Significance
Middle East respiratory syndrome coronavirus (MERS-CoV) recurrently infects humans from its dromedary camel reservoir, causing severe respiratory disease with an ∼35% fatality rate. The virus binds to the dipeptidyl peptidase 4 (DPP4) entry receptor on respiratory epithelial cells via its spike protein. We here report that the MERS-CoV spike protein selectively binds to sialic acid (Sia) and demonstrate that cell-surface sialoglycoconjugates can serve as an attachment factor. Our observations warrant further research into the role of Sia binding in the virus’s host and tissue tropism and transmission, which may be influenced by the observed Sia-binding fine specificity and by differences in sialoglycomes among host species.
Keywords: sialic acid, MERS-CoV, spike, attachment, receptor
Abstract
Middle East respiratory syndrome coronavirus (MERS-CoV) targets the epithelial cells of the respiratory tract both in humans and in its natural host, the dromedary camel. Virion attachment to host cells is mediated by 20-nm-long homotrimers of spike envelope protein S. The N-terminal subunit of each S protomer, called S1, folds into four distinct domains designated S1A through S1D. Binding of MERS-CoV to the cell surface entry receptor dipeptidyl peptidase 4 (DPP4) occurs via S1B. We now demonstrate that in addition to DPP4, MERS-CoV binds to sialic acid (Sia). Initially demonstrated by hemagglutination assay with human erythrocytes and intact virus, MERS-CoV Sia-binding activity was assigned to S subdomain S1A. When multivalently displayed on nanoparticles, S1 or S1A bound to human erythrocytes and to human mucin in a strictly Sia-dependent fashion. Glycan array analysis revealed a preference for α2,3-linked Sias over α2,6-linked Sias, which correlates with the differential distribution of α2,3-linked Sias and the predominant sites of MERS-CoV replication in the upper and lower respiratory tracts of camels and humans, respectively. Binding is hampered by Sia modifications such as 5-N-glycolylation and (7,)9-O-acetylation. Depletion of cell surface Sia by neuraminidase treatment inhibited MERS-CoV entry of Calu-3 human airway cells, thus providing direct evidence that virus–Sia interactions may aid in virion attachment. The combined observations lead us to propose that high-specificity, low-affinity attachment of MERS-CoV to sialoglycans during the preattachment or early attachment phase may form another determinant governing the host range and tissue tropism of this zoonotic pathogen.
The Middle East respiratory syndrome coronavirus (MERS-CoV) was identified in 2012 as a novel zoonotic virus, with cases largely confined to the Arabian Peninsula (1). Comparative sequence analysis identified MERS-CoV as a clade C Betacoronavirus (order Nidovirales, family Coronaviridae, subfamily Coronavirinae, genus Betacoronavirus) (2, 3), closely related to bat CoVs HKU4 and HKU5 and to a hedgehog CoV (4, 5). In humans, the virus primarily infects the lower respiratory tract and gives rise to acute pneumonia with clinical features remarkably similar to those associated with the severe acute respiratory syndrome (SARS)-CoV outbreak a decade earlier. As of July 6, 2017, 2,040 laboratory-confirmed cases of MERS have been reported, with 712 fatalities (34.9%) (6). Dromedary camels, the natural reservoir of MERS-CoV (7–9), are the main source for recurrent human infection. Human-to-human virus transmission is inefficient and occurs predominantly nosocomially (10–13).
CoV receptor recognition is mediated by the spike (S) glycoprotein. Hence, the receptor interaction of the S protein governs the zoonotic potential of CoVs (14). Recent elucidation of Betacoronavirus spike protein structures by cryoelectron microcopy revealed a multidomain architecture of the S1 receptor-binding subunit with four individually folded domains, designated S1A through S1D (15, 16). Depending on the Betacoronavirus species, S1A, S1B, or both have been implicated in receptor binding (17–22). CoVs may use both proteinaceous and sialoglycan-based receptor determinants for attachment (23, 24). Proteinaceous receptors are generally bound via S1B, as is the case for MERS-CoV attachment to entry receptor dipeptidyl peptidase 4 (DPP4) (25, 26). Conversely, for those CoVs binding to sialoglycan-based entry receptors, like the clade A beta-CoVs bovine CoV (BCoV) and human CoV (HCoV) OC43, these interactions are mediated by the S1 N-terminal domain (S1A) (21). Remarkably, some CoVs, among them two alphacoronaviruses [transmissible gastroenteritis virus (TGEV) and feline enteric CoV (FeCV)], use both protein- and sialoglycan-based receptor determinants, with binding to aminopeptidase N (APN) required for entry and cell surface sialic acids (Sias) serving as attachment factors (27–30). Although this phenomenon of dual-receptor binding as yet has received only modest attention in CoV research, it may be highly relevant to host and organ tropism and pathogenesis. Indeed, in the case of TGEV, binding to Sia is essential for enteropathogenicity (31). For MERS-CoV, host susceptibility correlates with DPP4 sequence conservation of the virus-binding motif and DPP4 respiratory tract distribution in most cases, but with notable exceptions (32). The question thus remains whether the zoonotic potential of MERS-CoV and its pathogenicity in the human host can be attributed to its specific binding to DPP4 exclusively, and whether other factors promoting or hampering cross-species transmission have to be taken into account.
In the present study, we observed hemagglutinating activity of MERS-CoV particles but were unable to confirm this interaction using recombinant soluble spike proteins. As virus–glycan interactions are inherently dynamic and characteristically of low affinity, we presented CoV S1 receptor-binding subunits on nanoparticles to increase the avidity through multivalent interactions, and thereby sensitivity of detection (33). This approach revealed a hitherto unknown, and potentially important, interaction of the MERS-CoV spike protein domain S1A with sialoglycoconjugates that may well contribute to the host and tissue tropism and transmission of this zoonotic pathogen.
Results
MERS-CoV Can Hemagglutinate Human Erythrocytes.
To identify cellular factors, other than DPP4, potentially involved in MERS-CoV cell attachment, we performed classical hemagglutination assays commonly used to detect virus–Sia interactions. Unlike SARS-CoV but similar to influenza A virus (IAV), MERS-CoV virions caused agglutination of human erythrocytes (Fig. 1). Because the spike glycoprotein is the only surface projection of clade C beta-CoVs, including MERS-CoV, we attempted to validate the observation in a hemagglutination assay using the recombinantly expressed S1 subunit of the MERS-CoV S protein that was transformed into a dimer by C-terminal fusion with the Fc part of human IgG. However, no hemagglutination was seen for MERS-CoV S1-Fc protein, not even when tested at concentrations of up to 500 μg/mL. We therefore considered the option that the observed hemagglutination by MERS-CoV was mediated through simultaneous low-affinity binding of multiple spike proteins on the virus particles with multiple receptors on the surface of human erythrocytes. Such low-affinity interactions may be augmented through multivalency-driven, high-avidity binding (34–36). Therefore, we opted to design a well-defined, self-assembling nanoparticle for arrayed presentation of S1 and S1 subdomains.
Fig. 1.
MERS-CoV particles display a hemagglutination phenotype. MERS-CoV (EMC strain), SARS-CoV (HKU39849 strain), and IAV (PR8 strain) stocks were serially diluted twofold (starting dilution of 107 TCID50 per milliliter) and then incubated with washed human erythrocytes. Hemagglutination was scored after 2 h of incubation at 4 °C. Mixing of erythrocytes with blocking buffer was used as a negative control (mock). Wells positive for hemagglutination are encircled. Hemagglutination assays were performed at least two times.
Design and Expression of Lumazine Synthase Nanoparticles Enabling Multivalent Presentation of Fc-Tagged Viral Receptor-Binding Proteins.
