Abstract
During embryonic development, mechanical forces are essential for cellular rearrangements driving tissue morphogenesis. Here, we show that in the early zebrafish embryo, friction forces are generated at the interface between anterior axial mesoderm (prechordal plate, ppl) progenitors migrating towards the animal pole and neurectoderm progenitors moving in the opposite direction towards the vegetal pole of the embryo. These friction forces lead to global rearrangement of cells within the neurectoderm and determine the position of the neural anlage. Using a combination of experiments and simulations, we show that this process depends on hydrodynamic coupling between neurectoderm and ppl as a result of E-cadherin-mediated adhesion between those tissues. Our data thus establish the emergence of friction forces at the interface between moving tissues as a critical force-generating process shaping the embryo.
Introduction
Throughout embryonic development, tissue morphogenesis depends on mechanical forces that drive cell rearrangements and global tissue shape changes1,2. In zebrafish gastrulation, epiboly, internalization, convergence and extension constitute the main cellular processes by which the embryo takes shape3. Although recent studies have unraveled key force-generating mechanisms mediating these different cellular processes3, how forces between neighboring tissues are generated, perceived and integrated is yet poorly understood.
Development of the central nervous system in vertebrates involves extensive morphogenetic movements within the embryonic neurectoderm4. The zebrafish nervous system organization becomes first apparent at gastrulation5, and morphogenesis of the neurectoderm is accompanied by neighboring tissues undergoing dynamic cellular reorganization6. Recent studies in zebrafish suggested that the formation of the mesoderm and endoderm (mesendoderm) germ layers is required for proper morphogenesis of the overlying neurectoderm during neural keel formation7,8. However, the mechanisms by which mesendoderm influences neurectoderm morphogenesis have only started to be unraveled.
Results
Anterior axial mesendoderm (prechordal plate) collective cell migration affects neurectoderm morphogenesis
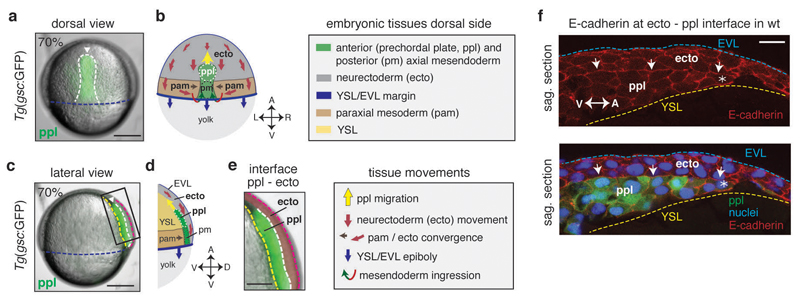
To investigate the role of mesendoderm in neurectoderm morphogenesis (for tissue organization within the gastrulating embryo, see Fig. 1), we turned to zebrafish maternal zygotic (MZ) one-eyed-pinhead (oep) mutant embryos 9, which lack much of the mesendoderm germ layers due to defective Nodal/TGFβ-signaling. Interestingly, when analyzing MZoep mutants at late stages of gastrulation, we found that the anterior neural anlage was positioned closer to the vegetal pole than in wild type (wt) embryos (Fig. 2a, b, i, j and Supplementary Fig. 2k-m). This points at the intriguing possibility that mesendoderm is required for proper positioning of the anterior neural anlage. To further test this possibility, we analyzed how the neurectoderm, which gives rise to the anterior neural anlage, interacts with the underlying anterior axial mesendoderm (prechordal plate, ppl) during gastrulation. Previous studies have suggested that the ppl moves as a migrating cell collective in a straight path towards the animal pole, while the neurectoderm moves in the opposite direction towards the vegetal pole (Fig. 1a-e)10. To understand how these in opposite directions moving tissues might influence each other, we first analyzed the localization of molecules involved in cell-cell and cell-extracellular matrix (ECM) adhesion at the neurectoderm-ppl interface. We found that the cell-cell adhesion receptor E-cadherin accumulated at the interface between ppl and neurectoderm during gastrulation (Fig. 1f), supporting previous observations that ppl and neurectoderm cells form E-cadherin mediated cell-cell contacts at this interface10. In contrast, ECM components, such as fibronectin, did not show any recognizable accumulations at the neurectoderm-ppl interface until late stages of gastrulation (Supplementary Fig. 1a-c), arguing against ECM playing an important role in mediating the interaction between ppl and neurectoderm cells during early stages of gastrulation11. Consistent with ppl and neurectoderm cells forming E-cadherin mediated cell-cell contacts, we also found interstitial fluid (IF) accumulations to be absent from places where E-cadherin accumulates at the neurectoderm-ppl interface (Supplementary Fig. 1d). Collectively, these observations suggest that neurectoderm and ppl constitute two directly adjacent tissues that globally move in opposite directions during gastrulation and contact each other directly at their interface via E-cadherin mediated cell-cell adhesions.
