Significance
Beetles are successful in the terrestrial ecosystem, which is attributable to, at least partly, their highly sclerotized exoskeleton. Here, we report a bacterial symbiont extremely specialized for underpinning the beetle’s hardness. The ancient endosymbiont Nardonella associated with weevils has an extremely small genome devoted to a single biological function, tyrosine provisioning, which is needed for insect’s cuticle formation and hardening. Notably, only the final step reaction of the tyrosine synthesis pathway is complemented by host-encoded aminotransferases up-regulated in the bacteriome, highlighting a highly focused aspect of the host–symbiont metabolic integrity. Both symbiont suppression by an antibiotic and RNA interference of the host aminotransferases induce reddish and soft weevils, verifying the pivotal role of the symbiosis for the beetle’s hardness.
Keywords: weevil, Nardonella, symbiont, genome, tyrosine
Abstract
Beetles, representing the majority of the insect species diversity, are characterized by thick and hard cuticle, which plays important roles for their environmental adaptation and underpins their inordinate diversity and prosperity. Here, we report a bacterial endosymbiont extremely specialized for sustaining beetle’s cuticle formation. Many weevils are associated with a γ-proteobacterial endosymbiont lineage Nardonella, whose evolutionary origin is estimated as older than 100 million years, but its functional aspect has been elusive. Sequencing of Nardonella genomes from diverse weevils unveiled drastic size reduction to 0.2 Mb, in which minimal complete gene sets for bacterial replication, transcription, and translation were present but almost all of the other metabolic pathway genes were missing. Notably, the only metabolic pathway retained in the Nardonella genomes was the tyrosine synthesis pathway, identifying tyrosine provisioning as Nardonella’s sole biological role. Weevils are armored with hard cuticle, tyrosine is the principal precursor for cuticle formation, and experimental suppression of Nardonella resulted in emergence of reddish and soft weevils with low tyrosine titer, confirming the importance of Nardonella-mediated tyrosine production for host’s cuticle formation and hardening. Notably, Nardonella’s tyrosine synthesis pathway was incomplete, lacking the final step transaminase gene. RNA sequencing identified host’s aminotransferase genes up-regulated in the bacteriome. RNA interference targeting the aminotransferase genes induced reddish and soft weevils with low tyrosine titer, verifying host’s final step regulation of the tyrosine synthesis pathway. Our finding highlights an impressively intimate and focused aspect of the host–symbiont metabolic integrity via streamlined evolution for a single biological function of ecological relevance.
Symbiotic associations with microorganisms are ubiquitously found in a variety of insects, which are rated among the important factors underpinning their adaptation, diversity, and prosperity (1–3). Many bacterial symbionts are indispensable for growth, survival, and reproduction of their insect hosts via, for example, provisioning of essential nutrients like amino acids and vitamins, where the host and the symbiont are integrated into an almost inseparable biological entity (4, 5). In such obligate symbiotic associations, the symbiont genomes tend to exhibit conspicuous structural degeneration, massive gene losses, and drastic size reduction, which are attributable to relaxed natural selection acting on many symbiont genes no longer necessary for the permanent intrahost lifestyle, and also to accumulation of deleterious mutations driven by attenuated natural selection acting on the symbiont genomes due to strong population bottlenecks and restricted horizontal gene acquisitions associated with continuous vertical symbiont transmission over evolutionary time (4, 6, 7). Some bacterial symbionts of plant-sucking insects like cicadas, leafhoppers, spittlebugs, psyllids, and mealybugs belonging to the order Hemiptera, which are associated with multiple endocellular bacterial cosymbionts within the symbiotic organ called the bacteriome, have experienced extreme genome reduction down to 0.2 Mb or smaller with 200 or less protein-coding genes (8–13), suggesting that metabolic complementation between the cosymbionts may have further facilitated the reductive genome evolution entailing losses of otherwise essential genes in either of the cosymbionts (7, 14–17).
In this study, we report another case of extremely reduced symbiont genome in a different insect group through a different evolutionary trajectory. Beetles, comprising the insect order Coleoptera, represent the majority of the biodiversity described (18–20), of which weevils comprise the most species-rich group, the superfamily Curculionoidea, with some 70,000 described species in the world (19, 21, 22). Many, if not all, weevils are associated with an ancient γ-proteobacterial endosymbiont lineage, Nardonella, in the bacteriome, whose evolutionary origin is estimated as older than 100 My (23–31). Despite the long-lasting host–symbiont coevolution, Nardonella’s biological role has been poorly understood (26). Previous studies have identified a number of weevil lineages in which Nardonella infections have been lost or replaced by different bacterial lineages, uncovering a strikingly dynamic aspect of the endosymbiotic evolution in the insect group (23, 24, 32–35).
Here, we report genomic, transcriptomic, and functional analyses of the Nardonella symbionts associated with diverse weevils, which unveiled their extremely reduced genomes down to as small as 0.2 Mb in the absence of any cosymbionts. The tiny genomes encode minimal but complete gene sets for bacterial replication, transcription, and translation, while lacking almost all of the other metabolic pathway genes, which indicate Nardonella’s near-complete dependence on host-derived metabolites toward a minimal cellular entity through the ancient coevolutionary history. Notably, a set of metabolic genes is conspicuously retained in the Nardonella genomes, namely synthesis pathway genes for a specific amino acid, tyrosine. Weevils are armored with hard cuticle, tyrosine is the principal precursor needed for cuticle formation, and the Nardonella genome has been streamlined for a single biological function, tyrosine provisioning, for sustaining the weevil’s highly sclerotized exoskeleton, which elucidates the general importance of endosymbiont-provisioned tyrosine for cuticle formation in weevils (36, 37), and potentially also in other beetles. Furthermore, we demonstrate that, reflecting the absence of transcriptional regulators in the tiny symbiont genome, Nardonella’s tyrosine provisioning is controlled by host’s final step regulation of the synthesis pathway.
Results and Discussion
Weevil-Nardonella Endosymbiotic System.
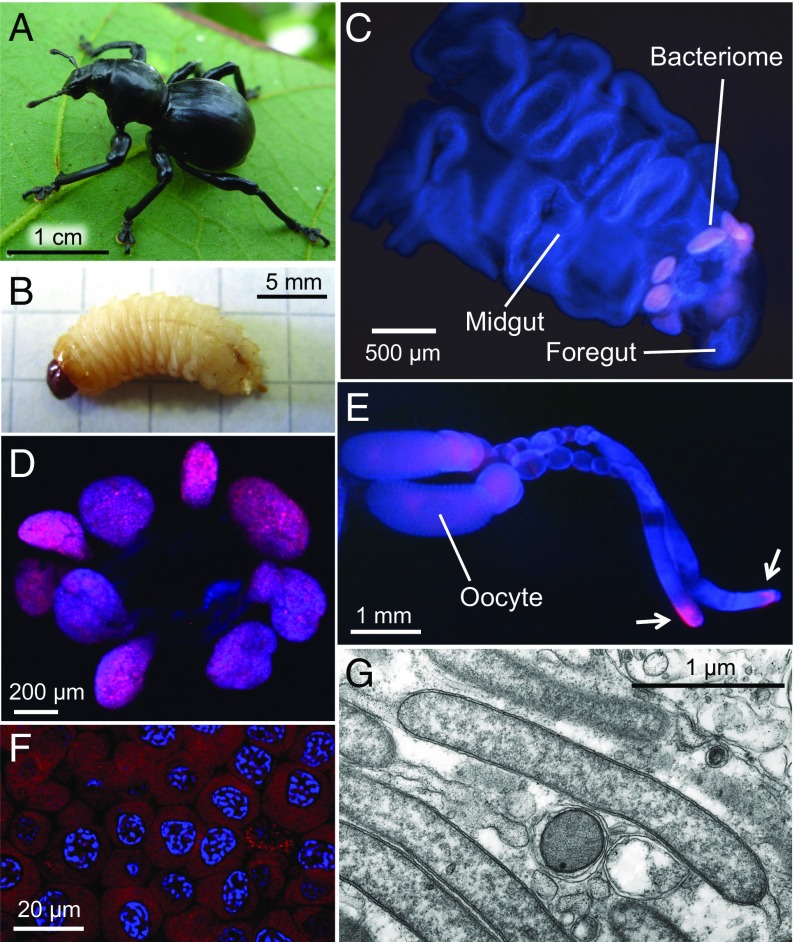
In the black hard weevil Pachyrhynchus infernalis (Fig. 1 A and B), Nardonella exhibited characteristic and specific endocellular localizations. In larvae, the symbiotic bacteria were localized within a conspicuous symbiotic organ, called the bacteriome, consisting of petal-like lobes surrounding the foregut-midgut junction (Fig. 1 C and D). In adult females, the larval bacteriome was not found, and the symbiotic bacteria were concentrated at a tip region of the ovarioles and also in developing oocytes (Fig. 1E). In the bacteriomes, the symbiotic bacteria were located within the cytoplasm of numerous bacteriocytes (Fig. 1F) as slender bacterial cells (Fig. 1G). Similar localization patterns of Nardonella were observed in the red palm weevil Rhynchopholus ferrugineus, the giant weevil Sipalinus gigas, the West Indian sweet poptato weevil Euscepes postfasciatus, and other weevil species, although size and shape of the bacteriomes as well as those of the bacterial cells were considerably different between the species (25, 29) (SI Appendix, Fig. S1).
Fig. 1.
P. infernalis and its bacterial symbiont Nardonella. (A) An adult. (B) A final instar larva. (C) A dissected larval gut with bacteriomes. Petal-like bacteriome lobes, each consisting of many bacteriocytes, are located around the foregut-midgut junction. (D) Isolated radial bacteriome lobes. (E) Detection of Nardonella in the ovarioles dissected from an adult female. Arrows highlight localization of Nardonella at the ovariole tips. (F) Visualization of Nardonella in the cytoplasm of larval bacteriocytes. (G) A transmission electron microscopic image of Nardonella cells in the larval bacteriocyte. In C–F, Nardonella 16S rRNA and host nuclear DNA are visualized in red and blue, respectively.
Extremely Reduced Nardonella Genomes.