We selected the 154-residue-long lumazine synthase (LS) protein of the hyperthermophile Aquifex aeolicus bacterium that self-assembles into 15-nm-wide 60-meric particles with icosahedral symmetry (37). To allow multivalent presentation of Fc-tagged MERS-CoV S1, the LS protein was extended N-terminally with the 59-residue-long domain B of protein A (pA) of Staphylococcus aureus (Fig. 2A), known for its capacity to bind immunoglobulins via their Fc region (38). The pA-LS protein was provided with an N-terminal signal sequence to allow secretion from mammalian cells, and a C-terminal streptavidin tag was appended for affinity purification (SI Appendix, Fig. S1). The pA-LS protein was affinity-purified from cell culture supernatant of transiently transfected HEK-293T cells and migrated according to its calculated molecular mass of 26 kDa (SI Appendix, Figs. S2 and S3). Analysis of purified pA-LS by electron microscopy revealed spherical particles of ±15 nm in diameter (Fig. 2B), and size exclusion chromatography coupled with multiangle light scattering (SEC-MALS) analysis of these pA-LS particles demonstrated that the mass of the main elution peak was close to that of the predicted 60-meric state of the nanoparticle (SI Appendix, Fig. S3). Binding of Fc-tagged S1 subunits (Fig. 2C) to the pA domain on pA-LS nanoparticles was confirmed by ELISA (SI Appendix, Fig. S4), and electron microscopy (Fig. 2B, Right).
Fig. 2.
(A) Design of self-assembling nanoparticles with Ig-binding potential. Surface representation of the 60-meric LS nanoparticle of A. aeolicus (N-terminal residues are labeled in red) and ribbon presentation of Ig-binding domain B of pA of S. aureus, including a schematic of LS, pA-LS, and pA-LS in complex with chimeric CoV S1 fused to Fc-part of human IgG1 (blue-purple). (B) Electron microscopic analysis of LS nanoparticles. Representative electron microscopic photographs of pA-LS (mean diameter of 15.21 ± 1.00) and pA-LS complexed with MERS-CoV S1-Fc (molar ratio of 1:1.2) are shown. (Scale bar, 50 nm.) (C) Schematic representation of the MERS-CoV spike protein sequence (drawn to scale). S1 and S2 subunits of the 1,353-aa-long MERS-CoV spike protein are indicated, as well as the four domains (A–D) within S1 and their respective boundaries: A (blue), B (green), C (yellow), and D (orange). The positions of the transmembrane domain (black bar; predicted by the TMHMM server) and of the predicted N-glycosylation sites (Ψ; predicted by the NetNGlyc server) are indicated.
MERS-CoV S1 Hemagglutinating Activity Is Sia-Dependent.
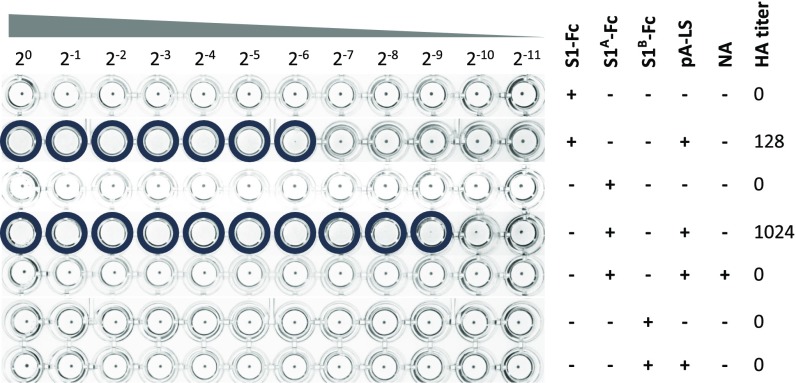
Nanoparticle-displayed MERS-CoV S1 exhibited hemagglutination activity (2.5 μg of S1-Fc combined with 0.5 μg of pA-LS; HA titer = 128) (Fig. 3A). Since no hemagglutination activity was observed for the (dimeric) MERS-CoV S1-Fc, multivalent presentation of S1-Fc appeared critical for the hemagglutination phenotype. No hemagglutination was observed with human erythrocytes desialylated by pretreatment with bacterial neuraminidase (NA; Arthrobacter ureafaciens), indicating that the observed MERS-CoV S1–erythrocyte interaction is strictly Sia-dependent (Fig. 3A).
Fig. 3.
(A) MERS-CoV S1-Fc hemagglutination phenotype is dependent on multivalent presentation on nanoparticles and on Sia-containing receptors on the erythrocyte surface. Human erythrocytes were mock-treated (PBS) or NA-treated, and incubated with a twofold serial dilution of MERS-CoV S1-Fc, pA-LS, or a combination. Hemagglutination was scored after 2 h of incubation at 4 °C. Wells positive for hemagglutination are encircled. Experiments were performed at least two times. (B) Defining the optimal pA-LS/S1-Fc ratio for hemagglutination. A fixed amount of MERS-CoV S1-Fc (2.5 μg) was preincubated with varying amounts of pA-LS (4, 2, 1, 0.5, 0.25, or 0.125 μg) serially diluted twofold and then incubated with washed human erythrocytes. Hemagglutination was scored after 2 h of incubation at 4 °C, and an optimal pA-LS/S1-Fc ratio was determined. Conditions lacking either pA-LS or MERS-CoV S1-Fc protein were used as a negative control. Wells positive for hemagglutination are encircled. Experiments were performed at least two times.
The pA-LS/S1-Fc ratio showing the highest hemagglutination titer was determined to be 1 μg per 2.5 μg (molar ratio = 1:0.6), indicating that ∼36 MERS-CoV S1 receptor-binding subunits, presented as 18 S1-Fc dimers, can be accommodated on the 60-meric pA-LS nanoparticle providing optimal hemagglutination efficiency (Fig. 3B and SI Appendix, Table S1). Aside from S1-Fc proteins of MERS-CoV and several previously documented Sia-interacting CoVs [TGEV, porcine epidemic diarrhea virus (PEDV), BCoV, HCoV-OC43, and infectious bronchitis virus (IBV) (39–43)], none of the tested S1 subunits, including those of the MERS-CoV–related Betacoronavirus clade C bat CoVs HKU4 and HKU5, displayed hemagglutination activity in the nanoparticle-based assay (SI Appendix, Table S2).
Sia Binding Site Locates Within Domain A of the MERS-CoV S1 Subunit.
To identify the S1 domain of the MERS-CoV spike protein involved in Sia binding, we expressed (SI Appendix, Fig. S2) and assessed the hemagglutination activity of Fc-tagged MERS-CoV S1A and S1B domains, as these are the two domains known to facilitate receptor interactions for Betacoronavirus. In contrast to the MERS-CoV S1B domain, nanoparticle-displayed MERS-CoV S1A-Fc was capable of mediating hemagglutination (Fig. 4). These data demonstrate that the Sia-binding capacity of the MERS-CoV S1 subunit resides in its domain A.
Fig. 4.
MERS-CoV S1-mediated Sia-binding activity resides in S1A. MERS-CoV S1-, S1A-, and S1B-Fc proteins alone or complexed with pA-LS were subjected to hemagglutination assay using human erythrocytes, after which HA titers were determined. MERS-CoV S1A-Fc hemagglutination activity was additionally tested with NA-treated human erythrocytes. Wells positive for hemagglutination are encircled. Hemagglutination assays were performed at least two times.
MERS-CoV S1A Preferentially Binds to Nonmodified α2,3-Linked N-Acetylneuraminic Acid.