Figure 1. Neurectoderm (ecto) and prechordal plate (ppl) morphogenesis during gastrulation.
(a,c) Bright-field/fluorescence images of a Tg(gsc:GFP) zebrafish embryo at 7.0hpf; GFP-labeled ppl leading edge cells are indicated (white arrowheads); rectangle in (c) marks magnified area in (e); dashed lines in (a) indicate axial mesendoderm (white), and in (c) ecto-to-ppl (white), yolk syncytial layer (YSL)-to-ppl (yellow), enveloping layer (EVL)-to-media (purple) and EVL-to-YSL (blue) interfaces; embryonic axes orientation as marked in (b,d) for same views.
(b,d) Illustration of embryonic [anterior (ppl) and posterior axial mesendoderm (pm), paraxial mesoderm (pam) and ecto] and extra-embryonic [YSL, EVL, yolk) tissues, and their respective direction of movement during gastrulation at the dorsal side of the zebrafish embryo; arrows in (b,d) indicate animal-vegetal (A-V), left-right (L-R), and dorsal-ventral (D-V) embryonic axes.
(e) Magnified view of the boxed area in (c) showing neighboring ppl (green) and overlying ecto (red pseudocolored) tissues; dashed lines as in (c).
(f) Immunofluorescence confocal images of sagittal sections of the ecto-to-ppl interface at 7.5hpf stained for E-cadherin (upper panel) and merged with ppl progenitors expressing gsc:GFP and DAPI-stained nuclei (lower panel); arrows highlight E-cadherin accumulations at ecto-to-ppl interface, and asterisks mark ppl leading edge cells; blue dashed line indicates EVL-to-media interface, and yellow dashed line outlines ppl- and ecto-to-YSL interfaces; animal pole to the right.
All embryos animal pole up; dorsal (a,b) and lateral (c,d,e,f) views with dorsal right; scale bars, 200µm (a,c), 100µm (e), and 20µm (f).
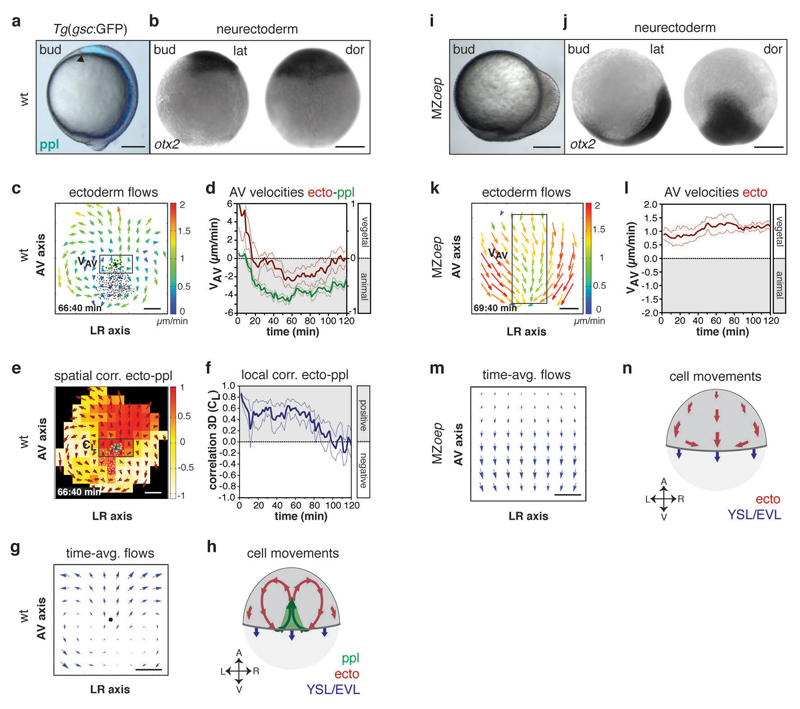
Figure 2. Defective neurectoderm (ecto) morphogenesis in MZoep mutants.
