Significance
Studies using experimental models have indicated that multiple sclerosis (MS)-like disease can be triggered in the gut following interactions of brain autoimmune T lymphocytes with local microbiota. Here we studied the gut microbiota from monozygotic human twin pairs discordant for multiple sclerosis. When we transferred human-derived microbiota into transgenic mice expressing a myelin autoantigen-specific T cell receptor, we found that gut microbiota from multiple sclerosis-affected twins induced CNS-specific autoimmunity at a higher incidence than microbiota from healthy co-twins. Our results offer functional evidence that human microbiome components contribute to CNS-specific autoimmunity.
Keywords: gut microbiome, multiple sclerosis, experimental autoimmune encephalomyelitis, twin study, germ-free mice
Abstract
There is emerging evidence that the commensal microbiota has a role in the pathogenesis of multiple sclerosis (MS), a putative autoimmune disease of the CNS. Here, we compared the gut microbial composition of 34 monozygotic twin pairs discordant for MS. While there were no major differences in the overall microbial profiles, we found a significant increase in some taxa such as Akkermansia in untreated MS twins. Furthermore, most notably, when transplanted to a transgenic mouse model of spontaneous brain autoimmunity, MS twin-derived microbiota induced a significantly higher incidence of autoimmunity than the healthy twin-derived microbiota. The microbial profiles of the colonized mice showed a high intraindividual and remarkable temporal stability with several differences, including Sutterella, an organism shown to induce a protective immunoregulatory profile in vitro. Immune cells from mouse recipients of MS-twin samples produced less IL-10 than immune cells from mice colonized with healthy-twin samples. IL-10 may have a regulatory role in spontaneous CNS autoimmunity, as neutralization of the cytokine in mice colonized with healthy-twin fecal samples increased disease incidence. These findings provide evidence that MS-derived microbiota contain factors that precipitate an MS-like autoimmune disease in a transgenic mouse model. They hence encourage the detailed search for protective and pathogenic microbial components in human MS.
The risk of developing multiple sclerosis (MS) is driven by both genetic factors and environmental exposures (1). Risk genes have been determined by large-scale genome-wide association studies (GWAS), which identified more than 200 different DNA variants associated with disease susceptibility (2). Environmental risk factors include smoking, reduced exposure to sunlight, and infection with Epstein–Barr virus (3). Very recently, the intestinal microbiota emerged as an additional potential triggering factor (4, 5).
The notion that commensal gut bacteria are causally related to brain autoimmunity is supported by a transgenic mouse model of spontaneous experimental autoimmune encephalomyelitis (EAE). In this model, nearly all animals raised in specific pathogen-free (SPF) conditions develop a relapsing–remitting (RR) variant of the disease within months of age (6). Importantly, when kept in a germ-free environment, animals from the same strain remain disease-free. However, spontaneous disease promptly follows exposure of germ-free mice to SPF-derived fecal material (7). Taken together, these observations indicate that the encephalitogenic immune response observed in these mice is mediated by the intestinal microbiota.
Translation of these experimental observations into human MS poses considerable practical challenges. Complicating factors are, in particular, genetic diversity (8) and lifestyle, such as diet (9), both of which profoundly impact the individual gut microbiota. In addition, age, therapy, and neurological condition (10) might also affect the gut microbial composition. In an attempt to eliminate genetic variance and reduce environmental variance to a minimum, we identified and recruited 34 monozygotic (MZ) twin pairs, discordant for MS, for a microbiome study. All the probands were of Caucasian origin and had grown up together with their healthy twins to adulthood in Germany. We studied their gut microbiota in two tiers: first, intestinal microbial profiles of MS twins and healthy twins were compared by 16S ribosomal RNA (rRNA) amplicon and metagenomic shotgun sequencing. Second, we transplanted fecal samples from selected twin pairs to germ-free mice to assess functional differences in the human intestinal microbiota of MS and healthy twins.
Results
MZ Twin Cohorts Discordant for MS.
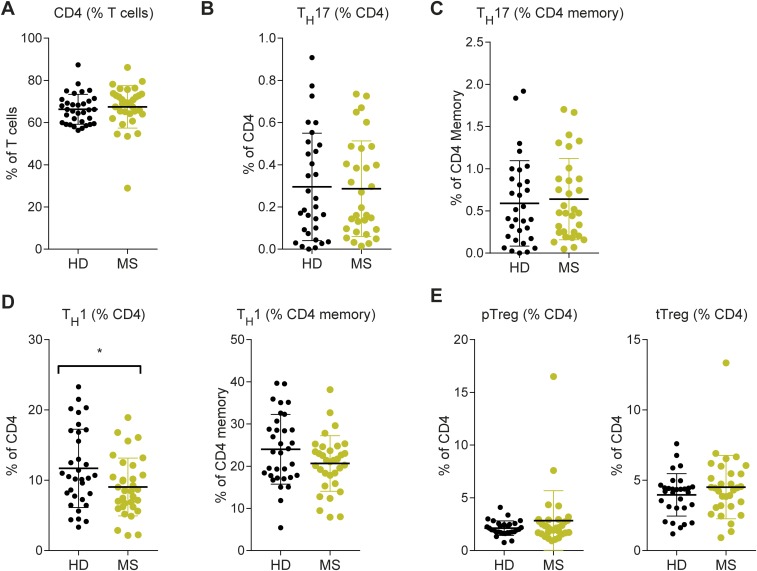
We assembled a cohort of 34 MZ twin pairs clinically discordant for MS. In each pair, one twin has clinically definite MS according to the current diagnostic criteria (11), whereas the co-twin is unaffected. Our MS twin cohort resembles the general MS population with respect to female preponderance, age distribution, age at onset, and distribution of clinical subtypes [RR MS, secondary progressive MS, primary progressive MS, and clinically isolated syndrome (CIS)] (Table 1) (12). The twin cohort is homogenous in terms of geographic environment and genetic background. Furthermore, immunophenotyping of peripheral blood mononuclear cells (PBMCs) from all twin pairs did not show relevant differences in the frequencies of CD4+ T cells, Th1 cells, Th17 cells, and Treg cells (Fig. S1).
Table 1.
Demographic and clinical characteristics of MZ twins discordant for MS
| Characteristic | MZ twins |
| No. of pairs | 34 |
| Gender, female/male | 26/8 |
| Age in years (range in years) | 41.3 ± 10.8 (21–63) |
| Age in years at disease onset (range in years) | 28.0 ± 9.0 (14–47) |
| Time in years clinically discordant for MS (range in years) | 13.2 ± 9.6 (1–33) |
| Pairs clinically discordant for MS longer than 10 y (%) | 18 (53) |
| Pairs with a positive family history of MS (%) | 11 (32) |
| MS type, n (%) | |
| CIS | 3 (9) |
| RR-MS | 22 (65) |
| SP-MS | 7 (21) |
| PP-MS | 2 (6) |
| Disease-modifying treatment, n (%) | |
| Untreated | 15 (44) |
| IFN-β | 13 (38) |
| Natalizumab | 4 (12) |
| Glatiramer acetate | 1 (3) |
| Azathioprine | 1 (3) |
Continuous data are expressed as mean ± SD. Categorical data are expressed as the number of observations (%). CIS, clinically isolated syndrome; PP-MS, primary progressive MS; RR-MS, relapsing–remitting MS; SP-MS, secondary progressive MS.
Fig. S1.