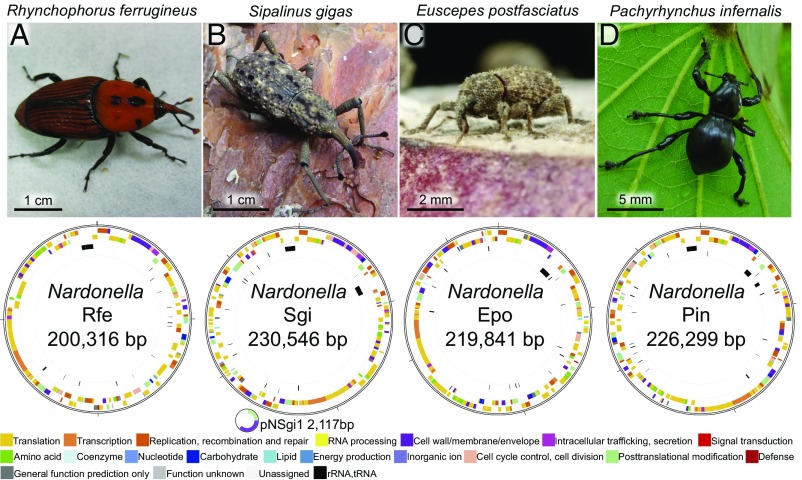
From these four weevil species, the larval bacteriomes were dissected, extracted for DNA preparation, and subjected to library construction, sequencing, and assembly, by which we obtained the complete Nardonella genome sequences (Fig. 2 and SI Appendix, Table S1). The main circular genomes of the Nardonella symbionts were strikingly small, ranging from 0.20 Mb to 0.23 Mb in size, with only 196–226 putative protein-coding ORFs. In the Nardonella Sgi genome, a 2.1-kb plasmid encoding only two genes was identified. Although extremely reduced, the Nardonella genomes retained a minimal but complete set of genes needed for replication, transcription, and translation: three or more ribosomal RNAs, 27 or more transfer RNAs, and all 20 aminoacyl tRNA synthetases (SI Appendix, Tables S1–S3). Besides some minor bacterial reads and contigs derived from contaminants, no other predominant bacterial contigs were identified, confirming the histological observations that these weevils harbor no major bacterial symbionts other than Nardonella in the bacteriome (25, 29) (Fig. 1 and SI Appendix, Fig. S1).
Fig. 2.
Weevils and their Nardonella genomes examined in this study. (A) Nardonella Rfe of the red palm weevil R. ferrugineus (Curculionidae: Dryophthorinae). (B) Nardonella Sgi of the giant weevil S. gigas (Curculionidae: Dryophthorinae). (C) Nardonella Epo of the West Indian sweet potato weevil E. postfasciatus (Curculionidae: Cryptorhynchinae). (D) Nardonella Pin of the black hard weevil P. infernalis (Curculionidae: Entiminae). Nardonella Sgi has a 2,117-bp plasmid encoding only two genes, mscS and ibpA-like. mscS, which is retained in all of the Nardonella genomes, encodes a channel for osmoregulation that exports water and ions upon hypotonic conditions (77). ibpA, which is found in Nardonella Epo but absent in Nardonella Rfe and Nardonella Pin, encodes a small chaperone Hsp20 (78). Note that the synteny of mscS and ibpA on the plasmid of Nardonella Sgi is conserved on the chromosome of Nardonella Epo.
Extreme Metabolic Capacity of Nardonella Specialized for Tyrosine Synthesis.
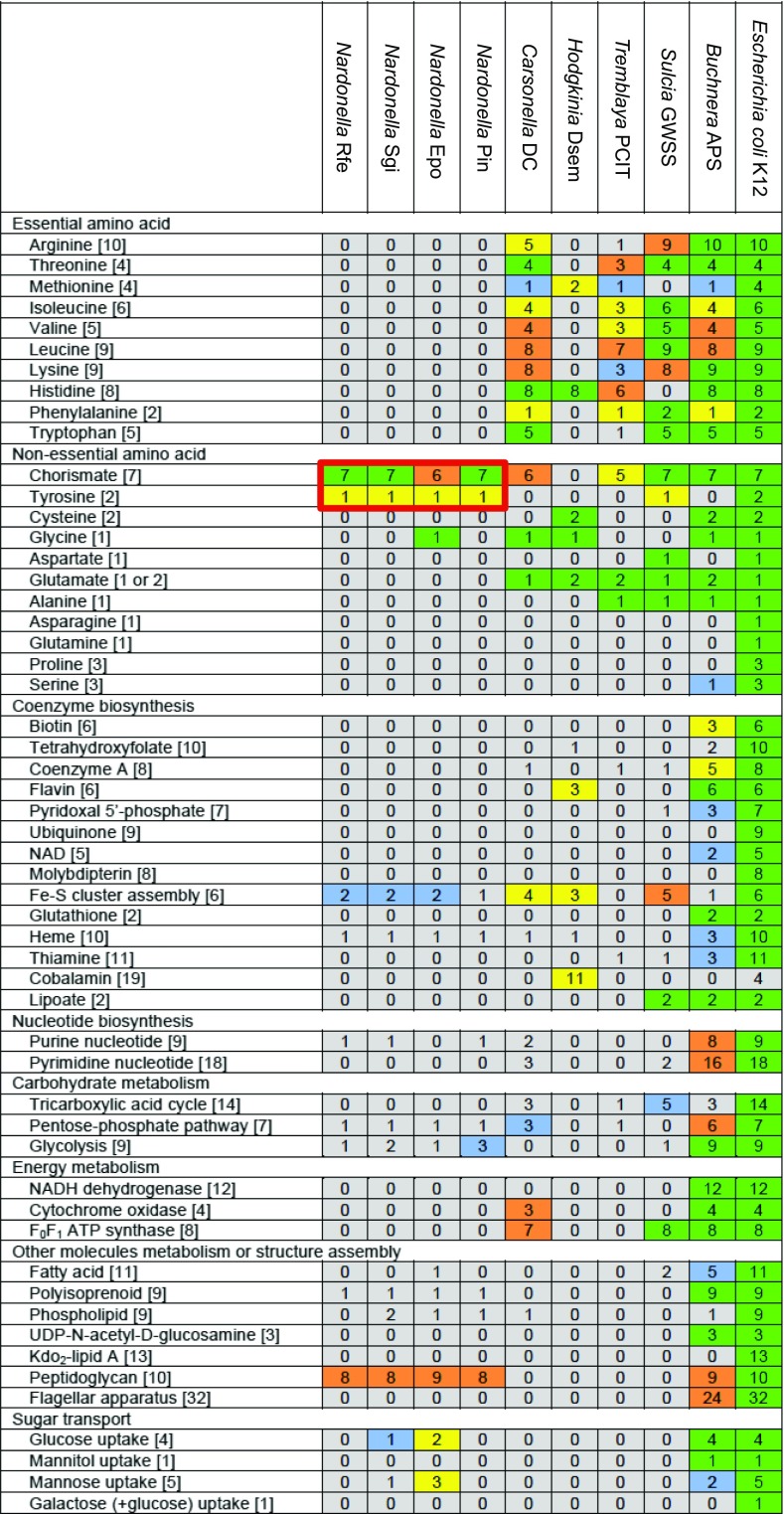
Reflecting their extreme genome reduction, the general metabolic capacity of the Nardonella symbionts was stringently limited: no synthesis genes for amino acids (aside from the exception involving tyrosine described later), no synthesis genes for B vitamins and cofactors (but a few genes for Fe-S cluster assembly), no functional nucleotide synthesis pathways, no TCA cycle genes, almost no pentose-phosphate pathway genes, no functional glycolysis pathway, and substantial absence of genes for energy metabolism, sugar transport, lipid synthesis, and flagellar apparatus (Fig. 3 and SI Appendix, Table S2). The only pathways that seemed to be complete were the synthesis pathway for tyrosine and the synthesis pathway for peptidoglycan (Fig. 3). Considering that the peptidoglycan synthesis is needed for cell wall construction to ensure the bacterial cell integrity, the Nardonella genome seems to be specialized toward production of a single amino acid, tyrosine.
Fig. 3.
Comparison of the metabolic gene repertoire between Nardonella genomes and other extremely reduced symbiont genomes. The minimal number of genes for a metabolic pathway is shown in each of the brackets. Each color indicates the ratio of retained genes to the minimal gene set for a metabolic pathway: green for 100%, orange for 75–99%, yellow for 50–74%, blue for 25–49%, and gray for 0–24%. Nardonella’s tyrosine synthesis pathway genes are highlighted in red. In the Nardonella Epo genome, aroE gene, located between def and sufE genes in the other Nardonella genomes, is lost and replaced by a 168-bp spacer sequence, which may be relevant to the fact that its host E. postfasciatus is smaller in size with thinner cuticle in comparison with the other large and hard weevil species R. ferrugineus, S. gigas, and P. infernalis.
In Vitro Assay of Nardonella’s Tyrosine Synthesis.
