Abstract
Human induced pluripotent stem cells (iPSCs) offer a novel, timely approach for investigating the etiology of neuropsychiatric disorders. Although we are starting to gain more insight into the specific mechanisms that cause Alzheimer’s disease and other forms of dementia, this has not resulted in therapies to slow the pathological processes. Animal models have been paramount in studying the neurobiological processes underlying psychiatric disorders. Nonetheless, these human conditions cannot be entirely recapitulated in rodents. Human cell models derived from patients’ cells now offer new hope for improving our understanding of the early molecular stages of these diseases, through to validating therapeutics. The impact of dementia is increasing, and a new model to investigate the early stages of this disease is heralded as an essential, new platform for translational research. In this article, we review current literature using iPSCs to study Alzheimer’s disease, describe drug discovery efforts using this platform, and discuss the future potential for this technology in psychiatry research.
Animal models vs. human models
The lack of predictive validity from preclinical animal studies for neuropsychiatric disorders is evidence of the need for new models in psychiatric research and neuroscience drug discovery to supplement these existing models (Markou et al., 2009). Alzheimer’s disease (AD) is an example of this, where the complex genetics is a major obstacle in successfully studying the disease. Established transgenic rodent models of AD have successfully recapitulated certain components of the neuropathology and cognitive deficits experienced by patients. However, even with known human AD mutations transgenic models do not exhibit all aspects of AD, such as extensive neuronal loss or distinct neurofibrillary tangle pathology (Duff, 2001; Saraceno et al., 2013). Moreover, inserting the right genetic mutation may cause some extent of AD pathology, but it may still not be applicable to the majority of patients who have no known inherited mutations. Post mortem tissue and immortalised cell lines offer opportunities for studying the AD process in human tissue. Donated post mortem tissue may be the best way to study pathological features of the disease, but this is largely limited to looking at the end stage of the disease, it being difficult to access the brains of patients at earlier stages.
Potential for human iPSCs in disease modelling
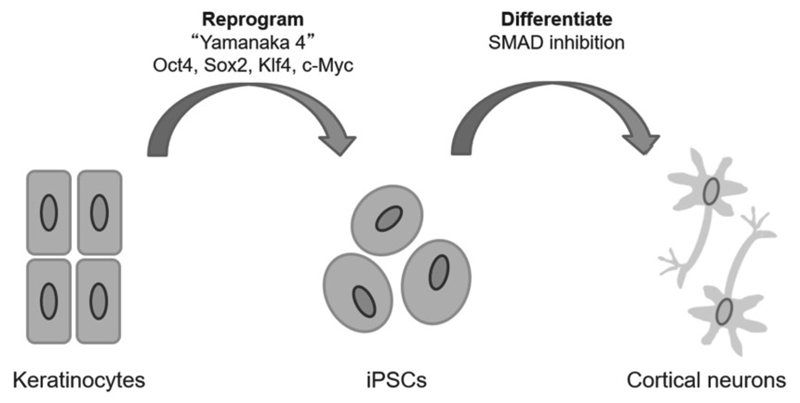
Induced pluripotent stem cells (iPSCs) allow disease processes to be modelled for the first time in cells directly from patients. Adult somatic cell biopsies can be obtained, reprogrammed into iPSCs and then differentiated into the cell type of interest in vitro, as summarised in Figure 1. For disorders of the brain, where tissue from patients is inaccessible, iPSC technology offers the opportunity to study pathology as it develops in brain cells, whilst retaining the genetic background of the patient.
Figure 1.
Adult somatic cells obtained from the root shaft of scalp hair are reprogrammed into iPSCs, which can then be differentiated into specific cell types. Cells from multiple sources can be utilised for reprogramming with the ‘Yamanaka factors’, with keratinocytes providing a non-invasive option. iPSCs can be differentiated into any cell type, with the SMAD inhibition protocol developed by Shi et al. (2012a) producing cortical neurons. These neurons can be used for studying disease processes or drug screening.
The development of iPSCs was a game changing technology by Takahashi and Yamanaka, winning Yamanaka the Nobel Prize in Medicine in 2012. They demonstrated that adult somatic cells could be restored to a pluripotent state by transducing the cells in vitro with four transcription factors: Oct4, Sox2, Klf4, and c-Myc (or the ‘Yamanaka factors’). These iPSCs express markers exclusive to embryonic stem cells, mimic their morphology and growth properties, and can be differentiated into any specified cell type when in the presence of the molecular signals that would typically be present during development (Takahashi and Yamanaka, 2006).
