Significance
We have experimentally investigated the immunoregulatory effects of human gut microbiota in multiple sclerosis (MS). We have identified specific bacteria that are associated with MS and demonstrated that these bacteria regulate T lymphocyte-mediated adaptive immune responses and contribute to the proinflammatory environment in vitro and in vivo. Thus, our results expand the knowledge of the microbial regulation of immunity and may provide a basis for the development of microbiome-based therapeutics in autoimmune diseases.
Keywords: multiple sclerosis, microbiome, autoimmunity
Abstract
The gut microbiota regulates T cell functions throughout the body. We hypothesized that intestinal bacteria impact the pathogenesis of multiple sclerosis (MS), an autoimmune disorder of the CNS and thus analyzed the microbiomes of 71 MS patients not undergoing treatment and 71 healthy controls. Although no major shifts in microbial community structure were found, we identified specific bacterial taxa that were significantly associated with MS. Akkermansia muciniphila and Acinetobacter calcoaceticus, both increased in MS patients, induced proinflammatory responses in human peripheral blood mononuclear cells and in monocolonized mice. In contrast, Parabacteroides distasonis, which was reduced in MS patients, stimulated antiinflammatory IL-10–expressing human CD4+CD25+ T cells and IL-10+FoxP3+ Tregs in mice. Finally, microbiota transplants from MS patients into germ-free mice resulted in more severe symptoms of experimental autoimmune encephalomyelitis and reduced proportions of IL-10+ Tregs compared with mice “humanized” with microbiota from healthy controls. This study identifies specific human gut bacteria that regulate adaptive autoimmune responses, suggesting therapeutic targeting of the microbiota as a treatment for MS.
A major role of the human gut microbiota is to regulate both innate and adaptive immune responses during health and disease (1). Most studies of the human microbiome to date have focused on analyzing microbial population structures. However, it is equally important to investigate how variability in microbial abundance and composition affects host functions (2, 3). Exposing primary human immune cells to microbes or microbial products in vitro allows functional investigation of immunomodulatory effects by the gut microbiota (4–6).
There is growing evidence of population differences in the gut microbiota in multiple human autoimmune diseases (7, 8), including multiple sclerosis (MS) (9–12). While these studies in MS were performed with small sample sizes and did not stratify patient groups by treatment with disease-modifying drugs, consistent patterns of modest dysbiosis appear to be emerging. Furthermore, microbiota have been shown to mediate the regulation of immune responses in experimental autoimmune encephalomyelitis (EAE), a mouse model of MS (13, 14). MS-like symptoms in EAE can be exacerbated by Th1 and Th17 responses and modulated by Tregs (15, 16).
This led us to investigate structural and functional changes in intestinal microbiota as a potential component of MS pathogenesis. Specifically, we identified differences in microbial abundance between MS patients and controls and investigated how particular MS-associated bacteria modulate T lymphocyte responses using both in vitro and in vivo model systems. Our results indicate that differences in specific gut bacteria are functionally associated with a shift toward a proinflammatory T cell profile that may exacerbate or perpetuate autoimmune responses, thus potentially identifying a previously unknown environmental contributor to MS pathogenesis.
Results
The MS Microbiome Elicits Differential Treg Responses and Shows Modest Dysbiosis at the Genus Level.
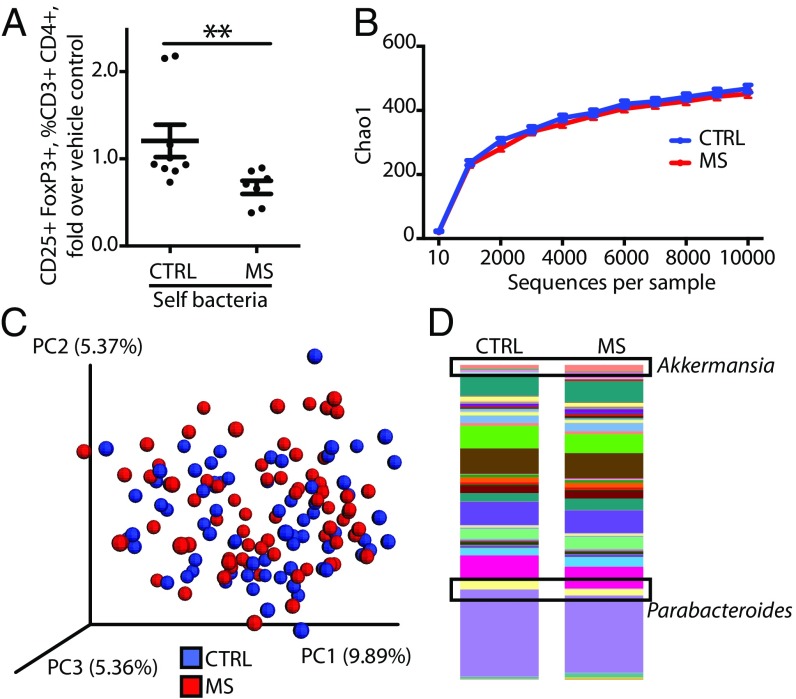
To investigate whether MS-associated bacteria affect immune functions in the host, we stimulated peripheral blood mononuclear cells (PBMCs) from MS patients or healthy controls, using extracts from total bacteria isolated from the stool samples of the same subjects who were PBMC donors (thus, “self” bacterial extracts). We observed that PBMCs from MS patients showed an impaired ability to differentiate or expand CD25+FoxP3+ Treg populations (Fig. 1A). The total CD3+CD4+ Th lymphocyte population was not altered by bacterial extract treatment, and the baseline proportion of CD25+FoxP3+ Tregs (in a population of CD3+CD4+ T cells) was not different between MS patients and healthy controls. These results suggest a specific immunoregulatory role of microbiota on PBMCs from MS patients.
Fig. 1.
