Abstract
Background
In-stent restenosis (ISR) remains a major cause of failure of contemporary percutaneous revascularization therapies. Invasive biomarkers to improve the prognosis of ISR should be considered. This study aimed to investigate the association between plasma ANRIL expression and ISR.
Material/Methods
A total of 444 patients were included in this research. Serial coronary angiography was performed at baseline (before and after intervention) and within 36 months’ follow-up. ISR was defined as >50% diameter stenosis at follow-up. ANRIL expression was quantified using reverse transcription-PCR. An area under the ROC curve (auROC) was generated to assess the diagnostic values of ANRIL. Logistic regression models were used to assess the independent risk factors for ISR.
Results
Plasma ANRIL expression was significantly increased in patients with ISR, as compared with that in patients without ISR (1.6 [1.1–2.5] vs. 0.9 [0.6–1.3], P<0.001). The auROC (95% confidence interval [CI]) of plasma ANRIL in diagnosing ISR was 0.745 (0.687–0.811). Multiple logistic regression models indicated that drinking (odds ratio [OR]=2.09, 95% CI: 1.08–4.04, P=0.028), hypertension (OR=2.01, 95% CI: 1.14–3.57, P=0.017), diabetes (OR=3.15, 95% CI: 1.63–3.57, P<0.001), low-density lipoprotein (OR=3.14, 95% CI: 1.57–6.31, P=0.001), and ANRIL (OR=2.21, 95% CI: 1.68–2.92, P<0.001) were the independent risk factors for ISR.
Conclusions
We found that higher ANRIL expression is associated with ISR, indicating that ANRIL may be an optimal prognostic factor for ISR.
MeSH Keywords: Coronary Restenosis; Prognosis; RNA, Long Noncoding
Background
Coronary interventions have revolutionized the treatment of acute and stable forms of coronary artery disease (CAD) [1]. However, the occurrence of in-stent restenosis (ISR), the re-narrowing of a coronary artery at the stented segment, remains the major reason for the failure of angioplasty therapies [2]. The use of drug-eluting stents (DESs) has drastically reduced the incidence of ISR, as compared with bare metal stents [3,4]. Nevertheless, DESs are not immune to restenosis. In fact, routine angiographic surveillance after the unrestricted use of these newer-generation devices have shown rates of angiographic restenosis of approximately 12% [5]. Thus, a sensitive, reliable biomarker to predict ISR would be ideal.
For decades, it has been known that non-protein-coding RNAs (ncRNAs) have important cellular functions [6]. Long ncRNAs (lncRNAs) belong to a novel heterogeneous class of ncRNAs that include thousands of different species, and have key roles in gene expression control during both developmental and differentiation processes [7]. Recent studies have clarified that some lncRNAs are expressed differentially in cardiovascular diseases, and that the aberrant changes of those lncRNAs are involved in the development of heart disorders. Hu et al. [8] found that an lncRNA called RP5-833A20.1 could regulate a gene called nuclear factor IA (NFIA), which is a primary regulator of adipocyte formation and lipid development. Their study also indicated that the RP5-833A20.1/miR-382-5p/NFIA pathway was essential to the regulation of cholesterol homeostasis and inflammatory reactions, and suggested that NFIA may represent a therapeutic target for ameliorating cardiovascular disease.
However, fewer studies have focused on the role of lncRNAs in ISR. In the present study, we determined the expression of lncRNA ANRIL in blood and assessed its diagnostic value for ISR screening by comparing the expression level in patients with and without ISR.
Material and Methods
Subjects
The clinical protocol of this study was approved by the ethics committee of Wuhan Asia Heart Hospital, and conducted according to the ethical guidelines outlined in the Declaration of Helsinki. Written informed consent was obtained from all subjects before their participation in the study.