Human iPSC technology has offered a new way of studying human cells throughout disease development without the use of embryonic or immortalised cells. There are now numerous protocols for converting different cell types into stem cells, including cells that can be acquired relatively non-invasively such as keratinocytes or blood cells (Cocks et al., 2014). iPSCs can be passaged multiple times with minimal impact on genomic integrity and can be differentiated into, in theory, any cell type (Musunuru, 2013). Efficient protocols for cortical neurons based on developmental neurobiology studies are now well established and can produce neuronal cells with functional excitatory synapses, providing an unprecedented opportunity to develop live patient-specific models for disease in vitro. (Shi et al., 2012a; Zhang et al., 2013; Qi et al., 2017).
iPSC-derived models of Alzheimer’s disease
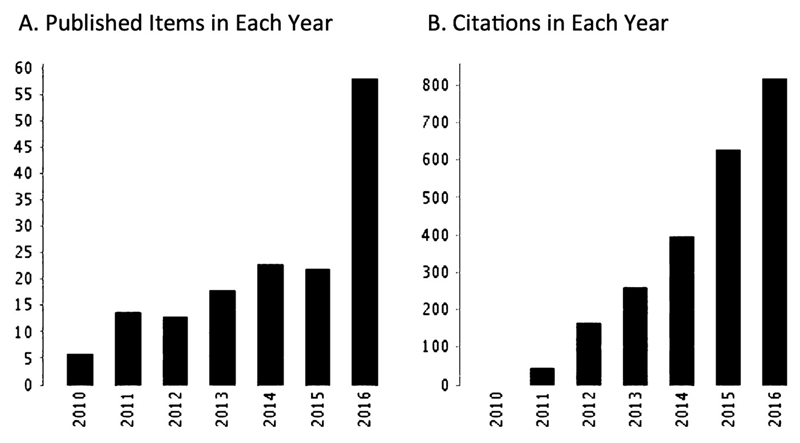
Dementia is an age-related neurodegenerative disorder, characterised by cognitive decline. Alzheimer’s disease (AD), the most common type of dementia, accounts for up to 80% of all cases, with prevalence sharply increasing with age (Lobo et al., 2000). AD is characterised by the presence of amyloid plaques, made up of β-amyloid (Aβ) protein aggregates, and neurofibrillary tangles, resulting from accumulated tau protein, in the brain. As with many central nervous system disorders, complex genetics of AD is still a major obstacle in successfully modelling the disease. A model based on cells derived from patients could be pivotal to determining relevant pathogenic mechanisms in humans. The excitement caused by this technological innovation is demonstrated by the rapid uptake of this system by the research community, with the number of journal articles on Alzheimer’s disease and iPSCs growing steadily and the number of citations now over 2000 for all articles combined (Figure 2).
Figure 2.
The increase in journal articles on Alzheimer’s disease using iPSCs. Generated from Web of Sciene citation report using search from ‘alzheimer’s included pluripotent stem cell’. A. Number of published articles by year. B. Number of citations of all articles per year. Citation Report graphic is derived from Thomson Reuters Web of Science, Copyright THOMSON REUTERS ® 201_. All rights reserved. (Date accessed 20/03/17.)
Studies using neurons derived from iPSCs have already provided insights into mechanisms and evaluation of pre-clinical drugs (Bellin et al., 2012, Mertens et al., 2013, Ross & Akimov, 2014). Increased Aβ is secreted by neurons derived from patients with familial AD (fAD) mutations compared to cells from healthy controls in a period of weeks in vitro, despite this process taking years in humans in vivo (Yagi et al., 2011; Israel et al., 2012). iPSC-derived neurons from Down’s syndrome patients with increased expression of the amyloid precursor protein (APP) gene on chromosome 21 also generated increased Aβ peptides, as well as aggregates of intracellular and extracellular Aβ. These neurons also presented a tau dysfunction phenotype, including abnormal tau localisation to the cell bodies and dendrites, and increased levels of total tau and two forms of phosphorylated tau (Shi et al., 2012b). Tau phenotypes have also been identified in lines from patients with mutations in the microtubule-associated protein tau (MAPT) gene. Neurons derived from patients with N279K showed changes in the 3R:4R tau isoform ratio and hyperphosphorylated tau aggregates, while neurons from P301L patients exhibited abnormal processes containing 4R tau (Iovino et al., 2015)
Variability between patient lines has been reported in multiple studies, indicating difficulty in drawing conclusions about cellular processes in AD from a small number of iPSC lines. Some sporadic AD (sAD, or late-onset AD) lines did not show higher levels of Aβ in the neurons, and differences between intracellular and extracellular Aβ levels in patients with different APP mutations and sAD patient lines have also been described (Israel et al., 2012; Kondo et al., 2013). The complex interplay between genetic and environmental factors may mean that a large number of iPSC lines from sAD patients will be required to derive robust phenotypes.