MS patient microbiota alter self-Treg differentiation despite similar alpha and beta diversity to control microbiota. (A) Quantification of CD25+FoxP3+ lymphocytes within the CD3+CD4+ population in MS patients (n = 7) and healthy controls (n = 9), run in triplicate and results averaged, in response to extracts from total bacteria isolated from the stool samples of the same subjects who were PBMC donors (thus, self bacteria). CD25+FoxP3+ lymphocyte induction is expressed as the fold difference over no-bacteria control. **P < 0.01, two-tailed Mann–Whitney test. (B–D) Comparison of microbial community composition of untreated MS patients (n = 71) and healthy controls (n = 71). (B) Chao1 metric of alpha diversity. Data are presented as mean ± SEM. (C) Principal coordinate analysis (PCoA) plot of beta diversity (unweighted UniFrac). (D) Mean relative abundance of microbial genera. Akkermansia and Parabacteroides are outlined. Colors represent bacterial genera.
We subsequently analyzed the microbiome by 16S rRNA gene sequencing of stool samples from 71 untreated relapsing–remitting MS patients and 71 healthy controls (SI Appendix, Table S1). A subset of 79 subjects was sampled on two consecutive days to account for variability over time, and all subjects were used to compare the variability within and between subject groups (SI Appendix, Fig. S1). Consistent with similar findings in other autoimmune diseases (7, 8), we did not observe major global shifts in bacterial community structure in terms of alpha or beta diversity (Fig. 1 B and C). However, significant differences at the level of individual microbial taxa were observed between MS and control subjects (Figs. 1D and 2).
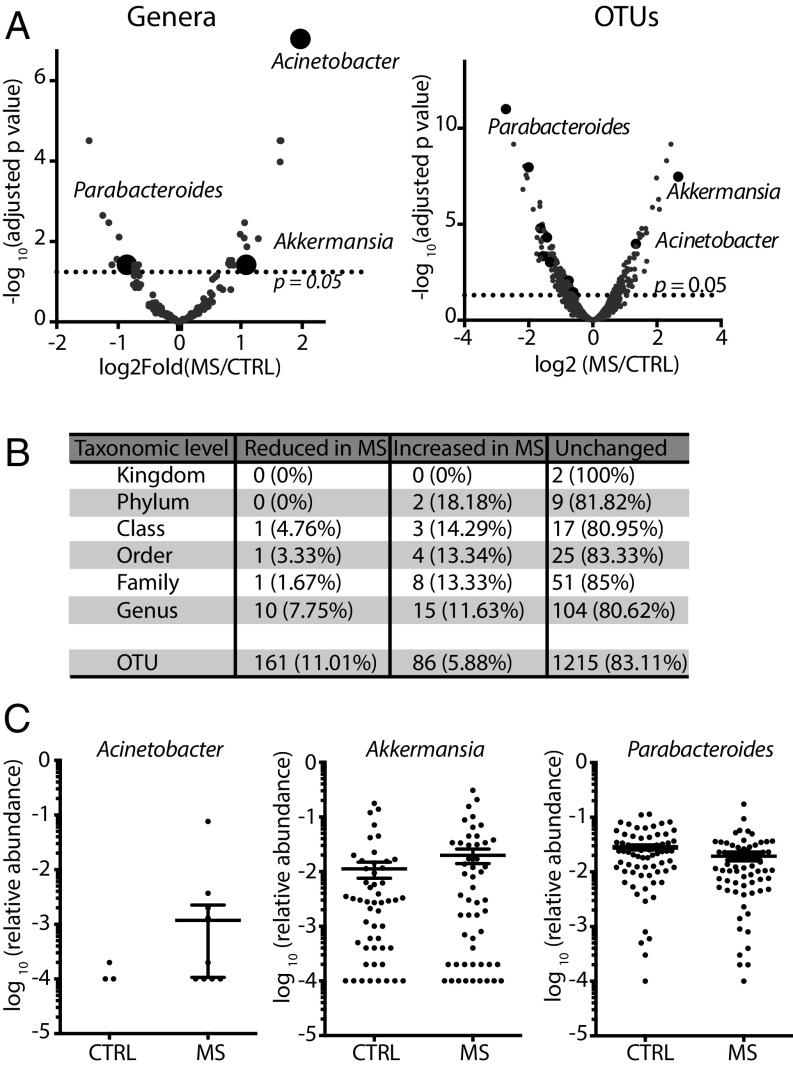
Fig. 2.
Relative abundances of individual microbial genera differ between MS patients and controls. (A) Volcano plots of the relative abundance distribution of microbial genera (Left) and OTUs (Right). The x axes show log twofold of relative abundance ratio between MS patients (n = 71) and controls (n = 71) after variance-stabilizing transformation. The y axes show negative log10 of P value (negative binomial Wald test with Benjamini–Hochberg correction for multiple comparisons). (B) Summary of taxonomic differences between MS and control microbiomes. (C) Relative abundance plots of selected microbial genera (highlighted in A) that were found to be significantly different between MS and controls. Data are shown as mean ± SEM.
Our analysis revealed significant differences in the relative abundance of 25 bacterial genera (19.38% of the total) (listed in SI Appendix, Table S2) and 247 operational taxonomic units (OTUs) (16.89% of the total) (listed in SI Appendix, Table S3) (Fig. 2 A and B).
We then selected individual significantly different taxa for functional studies to assess their potential contribution to autoimmune inflammation in MS. The specific taxa were selected based on the following criteria: (i) identifiable to species level or genus level with high overlap between species; (ii) culturable, to be able to study their functions in vitro and in vivo; (iii) type strains available from the ATCC to ensure reproducibility; and (iv) previously associated with immunoregulatory effects.
