Significance
The [PSI+] prion of baker’s yeast is a filamentous polymer (amyloid) of the Sup35 protein, producing readthrough of translation termination and different degrees of growth slowing, depending on the prion variant. We show that certain inositol polyphosphates and pyrophosphates promote the propagation of the [PSI+] prion and that an inositol pyrophosphate pyrophosphatase has an antiprion effect. Inositol poly-/pyrophosphates are intracellular signaling molecules not previously connected with any amyloidosis.
Keywords: prion, inositol polyphosphate, Arg82, Siw14, [PSI+]
Abstract
The yeast prions [PSI+] and [URE3] are folded in-register parallel β-sheet amyloids of Sup35p and Ure2p, respectively. In a screen for antiprion systems curing [PSI+] without protein overproduction, we detected Siw14p as an antiprion element. An array of genetic tests confirmed that many variants of [PSI+] arising in the absence of Siw14p are cured by restoring normal levels of the protein. Siw14p is a pyrophosphatase specifically cleaving the β phosphate from 5-diphosphoinositol pentakisphosphate (5PP-IP5), suggesting that increased levels of this or some other inositol polyphosphate favors [PSI+] propagation. In support of this notion, we found that nearly all variants of [PSI+] isolated in a WT strain were lost upon loss of ARG82, which encodes inositol polyphosphate multikinase. Inactivation of the Arg82p kinase by D131A and K133A mutations (preserving Arg82p’s nonkinase transcription regulation functions) resulted the loss of its ability to support [PSI+] propagation. The loss of [PSI+] in arg82Δ is independent of Hsp104’s antiprion activity. [PSI+] variants requiring Arg82p could propagate in ipk1Δ (IP5 kinase), kcs1Δ (IP6 5-kinase), vip1Δ (IP6 1-kinase), ddp1Δ (inositol pyrophosphatase), or kcs1Δ vip1Δ mutants but not in ipk1Δ kcs1Δ or ddp1Δ kcs1Δ double mutants. Thus, nearly all [PSI+] prion variants require inositol poly-/pyrophosphates for their propagation, and at least IP6 or 5PP-IP4 can support [PSI+] propagation.
There are a multitude of antiviral and antibacterial systems to deal with the variety of these infectious agents. Saccharomyces cerevisiae has at least nine proteins capable of forming prions, most based on amyloid filaments formed from normally nonamyloid proteins (reviewed in ref. 1). [PSI+] is a prion of the translation termination factor Sup35p, and [URE3] is a prion of the nitrogen catabolite repression factor Ure2p (2). These two prions are detected by phenotypes due to the partial deficiency of the active normal form of the protein. [PSI+] and [URE3] are based on amyloid filaments of Sup35p and Ure2p, respectively (3–11). Their folded in-register parallel β-sheet architecture (12–14) naturally suggests a mechanism by which the molecules in the filament transmit their conformation to monomers newly joining the chain by a type of templating (1, 15), in analogy to DNA transmitting its sequence to a newly forming chain. Distinct self-propagating amyloid conformations are believed to determine the many different prion variants that one can observe for a given prion protein sequence (9, 10, 16).
The newly formed [PSI+] and [URE3] prions are most often toxic or even lethal (17), and the infrequent occurrence of even their mildest forms (18–20) in wild strains indicates that they are, on the net, detrimental (19, 21; reviewed in ref. 22). One expects that there should be antiprion systems that prevent prion formation or cure them as they arise. Ssb1p and Ssb2p are ribosome-associated Hsp70 chaperones believed to assist the cotranslational folding of nascent proteins (23). In the absence of Ssb1/2, the frequency of [PSI+] generation is elevated (24). Restoring Ssb1 to the double mutant that has become [PSI+] does not cure the prion, indicating that the Ssb chaperones partially prevent [PSI+] from arising (24). The Hsp104 disaggregating chaperone is necessary for the propagation of most amyloid-based yeast prions but if overproduced can cure the [PSI+] prion (25–27). Mutation of the Hsp104 N-terminal domain eliminates its ability to cure [PSI+] by overproduction without affecting its prion propagation activity (28). Using this finding, we showed that this prion-curing activity of Hsp104 acts at normal levels of the protein to eliminate most spontaneous [PSI+] variants as they arise (29). Overproduction of Btn2p and Cur1p each cure the [URE3] prion (30), but normal levels of either protein cure most variants of [URE3] arising in their absence (31). Btn2p acts by collecting prion amyloid filaments at one place in the cell, so that one of the daughter cells is likely to be prion-free (30). These represent three (or four) antiprion systems working in normal cells to prevent prion generation or to cure newly arising prions. Mutation of each of these systems elevates spontaneous prion generation frequency by 10-fold or more.
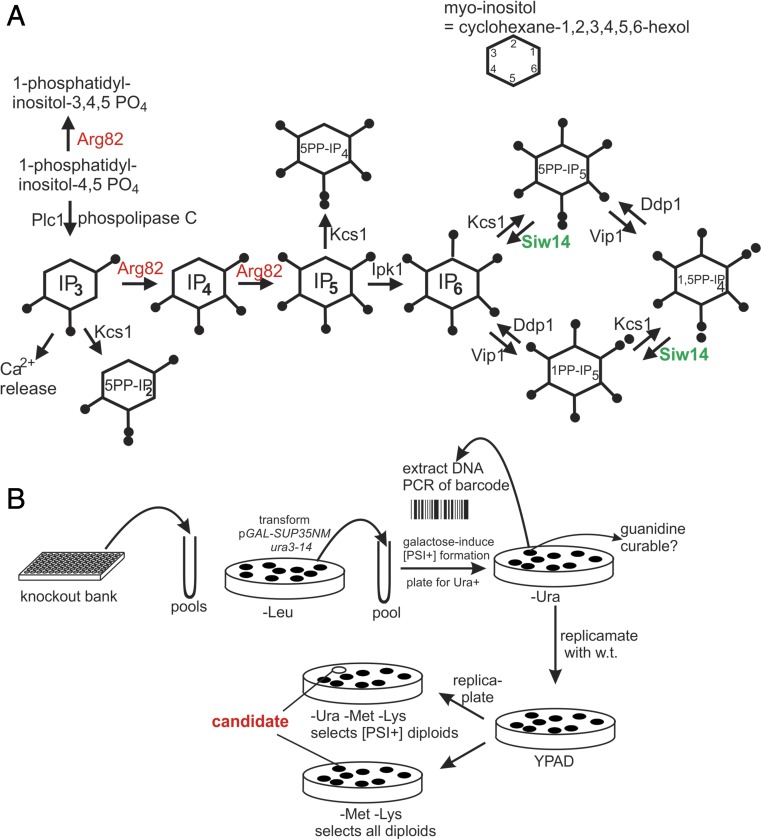
Based on this experience, we devised a general screen for antiprion systems and found that Siw14p acts as an antiprion element. Siw14 is a pyrophosphatase specific for 5PP-IP5 (5-diphosphoinositol pentakisphosphate) (32). Inositol polyphosphates (IPs) and pyrophosphates are signaling molecules regulating energy balance, phosphate uptake, DNA damage repair, telomere shortening, response to certain stress conditions, vesicle trafficking, and other functions (33). We further show that IPs are important for the propagation of most [PSI+] variants and that the Siw14 pyrophosphatase acts as a [PSI+]-curing factor by limiting the levels of some inositol poly-/pyrophosphates. The pathways of IP synthesis are shown in Fig. 1A (reviewed in refs. 3, 4, and 35).
Fig. 1.
(A) IP synthesis pathways. (B) Diagram of the isolation scheme for mutants in antiprion genes.
Results
Isolation of Anti-[PSI+]–Defective Mutants.
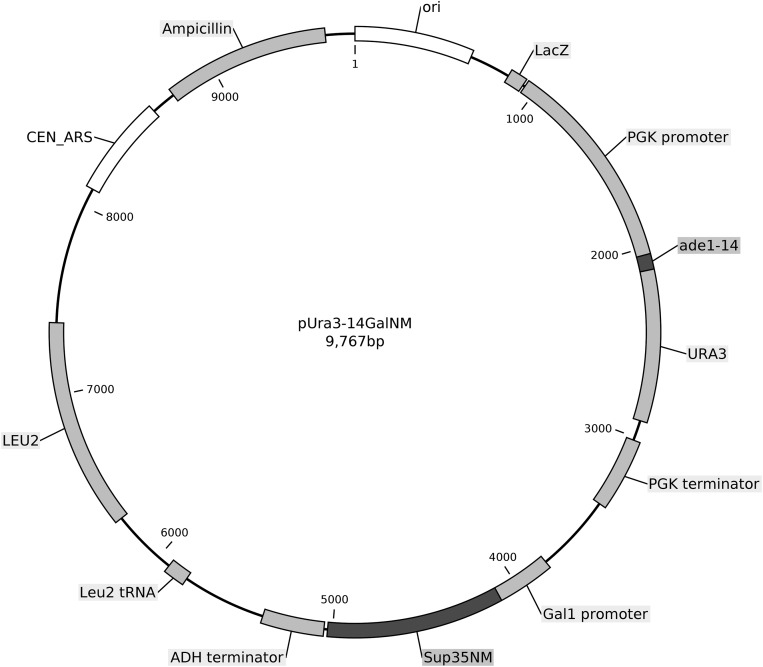
In designing a general screen to find [PSI+]-curing systems that are effective without protein overproduction, we assumed that such a system would be variant-specific because [PSI+] variants do arise despite such systems. Modeling our screen on the Btn2/Cur1 experiments, we generated [PSI+] variants in pools of subsets of the yeast knockout collection (36). To score [PSI+] in the knockout bank strains (all ura3Δ), we used ura3-14 (37), a [PSI+]-suppressible nonsense allele, on a CEN plasmid (p1520) that includes the part of SUP35 encoding the prion domain (NM) under control of the GAL1 promoter (Fig. S1 and Dataset S1). Overproduction of the amyloid-forming part of a prion protein (Sup35NM in this case) dramatically increases the frequency of prion formation (2). [PSI+] cells are Ura+, and [psi−] cells are Ura−. Pools of the MATa knockout bank were transformed en masse with p1520, grown for 24 h in galactose to induce the appearance of an array of [PSI+] variants, and plated on medium lacking Ura (−Ura) to select [PSI+] clones (Fig. 1B). These clones were replica-mated with an isogenic WT MATα strain to complement the knockout of each clone. We looked for clones that formed diploids but not [PSI+] (i.e., Ura+) diploids. [PSI+] is efficiently cured by growth in the presence of 5 mM guanidine (38), a specific inhibitor of the disaggregating chaperone Hsp104 (39–41). We confirmed that the candidate haploid clones were [PSI+] by showing they were curable by guanidine and, as shown below, are transferred by cytoplasmic mixing (cytoduction). To identify the gene deleted in candidate anti-[PSI+]–defective clones, we amplified by PCR and sequenced the bar-code region of the kanMX module (42). Two such isolates, PB7 and PB14, carried siw14::kanMX and are characterized here.
