but the nature of such cooperation remains unclear given the diversity of Syt1–SNARE complex binding modes observed in recently reported structures (23, 26). Moreover, the interactions observed in these structures involve primarily the Syt1 C2B domain and the key residues involved in binding are not conserved in the Syt7 C2A domain. Hence, the mechanisms of coupling of Syt1 and Syt7 with the SNAREs may be more complex than currently envisioned.
Materials and Methods
Recombinant Proteins.
N-terminal GST fusion proteins of rat Syt7 fragments 134–262 (C2A domain), 263–403 (C2B domain), and 134–403 (C2AB fragment) were expressed in Escherichia coli BL21 (DE3) cells. The bacteria were grown in Lennox L Broth media at 25 °C for 16 h and induced with 0.5 mM isopropyl β-d-1-thiogalactopyranoside. Cells were resuspended in a buffer containing 40 mM Tris⋅HCl (pH 8.0), 1 M NaCl, 2 mM DTT, 1% Triton and Sigma protease inhibitor mixture (P2714-1BTL), and lysed using an Avestin EmulsiFlex-C5 homogenizer. The soluble fraction of the cell lysate was collected after centrifugation at 48,000 × g for 30 min and incubated with 100 mg protamine sulfate (Sigma-Aldrich) per 35 mL supernatant for 1 h at room temperature. The mixture was spun again at 48,000 × g for 30 min and the soluble fraction was incubated with Glutathione Sepharose 4B (GE Healthcare) at 4 °C for 16 h. The resin was washed with buffer containing 40 mM Tris⋅HCl (pH 8.0) and 200 mM NaCl (buffer A), buffer A + 50 mM CaCl2, and buffer A + 50 mM CaCl2 + 1 M NaCl. Remaining nucleic acid contaminants bound to synaptotagmin fragments were then cleared with benzonase treatment (40 units per milliliter of solution, corresponding to ∼1,000 units per liter of cell culture) in 50 mM Tris (pH 8.0), 2 mM MgCl2 for 2 h at room temperature with gentle rotation of the beads. Thrombin cleavage of the GST tag was carried out at room temperature for 3 h in 10 mL thrombin cleavage buffer (50 mM Tris⋅HCl pH 8.0, 150 mM NaCl, 2.5 mM CaCl2) and 0.08 mg/mL thrombin (Sigma-Aldrich). Eighteen residues from the GST tag (SPGISGGGGGILDSMGRL) remained at the N terminus of Syt7 fragments after thrombin cleavage. Syt7 fragments were further purified with an ion exchange Source S column [buffers: 50 mM NaAc (pH 6.2), 5 mM CaCl2; and 50 mM NaAc (pH 6.2), 5 mM CaCl2, 1 M NaCl]. All proteins were further purified using size-exclusion chromatography on a Superdex 75 16/60 column using buffer containing 20 mM Hepes (pH 7.4), 125 mM KCl, and 1 mM TCEP for Syt7 C2A and C2B domains, and buffer containing 20 mM Hepes (pH 7.4), 500 mM NaCl, 1 mM TCEP for the Syt7 C2AB fragment. Following chromatography, the Syt7 C2AB fragment was buffer exchanged into 20 mM Hepes (pH 7.4), 125 mM KCl, and 1 mM TCEP by serial concentration and dilution. Syt1 C2A domain, C2B domain and C2AB fragment were purified as described previously (15, 20, 51, 52). All mutations were performed using the QuikChange site-directed mutagenesis kit (Stratagene). These included the cysteine mutations indicated in the text and the following mutations to disrupt the Ca2+-binding sites: for Syt7 C2A, D225A,D227A,D233A; for Syt7 C2B, D357A,D359A; for Syt1 C2A, D178A,D230A,D232A; for Syt1 C2B, D309A,D363A,D365A.
Crystallization and X-Ray Data Collection.
Purified Syt7 C2A domain and C2AB fragment were concentrated to 20 mg/mL in a 20 mM Hepes (pH 7.4), 125 mM KCl, 1 mM CaCl2 buffer, and used for crystal screening. Crystals of Syt7 C2A domain were grown in 21% t-butanol, 0.1 M Tris (pH 8.5), 0.1 M CaCl2 using the hanging-drop method at 20 °C, and the crystals were cryoprotected by transferring to a final solution of 15% (vol/vol) glycerol, 21% t-butanol, 0.1 M Tris (pH 8.5), 0.1 M CaCl2, then flash-cooled in liquid nitrogen. Crystals of Syt7 C2AB fragment were grown in 20% PEG 3350, 0.2 M Li2SO4, 0.1 M Bis⋅Tris (pH 6.0), 0.1 M CaCl2 using the hanging-drop method at 20 °C, and the crystals were cryoprotected by transferring to a final solution of 20% (vol/vol) ethylene glycol, 22% PEG 3350, 0.2 M Li2SO4, 0.1 M Bis⋅Tris (pH 6.0), 0.1 M CaCl2.