A total of 625 patients with single-vessel lesions who received a DES at the Wuhan Asia Heart Hospital between July 2011 and July 2013 were consecutively included in this retrospective study. The following patients were excluded: (i) patients with severe cardiac insufficiency, renal insufficiency, malignancy, severe infection, fever, acute pulmonary embolism and/or pulmonary heart disease; (ii) patients who had received a stent implantation before this study; (iii) patients with contraindications to aspirin or clopidogrel; (iv) patients with very early (<1 month) DES-ISR, those presenting with acute myocardial infarction, and/or those with obvious angiographic thrombus; and (v) patients who did not undergo a second angiographic scan within 3 years.
Percutaneous coronary intervention (PCI) procedure and follow-up
Premedication with aspirin 100 mg and clopidogrel 75 mg daily was commenced at least 2–3 days before the PCI procedure, and loading doses of aspirin (300 mg) and clopidogrel (450–600 mg) were readily provided to those who were not premedicated.
The PCI procedure and domestic rapamycin DES implantation were performed using conventional techniques. At discharge, aspirin 100 mg and clopidogrel 75 mg daily were recommended for at least 12 months. All study patients received standard cardioactive therapies, as indicated (e.g., beta-blockers, angiotensin-converting enzyme inhibitors, and statins). Serial coronary angiography was performed at baseline (before and after intervention) and within 36 months’ follow-up. ISR was defined as >50% diameter stenosis at follow-up [9].
Sample collection
Immediately after PCI, 5 mL whole blood was collected from each participant. The separation procedure was performed within 2 h of sample collection. Blood samples were centrifuged at 1000 g for 10 min at 4°C to separate the blood cells. The supernatant was then centrifuged at 13,000 g for 10 min at 4°C to completely remove cellular contaminants. The plasma was then aliquoted into microcentrifuge tubes, marked, and stored at −80°C for further analysis.
RNA extraction
Total RNA was isolated from 0.2 mL plasma using TRIzol reagent according to the manufacturer’s protocol (Invitrogen, Carlsbad, CA, USA). The concentration and purity of the RNA solution were measured by detecting its absorbance at 260/280 and at 260/230 nm with a NanoDrop 1000A spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). All of the purified RNA samples were stored at −80°C for further processing.
Reverse transcription and quantitative real-time PCR (qRT-PCR)
Reverse transcription for total RNA was performed using the TaqMan MicroRNA Reverse Transcription Kit (Thermo Fisher Scientific, Waltham, MA, USA). The reaction was performed on a PTC-200 Peltier thermocycler (MJ Research, Waltham, MA, USA) at 16°C for 30 min, 42°C for 40 min, and 85°C for 5 min, respectively.
Then, qRT-PCR was performed to quantify the expression levels of miR-155, miR-21, and miR-10b with SYBR Green PCR Master Mix (Thermo Fisher Scientific), following the manufacturer’s instructions. GAPDH was used as an intrinsic control. The following primers were used: ANRIL forward 5′-TGCTCTATCCGCCAATCAGG-3′, reverse 5′-GGGCCTCAGTGGCAC ATACC-3′; GAPDH forward 5′-AGCCA CATCGCTCAGACAC-3′ and reverse 5′-GCCCAATACGACCAAATCC-3′. The RT-PCR reaction was performed at 95°C for 10 min and in 40 cycles at 95°C for 15 s, and then at 60°C for 60 s on an ABI 7500 thermocycler (Applied Biosystems, Foster City, CA, USA). The relative lncRNA expression level was then calculated using the 2−ΔΔCt method. Each blood sample was analyzed in duplicate wells, and the analysis was repeated 3 times.
Statistical analysis
Continuous data were presented as mean ± standard deviation if normality was not rejected; otherwise, the data were presented as median and interquartile ranges (25th, 75th percentiles). Categorical values were expressed through absolute and relative frequencies. Plasma lncRNA ANRIL expression between patients with and without restenosis was compared using an unpaired Mann-Whitney test. To assess the diagnostic value of ANRIL, the area under the receiver operating characteristic curve (auROC), which is a measure of discrimination, was calculated. We also applied the generalized additive model (GAM) to explore the relationship between ANRIL and the risk of ISR. Logistic regression models adjusted for confounding variables were used to assess the independent risk factors for ISR. Interaction analysis was performed to investigate the interaction between ANRIL and other risk factors. All statistical analyses in this study were performed with R software version 2.8 (R Development Core Team 2013). P<0.05 (two-sided) was considered statistically significant.