Sialoglycans occur in an extraordinary structural variety, which arises from the composition and complexity of the glycan chain, differences in the glycosidic linkage through which the Sia is joined to the adjacent sugar residue, and differential modification of Sia at carbon 5 [either N-acetyl moiety [N-acetylneuraminic acid (Neu5Ac)] or N-glycolyl moiety [N-glycolylneuraminic (Neu5Gc)] in combination with modifications of any of the hydroxyl groups at C4, C7, C8, and C9, most often by acetate esters. To assess the specificity of MERS-CoV, glycan array analysis was performed with Fc-tagged MERS-S1A-loaded pA-LS nanoparticles against a library of 135 glycans. We used NA-treated MERS-CoV S1A as desialylation increased the Sia-binding potential of the MERS-CoV S1 subunit (SI Appendix, Table S3), similar to what has been reported for the Alphacoronavirus TGEV and Gammacoronavirus IBV (43, 44) for which Sia interaction has been shown to be biologically relevant (31, 45), consequently enhancing glycan array sensitivity. A recombinant Fc-tagged HA ectodomain of a human H1N1 IAV (A/California/04/2009) with a binding preference for α2,6-linked sialylated glycans (46) was taken along as a positive control. Unloaded nanoparticles, included as a negative control, displayed no binding on the glycan array (Fig. 5A). Indeed, nanoparticles loaded with dimeric HA-Fc displayed specific binding to α2,6-linked sialoglycoconjugates, as was previously reported the for trimeric HA ectodomain (46) (Fig. 5 and SI Appendix, Table S4). MERS-CoV spike protein domain A predominantly bound to short, sulfated, α2,3-linked mono-sialotrisaccharides (glycan nos. 11, 12, and 15), as well as to long, branched, α2,3-linked di-Sia and tri-Sia glycans with a minimum extension of 3 Galβ1–4GlcNAcβ1–3 (LacNAc) tandem repeats (Fig. 5 and Dataset S1). Generally, no or low binding was observed to α2,6-linked sialosides. Strikingly, MERS-CoV S1A did not bind Neu5Gc containing α2,3-linked glycans in contrast to the α2,3-linked Neu5Ac glycan counterparts (Fig. 5B). In accordance, nanoparticle-displayed MERS-CoV S1A did not agglutinate equine erythrocytes that exclusively contain Neu5Gc sialoglycans (47), irrespective of the presence of potentially interfering O-acetyl moieties on the C4 and C9 positions of erythrocyte sialoglycans, which were enzymatically removed using CoV hemagglutinin-esterase proteins (SI Appendix, Table S4). In contrast, the PEDV S1 protein that can bind to Neu5Ac and Neu5Gc (42) did agglutinate erythrocytes of horse origin.
Fig. 5.
(A) MERS-CoV S1A-loaded nanoparticles preferentially bind α2,3-linked sialoglycans. Glycan array screen of MERS-CoV S1A-Fc–loaded pA-LS nanoparticles (Upper), IAV HA-Fc–loaded pA-LS nanoparticles (Middle), or pA-LS nanoparticles alone (Lower). MERS-CoV S1A-Fc nanoparticles show specific binding toward several α2,3-linked Neu5Ac glycans (salmon, 11–77) and some α2,6-linked Neu5Ac glycans (blue, 78–130). IAV HA-Fc nanoparticles show specific binding to α2,6-linked Neu5Ac glycans. Neither of the viral lectins showed binding to glycans lacking terminal Sia residues (gray, 1–10) or to Neu5Gc sialoglycans (purple, 131–135). No binding was observed for pA-LS nanoparticles alone. Bars display the average fluorescence units of six measurements after removal of the highest and lowest signals. Error bars represent the corresponding SEM. The list of glycans imprinted on the array and binding scores are provided in SI Appendix, Table S4. Note that the panel for IAV HA is on a different scale due to the relatively low avidity of MERS-CoV S1A. (B) List of glycans bound by MERS-CoV S1A–loaded nanoparticles above the arbitrary cutoff value of 2,000 relative fluorescence units. Indicated are the glycan schematic structure, average relative fluorescence units (RFU), and glycan number. When present on the glycan array, the α2,3- or α2,6-linked Neu5Ac and Neu5Gc derivatives of each hit are shown for comparison.
Sia 9-O-Acetyl Moiety Prevents MERS-CoV S1A Binding.
The MERS-CoV spike protein appears to bind to nonmodified Neu5Ac sialosides, which suggests that, other than for beta 1 CoVs (40, 41, 48), Sia O-acetylation is not a prerequisite for binding. We wondered what effect Sia 9-O-acetylation would have on MERS-CoV spike–Sia interaction, and hence used a bovine submaxillary mucin (BSM)-based ELISA, as BSM is a well-characterized substrate rich in 9-O-acetylated Sias with only a minor population of unmodified Neu5Ac (49). While MERS-CoV S1A was unable to interact with mock-treated mucin of bovine origin (Fig. 6A), pretreatment of BSM with sialate–9-O-acetylesterase [i.e., BCoV hemagglutinin-esterase (HE)] enabled its interaction. Similarly, pretreatment of BSM with sialate–9-O-acetylesterase clearly enhanced binding of IAV HA to bovine mucin (Fig. 6A), for which it is known that the acetyl moiety at the Sia C9 position prevents recognition of the saccharide by HA (50). No binding of MERS-CoV S1A and IAV HA was observed upon pretreatment of BSM with NA, confirming that the spike–mucin interaction was Sia-dependent. Consistent with these observations, treatment of rat erythrocytes, which are rich in 9-O-acetylated sialo-glycoconjugates, with sialate–9-O-acetylesterase revealed receptors compatible with MERS-CoV S1A–mediated hemagglutination (SI Appendix, Fig. S5). We conclude that O-acetylation of the Sia glycerol side chain inhibits MERS-CoV S1A binding. Conclusively, we offer that MERS-CoV S1A comprises a Sia binding preference for α2,3-coupled, 5-N-acetylated neuraminic acid in which both 5-N-glycolylation and 9-O-acetylation are not tolerated.
Fig. 6.
(A) Sia-binding activity of MERS-CoV S1A nanoparticles is inhibited by the acetyl moiety of 9-O-acetylated Sias. BSM-coated ELISA plates were mock treated, de–9-O-acetylated using BCoV HE, or desialylated using NA before incubation with twofold serially diluted (pA-LS–complexed) viral proteins. Fc-tagged IAV HA and HCoV-OC43 S1 were taken along as control proteins for HE and NA treatment. BSM-bound pA-LS nanoparticles were detected by ELISA with HRP-conjugated StrepMAB antibody as described above. BSM pretreated with NA served as a negative control. The experiment was performed in triplicate and repeated at least two times. A representative experiment is shown. Error bars represent the corresponding SEM. (B) Binding of MERS-CoV S1A nanoparticles to human mucin. Equimolar amounts of Fc-tagged MERS-CoV S1A, IAV HA, and pAPN were coupled to pA-LS nanoparticles, serially diluted twofold, and incubated on plates coated with human mucin that were mock-pretreated or pretreated with NA. IAV HA-Fc and pAPN-Fc were taken along as controls. Mucin-bound pA-LS nanoparticles were detected as described in A. The experiment was performed in triplicate and repeated at least two times. A representative experiment is shown. Error bars represent the corresponding SEM. (C) Staining with M. amurensis lectin II (specific for α2,3-linked Sias) of camel and human nose and lung tissues. (Magnification: 400×.)
MERS-CoV S1A Is Suited to Bind Human Mucin.
Reportedly, 9-O-acetylated Sias are scarce in the respiratory tissue of humans (49), while glycolylated Sias are considered to be absent altogether (51, 52). Accordingly, we examined whether mucin derived from human respiratory epithelial cells could be bound by MERS-CoV S1A. Nanoparticle-displayed MERS-CoV S1A showed binding to human mucin in a concentration-dependent manner similar to IAV HA (Fig. 6B), whereas negative control pAPN did not. Pretreatment of human mucin with sialidase completely inhibited binding by MERS-CoV S1A and IAV HA, demonstrating the Sia-dependent nature of the observed interactions (Fig. 6B). Consistent with previous binding assays, MERS-CoV S1A binding to mucin was only detected when presented on nanoparticles, indicating that the multivalent lectin–carbohydrate interactions enhance binding affinity (SI Appendix, Fig. S6). In addition, MERS-CoV S1A binding to human mucin was observed to be temperature-sensitive and displayed weaker binding at higher temperatures (SI Appendix, Fig. S7). These results contrast with the observations made for BSM and suggest that the MERS-CoV S1A protein is particularly suited to bind human respiratory mucin.
α2,3-Linked Sias Colocalize with Site of MERS-CoV Replication in Vivo.
With the preferential binding of MERS-CoV S1A to α2,3-linked sialoglycans in mind, we next studied the distribution of these glycans in the upper (nose) and lower (lung) respiratory tissues of humans and the natural reservoir of MERS-CoV: dromedary camels. Lectin histochemistry on these tissues suggests that α2,3-linked sialoglycans are abundant in the camel nasal respiratory epithelium and the alveoli of the human lung (Fig. 6C), which coincides with DPP4 expression and the site of MERS-CoV replication in these mammals (53).