Among the genera significantly increased in MS samples were Acinetobacter and Akkermansia, while one of the most significantly reduced genera in MS patients was Parabacteroides, with the majority of OTUs mapping to Parabacteroides distasonis (Fig. 2C and SI Appendix, Fig. S3). Interestingly, P. distasonis was previously reported to induce a Treg phenotype in gnotobiotic mouse models (17, 18). All Acinetobacter species, including Acinetobacter baumannii, Acinetobacter calcoaceticus, and Acinetobacter lwoffii, are rare in the healthy human gut microbiome and share genome-wide homology (19, 20), making them indistinguishable by 16S amplicon sequencing. Thus, OTUs that mapped to the genus Acinetobacter did not allow species-level discrimination. Based on a previous report associating A. calcoaceticus with MS (21), we focused on this organism as a candidate for functional studies of immune regulation. Finally, all OTUs that mapped to the Akkermansia genus belonged to the species Akkermansia muciniphila, which has been studied mostly in the context of metabolism (22) but also contributes to inflammation during infection (12, 23).
MS-Associated Bacterial Species Reduce Tregs and Increase Th1 Lymphocyte Differentiation in Vitro.
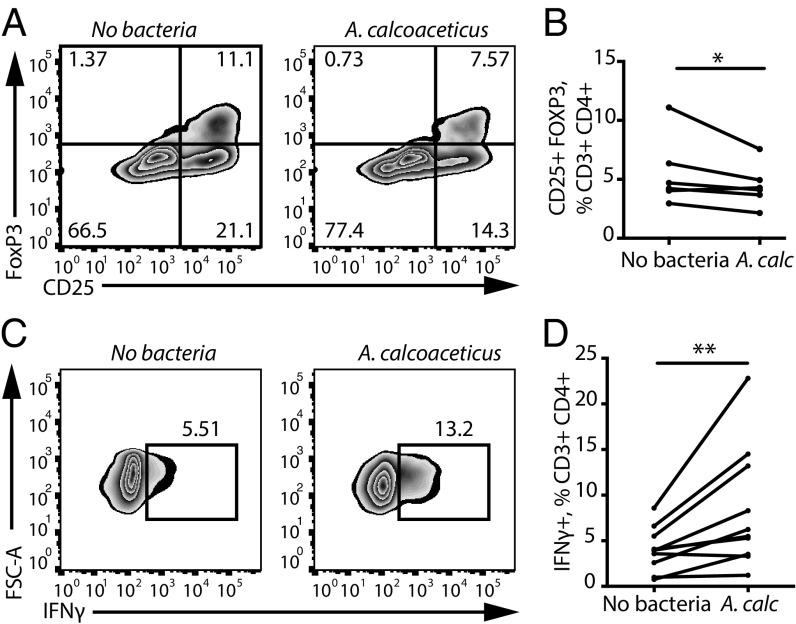
We hypothesized that bacterial taxa altered in MS patients play functional roles in regulating immune responses. To test this hypothesis, we established an in vitro model system by exposing PBMCs from healthy donors to a suspension of heat-killed and sonicated individual bacterial cultures (“bacterial extracts”) under different stimulating conditions (e.g., Treg, Th1, and so forth) and used flow cytometry to evaluate T lymphocyte differentiation and proliferation. We observed that extracts from A. calcoaceticus reduced the proportions of CD25+FoxP3+ Tregs among PBMCs (Fig. 3 A and B). These results suggest that intestinal A. calcoaceticus restrains immunoregulatory T cell development, as is consistent with its relative increase in the MS cohort. Furthermore, we observed that A. calcoaceticus increased the proportion of effector CD4+ lymphocytes that differentiated into IFNγ-producing Th1 cells (Fig. 3 C and D), thereby potentially exacerbating inflammation.
Fig. 3.
A. calcoaceticus inhibits Treg differentiation and stimulates Th1 differentiation in vitro. (A and B) Representative flow cytometry plots (A) and quantification (B) of CD25+FoxP3+ cell differentiation within the CD3+CD4+ population in response to A. calcoaceticus (A. calc) (n = 6 PBMC donors). (C and D) Representative flow cytometry plots (C) and quantification (D) of IFNγ+ Th1 lymphocytes within the CD3+CD4+ population in response to A. calcoaceticus (A. calc) (n = 11 PBMC donors). *P < 0.05, **P < 0.01, two-tailed repeated-measures t test. Data are shown as mean ± SEM.