Fig. S1.
Structure of p1520 carrying the ura3-14 gene and SUP35NM controlled by the GAL1 promoter.
PB7 was cured of [PSI+] and [PIN+] (a prion of Rnq1p needed for [PSI+] induction; see refs. 43–45) by growth on 5 mM guanidine, and then [PIN+] was replaced by cytoduction from strain 4457. [PSI+] generation was again induced by overproduction of Sup35NM in galactose medium, and Ura+ clones were isolated and again tested by guanidine curing and mating with the isogenic WT strain 4729. Nine of seventeen Ura+ guanidine-curable clones produced Ura− diploids on mating with 4729, suggesting that about half of the [PSI+] variants arising in the siw14Δ strain were cured by restoring normal amounts of Siw14p. We call such a Siw14p-sensitive prion variant “[PSI+ss].”
[PSI+ss] Is Stable in siw14::kanMX Strains and Is Lost When SIW14 Is Restored.
Eight new apparently [PSI+ss] isolates were obtained as above in the siw14Δ strain PB7 and were either subcloned on YPAD medium or were mated with isogenic WT strain 4729 and the diploids formed were subcloned on YPAD medium. Both the siw14Δ haploid and siw14Δ/+ diploid subclones were replica-plated to –Ura to test the stability of [PSI+] (Table 1). Each [PSI+ss] variant was more stable in the siw14Δ haploid than in the complemented (heterozygous) diploid.
Table 1.
[PSI+ss] is more stable in siw14Δ than in the siw14Δ/+ host
| Isolate no. | Ura+/total subclones | |
| siw14Δ | siw14Δ/+ | |
| 1 | 48/48 | 4/25 |
| 2 | 41/41 | 9/29 |
| 3 | 35/35 | 1/16 |
| 4 | 39/40 | 0/26 |
| 5 | 31/31 | 1/17 |
| 6 | 31/36 | 2/22 |
| 7 | 50/50 | 2/12 |
| 8 | 41/41 | 2/23 |
Ura+, guanidine-curable clones induced in strain 5255 (siw14Δ [PIN+]) were subcloned on YPAD medium or mated with strain 4729 (SIW14 [psi−]) and then subcloned on YPAD medium. Subclones were tested for [PSI+] by replica-plating on −Ura plates.
The Ura− Phenotype in SIW14 Hosts Is due to [PSI+] Loss.
If the Ura− phenotype of diploids formed by mating siw14Δ [PSI+ss] with a WT strain is due to the loss of [PSI+ss], then all meiotic segregants of such diploids should be Ura−, whether they are siw14Δ or SIW14. Indeed, Ura− diploids of PB7 × 4729 and PB14 × 4729 produced only Ura− meiotic segregants (12 tetrads each). However, if PB7 (siw14::kanMX [PSI+ss]) was mated with the isogenic WT [psi−] strain 4729 (= 4813) and sporulated immediately, before the loss of [PSI+ss] in the diploids could occur, the segregation was 2 Ura+ G418res: 2 Ura− G418sen (29 tetrads). These experiments show that siw14Δ does not simply affect the expression of the [PSI+] phenotype but determines the stability of prion propagation. The cosegregation of Ura+ and G418res also shows that it is indeed the siw14::kanMX allele that allows the propagation of [PSI+ss], and not some other incidental mutation in the strain.
Several [PSI+] variants (all guanidine-curable and mitotically stable) were isolated anew in 5255 (siw14Δ [PIN+] [psi−]) and cytoduced into isogenic WT and siw14Δ recipients (Table 2). Some variants were equally transmitted to both recipients, but the [PSI+ss] variants were better transmitted to the siw14Δ recipient. To confirm that cytoductants from siw14Δ [PSI+ss] into WT cells lost [PSI+ss], the Ura− cytoductants were used as donors to return the cytoplasm to a siw14Δ environment (strain 5255). These back cytoductants were uniformly Ura−, including experiments with three independent [PSI+ss]s (Table 3).
Table 2.
Cytoduction of Siw14p-sensitive and -insensitive [PSI+] isolates
| [PSI+] donor isolate no. | Recipient WT strain 5335 | Recipient siw14Δ strain 5337 | Stability of [PSI+] donor | |||
| [PSI+] | [psi−] | [PSI+] | [psi−] | [PSI+] | [psi−] | |
| 1 | 17 | 2 | 13 | 0 | 33 | 0 |
| 9 | 15 | 3 | 18 | 0 | 38 | 0 |
| 14* | 1 | 27 | 16 | 7 | 67 | 0 |
| 21* | 0 | 39 | 11 | 11 | 30 | 0 |
| 20* | 0 | 31 | 15 | 10 | 32 | 0 |
A series of [PSI+]s (each guanidine curable) was generated in strain 5255 (siw14 [PIN+] [psi−]). Cytoductions were performed into isogenic WT strain 5335 and siw14Δ strain 5337 recipients, and the original [PSI+] clones were subcloned to determine their stability.
Isolates 14, 20, and 21 are [PSI+ss].
Table 3.
Back-cytoduction proves [PSI+ss] is lost in a WT strain
| [PSI+ss] donor 1 | WT recipient 1 | siw14Δ recipient 2 |
| Cytoductants 1 = donor 2 | Cytoductants 2 | |
| PB7 siw14 [PSI+ss] | 2 Ura+, 9 Ura− | All Ura− (>17) |
| PB14 siw14 [PSI+ss] | 10 Ura− | All Ura− |
| 5355 siw14 [PSI+ss] | 15 Ura− | All Ura− |
| [psi−] control | 20 Ura− | All Ura− |
Recipient 1 is WT strain 5402, and recipient 2 is siw14Δ strain 5255, each isogenic with the donors but initially [psi−]. Ura− cytoductants of cytoduction 1 were used as donors in cytoduction 2.
To further confirm that it is the siw14Δ mutation that allows propagation of [PSI+ss], we transformed siw14Δ [PSI+ss] strains with a CEN plasmid carrying SIW14 driven by its own promoter and found curing by this plasmid (compared with the empty vector) (Table 4).
Table 4.
Transforming siw14Δ [PSI+ss] with SIW14 cures [PSI+ss]
| Plasmid | [PSI+ss] siw14Δ strain transformant phenotype: %Ura+ (total transformants) | ||
| PB7 | PB14 | 5355 | |
| Vector pH321 | 75 (57) | 97 (331) | 76 (238) |
| pSIW14 p1569 | 20 (570) | 36 (312) | 13 (686) |
siw14Δ Does Not Affect Translation Termination Readthrough.
The above experiments prove that SIW14 blocks propagation of [PSI+ss] variants. However, to determine whether Siw14p also affects translation termination readthrough, we used a dual luciferase plasmid with a 5′ Renilla luciferase gene separated by a UAA codon from an in-frame 3′ firefly luciferase gene (46). We examined two sets of isogenic siw14Δ [psi−] and WT [psi−] strains (Table 5). There were minor differences, but no consistent effect of siw14Δ on translational readthrough was observed.
Table 5.
Direct effects of siw14Δ and arg82Δ on translation termination efficiency
| Firefly luciferase/Renilla luciferase | |||
| Genotype (strain) | Experiment 1 | Experiment 2 | Experiment 3 |
| WT (4812) | 0.0030 | ||
| siw14Δ (5255) | 0.0040 | ||
| siw14Δ (5261) | 0.0026 | ||
| WT (4813) | 0.0010 | ||
| siw14Δ (5337) | 0.0032 | ||
| WT (4812) | 0.0053 | 0.0010 | 0.0012 |
| arg82Δ (5408) | 0.014 | 0.015 | 0.015 |
| WT (4813) | 0.015 | 0.0015 | 0.0019 |
| arg82Δ (5477) | 0.052 | 0.0020 | 0.044 |
Cells expressed Renilla luciferase upstream, separated by a UAA codon from firefly luciferase downstream from a single mRNA. The ratios show relative, not absolute, terminator readthrough rates because the enzyme activities are different. All strains tested were [psi−]. [PSI+] strains have values in the range 1.0–2.0.
Siw14p encodes a pyrophosphatase specific for 5PP-IP5 (Fig. 1) (32). A siw14Δ strain has substantially elevated levels of 5PP-IP5 and 1,5PP-IP4 (32), suggesting that one or both of these compounds may have effects favorable to the propagation of some [PSI+] variants.
Arg82p Is Necessary for Most [PSI+] Variants.