Immune-phenotyping of PBMCs from twin pairs did not reveal profound changes in T cell subset composition. Frozen PBMC specimens from all twin pairs (n = 34) were thawed and immediately subjected to 10-color flow cytometry for quantification of T cell subsets including CD4 T cells, Th17 cells (defined by simultaneous expression of CCR4+, CCR6+, CD161+, CD146+), Th17 memory cells, Th1 cells (defined as CCR4−, CCR6−, CD183+), Th1 memory cells, peripheral regulatory T cells (defined as FoxP3+ Helios−), and thymic regulatory T cells (defined as FoxP3+ Helios+). (A) CD4 T cells are depicted as percentage of T cells. (B–E) All other populations are depicted as percentage of CD4 T cells. Each dot represents an individual measurement, and horizontal lines and whiskers indicate mean ± SEM. *P < 0.05 (Mann–Whitney u test).
Microbial Profiling of MS-Discordant MZ Twins by 16S rRNA and Metagenomic Shotgun Sequencing.
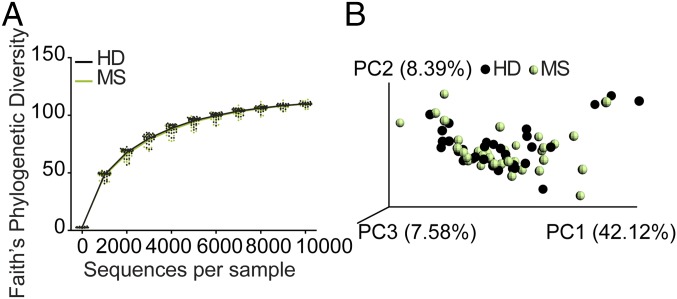
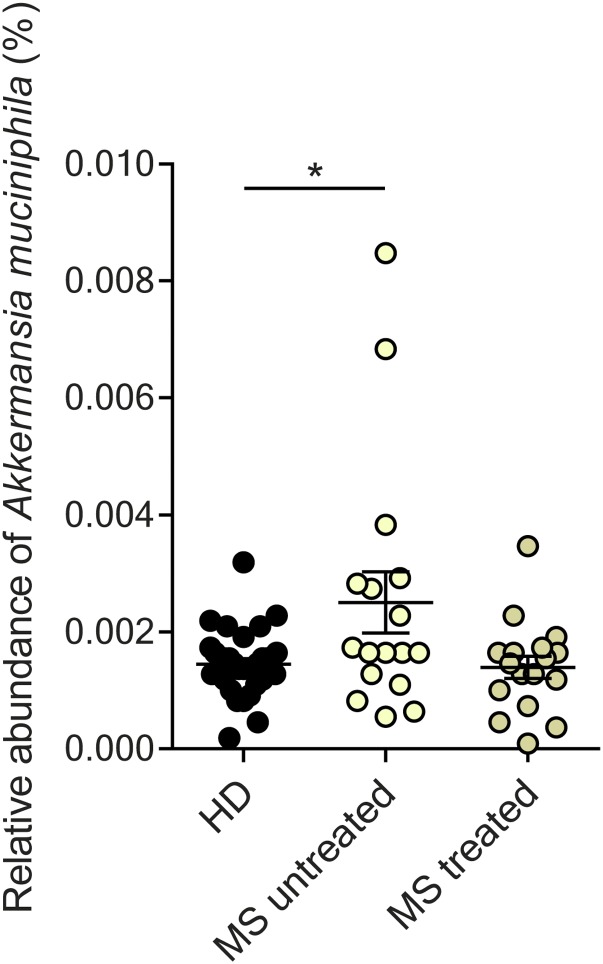
To search for differences between microbiomes of MS-affected versus nonaffected twins, we applied 16S rRNA sequencing and metagenomics shotgun sequencing. 16S rRNA amplicon sequencing revealed no major differences in microbial community structures. Specifically, MS twins and healthy twins exhibited comparable microbial community richness (alpha diversity) (Fig. 1A). A concomitant principal coordinate analysis (PCoA) also did not point to disease-dependent clustering of samples (Fig. 1B). Further, analysis at the level of individual taxa [up to genera or the operational taxonomic units (OTUs) level] did not indicate significant differences between MS and healthy twins. However, when patients were stratified for the use of disease-modifying therapy, several taxa (most notably Akkermansia muciniphila) were significantly increased in untreated MS twin siblings (Fig. S2).
Fig. 1.
No overt differences in alpha or beta diversity were detected by comparing the fecal microbial profiles of healthy twins with those of MS twins. (A) Phylogenetic (alpha) diversity of fecal microbiota in healthy twins (HD, black; n = 34) and MS twins (MS, green; n = 34). (B) PCoA of weighted UniFrac community distances for fecal microbiota of healthy twins (black; n = 34) and MS twins (green; n = 34).
Fig. S2.
Differential abundance of Akkermansia muciniphila in fecal samples of healthy twins (HD; n = 34) compared with co-twins with untreated MS (MS untreated; n = 17) or treated MS (MS treated; n = 17). Each dot represents an individual stool sample, and horizontal lines and whiskers indicate mean ± SEM. *P < 0.05 (Mann–Whitney u test).
Comparing the microbiome composition of MS and healthy twins is a first, purely descriptive step in exploring the role of the microbiota in MS. Disease-related functional differences in the microbiomes of healthy or MS twins might be revealed by shotgun metagenomic sequencing. Using Human Microbiome Project (HMP) Unified Metabolic Analysis Network (HUMAnN2) tool1, we assessed the microbial community, performed functional profiling on the bacterial DNA sequences, and identified 242 unique bacterial species across 98 genera, 1.2 million gene families, and 558 bacterial metabolic pathways. Interestingly, analysis of gut bacterial composition showed higher similarity between discordant twin siblings than among unrelated twin pairs (Fig. S3), thus confirming the influence of the host’s genetics in the composition of the gut microbiome. To determine possible associations of specific bacterial species, gene families, or metabolic pathways with MS, we performed logistic regression analysis. In harmony with 16S sequencing findings, after adjusting for twin pair, number of genome equivalents sequenced, and multiple comparisons, no specific bacterial species, gene families, or metabolic pathways were significantly associated with the MS disease phenotype.
Fig. S3.
Comparison of the metagenomic profiles of healthy and MS twins revealed a high similarity of the gut microbiomes between discordant twins. Pairwise analysis of gut microbial composition (sum of absolute differences in genus relative abundance between two individuals) shows that gut bacterial flora are more similar within twin pairs discordant for MS (discordant twins, n = 32) than within pairs of unrelated individuals with MS (unrelated cases; n = 16), unrelated healthy individuals (unrelated controls; n = 18) or unrelated healthy and MS individuals (unrelated individuals discordant phenotypes; n = 34).
RR Mice Colonized with Human MS Twin-Derived Microbiota.
To explore the functional role of human microbiota in CNS inflammation, we used our previously described spontaneous RR mouse model (6, 7). Several qualities render these mice a particularly promising reporter system. RR mice express a transgenic myelin oligodendrocyte glycoprotein (MOG)-specific T cell receptor in more than 70% of their CD4+ T cells. Beginning at 2 mo of age, more than 80% of the mice spontaneously develop an inflammatory demyelinating disease that recapitulates features of early human MS: The disease course is often relapsing–remitting, affects varying parts of the brain and spinal cord with lesions featuring round-cell infiltrates and large confluent areas of demyelination with axonal degeneration, and responds to B cell-depleting therapy (6). Most importantly, spontaneous disease critically depends on an intact commensal microbiome, as germ-free RR mice remain healthy (7).