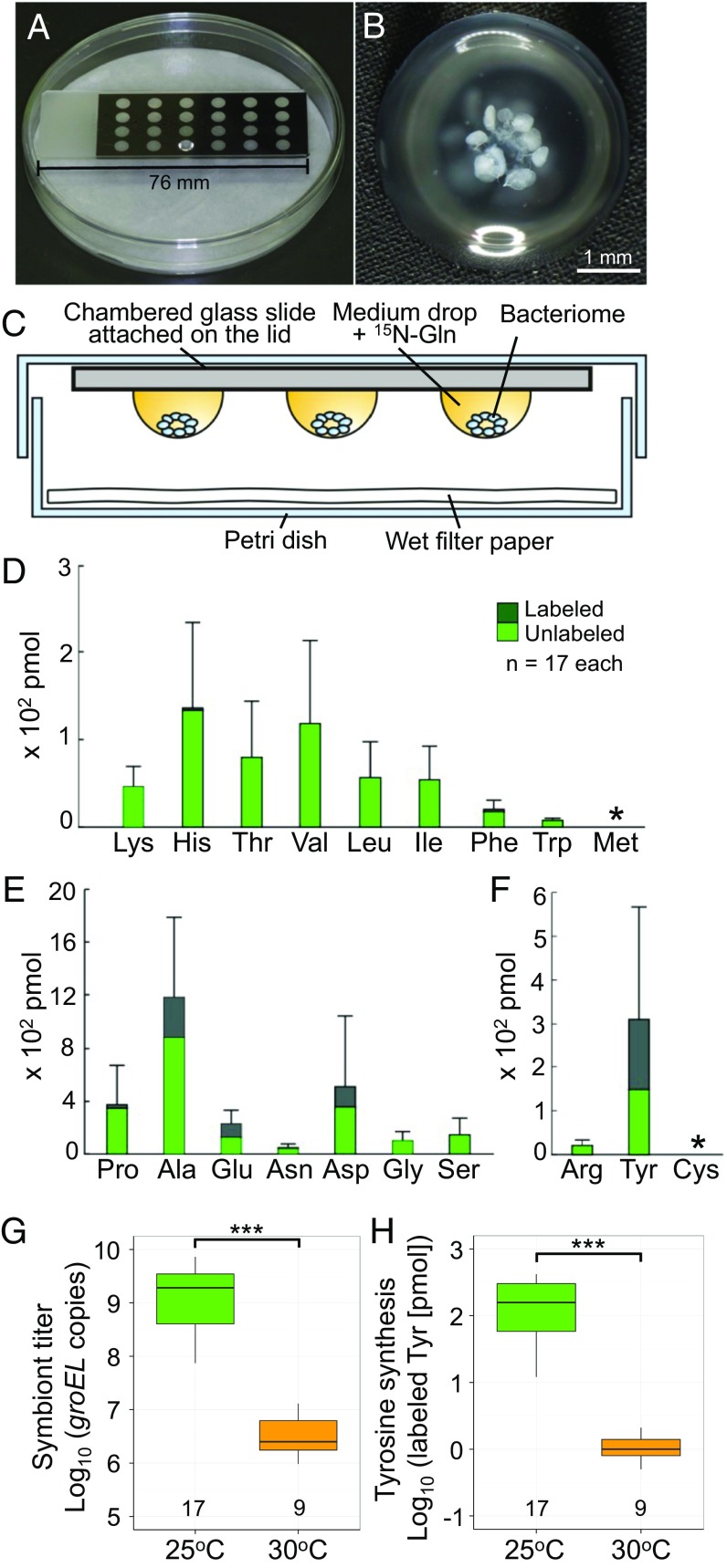
Hence, we constructed an in vitro assay system for testing whether Nardonella is capable of synthesizing tyrosine and other amino acids (Fig. 4 A–C). When bacteriomes dissected from mature larvae of P. infernalis were individually incubated with a culture medium containing 15N-labeled glutamine as a source of the amino group for newly synthesized amino acids, we observed that (i) essential amino acids were scarcely 15N-labeled, reflecting the absence of the synthesis pathways in both the host and the symbiont (Fig. 4D); (ii) some nonessential amino acids were 15N-labeled, likely owing to the synthesis pathways operating in the host bacteriocytes (Fig. 4E); and (iii) among semiessential amino acids, tyrosine exhibited a remarkably high proportion (about 50%) of 15N-labeling (Fig. 4F). These results suggest that the Nardonella symbiont in the bacteriome is preferentially synthesizing a substantial quantity of tyrosine.
Fig. 4.
In vitro assay of Nardonella’s tyrosine synthesis in P. infernalis. (A–C) The experimental system for hanging drop tissue culture. (A) An external view of the culture system. (B) An isolated larval bacteriome of P. infernalis in hanging drop medium. (C) A schematic view of the culture system. (D–F) Quantification of essential amino acids (D), nonessential amino acids (E), and semiessential amino acids (F) released from an isolated larval bacteriome of P. infernalis in the hanging drop medium supplemented with 15N-glutamine. Columns and bars show means and SDs, in which 15N-labeled and unlabeled amino acid fractions are depicted by dark green and light green, respectively. Asterisks indicate undetected amino acids, methionine, and cysteine. (G and H) Nardonella titers (G) and tyrosine synthesis activities (H) in the bacteriomes dissected from control larvae reared at 25 °C and heat-treated larvae reared at 30 °C. Asterisks indicate statistically significant differences [likelihood-ratio test of a generalized linear model (GLM) assuming a Gamma error distribution; ***P < 0.001]. Tukey box plots indicate the median (bold line), the 25th and 75th percentiles (box edges), the range (whiskers) and outliers, which are larger or smaller than 1.5 times the interquartile range from the box edge (dots), with sample sizes at the bottom.
High Temperature Eradication of Nardonella Infection in P. infernalis.
As previously described in a variety of insect-microbe symbiotic systems (38, 39), rearing of P. infernalis larvae at an elevated temperature, 30 °C, resulted in drastically suppressed symbiont titers in the bacteriomes (about 1/530 in terms of symbiont groEL gene copies) in comparison with control larvae reared at 25 °C (Fig. 4G). Considering that quantitative PCR also detects DNA molecules derived from dead bacterial cells, the level of symbiont suppression may actually be more severe. Accordingly, tyrosine synthesis by the dissected bacteriomes was also drastically suppressed in the larvae reared at 30 °C (about 1/150 in terms of 15N-labeled tyrosine titers) in comparison with the control larvae reared at 25 °C (Fig. 4H). These results indicate that the Nardonella symbiont is certainly involved in tyrosine synthesis and that the Nardonella-harboring bacteriome may function as a tyrosine-producing organ.
Antibiotic Suppression of Nardonella Infection in P. infernalis.
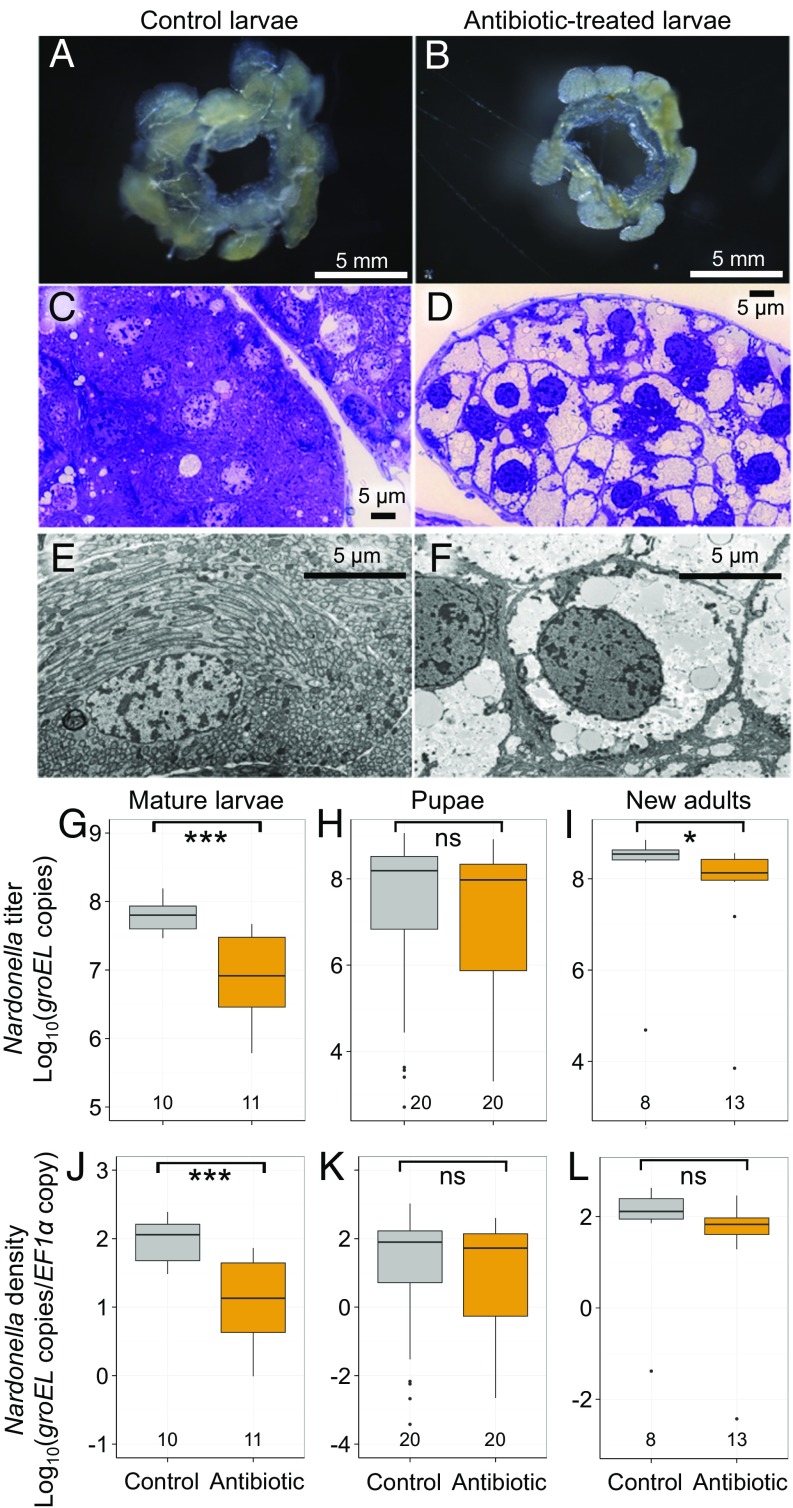
While the larvae of P. infernalis reared at 25 °C normally became adults, none of the larvae reared at 30 °C attained adulthood, suggesting that Nardonella is essential for pupal/adult survival or the high temperature condition is detrimental to pupal/adult survival. Hence, we attempted to develop a rearing condition for P. infernalis under which the Nardonella infection is not eradicated but suppressed significantly without high temperature. Finally, in addition to the standard rearing system on sweet potatoes (SI Appendix, Fig. S2 A–C), we established an agar-based artificial diet rearing system for P. infernalis, to which antibiotics can be conveniently supplemented, to generate and investigate normal and symbiont-suppressed insects (SI Appendix, Fig. S2 D–F). On the control artificial diet, the larvae grew normally with their bacteriomes full of Nardonella cells (Fig. 5 A, C, and E and SI Appendix, Table S4). On the antibiotic-supplemented artificial diet containing 0.003% rifampicin, the larvae also grew, but their bacteriomes were depleted of Nardonella cells (Fig. 5 B, D, and F and SI Appendix, Table S4). In mature larvae, the Nardonella titers in the antibiotic-treated insects were estimated by quantitative PCR to be around 10 times lower than those in the control insects (Fig. 5 G and J). In pupae and adults, notably, the Nardonella titers in the antibiotic-treated insects were still lower but recovering toward the levels of the control insects (Fig. 5 H, I, K, and L). Plausibly, intake of the antibiotic stopped upon the cessation of larval feeding before pupation, and remaining Nardonella cells proliferated and restored the infection density during the prepupal and pupal periods. The life tables of P. infernalis reared on the control diet and the antibiotic-supplemented diet revealed that the antibiotic treatment resulted in (i) lower mortality during the larval period (P < 0.05), (ii) higher mortality during the prepupal period (P < 0.05), (iii) higher mortality of pupae and newly emerged adults (although statistically not significant), and (iv) higher overall lifetime mortality (SI Appendix, Tables S4 and S5). These patterns were generally concordant with the results of fitness measurements: The antibiotic treatment resulted in greater larval body weight (P < 0.05) and prolonged pupal period (P < 0.05) (SI Appendix, Fig. S3). The superior performance of the antibiotic-treated larvae was likely relevant to the condition of the artificial diets. While the antibiotic-supplemented diet plates were usually clean throughout the rearing period (SI Appendix, Fig. S2D), the control diet plates suffered microbial contaminations, in particular bacterial ones, more frequently than the antibiotic-supplemented diet plates, which entailed change of color and smell of the diet (SI Appendix, Fig. S2E), and larvae on such contaminated diet plates tended to exhibit retarded growth or death. Meanwhile, the inferior performance of the antibiotic-treated prepupae, pupae, and adults seemed to be due to the antibiotic-induced symbiont depletion. It should be noted that almost all adult insects that successfully emerged were Nardonella-infected (Fig. 5 I and L) and a few Nardonella-deficient adult insects were frail and died early as described later, favoring the notion that Nardonella is essential for pupal/adult survival.