Protocols for human neural stem-cell-derived three-dimensional (3D) culture systems have been established that may support a more realistic cellular network and neural cell type representation, including GABAergic, glutamatergic, and dopaminergic neurons, as well as astrocytes and oligodendrocytes. The 3D neuronal cultures in this system exhibited extracellular deposition of Aβ, including Aβ plaques, and aggregates of phosphorylated tau in the cell bodies of neurons differentiated from fAD iPSC lines (Choi et al., 2014). Therefore, these 3D culture systems will be of particular interest in recapitulating disease phenotypes, as the 2D in vitro models did not report aggregates of amyloid or tau protein, the characteristic hallmarks of AD in the human brain.
Clinical Implications
Neurons derived from iPSCs are now being utilised for drug screens to test for chemical compounds that may prevent Aβ toxicity of neurons (Yahata et al., 2011; Xu et al., 2013). Gamma secretase inhibitors were found to modulate the increased Aβ found in fAD patients, and hence it may be beneficial to test the efficacy of compounds in reversing Aβ and tau-mediated toxicity in human neurons before clinical trials (Yagi et al., 2011). NSAID-based γ-secretase modulators were tested in iPSC-derived neurons from AD patients, modulators that had previously lowered Aβ levels in various model systems but had translated poorly into human clinical trials. Results showed no change of these drugs on Aβ, suggesting resistance of human neurons to NSAID-based γ-secretase modulation (Mertens et al.,2013). Another high-throughput Aβ toxicity screen revealed several small molecules that significantly blocked the toxic effects of Aβ (Xu et al., 2013).
These studies highlight the potential for iPSC models of AD as a pre-clinical tool for screening therapeutic compounds, testing early toxicity and efficacy, and developing a platform for drug development.Moreover, the advancement of this technology coincides with rapid progress in genetic techniques such as precise genome engineering and transcriptomic and proteomic analysis. The complement of these techniques could allow for human cells to model known pathogenic mutations in vitro, which could then be functionally validated and downstream targets could be confirmed. In the future these models could give rise to new preclinical model for drug discovery and even personalised therapeutics based on individuals genetics.
Genome engineering in in vitro models
Over the past decade, the field of genome editing has emerged whereby researchers can manipulate almost any gene in various cell types and organisms, through targeted cleavage of DNA. This is rapidly increasing our understanding of the genetic contribution of specific mutations to disease processes. Studies comparing iPSCs from patients with a monogenic disorder to healthy controls have not been able to account for differences in genetic background and epigenetic states of the iPSC lines. Precise genome editing allows for the specific effects of a mutation or gene to be studied in a cell line and the isogenic control. Correcting a single mutation, or creating one, removes all other variables, such as genetic alterations from reprogramming or irregularities with the epigenetic memory of an iPSC line (Musunuru, 2013).
Precise genome engineering technologies, such as the CRISPR/Cas system, have provided a breakthrough in genome-editing technology. CRISPR/Cas enables specific mutations to be introduced with no off-target effects to the rest of the genome. iPSC lines with fAD mutations have been generated, displaying genotype-dependent disease-associated phenotypes in the differentiated cortical neurons (Paquet et al., 2016). These cell lines are established from healthy controls, maintaining an identical genetic background in all lines. This technology facilitates the study of human disease by attributing certain phenotypes to specific genotypes, helping to decipher whether a single gene or a number of interacting genes is responsible for disease-associated phenotypes.