(a,i) Brightfield/fluorescence images of Tg(gsc:GFP) wt (a) and MZoep mutant embryos (i) at the end of gastrulation (bud stage, 10hpf); arrowhead in (a) marks anterior edge of GFP (blue)-labeled ppl.
(b,j) Anterior neurectoderm progenitor cells in a wt (b) and MZoep embryo (j) at bud stage (10hpf) visualized by whole-mount in situ hybridization of otx-2 mRNA.
(c,k) 2D tissue flow map indicating average velocities of ecto movements along the animal-vegetal (AV) and left-right (LR) axis at the dorsal side of a wt (c; 7.1hpf) and MZoep embryo (k; 7.2hpf); local average ecto velocities color-coded ranging from 0 (blue) to 2 (red) µm/min; positions of all/leading edge ppl cells marked by black/green dots; boxed areas are used for measurements in (d,l).
(d,l) Mean velocities along the AV axis (VAV) of ecto (red; right y-axis; boxed area in c,k) and underlying ppl leading edge cells (green, left y-axis) in wt (d; n=6 embryos) and MZoep embryos (l; n=4 embryos); 6-8hp; error bars, s.e.m..
(e) 3D directional correlation between ecto and ppl in a wt embryo at 7.1hpf; color-coded correlation ranging from 1 (red, highest) to -1 (white, lowest); red arrows indicate local averaged ecto velocities; boxed area was used for measurements in (f).
(f) 3D average directional correlation between leading edge ppl and adjacent neurectoderm cells (black boxed area in e) used for local correlation (CL) calculation in wt embryos (n=6 embryos); 6-8hpf; error bars, s.e.m.
(g,m) 2D tissue flow map of ecto cells showing time-averaged velocities (over 120min from 3 embryos) along the AV and LR axes at the dorsal side in wt (g) and MZoep embryos (m); black dot in (g) marks position of ppl leading edge.
(h,n) Schematic of ecto (red), ppl (green), and enveloping layer (EVL)/yolk syncytial layer (YSL) movements (blue) in wt (h) and MZoep embryos (n); arrows indicate AV and LR embryonic axes.
All embryos animal pole up; dorsal [b,j (dor) and h,n] and lateral [a,i and b,j(lat)] views with dorsal right; scale bars, 200µm (a,b,i,j) and 100µm (c,e,k,m).
Next we asked whether neurectoderm movements towards the vegetal pole might be influenced by the underlying ppl migrating in the opposite direction towards the animal pole of the embryo. To this end, we compared neurectoderm cell movements in wt embryos forming a ppl versus MZoep mutant embryos defective in ppl formation (Supplementary Fig. 2a, i and Supplementary Video 1, 2). For quantifying neurectoderm cell movements relative to the movement of ppl cells, we constructed 2D cellular flow maps of velocities and directions from 3D tracking data (Fig. 2c). Consistent with previous observations10,12,13, we found that ppl progenitors migrated in a highly coordinated and directed manner towards the animal pole of the wt gastrula (Supplementary Fig. 2b-d). Remarkably, while the bulk of neurectoderm cells in wt underwent epiboly movements directed towards the vegetal pole, cells located directly above and anterior of the leading edge of the ppl slowed down their vegetal-directed movement and reoriented their direction of motion from vegetal to animal (Fig. 2c, d and Supplementary Fig. 2e, f and Supplementary Video 3), resulting in high animal-directed movement alignment with the adjacent ppl progenitors (measured average local correlation (CL) over time interval (t, min) CL (t120) = 0.37 ± 0.03 (s.e.m., n=6 embryos); Fig. 2e, f and Supplementary Fig. 2g, h and Supplementary Video 4). This local reorientation of neurectoderm cell movements close to the leading edge of the ppl in wt was accompanied by the formation of characteristic large-scale cell flows within the neurectoderm resembling two counter-rotating vortices, which were mirrored along the embryo midline (Fig. 2c, g, h and Supplementary Video 3). Notably, there was only little influence on the movements of neurectoderm cells posterior of the ppl, likely due to posterior axial mesendoderm behind the ppl displaying much less pronounced animal-directed movements (Supplementary Fig. 1e, f). In contrast to the situation in wt embryos, neurectoderm cells in MZoep embryos lacking ppl progenitors (Supplementary Fig. 2i) showed none of the characteristic flow patterns found in wt and, instead, moved exclusively towards the vegetal pole (Fig. 1k-n and Supplementary Fig. 2j and Supplementary Video 5). Collectively, these observations point to the possibility that animal-directed migration of ppl cells reorient the vegetal-directed movement of adjacent neuroectoderm cells.