Fig. 5.
Antibiotic suppression of Nardonella infection in P. infernalis. (A and B) Dissected bacteriomes. (C and D) Light microscopic images of semiultrathin sections of the larval bacteriome stained with toluidine blue. (E and F) Transmission electron microscopic images of the larval bacteriocytes. (A, C, and E) Larvae reared on the control artificial diet. (B, D, and F) Larvae reared on the artificial diet containing 0.003% rifampicin. (G–I) Nardonella titers (in terms of bacterial groEL gene copies per insect) in mature larvae (G), pupae (H), and newly emerged adults (I). (J–L) Nardonella densities (in terms of bacterial groEL gene copies per host Elongation Factor 1α [EF1α] gene copy) in mature larvae (J), pupae (K), and newly emerged adults (L). In G–L, asterisks indicate statistically significant differences (likelihood-ratio test of GLM assuming a Gamma error distribution; *P < 0.05; ***P < 0.001; ns, no significant difference). Tukey box plots are as shown in Fig. 4 G and H.
Effects of Nardonella Suppression on Adult Color and Cuticle Formation in P. infernalis.
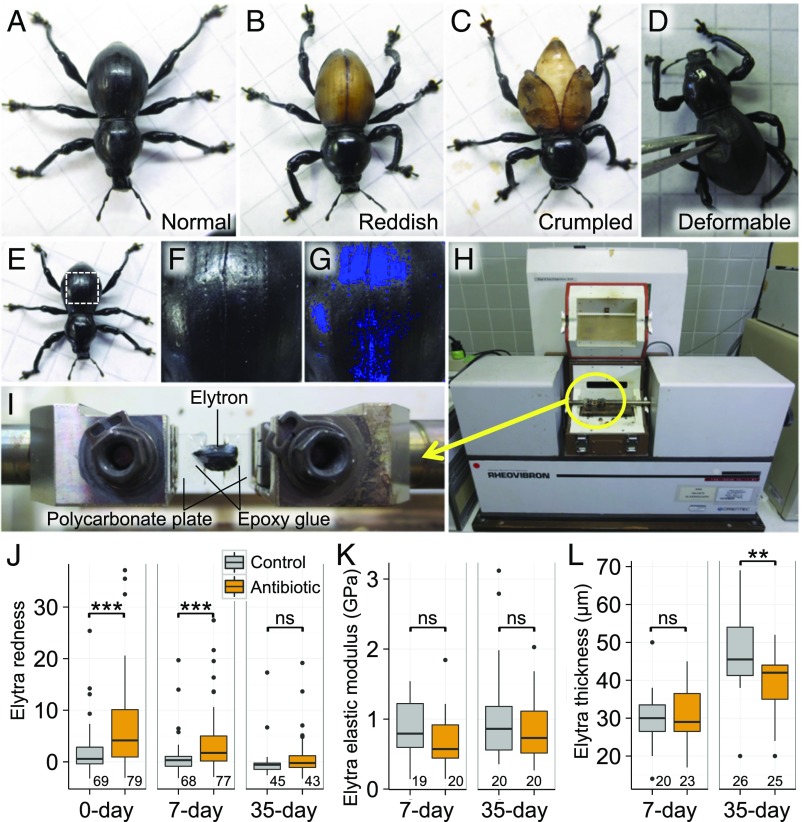
The majority of the adult insects that emerged from the control diet were morphologically normal with black and rigid elytra (Fig. 6A), whereas the Nardonella-suppressed adult insects that emerged from the antibiotic-supplemented diet frequently suffered morphological abnormalities with reddish, crumpled, and/or deformable elytra (Fig. 6 B–D and SI Appendix, Fig. S4). Elytra color analysis (Fig. 6 E–G and SI Appendix, Fig. S5) revealed that the antibiotic-treated adult insects exhibited significantly more reddish elytra in comparison with the control adult insects, while the reddish color turned into black as the insect age proceeded (Fig. 6J). The red-black color transition was partly because reddish frail adult insects, which were presumably suffering severe symbiosis deficiency, tended to die early, and also because surviving adult insects became darker in color. Physical property analysis of the elytra using a viscoelastometer (Fig. 6 H and I) revealed that, unexpectedly, the elastic modulus, a qualitative index of physical hardness, was not significantly different between the control insects and the antibiotic-treated insects irrespective of adult aging (Fig. 6K). However, elytra thickness increased as the insect age proceeded, and the control adult insects developed thicker elytra than the antibiotic-treated adult insects (Fig. 6L). These results indicate that the Nardonella-suppressed adult insects exhibit reddish and thinner elytra in contrast to black and thicker elytra of the control adult insects.
Fig. 6.
Effects of Nardonella suppression on adult color and cuticle formation in P. infernalis. (A) A control adult insect with black and hard elytra. (B–D) Antibiotic-treated adult insects with reddish elytra (B), crumpled fragile elytra (C), and soft and deformable elytra (D). (E–G) The process of quantifying the redness of elytra. From each of dorsal images of adult insects (E), a square area of maximal size (dotted square) was extracted. On the square image (F), the pixels whose brightness was either over top 10% or below bottom 10% were masked in blue (G) and excluded from the analysis to minimize the effects of highlights and shadows. Then, red-green-blue (RGB) values for all (=n) pixels were measured and averaged to obtain the redness index by Σ (R − mean [R, G, B])/n. (H and I) The system for measuring the viscoelasticity of elytra. On the stage of a viscoelastometer (H), each sample elytron was set by gluing onto two plastic plates with epoxy resin (I) to measure elastic modulus. (J–L) Elytra redness (J), elytra elastic modulus at 10 Hz (K), and elytra thickness (L) of the control and antibiotic-treated adult insects 0, 7, and 35 d after emergence. Asterisks indicate statistically significant differences [Wilcoxon rank sum test for (J), and t test for (K) and (L): ***P < 0.001; **P < 0.01; ns, no significant difference]. Tukey box plots are as shown in Fig. 4 G and H.
Effects on Nardonella Suppression on Levels of Tyrosine and l-DOPA During Pupal and Adult Development of P. infernalis.
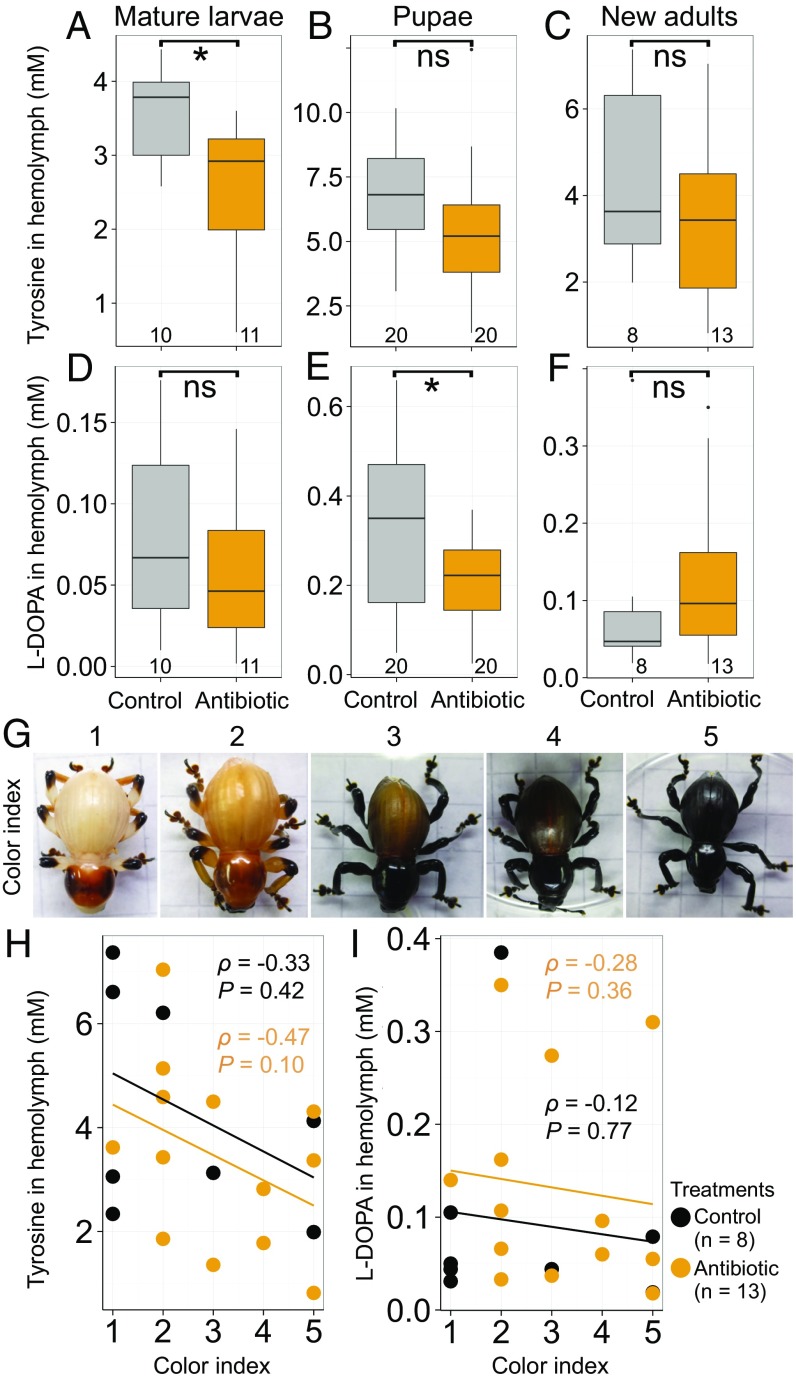
In the process of cuticular pigmentation and hardening in beetles and other insects, tyrosine and its derivative l-dopa (l-DOPA) play essential roles as principal substrates for initiating a series of chemical reactions toward cuticle tanning, polymerization, and melanization (40, 41) (SI Appendix, Fig. S6). Quantification of the levels of tyrosine and l-DOPA in hemolymph of mature larvae, pupae, and newly emerged adults of P. infernalis revealed that (i) the levels of tyrosine were consistently 10–100 times higher than the levels of l-DOPA irrespective of the developmental stages and the symbiotic conditions (Fig. 7 A–F), probably reflecting the fact that l-DOPA is a transient intermediate in the cuticle formation process (SI Appendix, Fig. S6); (ii) both the levels of tyrosine and l-DOPA were the highest at the pupal stage (Fig. 7 A–F), suggesting active mobilization of these metabolites at the developmental stage of active adult body formation; (iii) at the mature larval stage, the levels of tyrosine were significantly higher in the control insects than in the antibiotic-treated insects (Fig. 7A), suggesting that the normal symbiotic larvae are able to synthesize and accumulate more tyrosine than the symbiosis-deficient larvae; and (iv) at the pupal stage, the levels of l-DOPA were significantly higher in the control insects than in the antibiotic-treated insects (Fig. 7E), suggesting the possibility that the normal symbiotic pupae are recruiting l-DOPA and constructing adult cuticles more actively than the symbiosis-deficient pupae. Moreover, by carefully monitoring plastic sandboxes in which pupae were observable in pupal chambers adjacent to a transparent wall (SI Appendix, Fig. S2 G–I), we collected teneral adult insects newly eclosed within 24 h (Fig. 7G) and measured the levels of tyrosine and l-DOPA in their hemolymph (Fig. 7 H and I). As the cuticle pigmentation in the teneral adult insects proceeded, the tyrosine levels tended to decrease in both the control adult insects and the antibiotic-treated adult insects (Fig. 7H).