Results
Clinical characteristics of the study population
A total of 625 patients with CAD who received DESs were enrolled in this study. As Figure 1 shows, after the exclusion of participants who did not meet the inclusive criteria, 444 patients were finally included. The average age was 62.0±6.6 years, and 262 (59.0%) of the patients were men. Of the 444 patients, 76 (17.1%) patients had ISR within 2 years’ follow-up. The main characteristics of the patients with restenosis (n=75) are reported in Table 1. Briefly, these patients were more likely to have hypertension (50% vs. 32.1%, P=0.003); have diabetes (28.9% vs. 12.0%, P<0.001) or be drinkers (28.9% vs. 15.2%, P<0.001); and/or have higher systolic blood pressure (SBP) (145.0±10.5 vs. 139.6±10.9 mm Hg, P<0.001), low-density lipoprotein (LDL) (2.7±0.3 vs. 2.5±0.4 mmol/L, P=0.001), and/or lower high-density lipoprotein (HDL) (1.3±0.2 vs. 1.5±0.4 mmol/L, P=0.011) than patients without restenosis.
Figure 1.
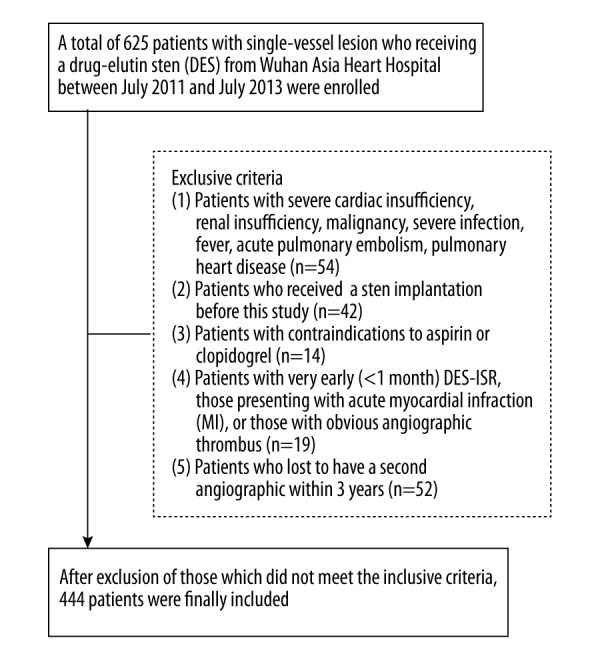
Flow diagram of study patients.
Table 1.
Baseline characteristics of patients.
| Total | Non-restenosis (n=368) | Restenosis (n=76) | P-value | |
|---|---|---|---|---|
| Male (kg/m2) | 262 (59.0%) | 220 (59.8%) | 42 (55.3%) | 0.446 |
| Age (mmHg) | 62.0±6.6 | 61.8±6.1 | 63.0±8.7 | 0.165 |
| BMI (mmHg) | 21.9±2.0 | 21.9±1.9 | 21.7±2.2 | 0.343 |
| SBP | 140.5±11.1 | 139.6±10.9 | 145.0±10.5 | <0.001 |
| DBP | 80.8±7.6 | 80.6±7.2 | 81.7±9.6 | 0.277 |
| History of: | ||||
| Smoking | 100 (22.5%) | 78 (21.2%) | 22 (28.9%) | 0.141 |
| Drinking | 78 (17.6%) | 56 (15.2%) | 22 (28.9%) | 0.004 |
| Hypertension | 156 (35.1%) | 118 (32.1%) | 38 (50.0%) | 0.003 |
| Diabetes | 66 (14.9%) | 44 (12.0%) | 22 (28.9%) | <0.001 |
| Stroke | 50 (11.3%) | 38 (10.3%) | 12 (15.8%) | 0.17 |
| HDL (mmol/L) | 1.4±0.4 | 1.5±0.4 | 1.3±0.2 | 0.011 |
| LDL (mmol/L) | 2.6±0.4 | 2.5±0.4 | 2.7±0.3 | 0.001 |
| Creatinine (μmol/L) | 85.5±24.0 | 85.2±24.2 | 87.1±23.3 | 0.740 |
| LVEF (%) | 51.4±6.4 | 51.3±6.5 | 51.7±5.6 | 0.632 |
BMI – body mass index; SBP – systolic blood pressure; DBP – diastolic blood pressure; HDL – high density lipoprotein; LDL – low density lipoprotein; LVEF – left ventricular ejection fraction.