Sia Serves as an Attachment Receptor for MERS-CoV.
To study whether the MERS-CoV Sia-binding activity may aid virus cell entry, African green monkey kidney Vero cells and human airway epithelial Calu-3 cells were depleted for cell surface Sias by NA treatment or mock-treated. The cells were then inoculated with MERS-CoV and IAV [multiplicity of infection (MOI) = 0.1], and infection levels were assessed by immunostaining. As expected, NA treatment rendered Vero and Calu-3 cells almost completely resistant to the Sia-dependent IAV. In Vero cells, NA treatment did not affect MERS-CoV entry efficiency. However, Sia depletion of Calu-3 cells reduced MERS-CoV entry by more than 70% compared with control-treated cells (Fig. 7 A and B). The difference between Vero and Calu-3 cells may be explained by a difference in the levels of Sia cell surface expression. As shown by flow cytometry, both cell types display similar levels of the entry receptor DPP4 (Fig. 7C), but in Calu-3 cells, those of cell surface MERS-CoV S1A glycotopes are fivefold higher (Fig. 7D). Our findings provide direct evidence that sialylated proteins or glycolipids on the surface of human airway epithelial cells can be used as an attachment receptor by MERS-CoV, and thereby increase infection efficiency.
Fig. 7.
(A) Sia depletion of Calu-3 cells inhibits MERS-CoV infection. Vero and Calu-3 cells were treated with or without NA and inoculated with MERS-CoV or IAV for 1 h at 37 °C. Cells were subsequently washed once, and fresh medium was added. Following incubation at 37 °C for 8 h, cells were fixed and the percentage of infected cells was determined by immunostaining and counting. Data are presented as mean ± SE and statistically analyzed with a t test. (B) Immunofluorescence staining on MERS-CoV–infected Vero and Calu-3 cells with and without NA pretreatment with the MERS-CoV N protein visualized in green. (Magnification: 100×.) (C) Surface expression of DPP4 on Vero and Calu-3 cells measured by flow cytometry. (D) Binding of nanoparticle-displayed MERS-CoV spike domain S1A to Vero and Calu-3 cells with and without NA pretreatment measured by flow cytometry. FACS data are presented as the geometric mean fluorescence intensity (gMFI) ratio between samples and isotype controls at a 95% confidence interval. The dotted line indicates the background level, equal to the isotype control. The data were analyzed with a nonparametric Mann–Whitney test (*P < 0.05; **P < 0.01; ***P < 0.001). All experiments were performed at least in triplicate and repeated three times. n.s., not significant.
Discussion
Our data demonstrate that MERS-CoV carries dual-binding specificity for host molecules and engages both sialoglycans and DPP4 via distinct domains of its spike protein. The multivalent presentation of S1 receptor-binding subunits onto 60-meric nanoparticles enabled us to detect a low-affinity interaction of the MERS-CoV spike protein with Sias via its domain A. The existence of CoV spike–Sia interactions has been reported for several CoVs, including clade A betacoronaviruses; the alphacoronaviruses TGEV, PEDV, and FeCV; and Gammacoronavirus IBV (27, 39–43), all of which interact with Sia via their N-terminal spike domains. Although the structural conservation of these domains is not obvious from the primary protein sequence, available cryoelectron microscopy structures demonstrated that the spike N-terminal domains of the Alphacoronavirus HCoV-NL63 and betacoronaviruses murine hepatitis virus (MHV), HCoV-HKU1 and MERS-CoV display structural similarity (54, 55). Despite the apparent structural conservation of this domain, the observation that genetically distant CoVs comprise Sia-binding activity implies that this feature has independently evolved several times throughout CoV history.
Viral interactions with Sias are typically of a reversible nature so as to prevent nonproductive binding to the abundance of sialoglycans on off-target host cells and non–cell-associated structures, as well as for release of viral progeny from infected cells. For instance, clade A betacoronaviruses and ICV encode sialate–O-acetyl-esterases to mediate their reversible binding to O-acetylated Sias (56), while IAVs and IBVs are provided with the sialidase NA to facilitate reversible binding to Sia decoys. By contrast, reversible interaction with Sia by viruses that lack Sia-destroying enzymes, such as several noroviruses, polyoma viruses, and CoVs, including MERS-CoV, is merely governed by the binding equilibrium (57). Because of the inherent low affinity for such receptors by viruses lacking a receptor-destroying enzyme, detection of these weak interactions, particularly by isolated viral lectin domains, may prove difficult. This is illustrated by our observation that only nanoparticle-based assays constituting enhanced avidity through multivalency proved sensitive enough to detect the direct interaction between MERS-CoV S1 and sialoglycans.
Our data indicate that sialoglycans may aid MERS-CoV entry on DPP4-positive cells. Absence of a Sia-dependent effect on Vero cell infection correlated with a low abundancy of MERS-CoV S1A receptors. By contrast, infection efficiency of a more relevant cell type, namely, the human airway Calu-3 epithelial cells, was reduced significantly after depletion of cell surface Sias. These cells, derived from bronchial submucosal glands that are targeted by MERS-CoV (58), showed high abundancy of MERS-CoV–receptive glycans. These findings provide formal evidence that sialoglycans may serve as an attachment receptor for virus particles, and thereby aid viral entry. Presumably, binding to host cell sialoglycans mediates virion concentration at the cell surface, and thus may increase the likelihood of MERS-CoV spike engagement with the DPP4 entry receptor.
The receptor use of MERS-CoV resembles that of the Alphacoronavirus TGEV and Betacoronavirus MHV, for which binding to cell surface-sialylated glycans was not required for infection of cultured cells but contributes to the efficiency of binding (30, 59–62). In addition to Sias, TGEV is able to interact with porcine APN (29) and MHV engages with CEACAM1a (63), both of which serve as a cellular receptor. High-affinity binding to proteinaceous receptors by MERS-CoV, TGEV, and MHV appears sufficient for entry in cultured cells, and primary attachment to sialylated proteins or lipids on the cell surface may aid the virus to get into contact with its protein receptor. This is in sharp contrast to other Sia-binding CoVs such as BCoV and IBV, for which no protein receptor has been identified and which critically depend on (O-acetylated) Sias during cell entry (45, 48, 64).
Our data indicate that MERS-CoV has evolved a binding site of low affinity but high selectivity for Neu5Ac that excludes Neu5Gc. Likewise, binding to Sias modified by the addition of an acetyl group at the C9 position is not permitted. The selection on Sia linkage types to the penultimate sugar in the glycan chain appears less stringent, although a distinct preference for α2,3-linked sialoglycans (over α2,6 linkages) is observed, the distribution of which correlates with the predominant sites of MERS-CoV replication in the upper and lower respiratory tracts of camels and humans, respectively. In particular, long bi- and triantennary α2,3-linked, sialylated poly-LacNAc structures are recognized, indicating that multivalency of the ligand is important for binding.
Virus receptor-binding specificity and variation in receptor distribution can be critical for host and tissue tropism, pathogenesis, and (cross-species) transmissibility of viruses (65, 66). For the zoonotic MERS-CoV, the spike–DPP4 interaction has been a focus of study to understand these aspects of the virus biology and epidemiology. The high sequence identity of spike-interacting residues on dromedary and human DPP4 facilitates cross-species transmission without the need for adaptive mutations (17, 67). The reported differential receptor distribution in the respiratory tract may account for the observed dissimilar MERS-CoV–induced pathology in dromedaries and humans, as well as the apparent inefficient interhuman transmission (53). Although receptor interaction and distribution are key determinants for virus tropism, they cannot solely explain the species’ susceptibility to MERS-CoV infection, as illustrated by the resistance of horses to MERS-CoV infection (32). MERS-CoV–contacting residues on equine and human DPP4 are identical (68), and equine DPP4 supports MERS-CoV infection in cultured cells (69). In addition, DPP4 is highly and widely expressed along the respiratory tract of horses (32). Nevertheless, horses appear to be resistant to experimental infection with MERS-CoV (32), an observation that is supported by the lack of MERS-CoV antibodies in horse sera collected in the Middle East region (70), suggesting that host factors other than DPP4 contribute to viral tropism. For influenza viruses, it has been well established that Sia-binding preference and distribution of Sia species in animals and humans are critical determinants for host and tissue tropism (65, 66, 71, 72). Intriguingly, the epithelial cells of the horse trachea are highly enriched in N-glycolylneuraminic acid (71), and equine nose epithelia are reportedly covered with a particularly thick mucus layer (32). Whether and how the MERS-CoV Sia-binding fine specificity and the differences in sialoglycomes among species affect virus host and tissue tropism clearly warrant further investigation. Detailed structural insight into the binding mode of the S1A domain with Sias would enable generation of MERS-CoV mutants incapable of Sia interaction that could be instrumental in determining the role of Sia interaction in vivo.