Fig. 7.
Effects of Nardonella suppression on levels of tyrosine and l-DOPA during pupal and adult development of P. infernalis. (A–C) Tyrosine levels in the hemolymph of mature larvae (A), pupae (B), and newly emerged adults (C). (D–F) l-DOPA levels in the hemolymph of mature larvae (D), pupae (E), and newly emerged adults (F). Asterisks indicate statistically significant differences (t test; *P < 0.05; ns, no significant difference). Tukey box plots are as shown in Fig. 4 G and H. (G) Color indices of newly eclosed (within 24 h) adult insects defined by the levels of cuticle pigmentation. The younger the insects, the paler their color is. (H and I) Relationships between the levels of cuticle pigmentation and the titers of hemolymph tyrosine (H) or hemolymph l-DOPA (I) in the control insects and the antibiotic-treated insects. Regression lines, Spearman’s rank correlation coefficients (ρ), and P values (P) are depicted.
Nardonella as an Ancient Symbiont Specialized for Provisioning Tyrosine That Underpins Hard Cuticle of Weevils.
All these results taken together, we conclude that the ancient endosymbiont Nardonella has experienced an extremely streamlined genome evolution toward a specific biological function, namely tyrosine provisioning, which underpins the formation of the hard cuticle of weevils. Weevils constitute the most species-rich animal group with some 70,000 described species, including many highly sclerotized lineages such as Pachyrhynchus spp., Eupholus spp., Rhynchophorus spp., and Trigonopterus spp. (19, 21, 22, 42–44). The hard cuticle of these and other weevils has been shown to confer mechanical strength, antipredator effects, tolerance to desiccation, and other beneficial consequences, which facilitate their survival and adaptation (45–48). In this context, the Nardonella-mediated tyrosine production and cuticle sclerotization are likely to play pivotal roles in the biology of weevils, which potentially contribute to their diversity and prosperity. Whether such symbiont-assisted cuticle hardening is also found in other insect groups deserves future survey of diverse beetle–microbe associations. Recent detailed structural studies unveiled that, among diverse beetles, weevils are prominent in that their cuticle exhibits a peculiar microstructure of densely interlocked exocuticle and endocuticle, which may be relevant to the mechanical strength of their exoskeleton (48, 49). Whether the symbiont-provisioned tyrosine contributes to formation of the cuticular structure, and if so, how, is of interest and open to future investigation.
Insights into Symbiont Replacements and Diversification in Evolution of Weevils.
While the Nardonella symbionts have been conserved among diverse weevils and cospeciated with the weevil hosts for over 100 My (23–31), previous studies have identified a number of weevil lineages in which the Nardonella symbiont had been either lost or replaced by novel bacterial symbionts (23, 24, 32–35). The extremely reduced Nardonella genome down to 0.2 Mb (Fig. 2 and SI Appendix, Table S1) must be relevant to the recurrent symbiont losses and replacements as a consequence of genomic decay theoretically expected and empirically observed in ancient symbiont lineages subjected to strict vertical transmission and strong population bottleneck (4, 6, 7, 14–17). The highly specific Nardonella’s function, tyrosine provisioning only, is also likely to have facilitated the symbiont losses and replacements. For example, in weevil lineages that had evolved feeding habits on tyrosine-rich food sources, it is expected that evolutionary losses of Nardonella may proceed easily. Considering that many bacteria are capable of synthesizing amino acids including tyrosine, any bacterial infections with secondary facultative symbionts or gut microbial associates may potentially result in complementation of Nardonella’s biological function. Such a symbiont replacement has been presumed and best documented for grain weevils of the genus Sitophilus, in which Nardonella was replaced by a γ-proteobacterial lineage Sodalis pierantonius (23, 24, 32, 50). The weevil-associated Sodalis genome was determined as 4.5 Mb in size, which was much larger than the Nardonella genome and retaining many metabolic pathways intact; however, the Sodalis genome was full of amplified IS elements and pseudogenes (50, 51), representing an early stage of the degenerative genome evolution after replacing the original Nardonella symbiont. A number of classic and recent studies have documented a variety of biological roles of the Sodalis symbiont for the grain weevils: at phenotypic levels, enhanced growth, survival, and fecundity (37, 52, 53), improved flight activity (52, 54) and facilitated cuticular tanning and hardening (36, 37, 52); and at biochemical and metabolic levels, provisioning of B vitamins such as pantothenic acid, biotin, and riboflavin (55), supply of aromatic amino acids phenylalanine and tyrosine (36, 37), metabolism of methionine and sarcosine (56, 57), and involvement in mitochondrial energy metabolism (58, 59). Our results strongly suggest that the tyrosine provisioning, which underpins the cuticle hardening and the fitness improvement, is the original essential role of the weevil-bacterium endosymbiosis, and the other biological functions are likely acquired following the symbiont replacement from Nardonella to Sodalis in an ancestor of the grain weevils.
Incomplete Tyrosine Synthesis Pathway of Nardonella.
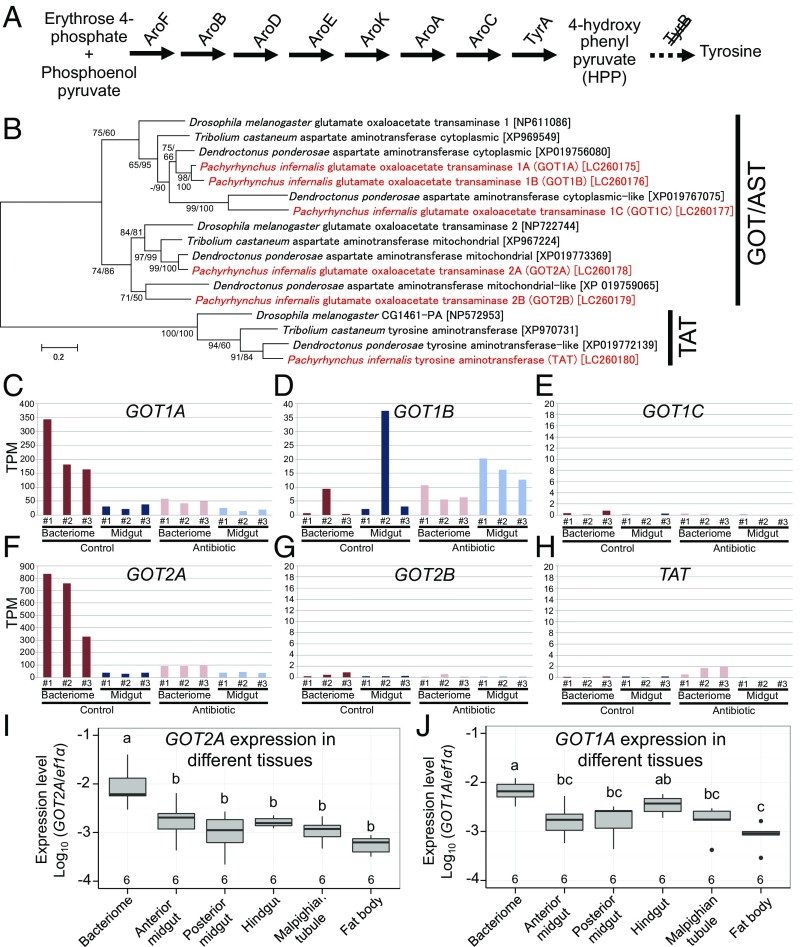
It should be noted that, despite the importance of tyrosine provisioning for the host weevil, Nardonella’s tyrosine synthesis pathway is incomplete, lacking the final step gene, tyrB, that encodes a tyrosine transaminase (Fig. 8A and SI Appendix, Fig. S7). We expected that the final step reaction of converting 4-hydroxyphenylpyruvate (HPP) to tyrosine may be catalyzed by some gene(s) expressed in the host bacteriome. In the genome of Drosophila melanogaster (60), there are three aminotransferase genes, namely glutamate oxaloacetate transaminase 1 (GOT1), glutamate oxaloacetate transaminase 2 (GOT2), and tyrosine aminotransferase (TAT), which are potentially capable of converting HPP to tyrosine by transamination. Hence, we surveyed the transcriptomes of P. infernalis for these aminotransferase genes.