Association between ANRIL and incidence of ISR
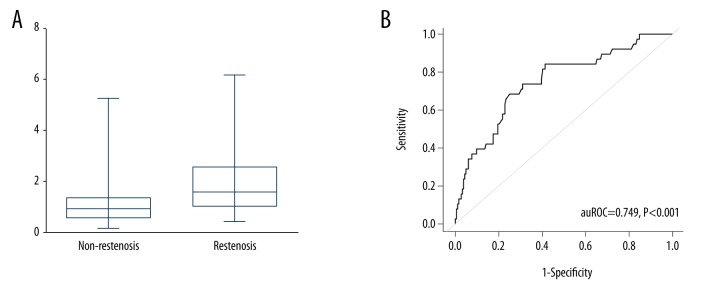
To detect the expression level of lncRNA ANRIL, RT-PCR was performed. As shown in Figure 2A, the relative expression of ANRIL was 1.6 (1.1, 2.5) and 0.9 (0.6, 1.3) in patients with and without restenosis, respectively. ROC curves were constructed, and the auROC was evaluated to assess the diagnostic value. An auROC of 0.749 (0.687, 0.811) was obtained (Figure 2B). Additionally, when using a cutoff value of 1.34 for ANRIL, sensitivity was found to be 68.4%; specificity, 75.0%; positive likelihood ratio, 2.74; negative likelihood ratio, 0.42; positive predictive value, 0.36; and negative predictive value, 0.92.
Figure 2.
Diagnostic performance of lncRNA ANRIL for in-stent restenosis. (A) Relative expression levels of ANRIL in patients with and without ISR. (B) ROC curve analysis of ANRIL in distinguishing ISR from non-ISR. ROC, receiver operating characteristic curve; ISR, in-stent restenosis.
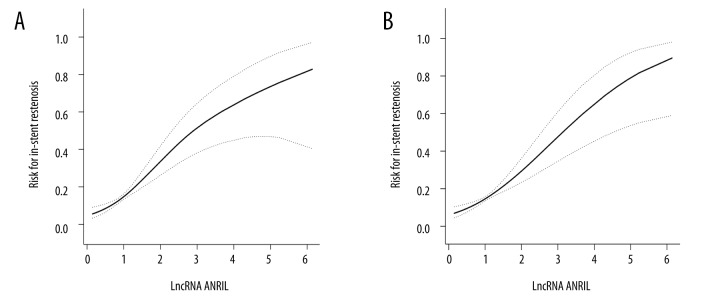
A GAM was applied to explore the relationship between ANRIL expression and the risk of ISR. Figure 3 shows curve fitting for ANRIL expression and the incidence of ISR. ANRIL was found to have had a positive association with the incidence of restenosis, in spite of whether the confounding variables were adjusted or not.
Figure 3.
Curve fitting between ANRIL and risk of ISR. (A) Unadjusted; (B) adjusted for sex, age, BMI, smoking, drinking, hypertension, diabetes, stroke, HDL, LDL, creatinine, and LVEF. BMI – body mass index; HDL – high-density lipoprotein; LDL – low-density lipoprotein; LVEF – left ventricular ejection fraction.