Considerations in iPSC modelling
The heterogeneity of iPSC lines due to variation in the genetic background of individuals is often discussed as a concern within the iPSC field. For example, Kondo et al. (2013) reported different phenotypes amongst cell lines from 4 AD patients (2 sAD and 2 fAD). This has made it impossible to set guidleines for the number of patient and control lines that must be generated in order to validate specific clinical phenotypes in these models. Using fAD patients, in which a single mutation can cause a phenotype, reduces the number of lines required, however, cells from sAD patients may be more relevant to advancing the understanding of AD development in the majority of people living with dementia. In both these experimental designs the careful selection of gender and age-matched controls, including obtaining cells from unaffected siblings, can overcome such issues (Brennand et al., 2015; Santostefano et al., 2015). The generation of cell lines with disease-relevant mutations by genome editing methods also produces a perfect isogenic control, removing the need for numerous cell lines. A more general model for AD may be to treat wildtype neurons with exogenous Aβ, as done in Aβ toxicity screening by Xu et al. (2013), reducing the need for multiple control lines. However, this strategy also has issues due to the lack of consensus on the biophysical and biological behaviour of A β in vivo, and the instability and irreproducibility of oligomeric Aβ preparations (Roychaudhuri et al., 2009; Ryan et al., 2010). The different rates of Aβ and tau production in iPSC-derived neurons and the range of neuronal responses to Aβ reflects the heterogeneity of patients with AD, and make iPSC models a representative tool for studying the multiple biological phenotypes of the disease.
Another potential limitation for modelling disorders associated with ageing is the preservation of age in iPSCs reprogrammed from donors. Studies indicate that during reprogramming to an embryo-like iPSC state the original age of the somatic cells is lost (Lapasset et al., 2011, Meissner et al., 2008). The reprogramming phase resets many important hallmarks of ageing, including: extension of telomere length, restoration of mitochondrial function and reduced oxidative stress (Rohani et al., 2014). iPSCs generated from donors aged 0 to 89 years did not retain ageing-associated molecular signatures. However, the neurons they made directly from fibroblasts, induced neurons (iNs), preserved these signatures (Mertens et al., 2015). Therefore, iNs may be a useful technique in the future for modelling AD, and other diseases where ageing is an important risk factor, in vitro.
The future of iPSC models in dementia
One of the greatest aspects of iPSC-derived neurons is that they are generated from individuals, and so the disease process of patients with different genetic backgrounds can be investigated. This is interesting from the aspect of pharmacogenomics, where patients can be divided into subgroups, depending on factors such as sex and genotype, to identify successful treatments. This may be beneficial in heterogeneous neuropsychiatric disorders, such as AD, as drugs that have offered poor results in large trials may be effective in specific patient groups.
Protocols for rapidly generating different glial cell types are emerging, together with 3D cell cultures, providing more realistic in vitro systems incorporating cell types such as astrocytes and microglia that are known to be important in neurodegenerative disorders. Indeed, stem cell-derived neurons have increased synaptic activity in the presence of astrocytes (Johnson et al., 2007), and co-culture models could produce more relevant AD phenotypes. With genetic techniques rapidly advancing, iPSC-derived model systems allow risk mutations identified in dementia patients through genomics to be functionally validated. The human in vitro models discussed here with relevant AD pathologies offer hope for identifying novel therapeutic strategies and improving translation of therapies from preclinical to clinical trials.
Conclusion
In summary, iPSC technology offers a new method of studying the human brain and its pathologies, with exciting implications for basic research, as well as translational. iPSC modelling together with precise genome editing is already starting to complement existing models of psychiatric disorders and other human diseases. Cellular and molecular phenotypes can inform animal studies, and pathways from animal models can be confirmed in iPSC models. Human cell models could rapidly advance understanding of disease mechanisms, leading to potential therapeutics, and novel targets can undergo safety and efficacy testing in a human in vitro model prior to clinical trials. The revolutionary technologies described in this article could lead to the advancement in understanding of AD mechanisms and the refinement in drug discovery for neuropsychiatric disorders desperately needed for novel AD therapies.
Acknowledgements
We acknowledge the funding provided by the Medical Research Council, project 1246016. We acknowledge the financial support of the National Institute for Health Research (NIHR) Biomedical Research Centre for Mental Health at South London and Maudsley NHS Foundation Trust and Institute of Psychiatry, Psychology and Neuroscience, King’s College London. The research leading to these results has received support from the Innovative Medicines Initiative Joint Undertaking under grant agreement no. 115300 and agreement no. 115439, resources of which are composed of financial contribution from the European Union’s Seventh Framework Programme (FP7/2007–2013) and EFPIA companies’ in kind contribution, and from and the Mortimer D Sackler Foundation. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Declaration of Interest
None declared.