Data were collected at the APS beamline 19-ID (SBC-CAT) at the Advanced Photon Source (Argonne National Laboratory, Argonne, IL) at 100 K, and were indexed, integrated and scaled using HKL-3000 (53), with applied corrections for effects resulting from absorption in a crystal and for radiation damage (54, 55), the calculation of an optimal error model, and corrections to compensate the phasing signal for a radiation-induced increase of nonisomorphism within the crystal (56, 57). Syt7 C2A crystals exhibited the symmetry of space group P65 with unit cell parameters of a = 55.30 Å, c = 89.81 Å, and contained one molecule of Syt7 C2A per asymmetric unit, with a solvent content of 50%. Syt7 C2AB fragment crystals belonged to space group P21 with unit cell parameters of a = 60.64 Å, b = 75.06 Å, c = 70.98 Å, β = 114.5°, and contained two molecules each of Syt7 C2AB fragment per asymmetric unit, with a solvent content of 45%. Syt7 C2A crystals diffracted isotropically to a dmin of 1.70 Å when exposed to synchrotron radiation. Syt7 C2AB fragment crystals diffracted anisotropically to a dmin of 2.25 Å when exposed to synchrotron radiation, and displayed strong translational noncrystallographic symmetry characteristic of a pseudo B-centered lattice, as evidenced by a peak in the Patterson function at approximately (1/2, 0, 1/2) with a height ≈49.9% of the origin peak. This resulted in numerous systematically weak reflections in the diffraction pattern, namely for the following reflection conditions: hkl, h + l = 2n + 1; hk0, h = 2n + 1; 0kl, l = 2n + 1; h0l, h + l = 2n + 1; 00l, l = 2n + 1; and h00, h = 2n + 1. Data collection statistics are provided in Table S1.
Phase Determination and Structure Refinement.
Phases for the Syt7 C2A domain were obtained via molecular replacement in the program Phaser (58) using a search model derived from the NMR structure of the Ca2+-free Syt7 C2A domain (PDB ID code 2D8K). Phases for the Syt7 C2AB fragment were obtained via molecular replacement in the program Phaser using search models derived from the structure of the Syt7 C2A domain described herein and the crystal structure of the Syt7 C2B domain (PDB ID code 3N5A) (29). Additional protein residues and calcium ions were manually modeled into the electron density maps via the program COOT (59). Refinement was performed using the program Phenix (60) with a random 5% of all data set aside for an Rfree calculation. The final model for Syt7 C2A (Rwork = 14.4%, Rfree = 18.1%) contained 138 residues and three Ca2+ ions. The final model for Syt7 C2AB fragment (Rwork = 21.4%, Rfree = 25.6%) contained 262 residues in chain A, 263 residues in chain B, and 8 Ca2+ ions, or 2 Ca2+ ions per C2 domain. The presence of the pseudocentering operator in the Syt7 C2AB fragment lattice translates into a systematic modulation of the observed intensities (61, 62) and thus resulted in lower than 100% completeness in the highest resolution shells between 2.70 and 2.25 Å. The L-value twinning test (63), which is not influenced by the presence of pseudocentering, shows no significant deviation from the expected values for untwinned data. Ramachandran plots generated with MolProbity (64) indicated no outliers in the Ramachandran plot for Syt7 fragments. Data collection and structure refinement statistics are summarized in Table S1. Coordinates and structure factors have been deposited in the Protein Data Bank with ID codes 6ANJ (Syt7 C2A domain) and 6ANK (Syt7 C2AB fragment).
Isothermal Titration Calorimetry.