Arg82 is an IP multikinase converting IP3 to IP4 and then to IP5 (47, 48). The Arg82p kinase activity is necessary for the synthesis of 5PP-IP5 and all other inositol poly/pyrophosphates with more than three phosphates (47, 48). Our finding that Siw14p, which lowers the levels of certain inositol poly/pyrophosphates, antagonizes the propagation of certain [PSI+] variants suggests that other genetic modifications that lower the levels of these compounds may have a similar effect. We therefore tested whether arg82Δ has a similar effect. We prepared an arg82Δ lys2 strain carrying a single-copy LYS2 plasmid with ARG82 under its own promoter (p1574) as well as p1520 carrying ura3-14 and GAL1-SUP35NM on a LEU2 CEN plasmid for [PSI+] induction and detection.
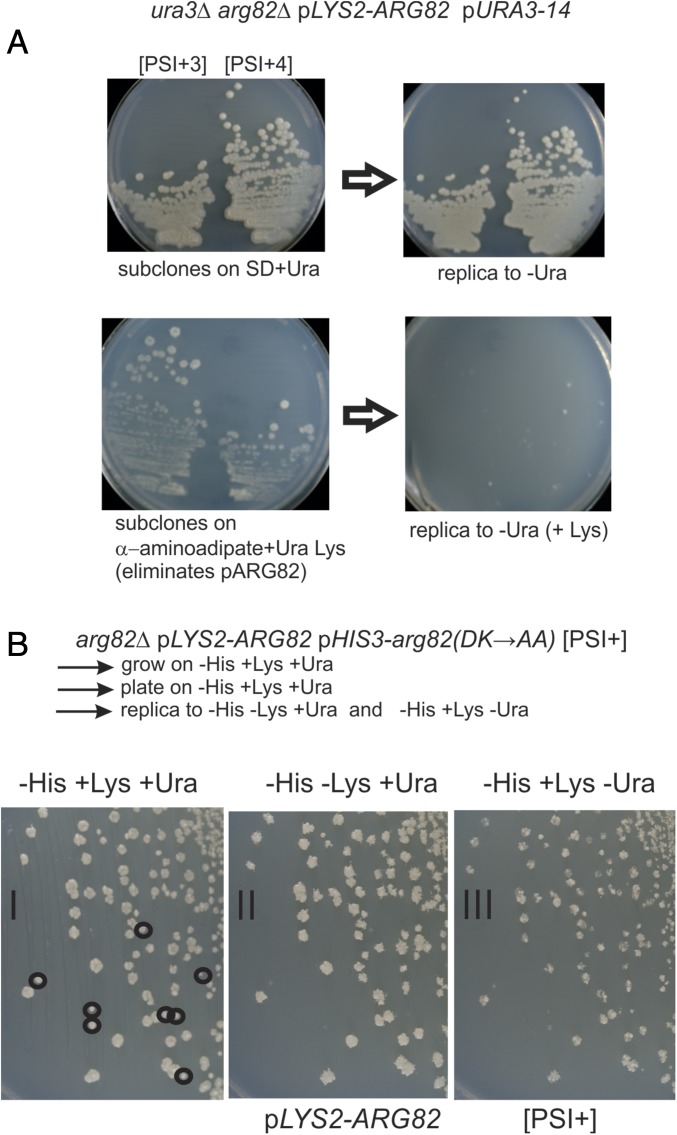
We induced [PSI+] formation in this essentially WT strain (5478) by growth in galactose for 24 h and selected [PSI+] clones by plating on −Ura −Leu −Lys medium. Guanidine-curable clones were identified as [PSI+] (called “[PSI+1],” “[PSI+2],” and so forth), and the loss of the ARG82 plasmid from such clones was then selected by plating on α-aminoadipate plates (49) or was screened for among colonies growing on rich medium by replica-plating to –Lys medium (Fig. 2A). For each of the 16 [PSI+] isolates tested, loss of pLYS2-ARG82 resulted in all cells becoming Ura−, but nearly all clones retaining pLYS2-ARG82 remained Ura+. Retransformation of the arg82Δ Ura− clones with p1574 (pLYS2-ARG82) resulted in most cells remaining Ura−. A minority of cells (13 of 64 for [PSI+3], 27 of 88 for [PSI+1]) became Ura+ again, indicating that loss of ARG82 did not eliminate the prion completely in all cells. However, essentially all arg82Δ cells were Ura−. We suggest that the minority arg82Δ Ura− [PSI+] cells had seed number so low that there was no phenotype, while the majority of cells had completely lost [PSI+].
Fig. 2.
(A) [PSI+] requires Arg82p for its propagation. [PSI+] cells with chromosomal arg82Δ and pLYS2-ARG82 become Ura− ([psi−]) when loss of the plasmid is selected on α-aminoadipate medium (selects lys2−). (B) The kinase activity of Arg82p is required for propagation of [PSI+]. The circled colonies in the left panel have lost pLYS2-ARG82 (i.e., fail to grow on −Lys in the middle panel) but retain pHIS3-arg82(D131A K133A) expressing the kinase-dead Arg82p. All such colonies are Ura− (i.e., fail to grow on −Ura in the right panel), showing that they cannot maintain [PSI+].
To test whether the arg82Δ mutation might directly affect translation terminator readthrough efficiency, we used the dual luciferase system mentioned above (Table 5). We found that arg82Δ resulted in a significant increase in readthrough efficiency in [psi−] strains, a result previously reported using a different tandem reporter plasmid (50). This result is the opposite of what would be expected if a direct effect on translation were to explain the Ura− phenotype produced from [PSI+] arg82Δ pARG82 cells on loss of pARG82. We infer that most arg82Δ cells have become [psi−] and that the minority that are not [psi−] by the retransformation test have a very low seed number.
Similar experiments were carried out in the 779-6A background using the [PSI+]-suppressible ade2-1 as the reporter, with similar results. The arg82Δ mutation and pARG82 (p1574) were introduced, [PSI+] clones were induced, and loss of pARG82 resulted in the loss of [PSI+] in each case (eight variants were tested).
The Kinase Activity of Arg82p Is Necessary for Propagation of [PSI+].
In addition to its inositol multikinase activity (47, 48), Arg82p is known to stabilize the essential transcription factor Mcm1p (51), facilitating mating, cell-cycle events, osmotolerance, and arginine metabolism (52, 53). The Mcm1p stabilization does not require the kinase activity, as inactivation of the kinase by the mutations D131A and K133A does not impair these activities (52, 54). To determine the role of the Arg82 kinase activity in [PSI+] propagation, we constructed an arg82Δ [PSI+] strain carrying p1574 (CEN LYS2 ARG82) as well as p1585 [CEN HIS3 arg82(D131A K133A)] and p1520 (CEN LEU2 ura3-14 Gal1p-NM). Upon loss of the LYS2 ARG82 plasmid, only the kinase-defective Arg82pD131A K133A is available, and [PSI+] is uniformly lost (Fig. 2B). This shows that the IP multikinase activity of Arg82p is responsible for the propagation of the [PSI+] prion.
ARG82 is immediately adjacent to SUP35 on chromosome IV with converging transcription and overlapping 3′ UTRs. One could suggest that the direct effect of arg82Δ on terminator readthrough (Table 5) and the loss of [PSI+] might both be a result of decreased SUP35 expression. However, the fact that ARG82 on a plasmid supports [PSI+] while the arg82(D131A K133A) mutant on a plasmid does not argues strongly against this interpretation. Moreover, as shown below, other mutants in the IP pathway in genes not located near SUP35 also lose [PSI+].
[PSI+] Propagation in Inositol Poly/Pyrophosphate Mutants.
To further narrow the range of possible IP species that may be involved in [PSI+] propagation, we used cytoduction to pass each of two Arg82p-dependent [PSI+] variants to an array of single and double mutants with altered IP metabolism (Fig. 1A and Table 6). The arg82Δ strains are deficient in all IPs above IP3 but accumulate elevated levels of its substrate, IP3 (48, 55). If IP3 were an inhibitor of [PSI+] propagation, then plc1Δ strains, unable to make IP3, should be able to propagate the prion (Fig. 1A). However, like arg82Δ mutants, plc1Δ strains could not propagate either [PSI+] variant (Table 6), showing that it is a product of Arg82 that is needed for [PSI+] rather than inhibition by its accumulated substrate.
Table 6.
Propagation of [PSI+] in IP mutants
| Donor arg82Δ pARG82 [PSI+x] | Recipient strain no. | % Ura+ cytoductants (total) | Donor arg82Δ pARG82 [PSI+x] | Recipient strain no. | % Ura+ cytoductants (total) |
| [PSI+1] | WT 4812 | 100 (19) | [PSI+1] | plc1Δ 5526 | 0 (10) |
| [PSI+2] | 100 (6) | [PSI+2] | 0 (14) | ||
| [PSI+1] | WT 4445 | 76 (25) | [PSI+4] | kcs1Δ ipk1Δ | 0 (31) |
| [PSI+2] | 94 (33) | [PSI+6] | 5538 | 0 (21) | |
| [PSI+1] | WT 5506 | 100 (13) | [PSI+1] | kcs1Δ ipk1Δ | 0 (14) |
| [PSI+1] | gal7Δ 4866 | 100 (19) | [PSI+2] | 5536 | 0 (5) |
| [PSI+2] | 94 (17) | [PSI+1] | kcs1Δ ipk1Δ | 0 (15) | |
| [PSI+1] | siw14Δ 5498 | 94 (17) | [PSI+2] | 5538 | 0 (16) |
| [PSI+2] | 100 (30) | [PSI+1] | kcs1Δ ddp1Δ | 0 (10) | |
| [PSI+1] | siw14Δ 5505 | 92 (26) | [PSI+2] | 5525 | 0 (8) |
| [PSI+2] | 88 (17) | [PSI+4] | kcs1Δ ddp1Δ | 0 (36) | |
| [PSI+1] | ipk1Δ 5503 | 95 (19) | [PSI+6] | 5525 | 0 (17) |
| [PSI+2] | 96 (26) | [PSI+1] | kcs1Δ vip1Δ | 88 (24) | |
| [PSI+1] | ipk1Δ 5492 | 89 (28) | [PSI+2] | 5440 | 83 (30) |
| [PSI+2] | 88 (8) | [PSI+1] | siw14Δ vip1Δ | 98 (85) | |
| [PSI+1] | kcs1Δ 5502 | 100 (27) | [PSI+2] | 5522 | 100 (68) |
| [PSI+2] | 96 (25) | [PSI+1] | siw14Δ kcs1Δ | 100 (20) | |
| [PSI+1] | kcs1Δ 5495 | 100 (15) | [PSI+2] | 5497 | 95 (38) |
| [PSI+2] | 100% (19) | [PSI+1] | siw14Δ ddp1Δ | 100 (20) | |
| [PSI+1] | vip1Δ 5504 | 100% (22) | [PSI+2] | 5457 | 100 (10) |
| [PSI+2] | 100% (43) | [PSI+1] | arg82Δ pARG82 | 100 (15) | |
| [PSI+1] | vip1Δ 5496 | 100% (33) | [PSI+2] | 5499 | 100 (20) |
| [PSI+2] | 87% (30) | [PSI+1] | ipk1Δ vip1Δ | 100 (29) | |
| [PSI+1] | ddp1Δ 5494 | 100% (12) | [PSI+2] | 5197-2D | 100 (32) |
| [PSI+2] | 100% (16) | [PSI+1] | ipk1Δ vip1Δ | 100 (30) | |
| [PSI+2] | 5197-5B | 97 (41) |
Each donor was 5478 kar1Δ15 carrying one of four ARG82-dependent [PSI+] variants. As a control, the cytoductants into 5499 (arg82Δ pARG82) were [PSI+] but became [psi−] on the loss of pARG82.