Movement speed of neurectoderm and prechordal plate (ppl) controls neurectoderm morphogenesis
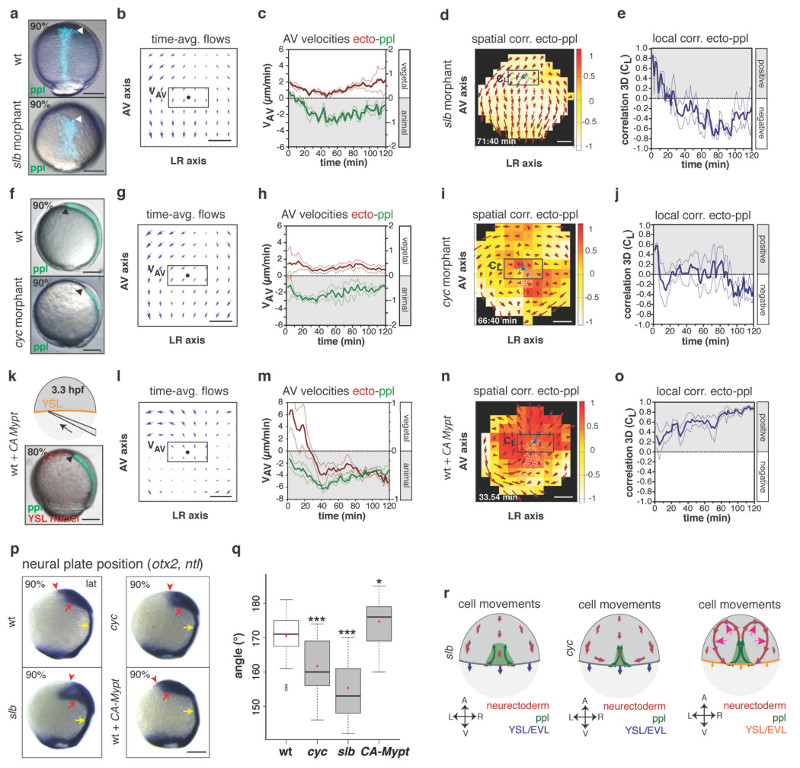
To determine whether changing ppl cell movement would affect neurectoderm cell rearrangements, we turned to slb/wnt11 morphant embryos (Fig. 3a), in which ppl cells move less coordinated and slower towards the animal pole due to compromised expression of the non-canonical Wnt ligand Wnt11 (Supplementary Fig. 3e-h)14,15. When analyzing cell movements in slb morphants, we found that neurectoderm cells located above and ahead of the ppl displayed increased vegetal-directed movements, and that the characteristic vortex movements within the neurectoderm were largely lost (Fig. 3b, c). Moreover, the alignment of neurectoderm with ppl cell movements was strongly diminished (CL (t120) = -0.24 ± 0.04 (s.e.m., n=4 embryos); Fig. 3d, e) and, importantly, the anterior neural anlage was positioned closer to the vegetal pole compared to wt embryos (Fig. 3p, q and Supplementary Fig. 3j, k). Notably, similar effects were observed in cyclops/ndr2 (cyc) mutant embryos (Fig. 3f), in which a diminished number of ppl cells due to compromised expression of the Nodal signal Ndr216,17 displayed reduced velocity and less coordinated movements (Supplementary Fig. 3a-d). This resulted in increased vegetal-directed movements of neurectoderm cells close to the ppl (Fig. 3g, h), decreased movement alignment between ppl and the neurectoderm (CL (t120) = -0.035 ± 0.027 (s.e.m., n=3 embryos); Fig. 3i, j) and a more vegetal localization of the neural anlage along the animal-vegetal (AV) axis compared to wt embryos (Fig. 3p, q and Supplementary Fig. 3i, k). Collectively, these observations strongly support the notion that proper animal-directed collective ppl cell migration is critical for normal neurectoderm cell movements and positioning of the neural anlage (Fig. 3r).