Materials and Methods
Protein Design and Expression.
Genes encoding the 6,7-dimethyl-8-ribityllumazine synthase (LS; GenBank accession no. WP_010880027.1) of A. aeolicus and domain B of pA (UniProt accession no. P38507; amino acids 212–270) of S. aureus were synthesized using human-preferred codons obtained from GenScript USA, Inc. The cysteine at position 37 and asparagine at position 102 of LS were mutated to alanine and glutamine, respectively. A pA-LS expression vector was generated by ligating the pA-encoding sequence in-frame with the N terminus of the LS-encoding sequence via a Gly-Ser linker and subsequent cloning into the pCAGGS mammalian expression vector. In addition, LS ORFs in the LS and pA-LS expression vectors were provided with a sequence encoding an N-terminal CD5 signal peptide, and a streptomycin tag purification tag sequence (IBA) (SI Appendix, Fig. S1). To generate the MERS-CoV S1A-LS and S1B-LS expression vectors, the pA sequence in the pA-LS expression vector was replaced by a sequence encoding the MERS-CoV S1A domain (residues 19–357) (protein sequences are provided in SI Appendix, Fig. S1). Plasmids encoding pA-LS were polyethylenimine (PEI)-transfected into 60% confluent HEK-293T cells for 6 h, after which transfections were removed and medium was replaced with 293 SFM II-based expression medium (Gibco Life Technologies) and incubated at 37 °C in 5% CO2. Tissue culture supernatants were harvested 5–6 d posttransfection, and expressed proteins were purified using StrepTactin Sepharose beads (IBA) according to the manufacturer’s instruction. Purified proteins were stored at 4 °C until further use.
Expression of Fc-tagged S1 subunits of spike proteins of TGEV (GB: ABG89325.1), HCoV-229E (GB: NP_073551.1), HCoV-NL63 (GB: YP_003767.1), PEDV (GB: AOG30832.1), HCoV-HKU1 (GB: YP_173238.1), BCoV (GB: P15777.1), HCoV-OC43 (GB: AAR01015.1), SARS-CoV (GB: AAX16192.1), MERS-CoV (GB: YP_009047204.1), bat CoV HKU4.2 (GB: ABN10848.1), bat CoV HKU5 (GB: ABN10893.1), IBV (GB: AAW33786.1), and porcine delta coronavirus (PDCoV) (AML40790.1) was performed as described previously (26). Briefly, plasmids encoding the CoV-S1 subunit (or a domain thereof), C-terminally fused to the Fc part of human IgG1, were generated, and proteins were expressed in HEK-293T cells as described above. Similarly, an Fc-tagged version of the HA ectodomain of IAV [A/California/04/2009 (H1N1)] containing T200A and E227A substitutions in the Sia-binding site enhancing Sia affinity (46) and of the ectodomain of porcine APN (GB: XP_005653580.1) were generated. Fc-tagged proteins were purified from tissue culture supernatants 5–6 d posttransfection by pA affinity chromatography (17-0780-01; GE Healthcare), eluted using acid solution (0.1 M citric acid at pH 3.0), and neutralized immediately using Tris at pH 8.8 (0.2 M final concentration), with the exception of HA-Fc, which was purified via a C-terminal streptomycin tag and StrepTactin Sepharose beads and eluted using StrepTactin elution buffer (100 mM Tris⋅HCl, 150 mM NaCl, 1 mM EDTA, 2.5 mM biotin). Purified proteins were analyzed on a 12% SDS/PAGE gel under reducing conditions and stained with GelCodeBlue stain reagent (Thermo Scientific). Purified proteins were stored at 4 °C until further use.
Electron Microscopic Analysis of LS Nanoparticles.
Before the application of samples, 400-mesh copper grids with a pure carbon film were exposed to a glow discharge in air for 20 s to make them hydrophilic. Ten microliters of purified LS, pA-LS, or pA-LS + MERS-CoV S1-Fc (premixed at a 1:1.2 ratio) at 0.2–0.3 mg/mL was applied to the grids and incubated for 1 min. Excess sample was blotted with a filter paper, and for negative staining, 10 μL of 1% phosphotungstic acid at pH 6.8 was applied. After 1 min, excess stain was blotted and grids were left to dry. The specimens were examined in a JEOL JEM2100 transmission electron microscope at 200 kV, and images were taken at a magnification of 80,000× with a Gatan US4000 4K × 4K camera. At least 50 particles were measured to calculate the mean diameter and SD of the pA-LS particles.
SEC-MALS.
For SEC-MALS analysis, purified LS nanoparticles [150 μg in 20 mM Tris⋅HCl (pH 8.0), 150 mM NaCl] were loaded onto a Superose 6 10/300 GL column (0.4 mL⋅min−1; GE Life Sciences) and passed through a Wyatt DAWN Heleos II EOS 18-angle laser photometer coupled to a Wyatt Optilab TrEX differential refractive index detector. Data were analyzed, and weight-averaged molecular masses and mass distributions (polydispersity) for each sample were calculated using Astra 6 software (Wyatt Technology Corp.).
MERS-CoV S1-Fc–Based ELISA.
Nunc Maxisorp plates were coated with 100 μL of D-PBS (PBS with Ca2+/Mg2+) containing 50 ng of MERS-CoV S1-Fc per well and incubated for 4 h at room temperature. Plates were washed three times with washing buffer (PBS + 0.05% Tween-20) and subsequently blocked overnight at 4 °C with 200 μL of blocking buffer (PBS + 0.1% Tween-20, containing 2% BSA). Plates were put at room temperature and washed three times with washing buffer before use. Streptomycin-tagged LS and pA-LS were threefold serially diluted in blocking buffer and incubated for 1 h at 37 °C. Plates were washed three times with washing buffer and once with PBS + 2% BSA, and then incubated for 1 h at 37 °C with 100 μL of a 1:10,000 StrepMAB-classic HRP (IBA) dilution in PBS + 2% BSA. Plates were washed three times with washing buffer. One hundred microliters of TMB Super Slow One Component HRP Microwell Substrate (SurModics) was added to the wells and developed for 4 min. Reaction was stopped with 100 μL of 12.5% sulfuric acid per well, and absorbance was measured at 450 nm using an ELx808 Absorbance Microplate Reader (Bio-Tek Instruments).
Hemagglutination Assays.
The classical hemagglutination assay was performed according to standard procedures. Briefly, twofold dilutions of the MERS-CoV (EMC strain), SARS-CoV (HKU39849 strain), and IAV (PR8 strain) [starting dilution of 107 TCID50 (tissue culture infectious dose at which 50% of cells display cytopathogenic effects) per milliliter] were incubated with 0.5% washed human erythrocytes (diluted in PBS) in V-bottom, 96-well plates (Greiner Bio-One) for 30 min at 4 °C. Plates were incubated for 2 h at 4 °C, after which hemagglutination titer was scored. For the nanoparticle-based hemagglutination assay, Fc-tagged chimeric proteins were complexed with pA-LS at a 0.6:1 molar ratio (unless stated otherwise) for 30 min at 4 °C in blocking buffer (PBS + 0.1% BSA) and subjected to hemagglutination assay as described above. In some experiments, human, rat, or equine erythrocytes were pretreated for 3 h at 37 °C with NA from A. ureafaciens (Roche; diluted to 20 mU/mL in PBS), Fc-tagged ectodomains of the bovine CoV-LUN hemagglutinin-esterase (20 μg/mL), or Fc-tagged ectodomains of MHV-S HE (4 μg/mL), the latter two of which were produced and purified as described by Zeng et al. (73). In some instances, MERS-S1A-Fc protein was treated with 20 mU/mL NA in PBS for 3 h at 37 °C before complexation to pA-LS nanoparticles.