Multivariate predictors of ISR
A univariate logistic regression model was used to assess the risk factors for ISR. Table 2 shows that SBP (odds ratio [OR]=1.05, 95% confidence interval [CI]: 1.02–1.08, P<0.001), drinking (OR=2.27, 95% CI: 1.28–4.02, P=0.005), hypertension (OR=2.12, 95% CI: 1.28–3.49, P=0.003), diabetes (OR=3.00, 95% CI: 1.67–5.40, P<0.001), HDL (OR=0.43, 95% CI: 0.22–0.83, P=0.012), LDL (OR=2.65, 95% CI: 1.46–4.83, P=0.001), and ANRIL (OR=2.35, 95% CI: 1.80–3.06, P < 0.001) were statistically significant risk factors affecting restenosis. The above variables were entered into the multivariate logistic regression analyses. As Table 2 shows, drinking (OR=2.09, 95% CI: 1.08–4.04, P=0.028), hypertension (OR=2.01, 95% CI: 1.14–3.57, P=0.017), diabetes (OR=3.15, 95% CI: 1.63–3.57, P < 0.001), LDL (OR=3.14 95% CI: 1.57–6.31, P=0.001), and ANRIL (OR=2.21, 95% CI: 1.68–2.92, P < 0.001) were found to be the independent risk factors.
Table 2.
Univariate and multivariate analysis of risk factors for in-stent restenosis.
| Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|
| OR | 95CI% | P-value | OR | 95CI% | P-value | |
| Male | 0.83 | 0.51–1.37 | 0.466 | |||
| Age | 1.03 | 0.99–1.07 | 0.166 | |||
| BMI | 0.94 | 0.83–1.07 | 0.343 | |||
| SBP | 1.05 | 1.02–1.08 | <0.001 | |||
| DBP | 1.02 | 0.99–1.05 | 0.277 | |||
| History of: | ||||||
| Smoking | 1.51 | 0.87–2.64 | 0.143 | |||
| Drinking | 2.27 | 1.28–4.02 | 0.005 | 2.09 | 1.08–4.04 | 0.028 |
| Hypertension | 2.12 | 1.28–3.49 | 0.003 | 2.01 | 1.14–3.57 | 0.017 |
| Diabetes | 3.00 | 1.67–5.40 | <0.001 | 3.15 | 1.63–3.57 | <0.001 |
| Stroke | 1.63 | 0.81–3.29 | 0.174 | |||
| HDL (mmol/L) | 0.43 | 0.22–0.83 | 0.012 | 0.52 | 0.24–1.15 | 0.100 |
| LDL (mmol/L) | 2.65 | 1.46–4.83 | 0.001 | 3.14 | 1.57–6.31 | 0.001 |
| Cr (μmol/L) | 1.00 | 0.99–1.01 | 0.526 | |||
| LVEF(%) | 1.01 | 0.97–1.05 | 0.631 | |||
| ANRIL | 2.35 | 1.80–3.06 | <0.001 | 2.21 | 1.68–2.92 | <0.001 |
BMI – body mass index; SBP – systolic blood pressure; DBP – diastolic blood pressure; HDL – high density lipoprotein cholesterin; LDL-c – low density lipoprotein cholesterin; Cr – Creatinine; LVEF – left ventricular ejection fraction.
As shown in Figure 4, similar ORs were observed in different sexes; ages; and status of alcohol use, diabetes, and/or LDL (all P for interaction >0.05). Moreover, a significant P for interaction between ANRIL and hypertension was observed, with OR (95% CI) of 1.94 (1.39–2.71) in patients without hypertension and 4.38 (2.42–7.93) in patients with hypertension (P for interaction=0.013).
Figure 4.