Fig. 8.
Identification of GOT1, GOT2, and TAT genes of P. infernalis that are potentially involved in tyrosine synthesis in place of Nardonella’s tyrB gene. (A) Tyrosine synthesis pathway encoded by the Nardonella genome. Also see SI Appendix, Fig. S7. (B) Phylogenetic relationship of GOT1, GOT2, and TAT transcripts identified from P. infernalis to those from the fruit fly D. melanogaster, the flour beetle T. castaneum, and the bark beetle D. ponderosae. A maximum-likelihood phylogeny inferred from 515 aligned amino acid sites is shown with bootstrap values on the nodes in the order of maximum likelihood/neighbor joining. Note that three GOT1 (GOT1A, GOT1B, and GOT1C), two GOT2 (GOT2A and GOT2B) and one TAT sequences were obtained from P. infernalis, which are highlighted in red. In brackets are sequence accession numbers. (C–H) TPM values for GOT1A (C), GOT1B (D), GOT1C (E), GOT2A (F), GOT2B (G), and TAT (H) genes transcribed in bacteriomes and midguts dissected from mature larvae reared on the control diet and the antibiotic-supplemented diet. (I and J) Quantitative RT-PCR evaluating the expression levels of GOT2A (I) and GOT1A (J) in tissues dissected from mature larvae. Gene copy numbers evaluated by quantitative RT-PCR are normalized as per EF1α gene copy. Different letters (a, b, c) indicate statistically significant differences (likelihood-ratio test of GLM and post hoc multiple comparisons; P < 0.05).
Genes of P. infernalis Up-Regulated in the Bacteriome and Complementing Nardonella’s Tyrosine Synthesis Pathway.
We performed RNA extraction of bacteriomes and midguts dissected from mature larvae of P. infernalis reared on the control diet and the antibiotic-supplemented diet, and the RNA samples were subjected to RNA sequencing analysis (SI Appendix, Table S6). From the assembled contigs, we identified three GOT1 (designated as GOT1A, GOT1B, and GOT1C), two GOT2 (GOT2A and GOT2B) and one TAT gene sequences (accession numbers LC260175–LC260180), which were orthologous to those of D. melanogaster and other insects based on molecular phylogenetic analyses (Fig. 8B). TPM (transcripts per million reads) values indicated that, among them, GOT1A and GOT2A represented the overwhelmingly predominant transcripts in the bacteriomes of Nardonella-infected control insects (Fig. 8 C–H). Quantitative RT-PCR analysis of dissected tissues confirmed preferential expression of GOT2A and GOT1A in the larval bacteriomes (Fig. 8 I and J).
Host’s Aminotransferase Genes Involved in Nardonella-Mediated Tyrosine Production and Cuticle Formation.
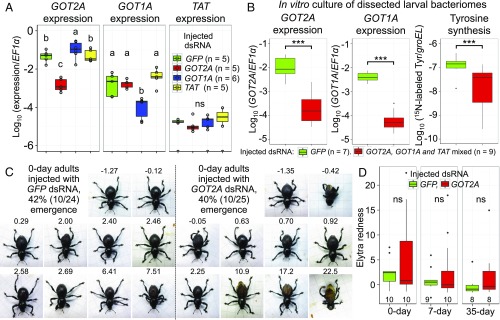
Based on these results, we attempted to suppress the expression levels of GOT2A and GOT1A (and additionally TAT) by RNAi. Injection of dsRNA into mature larvae resulted in significant suppression of GOT2A and GOT1A, while we could not detect RNAi effects on TAT because of the originally very low expression level (Fig. 9A). In vitro culturing of bacteriomes dissected from mature larvae, which had been injected with dsRNAs targeting GOT2A, GOT1A, and TAT, revealed significantly reduced tyrosine production in association with drastically reduced expression levels of GOT2A and GOT1A (Fig. 9B). To observe phenotypic consequences of the mixed dsRNA injection, some of the injected larvae were reared to adulthood but in vain: Most died as pupae and a few died as teneral adults in pupal chambers without cuticle sclerotization. Hence, we performed another experiment in which only GOT2A was suppressed by RNAi. The treated larvae injected with dsRNA targeting GOT2A attained 40% (10/25) adult emergence rate, which was similar to the 42% (10/24) adult emergence rate of the control larvae injected with dsRNA targeting GFP (Fig. 9C). Notably, the GOT2A-suppressed adult insects tended to exhibit morphological abnormalities, represented by reddish, crumpled, and/or deformed elytra, in comparison with the control adult insects that were mostly with normally sclerotized black elytra (Fig. 9C). Elytra color analysis confirmed that the GOT2A-suppressed adult insects were more reddish in comparison with the control adult insects (although the differences were statistically not significant) and the reddish color became darker as the insect age proceeded (Fig. 9D). These results strongly suggest that the host’s aminotransferase genes up-regulated in the bacteriome are needed for Nardonella-mediated tyrosine production in the bacteriome and involved in formation and pigmentation of adult cuticle.
Fig. 9.
RNA interference (RNAi) suppression of host’s tyrosine synthesis genes up-regulated in the bacteriome of P. infernalis, and its effects on cuticle formation and pigmentation. (A) RNAi targeting GOT2, GOT1, and TAT by injecting double-stranded RNA (dsRNA) into mature larvae. Expression levels of GOT2A, GOT1A, and TAT were measured by quantitative RT-PCR 3 d after the injection. Different letters (a, b, c) indicate statistically significant differences (likelihood-ratio test of GLM and post hoc multiple comparisons; P < 0.05). (B) Tyrosine synthesis activity of dissected larval bacteriomes suppressed by RNAi of GOT2 and GOT1. Mature larvae were injected with dsRNAs, their bacteriomes were dissected 7 d after the injection and cultured in a medium containing 15N-labeled glutamine for 2 h, and the bacteriomes were subjected to quantitative RT-PCR, whereas the culture media were analyzed by LC-MS for quantification of 15N-labeled tyrosine. Asterisks indicate statistically significant differences (likelihood-ratio test of GLM assuming a gamma error distribution; ***P < 0.001). (C) Images of adult insects on the day of emergence, which were subjected to larval injection with either GFP dsRNA or GOT2A dsRNA. Numbers on the top of the images indicate the values of elytra redness. (D) Comparison of elytra redness between the adult insects 0, 7, and 35 d after emergence. “ns” indicates no statistically significant difference (Wilcoxon rank sum test; P > 0.05). Tukey box plots are as shown in Fig. 4 G and H. Note that the primers for quantitative RT-PCR are highly specific, whereas dsRNAs for RNAi may potentially cause some cross-suppressions: dsRNA for GOT2A may also recognize GOT2B, and dsRNA for GOT1A may also target GOT1B and GOT1C (SI Appendix, Fig. S8).
Host’s Final Step Control over Symbiont’s Metabolic Pathway.
All these results taken together, we conclude that the Nardonella’s sole and most important role, tyrosine synthesis, is subjected to the final step regulation by host’s enzymes up-regulated in the symbiotic organ, which highlights an impressively intimate and focused aspect of the host–symbiont metabolic integrity. We suggest that the regulation by the host side may have been evolutionarily facilitated by, on one hand, the extremely reduced Nardonella genome that has purged out transcriptional and translational regulator genes, and on the other hand, the drastic change in demand for tyrosine, much more for highly sclerotized adults than for nonsclerotized larvae, in the developmental course of the weevil host. In the aphid-Buchnera endosymbiosis, similar patterns are observed in metabolic pathway genes for synthesis of essential amino acids isoleucine, leucine, valine, and phenylalanine, and also tyrosine. Specifically, the Buchnera genome encodes almost all of the synthesis pathway enzymes for these amino acids except for the final step enzymes, which are complemented by host-encoded enzymes, namely, branched-chain aminotransferase for isoleucine, leucine, and valine; aspartate aminotransferase for phenylalanine; and phenylalanine 4-monooxigenase for tyrosine (61–68). Notably, in contrast to the synthesis pathway for tyrosine via phenylalanine in the aphid-Buchnera system, the synthesis pathway for tyrosine in the weevil-Nardonella system circumvents phenylalanine (SI Appendix, Fig. S7), which is probably relevant to the facts that (i) distinct from the Buchnera of aphids, Nardonella of weevils is incapable of synthesizing phenylalanine; (ii) weevils require incomparably more tyrosine for building their hard cuticle than aphids; (iii) thus, if Nardonella could synthesize tyrosine from phenylalanine, it would readily end up with depletion of the essential amino acid phenylalanine in weevils; and (iv) this is probably the reason why Nardonella has evolved the phenylalanine-independent synthesis pathway for tyrosine. In the aphid-Buchnera system, amino acid transporter proteins located at the host symbiosomal membrane in the bacteriocyte play pivotal roles for metabolite exchange and host–symbiont metabolic integration (69, 70). In the weevil-Nardonella system, it is of interest but unknown how the precursor of tyrosine, 4-HPP, produced by the symbiont is transported to the host cytoplasm within the bacteriome (SI Appendix, Fig. S7), which is open to future investigation.
Conclusion and Perspective
The weevil-Nardonella system represents an unprecedented form of nutritional symbiosis. The extremely reduced symbiont genome, around 0.2 Mb in size, encodes minimal but complete gene sets for bacterial cell replication, transcription, and translation, while lacking almost all of the other metabolic pathway genes, which is indicative of Nardonella’s near-complete dependence on host-derived metabolites toward a minimal cellular entity through the ancient endosymbiotic evolution. The only functional metabolism retained in the Nardonella genome is the tyrosine synthesis pathway, uncovering the sole biological role of Nardonella as a tyrosine supplier. Experimental suppression of Nardonella infection results in emergence of reddish and soft weevils, verifying the importance of Nardonella for host’s cuticle formation and hardening. In accordance with the stringently limited gene repertoire of Nardonella lacking transcriptional and translational regulators, the Nardonella-mediated tyrosine synthesis is regulated by host aminotransferases up-regulated in the bacteriome at the final step of the synthesis pathway, highlighting an intricate host–symbiont metabolic integration that underpins the bacteriome function as a tyrosine-producing organ. In an evolutionary perspective, the extremely reduced Nardonella genome specialized for a single metabolic function may have facilitated the recurrent symbiont losses and replacements observed in weevils. In an ecological perspective, the symbiont-assisted formation of hard cuticle may have contributed to, at least to some extent, the adaptation, diversity, and prosperity of weevils. The weevil-Nardonella endosymbiotic system presents an impressive example as to how far such an intimate and ancient host–symbiont association can go over evolutionary time and why such a highly sophisticated symbiotic system inherently entails evolutionary consequences of instability, losses, and replacements.