Glycan Array Analysis.
Glycans containing Neu5Gc were synthesized essentially as described for the corresponding glycans terminating in Neu5Ac (74), except that enzymatic addition of Sia in the last step was done with the donor substrate CMP-Neu5Gc instead of CMP-Neu5Ac. Microarrays were printed as described previously (75). Microarray slides were blocked with PBS + 0.1% Tween-20 and 5% Protifar skimmed milk. Empty pA-LS nanoparticles or nanoparticles complexed with A. ureafaciens NA-treated MERS-S1A-Fc or IAV HA-Fc at a 0.6:1 molar ratio (as described above) were diluted in PBS + 1% BSA to a concentration of 500 μM (based on the pA-LS concentration) and incubated for 120 min at 4 °C on the microarray slides under a microscope cover glass in a humidified chamber at room temperature. Microarray slides were subsequently washed by successive rinses with PBS + 0.05% Tween-20 and overlaid with a Chromeo488-conjugated secondary StrepMAB antibody (IBA). Slides were repeatedly washed with PBS + 0.05% Tween-20, PBS, and deionized water, and immediately subjected to imaging as described elsewhere (75).
Immunohistochemistry.
The α2,3-Sia expression on human and dromedary camel nose and lung tissues was assessed by lectin histochemistry. Human and camel tissue samples used in this study were similar to those described in our previous study (53). The staining was performed as previously described using biotinylated Maackia amurensis lectin II (76). The use of human materials was approved by the local medical ethical committee (MEC approval: 2014-414). Human tissues were residual human biomaterials, which are collected, stored, and issued by the Erasmus MC Tissue Bank under ISO 15189:2007 standard operating procedures. Use of these materials for research purposes is regulated according to the Human Tissue and Medical Research code of conduct for responsible use (https://www.federa.org/sites/default/files/digital_version_first_part_code_of_conduct_in_uk_2011_12092012.pdf.). Experimental procedures using dromedaries were approved by the local Ethical Committee of the Autonomous University of Barcelona (no. 8003).
Solid-Phase Lectin-Binding Assay with Human or Bovine Mucin.
Maxisorp flat-bottom, 96-well plates were coated per well with 100 μL of PBS containing 1% (vol/vol) mucin from human airway epithelium (Epithelix) or with 100 μL of D-PBS containing 1 μg of BSM, and incubated for 4 h at room temperature. Plates were washed three times with washing buffer (PBS + 0.05% Tween-20) and subsequently blocked for 2 h at 37 °C with 200 μL of blocking buffer (PBS + 0.1% Tween-20, containing 5% Protifar skimmed milk). Plates were put at room temperature and washed three time with washing buffer before use. In some instances, mucin-coated plates were mock-treated (with PBS), treated with 20 μg/mL sialate–9-O-acetylesterase (BCoV-HE), or treated with 20 mU/mL NA (A. ureafaciens; Roche) (diluted in PBS, 100 μL per well) for 3 h at 37 °C. MERS-CoV S1A-Fc, IAV HA-Fc [A/California/04/2009 (H1N1)] and pAPN-Fc were mock-treated or treated with 20 mU/mL NA (A. ureafaciens; Roche) in PBS for 3 h at 37 °C. Fc-tagged proteins were complexed with pA-LS (2:1 molar ratio) for 30 min at 4 °C, and twofold serial dilutions of mixtures were made using ice-cold blocking buffer. Complexed nanoparticles were incubated on mucin-coated plates for 2 h at 4 °C, after which bound LS nanoparticles were detected by ELISA using the HRP-conjugated anti-StrepMAB antibody that recognizes their C-terminally appended streptomycin tag.
MERS-CoV Infection Experiments.
Confluent layers of Vero or Calu-3 cells were mock-treated or treated with NA (A. ureafaciens) for 2 h at 37 °C, followed by inoculation with MERS-CoV (EMC isolate) or IAV (PR8 strain) (MOI = 0.1). Inoculum was removed after 1 h, and cells were washed once before addition of fresh culture medium. At 8 h postinfection, cells were fixed with formaldehyde, permeabilized with 70% ethanol, and stained using monoclonal antibody against MERS-CoV Nucleoprotein (Immunosource) (32) or a monoclonal antibody against influenza nucleoprotein (IgG2A, Clone Hb65; American Type Culture Collection) according to standard protocols using Alexa Fluor 488-conjugated goat α-rabbit or goat α-mouse antibodies as a second step (25). The percentage of infected cells was determined by immunostaining and counting.
Flow Cytometric Analysis of Cell Surface DPP4 and Sia Expression.
To measure DPP4 expression, Vero and Calu-3 cells were incubated with 5 μg/mL polyclonal goat anti-hDPP4 antiserum (clone AF1180; R&D Systems), washed three times with FACS buffer (2 mM EDTA and 0.05% BSA in PBS), and then subsequently stained with rabbit anti-goat IgG antibodies conjugated with Alexa Fluor 488 in a 1:250 dilution (Life Technologies). To measure Sia expression, Vero and Calu-3 cells were incubated with 7.5 μg/mL S1A-Fc–loaded pA-LS nanoparticles, followed by rabbit antistreptomycin antibodies (1:200), after which cells were washed three times with FACS buffer and stained with Alexa Fluor 633-conjugated secondary anti-rabbit IgG in a 1:250 dilution (Invitrogen). Cells were washed and resuspended for flow cytometric analysis using a BD FACS Canto II (BD Biosciences). Data were analyzed using FlowJo software.
Supplementary Material
Acknowledgments
We thank Herman Egberink and Floor van der Vegt for technical assistance. We thank Monique Spronken and Debby van Riel (Department of Viroscience, Erasmus Medical Center) for providing influenza virus strain and human nose and lung tissues, respectively. This study is supported by TOP Project Grant 40-00812-98-13066 (to B.L.H., F.J.M.v.K., and B.-J.B.), which is funded by ZonMw (Dutch Organization for Health Research and Innovation). Research was performed as part of the Zoonoses Anticipation and Preparedness Initiative [IMI (Innovative Medicines Initiative) Grant Agreement 115760], with the assistance and financial support of the IMI and European Commission, and in-kind contributions from EFPIA (European Federation of Pharmaceutical Industries and Associations) partners. The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1712592114/-/DCSupplemental.
References
- 1.Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus ADME, Fouchier RAM. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med. 2012;367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 2.de Groot RJ, et al. Middle East respiratory syndrome coronavirus (MERS-CoV): Announcement of the Coronavirus Study Group. J Virol. 2013;87:7790–7792. doi: 10.1128/JVI.01244-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Boheemen S, et al. Genomic characterization of a newly discovered coronavirus associated with acute respiratory distress syndrome in humans. MBio. 2012;3:1–9. doi: 10.1128/mBio.00473-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lau SKP, et al. Genetic characterization of Betacoronavirus lineage C viruses in bats reveals marked sequence divergence in the spike protein of pipistrellus bat coronavirus HKU5 in Japanese pipistrelle: Implications for the origin of the novel Middle East respiratory syndrome coronavirus. J Virol. 2013;87:8638–8650. doi: 10.1128/JVI.01055-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corman VM, et al. Characterization of a novel betacoronavirus related to MERS-CoV in European hedgehogs. J Virol. 2013;88:717–724. doi: 10.1128/JVI.01600-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.WHO 2015 Middle East respiratory syndrome coronavirus (MERS-CoV) – Saudi Arabia. Available at www.who.int/csr/don/06-july-2017-mers-saudi-arabia/en/. Accessed July 13, 2017.