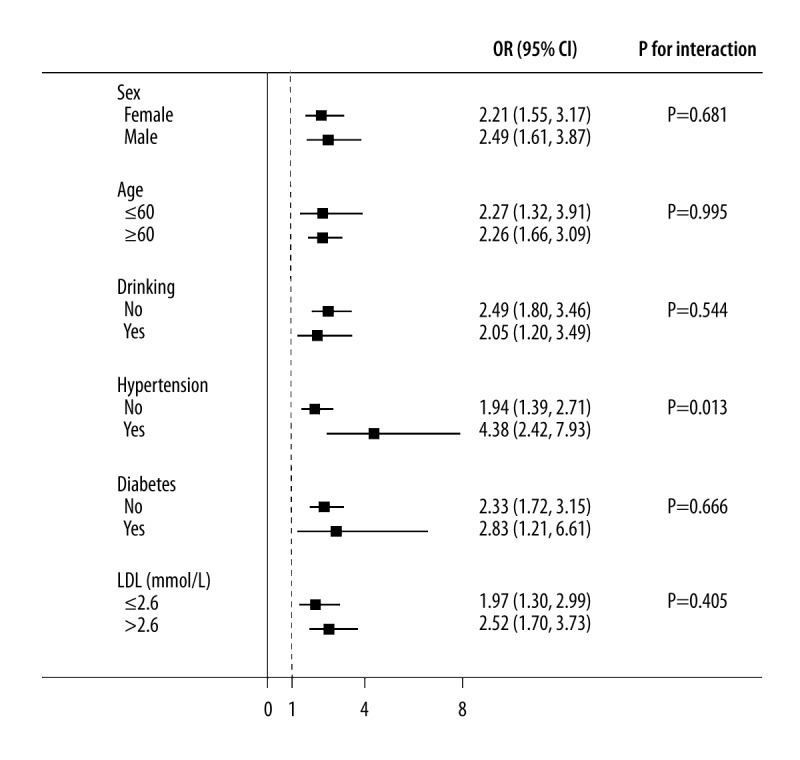
Interaction between ANRIL and other risk factors with regard to the risk of in-stent restenosis. Adjusted for sex, age, body mass index, smoking, drinking, hypertension, diabetes, stroke, high-density lipoprotein, low-density lipoprotein, creatinine, and left ventricular ejection fraction.
Discussion
In this study, we found that lncRNA ANRIL expression is significantly increased in the plasma of patients with ISR as compared with that in the plasma of patients without ISR. We confirmed that high ANRIL expression predicts a high risk of ISR, with an auROC of 0.745. Furthermore, a GAM showed that ANRIL had a positive association with the incidence of restenosis, whether the confounding variables were adjusted or not. Additionally, multiple logistic regression demonstrated that ANRIL is an independent risk factor for the incidence of ISR, which suggests that it may be an optimal prognostic factor for ISR.
Recently, with the emergence of sequencing techniques, the widespread existence of lncRNAs in cardiovascular development and diseases has been confirmed, and their action mechanisms and important biological functions have gradually been elucidated [10,11]. In fact, lncRNAs play crucial roles in regulating cell transformation and organogenesis, as well as in mammalian heart development and cellular lineage [12–14]. In a study conducted in mice, Braveheart (Bvht), known as a novel long noncoding RNA, was identified as a critical regulator of cardiovascular-associated cell differentiation and revealed to be required for the development of nascent mesoderm [15].
Given the emerging role of lncRNAs in the large spectrum of biological systems examined, it is not surprising that several recent studies have identified many lncRNAs associated with diseased hearts. In the study by Kumarswamy et al., regulated candidates, screened using global transcriptomic analyses, were validated in 788 patients with developing cardiac remodeling and heart failure; the study also demonstrated that the mitochondrial long noncoding RNA uc022bqs.1 (LIPCAR) is a novel biomarker of cardiac remodeling, and could help predict future death in patients with heart failure [16].
Additionally, deep sequencing of RNA isolated from paired nonischemic (NICM) and ischemic (ICM) human failing left ventricular samples indicated that the expression signature of lncRNAs, but not miRNAs or mRNAs, distinguishes ICM from NICM, suggesting an important role for lncRNAs in the pathogenesis of heart failure and in the reverse remodeling observed with mechanical support [17]. However, few studies have focused on lncRNAs and ISR. Lnc-Ang362 and lincRNA-p21 are expressed in both vascular smooth muscle cells (VSMCs) and endothelial cells, and in turn are able to modulate VSMC proliferation, vascular remodeling, and neointimal hyperplasia, which may affect ISR [18].