Materials and Methods
Weevil samples used in this study are as listed (SI Appendix, Table S7). P. infernalis was reared in the laboratory for experiments (SI Appendix, Fig. S2). Histological analyses were conducted as described (28, 71). Symbiont genome sequencing and annotation were performed as described (72–75). Transcriptomic data of the host bacteriomes were obtained by RNA sequencing using TruSeq RNA Sample Preparation Kit v2 (Illumina) and HiSeq2000 sequencer (Illumina), and analyzed using the program Trinity (76) implemented in the MASER pipeline (cell-innovation.nig.ac.jp/). Symbiont visualization by fluorescence in situ hybridization, symbiont quantification by quantitative PCR, and evaluation of host gene expression by quantitative RT-PCR were conducted using the probes and primers listed (SI Appendix, Table S8). Tracer experiments with [2-15N] glutamine (Cambridge Isotope Laboratories) using tissue culture and amino acid analyses were performed using a high-performance liquid chromatography system (Prominence, Shimadzu) with a Shim-pack FC-ODS column (150 mm × 2 mm i.d., Shimadzu) and an electrospray ionization mass spectrometry system (LCQduo; Thermo). Elytra color analysis was conducted using the software Natsumushi version 0.99. An automatic dynamic viscoelastometer (Rheovibron DDV-25FP; A&D) was used to measure mechanical properties of elytra.
See SI Appendix, SI Materials and Methods for complete details on the materials and methods.
Supplementary Material
Acknowledgments
We thank Takuya Aikawa, Katsunori Nakamura, Takuma Takanashi, Wataru Toki, and Yosuke Usui for insect samples; Norikuni Kumano, Munetoshi Maruyama, Katsunori Nakamura, and Katsushi Yamaguchi for insect photos; Akira Oyafuso for artificial diet ingredients; Katsushi Yamaguchi and Tomoko F. Shibata for supporting genomic and transcriptomic data processing; Hiroshi Shimizu and Hideyuki Tsukada for supporting measurement of physical properties of insect cuticle; and Wakana Kikuchi, Junko Makino, and Kaoru Nikoh for technical and secretarial assistance. This study was supported by the Japan Society for the Promotion of Science Grant-in-Aid for Scientific Research Grants JP25221107 (to T.F., N.N., T.H., and R.K.), JP22128001, and JP22128007 (to T.F., N.N. and S.S.); the Program for Promotion of Basic and Applied Researches for Innovations in Bio-Oriented Industry (to T.F.); and National Institute for Basic Biology Collaborative Research Projects for Integrative Genomics (to T.F. and S.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the DNA Data Bank Japan Read Archive, www.ddbj.nig.ac.jp (accession nos. AP018159–AP018162, LC260175–LC260180, LC260491, and DRR095964–DRR095975).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1712857114/-/DCSupplemental.
References
- 1.Buchner P. Endosymbiosis of Animals with Plant Microorganisms. Interscience; New York: 1965. [Google Scholar]
- 2.Boutzis K, Miller TA. Insect Symbiosis. CRC; Boca Raton, FL: 2003. [Google Scholar]
- 3.Zchori-Fein E, Miller TA. Manipulative Tenants: Bacteria Associated with Arthropods. CRC; Boca Raton, FL: 2011. [Google Scholar]
- 4.Moran NA, McCutcheon JP, Nakabachi A. Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet. 2008;42:165–190. doi: 10.1146/annurev.genet.41.110306.130119. [DOI] [PubMed] [Google Scholar]
- 5.Douglas AE. The microbial dimension in insect nutritional ecology. Funct Ecol. 2009;23:38–47. [Google Scholar]
- 6.Wernegreen JJ. Genome evolution in bacterial endosymbionts of insects. Nat Rev Genet. 2002;3:850–861. doi: 10.1038/nrg931. [DOI] [PubMed] [Google Scholar]
- 7.McCutcheon JP, Moran NA. Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol. 2011;10:13–26. doi: 10.1038/nrmicro2670. [DOI] [PubMed] [Google Scholar]
- 8.Nakabachi A, et al. The 160-kilobase genome of the bacterial endosymbiont Carsonella. Science. 2006;314:267. doi: 10.1126/science.1134196. [DOI] [PubMed] [Google Scholar]
- 9.McCutcheon JP, Moran NA. Parallel genomic evolution and metabolic interdependence in an ancient symbiosis. Proc Natl Acad Sci USA. 2007;104:19392–19397. doi: 10.1073/pnas.0708855104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCutcheon JP, McDonald BR, Moran NA. Convergent evolution of metabolic roles in bacterial co-symbionts of insects. Proc Natl Acad Sci USA. 2009;106:15394–15399. doi: 10.1073/pnas.0906424106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McCutcheon JP, Moran NA. Functional convergence in reduced genomes of bacterial symbionts spanning 200 My of evolution. Genome Biol Evol. 2010;2:708–718. doi: 10.1093/gbe/evq055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCutcheon JP, von Dohlen CD. An interdependent metabolic patchwork in the nested symbiosis of mealybugs. Curr Biol. 2011;21:1366–1372. doi: 10.1016/j.cub.2011.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bennett GM, Moran NA. Small, smaller, smallest: The origins and evolution of ancient dual symbioses in a Phloem-feeding insect. Genome Biol Evol. 2013;5:1675–1688. doi: 10.1093/gbe/evt118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCutcheon JP. The bacterial essence of tiny symbiont genomes. Curr Opin Microbiol. 2010;13:73–78. doi: 10.1016/j.mib.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moran NA, Bennett GM. The tiniest tiny genomes. Annu Rev Microbiol. 2014;68:195–215. doi: 10.1146/annurev-micro-091213-112901. [DOI] [PubMed] [Google Scholar]
- 16.Bennett GM, Moran NA. Heritable symbiosis: The advantages and perils of an evolutionary rabbit hole. Proc Natl Acad Sci USA. 2015;112:10169–10176. doi: 10.1073/pnas.1421388112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Douglas AE. How multi-partner endosymbioses function. Nat Rev Microbiol. 2016;14:731–743. doi: 10.1038/nrmicro.2016.151. [DOI] [PubMed] [Google Scholar]
- 18.Grimardi D, Engel MS. Evolution of the Insects. Cambridge Univ Press; New York: 2005. [Google Scholar]
- 19.Hunt T, et al. A comprehensive phylogeny of beetles reveals the evolutionary origins of a superradiation. Science. 2007;318:1913–1916. doi: 10.1126/science.1146954. [DOI] [PubMed] [Google Scholar]
- 20.Stork NE, McBroom J, Gely C, Hamilton AJ, Hamilton AJ. New approaches narrow global species estimates for beetles, insects, and terrestrial arthropods. Proc Natl Acad Sci USA. 2015;112:7519–7523. doi: 10.1073/pnas.1502408112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oberprieler RG, Marvaldi AE, Anderson RS. Weevils, weevils, weevils everywhere. Zootaxa. 2007;1668:491–520. [Google Scholar]
- 22.McKenna DD, Sequeira AS, Marvaldi AE, Farrell BD. Temporal lags and overlap in the diversification of weevils and flowering plants. Proc Natl Acad Sci USA. 2009;106:7083–7088. doi: 10.1073/pnas.0810618106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lefèvre C, et al. Endosymbiont phylogenesis in the dryophthoridae weevils: Evidence for bacterial replacement. Mol Biol Evol. 2004;21:965–973. doi: 10.1093/molbev/msh063. [DOI] [PubMed] [Google Scholar]
- 24.Conord C, et al. Long-term evolutionary stability of bacterial endosymbiosis in Curculionoidea: Additional evidence of symbiont replacement in the Dryophthoridae family. Mol Biol Evol. 2008;25:859–868. doi: 10.1093/molbev/msn027. [DOI] [PubMed] [Google Scholar]
- 25.Hosokawa T, Fukatsu T. Nardonella endosymbiont in the West Indian sweet potato weevil Euscepes postfasciatus (Coleoptera: Curculionidae) Appl Entomol Zool (Jpn) 2010;45:115–120. [Google Scholar]
- 26.Kuriwada T, et al. Biological role of Nardonella endosymbiont in its weevil host. PLoS One. 2010;5:e13101. doi: 10.1371/journal.pone.0013101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rinke R, et al. Microbial diversity in the larval gut of field and laboratory populations of the sugarcane weevil Sphenophorus levis (Coleoptera, Curculionidae) Genet Mol Res. 2011;10:2679–2691. doi: 10.4238/2011.November.1.1. [DOI] [PubMed] [Google Scholar]
- 28.Hirsch J, Strohmeier S, Pfannkuchen M, Reineke A. Assessment of bacterial endosymbiont diversity in Otiorhynchus spp. (Coleoptera: Curculionidae) larvae using a multitag 454 pyrosequencing approach. BMC Microbiol. 2012;12(Suppl 1):S6. doi: 10.1186/1471-2180-12-S1-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hosokawa T, et al. Nardonella endosymbionts of Japanese pest and non-pest weevils (Coleoptera: Curculionidae) Appl Entomol Zool (Jpn) 2015;50:223–229. [Google Scholar]
- 30.White JA, et al. Endosymbiotic candidates for parasitoid defense in exotic and native New Zealand weevils. Microb Ecol. 2015;70:274–286. doi: 10.1007/s00248-014-0561-8. [DOI] [PubMed] [Google Scholar]
- 31.Huang X, et al. The symbiotic bacteria Nardonella in rice water weevil (Coleoptera: Curculionidae): Diversity, density, and associations with host reproduction. Ann Entomol Soc Am. 2016;109:415–423. [Google Scholar]
- 32.Heddi A, Nardon P. Sitophilus oryzae L.: A model for intracellular symbiosis in the Dryophthoridae weevils (Coleoptera) Symbiosis. 2005;39:1–11. [Google Scholar]
- 33.Toju H, et al. “Candidatus Curculioniphilus buchneri,” a novel clade of bacterial endocellular symbionts from weevils of the genus Curculio. Appl Environ Microbiol. 2010;76:275–282. doi: 10.1128/AEM.02154-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Toju H, Fukatsu T. Diversity and infection prevalence of endosymbionts in natural populations of the chestnut weevil: Relevance of local climate and host plants. Mol Ecol. 2011;20:853–868. doi: 10.1111/j.1365-294X.2010.04980.x. [DOI] [PubMed] [Google Scholar]