The protein solutions were dialyzed twice against the ITC buffer (20 mM Hepes, pH 7.4, 125 mM KCl, 1 mM TCEP). The buffer was pretreated with Chelex-100 (Bio-Rad; calcium binding constant ≈4.6 × 103 M−1) to remove residual calcium ions bound with moderate affinity. The Chelex-100 beads were first washed with water and then added directly to the dialysis buffer for 2 h. The Chelex-100 beads were removed by filtration. The buffer pH was adjusted to 7.4 after filtration. ITC experiments were performed using a VP-ITC system (MicroCal) at 37 °C in ITC buffer. The protein solution (200 µM) was loaded into the sample cell, and CaCl2 solution (20 mM for the Syt7 C2A domain and 40 mM for the Syt7 C2B domain) was loaded in the syringe. The CaCl2 solution was prepared by diluting a 1 M stock solution with ITC buffer to the appropriate concentration. CaCl2 was injected at 3-μL volumes 100 times, and the heat evolved per injection was measured. All ITC data were analyzed using the Microcal Origin ITC software packet.
Labeling Proteins with BODIPY-FL Maleimide.
Purified proteins at a concentration of 100 µM were incubated with 1 mM BODIPY FL N-(2-aminoethyl) maleimide (Molecular Probes) for 2 h at room temperature or for 16 h at 4 °C with rotation. The reaction was quenched by adding 10 mM DTT to the mixture. The unlabeled dye was separated from the labeled protein by ion exchange chromatography on a Source S column (GE Healthcare) [buffers: 50 mM NaAc (pH 6.2), 5 mM CaCl2; and 50 mM NaAc (pH 6.2), 5 mM CaCl2, 1 M NaCl].
Preparation of Phospholipid Vesicles.
For preparation of phospholipid vesicles, 1,2-dioleoyl-sn-glycero-3-[phospho-L-serine] (DOPS), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), Cholesterol, PIP2, l-α-phosphatidylinositol (PI), and 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N- (lissamine rhodamine B sulfonyl) (ammonium salt; Liss Rho PE; Avanti Polar Lipids) in chloroform were mixed in a glass test tube in a desired ratio, and chloroform was evaporated using a dry nitrogen stream. The lipids were placed in a vacuum chamber overnight for complete removal of organic solvent. Lipid films were hydrated with 20 mM Hepes (pH 7.4) 150 mM KCl buffer in an appropriate volume yielding 10 mM lipids. Lipids were vortexed for >5 min, then frozen and thawed five times. Large unilamellar vesicles were prepared by extruding the hydrated lipid solution through 80 nm polycarbonate membranes 23 times using an Avanti Mini-Extruder. For most FRET assays, we used liposomes containing 41% POPC, 31% POPE, 1% Liss Rho PE, 1% cholesterol, 12% DOPS, and 5% PI. PIP2-containing liposomes were made by mixing 40% POPC, 31% POPE, 1% Liss Rho PE, 1% cholesterol, 12% DOPS, 1% PIP2, and 5% PI.
Phospholipid Binding Assays.
The FRET experiments were performed at 22 °C on a PTI spectrofluorimeter. All of the experiments were carried out in a buffer containing 20 mM Hepes (pH 7.4) and 150 mM KCl. The labeled protein (20 nM in case of Syt7 fragments and 100 nM in the case of Syt1 fragments, unless otherwise indicated) was mixed with variable amounts of liposomes and 1 mM CaCl2. Note that, although the intrinsic Ca2+ affinities of the Syt1 and Syt7 C2 domains are weak, the apparent Ca2+ affinities in the presence of phospholipids are much higher (6, 40), and hence 1 mM Ca2+ is sufficient for Ca2+ saturation. These samples were excited at 485 nm and the emission spectra from 500 to 600 nm were acquired. A fluorescence spectrum of the same sample after the addition of 2 mM EDTA was also collected. To correct for the bleed-through from Rhodamine at the emission maxima of BODIPY-FL, a spectrum of liposomes alone was collected and subtracted from the corresponding spectrum acquired in the presence of the protein. The FRET efficiency (E) was calculated with the formula: E = (IEDTA − ICa)/IEDTA, where ICa is the fluorescence intensity of the protein-liposome sample at the emission maxima of BODIPY-FL (512 nm) in the presence of 1 mM Ca2+ and IEDTA is the fluorescence intensity of the protein-liposome sample at 512 nm after the addition of 2 mM EDTA. The binding data were fitted to a Hill function using GraphPad PRISM7.
Dynamic Light Scattering.
The clustering ability of Syt7 C2A and C2B domains was measured by DLS using a DynaPro (Wyatt Technology) instrument equipped with a temperature controlled Microsampler. Liposomes containing 70% POPC and 30% DOPS were used for the DLS experiments. Samples were prepared by mixing 100 μM liposomes and 2 μM protein in 20 mM Hepes, pH 7.4, 125 mM KCl buffer. DLS measurements were made at 22 °C.