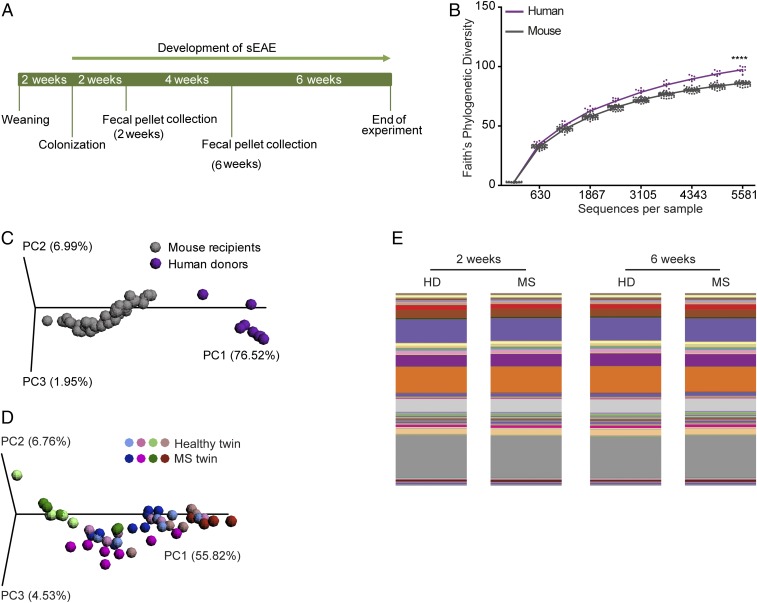
We reasoned that if the gut microbiota is a determinant factor contributing to the severity and course of human MS, transplantation of human fecal microbial communities from MS twins should result in a higher frequency of spontaneous EAE than the transfer of fecal material from healthy twins. We selected a subgroup of five twin pairs who were all discordant for RR MS and in which the affected twins had different disease durations and were either without therapy or were treated with IFN-β (for further details refer to Table S1). Each donor fecal sample was diluted in prereduced PBS and introduced into a group of 6-wk-old, germ-free RR mice by oral gavage. Mouse fecal pellets were collected 2 and 6 wk after transplantation to assess the stability as well as composition of the intestinal microbiota by 16S rRNA amplicon sequencing (Fig. 2A).
Table S1.
Clinical characteristics of the five MZ twin pairs discordant for MS used for colonization experiments
| Twin pair | Age in years | Gender | Disease duration in years | Disease course | EDSS | DMD |
| 1 | 40 | Female | 14 | RR-MS | 2.0 | None in 3 mo prior |
| 40 | Female | N.a. | N.a. | N.a. | N.a. | |
| 2 | 20 | Female | 6 | RR-MS | 2.0 | IFN-β |
| 20 | Female | N.a. | N.a. | N.a. | N.a. | |
| 3 | 21 | Female | 1 | RR-MS | 2.0 | IFN-β |
| 21 | Female | N.a. | N.a. | N.a. | N.a. | |
| 4 | 29 | Female | 15 | RR-MS | 4.5 | None in 3 mo prior |
| 29 | Female | N.a. | N.a. | N.a. | N.a. | |
| 5 | 40 | Female | 2 | RR-MS | 1.5 | IFN-β |
| 40 | Female | N.a. | N.a. | N.a. | N.a. |
DMD, disease-modifying drugs; EDSS, Expanded Disability Status Scale; N.a., not applicable; RR-MS, relapsing-remitting MS.
Fig. 2.
Human microbiota can be efficiently transferred to mouse recipients. (A) Design of humanized gnotobiotic mouse experiment: 6-wk-old, germ-free RR SJL/J mice were gavaged with fecal samples from the healthy twin or the MS twin from a MZ twin pair. Fecal samples for 16S rRNA sequencing analysis were harvested at 2 and 6 wk after colonization. Humanized gnotobiotic RR mice were observed for the development of clinical signs of EAE for 12 wk. (B) Phylogenetic (alpha) diversity of fecal microbiota in human twin pairs (purple; n = 8) and mouse recipients (gray; n = 47). ****P < 0.0001 (Mann–Whitney u test). (C) PCoA of weighted UniFrac community distances comparing fecal microbiota of human donors (purple; n = 8) and mouse recipients (gray; n = 47). (D) PCoA of weighted UniFrac community distances for fecal microbiota of mice transplanted with microbiota from healthy (n = 23) or MS (n = 24) twins. (E) Mean relative abundances of bacterial genera in fecal samples of mice that received microbiota of healthy donors (HD; n = 20–26) or MS patients (MS; n = 22–26) at 2 and 6 wk after transplantation.
Unsurprisingly, we detected significantly reduced alpha diversity (∼10%) in colonized mice compared with the donor humans (Fig. 2B). PCoA confirmed the divergent gut microbial community structures in human and mouse samples (Fig. 2C), indicating that only a subset of microbial species from human donors was able to colonize mouse recipients. The microbiota transfer rate was consistent across samples and independent of the disease state of the donor. Since the composition of the intestinal microbiota is profoundly imprinted by host genetics, diet, and other environmental factors (13), it was not surprising that the composition of gnotobiotic mouse fecal samples reflected incomplete colonization by the human donor microbiota. Similar changes were noted in previous human-to-mouse transplant experiments (14, 15). PCoA of mouse samples demonstrated clustering by donor and twin pair but not by EAE disease state (Fig. 2D). In addition, comparison of the percentage of microbial genera that was transferred from the human donor to recipient mice did not indicate any disease-based trends and highlighted a comparatively high mean transfer rate of 90.3% (Table S2). Finally, the recipient mouse microbiome remained highly stable between 2 and 6 wk after colonization, as indicated by representative bar plots of mean relative abundance of bacterial genera and corresponding analysis (Fig. 2E).
Table S2.
Efficiency of transfer of human microbiota to mouse recipients
| Donor | Transferred genera, n (%) | Nontransferred genera n (%) |
| HD | 122.5 (90.725) | 12.5 (9.275) |
| MS | 118.5 (89.95) | 13.25 (10.05) |
HD, healthy twins; MS, MS twins.
Increased Incidence of Spontaneous EAE in RR Mice Colonized with Microbiota from MS-Affected Twins.
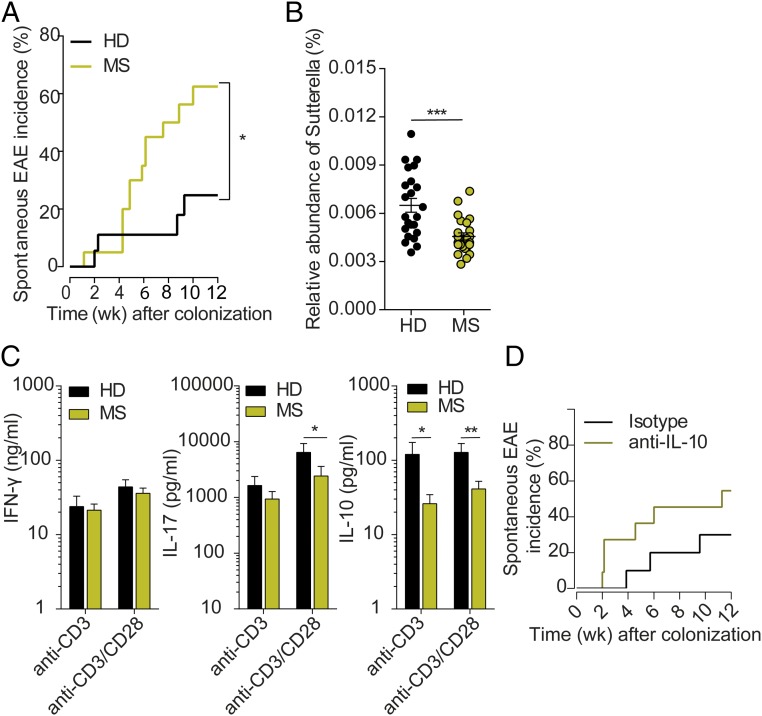
Remarkably, RR mice transplanted with MS patient-derived microbiota developed spontaneous EAE at a higher frequency than animals colonized with intestinal bacteria from healthy twins (Fig. 3A). The triggering of EAE in germ-free RR mice by human microbial transfer per se is notable, considering that so far this has been achieved only with some mouse-associated bacteria (7). However, monocolonization with experimental bacterial consortia (such as altered Schaedler flora) and segmented filamentous bacteria did not induce spontaneous EAE (7).