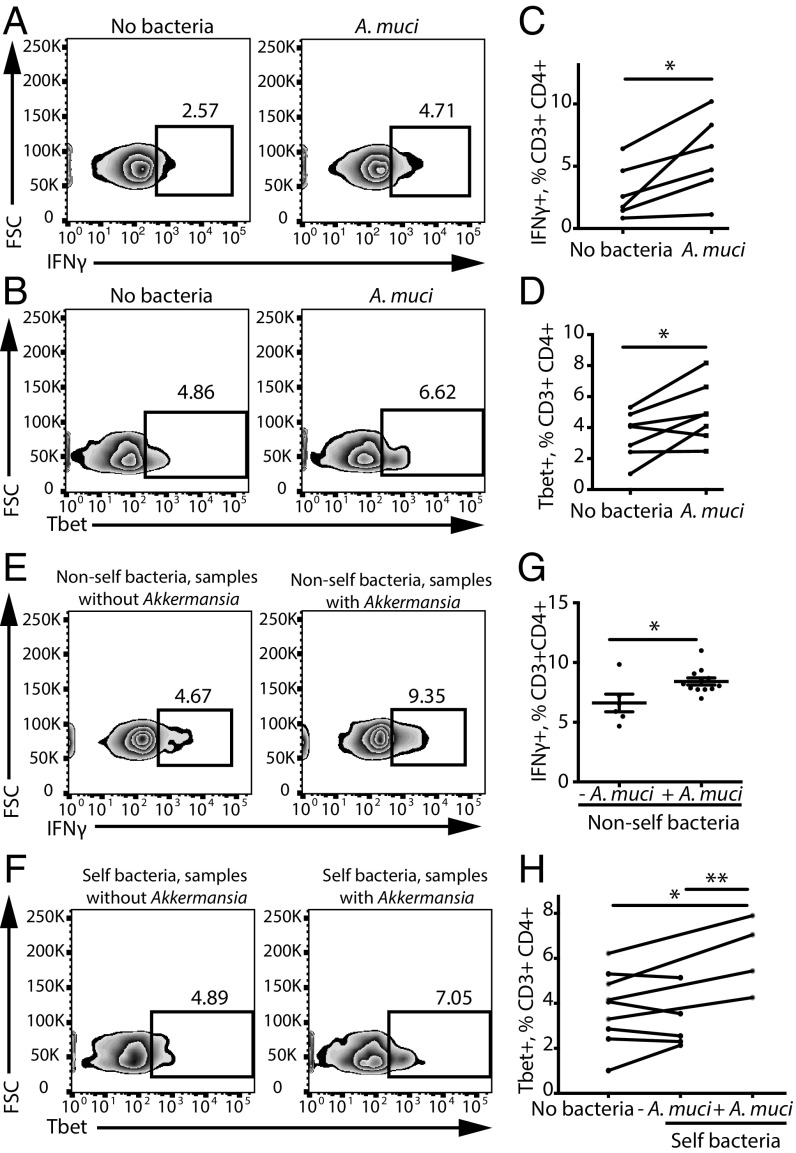
Analysis of A. muciniphila, another bacterial species increased in the MS microbiome, revealed an even more pronounced effect on stimulating Th1 differentiation. Extracts from A. muciniphila significantly increased healthy donor PBMC differentiation into Th1 lymphocytes (Fig. 4 A–D). Furthermore, we discovered that the mere presence of A. muciniphila in total stool bacteria was sufficient to increase Th1 lymphocyte differentiation in vitro. Specifically, exposing PBMCs to total bacterial extracts isolated from unrelated subjects with detectable levels of A. muciniphila increased the differentiation of IFNγ+ Th1 lymphocytes compared with bacterial extracts that did not have A. muciniphila (Fig. 4 E and G). Similarly, subjects with detectable A. muciniphila showed a significant increase in IFNγ+ Th1 differentiation in response to extracts of their own bacteria (Fig. 4 F and H). In summary, we have identified A. muciniphila and A. calcoaceticus as examples of common and rare MS-associated bacterial species that favor proinflammatory T lymphocyte responses in vitro.
Fig. 4.
A. muciniphila increases Th1 lymphocyte differentiation in vitro. (A–D) Representative flow cytometry plots (A and B) and quantification (C and D) of IFNγ+ and of Tbet+ Th1 lymphocytes within the CD3+CD4+ population in response to A. muciniphila (A. muci). n = 6 PBMC donors for the IFNγ experiment; n = 7 PBMC donors for the Tbet experiment. *P < 0.05, two-tailed repeated-measures t test. (E–H) Representative flow cytometry plots (E and F) and quantification (G and H) of IFNγ+ Th1 lymphocytes within the CD3+CD4+ population in response to nonself or self bacteria from subjects with or without detected A. muciniphila. n = 6 subjects without A. muciniphila; n = 12 subjects with A. muciniphila. *P < 0.05, two-tailed t test; **P < 0.01, two-way ANOVA for repeated measures. Data are shown as mean ± SEM.
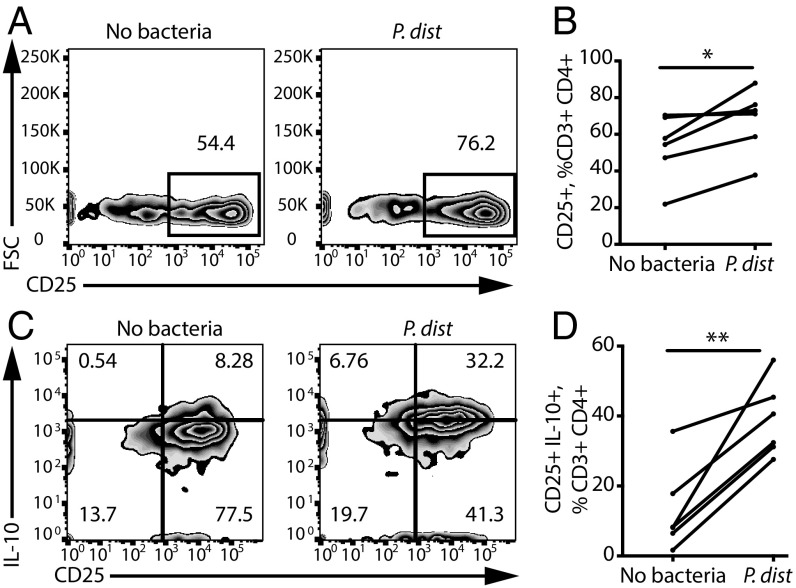
We next explored whether individual taxa that are less abundant in MS patients could promote immunoregulatory responses. Exposing healthy donor PBMCs to extracts from P. distasonis significantly increased the percentage of CD25+ T lymphocytes among the CD3+CD4+ T cell population (Fig. 5 A and B). Furthermore, we observed an enrichment of CD25+IL-10+ cells (Fig. 5 C and D), including CD25+IL-10+FoxP3− Tr1 cells (but not CD25+FoxP3+ Tregs), which have been associated with strong immunoregulatory properties (14, 24). Thus, our results demonstrate that P. distasonis is sufficient to skew T lymphocytes toward a regulatory profile in vitro.
Fig. 5.
P. distasonis stimulates IL-10+ Treg differentiation in vitro. (A–D) Representative flow cytometry plots (A and B) and quantification (C and D) of CD25+ and CD25+IL-10+ lymphocytes within the CD3+CD4+ population in response to P. distasonis (P. dist). n = 6 PBMC donors. *P < 0.05, **P < 0.01, two-tailed repeated measures t test. Data are shown as mean ± SEM.