NMR Spectroscopy.
1H-15N HSQC spectra were acquired at 25 °C on Agilent DD2 spectrometers operating at 800 or 600 MHz. Samples contained 30–50 μM uniformly 15N-labeled proteins dissolved in buffer with 20 mM Hepes (pH 7.4), 125 mM KCl containing 5% D2O. All data were processed with NMRpipe (65) and analyzed with NMRView (66).
Acknowledgments
We thank Jefferson Knight for fruitful discussions and valuable comments on the mansucript; Thomas Scheuermann for assistance in acquiring the ITC data; and Junjie Xu for help fitting the ITC data. The structures in this report are derived from work performed on beamline 19-ID at the Argonne National Laboratory, Structural Biology Center at the Advanced Photon Source, operated by University of Chicago Argonne, LLC, for the US Department of Energy, Office of Biological and Environmental Research under Contract DE-AC02-06CH11357. The Agilent DD2 consoles of the 800-MHz spectromenter and one of the 600-MHz spectrometers used for the research presented here were purchased with shared instrumentation grants from the NIH: Grants S10OD018027 (to J.R.) and S10RR026461 (to Michael K. Rosen). R.V. was supported by a fellowship from the Howard Hughes Medical Institute. This work was supported by Grant I-1304 from the Welch Foundation (to J.R.) and by NIH Research Project Award R35 NS097333 (to J.R.), which continues work performed under NIH Grants NS037200 and NS049044 (to J.R.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org [PDB ID codes 6ANJ (Syt7 C2A domain) and 6ANK (Syt7 C2AB fragment)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1710708114/-/DCSupplemental.
References
- 1.Südhof TC. Neurotransmitter release: The last millisecond in the life of a synaptic vesicle. Neuron. 2013;80:675–690. doi: 10.1016/j.neuron.2013.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Südhof TC, Rothman JE. Membrane fusion: Grappling with SNARE and SM proteins. Science. 2009;323:474–477. doi: 10.1126/science.1161748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rizo J, Xu J. The synaptic vesicle release machinery. Annu Rev Biophys. 2015;44:339–367. doi: 10.1146/annurev-biophys-060414-034057. [DOI] [PubMed] [Google Scholar]
- 4.Brunger AT, Cipriano DJ, Diao J. Towards reconstitution of membrane fusion mediated by SNAREs and other synaptic proteins. Crit Rev Biochem Mol Biol. 2015;50:231–241. doi: 10.3109/10409238.2015.1023252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Südhof TC. Synaptotagmins: Why so many? J Biol Chem. 2002;277:7629–7632. doi: 10.1074/jbc.R100052200. [DOI] [PubMed] [Google Scholar]
- 6.Fernández-Chacón R, et al. Synaptotagmin I functions as a calcium regulator of release probability. Nature. 2001;410:41–49. doi: 10.1038/35065004. [DOI] [PubMed] [Google Scholar]
- 7.Pang ZP, Sun J, Rizo J, Maximov A, Südhof TC. Genetic analysis of synaptotagmin 2 in spontaneous and Ca2+-triggered neurotransmitter release. EMBO J. 2006;25:2039–2050. doi: 10.1038/sj.emboj.7601103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu J, Mashimo T, Südhof TC. Synaptotagmin-1, -2, and -9: Ca(2+) sensors for fast release that specify distinct presynaptic properties in subsets of neurons. Neuron. 2007;54:567–581. doi: 10.1016/j.neuron.2007.05.004. [DOI] [PubMed] [Google Scholar]
- 9.Bacaj T, et al. Synaptotagmin-1 and synaptotagmin-7 trigger synchronous and asynchronous phases of neurotransmitter release. Neuron. 2013;80:947–959. doi: 10.1016/j.neuron.2013.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma C, Su L, Seven AB, Xu Y, Rizo J. Reconstitution of the vital functions of Munc18 and Munc13 in neurotransmitter release. Science. 2013;339:421–425. doi: 10.1126/science.1230473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sutton RB, Davletov BA, Berghuis AM, Südhof TC, Sprang SR. Structure of the first C2 domain of synaptotagmin I: A novel Ca2+/phospholipid-binding fold. Cell. 1995;80:929–938. doi: 10.1016/0092-8674(95)90296-1. [DOI] [PubMed] [Google Scholar]