- 7.Briese T, et al. Middle East respiratory syndrome coronavirus quasispecies that include homologues of human isolates revealed through whole-genome analysis and virus cultured from dromedary camels in Saudi Arabia. MBio. 2014;5:e01146–e14. doi: 10.1128/mBio.01146-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haagmans BL, et al. Middle East respiratory syndrome coronavirus in dromedary camels: An outbreak investigation. Lancet Infect Dis. 2014;14:140–145. doi: 10.1016/S1473-3099(13)70690-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reusken CBEM, et al. Middle East respiratory syndrome coronavirus neutralising serum antibodies in dromedary camels: A comparative serological study. Lancet Infect Dis. 2013;13:859–866. doi: 10.1016/S1473-3099(13)70164-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Assiri A, et al. KSA MERS-CoV Investigation Team Hospital outbreak of Middle East respiratory syndrome coronavirus. N Engl J Med. 2013;369:407–416. doi: 10.1056/NEJMoa1306742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Memish ZA, et al. Human infection with MERS coronavirus after exposure to infected camels, Saudi Arabia, 2013. Emerg Infect Dis. 2014;20:1012–1015. doi: 10.3201/eid2006.140402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cotten M, et al. Transmission and evolution of the Middle East respiratory syndrome coronavirus in Saudi Arabia: A descriptive genomic study. Lancet. 2013;382:1993–2002. doi: 10.1016/S0140-6736(13)61887-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Azhar EI, et al. Evidence for camel-to-human transmission of MERS coronavirus. N Engl J Med. 2014;370:2499–2505. doi: 10.1056/NEJMoa1401505. [DOI] [PubMed] [Google Scholar]
- 14.Hulswit RJG, de Haan CAM, Bosch B-J. Coronavirus spike protein and tropism changes. Adv Virus Res. 2016;96:29–57. doi: 10.1016/bs.aivir.2016.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walls AC, et al. Cryo-electron microscopy structure of a coronavirus spike glycoprotein trimer. Nature. 2016;531:114–117. doi: 10.1038/nature16988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kirchdoerfer RN, et al. Pre-fusion structure of a human coronavirus spike protein. Nature. 2016;531:118–121. doi: 10.1038/nature17200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu G, et al. Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26. Nature. 2013;500:227–231. doi: 10.1038/nature12328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li F, Li W, Farzan M, Harrison SC. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 19.Peng G, et al. Crystal structure of mouse coronavirus receptor-binding domain complexed with its murine receptor. Proc Natl Acad Sci USA. 2011;108:10696–10701. doi: 10.1073/pnas.1104306108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu K, Li W, Peng G, Li F. Crystal structure of NL63 respiratory coronavirus receptor-binding domain complexed with its human receptor. Proc Natl Acad Sci USA. 2009;106:19970–19974. doi: 10.1073/pnas.0908837106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peng G, et al. Crystal structure of bovine coronavirus spike protein lectin domain. J Biol Chem. 2012;287:41931–41938. doi: 10.1074/jbc.M112.418210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reguera J, et al. Structural bases of coronavirus attachment to host aminopeptidase N and its inhibition by neutralizing antibodies. PLoS Pathog. 2012;8:e1002859. doi: 10.1371/journal.ppat.1002859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwegmann-Wessels C, Herrler G. Sialic acids as receptor determinants for coronaviruses. Glycoconj J. 2006;23:51–58. doi: 10.1007/s10719-006-5437-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wickramasinghe INA, de Vries RP, Gröne A, de Haan CAM, Verheije MH. Binding of avian coronavirus spike proteins to host factors reflects virus tropism and pathogenicity. J Virol. 2011;85:8903–8912. doi: 10.1128/JVI.05112-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raj VS, et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mou H, et al. The receptor binding domain of the new Middle East respiratory syndrome coronavirus maps to a 231-residue region in the spike protein that efficiently elicits neutralizing antibodies. J Virol. 2013;87:9379–9383. doi: 10.1128/JVI.01277-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Desmarets LMB, Theuns S, Roukaerts IDM, Acar DD, Nauwynck HJ. Role of sialic acids in feline enteric coronavirus infections. J Gen Virol. 2014;95:1911–1918. doi: 10.1099/vir.0.064717-0. [DOI] [PubMed] [Google Scholar]
- 28.Tresnan DB, Levis R, Holmes KV. Feline aminopeptidase N serves as a receptor for feline, canine, porcine, and human coronaviruses in serogroup I. J Virol. 1996;70:8669–8674. doi: 10.1128/jvi.70.12.8669-8674.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Delmas B, et al. Aminopeptidase N is a major receptor for the entero-pathogenic coronavirus TGEV. Nature. 1992;357:417–420. doi: 10.1038/357417a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwegmann-Wessels C, Zimmer G, Laude H, Enjuanes L, Herrler G. Binding of transmissible gastroenteritis coronavirus to cell surface sialoglycoproteins. J Virol. 2002;76:6037–6043. doi: 10.1128/JVI.76.12.6037-6043.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krempl C, Schultze B, Laude H, Herrler G. Point mutations in the S protein connect the sialic acid binding activity with the enteropathogenicity of transmissible gastroenteritis coronavirus. J Virol. 1997;71:3285–3287. doi: 10.1128/jvi.71.4.3285-3287.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vergara-Alert J, et al. Livestock susceptibility to infection with Middle East respiratory syndrome coronavirus. Emerg Infect Dis. 2017;23:232–240. doi: 10.3201/eid2302.161239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee RT, Lee YC. Affinity enhancement by multivalent lectin-carbohydrate interaction. Glycoconj J. 2000;17:543–551. doi: 10.1023/a:1011070425430. [DOI] [PubMed] [Google Scholar]
- 34.Bakkers MJG, et al. Betacoronavirus adaptation to humans involved progressive loss of hemagglutinin-esterase lectin activity. Cell Host Microbe. 2017;21:356–366. doi: 10.1016/j.chom.2017.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jardine J, et al. Rational HIV immunogen design to target specific germline B cell receptors. Science. 2013;340:711–716. doi: 10.1126/science.1234150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carlson CB, Mowery P, Owen RM, Dykhuizen EC, Kiessling LL. Selective tumor cell targeting using low-affinity, multivalent interactions. ACS Chem Biol. 2007;2:119–127. doi: 10.1021/cb6003788. [DOI] [PubMed] [Google Scholar]
- 37.Wörsdörfer B, Woycechowsky KJ, Hilvert D. Directed evolution of a protein container. Science. 2011;331:589–592. doi: 10.1126/science.1199081. [DOI] [PubMed] [Google Scholar]
- 38.Löfkvist T, Sjöquist J. Chemical and serological analysis of antigen preparations from Staphylococcus aureus. Acta Pathol Microbiol Scand. 1962;56:295–304. [Google Scholar]
- 39.Bingham RW, Madge MH, Tyrrell DA. Haemagglutination by avian infectious bronchitis virus-a coronavirus. J Gen Virol. 1975;28:381–390. doi: 10.1099/0022-1317-28-3-381. [DOI] [PubMed] [Google Scholar]
- 40.Künkel F, Herrler G. Structural and functional analysis of the surface protein of human coronavirus OC43. Virology. 1993;195:195–202. doi: 10.1006/viro.1993.1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schultze B, Gross H-J, Brossmer R, Herrler G. The S protein of bovine coronavirus is a hemagglutinin recognizing 9-O-acetylated sialic acid as a receptor determinant. J Virol. 1991;65:6232–6237. doi: 10.1128/jvi.65.11.6232-6237.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu C, et al. Receptor usage and cell entry of porcine epidemic diarrhea coronavirus. J Virol. 2015;89:6121–6125. doi: 10.1128/JVI.00430-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schultze B, et al. Transmissible gastroenteritis coronavirus, but not the related porcine respiratory coronavirus, has a sialic acid (N-glycolylneuraminic acid) binding activity. J Virol. 1996;70:5634–5637. doi: 10.1128/jvi.70.8.5634-5637.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schultze B, Cavanagh D, Herrler G. Neuraminidase treatment of avian infectious bronchitis coronavirus reveals a hemagglutinating activity that is dependent on sialic acid-containing receptors on erythrocytes. Virology. 1992;189:792–794. doi: 10.1016/0042-6822(92)90608-R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Winter C, Schwegmann-Wessels C, Cavanagh D, Neumann U, Herrler G. Sialic acid is a receptor determinant for infection of cells by avian infectious bronchitis virus. J Gen Virol. 2006;87:1209–1216. doi: 10.1099/vir.0.81651-0. [DOI] [PubMed] [Google Scholar]