All immunoregulatory effects described here were at least partially specific to selected bacterial extracts. To show that exposure to any differentially abundant bacteria did not elicit similar effects, we analyzed the immune effects of Eggerthella lenta, which is significantly increased in MS patients, and found that it did not alter Th1 or Treg differentiation (SI Appendix, Fig. S3 A and B). In addition, the specificity of P. distasonis, A. calcoaceticus, and A. muciniphila functions is emphasized by the fact that these bacteria did not alter the differentiation of all lymphocyte populations indiscriminately; for example, P. distasonis had no effect on Th1 cells, and A. muciniphila had no effect on CD25+FoxP3+ Tregs (SI Appendix, Fig. S3 C–E).
While regulation of immune functions by gut microbiota are likely multifaceted and complex, we speculate that the observed P. distasonis-associated reduction in immunoregulatory T cells could act in concert with the described increases in A. calcoaceticus and A. muciniphila and contribute to create an overall proinflammatory environment in MS patients.
Colonization of Mice with Single Species of MS-Associated Bacteria Recapitulates in Vitro T Lymphocyte Differentiation Profiles.
To elucidate the role of individual MS-associated bacteria in vivo, we colonized antibiotic-treated or germ-free (GF) mice with a single species: A. calcoaceticus, A. muciniphila, or P. distasonis. Following colonization, we measured T lymphocyte differentiation in multiple peripheral lymphoid tissues. We were unable to observe an effect of A. muciniphila in monocolonized mice as described in our in vitro experiments, and we hypothesize that this discrepancy is likely due to differences in host (e.g., mice vs. human). However, we were able to replicate our key findings with the other two species analyzed.
In antibiotic-treated mice A. calcoaceticus inhibited FoxP3+ Treg differentiation, while P. distasonis stimulated CD4+IL-10+ lymphocyte differentiation (SI Appendix, Fig. S4 A and B). Furthermore, splenocytes from mice colonized with P. distasonis also displayed induction of CD4+IL10+ lymphocytes in response to their bacterial extracts, while this was not observed when splenocytes from control SPF mice were exposed to their own bacterial extracts (SI Appendix, Fig. S4C).
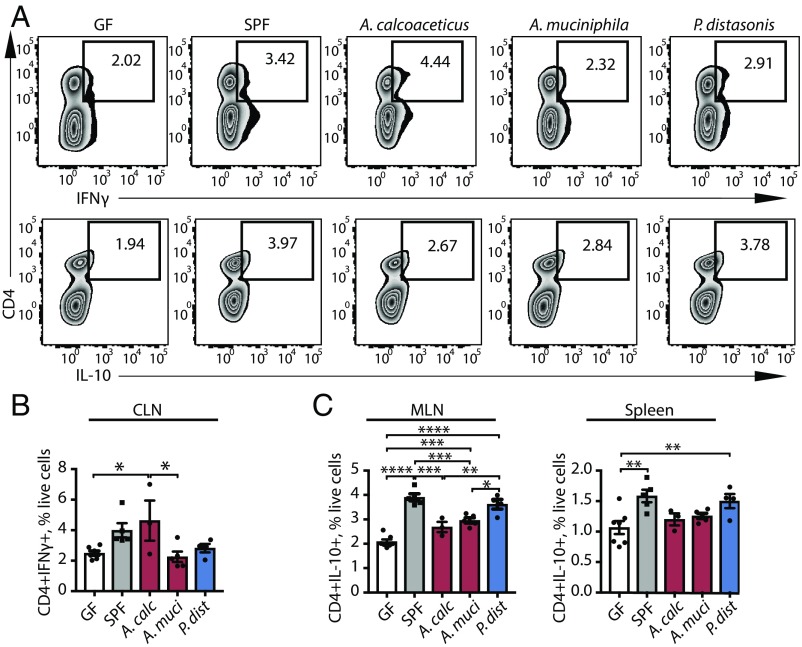
Furthermore, monocolonization of GF mice with A. calcoaceticus increased T lymphocyte differentiation into the IFNγ+ Th1 phenotype in cervical lymph nodes (Fig. 6 A and B), while monocolonization with P. distasonis led to significant increases in the CD4+IL-10+ T lymphocyte population in mesenteric lymph nodes and spleens (Fig. 6C and SI Appendix, Fig. S5). Taken together, the in vivo monocolonization results form a consistent complement to our in vitro data.
Fig. 6.
Monocolonization of GF mice with MS-associated bacteria mediates T lymphocyte differentiation. (A–C) Representative flow cytometry plots (A) and quantification (B and C) of CD4+IFNγ+ lymphocytes (B) and CD4+IL-10+ lymphocytes (C) within the live cell population in GF mice colonized with A. calcoaceticus, A. muciniphila, and P. distasonis. GF mice and specific pathogen-free (SPF) mice are used as controls. n = 3–8 mice per group. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, one-way ANOVA with Tukey adjustment for multiple comparisons. Data are shown as mean ± SEM. CLN, cervical lymph nodes; MLN, mesenteric lymph nodes.
Colonization of Mice with MS Donor Microbiota Inhibits Treg Differentiation and Exacerbates Disease Severity in EAE.