Fig. 3.
Intestinal microbiota plays a pivotal role in CNS-specific autoimmunity. (A) Incidence of spontaneous EAE in humanized gnotobiotic RR SJL/J mice. Germ-free RR SJL/J mice were gavaged with fecal material from healthy (HD; n = 18) or MS (MS; n = 20) twins using in total five MZ twin pairs. *P < 0.05 (Gehan–Breslow–Wilcoxon test). (B) Relative abundance of Sutterella in fecal samples of mice that received the microbiota of healthy donor (HD; n = 23) or an MS patient (MS; n = 24) using in total five MZ twin pairs. Each circle represents an individual stool sample, and the horizontal line and whiskers indicate mean ± SEM. ***P < 0.001 (Mann–Whitney u test). (C) Cytokine production by splenocytes from humanized gnotobiotic SJL/J mice. Spleen cells were stimulated for 72 h with 1 µg/mL anti-CD3 antibody or 1 µg/mL anti-CD3 and 0.5 µg/mL anti-CD28 antibody. Levels of IFN-γ, IL-17, and IL-10 in the supernatants were measured by ELISA. Bars display mean ± SEM; n = 12–16 mice per group. Data were pooled from three independent experiments. *P < 0.05; **P = 0.0069 (Mann–Whitney u test). (D) IL-10 neutralization increases spontaneous EAE in humanized gnotobiotic RR SJL/J mice. Germ-free RR SJL/J mice were gavaged with fecal material from healthy twins and were treated weekly with either isotype control antibodies (n = 10) or neutralizing anti–IL-10 antibodies (n = 11). Data were pooled from two independent experiments.
We next set out to determine the differences that made MS-derived microbiota more pathogenic than healthy-derived samples. Analysis of the mouse microbiome at the level of individual taxa revealed significant differences between MS- and healthy twin-colonized mice, both at the genus and OTU taxonomical levels. While the genera Adlercreutzia and Tannerella were more abundant in the feces of mice transplanted with the healthy twin fecal material, fewer bacteria of the genera Ruminococcus were present in these mice. The most significant difference, however, was a reduced abundance of the genus Sutterella in mice colonized with microbiota from the MS twins compared with recipients of fecal material from healthy twins (Fig. 3B). Consistent with our results, Sutterella has previously been associated with better outcomes in patients with inflammatory bowel disease (16) as well as with resistance to EAE development in male TNFR2−/− mice (17).
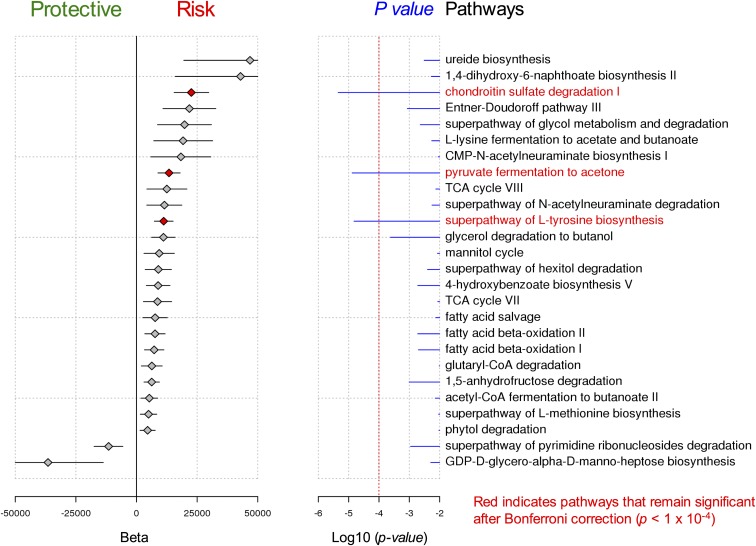
The gut flora may also affect CNS autoimmune responses by changing the metabolic environment of the gastrointestinal tract to produce metabolites such as short-chain fatty acids and aryl hydrocarbon receptor (Ahr) ligands, which are known to affect microglia and astrocytes (18–20). To identify potential metabolic pathways that are altered in recipient mice, we performed shotgun metagenomic sequencing on total stool DNA. Interestingly, shotgun metagenomic sequencing of mouse fecal pellets uncovered several metabolic pathways that were associated with the MS disease status of the donor and thus were up-regulated in the intestines of mouse recipients of MS microbiota. Pathways associated with the disease phenotype included pyruvate fermentation, l-tyrosine biosynthesis, and chondroitin sulfate degradation (Fig. S4). While the functional significance of these pathways to MS pathogenesis remains unclear, these results indicate that the gut microbiome influences MS disease risk not only through specific bacterial composition, as suggested by other studies, but also by potentially changing the metabolic environment of the human gastrointestinal tract. Additional studies are required to validate these findings as well as to identify similar associations in a larger cohort of human subjects.
Fig. S4.
Shotgun metagenomic sequencing of mouse fecal pellets uncovered several metabolic pathways associated with MS. Examination of gut microbiome metabolic pathways identified by metagenomic sequencing in 25 germ-free mice colonized by stool from four twins discordant for MS. Pathways that remained significantly associated with MS after multiple testing correction included chondroitin sulfate degradation (beta = 22,600, P = 4.5 × 10−6), pyruvate fermentation to acetone (beta = 13,400, P = 1.3 × 10−5), and superpathway of l-tyrosine biosynthesis (beta = 11,200, P = 1.5 × 10−5).
Reduced IL-10 Production in RR Mice Colonized with Microbiota from MS-Affected Twins.
A search for relevant immunological changes possibly responsible for the differential EAE induction pointed to IL-10 as a key factor. While the global composition of immune tissues (dendritic cells, macrophages, and B cells, as well as T cell subsets, including Foxp3+ regulatory T cells) remained unaffected by fecal transfers (Fig. S5), there were functional dissimilarities in the production of antibodies and cytokines. Serum anti-MOG autoantibody titers of the IgG1 class were slightly increased in MS-colonized RR mice, while IgG2a levels were decreased (Fig. S6). In addition, we performed a comprehensive qPCR-based screening for cytokines and barrier and tight junction proteins in the small intestine and colon of mouse recipients. We did not find significant differences in the expression levels of IL-1α, IL-1β, IL-6, IL-9, IL-11, IL-18, IL-21, GM-CSF, IFN-γ, TNF-α, Claudin-1, Claudin-2, Claudin-4, Claudin-5, Myo9b, Occludin, RegIIIγ, Tjp-1, Tjp-2, or Tjp-3 between mice transplanted with control or MS twin fecal samples (Fig. S7). The expression levels of IL-4, IL-5, IL-13, IL-22, and IL-27 were below the detection limit.
Fig. S5.