- 46.de Vries RP, et al. Only two residues are responsible for the dramatic difference in receptor binding between swine and new pandemic H1 hemagglutinin. J Biol Chem. 2011;286:5868–5875. doi: 10.1074/jbc.M110.193557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Suzuki Y, Matsunaga M, Matsumoto M. N-Acetylneuraminyllactosylceramide, GM3-NeuAc, a new influenza A virus receptor which mediates the adsorption-fusion process of viral infection. Binding specificity of influenza virus A/Aichi/2/68 (H3N2) to membrane-associated GM3 with different molecular species of sialic acid. J Biol Chem. 1985;260:1362–1365. [PubMed] [Google Scholar]
- 48.Huang X, et al. Human coronavirus HKU1 spike protein uses O-acetylated sialic acid as an attachment receptor determinant and employs hemagglutinin-esterase protein as a receptor-destroying enzyme. J Virol. 2015;89:7202–7213. doi: 10.1128/JVI.00854-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Langereis MA, et al. Complexity and diversity of the mammalian sialome revealed by nidovirus virolectins. Cell Rep. 2015;11:1966–1978. doi: 10.1016/j.celrep.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rogers GN, Herrler G, Paulson JC, Klenk HD. Influenza C virus uses 9-O-acetyl-N-acetylneuraminic acid as a high affinity receptor determinant for attachment to cells. J Biol Chem. 1986;261:5947–5951. [PubMed] [Google Scholar]
- 51.Chou H-H, et al. A mutation in human CMP-sialic acid hydroxylase occurred after the Homo-Pan divergence. Proc Natl Acad Sci USA. 1998;95:11751–11756. doi: 10.1073/pnas.95.20.11751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Irie A, Koyama S, Kozutsumi Y, Kawasaki T, Suzuki A. The molecular basis for the absence of N-glycolylneuraminic acid in humans. J Biol Chem. 1998;273:15866–15871. doi: 10.1074/jbc.273.25.15866. [DOI] [PubMed] [Google Scholar]
- 53.Widagdo W, et al. Differential expression of the Middle East respiratory syndrome coronavirus receptor in the upper respiratory tracts of humans and dromedary camels. J Virol. 2016;90:4838–4842. doi: 10.1128/JVI.02994-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Walls AC, et al. Glycan shield and epitope masking of a coronavirus spike protein observed by cryo-electron microscopy. Nat Struct Mol Biol. 2016;23:899–905. doi: 10.1038/nsmb.3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yuan Y, et al. Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nat Commun. 2017;8:15092. doi: 10.1038/ncomms15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.de Groot RJ. Structure, function and evolution of the hemagglutinin-esterase proteins of corona- and toroviruses. Glycoconj J. 2006;23:59–72. doi: 10.1007/s10719-006-5438-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Neu U, Bauer J, Stehle T. Viruses and sialic acids: Rules of engagement. Curr Opin Struct Biol. 2011;21:610–618. doi: 10.1016/j.sbi.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ng DL, et al. Clinicopathologic, immunohistochemical, and ultrastructural findings of a fatal case of Middle East respiratory syndrome coronavirus infection in the United Arab Emirates, April 2014. Am J Pathol. 2016;186:652–658. doi: 10.1016/j.ajpath.2015.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Krempl C, et al. Characterization of the sialic acid binding activity of transmissible gastroenteritis coronavirus by analysis of haemagglutination-deficient mutants. J Gen Virol. 2000;81:489–496. doi: 10.1099/0022-1317-81-2-489. [DOI] [PubMed] [Google Scholar]
- 60.Yokomori K, Banner LR, Lai MMC. Heterogeneity of gene expression of the hemagglutinin-esterase (HE) protein of murine coronaviruses. Virology. 1991;183:647–657. doi: 10.1016/0042-6822(91)90994-M. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Luytjes W, Bredenbeek PJ, Noten AFH, Horzinek MC, Spaan WJM. Sequence of mouse hepatitis virus A59 mRNA 2: Indications for RNA recombination between coronaviruses and influenza C virus. Virology. 1988;166:415–422. doi: 10.1016/0042-6822(88)90512-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Langereis MA, van Vliet ALW, Boot W, de Groot RJ. Attachment of mouse hepatitis virus to O-acetylated sialic acid is mediated by hemagglutinin-esterase and not by the spike protein. J Virol. 2010;84:8970–8974. doi: 10.1128/JVI.00566-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Williams RK, Jiang GS, Holmes KV. Receptor for mouse hepatitis virus is a member of the carcinoembryonic antigen family of glycoproteins. Proc Natl Acad Sci USA. 1991;88:5533–5536. doi: 10.1073/pnas.88.13.5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schultze B, Herrler G. Bovine coronavirus uses N-acetyl-9-O-acetylneuraminic acid as a receptor determinant to initiate the infection of cultured cells. J Gen Virol. 1992;73:901–906. doi: 10.1099/0022-1317-73-4-901. [DOI] [PubMed] [Google Scholar]
- 65.Matrosovich M, Herrler G, Klenk HD. Sialic acid receptors of viruses. Top Curr Chem. 2015;367:1–28. doi: 10.1007/128_2013_466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Böttcher-Friebertshäuser E, Garten W, Matrosovich M, Klenk HD. The hemagglutinin: A determinant of pathogenicity. Curr Top Microbiol Immunol. 2014;385:3–34. doi: 10.1007/82_2014_384. [DOI] [PubMed] [Google Scholar]
- 67.Wang N, et al. Structure of MERS-CoV spike receptor-binding domain complexed with human receptor DPP4. Cell Res. 2013;23:986–993. doi: 10.1038/cr.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bosch BJ, Raj VS, Haagmans BL. Spiking the MERS-coronavirus receptor. Cell Res. 2013;23:1069–1070. doi: 10.1038/cr.2013.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Barlan A, et al. Receptor variation and susceptibility to Middle East respiratory syndrome coronavirus infection. J Virol. 2014;88:4953–4961. doi: 10.1128/JVI.00161-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Meyer B, et al. Serologic assessment of possibility for MERS-CoV infection in equids. Emerg Infect Dis. 2015;21:181–182. doi: 10.3201/eid2101.141342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Suzuki Y, et al. Sialic acid species as a determinant of the host range of influenza A viruses. J Virol. 2000;74:11825–11831. doi: 10.1128/jvi.74.24.11825-11831.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shinya K, et al. Avian flu: Influenza virus receptors in the human airway. Nature. 2006;440:435–436. doi: 10.1038/440435a. [DOI] [PubMed] [Google Scholar]
- 73.Zeng Q, Langereis MA, van Vliet ALW, Huizinga EG, de Groot RJ. Structure of coronavirus hemagglutinin-esterase offers insight into corona and influenza virus evolution. Proc Natl Acad Sci USA. 2008;105:9065–9069. doi: 10.1073/pnas.0800502105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Peng W, et al. Recent H3N2 viruses have evolved specificity for extended, branched human-type receptors, conferring potential for increased avidity. Cell Host Microbe. 2017;21:23–34. doi: 10.1016/j.chom.2016.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Blixt O, et al. Printed covalent glycan array for ligand profiling of diverse glycan binding proteins. Proc Natl Acad Sci USA. 2004;101:17033–17038. doi: 10.1073/pnas.0407902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.van Riel D, Leijten LM, Kochs G, Osterhaus ADME, Kuiken T. Decrease of virus receptors during highly pathogenic H5N1 virus infection in humans and other mammals. Am J Pathol. 2013;183:1382–1389. doi: 10.1016/j.ajpath.2013.07.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.