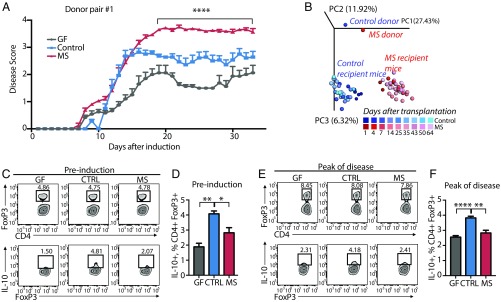
To investigate whether the proinflammatory environment established by MS-associated bacteria is physiologically relevant, we randomly selected three MS and control donor pairs (each composed of an untreated relapsing–remitting MS patient and a household control) to perform fecal microbiota transplants into groups of GF C57BL/6 mice (n = 6–8 mice per group). Six weeks after transplantation, mice were immunized with myelin oligodendrocyte glycoprotein (MOG35–55) to induce EAE. Remarkably, EAE disease scores were significantly increased in mice colonized with microbiota from MS patients compared with animals colonized with microbiota from healthy controls or GF mice (Fig. 7A). This result was recapitulated across all three donor pairs tested (SI Appendix, Fig. S6) and was accompanied by a lack of IL-10+ Treg induction in mesenteric lymph nodes from MS microbiota-colonized mice (Fig. 7 C–F and SI Appendix, Fig. S7). RNA-sequencing (RNA-seq) was performed on spinal cord samples derived from GF mice humanized with MS (n = 11) or control (n = 9) microbiota before and after EAE induction. Analysis of transcripts with differential expression in the two groups before and after EAE identified the up-regulation of several genes identified as the “immune response gene” category. Interestingly, when this dataset was used to infer the enrichment of specific cell types in the CNS, we detected a noticeable enrichment toward genes expressed by microglia in mice colonized with MS microbiota compared with controls (SI Appendix, Fig. S8).
Fig. 7.
Transfer of healthy control microbiota protects against EAE and mediates Treg induction in mouse mesenteric lymph nodes compared with transfer of MS patient microbiota. (A) Clinical EAE scores of mice that had been colonized with healthy control or MS patient microbiota for at least 5 wk or kept GF before the induction of EAE at 9–10 wk of age. Asterisks indicate significance between both the MS vs. control and the MS vs. GF groups. n = 6–8 mice per group. ****P < 0.0001, two-way ANOVA with Tukey adjustment for multiple comparisons. Data are shown as mean ± SEM. (B) PCoA of mouse microbiota at different time points after colonization with fecal microbiota from donor pair #1. n = 3–5 mice per group. EAE induction occurs at 35 d after transplantation. PC1, -2, -3, principal components 1, 2, and 3. (C and E) Representative flow cytometry plots of FoxP3+ lymphocytes within CD4+ populations and IL-10+ lymphocytes within CD4+FoxP3+ populations before EAE induction (C) and at peak of EAE disease (E). (D and F) Frequencies of IL-10+ lymphocytes within CD4+FoxP3+ populations in mesenteric lymph nodes of mice killed before EAE induction (D) and at peak of EAE progression (22 d after immunization) (F). *P < 0.05, **P < 0.01, ****P < 0.0001, one-way ANOVA with Tukey adjustment for multiple comparisons. Data are shown as mean ± SEM.
The inability of fecal bacteria from MS patients to promote Treg responses was observed both pre- and post-EAE induction, consistent with the microbiota showing no major differences in beta diversity at time points before and after the disease (Fig. 7B).
Principal component analysis of beta diversity of the microbiota in recipient animals showed a significant separation by donor that was stabilized as early as 7 d after transplantation (Fig. 7B and SI Appendix, Fig. S9A). This separation was recapitulated by metrics of alpha diversity (SI Appendix, Fig. S9B). Interestingly, some of the changes in relative abundance of individual bacterial genera, including a decrease in Sutterella and an increase in Ruminococcus, were also observed in mice colonized with microbiota from MS-discordant twins (SI Appendix, Fig. S9C) (25). Although human and mouse microbiota are not directly comparable, the biological pathways represented by MS-associated taxa largely overlap in the two groups (SI Appendix, Tables S3 and S4).
Collectively, the MS microbial community in vivo enhances EAE disease progression and fails to induce IL-10+ Tregs relative to gut bacteria from healthy controls, suggesting a functional role for the microbiota in autoimmunity that may be independent of host factors.
Discussion
Here we present a comparative structural analysis of the gut microbiome from patients with MS followed by functional studies of MS-associated microbiota. Our results reinforce the concept of MS pathogenesis as a multihit model that combines genetic predisposition and environmental factors, one of which is the microbiota. Based on our findings, we hypothesize that gut microbiota contribute to creating a sustained proinflammatory environment, which, in combination with genetic and other environmental factors, may crystallize the pathogenic autodestructive process of myelin.
In our admittedly simplistic experimental setup, we used crude bacterial extracts without prior fractionation; thus the relevant active components may consist of any bacterial products, either secreted or intracellular. Known bacterial metabolites with immunoregulatory effects fall into multiple categories, including polysaccharides (26), short-chain fatty acids (27), and aryl hydrocarbons (28). In addition, although we analyzed the effects of bacterial extracts on peripheral immune cells, some bacterial metabolites are able to cross the blood–CNS barrier and directly regulate CNS inflammation via microglia (29, 30) or astrocytes (28). Future research will likely be directed toward identifying the therapeutic potential of such products in MS and other complex diseases.
Although intestinal A. muciniphila has been extensively studied in the context of diet and obesity (22, 31), its role in regulating immune responses is less well understood. Here we provide in vitro evidence that A. muciniphila promotes Th1 lymphocyte differentiation. Consistent with our observations, A. muciniphila has been reported to exacerbate inflammation during infection (23). In contrast, a recent study reported that EAE-resistant male TNFR2−/− mice harbor more A. muciniphila than do disease-susceptible TNFR2−/− females (32). However, it remains to be addressed whether EAE susceptibility in this genotype is driven by the gut microbiome or by other factors that stem from gender and genetic differences. Notably, in a recently published study (12), as well as in companion the MS-discordant twin study in this issue of PNAS (25), Akkermansia was reported to be elevated in untreated MS patients.
The higher prevalence of Acinetobacter within MS subjects is consistent with previous reports of increased serum antibody responses (21, 33). Strikingly, A. calcoaceticus has also been shown to encode peptides that mimic the amino acid sequences of myelin basic protein (MBP) and MOG (21), both of which are myelin components (34). This suggests that molecular mimicry could potentially convert a normal immune response toward Acinetobacter into autoimmunity against myelin. Recently, another model of molecular mimicry-mediated CNS autoimmunity was proposed when aquaporin four-specific T lymphocytes from neuromyelitis optica patients were found to recognize a peptide from Clostridium perfringens and induce a Th17 bias (6), and this organism was found to be overabundant in neuromyelitis optica patients compared with healthy controls (35).