References
- Bellin M, Marchetto MC, Gage FH, Mummery CL. Induced pluripotent stem cells: the new patient? Nature Reviews Molecular Cell Biology. 2012;13:713–726. doi: 10.1038/nrm3448. [DOI] [PubMed] [Google Scholar]
- Brennand K, Marchetto M, Benvenisty N, Brüstle O, Ebert A, Izpisua Belmonte J, Kaykas A, Lancaster M, Livesey F, McConnell M, McKay R, et al. Creating Patient-Specific Neural Cells for the In Vitro Study of Brain Disorders. Stem Cell Reports. 2015;5:933–945. doi: 10.1016/j.stemcr.2015.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SH, Kim YH, Hebisch M, Sliwinski C, Lee S, D'Avanzo C, Chen H, Hooli B, Asselin C, Muffat J, Klee JB, et al. A three-dimensional human neural cell culture model of Alzheimer/'s disease. Nature. 2014;515:274–278. doi: 10.1038/nature13800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocks G, Curran S, Gami P, Uwanogho D, Jeffries AR, Kathuria A, Lucchesi W, Wood V, Dixon R, Ogilvie C, Steckler T, et al. The utility of patient specific induced pluripotent stem cells for the modelling of Autistic Spectrum Disorders. Psychopharmacology. 2014;231:1079–1088. doi: 10.1007/s00213-013-3196-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duff K. Transgenic mouse models of Alzheimer's disease: phenotype and mechanisms of pathogenesis. Biochemical Society symposium. 2001;67:195–202. doi: 10.1042/bss0670195. [DOI] [PubMed] [Google Scholar]
- Iovino M, Agathou S, Gonzalez-Rueda A, Del Castillo Velasco-Herrera M, Borroni B, Alberici A, Lynch T, O'Dowd S, Geti I, Gaffney D, Vallier L, et al. Early maturation and distinct tau pathology in induced pluripotent stem cell-derived neurons from patients with MAPT mutations. Brain. 2015;138:3345–3359. doi: 10.1093/brain/awv222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Israel MA, Yuan SH, Bardy C, Reyna SM, Mu Y, Herrera C, Hefferan MP, Van Gorp S, Nazor KL, Boscolo FS, Carson CT, et al. Probing sporadic and familial Alzheimer/'s disease using induced pluripotent stem cells. Nature. 2012;482:216–220. doi: 10.1038/nature10821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MA, Weick JP, Pearce RA, Zhang SC. Functional neural development from human embryonic stem cells: accelerated synaptic activity via astrocyte coculture. The Journal of Neuroscience. 2007;27:3069–3077. doi: 10.1523/JNEUROSCI.4562-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T, Asai M, Tsukita K, Kutoku Y, Ohsawa Y, Sunada Y, Imamura K, Egawa N, Yahata N, Okita K, Takahashi K, et al. Modeling Alzheimer's disease with iPSCs reveals stress phenotypes associated with intracellular Abeta and differential drug responsiveness. Cell Stem Cell. 2013;12:487–496. doi: 10.1016/j.stem.2013.01.009. [DOI] [PubMed] [Google Scholar]
- Lapasset L, Milhavet O, Prieur A, Besnard E, Babled A, Ait-Hamou N, Leschik J, Pellestor F, Ramirez JM, De Vos J, Lehmann S, et al. Rejuvenating senescent and centenarian human cells by reprogramming through the pluripotent state. Genes & Development. 2011;25:2248–2253. doi: 10.1101/gad.173922.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo A, Launer LJ, Fratiglioni L, Andersen K, Di Carlo A, Breteler MM, Copeland JR, Dartigues JF, Jagger C, Martinez-Lage J, Soininen H, et al. Prevalence of dementia and major subtypes in Europe: A collaborative study of population-based cohorts. Neurologic Diseases in the Elderly Research Group. Neurology. 2000;54:S4–9. [PubMed] [Google Scholar]
- Markou A, Chiamulera C, Geyer MA, Tricklebank M, Steckler T. Removing obstacles in neuroscience drug discovery: the future path for animal models. Neuropsychopharmacology. 2009;34:74–89. doi: 10.1038/npp.2008.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertens J, Paquola Apuã CM, Ku M, Hatch E, Böhnke L, Ladjevardi S, McGrath S, Campbell B, Lee H, Herdy Joseph R, Gonçalves JT, et al. Directly Reprogrammed Human Neurons Retain Aging-Associated Transcriptomic Signatures and Reveal Age-Related Nucleocytoplasmic Defects. Cell Stem Cell. 2015;17:705–718. doi: 10.1016/j.stem.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertens J, Stüber K, Wunderlich P, Ladewig J, Kesavan Jaideep C, Vandenberghe R, Vandenbulcke M, van Damme P, Walter J, Brüstle O, Koch P. APP Processing in Human Pluripotent Stem Cell-Derived Neurons Is Resistant to NSAID-Based γ-Secretase Modulation. Stem Cell Reports. 