Figure 3. Prechordal plate (ppl) velocity determines the effect of ppl on neurectoderm (ecto) morphogenesis.
(a,f,k) Brightfield/fluorescence images of a Tg(gsc:GFP) wt (a,f; top panel), slb (a; bottom panel) and cyc morphant embryo (f, bottom panel) at 9hpf; Tg(gsc:GFP) embryo (k) injected at 3.3hpf with CA-Mypt and H2A-mCherry mRNAs into the YSL (top panel, schematic) at 8hpf; arrowheads mark GFP (green/blue)-labeled ppl leading edge.
(b,g,l) 2D tissue flow map of time-averaged velocities of ecto cells (over 120min from 3 embryos) along animal-vegetal (AV) and left-right (LR) axes at the dorsal side of slb (b), cyc (g) and CA-Mypt injected embryos (l); black dots; ppl leading edge positions; boxed areas used for measurements in (c,h,m).
(c,h,m) Mean velocities along the AV axis (VAV) of ecto (red; right y-axis; boxed areas in b,g,l) and underlying leading edge ppl cells (green, left y-axis) in slb (c; n=4 embryos), cyc (h; n=3 embryos) and CA-Mypt injected embryos (m; n=4 embryos); 6-8hpf; error bars (dashed lines), s.e.m.
(d,i,n) 3D directional correlation map between ecto and ppl cells in a slb (d; 7.1hpf), cyc (i; 7.2hpf) and CA-Mypt-injected embryo (n; 6.6hpf); color-coded correlation ranging from 1 (red, highest) to -1 (white, lowest); red arrows; local averaged ecto velocities; boxed areas used for measurements in (e,j,o).
(e,j,o) 3D average directional correlation between leading edge ppl and ecto cells (boxed areas in d,I,n) in slb (e; n=4 embryos), cyc (j; n=3 embryos) and CA-Mypt-injected embryos (o; n=4 embryos); 6-8hpf; error bars (dashed lines), s.e.m.
(p) Anterior neural anlage and notochord labeled by otx2 (red arrows) and ntl expression (yellow arrows), respectively, in wt, slb, cyc and CA-Mypt-injected embryos at 9hpf; arrowheads denote anterior neural plate edge.
(q) Angle (°) between the vegetal pole and neural plate anterior edge in wt, slb, cyc and CA-Mypt-injected embryos at 9hpf; student’s t-test (P value indicated) for all graphs; ***, P<0.001; *, P<0.05; n (embryos from 4 independent experiments) wt/cyc/slb/CA-Mypt=36(<0.001)/39(<0.001)/17(<0.0001)/22(0.0194); box plot centre, median; red dot, mean; upper whisker, maximum; lower whisker, minimum.
(r) Schematic of ecto (red), ppl (green) and enveloping layer (EVL)/yolk syncytial layer (YSL) (blue/orange) movements in slb (f), cyc (l) and CA-Mypt-injected (r; magenta arrows;increased vortex flow) embryos; arrows;AV and LR axes.
All embryos animal pole up; dorsal (f) and lateral (a,k,p) views with dorsal right; scale bars, 200µm (a,f,k,p) or 100µm (b,g,l,d,i,n).