A growing body of literature has associated both CD25+FoxP3+ Tregs and IL-10–expressing T lymphocytes with alterations in gut microbiota. For example, monocolonization of GF mice with specific bacterial species is sufficient to drive CD25+FoxP3+ Treg differentiation and alter disease phenotype (3). Of interest, P. distasonis has also been shown to induce Treg differentiation in GF mice (2). In addition, our findings suggest that in vivo exposure to pure P. distasonis is associated with a subsequent immunoregulatory response to this bacterium in vitro. This observation is supported by the result that the immune cells of MS patients have impaired Treg differentiation in response to autologous (self) bacteria. Thus, the initial exposure to P. distasonis or other “beneficial” bacteria found in healthy subjects may contribute to expanding regulatory T lymphocyte precursor populations, thus promoting antiinflammatory responses upon subsequent exposure to the same bacteria.
While previous studies using GF mouse models have identified that the absence of gut bacteria ameliorates EAE (13, 14), here we show that gut bacteria transplanted from MS patients promotes more severe EAE symptoms than seen in mice that were transplanted with fecal bacteria from household controls. Similar results of microbiota being sufficient to transfer a human donor phenotype to GF mice have been reported in the context of obesity (36) and inflammatory bowel disease (37), and rheumatoid arthritis-associated bacteria were shown to exacerbate the disease in a mouse model of colitis (7). However, our study shows that the gut microbiota is able to transfer the phenotype in a disease model unrelated to the digestive system and suggests a potentially causal role for the gut microbiota in MS.
We consider GF mouse monocolonization as a valid experimental model to study microbial functions in vivo. However, we also recognize this approach has caveats, as monocolonization may not represent bacterial functions within the entire microbial community of the gut, and it requires using a mouse host for bacteria that presumably have a function in human disease. Therefore, it is not surprising to find that in vivo studies using monocolonized mice do not completely replicate in vitro results from human cells. We interpret our in vitro and in vivo findings as the first step toward future studies to identify pathways and metabolites that modulate Th1 and IL-10+ regulatory T lymphocytes. Such studies will likely open new avenues for the development of novel, microbiome-based therapeutic approaches for autoimmunity.
Materials and Methods
Animal work was approved by the Institutional Animal Care and Use Committee office at the California Institute of Technology. All human participants signed a written informed consent approved by the Institutional Review Boards of the University of California, San Francisco and the Icahn School of Medicine at Mount Sinai. Details about human fecal sample collection, 16S rRNA amplicon sequencing, and computational analysis of human and mouse microbiome samples are provided in the SI Appendix. Similarly, comparison of functional pathways expressed by microbiota, bacterial extract preparation for stimulation of human PBMCs, mouse colonization with microbiota, and induction of EAE can be found in the SI Appendix.
Supplementary Material
Acknowledgments
We thank the patients who participated in this study and M. Fischbach, S. S. Zamvil, and J. R. Oksenberg for critically reading the manuscript. We also thank the international multiple sclerosis microbiome consortium (iMSMS) for helpful discussions and feedback. This work was supported by the US National Multiple Sclerosis Society, a NIH Institutional Research and Academic Career Development Award Postdoctoral Fellowship, the US Department of Defense, the Valhalla Charitable Foundation, the Emerald Foundation, and Heritage Medical Research Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Normalized datasets related to this paper are available from the UCSF Data Sharing Service (Dash) at https://doi.org/10.7272/Q6N58JH2 and https://doi.org/10.7272/Q6RX997G. Raw data are available upon request.
See Commentary on page 10528.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1711235114/-/DCSupplemental.
References
- 1.Lee YK, Mazmanian SK. Has the microbiota played a critical role in the evolution of the adaptive immune system? Science. 2010;330:1768–1773. doi: 10.1126/science.1195568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Faith JJ, Ahern PP, Ridaura VK, Cheng J, Gordon JI. Identifying gut microbe-host phenotype relationships using combinatorial communities in gnotobiotic mice. Sci Transl Med. 2014;6:220ra11. doi: 10.1126/scitranslmed.3008051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci USA. 2010;107:12204–12209. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lozupone CA, et al. Alterations in the gut microbiota associated with HIV-1 infection. Cell Host Microbe. 2013;14:329–339. doi: 10.1016/j.chom.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chu H, et al. Gene-microbiota interactions contribute to the pathogenesis of inflammatory bowel disease. Science. 2016;352:1116–1120. doi: 10.1126/science.aad9948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Varrin-Doyer M, et al. Aquaporin 4-specific T cells in neuromyelitis optica exhibit a Th17 bias and recognize Clostridium ABC transporter. Ann Neurol. 2012;72:53–64. doi: 10.1002/ana.23651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Scher JU, et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. Elife. 2013;2:e01202. doi: 10.7554/eLife.01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gevers D, et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe. 2014;15:382–392. doi: 10.1016/j.chom.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cantarel BL, et al. Gut microbiota in multiple sclerosis: Possible influence of immunomodulators. J Investig Med. 2015;63:729–734. doi: 10.1097/JIM.0000000000000192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Miyake S, et al. Dysbiosis in the gut microbiota of patients with multiple sclerosis, with a striking depletion of species belonging to Clostridia XIVa and IV clusters. PLoS One. 2015;10:e0137429. doi: 10.1371/journal.pone.0137429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tremlett H, et al. US Network of Pediatric MS Centers Gut microbiota composition and relapse risk in pediatric MS: A pilot study. J Neurol Sci. 2016;363:153–157. doi: 10.1016/j.jns.2016.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jangi S, et al. Alterations of the human gut microbiome in multiple sclerosis. Nat Commun. 2016;7:12015. doi: 10.1038/ncomms12015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berer K, et al. Commensal microbiota and myelin autoantigen cooperate to trigger autoimmune demyelination. Nature. 2011;479:538–541. doi: 10.1038/nature10554. [DOI] [PubMed] [Google Scholar]