Recent studies have also linked other lncRNAs to heart diseases. The first evidence for a putative role of lncRNAs in vascular disease came from several genome-wide association studies that independently identified a susceptibility locus of CAD on the human chromosome 9p21.53 [19–21], which is adjacent to the last exon of an lncRNA known as an antisense noncoding RNA in the INK4 locus (i.e., ANRIL, also known as CDKN2BAS). Subsequent studies showed that ANRIL is expressed in endothelial, smooth muscle, and inflammatory cells, and is associated with the risk for coronary atherosclerosis, carotid arteriosclerosis, and other vascular diseases [22–24]. In our study, we found that ANRIL is an independent risk factor for the incidence of ISR, and that it may be an optimal prognostic factor for ISR.
To our knowledge, this is the first study to focus on lncRNAs and ISR, especially in which an ideal result was observed. However, the limitations of this study require further comments. First, the sample size of enrolled patients was relatively small. Further examination with a larger sample and a longer follow-up is needed to confirm our results. Second, the patients involved in this study all had a second coronary angiography within 3 years; however, perhaps an annual coronary angiography should be taken. Thirdly, many factors may influence the incidence of ISR, because it is a complex disease, and we could not consider all the potentially confounding factors in our study, such as lifestyle and dietary factors. Last, but not least, a prospective study should be designed and performed to demonstrate the relationship between ANRIL and ISR.
Conclusions
This study found that higher ANRIL expression is associated with ISR and that it may be an optimal prognostic factor for ISR. However, whether this correlation is exactly proportional or not requires further careful scrutiny.
Footnotes
Source of support: Departmental sources
Conflicts of interest
The authors report that there are no conflicts of interest.
References
- 1.Gruntzig AR, Senning A, Siegenthaler WE. Nonoperative dilatation of coronary-artery stenosis: Percutaneous transluminal coronary angioplasty. N Engl J Med. 1979;301(2):61–68. doi: 10.1056/NEJM197907123010201. [DOI] [PubMed] [Google Scholar]
- 2.Alfonso F, Byrne RA, Rivero F, Kastrati A. Current treatment of in-stent restenosis. J Am Coll Cardiol. 2014;63(24):2659–73. doi: 10.1016/j.jacc.2014.02.545. [DOI] [PubMed] [Google Scholar]
- 3.Stefanini GG, Holmes DR., Jr Drug-eluting coronary-artery stents. N Engl J Med. 2013;368(3):254–65. doi: 10.1056/NEJMra1210816. [DOI] [PubMed] [Google Scholar]
- 4.Palmerini T, Biondi-Zoccai G, Della Riva D, et al. Stent thrombosis with drug-eluting and bare-metal stents: Evidence from a comprehensive network meta-analysis. Lancet. 2012;379(9824):1393–402. doi: 10.1016/S0140-6736(12)60324-9. [DOI] [PubMed] [Google Scholar]
- 5.Cassese S, Byrne RA, Tada T, et al. Incidence and predictors of restenosis after coronary stenting in 10 004 patients with surveillance angiography. Heart. 2014;100(2):153–59. doi: 10.1136/heartjnl-2013-304933. [DOI] [PubMed] [Google Scholar]
- 6.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: Insights into functions. Nat Rev Genet. 2009;10(3):155–59. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 7.Fatica A, Bozzoni I. Long non-coding RNAs: New players in cell differentiation and development. Nat Rev Genet. 2014;15(1):7–21. doi: 10.1038/nrg3606. [DOI] [PubMed] [Google Scholar]
- 8.Hu YW, Zhao JY, Li SF, et al. RP5-833A20.1/miR-382-5p/NFIA-dependent signal transduction pathway contributes to the regulation of cholesterol homeostasis and inflammatory reaction. Arterioscler Thromb Vasc Biol. 2015;35(1):87–101. doi: 10.1161/ATVBAHA.114.304296. [DOI] [PubMed] [Google Scholar]