- 12.Shao X, Fernandez I, Südhof TC, Rizo J. Solution structures of the Ca2+-free and Ca2+-bound C2A domain of synaptotagmin I: Does Ca2+ induce a conformational change? Biochemistry. 1998;37:16106–16115. doi: 10.1021/bi981789h. [DOI] [PubMed] [Google Scholar]
- 13.Fernandez I, et al. Three-dimensional structure of the synaptotagmin 1 C2B-domain: Synaptotagmin 1 as a phospholipid binding machine. Neuron. 2001;32:1057–1069. doi: 10.1016/s0896-6273(01)00548-7. [DOI] [PubMed] [Google Scholar]
- 14.Shao X, Davletov BA, Sutton RB, Südhof TC, Rizo J. Bipartite Ca2+-binding motif in C2 domains of synaptotagmin and protein kinase C. Science. 1996;273:248–251. doi: 10.1126/science.273.5272.248. [DOI] [PubMed] [Google Scholar]
- 15.Ubach J, Zhang X, Shao X, Südhof TC, Rizo J. Ca2+ binding to synaptotagmin: How many Ca2+ ions bind to the tip of a C2-domain? EMBO J. 1998;17:3921–3930. doi: 10.1093/emboj/17.14.3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shao X, et al. Synaptotagmin-syntaxin interaction: The C2 domain as a Ca2+-dependent electrostatic switch. Neuron. 1997;18:133–142. doi: 10.1016/s0896-6273(01)80052-0. [DOI] [PubMed] [Google Scholar]
- 17.Zhang X, Rizo J, Südhof TC. Mechanism of phospholipid binding by the C2A-domain of synaptotagmin I. Biochemistry. 1998;37:12395–12403. doi: 10.1021/bi9807512. [DOI] [PubMed] [Google Scholar]
- 18.Chapman ER, Davis AF. Direct interaction of a Ca2+-binding loop of synaptotagmin with lipid bilayers. J Biol Chem. 1998;273:13995–14001. doi: 10.1074/jbc.273.22.13995. [DOI] [PubMed] [Google Scholar]
- 19.Rhee JS, et al. Augmenting neurotransmitter release by enhancing the apparent Ca2+ affinity of synaptotagmin 1. Proc Natl Acad Sci USA. 2005;102:18664–18669. doi: 10.1073/pnas.0509153102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Araç D, et al. Close membrane-membrane proximity induced by Ca(2+)-dependent multivalent binding of synaptotagmin-1 to phospholipids. Nat Struct Mol Biol. 2006;13:209–217. doi: 10.1038/nsmb1056. [DOI] [PubMed] [Google Scholar]
- 21.Xue M, Ma C, Craig TK, Rosenmund C, Rizo J. The Janus-faced nature of the C(2)B domain is fundamental for synaptotagmin-1 function. Nat Struct Mol Biol. 2008;15:1160–1168. doi: 10.1038/nsmb.1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seven AB, Brewer KD, Shi L, Jiang QX, Rizo J. Prevalent mechanism of membrane bridging by synaptotagmin-1. Proc Natl Acad Sci USA. 2013;110:E3243–E3252. doi: 10.1073/pnas.1310327110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brewer KD, et al. Dynamic binding mode of a synaptotagmin-1-SNARE complex in solution. Nat Struct Mol Biol. 2015;22:555–564. doi: 10.1038/nsmb.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bai J, Tucker WC, Chapman ER. PIP2 increases the speed of response of synaptotagmin and steers its membrane-penetration activity toward the plasma membrane. Nat Struct Mol Biol. 2004;11:36–44. doi: 10.1038/nsmb709. [DOI] [PubMed] [Google Scholar]
- 25.Li L, et al. Phosphatidylinositol phosphates as co-activators of Ca2+ binding to C2 domains of synaptotagmin 1. J Biol Chem. 2006;281:15845–15852. doi: 10.1074/jbc.M600888200. [DOI] [PubMed] [Google Scholar]
- 26.Zhou Q, et al. Architecture of the synaptotagmin-SNARE machinery for neuronal exocytosis. Nature. 2015;525:62–67. doi: 10.1038/nature14975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dai H, et al. Structural basis for the evolutionary inactivation of Ca2+ binding to synaptotagmin 4. Nat Struct Mol Biol. 2004;11:844–849. doi: 10.1038/nsmb817. [DOI] [PubMed] [Google Scholar]