[PSI+1] and [PSI+2] were efficiently transmitted by cytoduction to several single mutants in IP metabolism, including vip1Δ, kcs1Δ, ipk1Δ, siw14Δ, and ddp1Δ. However, ipk1Δ kcs1Δ or kcs1Δ ddp1Δ double mutants uniformly lost [PSI+] (Fig. 1A and Table 6). Mating the Ura− ipk1Δ kcs1Δ or kcs1Δ ddp1Δ double mutants with an isogenic WT strain produced all Ura− diploids except for a very rare Ura+ diploid. This shows that [PSI+] was indeed lost from the ipk1Δ kcs1Δ or kcs1Δ ddp1Δ double mutants and that the mutations did not simply affect the phenotype. Note that [PSI+] can propagate in the vip1Δ kcs1Δ double mutant, lacking all known inositol pyrophosphate-synthesizing enzymes. This indicates either that another inositol pyrophosphate-synthesizing enzyme exists or that some IP, presumably IP6, made by Ipk1p, can help [PSI+] (Discussion).
In addition to acting on IP3, Arg82p can phosphorylate phosphatidylinositol-4,5-diphosphate [PI(4,5)P2] to form phosphatidylinositol-3,4,5-triphosphate [PI(3,4,5)P3] (Fig. 1A) (56). If this were sufficient for prion propagation, then the double mutant ipk1Δ kcs1Δ would be able to propagate [PSI+] because neither protein is involved in making PI(3,4,5)P3. However, the double mutant cannot propagate [PSI+]. The inability of the plc1Δ strain to propagate [PSI+] supports this conclusion as well, because this mutant should not be impaired in making PI(3,4,5)P3.
Cytoduction of [PSI+1] or [PSI+2] into another arg82Δ pARG82 strain produced only Ura+ cytoductants that again lost [PSI+] on loss of pARG82 (Table 6). This important control shows that the [PSI+] variants had not changed since their initial isolation.
Overproduction of Siw14p Does Not Cure [PSI+].
The [PSI+] strain 779-6A (assaying [PSI+] using the suppressible allele ade2-1) was transformed with p1534 (GAL1 promoter-SIW14) or the vector (pH773), and transformants were grown on galactose for 2 d and then plated on 1/2 yeast extract/peptone/dextrose (YPD) medium to detect [PSI+] loss by the red pigment that accumulates in unsuppressed ade2 mutants. There was no increase in [psi−] clones. In another experiment, strain 74D-694 (ade1-14 [PIN+] [psi−]) carrying pSL1066 (CUP1 promoter, SUP35NM) and p1534 (GAL1 promoter, SIW14) was grown with copper to induce [PSI+] appearance, and 12 variants were tested for increased loss of [PSI+] after growth in galactose for 3 d. None showed greater instability than with the glucose control. Thus, this overproduction of SIW14p does not cure [PSI+] variants isolated at normal levels of the protein.
[PSI+] Variants Independent of ARG82.
Although the loss of the pARG82 from arg82Δ pARG82 [PSI+] Ura+ strains results in apparently uniformly Ura− cells, incubation for >1 wk results in growth of rare Ura+ clones. Each is guanidine-curable and produces frequent Ura− subclones in the absence of guanidine, suggesting that these are unstable, Arg82p-independent [PSI+] variants.
Is the Loss of [PSI+] in arg82Δ Cells a Result of Impairment of the Environmental Stress Response?
Cells exposed to a variety of stresses, including high salt, oxidation, and heat shock, respond by shutting down translation and turning on stress-response genes. This response is controlled by inositol pyrophosphates, and in arg82Δ strains the environmental stress response (ESR) is not effective (57). To test whether this system is responsible for our observation of the involvement of IPs in [PSI+] prion propagation, we tested cytoduction of two ARG82-requiring [PSI+] variants to strains carrying kcs1Δ vip1Δ. In kcs1Δ vip1Δ strains inositol pyrophosphate synthesis is blocked, and there is little or no ESR (57), but this mutant combination has no effect on [PSI+] propagation (Table 6). In addition, in our experiments, we do not subject the cells to any of the known inducers of the ESR. Thus [PSI+] propagation is not dependent on the ESR.
[URE3] Is Not Lost from arg82Δ Strains.
DAL5 is strongly repressed by active Ure2p in medium with a good nitrogen source, such as ammonia (58). Placing the ADE2 gene under the DAL5 promoter enables assay of the loss of Ure2p activity in a [URE3] strain as an Ade+ phenotype (11, 59). Strain BY241 (DAL5:ADE2) was made arg82Δ lys2, and p1330 (GAL-URE2N) and p1574 (pLYS2 ARG82) were introduced. [URE3] prion formation was induced by overproduction of the Ure2p prion domain (Ure2N). Sixteen Ade+ clones were tested for curing on 5 mM guanidine, and 15 were found to be curable. These 15 [URE3] isolates were grown to single colonies on either SD medium without lysine, to ensure they remain ARG82+, or on α-aminoadipate medium to select for loss of the ARG82 plasmid. For these 15 [URE3] isolates, there was no increase in loss of the prion following loss of ARG82.
Relation of Inositol Poly/Pyrophosphate Effects on [PSI+] to the Hsp104 [PSI+]-Curing Activity.
The disaggregating chaperone Hsp104, when overproduced, cures [PSI+] (25, 26), but Hsp104T160M lacks this activity (28). Normal levels of Hsp104 cure more than half of [PSI+] variants arising in the hsp104T160M mutant, showing that this constitutes an antiprion system (29). Two [PSI+] variants dependent on ARG82 for their propagation were cytoduced into isogenic arg82Δ hsp104T160M pLYS2-ARG82 and arg82Δ HSP104 pLYS2-ARG82 hosts. Loss of the pLYS2-ARG82 plasmid resulted in the uniform loss of each [PSI+] variant in both hosts (Table 7). This shows that the inositol poly/pyrophosphate requirement for [PSI+] does not operate through Hsp104’s [PSI+]-curing activity.
Table 7.
Loss of [PSI+] from arg82Δ cells does not require the Hsp104 antiprion activity
| Strain | Genotype | Lys− clones% Ade+ (no.) | Lys+ clones% Ade+ (>50) |
| 5565 | arg82Δ pLYS2-ARG82 [PSI+1] HSP104 | 0 (25) | 100 |
| 5571 | arg82Δ pLYS2-ARG82 [PSI+1] hsp104T160M | 0 (18) | 100 |
| 5565 | arg82Δ pLYS2-ARG82 [PSI+2] HSP104 | 0 (18) | 100 |
| 5571 | arg82Δ pLYS2-ARG82 [PSI+2] hsp104T160M | 0 (12) | 100 |
Strains 5565 (HSP104) and 5571 (hsp104T160M) were used as cytoduction recipients for [PSI+1] and [PSI+2] from strains 5552 and 5553. All cytoductants were Ade+ ([PSI+]). Single cytoductants (two for each genotype) were subcloned in the presence of lysine and adenine to allow plasmid loss. Clones were replica-plated to Lys− and Ade− plates. The hsp104T160M allele, which eliminates the Hsp104 antiprion activity, does not affect the requirement of [PSI+1] or [PSI+2] for ARG82.
Possible Targets of IP Action in Controlling [PSI+].
Among the IP6/5PP-IP5–binding proteins identified by Wu et al. were two chaperones, Sse1p and Hsp26p (60). Sse1p overproduction or deficiency cures [URE3], and sse1Δ strains lose a weak [PSI+] (61). In addition, overproduction of Sse1p stimulates [PSI+] generation, and Sse1p deficiency deters the formation of this prion (62). Deficiency of Hsp26p destabilizes [URE3] (31). Hsp42p is necessary for Btn2p overproduction curing of [URE3-1], and overproduction of Hsp42p itself can cure [URE3-1] (31). Thus, Hsp42p is also a candidate for involvement in [PSI+] curing. One group reports that overproduction of Hsp26 or Hsp42 cures [PSI+] (63), but we did not find that either cures [PSI+] (31). In this work, we also found that CEN plasmids expressing Hsp26 or Hsp42 from the very strong GPD1 promoter failed to produce curing of [PSI+1] or [PSI+2]. We also find here that Sse1p overproduction sufficient to cure [URE3-1] did not prevent the loss of [PSI+1] or [PSI+2] on the loss of Arg82p. Likewise, expression of Hsp26 from a high-copy plasmid (p1606) did not affect the loss of these [PSI+] variants in arg82 cells. Neither hsp26Δ nor hsp42Δ results in loss of [PSI+1] or [PSI+2] from an otherwise WT host, as shown by efficient cytoduction of either variant from strains 5548 or 5549 into the respective knockout mutant (Table 8). Further work will be required to identify the target of IP action affecting prion propagation.