2013;1:491–498. doi: 10.1016/j.stemcr.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musunuru K. Genome editing of human pluripotent stem cells to generate human cellular disease models. Disease Models and Mechanisms. 2013;6:896–904. doi: 10.1242/dmm.012054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquet D, Kwart D, Chen A, Sproul A, Jacob S, Teo S, Olsen KM, Gregg A, Noggle S, Tessier-Lavigne M. Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature. 2016;533:125–129. doi: 10.1038/nature17664. [DOI] [PubMed] [Google Scholar]
- Qi Y, Zhang X, Renier N, Wu Z, Atkin T, Sun Z, Ozair MZ, Tchieu J, Zimmer B, Fattahi F, Ganat Y, et al. Combined small-molecule inhibition accelerates the derivation of functional cortical neurons from human pluripotent stem cells. Nature Biotechnology. 2017;35:154–163. doi: 10.1038/nbt.3777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohani L, Johnson AA, Arnold A, Stolzing A. The aging signature: a hallmark of induced pluripotent stem cells? Aging Cell. 2014;13:2–7. doi: 10.1111/acel.12182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross CA, Akimov SS. Human-induced pluripotent stem cells: potential for neurodegenerative diseases. Human Molecular Genetics. 2014;23:17–26. doi: 10.1093/hmg/ddu204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roychaudhuri R, Yang M, Hoshi MM, Teplow DB. Amyloid β-Protein Assembly and Alzheimer Disease. Journal of Biological Chemistry. 2009;284:4749–4753. doi: 10.1074/jbc.R800036200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan DA, Narrow WC, Federoff HJ, Bowers WJ. An improved method for generating consistent soluble amyloid-beta oligomer preparations for in vitro neurotoxicity studies. Journal of Neuroscience Methods. 2010;190:171–179. doi: 10.1016/j.jneumeth.2010.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santostefano KE, Hamazaki T, Biel NM, Jin S, Umezawa A, Terada N. A practical guide to induced pluripotent stem cell research using patient samples. Laboratory Investigation. 2015;95:4–13. doi: 10.1038/labinvest.2014.104. [DOI] [PubMed] [Google Scholar]
- Saraceno C, Musardo S, Marcello E, Pelucchi S, Di Luca M. Modeling Alzheimer’s disease: from past to future. Frontiers in Pharmacology. 2013;4:77. doi: 10.3389/fphar.2013.00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Kirwan P, Smith J, MacLean G, Orkin SH, Livesey FJ. A human stem cell model of early Alzheimer's disease pathology in Down syndrome. Science Translational Medicine. 2012a;4:124–129. doi: 10.1126/scitranslmed.3003771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Kirwan P, Smith J, Robinson HPC, Livesey FJ. Human cerebral cortex development from pluripotent stem cells to functional excitatory synapses. Nature Neuroscience. 2012b;15:477–486. doi: 10.1038/nn.3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Xu X, Lei Y, Luo J, Wang J, Zhang S, Yang XJ, Sun M, Nuwaysir E, Fan G, Zhao J, Lei L, et al. Prevention of beta-amyloid induced toxicity in human iPS cell-derived neurons by inhibition of Cyclin-dependent kinases and associated cell cycle events. Stem Cell Research. 2013;10:213–227. doi: 10.1016/j.scr.2012.11.005. [DOI] [PubMed] [Google Scholar]
- Yagi T, Ito D, Okada Y, Akamatsu W, Nihei Y, Yoshizaki T, Yamanaka S, Okano H, Suzuki N. Modeling familial Alzheimer's disease with induced pluripotent stem cells. Human Molecular Genetics. 2011;20:4530–4539. doi: 10.1093/hmg/ddr394. [DOI] [PubMed] [Google Scholar]
- Yahata N, Asai M, Kitaoka S, Takahashi K, Asaka I, Hioki H, Kaneko T, Maruyama K, Saido TC, Nakahata T, Asada T, et al. Anti-Aβ Drug Screening Platform Using Human iPS Cell-Derived Neurons for the Treatment of Alzheimer's Disease. PLoS ONE. 2011;6:e25788. doi: 10.1371/journal.pone.0025788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Pak CH, Han Y, Ahlenius H, Zhang Z, Chanda S, Marro S, Patzke C, Acuna C, Covy J, Xu W, et al. Rapid Single-Step Induction of Functional Neurons from Human Pluripotent Stem Cells. Neuron. 2013;78:785–798. doi: 10.1016/j.neuron.2013.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]