- 35.Toju H, Tanabe AS, Notsu Y, Sota T, Fukatsu T. Diversification of endosymbiosis: Replacements, co-speciation and promiscuity of bacteriocyte symbionts in weevils. ISME J. 2013;7:1378–1390. doi: 10.1038/ismej.2013.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wicker C, Nardon P. Development responses of symbiotic and aposymbiotic weevils Sitophilus oryzae L. (Coleoptera, Curculionidae) to a diet supplemented with aromatic amino acids. J Insect Physiol. 1982;28:1021–1024. [Google Scholar]
- 37.Vigneron A, et al. Insects recycle endosymbionts when the benefit is over. Curr Biol. 2014;24:2267–2273. doi: 10.1016/j.cub.2014.07.065. [DOI] [PubMed] [Google Scholar]
- 38.Wernegreen JJ. Mutualism meltdown in insects: Bacteria constrain thermal adaptation. Curr Opin Microbiol. 2012;15:255–262. doi: 10.1016/j.mib.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kikuchi Y, et al. Collapse of insect gut symbiosis under simulated climate change. MBio. 2016;7:e01578–e16. doi: 10.1128/mBio.01578-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.True JR. Insect melanism: The molecules matter. Trends Ecol Evol. 2003;18:640–647. [Google Scholar]
- 41.Noh MY, Muthukrishnan S, Kramer KJ, Arakane Y. Cuticle formation and pigmentation in beetles. Curr Opin Insect Sci. 2016;17:1–9. doi: 10.1016/j.cois.2016.05.004. [DOI] [PubMed] [Google Scholar]
- 42.Faleiro JR. A review of the issues and management of the red palm weevil Rhynchophorus ferrugineus (Coleoptera: Rhynchophoridae) in coconut and date palm during the last one hundred years. Int J Trop Insect Sci. 2006;26:135–154. [Google Scholar]
- 43.Seago AE, Brady P, Vigneron JP, Schultz TD. Gold bugs and beyond: A review of iridescence and structural colour mechanisms in beetles (Coleoptera) J R Soc Interface. 2009;6(Suppl 2):S165–S184. doi: 10.1098/rsif.2008.0354.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Riedel A, Sagata K, Surbakti S, Rene Tänzler, Michael Balke (2013) One hundred and one new species of Trigonopterus weevils from New Guinea. ZooKeys 280:1–150. [DOI] [PMC free article] [PubMed]
- 45.Crowson RA. The Biology of the Coleoptera. Academic; London: 1981. [Google Scholar]
- 46.Weissling TJ, Giblin-Davis RM. Water loss dynamics and humidity preference of Rhynchophorus cruentatus (Coleoptera: Curculionidae) adults. Environ Entomol. 1993;22:93–98. [Google Scholar]
- 47.Tseng HY, Lin CP, Hsu JY, Pike DA, Huang WS. The functional significance of aposematic signals: Geographic variation in the responses of widespread lizard predators to colourful invertebrate prey. PLoS One. 2014;9:e91777. doi: 10.1371/journal.pone.0091777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van de Kamp T, Riedel A, Greven H. Micromorphology of the elytral cuticle of beetles, with an emphasis on weevils (Coleoptera: Curculionoidea) Arthropod Struct Dev. 2016;45:14–22. doi: 10.1016/j.asd.2015.10.002. [DOI] [PubMed] [Google Scholar]
- 49.van de Kamp T, et al. Comparative thorax morphology of death-feigning flightless cryptorhynchine weevils (Coleoptera: Curculionidae) based on 3D reconstructions. Arthropod Struct Dev. 2015;44:509–523. doi: 10.1016/j.asd.2015.07.004. [DOI] [PubMed] [Google Scholar]
- 50.Oakeson KF, et al. Genome degeneration and adaptation in a nascent stage of symbiosis. Genome Biol Evol. 2014;6:76–93. doi: 10.1093/gbe/evt210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gil R, et al. Massive presence of insertion sequences in the genome of SOPE, the primary endosymbiont of the rice weevil Sitophilus oryzae. Int Microbiol. 2008;11:41–48. [PubMed] [Google Scholar]
- 52.Nardon P. Obtention d’une souche asymbiotique chez le charançon Sitophilus sasakii: Différentes méthodes et comparaison avec la souche symbiotique d’origine. C R Acad Sci Paris D. 1973;3:65–67. [Google Scholar]
- 53.Grenier AM, Nardon P, Bonnot G. Importance de la symbiose dans la croissance des populations de Sitophilus oryzae L. (Coleoptère Curculionidae) Oecol Appl. 1986;7:93–110. [Google Scholar]
- 54.Grenier AM, Nardon C, Nardon P. The role of symbiotes in flight activity of Sitophilus weevils. Entomol Exp Appl. 1994;70:201–208. [Google Scholar]
- 55.Wicker C. Differential vitamin and choline requirements of symbiotic and aposymbiotic S. oryzae (Coleoptera: Curculionidae) Comp Biochem Physiol A. 1983;76:177–182. [Google Scholar]
- 56.Gasnier-Fauchet F, Gharib A, Nardon P. Comparison of methionine metabolism in symbiotic and aposymbiotic larvae of Sitophilus oryzae L. (Coleoptera: Curculionidae)—I. Evidence for a glycine N-methyltransferase-like activity in the aposymbiotic larvae. Comp Biochem Physiol B. 1986;85:245–250. [Google Scholar]
- 57.Gasnier-Fauchet F, Nardon P. Comparison of methionine metabolism in symbiotic and aposymbiotic larvae of Sitophilus oryzae L. (Coleoptera: Curculionidae)—II. involvement of the symbiotic bacteria in the oxidation of methionine. Comp Biochem Physiol B. 1986;85:251–254. [Google Scholar]
- 58.Heddi A, Lefebvre F, Nardon P. Effect of endocytobiotic bacteria on mitochondrial enzymatic activities in the weevil Sitophilus oryzae (Coleoptera: Curculionidae) Insect Biochem Mol Biol. 1993;23:403–411. [Google Scholar]
- 59.Heddi A, Nardon P. Mitochondrial DNA expression in symbiotic and aposymbiotic strains of Sitophilus oryzae. J Stored Prod Res. 1993;29:243–252. [Google Scholar]
- 60.Adams MD, et al. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 61.Shigenobu S, Watanabe H, Hattori M, Sakaki Y, Ishikawa H. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature. 2000;407:81–86. doi: 10.1038/35024074. [DOI] [PubMed] [Google Scholar]
- 62.International Aphid Genomics Consortium Genome sequence of the pea aphid Acyrthosiphon pisum. PLoS Biol. 2010;8:e1000313. doi: 10.1371/journal.pbio.1000313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wilson ACC, et al. Genomic insight into the amino acid relations of the pea aphid, Acyrthosiphon pisum, with its symbiotic bacterium Buchnera aphidicola. Insect Mol Biol. 2010;19:249–258. doi: 10.1111/j.1365-2583.2009.00942.x. [DOI] [PubMed] [Google Scholar]
- 64.Shigenobu S, Wilson ACC. Genomic revelations of a mutualism: The pea aphid and its obligate bacterial symbiont. Cell Mol Life Sci. 2011;68:1297–1309. doi: 10.1007/s00018-011-0645-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hansen AK, Moran NA. Aphid genome expression reveals host-symbiont cooperation in the production of amino acids. Proc Natl Acad Sci USA. 2011;108:2849–2854. doi: 10.1073/pnas.1013465108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rabatel A, et al. Tyrosine pathway regulation is host-mediated in the pea aphid symbiosis during late embryonic and early larval development. BMC Genomics. 2013;14:235. doi: 10.1186/1471-2164-14-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Russell CW, Bouvaine S, Newell PD, Douglas AE. Shared metabolic pathways in a coevolved insect-bacterial symbiosis. Appl Environ Microbiol. 2013;79:6117–6123. doi: 10.1128/AEM.01543-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Simonet P, et al. Disruption of phenylalanine hydroxylase reduces adult lifespan and fecundity, and impairs embryonic development in parthenogenetic pea aphids. Sci Rep. 2016;6:34321. doi: 10.1038/srep34321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Price DRG, et al. Aphid amino acid transporter regulates glutamine supply to intracellular bacterial symbionts. Proc Natl Acad Sci USA. 2014;111:320–325. doi: 10.1073/pnas.1306068111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Douglas AE. Molecular dissection of nutrient exchange at the insect-microbial interface. Curr Opin Insect Sci. 2014;4:23–28. doi: 10.1016/j.cois.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 71.Koga R, Tsuchida T, Fukatsu T. Quenching autofluorescence of insect tissues for in situ detecton of endosymbionts. Appl Entomol Zool (Jpn) 2009;44:281–291. [Google Scholar]
- 72.Nikoh N, Hosokawa T, Oshima K, Hattori M, Fukatsu T. Reductive evolution of bacterial genome in insect gut environment. Genome Biol Evol. 2011;3:702–714. doi: 10.1093/gbe/evr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nikoh N, et al. Evolutionary origin of insect-Wolbachia nutritional mutualism. Proc Natl Acad Sci USA. 2014;111:10257–10262. doi: 10.1073/pnas.1409284111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kaiwa N, et al. Symbiont-supplemented maternal investment underpinning host’s ecological adaptation. Curr Biol. 2014;24:2465–2470. doi: 10.1016/j.cub.2014.08.065. [DOI] [PubMed] [Google Scholar]
- 75.Hosokawa T, et al. Obligate bacterial mutualists evolving from environmental bacteria in natural insect populations. Nat Microbiol. 2016;1:15011. doi: 10.1038/nmicrobiol.2015.11. [DOI] [PubMed] [Google Scholar]
- 76.Grabherr MG, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29:644–652. doi: 10.1038/nbt.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kung C, Martinac B, Sukharev S. Mechanosensitive channels in microbes. Annu Rev Microbiol. 2010;64:313–329. doi: 10.1146/annurev.micro.112408.134106. [DOI] [PubMed] [Google Scholar]
- 78.Narberhaus F. α-crystallin-type heat shock proteins: Socializing minichaperones in the context of a multichaperone network. Microbiol Mol Biol Rev. 2002;66:64–93. doi: 10.1128/MMBR.66.1.64-93.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.