Table 8.
Hsp26 and Hsp42 are dispensable for [PSI+] propagation
| Donor arg82Δ pARG82 [PSI+x] | Recipient strain no. (ρo p1520 pRS313) | %Ura+ cytoductants (total) |
| 5548 [PSI+1] | WT, 4812 | 88 (25) |
| 5549 [PSI+2] | WT, 4812 | 86 (14) |
| 5548 [PSI+1] | ipk1Δ kcs1Δ, 5536 | 0 (15) |
| 5549 [PSI+2] | ipk1Δ kcs1Δ, 5536 | 0 (22) |
| 5548 [PSI+1] | hsp26Δ, 5558 | 98 (40) |
| 5549 [PSI+2] | hsp26Δ, 5558 | 100 (23) |
| 5548 [PSI+1] | hsp42Δ, 5559 | 100 (25) |
| 5549 [PSI+2] | hsp42Δ, 5559 | 90 (21) |
Cytoduction donors were ρ+, and recipients were ρo. Cytoductants were identified as those with recipient nuclear genotype and donor cytoplasm (ρ+). The recipients carried p1520 bearing the ura3-14 allele, enabling scoring of [PSI+] as Ura+.
Discussion
Our search for antiprion systems has found that elevated inositol poly/pyrophosphates in the siw14Δ mutant allows propagation of many [PSI+] variants that cannot propagate in a WT strain. In this sense, Siw14p is an antiprion factor. Our search method has the virtue of detecting prion curing in a WT strain not overexpressing or deficient for any proteins. The prion variants arose in a mutant, but probably not because of the mutation, as prion formation occurs readily in the absence of any other proteins in vitro (9–11, 64) and probably in vivo. We also find that elimination of most of the inositol poly/pyrophosphates by an arg82Δ mutation results in the loss of almost all [PSI+] variants.
What Inositol Pyro-/Polyphosphates Allow [PSI+] to Propagate?
Because arg82Δ cells lose [PSI+], the IPs necessary for [PSI+] must be downstream of Arg82’s kinase steps, unless IP3, the substrate of Arg82, is inhibiting [PSI+] propagation. However, Plc1p, which produces IP3, is also necessary for [PSI+], indicating that [PSI+] is not lost from inhibition by IP3. The loss of [PSI+] in plc1Δ strains also argues that 1-phosphatidylinositol-3,4,5 phosphate, also produced by Arg82p, is not sufficient to support [PSI+] (Fig. 1A).
Although the only known effect of siw14Δ on the IPs is elevation of 5PP-IP5 (and possibly 1,5PP-IP4 or 5PP-IP4) (32), both kcs1Δ and kcs1Δ vip1Δ mutants can propagate [PSI+], suggesting that inositol pyrophosphates are not the only molecules capable of supporting [PSI+]. The failure of kcs1Δ ipk1Δ double mutants to propagate [PSI+] indicates that IP6 can also fulfill the [PSI+]-promoting role. IP6 is the only species missing in the kcs1Δ ipk1Δ double mutants that is present in the kcs1Δ vip1Δ strains. However, ipk1Δ single mutants propagate [PSI+] well, so IP6 is not the only species capable of supporting [PSI+]. These results suggest that 5PP-IP4 can help [PSI+]. If 5PP-I-1,4P (5PP-IP2), another product of Kcs1p action, were sufficient to help [PSI+], then the arg82Δ strain would not lose [PSI+]. Assuming the current model of inositol poly/pyrophosphate synthesis is correct, our results prove that IP6 and 5PP-IP4 are each sufficient to support [PSI+] propagation and that 5PP-IP5 and 1,5PP-IP4 are likely also capable of enabling [PSI+] propagation.
The above inferences do not provide an explanation of the loss of [PSI+] in the kcs1Δ ddp1Δ double mutants but not in either single mutant. If 1PP-IP5 were an inhibitor of [PSI+], its elevation in a ddp1Δ mutant, coupled with a loss of the [PSI+]-helping 5PP-IP5, would explain this result. However, expression of VIP1 from a GAL1 promoter in a kcs1Δ [PSI+] strain did not produce increased loss of the prion.
What is the Target of Inositol Poly/Pyrophosphates in Enabling [PSI+] Propagation?
Three components previously known to be involved in mRNA export from the nucleus, Dbp5p, Gle1p, and IP6, are now known to be necessary for efficient translation termination (50, 65, 66). IP6 (inositol hexakisphosphate) binds to Gle1p, and both Gle1p and Dbp5p bind to Sup45p, the partner of Sup35p in the translation termination complex. Thus, IP-deficient ipk1 mutants show inefficient translation termination, the opposite of the increased translation termination efficiency we see in ipk1Δ kcs1Δ strains due to the loss of [PSI+]. Thus, we cannot explain our results as simply the effect of IP6 binding to Gle1. However, further work will be needed to determine whether there is some indirect relation of these two inositol poly/pyrophosphate effects.
Recently, Wu et al. prepared affinity reagents to capture proteins binding to IP6 or 5PP-IP5 and isolated proteins from extracts of S. cerevisiae that were specifically bound (60). Among those identified were Sse1p, Hsp26p, and Ssb1,2p. Remarkably, most proteins isolated had similar affinity for the IP6 and the 5PP-IP5 affinity substrates (60), similar to the apparent ability, in our experiments, for either IP6 or a Kcs1p product (presumably 5PP-IP4 or 5PP-IP5) to support [PSI+] propagation. As discussed above, Ssbs lower the frequency of [PSI+] arising but do not cure [PSI+] variants produced in their absence (24). Sse1p is known to be critical for propagation of the [URE3] prion, with either overproduction or deficiency resulting in curing (61). It was found that a weak [PSI+] prion was lost from an sse1Δ strain, but a strong [PSI+] was maintained with a weakened phenotype (61). While overproduction of Sse1p enhanced [PSI+] generation, deficiency severely restricted it with a limited range of variants found (62). This makes Sse1p a candidate for mediating IP effects on [PSI+]. However, while all of the 24 [PSI+] tested here (including many strong [PSI+]) were lost from an arg82Δ, a strong [PSI+] was not lost from an Sse1Δ strain (61).
Hsp26p and Hsp42p are small heat-shock proteins, oligomeric inhibitors of protein aggregation with α-crystallin domains characteristic of this group (67). Hsp26 and Hsp42 are reported to block Sup35p amyloid formation and propagation in vitro (63). However, we find that neither deficiency nor overproduction of either of these proteins affects the propagation of either of two Arg82p-requiring [PSI+] variants in a WT strain. Further work will be needed to detail the mechanism of inositol poly/pyrophosphates on prion propagation. The IP may be stimulating, or inhibiting, or change the specificity of one or more of the components involved in prion propagation. IPs and pyrophosphates affect a wide array of cellular processes, most of them seemingly not likely to impinge on the process of prion propagation. In most cases, the direct target of the inositol poly/pyrophosphate is not known. Further work will be required to trace the pathway of prion control by these signal transducers. The known parallel of IP pathways between yeast and humans (33) and our finding of a previously unrecognized mode of control of prion propagation open possible avenues for control of a range of amyloid-based diseases.
Methods
Media and Strains.
Rich medium (YPAD), minimal medium (SD), and sporulation medium were as described (68). Low-adenine rich medium (1/2 YPD) is 0.5% yeast extract, 2% peptone, 2% dextrose, and 2% agar. Most strains used were isogenic to BY4741 (69). Most knockouts were made by PCR amplification of the gene::kanMX module from the knockout collection including 200 bp on each side, transformation selecting for G418 resistance, and confirmation by PCR with internal primers (kanB and kanC inside kanMX or primers inside the normal gene) and primers 300 bp 5′ or 3′ to the ORF being disrupted. The presence of the disruption and the absence of the normal gene was confirmed in each case. Double-mutant strains were constructed by meiotic crosses of two (isogenic) knockout bank strains and scoring of the segregants by PCR. Strains with the kar1Δ15 allele (70) were made as described (71) (Table S1).
Table S1.