No differences were seen in immune cell populations of mice receiving control or MS patient fecal material. Bar graphs display frequencies (± SEM) of dendritic cells (CD45+CD11c+), macrophages (CD45+CD11b+), B cells (CD45+B220+), as well as regulatory T cells (CD4+FoxP3+) in spleen and small intestinal lamina propria (siLP) of germ-free SJL/J mice receiving fecal material from healthy twins (HD) or MS patients (MS). n = 10–13 mice per group. Data were pooled from three independent experiments.
Fig. S6.
No differences were seen in serum autoantibody titers in recipients of healthy twin (HD; n = 16) or MS twin (MS; n = 19) microbiota. Serum MOG-specific IgG1 or IgG2a antibodies were measured by sandwich ELISA. Values shown are the absorbances at 405 nm. Bars depict mean ± SEM.
Fig. S7.
No differences were seen in the expression of cytokines and tight junction proteins in recipients of healthy (HD) or MS twin microbiota. Expression levels of tight junction proteins (A) and cytokines (B) were measured by real-time qPCR. (Left) Expression levels in small intestine (ileum). (Right) Expression levels in colon. n = 3 mice per group. Bars depict mean ± SEM.
Most informative was the pattern of cytokines released by activated splenic T cells. There was a modest change in IL-17 release, with IFN-γ remaining unaltered (Fig. 3C). More strikingly, spleen cells from recipients of control microbiota produced much higher amounts of IL-10 upon stimulation with anti-CD3/anti-CD28 monoclonal antibodies than cells from recipients of MS microbiota (Fig. 3C). This difference was of functional relevance. We colonized germ-free RR mice with healthy donor fecal material and subsequently treated them with anti–IL-10 neutralizing or isotype control antibodies. Blocking IL-10 increased disease incidence compared with isotype control-treated mice (from 30 to 55%; P = 0.0006, Fisher’s exact test) (Fig. 3D).
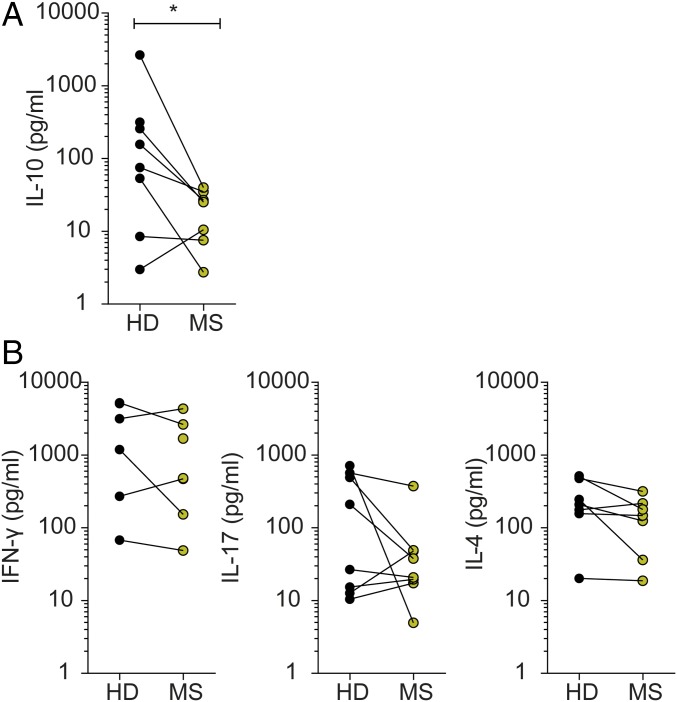
The immunological profiles observed in the colonized RR mice were consistent with the patterns observed in PBMCs from a subgroup of eight twin pairs which included several donors used for the mouse colonization experiments. There, anti-CD3/anti-CD28 stimulation induced stronger production of anti-inflammatory IL-10 in T cells from healthy than from MS donors (Fig. 4A). In contrast, we found no differences in the frequencies of CD4+ T cells, Th1 cells, Th17 cells, and Treg cells (Fig. S1). Also, production of IFN-γ, IL-17, and IL-4 cytokines in response to in vitro stimulation with phytohemagglutinin (PHA) or anti-CD3/anti-CD28 was largely indistinguishable between T cells from MS-affected and healthy twins (Fig. 4B).
Fig. 4.
Gut bacteria from healthy twins trigger an antiinflammatory T cell response. (A) IL-10 production of CD4+ T cells isolated from PBMCs of selected twin pairs. T cells were stimulated for 96 h with 1 µg/mL anti-CD3 and anti-CD28 antibodies. Levels of IL-10 (seven pairs, two singletons) in the supernatants were measured by ELISA. *P < 0.05 (Wilcoxon test). (B) Cytokine profiles of CD4+ T cells isolated from PBMCs of selected twin pairs Depending on PBMC availability and quality, eight twin pairs were selected for further in vitro stimulation assays, and data are depicted for each cytokine where stimulation-dependent production above the individual detection level could be documented. T cells were stimulated for 48 h with 5 µg/mL PHA. Levels of IFN-γ (five pairs, three singletons), IL-17 (eight pairs), and IL-4 (seven pairs, one singleton) in the supernatants were measured by the Luminex Bead-based Multiplex Assay.
Discussion
Chronic inflammatory diseases have been recently associated with altered profiles of the intestinal bacterial flora (21). These changes were revealed by studies making use of new sequencing technologies, which, however, cannot per se resolve whether the microbial changes were primary or secondary to cognate diseases. Causal relations were established in metabolic conditions such as obesity by transplanting human-derived microbiota to rodents; these recipients of disease-derived fecal samples developed an equivalent metabolic condition (22). Applying similar strategies to studies of chronic autoimmune or degenerative diseases (23–25) has remained more challenging. This definitely applies to exploring links between MS and changes of intestinal microbiota.
Here we took a two-tiered approach to address this question. First, we compared the intestinal microbial profiles of MS twins and healthy twins by microbial 16S rRNA amplicon and metagenomic shotgun sequencing. Second, we transplanted fecal samples from MS-affected and healthy twins into germ-free mice. We did not detect significant differences in MS-related microbial profiles between MS-affected and healthy twins. In this respect, our results differ from findings reported by others. A recent study using pyrosequencing of 16S rRNA gene in 20 Japanese patients with RR MS detected associations with Clostridium and Bacteroidetes species (26). These signatures were not seen in our cohort, nor were they reported in another independent screening (27). However, it should be noted that these studies (including our own) are relatively limited in size. Second, it is likely that microbial associations vary according to geographical region, human genetic background, and many other factors, including treatment. However, it is worth noting that, after adjusting for treatment, we found Akkermansia species were increased in untreated MS twins, which was also found to be increased in another independent cohort of MS patients [see ref. 27 and the companion paper by Cekanaviciute et al. (28)]. Third, fecal material may be suboptimal for studying processes possibly restricted to circumscribed intestinal segments. Gut microbiota differ radically in their functional potential, density, and composition along different segments of the small and large intestine (29). Thus, in the feces, pathogenic organisms populating the small intestine at low density could be eclipsed by high-density colonic communities. Obviously, more detailed studies of much larger cohorts are needed to further explore the complexities of MS-related microbial associations.
The second tier of our strategy, transfer of human microbiota into germ-free RR mice, led to a particularly interesting observation. We found that not only could human microbiota trigger classic RR EAE, but, even more significantly, transplants from MS-affected donors triggered EAE at higher rates than did transplants from their healthy control-derived counterparts. This difference was not large in magnitude but was significant and reproducible. A detailed analysis of 16S rRNA sequences revealed significant differences between MS- and healthy twin-colonized mice at the level of individual taxa (genera and OTUs). The most prominent MS-dependent change was a reduction in the bacterial genus Sutterella.