- 14.Lee YK, Menezes JS, Umesaki Y, Mazmanian SK. Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc Natl Acad Sci USA. 2011;108:4615–4622. doi: 10.1073/pnas.1000082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simmons SB, Pierson ER, Lee SY, Goverman JM. Modeling the heterogeneity of multiple sclerosis in animals. Trends Immunol. 2013;34:410–422. doi: 10.1016/j.it.2013.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carbajal KS, et al. Th cell diversity in experimental autoimmune encephalomyelitis and multiple sclerosis. J Immunol. 2015;195:2552–2559. doi: 10.4049/jimmunol.1501097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kverka M, et al. Oral administration of Parabacteroides distasonis antigens attenuates experimental murine colitis through modulation of immunity and microbiota composition. Clin Exp Immunol. 2011;163:250–259. doi: 10.1111/j.1365-2249.2010.04286.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geuking MB, et al. Intestinal bacterial colonization induces mutualistic regulatory T cell responses. Immunity. 2011;34:794–806. doi: 10.1016/j.immuni.2011.03.021. [DOI] [PubMed] [Google Scholar]
- 19.Peleg AY, Seifert H, Paterson DL. Acinetobacter baumannii: Emergence of a successful pathogen. Clin Microbiol Rev. 2008;21:538–582. doi: 10.1128/CMR.00058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Almeida LA, Araujo R. Highlights on molecular identification of closely related species. Infect Genet Evol. 2013;13:67–75. doi: 10.1016/j.meegid.2012.08.011. [DOI] [PubMed] [Google Scholar]
- 21.Hughes LE, et al. Cross-reactivity between related sequences found in Acinetobacter sp., Pseudomonas aeruginosa, myelin basic protein and myelin oligodendrocyte glycoprotein in multiple sclerosis. J Neuroimmunol. 2003;144:105–115. doi: 10.1016/s0165-5728(03)00274-1. [DOI] [PubMed] [Google Scholar]
- 22.Everard A, et al. Microbiome of prebiotic-treated mice reveals novel targets involved in host response during obesity. ISME J. 2014;8:2116–2130. doi: 10.1038/ismej.2014.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ganesh BP, Klopfleisch R, Loh G, Blaut M. Commensal Akkermansia muciniphila exacerbates gut inflammation in Salmonella Typhimurium-infected gnotobiotic mice. PLoS One. 2013;8:e74963. doi: 10.1371/journal.pone.0074963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hua J, Davis SP, Hill JA, Yamagata T. Diverse gene expression in human regulatory T cell subsets uncovers connection between regulatory T cell genes and suppressive function. J Immunol. 2015;195:3642–3653. doi: 10.4049/jimmunol.1500349. [DOI] [PubMed] [Google Scholar]
- 25. Berer K, et al. (2017) Gut microbiota from multiple sclerosis patients enables spontaneous autoimmune encephalomyelitis in mice. Proc Nat Acad Sci USA 10.1073/pnas1711233114. [DOI] [PMC free article] [PubMed]
- 26.Wang Y, et al. A commensal bacterial product elicits and modulates migratory capacity of CD39(+) CD4 T regulatory subsets in the suppression of neuroinflammation. Gut Microbes. 2014;5:552–561. doi: 10.4161/gmic.29797. [DOI] [PubMed] [Google Scholar]
- 27.Arpaia N, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rothhammer V, et al. Type I interferons and microbial metabolites of tryptophan modulate astrocyte activity and central nervous system inflammation via the aryl hydrocarbon receptor. Nat Med. 2016;22:586–597. doi: 10.1038/nm.4106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Erny D, et al. Host microbiota constantly control maturation and function of microglia in the CNS. Nat Neurosci. 2015;18:965–977. doi: 10.1038/nn.4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sampson TR, et al. Gut microbiota regulate motor deficits and neuroinflammation in a model of Parkinson’s disease. Cell. 2016;167:1469–1480.e1412. doi: 10.1016/j.cell.2016.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Derrien M, Belzer C, de Vos WM. Akkermansia muciniphila and its role in regulating host functions. Microb Pathog. 2016;106:171–181. doi: 10.1016/j.micpath.2016.02.005. [DOI] [PubMed] [Google Scholar]
- 32.Miller PG, Bonn MB, Franklin CL, Ericsson AC, McKarns SC. TNFR2 deficiency acts in concert with gut microbiota to precipitate spontaneous sex-biased central nervous system demyelinating autoimmune disease. J Immunol. 2015;195:4668–4684. doi: 10.4049/jimmunol.1501664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hughes LE, et al. Antibody responses to Acinetobacter spp. and Pseudomonas aeruginosa in multiple sclerosis: Prospects for diagnosis using the myelin-acinetobacter-neurofilament antibody index. Clin Diagn Lab Immunol. 2001;8:1181–1188. doi: 10.1128/CDLI.8.6.1181-1188.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Derfuss T, Meinl E. Identifying autoantigens in demyelinating diseases: Valuable clues to diagnosis and treatment? Curr Opin Neurol. 2012;25:231–238. doi: 10.1097/WCO.0b013e3283533a64. [DOI] [PubMed] [Google Scholar]
- 35.Cree BA, Spencer CM, Varrin-Doyer M, Baranzini SE, Zamvil SS. Gut microbiome analysis in neuromyelitis optica reveals overabundance of Clostridium perfringens. Ann Neurol. 2016;80:443–447. doi: 10.1002/ana.24718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ridaura VK, et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science. 2013;341:1241214. doi: 10.1126/science.1241214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sokol H, et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci USA. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.