Strains, plasmids, and primers
| Strains, plasmids, and oligonucleotides | Description | Source |
| Strains | ||
| BY4741/4812/4445 | MATa his3 ura3 leu2 met15 [PIN+][psi−] | (69) |
| BY4742/4813/4729 | MATα his3 ura3 leu2 lys2 [PIN+] [psi−] | (69) |
| 4457 | MATα trp1 ura2 leu2 kar1-1 [PIN+] | |
| PB7 | MATa siw14::kanMX his3 ura3 leu2 met15 [PIN+][PSI+ss] p1520 | This work |
| PB14 | MATa siw14::kanMX his3 ura3 leu2 met15 [PIN+][PSI+ss] p1520 | This work |
| 5255 | PB7 cured of [PSI+] and [PIN+], and [PIN+] then restored from strain 4457 | This work |
| 5335 | 4813 kar1Δ15 | This work |
| 5337 | 5335 siw14::kanMX | This work |
| 5355 | 5255 with a new [PSI+ss15] | This work |
| 5402 | MATα ade1-14 kar1Δ15 his3 ura3 leu2 lys2 p1520 | This work |
| 5261 | PB14 cured of [PSI+] and [PIN+], and [PIN+] then restored from strain 4457 | This work |
| 5408 | BY4741 arg82::kanMX | |
| 5477 | BY4742 arg82::kanMX | |
| 5478 | 5477 p1520 p1574 [PIN+] [psi−] | This work |
| 5506 | MATa ade1-14 kar1Δ15 his3 ura3 leu2 met15 p1520 pRS313 | This work |
| 4866 | BY4741 gal7::kanMX | |
| 5498, 5505 | MATa ade1-14 kar1Δ15 siw14::kanMX his3 ura3 leu2 met15 p1520 pRS313 ρo | This work |
| 5503, 5492 | MATa ipk1::kanMX his3 ura3 leu2 met15 p1520 pRS313 ρo | |
| 5502, 5495 | MATa kcs1::kanMX his3 ura3 leu2 met15 p1520 pRS313 ρo | |
| 5504, 5496 | MATa vip1::kanMX his3 ura3 leu2 met15 p1520 pRS313 ρo | |
| 5494 | MATa ddp1::kanMX his3 ura3 leu2 met15 p1520 pRS313 ρo | |
| 5538 | MATa ipk1::kanMX kcs1::kanMX his3 ura3 leu2 met15 kar1Δ15 p1520 pRS313 ρo | This work |
| 5536 | MATa ipk1::kanMX kcs1::kanMX his3 ura3 leu2 met15 kar1Δ15 p1520 pRS313 ρo | This work |
| 5526 | MATa plc1::kanMX his3 ura3 leu2 met15 p1520 pRS313 ρo | |
| 5525 | MATa kcs1::kanMX ddp1::kanMX his3 ura3 leu2 met15 kar1Δ15 p1520 pRS313 ρo | This work |
| 5440 | MATa vipk1::kanMX kcs1::kanMX his3 ura3 leu2 met15 p1520 pRS313 ρo | This work |
| 5522 | MATa vip1::kanMX siw14::kanMX ade1-14 his3 ura3 leu2 met15 p1520 pRS313 ρo | This work |
| 5497 | MATa kcs1::kanMX siw14::kanMX ade1-14 his3 ura3 leu2 met15 p1520 pRS313 ρo | This work |
| 5457 | MATa ddp1::kanMX siw14::kanMX ade1-14 his3 ura3 leu2 met15 p1520 pRS313 ρo | This work |
| 5197-2D | MATa vip1::kanMX ipk1::kanMX leu2 ura3 his3 lys2 | This work |
| 5197-5B | MATa vip1::kanMX ipk1::kanMX leu2 ura3 his3 lys2 | This work |
| 5197-7B | MATa vip1::kanMX ipk1::kanMX leu2 ura3 his3 lys2 met15 | This work |
| 74D-694 | MATa ade1-14 trp1-289 his3-200 ura3-52 leu2= [PIN+][psi−] | (24) |
| BY241 | MATa leu2 trp1 ura3 PDAL5:ADE2 PDAL5:CAN1 kar1 | (11) |
| 5560 | MATa leu2 trp1 ura3 PDAL5:ADE2 PDAL5:CAN1 kar1 arg82::kanMX lys2 | This work |
| 779-6A | MATa ade2-1 SUQ5 ura3 leu2 trp1 his3 [psi−] or [PSI+] | (39) |
| AG571 | MATα ade2-1 SUQ5 ura3 leu2 trp1 his3 [pin−] [psi−] ρo, isogenic with 779-6A | (29) |
| AG537 | MATα ade2-1 SUQ5 ura3 leu2 trp1 his3 hsp104T160M loxP-URA3-loxP [pin−] [psi−] ρo, isogenic with 779-6A | (29) |
| 5565 | AG571 lys2 arg82Δ pLYS2-ARG82 [PSI+1] HSP104 | This work |
| 5571 | AG537 lys2 arg82Δ pLYS2-ARG82 [PSI+1] hsp104T160M | This work |
| 5201-7B | MATa met15 lys2 leu2 his3 ura3 arg82::kanMX hsp26::kanMX | This work |
| Plasmids | ||
| p1520 | LEU2 CEN ura3-14 GAL1 promoter-SUP35NM | This work |
| p1574 | pRS317-ARG82 promoter-ARG82 LYS2 CEN | This work |
| pH773 | CEN HIS3 GAL1 promoter | H.K.E. |
| pH321 | pRS313 (CEN HIS3) with the HindIII and NheI sites removed from HIS3 | H.K.E. |
| p1569 | pH321 with SIW14 + 500 bp upstream of the ORF | This work |
| pSC4 | CEN LEU2 Renilla luciferase UAA firefly luciferase | (46) |
| pRS315-SIW14 | SIW14 with 500 bp of upstream sequence inserted as BamHI-SalI fragment in pRS315 | This work |
| pSIW14 p1569 | SIW14 with 500 bp of upstream sequence as BamHI-SalI fragment from pRS315-SIW14 inserted in pH321 | This work |
| p1534 | pH773 expressing the SIW14 orf from the GAL1 promoter | This work |
| pSL1066 | CEN URA3 CUP1promoter-SUP35NM-GFP | (74) |
| Oligonucleotides | ||
| U1 | GATGTCCACGAGGTCTCT | (42) |
| KanB | CTGCAGCGAGGAGCCGTAAT | (42) |
| 978 | CCCAAGCTTGAGACTAGTTACGTAAAGGTAATCAC | This work |
| 979 | CCCGGATCCCAGCGGCATTGAGTCTTGCGGC | This work |
| 1092 | CCTGAGGGCCCGCTGGGACTAATTGG | This work |
| 1093 | CCCGAGCTCGGTAAACTTCACCTCTCAATATATCTAGAATTTC | This work |
In most experiments [PSI+] is measured using the ura3-14 allele, a nonsense allele suppressible by the partial deficiency of the translation termination factor Sup35p resulting from its being largely sequestered in amyloid filaments. In some experiments (e.g., Table 7) [PSI+] is measured using suppression of ade2-1 with SUQ5. [URE3] was assayed using a DAL5 promoter-ADE2 fusion since normal Ure2p represses DAL5 transcription (58).
pGAL1-SUP35NM-URA3-14.
We combined the URA3-14 plasmid useful for detecting [PSI+] (37) with the GAL1 promoter/SUP35NM/ADH1 terminator from pHK006 (72). The GAL1 promoter/Sup35NM/ADH1 terminator from pHK006 was amplified using primers with 40-bp ends that were homologous to regions of pLEU2Ura3-14 surrounding the NaeI restriction site, between the PGK terminator of URA3-14 and the leucine tRNA gene adjacent to LEU2. pLEU2Ura3-14 was cut with NaeI and transformed into yeast strain YB4741 (MATa his3 met5 leu2 ura3) with the amplified purified Gal-SUP35NM fragment, selecting Leu+. Colony PCR identified clones with the desired homologous recombination event. The plasmid (p1520) was isolated and sequenced (see Supporting Information).
pSIW14 (p1569).
SIW14 and 500 bp of the upstream genomic sequence were amplified with oligos 978 and 979 including 5′ BamHI and 3′ HindIII sites; the product cut with these enzymes was inserted into pRS315 cut with the same enzymes. The SIW14 gene was excised with the same enzymes and ligated to pH321 (pRS313 from which the HindIII and NheI sites were removed from the HIS3 gene by site-directed mutagenesis), forming p1569.
pLYS2-ARG82 (p1574).
ARG82, including 408 bp of its 5′ upstream sequence, including all but 68 bp of the region between ARG82 and the adjoining gene (HMO1), was amplified by PCR using oligos 1092 and 1093, cut with ApaI (a site present in the genome 408 bp upstream of the ORF) and SacI, and inserted into pRS317 (CEN LYS2) cut with the same enzymes, forming p1574.
Isolation of Antiprion Mutants.
Pools of the S. cerevisiae MATa knockout bank (36) were made and transformed with p1520 carrying GAL1-SUP35NM and ura3-14, the latter suppressible by [PSI+] (Fig. 1B) (37). Thousands of transformant colonies from each pool were pooled, and an aliquot was grown in SGal medium (identical to SD medium but with galactose in place of dextrose) supplemented with uracil, histidine, and methionine for 24 h at 30 °C. Dilutions were plated on SD medium supplemented with histidine and methionine (i.e., −Ura) and grown for 6 d at 30 °C. These plates were replica-plated to YPAD with a seeded lawn of the isogenic WT MATα strain 4729. The mating on YPAD was allowed to proceed for 18 h, and then the plate was replica-plated to SD+His+Ura medium to confirm that mating had occurred and to SD+His medium to select [PSI+] diploids. Clones that formed diploids on SD+His+Ura but not on SD+His medium were candidates for antiprion mutants.
To confirm that candidates were [PSI+], each was streaked for single colonies on 1/2 YPD medium and on 1/2 YPD medium containing 5 mM guanidine hydrochloride. Colonies were replica-plated to SD+His+Met medium (i.e., −Ura). Only candidates becoming Ura− following exposure to guanidine but remaining Ura+ in its absence were examined further. Mating with WT strain 4729 was repeated to confirm that diploids were Ura−. DNA from candidates was extracted using the YeaStar Genomic DNA Kit (Zymo Research), and the gene-specific barcode sequences embedded in each KanMX-knockout cassette were amplified using primers U1 (73) and KanB.
Cytoduction.
Recipients were made ρo by growth on rich medium containing 30 μg/mL of ethidium bromide, and donors were ρ+. In some experiments, recipients also carried pRS313, a CEN HIS3 vector, to enable selecting against donor cells after the mating period. Mating mixtures were incubated for 7 h on YPAD medium and then were streaked for single colonies on media selecting against the donor. Clones were replica-plated to medium allowing only diploids to grow, to glycerol medium (to check for transfer of cytoplasm), and to –Ura or –Ade medium as appropriate for transfer and propagation of the prion.
Using Dual Luciferase Vectors to Measure Translation Termination Efficiency.