The increased incidence of spontaneous EAE in MS sample-recipient gnotobiotic RR mice could be due to either increased activation of autoimmune effector T cells or a weakened regulatory mechanism. Our results seem to favor the latter. Indeed, functional screening of peripheral blood lymphocytes from a subgroup of monozygotic twins indicated decreased IL-10 production in MS-derived samples. This is in accord with previous work, which described decreases of Tr1-like, IL-10–producing T cells (30–32). The differential IL-10 response noted in human PBMCs was mirrored in gnotobiotic RR mice. In germfree RR mice recolonized with MS-derived fecal samples, spleen T cells produce less IL-10 than their equivalents from control sample recipients. This recalls previous reports describing enhanced IL-10 production in the intestinal immune system driven by bacterial polysaccharide A (33) and protection from actively induced CNS autoimmune disease (34). A relative lack of protective factors rather than an increase in disease-promoting factors is also supported by our observation that in vivo treatment with anti–IL-10 antibody resulted in an increased incidence of spontaneous EAE in our RR model. Although there is currently no conclusive evidence of a genetic association of MS with the IL-10 gene or IL-10–related pathways, this by no means excludes the possibility that this crucially important pathway is modulated by external factors (e.g., microbiota).
In conclusion, we here provide evidence that human MS-derived microbiota contain factors that precipitate an MS-like autoimmune disease in a humanized transgenic mouse model. This observation provides possibilities for characterizing the precise role and functional mechanisms by which the human intestinal microbiota contributes to the pathogenesis of neuroinflammatory diseases. The findings may eventually have important implications not only for the pathogenesis but also for the therapy and potentially even prevention of human MS.
Methods
Details of cohort recruitment, sample acquisition, and information collection are provided in SI Methods. The study was approved by the local Ethics Committee of the Ludwig-Maximilians University Munich, and all participants gave written informed consent. All animal procedures were in accordance with the guidelines of the Committee on Animals of the Max Planck Institute of Neurobiology and the Max Planck Institute of Immunobiology and Epigenetics with a license from the Regierung von Oberbayern as well as the Regierungspräsidium Freiburg. Procedures for 16S and metagenomic sequencing, data analysis, mouse colonization, flow cytometry, ELISA, cytokine profiling, and real-time qPCR are described in detail in SI Methods.
SI Methods
Study Design.
MZ twins were recruited by launching a national televised appeal as well as internet notification in Germany with support from the German Multiple Sclerosis Society (DMSG). Inclusion criteria for study participation were MZ twins with an MS diagnosis according to the revised McDonald criteria or CIS in one twin only. Exclusion criteria were antibiotic, glucocorticosteroidal, or immunosuppressive treatment, gastrointestinal infection, or diet irregularities in the 3 mo before study entry. In total, 34 pairs (Table 1) visited the outpatient department at the Institute of Clinical Neuroimmunology in Munich for a detailed interview on past and present medical, family, and social history, a neurological examination, and a nutrition questionnaire. To confirm the MS diagnosis, medical records including MRI scans were obtained and reviewed. Fecal samples were directly collected in hospital or were taken at home, stored at −20 °C, and transferred to the hospital in cooling bags. Finally, all samples were stored at −80 °C. Buccal swabs for zygosity testing were taken from all participants. The study was approved by the local Ethics Committee of the Ludwig-Maximilians University Munich, and all participants gave written informed consent.
16S rRNA Sequencing and Analysis.
The V3–V5 region of the bacterial 16S rRNA gene was amplified using the universal forward (5′-CCGTCAATTCMTTTGAGTTT-3′) and reverse (5′-ACTCCTA CGGGAGGCAGCAG-3′) primers incorporating the FLX Titanium adapters and a unique barcode sequence. PCR products were sequenced on a 454 GS FLX titanium pyrosequencer (Life Sciences) at BGI-Shenzhen. Analysis was performed using QIIME v1.9 as described (35). Essentially, amplicon sequences were quality-filtered and grouped by OTUs using the SortMeRNA method (36) using the Greengenes version 13.8 97% dataset for closed reference. Sequences that did not match reference sequences in the Greengenes database were dropped from the analysis. Taxonomy was assigned to the retained OTUs based on the Greengenes reference sequence, and the Greengenes tree was used for all downstream phylogenetic community comparisons. Samples were filtered to at least 10,000 sequences per sample, and OTUs were filtered to retain only OTUs present in at least 5% of samples, covering at least 0.01% of total reads. After filtering, sequences were rarefied to the lowest number of sequences per sample: 10,975 sequences in human samples and 8,137 sequences in mouse samples. For comparison between human and mouse samples, the human and mouse datasets were combined before OTU filtering and rarefaction. The resulting OTUs were filtered as described above, and samples were rarefied to 6,200 sequences per sample. Alpha diversity was calculated using the phylogenetic diversity index method (37). For analysis of beta diversity, pairwise distance matrices were generated by phylogenetic metric of weighted UniFrac (38) and used for PCoA. For comparison of individual taxa, samples were not rarefied. Instead, OTU abundances were normalized using variance-stabilizing transformation, and taxa distributions were compared using the Wald negative binomial test from the R software package DESeq2 (as described in refs. 38 and 39) with Benjamini–Hochberg correction for multiple comparisons. All statistical analyses of differences between individual bacterial species were performed using QIIME v.1.9 or R (packages DESeq2 and phyloseq).
Metagenomic Analysis of Human and Mouse Samples.
We performed metagenomic sequencing of the gut microbiome in 16 pairs of identical twins, each composed of a sibling affected by MS and one unaffected sibling. In addition, stool samples from 25 germ-free mice that were colonized with four twin pair samples also underwent metagenomic sequencing. Each human and mouse fecal sample produced at least 30 million paired-end DNA reads 100 bp in length. Sequence quality, evaluated using FastQC, was high in the majority of sequences across all samples. We used the HMP Unified Metabolic Analysis Network (HUMAnN2) tool to calculate the relative abundance of specific microbes, gene families, and metabolic pathways. This software pipeline uses MetaPhlAn2 to obtain a list of abundant organisms by aligning sequences to genes unique to known bacterial species. DNA sequences are subsequently aligned to genomes of the identified organisms using the Bowtie 2 aligner and an annotated pangenome database, ChocoPhlAn. Unmapped DNA reads undergo translated alignment to the bacterial proteome using the software Diamond and a large protein database, UniRef50. The product of sequence alignment is a quantitative relative abundance of specific protein families. The HUMAnN2 software subsequently uses this information to determine the number complete copies of specific metabolic pathways using MetaCyc, a database mapping metabolic reactions to pathways. After obtaining quantitative measurements of gut bacterial abundance, gene families, and metabolic pathways, we performed association testing to identify pathogenic and protective factors in MS. We calculated the pairwise sum of absolute differences of microbial relative abundance between two individuals to determine if gut bacterial flora are more similar between twins with discordant phenotypes than between pairs of unrelated individuals. We used logistic regression (adjusting for twin pair and number of genome equivalents sequenced per sample) to examine associations between each gut bacterial variable and MS phenotype. We corrected for multiple comparisons and adjusted P values using a false-discovery rate of 5%. We applied the same rigorous statistical approach to mouse samples and also adjusted for the twin pair from which mice were colonized.
Colonization of Germ-Free RR Mice with Human MS Twin-Derived Fecal Samples.