- 28.Schonn JS, Maximov A, Lao Y, Südhof TC, Sørensen JB. Synaptotagmin-1 and -7 are functionally overlapping Ca2+ sensors for exocytosis in adrenal chromaffin cells. Proc Natl Acad Sci USA. 2008;105:3998–4003. doi: 10.1073/pnas.0712373105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xue M, et al. Structural and mutational analysis of functional differentiation between synaptotagmins-1 and -7. PLoS One. 2010;5:e12544. doi: 10.1371/journal.pone.0012544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mackler JM, Drummond JA, Loewen CA, Robinson IM, Reist NE. The C(2)B Ca(2+)-binding motif of synaptotagmin is required for synaptic transmission in vivo. Nature. 2002;418:340–344. doi: 10.1038/nature00846. [DOI] [PubMed] [Google Scholar]
- 31.Robinson IM, Ranjan R, Schwarz TL. Synaptotagmins I and IV promote transmitter release independently of Ca(2+) binding in the C(2)A domain. Nature. 2002;418:336–340. doi: 10.1038/nature00915. [DOI] [PubMed] [Google Scholar]
- 32.Nishiki T, Augustine GJ. Dual roles of the C2B domain of synaptotagmin I in synchronizing Ca2+-dependent neurotransmitter release. J Neurosci. 2004;24:8542–8550. doi: 10.1523/JNEUROSCI.2545-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shin OH, Xu J, Rizo J, Südhof TC. Differential but convergent functions of Ca2+ binding to synaptotagmin-1 C2 domains mediate neurotransmitter release. Proc Natl Acad Sci USA. 2009;106:16469–16474. doi: 10.1073/pnas.0908798106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu D, et al. Postsynaptic synaptotagmins mediate AMPA receptor exocytosis during LTP. Nature. 2017;544:316–321. doi: 10.1038/nature21720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rizo J, Südhof TC. C2-domains, structure and function of a universal Ca2+-binding domain. J Biol Chem. 1998;273:15879–15882. doi: 10.1074/jbc.273.26.15879. [DOI] [PubMed] [Google Scholar]
- 36.Maximov A, et al. Genetic analysis of synaptotagmin-7 function in synaptic vesicle exocytosis. Proc Natl Acad Sci USA. 2008;105:3986–3991. doi: 10.1073/pnas.0712372105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fuson KL, Montes M, Robert JJ, Sutton RB. Structure of human synaptotagmin 1 C2AB in the absence of Ca2+ reveals a novel domain association. Biochemistry. 2007;46:13041–13048. doi: 10.1021/bi701651k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sutton RB, Ernst JA, Brunger AT. Crystal structure of the cytosolic C2A-C2B domains of synaptotagmin III. Implications for Ca(+2)-independent snare complex interaction. J Cell Biol. 1999;147:589–598. doi: 10.1083/jcb.147.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Radhakrishnan A, Stein A, Jahn R, Fasshauer D. The Ca2+ affinity of synaptotagmin 1 is markedly increased by a specific interaction of its C2B domain with phosphatidylinositol 4,5-bisphosphate. J Biol Chem. 2009;284:25749–25760. doi: 10.1074/jbc.M109.042499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shin OH, Rizo J, Südhof TC. Synaptotagmin function in dense core vesicle exocytosis studied in cracked PC12 cells. Nat Neurosci. 2002;5:649–656. doi: 10.1038/nn869. [DOI] [PubMed] [Google Scholar]
- 41.Brandt DS, Coffman MD, Falke JJ, Knight JD. Hydrophobic contributions to the membrane docking of synaptotagmin 7 C2A domain: Mechanistic contrast between isoforms 1 and 7. Biochemistry. 2012;51:7654–7664. doi: 10.1021/bi3007115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hui E, et al. Three distinct kinetic groupings of the synaptotagmin family: Candidate sensors for rapid and delayed exocytosis. Proc Natl Acad Sci USA. 2005;102:5210–5214. doi: 10.1073/pnas.0500941102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Frazier AA, Roller CR, Havelka JJ, Hinderliter A, Cafiso DS. Membrane-bound orientation and position of the synaptotagmin I C2A domain by site-directed spin labeling. Biochemistry. 2003;42:96–105. doi: 10.1021/bi0268145. [DOI] [PubMed] [Google Scholar]
- 44.Rufener E, Frazier AA, Wieser CM, Hinderliter A, Cafiso DS. Membrane-bound orientation and position of the synaptotagmin C2B domain determined by site-directed spin labeling. Biochemistry. 2005;44:18–28. doi: 10.1021/bi048370d. [DOI] [PubMed] [Google Scholar]