We used a plasmid (pSC5) constructed by Harger and Dinman in which the upstream Renilla luciferase was fused in frame to the firefly luciferase but with a UAA stop codon at the sixth codon of the downstream firefly luciferase section (46). The two luciferase activities were separately assayed in the same preparation using the Dual-Glo Luciferase Assay System (Promega) with a Berthold luminometer (Titertek Berthold). Cells were grown to late log phase in media selective for retention of pSC5, washed with water, and lysed with the Dual-Glo Luciferase Reagent, and firefly luciferase activity was measured. Then the Stop & Glo reagent was added, which inhibits the firefly luciferase ∼10,000-fold but allows the Renilla luciferase reaction to proceed. The two luciferases are expressed as a fusion protein from the same mRNA, providing an internal control for mRNA amounts.
Supplementary Material
Acknowledgments
We thank Sue Liebman, Mike Reidy, and Dan Masison for plasmids. This work was supported by the Intramural Program of the National Institute of Diabetes and Digestive and Kidney Diseases.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1714361114/-/DCSupplemental.
References
- 1.Wickner RB, et al. Yeast prions: Structure, biology, and prion-handling systems. Microbiol Mol Biol Rev. 2015;79:1–17. doi: 10.1128/MMBR.00041-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wickner RB. [URE3] as an altered URE2 protein: Evidence for a prion analog in Saccharomyces cerevisiae. Science. 1994;264:566–569. doi: 10.1126/science.7909170. [DOI] [PubMed] [Google Scholar]
- 3.Masison DC, Wickner RB. Prion-inducing domain of yeast Ure2p and protease resistance of Ure2p in prion-containing cells. Science. 1995;270:93–95. doi: 10.1126/science.270.5233.93. [DOI] [PubMed] [Google Scholar]
- 4.King C-Y, et al. Prion-inducing domain 2-114 of yeast Sup35 protein transforms in vitro into amyloid-like filaments. Proc Natl Acad Sci USA. 1997;94:6618–6622. doi: 10.1073/pnas.94.13.6618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paushkin SV, Kushnirov VV, Smirnov VN, Ter-Avanesyan MD. In vitro propagation of the prion-like state of yeast Sup35 protein. Science. 1997;277:381–383. doi: 10.1126/science.277.5324.381. [DOI] [PubMed] [Google Scholar]
- 6.Glover JR, et al. Self-seeded fibers formed by Sup35, the protein determinant of [PSI+], a heritable prion-like factor of S. cerevisiae. Cell. 1997;89:811–819. doi: 10.1016/s0092-8674(00)80264-0. [DOI] [PubMed] [Google Scholar]
- 7.Edskes HK, Gray VT, Wickner RB. The [URE3] prion is an aggregated form of Ure2p that can be cured by overexpression of Ure2p fragments. Proc Natl Acad Sci USA. 1999;96:1498–1503. doi: 10.1073/pnas.96.4.1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Taylor KL, Cheng N, Williams RW, Steven AC, Wickner RB. Prion domain initiation of amyloid formation in vitro from native Ure2p. Science. 1999;283:1339–1343. doi: 10.1126/science.283.5406.1339. [DOI] [PubMed] [Google Scholar]
- 9.King CY, Diaz-Avalos R. Protein-only transmission of three yeast prion strains. Nature. 2004;428:319–323. doi: 10.1038/nature02391. [DOI] [PubMed] [Google Scholar]
- 10.Tanaka M, Chien P, Naber N, Cooke R, Weissman JS. Conformational variations in an infectious protein determine prion strain differences. Nature. 2004;428:323–328. doi: 10.1038/nature02392. [DOI] [PubMed] [Google Scholar]
- 11.Brachmann A, Baxa U, Wickner RB. Prion generation in vitro: Amyloid of Ure2p is infectious. EMBO J. 2005;24:3082–3092. doi: 10.1038/sj.emboj.7600772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shewmaker F, Wickner RB, Tycko R. Amyloid of the prion domain of Sup35p has an in-register parallel β-sheet structure. Proc Natl Acad Sci USA. 2006;103:19754–19759. doi: 10.1073/pnas.0609638103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baxa U, et al. Characterization of β-sheet structure in Ure2p1-89 yeast prion fibrils by solid-state nuclear magnetic resonance. Biochemistry. 2007;46:13149–13162. doi: 10.1021/bi700826b. [DOI] [PubMed] [Google Scholar]
- 14.Gorkovskiy A, Thurber KR, Tycko R, Wickner RB. Locating folds of the in-register parallel β-sheet of the Sup35p prion domain infectious amyloid. Proc Natl Acad Sci USA. 2014;111:E4615–E4622. doi: 10.1073/pnas.1417974111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wickner RB, Edskes HK, Shewmaker F, Nakayashiki T. Prions of fungi: Inherited structures and biological roles. Nat Rev Microbiol. 2007;5:611–618. doi: 10.1038/nrmicro1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bessen RA, Marsh RF. Biochemical and physical properties of the prion protein from two strains of the transmissible mink encephalopathy agent. J Virol. 1992;66:2096–2101. doi: 10.1128/jvi.66.4.2096-2101.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McGlinchey RP, Kryndushkin D, Wickner RB. Suicidal [PSI+] is a lethal yeast prion. Proc Natl Acad Sci USA. 2011;108:5337–5341. doi: 10.1073/pnas.1102762108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chernoff YO, et al. Evolutionary conservation of prion-forming abilities of the yeast Sup35 protein. Mol Microbiol. 2000;35:865–876. doi: 10.1046/j.1365-2958.2000.01761.x. [DOI] [PubMed] [Google Scholar]
- 19.Nakayashiki T, Kurtzman CP, Edskes HK, Wickner RB. Yeast prions [URE3] and [PSI+] are diseases. Proc Natl Acad Sci USA. 2005;102:10575–10580. doi: 10.1073/pnas.0504882102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Halfmann R, et al. Prions are a common mechanism for phenotypic inheritance in wild yeasts. Nature. 2012;482:363–368. doi: 10.1038/nature10875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kelly AC, Shewmaker FP, Kryndushkin D, Wickner RB. Sex, prions, and plasmids in yeast. Proc Natl Acad Sci USA. 2012;109:E2683–E2690. doi: 10.1073/pnas.1213449109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wickner RB, Kelly AC. Prions are affected by evolution at two levels. Cell Mol Life Sci. 2016;73:1131–1144. doi: 10.1007/s00018-015-2109-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pfund C, et al. The molecular chaperone Ssb from Saccharomyces cerevisiae is a component of the ribosome-nascent chain complex. EMBO J. 1998;17:3981–3989. doi: 10.1093/emboj/17.14.3981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chernoff YO, Newnam GP, Kumar J, Allen K, Zink AD. Evidence for a protein mutator in yeast: Role of the Hsp70-related chaperone ssb in formation, stability, and toxicity of the [PSI] prion. Mol Cell Biol. 1999;19:8103–8112. doi: 10.1128/mcb.19.12.8103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chernoff YO, Ono B-I. Dosage-dependent modifiers of PSI-dependent omnipotent suppression in yeast. In: Brown AJP, Tuite MF, McCarthy JEG, editors. Protein Synthesis and Targeting in Yeast. Springer; Berlin: 1992. pp. 101–107. [Google Scholar]
- 26.Chernoff YO, Lindquist SL, Ono B, Inge-Vechtomov SG, Liebman SW. Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+] Science. 1995;268:880–884. doi: 10.1126/science.7754373. [DOI] [PubMed] [Google Scholar]
- 27.Moriyama H, Edskes HK, Wickner RB. [URE3] prion propagation in Saccharomyces cerevisiae: Requirement for chaperone Hsp104 and curing by overexpressed chaperone Ydj1p. Mol Cell Biol. 2000;20:8916–8922. doi: 10.1128/mcb.20.23.8916-8922.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hung GC, Masison DC. N-terminal domain of yeast Hsp104 chaperone is dispensable for thermotolerance and prion propagation but necessary for curing prions by Hsp104 overexpression. Genetics. 2006;173:611–620. doi: 10.1534/genetics.106.056820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gorkovskiy A, Reidy M, Masison DC, Wickner RB. Hsp104 disaggregase at normal levels cures many [PSI+] prion variants in a process promoted by Sti1p, Hsp90, and Sis1p. Proc Natl Acad Sci USA. 2017;114:E4193–E4202. doi: 10.1073/pnas.1704016114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kryndushkin DS, Shewmaker F, Wickner RB. Curing of the [URE3] prion by Btn2p, a Batten disease-related protein. EMBO J. 2008;27:2725–2735. doi: 10.1038/emboj.2008.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wickner RB, Bezsonov E, Bateman DA. Normal levels of the antiprion proteins Btn2 and Cur1 cure most newly formed [URE3] prion variants. Proc Natl Acad Sci USA. 2014;111:E2711–E2720. doi: 10.1073/pnas.1409582111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Steidle EA, et al. A novel inositol pyrophosphate phosphatase in Saccharomyces cerevisiae: Siw14 protein selectively cleaves the β-phosphate from 5-diphosphoinositol pentakisphosphate (5PP-IP5) J Biol Chem. 2016;291:6772–6783. doi: 10.1074/jbc.M116.714907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wundenberg T, Mayr GW. Synthesis and biological actions of diphosphoinositol phosphates (inositol pyrophosphates), regulators of cell homeostasis. Biol Chem. 2012;393:979–998. doi: 10.1515/hsz-2012-0133. [DOI] [PubMed] [Google Scholar]
- 34.Monserrate JP, York JD. Inositol phosphate synthesis and the nuclear processes they affect. Curr Opin Cell Biol. 2010;22:365–373. doi: 10.1016/j.ceb.2010.03.006. [DOI] [PubMed] [Google Scholar]
- 35.Livermore TM, Azevedo C, Kolozsvari B, Wilson MS, Saiardi A. Phosphate, inositol and polyphosphates. Biochem Soc Trans. 2016;44:253–259. doi: 10.1042/BST20150215. [DOI] [PubMed] [Google Scholar]