For the human-to-mouse fecal transfer experiments we selected a subgroup of five discordant twin pairs, mainly based on pragmatic criteria such as relatively young age (20–40 y), female sex, and either no treatment or treatment only with IFN-β (Table S1). One gram of human fecal material was suspended in 15 mL prereduced PBS (PBS supplemented with 0.1% l-cysteine hydrochloride monohydrate) and was vortexed at room temperature for 5 min. Large insoluble particles were allowed to settle by gravity for 5 min. The supernatant was transferred to an anaerobic crimped tube (Sigma-Aldrich). Prereduced glycerol (containing 0.1% l-cysteine hydrochloride monohydrate) was added to a final concentration of 20%, and tubes were frozen at −80 °C. Tubes were sprayed thoroughly with Virkon (V.P. Produkte) before being transferred to the gnotobiotic isolators. Germ-free RR mice were gavaged with ≈300 µL of fecal bacterial suspension. In addition, mice colonized with healthy twin fecal material were injected with 250 µg anti–IL-10 (JES5-2A5; BioXcell) or isotype control antibodies once every week. All animal procedures were in accordance with the guidelines of the Committee on Animals of the Max Planck Institute of Neurobiology and the Max Planck Institute of Immunobiology and Epigenetics with a license from the Regierung von Oberbayern as well as the Regierungspräsidium Freiburg.
Cell Isolation and Flow Cytometry.
Isolation and phenotyping of immune cells by flow cytometry were done as previously described (7). Briefly, single-cell suspensions were prepared from spleens by mechanical disruption via forcing through 40-µm cell strainers (Thermo Fisher Scientific). For the isolation of lymphocytes from the small intestine, the intestine was collected in ice-cold HBSS buffered with 15 mM Hepes. After careful removal of fatty tissue and fecal contents, the intestine was opened longitudinally and cut into small pieces. The intestinal fragments were washed three times for 15 min with stirring in HBSS containing 5 mM EDTA, 15 mM Hepes, and 10% FBS. Next, intestinal pieces were washed once for 5 min with stirring in RPMI medium containing 15 mM Hepes and 10% FBS, followed by an incubation step at 37 °C with stirring in RPMI medium with 15 mM Hepes, 10% FBS, and 100 U/mL collagenase D (Roche Diagnostics). The digested tissue was washed twice in HBSS containing 5 mM EDTA before the lymphocytes of the small intestine were resuspended in 5 mL of 40% Percoll (Sigma-Aldrich) and overlaid on 2.5 mL of 80% Percoll. Percoll gradient separation was performed by centrifugation at 780 × g for 20 min at room temperature. Small intestinal lamina propria lymphocytes were harvested from the interphase of the Percoll gradient and washed once in RPMI medium containing 15 mM Hepes and 10% FBS. For detection of cell-surface markers, cells were stained in FACS buffer (PBS containing 1% BSA and 0.1% NaN3) with fluorochrome-labeled antibodies: PerCP-Cy5.5–conjugated anti-CD4 (RM4-5); eFluor 450-conjugated anti-CD45 (30-F11); PerCP-Cy5.5–conjugated anti-B220 (RA3-6B2); FITC-conjugated anti-CD11c (HL3), and PE-Cyanine 7–conjugated anti-CD11b (M1/70). For intracellular cytokine staining, 2 × 106 cells/mL were stimulated for 16 h with anti-CD3 antibody (1 µg/mL) (BD Pharmingen). Brefeldin A (5 µg/mL) (Sigma-Aldrich) was added for the last 5 h. After surface staining, cells were fixed and permeabilized using the Transcription Factor Staining Buffer Set (eBioscience) and stained intracellularly using the following antibodies: PE-conjugated anti-IL17 (TC11-18H10), FITC-conjugated anti–IFN-γ (XMG1.2), and APC-conjugated anti-FoxP3 (FJK-16s). All antibodies were purchased from BD Pharmingen, eBioscience, or BioLegend. Cells were acquired on a FACSVerse flow cytometer (BD Biosciences), and analysis was performed using FlowJo (TreeStar) software.
ELISA.
For the measurement of cytokine production by T cells, 2 × 105 splenocytes were cultured in the presence of 1 µg/mL anti-CD3 antibody (BD Pharmingen) or 1 µg/mL anti-CD3 and 0.5 µg/mL anti-CD28 antibody (BD Pharmingen). Cytokines in cell-culture supernatants were measured by ELISA with antibody pairs for IFN-γ (BD Biosciences), IL-17 (eBioscience), and IL-10 (R&D Systems). Serum titers of anti-MOG antibodies were quantified as previously described (6).
Cytokine Profiling of Human CD4+ T Cells.
From a subgroup of eight twin pairs, including three of the donors of fecal samples, we used available frozen PBMC specimens for in vitro cytokine production analyses. Frozen PBMC specimens from these pairs were thawed, and CD4+ T cells were isolated using the RosetteSep Kit (Stemcell Technologies). For assessment of IFN-γ, IL-4, and IL-17A secretion, 1 × 105 CD4+ T cells per well in triplicate were stimulated for 48 h with 5 µg/mL PHA. Levels of IFN-γ, IL-4, and IL-17A in the supernatants were measured by Luminex Bead-based Multiplex Assay (R&D systems) using a Bio-Plex MAGPIX Multiplex Reader (Bio-Rad). For assessment of IL-10 secretion, 1 × 105 CD4+ T cells per well in triplicate were stimulated for 96 h with either 5 µg/mL PHA or 1 µg/mL anti-CD3 (clone OKT3; BioLegend) and anti-CD28 (clone: CD28.2; eBioscience) antibodies, respectively. Levels of IL-10 in the supernatants were measured by ELISA according to the manufacturer’s instructions (eBioscience).
Real-Time qPCR.
RNA was prepared from ileum and colon of gnotobiotic mice using the Qiagen RNeasy mini kit (Qiagen) followed by conversion to cDNA using a verso cDNA synthesis kit (Thermo Scientific). Triplicate wells of PCR with gene-specific primers were performed using the Absolute QPCR SYBR Green Mix or Absolute QPCR mix with ROX (Thermo Fisher Scientific) following the manufacturer’s instructions and were measured using a 7900HT Real-Time PCR System (Applied Biosystems).
Statistical Analysis.
GraphPad Prism 6 (GraphPad Software, Inc.) was used for all statistical analysis. P values < 0.05 were considered to be significant.
Acknowledgments
We thank Prof. Thomas Boehm for supervising the maintenance of our germ-free mouse colony; Birgit Kunkel for technical assistance; Norbert Joswig, Melanie Pfunder, Manuela Schätzle, and Christian Schauerte for maintaining our germ-free colony and for technical support; Angelika Bamberger for support in patient care; Dr. Klaus Dornmair and Dr. Naoto Kawakami for helpful suggestions and comments; and Dr. Nicolaus König for support in the recruitment of the twin cohort. We also thank the international multiple sclerosis microbiome consortium (iMSMS) for helpful discussions and feedback. This work was funded by the Hertie Foundation, a DFG Koselleck Award, DFG Grant TR-128, the German Competence Network on Multiple Federal and Bavarian Divisions of the German MS Society, Verein Therapieforschung für MS Kranke e.V., Cyliax Stiftung, and the Max Planck Society. K.B. was supported by the James Heineman Research Award of the Minerva Stiftung. G.K. is supported by European Research Council Starting Grant GAMES 635617. S.E.B. is supported by grants from the US Department of Defense and the US National MS Society. E.C. is supported by a NIH/Institutional Research and Academic Career Development Award.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Normalized datasets related to this paper are available from the UCSF Data Sharing Service (Dash) at https://doi.org/10.7272/Q6N58JH2 and raw data have been deposited in the European Bioinformatics Institute (EMBL-EBI) database, https://www.ebi.ac.uk (accession no. ERP101460).