- 45.Rizo J, Rosenmund C. Synaptic vesicle fusion. Nat Struct Mol Biol. 2008;15:665–674. doi: 10.1038/nsmb.1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chapman ER. How does synaptotagmin trigger neurotransmitter release? Annu Rev Biochem. 2008;77:615–641. doi: 10.1146/annurev.biochem.77.062005.101135. [DOI] [PubMed] [Google Scholar]
- 47.van den BG, Meyenberg K, Diederichsen U, Jahn R. Phosphatidylinositol 4,5-bisphosphate increases Ca2+ affinity of synaptotagmin-1 by 40-fold. J Biol Chem. 2012;287:16447–16453. doi: 10.1074/jbc.M112.343418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tang J, et al. A complexin/synaptotagmin 1 switch controls fast synaptic vesicle exocytosis. Cell. 2006;126:1175–1187. doi: 10.1016/j.cell.2006.08.030. [DOI] [PubMed] [Google Scholar]
- 49.Martens S, Kozlov MM, McMahon HT. How synaptotagmin promotes membrane fusion. Science. 2007;316:1205–1208. doi: 10.1126/science.1142614. [DOI] [PubMed] [Google Scholar]
- 50.Dai H, Shen N, Araç D, Rizo J. A quaternary SNARE-synaptotagmin-Ca2+-phospholipid complex in neurotransmitter release. J Mol Biol. 2007;367:848–863. doi: 10.1016/j.jmb.2007.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ubach J, et al. The C2B domain of synaptotagmin I is a Ca2+-binding module. Biochemistry. 2001;40:5854–5860. doi: 10.1021/bi010340c. [DOI] [PubMed] [Google Scholar]
- 52.Zhou A, Brewer KD, Rizo J. Analysis of SNARE complex/synaptotagmin-1 interactions by one-dimensional NMR spectroscopy. Biochemistry. 2013;52:3446–3456. doi: 10.1021/bi400230u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: The integration of data reduction and structure solution—From diffraction images to an initial model in minutes. Acta Crystallogr D Biol Crystallogr. 2006;62:859–866. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- 54.Borek D, Minor W, Otwinowski Z. Measurement errors and their consequences in protein crystallography. Acta Crystallogr D Biol Crystallogr. 2003;59:2031–2038. doi: 10.1107/s0907444903020924. [DOI] [PubMed] [Google Scholar]
- 55.Otwinowski Z, Borek D, Majewski W, Minor W. Multiparametric scaling of diffraction intensities. Acta Crystallogr A. 2003;59:228–234. doi: 10.1107/s0108767303005488. [DOI] [PubMed] [Google Scholar]
- 56.Borek D, Cymborowski M, Machius M, Minor W, Otwinowski Z. Diffraction data analysis in the presence of radiation damage. Acta Crystallogr D Biol Crystallogr. 2010;66:426–436. doi: 10.1107/S0907444909040177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Borek D, Dauter Z, Otwinowski Z. Identification of patterns in diffraction intensities affected by radiation exposure. J Synchrotron Radiat. 2013;20:37–48. doi: 10.1107/S0909049512048807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 60.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zwart PH, Grosse-Kunstleve RW, Lebedev AA, Murshudov GN, Adams PD. Surprises and pitfalls arising from (pseudo)symmetry. Acta Crystallogr D Biol Crystallogr. 2008;64:99–107. doi: 10.1107/S090744490705531X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chook YM, Lipscomb WN, Ke H. Detection and use of pseudo-translation in determination of protein structures. Acta Crystallogr D Biol Crystallogr. 1998;54:822–827. doi: 10.1107/s0907444997020064. [DOI] [PubMed] [Google Scholar]
- 63.Padilla JE, Yeates TO. A statistic for local intensity differences: Robustness to anisotropy and pseudo-centering and utility for detecting twinning. Acta Crystallogr D Biol Crystallogr. 2003;59:1124–1130. doi: 10.1107/s0907444903007947. [DOI] [PubMed] [Google Scholar]
- 64.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Delaglio F, et al. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 66.Johnson BA, Blevins RA. NMR View: A computer program for the visualization and analysis of NMR data. J Biomol NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]