- 36.Winzeler EA, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 37.Manogaran AL, Kirkland KT, Liebman SW. An engineered nonsense URA3 allele provides a versatile system to detect the presence, absence and appearance of the [PSI+] prion in Saccharomyces cerevisiae. Yeast. 2006;23:141–147. doi: 10.1002/yea.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tuite MF, Mundy CR, Cox BS. Agents that cause a high frequency of genetic change from [psi+] to [psi-] in Saccharomyces cerevisiae. Genetics. 1981;98:691–711. doi: 10.1093/genetics/98.4.691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jung G, Masison DC. Guanidine hydrochloride inhibits Hsp104 activity in vivo: A possible explanation for its effect in curing yeast prions. Curr Microbiol. 2001;43:7–10. doi: 10.1007/s002840010251. [DOI] [PubMed] [Google Scholar]
- 40.Ferreira PC, Ness F, Edwards SR, Cox BS, Tuite MF. The elimination of the yeast [PSI+] prion by guanidine hydrochloride is the result of Hsp104 inactivation. Mol Microbiol. 2001;40:1357–1369. doi: 10.1046/j.1365-2958.2001.02478.x. [DOI] [PubMed] [Google Scholar]
- 41.Jung G, Jones G, Masison DC. Amino acid residue 184 of yeast Hsp104 chaperone is critical for prion-curing by guanidine, prion propagation, and thermotolerance. Proc Natl Acad Sci USA. 2002;99:9936–9941. doi: 10.1073/pnas.152333299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Eason RG, et al. Characterization of synthetic DNA bar codes in Saccharomyces cerevisiae gene-deletion strains. Proc Natl Acad Sci USA. 2004;101:11046–11051. doi: 10.1073/pnas.0403672101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Derkatch IL, Bradley ME, Zhou P, Chernoff YO, Liebman SW. Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics. 1997;147:507–519. doi: 10.1093/genetics/147.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sondheimer N, Lindquist S. Rnq1: An epigenetic modifier of protein function in yeast. Mol Cell. 2000;5:163–172. doi: 10.1016/s1097-2765(00)80412-8. [DOI] [PubMed] [Google Scholar]
- 45.Derkatch IL, Bradley ME, Hong JY, Liebman SW. Prions affect the appearance of other prions: The story of [PIN(+)] Cell. 2001;106:171–182. doi: 10.1016/s0092-8674(01)00427-5. [DOI] [PubMed] [Google Scholar]
- 46.Harger JW, Dinman JD. An in vivo dual-luciferase assay system for studying translational recoding in the yeast Saccharomyces cerevisiae. RNA. 2003;9:1019–1024. doi: 10.1261/rna.5930803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saiardi A, Erdjument-Bromage H, Snowman AM, Tempst P, Snyder SH. Synthesis of diphosphoinositol pentakisphosphate by a newly identified family of higher inositol polyphosphate kinases. Curr Biol. 1999;9:1323–1326. doi: 10.1016/s0960-9822(00)80055-x. [DOI] [PubMed] [Google Scholar]
- 48.York JD, Odom AR, Murphy R, Ives EB, Wente SR. A phospholipase C-dependent inositol polyphosphate kinase pathway required for efficient messenger RNA export. Science. 1999;285:96–100. doi: 10.1126/science.285.5424.96. [DOI] [PubMed] [Google Scholar]
- 49.Chattoo BB, et al. Selection of lys2 mutants in the yeast Saccharomyces cerevisiae by the utilization of alpha-aminoadipate. Genetics. 1979;93:51–65. doi: 10.1093/genetics/93.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bolger TA, Folkmann AW, Tran EJ, Wente SR. The mRNA export factor Gle1 and inositol hexakisphosphate regulate distinct stages of translation. Cell. 2008;134:624–633. doi: 10.1016/j.cell.2008.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.El Bakkoury M, Dubois E, Messenguy F. Recruitment of the yeast MADS-box proteins, ArgRI and Mcm1 by the pleiotropic factor ArgRIII is required for their stability. Mol Microbiol. 2000;35:15–31. doi: 10.1046/j.1365-2958.2000.01665.x. [DOI] [PubMed] [Google Scholar]
- 52.El Alami M, Messenguy F, Scherens B, Dubois E. Arg82p is a bifunctional protein whose inositol polyphosphate kinase activity is essential for nitrogen and PHO gene expression but not for Mcm1p chaperoning in yeast. Mol Microbiol. 2003;49:457–468. doi: 10.1046/j.1365-2958.2003.03562.x. [DOI] [PubMed] [Google Scholar]
- 53.Hatch AJ, Odom AR, York JD. Inositol phosphate multikinase dependent transcriptional control. Adv Biol Regul. 2017;64:9–19. doi: 10.1016/j.jbior.2017.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bosch D, Saiardi A. Arginine transcriptional response does not require inositol phosphate synthesis. J Biol Chem. 2012;287:38347–38355. doi: 10.1074/jbc.M112.384255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Azevedo C, Saiardi A. Extraction and analysis of soluble inositol polyphosphates from yeast. Nat Protoc. 2006;1:2416–2422. doi: 10.1038/nprot.2006.337. [DOI] [PubMed] [Google Scholar]
- 56.Resnick AC, et al. Inositol polyphosphate multikinase is a nuclear PI3-kinase with transcriptional regulatory activity. Proc Natl Acad Sci USA. 2005;102:12783–12788. doi: 10.1073/pnas.0506184102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Worley J, Luo X, Capaldi AP. Inositol pyrophosphates regulate cell growth and the environmental stress response by activating the HDAC Rpd3L. Cell Reports. 2013;3:1476–1482. doi: 10.1016/j.celrep.2013.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rai R, Genbauffe F, Lea HZ, Cooper TG. Transcriptional regulation of the DAL5 gene in Saccharomyces cerevisiae. J Bacteriol. 1987;169:3521–3524. doi: 10.1128/jb.169.8.3521-3524.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schlumpberger M, Prusiner SB, Herskowitz I. Induction of distinct [URE3] yeast prion strains. Mol Cell Biol. 2001;21:7035–7046. doi: 10.1128/MCB.21.20.7035-7046.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu M, Chong LS, Perlman DH, Resnick AC, Fiedler D. Inositol polyphosphates intersect with signaling and metabolic networks via two distinct mechanisms. Proc Natl Acad Sci USA. 2016;113:E6757–E6765. doi: 10.1073/pnas.1606853113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kryndushkin D, Wickner RB. Nucleotide exchange factors for Hsp70s are required for [URE3] prion propagation in Saccharomyces cerevisiae. Mol Biol Cell. 2007;18:2149–2154. doi: 10.1091/mbc.E07-02-0128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fan Q, Park K-W, Du Z, Morano KA, Li L. The role of Sse1 in the de novo formation and variant determination of the [PSI+] prion. Genetics. 2007;177:1583–1593. doi: 10.1534/genetics.107.077982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Duennwald ML, Echeverria A, Shorter J. Small heat shock proteins potentiate amyloid dissolution by protein disaggregases from yeast and humans. PLoS Biol. 2012;10:e1001346. doi: 10.1371/journal.pbio.1001346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Maddelein ML, Dos Reis S, Duvezin-Caubet S, Coulary-Salin B, Saupe SJ. Amyloid aggregates of the HET-s prion protein are infectious. Proc Natl Acad Sci USA. 2002;99:7402–7407. doi: 10.1073/pnas.072199199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gross T, et al. The DEAD-box RNA helicase Dbp5 functions in translation termination. Science. 2007;315:646–649. doi: 10.1126/science.1134641. [DOI] [PubMed] [Google Scholar]
- 66.Alcázar-Román AR, Bolger TA, Wente SR. Control of mRNA export and translation termination by inositol hexakisphosphate requires specific interaction with Gle1. J Biol Chem. 2010;285:16683–16692. doi: 10.1074/jbc.M109.082370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mogk A, Bukau B. Role of sHsps in organizing cytosolic protein aggregation and disaggregation. Cell Stress Chaperones. 2017;22:493–502. doi: 10.1007/s12192-017-0762-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sherman F. 1991. Getting Started with Yeast. Guide to Yeast Genetics and Molecular Biology, Methods in Enzymology, eds Guthrie C, Fink GR (Academic, San Diego), Vol 194, pp 3–21.
- 69.Brachmann CB, et al. Designer deletion strains derived from Saccharomyces cerevisiae S288C: A useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 70.Vallen EA, Hiller MA, Scherson TY, Rose MD. Separate domains of KAR1 mediate distinct functions in mitosis and nuclear fusion. J Cell Biol. 1992;117:1277–1287. doi: 10.1083/jcb.117.6.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Edskes HK, McCann LM, Hebert AM, Wickner RB. Prion variants and species barriers among Saccharomyces Ure2 proteins. Genetics. 2009;181:1159–1167. doi: 10.1534/genetics.108.099929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Edskes HK, et al. Sporadic distribution of prion-forming ability of Sup35p from yeasts and fungi. Genetics. 2014;198:605–616. doi: 10.1534/genetics.114.166538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shoemaker DD, Lashkari DA, Morris D, Mittmann M, Davis RW. Quantitative phenotypic analysis of yeast deletion mutants using a highly parallel molecular bar-coding strategy. Nat Genet. 1996;14:450–456. doi: 10.1038/ng1296-450. [DOI] [PubMed] [Google Scholar]
- 74.Zhou P, Derkatch IL, Liebman SW. The relationship between visible intracellular aggregates that appear after overexpression of Sup35 and the yeast prion-like elements [PSI(+)] and [PIN(+)] Mol Microbiol. 2001;39:37–46. doi: 10.1046/j.1365-2958.2001.02224.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.