See Commentary on page 10528.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1711233114/-/DCSupplemental.
References
- 1.Wekerle H. Nature plus nurture: The triggering of multiple sclerosis. Swiss Med Wkly. 2015;145:w14189. doi: 10.4414/smw.2015.14189. [DOI] [PubMed] [Google Scholar]
- 2.Sawcer S, Franklin RJ, Ban M. Multiple sclerosis genetics. Lancet Neurol. 2014;13:700–709. doi: 10.1016/S1474-4422(14)70041-9. [DOI] [PubMed] [Google Scholar]
- 3.Simon KC, Munger KL, Ascherio A. XVI European Charcot Foundation lecture: Nutrition and environment: Can MS be prevented? J Neurol Sci. 2011;311:1–8. doi: 10.1016/j.jns.2011.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berer K, Krishnamoorthy G. Microbial view of central nervous system autoimmunity. FEBS Lett. 2014;588:4207–4213. doi: 10.1016/j.febslet.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 5.Wekerle H, Hohlfeld R. Gut Microbiota in Multiple Sclerosis: A Bioreactor Driving Brain Autoimmunity. Elsevier; Amsterdam: 2016. pp. 113–123. [Google Scholar]
- 6.Pöllinger B, et al. Spontaneous relapsing-remitting EAE in the SJL/J mouse: MOG-reactive transgenic T cells recruit endogenous MOG-specific B cells. J Exp Med. 2009;206:1303–1316. doi: 10.1084/jem.20090299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berer K, et al. Commensal microbiota and myelin autoantigen cooperate to trigger autoimmune demyelination. Nature. 2011;479:538–541. doi: 10.1038/nature10554. [DOI] [PubMed] [Google Scholar]
- 8.Goodrich JK, et al. Human genetics shape the gut microbiome. Cell. 2014;159:789–799. doi: 10.1016/j.cell.2014.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goodrich JK, Davenport ER, Waters JL, Clark AG, Ley RE. Cross-species comparisons of host genetic associations with the microbiome. Science. 2016;352:532–535. doi: 10.1126/science.aad9379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sampson TR, Mazmanian SK. Control of brain development, function, and behavior by the microbiome. Cell Host Microbe. 2015;17:565–576. doi: 10.1016/j.chom.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Polman CH, et al. Diagnostic criteria for multiple sclerosis: 2010 revisions to the McDonald criteria. Ann Neurol. 2011;69:292–302. doi: 10.1002/ana.22366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lublin FD, et al. Defining the clinical course of multiple sclerosis: The 2013 revisions. Neurology. 2014;83:278–286. doi: 10.1212/WNL.0000000000000560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seedorf H, et al. Bacteria from diverse habitats colonize and compete in the mouse gut. Cell. 2014;159:253–266. doi: 10.1016/j.cell.2014.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Turnbaugh PJ, et al. The effect of diet on the human gut microbiome: A metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009;1:6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chung H, et al. Gut immune maturation depends on colonization with a host-specific microbiota. Cell. 2012;149:1578–1593. doi: 10.1016/j.cell.2012.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morgan BP, Harris CL. Complement, a target for therapy in inflammatory and degenerative diseases. Nat Rev Drug Discov. 2015;14:857–877. doi: 10.1038/nrd4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller PG, Bonn MB, Franklin CL, Ericsson AC, McKarns SC. TNFR2 deficiency acts in concert with gut microbiota to precipitate spontaneous sex-biased central nervous system demyelinating autoimmune disease. J Immunol. 2015;195:4668–4684. doi: 10.4049/jimmunol.1501664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Erny D, et al. Host microbiota constantly control maturation and function of microglia in the CNS. Nat Neurosci. 2015;18:965–977. doi: 10.1038/nn.4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sampson TR, et al. Gut microbiota regulate motor deficits and neuroinflammation in a model of Parkinson’s disease. Cell. 2016;167:1469–1480.e12. doi: 10.1016/j.cell.2016.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rothhammer V, et al. Type I interferons and microbial metabolites of tryptophan modulate astrocyte activity and central nervous system inflammation via the aryl hydrocarbon receptor. Nat Med. 2016;22:586–597. doi: 10.1038/nm.4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kamada N, Seo SU, Chen GY, Núñez G. Role of the gut microbiota in immunity and inflammatory disease. Nat Rev Immunol. 2013;13:321–335. doi: 10.1038/nri3430. [DOI] [PubMed] [Google Scholar]
- 22.Ridaura VK, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341:1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang X, et al. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat Med. 2015;21:895–905. doi: 10.1038/nm.3914. [DOI] [PubMed] [Google Scholar]
- 24.de Goffau MC, et al. Fecal microbiota composition differs between children with β-cell autoimmunity and those without. Diabetes. 2013;62:1238–1244. doi: 10.2337/db12-0526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scheperjans F, et al. Gut microbiota are related to Parkinson’s disease and clinical phenotype. Mov Disord. 2015;30:350–358. doi: 10.1002/mds.26069. [DOI] [PubMed] [Google Scholar]
- 26.Miyake S, et al. Dysbiosis in the gut microbiota of patients with multiple sclerosis, with a striking depletion of species belonging to Clostridia XIVa and IV clusters. PLoS One. 2015;10:e0137429. doi: 10.1371/journal.pone.0137429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jangi S, et al. Alterations of the human gut microbiome in multiple sclerosis. Nat Commun. 2016;7:12015. doi: 10.1038/ncomms12015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cekanaviciute E, et al. Gut bacteria from multiple sclerosis patients modulate human T cells and exacerbate symptoms in mouse models. Proc Nat Acad Sci USA. 2017 doi: 10.1073/pnas1711235114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walter J, Ley R. The human gut microbiome: Ecology and recent evolutionary changes. Annu Rev Microbiol. 2011;65:411–429. doi: 10.1146/annurev-micro-090110-102830. [DOI] [PubMed] [Google Scholar]
- 30.Astier AL, Meiffren G, Freeman S, Hafler DA. Alterations in CD46-mediated Tr1 regulatory T cells in patients with multiple sclerosis. J Clin Invest. 2006;116:3252–3257. doi: 10.1172/JCI29251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martinez-Forero I, et al. IL-10 suppressor activity and ex vivo Tr1 cell function are impaired in multiple sclerosis. Eur J Immunol. 2008;38:576–586. doi: 10.1002/eji.200737271. [DOI] [PubMed] [Google Scholar]
- 32.Cao Y, et al. Functional inflammatory profiles distinguish myelin-reactive T cells from patients with multiple sclerosis. Sci Transl Med. 2015;7:287ra74. doi: 10.1126/scitranslmed.aaa8038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci USA. 2010;107:12204–12209. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Y, et al. An intestinal commensal symbiosis factor controls neuroinflammation via TLR2-mediated CD39 signalling. Nat Commun. 2014;5:4432. doi: 10.1038/ncomms5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Navas-Molina JA, et al. Advancing our understanding of the human microbiome using QIIME. Methods Enzymol. 2013;531:371–444. doi: 10.1016/B978-0-12-407863-5.00019-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kopylova E, et al. Open-source sequence clustering methods improve the state of the art. mSystems. 2016;1:e00003–e00015. doi: 10.1128/mSystems.00003-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Faith DP, Baker AM. Phylogenetic diversity (PD) and biodiversity conservation: Some bioinformatics challenges. Evol Bioinform Online. 2007;2:121–128. [PMC free article] [PubMed] [Google Scholar]
- 38.Lozupone C, Knight R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]