- 9.Alfonso F, Perez-Vizcayno MJ, Garcia Del Blanco B, et al. Everolimus-eluting stents in patients with bare-metal and drug-eluting in-stent restenosis: Results from a patient-level pooled analysis of the RIBS IV and V trials. Circ Cardiovasc Interv. 2016;9(7) doi: 10.1161/CIRCINTERVENTIONS.115.003479. pii: e003479. [DOI] [PubMed] [Google Scholar]
- 10.Uchida S, Dimmeler S. Long noncoding RNAs in cardiovascular diseases. Circ Res. 2015;116(4):737–50. doi: 10.1161/CIRCRESAHA.116.302521. [DOI] [PubMed] [Google Scholar]
- 11.Kataoka M, Wang DZ. Non-coding RNAs including miRNAs and lncRNAs in cardiovascular biology and disease. Cells. 2014;3(3):883–98. doi: 10.3390/cells3030883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rayner KJ, Liu PP. Long noncoding RNAs in the heart: The regulatory roadmap of cardiovascular development and disease. Circ Cardiovasc Genet. 2016;9(2):101–3. doi: 10.1161/CIRCGENETICS.116.001413. [DOI] [PubMed] [Google Scholar]
- 13.Thum T, Condorelli G. Long noncoding RNAs and microRNAs in cardiovascular pathophysiology. Circ Res. 2015;116(4):751–62. doi: 10.1161/CIRCRESAHA.116.303549. [DOI] [PubMed] [Google Scholar]
- 14.Bar C, Chatterjee S, Thum T. Long noncoding RNAs in cardiovascular pathology, diagnosis, and therapy. Circulation. 2016;134(19):1484–99. doi: 10.1161/CIRCULATIONAHA.116.023686. [DOI] [PubMed] [Google Scholar]
- 15.Klattenhoff CA, Scheuermann JC, Surface LE, et al. Braveheart, a long noncoding RNA required for cardiovascular lineage commitment. Cell. 2013;152(3):570–83. doi: 10.1016/j.cell.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumarswamy R, Bauters C, Volkmann I, et al. Circulating long noncoding RNA, LIPCAR, predicts survival in patients with heart failure. Circ Res. 2014;114(10):1569–75. doi: 10.1161/CIRCRESAHA.114.303915. [DOI] [PubMed] [Google Scholar]
- 17.Yang KC, Yamada KA, Patel AY, et al. Deep RNA sequencing reveals dynamic regulation of myocardial noncoding RNAs in failing human heart and remodeling with mechanical circulatory support. Circulation. 2014;129(9):1009–21. doi: 10.1161/CIRCULATIONAHA.113.003863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deng L, Bradshaw AC, Baker AH. Role of noncoding RNA in vascular remodelling. Curr Opin Lipidol. 2016;27(5):439–48. doi: 10.1097/MOL.0000000000000336. [DOI] [PubMed] [Google Scholar]
- 19.Samani NJ, Erdmann J, Hall AS, et al. WTCCC and the Cardiogenics Consortium. Genomewide association analysis of coronary artery disease. N Engl J Med. 2007;357(5):443–53. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gilad GM, Dahl D, Gilad VH. Effects of glycosaminoglycans and proteinase inhibitors on astroglia-induced detachment of cultured rat cerebellar neurons. Int J Dev Neurosci. 1989;7(2):133–43. doi: 10.1016/0736-5748(89)90064-6. [DOI] [PubMed] [Google Scholar]
- 21.Helgadottir A, Thorleifsson G, Manolescu A, et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316(5830):1491–93. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- 22.Holdt LM, Beutner F, Scholz M, et al. ANRIL expression is associated with atherosclerosis risk at chromosome 9p21. Arterioscler Thromb Vasc Biol. 2010;30(3):620–27. doi: 10.1161/ATVBAHA.109.196832. [DOI] [PubMed] [Google Scholar]
- 23.Tsai PC, Liao YC, Lin TH, et al. Additive effect of ANRIL and BRAP polymorphisms on ankle-brachial index in a Taiwanese population. Circ J. 2012;76(2):446–52. doi: 10.1253/circj.cj-11-0925. [DOI] [PubMed] [Google Scholar]
- 24.Burd CE, Jeck WR, Liu Y, et al. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet. 2010;6(12):e1001233. doi: 10.1371/journal.pgen.1001233. [DOI] [PMC free article] [PubMed] [Google Scholar]