Next, we asked whether epiboly movements of neurectoderm cells towards the vegetal pole are also important for ppl cells to control neurectoderm morphogenesis. To reduce epiboly movements, we overexpressed a constitutively active version of the myosin-II phosphatase (CA-Mypt) specifically within the yolk syncytial layer (YSL) (Fig. 3k; Supplementary Fig. 4a-c)18. In CA-Mypt overexpressing embryos, animal-directed movements of neurectoderm cells were more pronounced (Fig. 3l, m), whereas ppl cell migration remained unchanged (Supplementary Fig. 4d-h). Moreover, the degree of alignment between ppl and neurectoderm movements was enhanced (CL (t120) = 0.61 ± 0.02 (s.e.m., n=4 embryos); Fig. 3n, o and Supplementary Video 6), and the anterior neural anlage was positioned closer to the animal pole (Fig. 3p, q and Supplementary Fig. 4i, j). This suggests that the effect of ppl cell movements on neurectoderm morphogenesis becomes more apparent when neurectoderm epiboly movements are reduced (Fig. 3r).
Continuous mesendoderm cell ingression is required for ppl cells affecting neurectoderm morphogenesis
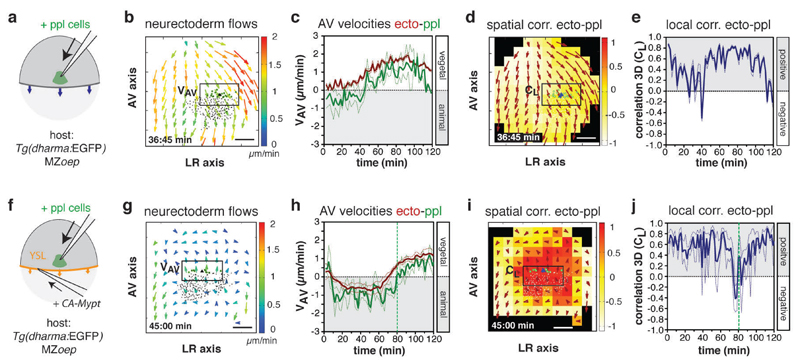
Movement of ppl cells towards the animal pole is initiated by the synchronized ingression of ppl progenitors at the dorsal germ ring margin10. To test whether continuous ingression of mesendoderm progenitors is required for animal-directed ppl cell migration and, consequently, their effect on neurectoderm cell movements, we analyzed ppl and neurectoderm cell movements in the absence of mesendoderm cell ingression. To this end, we transplanted 100-150 induced ppl progenitor cells19 below the neurectoderm close to the dorsal germ ring margin of MZoep embryos lacking endogenous mesendoderm cell ingression9 and monitored their movement relative to adjacent neurectoderm cells (Fig. 4a and Supplementary Fig. 5a-c, e). Although most of the transplanted cells displayed protrusions directed towards the animal pole (Supplementary Fig. 5d), their movement coordination and directed migration were severely impaired (Supplementary Fig. 5f, g). This is consistent with previous suggestions that anchorage of ppl progenitors to newly internalizing cells at their rear is required for their animal-directed migration20. Notably, neurectoderm cells adjacent to the transplanted ppl progenitors showed vegetal-directed movements indistinguishable from neurectoderm cells in non-transplanted MZoep mutant embryos (Fig. 4b, c and Supplementary Video 7). Moreover, as gastrulation proceeded, transplanted ppl cells started to move towards the vegetal pole in the same direction as the overlying neurectoderm cells, resulting in highly aligned vegetal-directed movements of neurectoderm and transplanted ppl cells (CL = 0.48 ± 0.04 (s.e.m. n=3 embryos); Fig. 4d, e).
Figure 4. Mesendoderm cell ingression is required for prechordal plate (ppl) affecting neurectoderm (ecto) cell movements.
(a,f) Schematic of ppl cells (green) transplanted at 6hpf into the dorsal side of a MZoep (a) or MZoep embryos injected with CA-Mypt mRNA into the YSL (f).
(b,g) 2D tissue flow map indicating average velocities of ecto movements along the animal-vegetal (AV) (VAV) and left-right (LR) (VLR) axis at the dorsal side of a transplanted MZoep mutant (b; 6.6hpf) and transplanted MZoep embryo overexpressing CA-Mypt within the YSL (g; 6.7hpf); local average ecto velocites indicated by arrows color-coded ranging from 0 (blue) to 2 (red) µm/min; positions of all/leading edge transplanted ppl cells marked by black/green dots; boxed areas were used for measurements in (c,h).
(c,h) Mean velocities along the AV axis (VAV) of ecto (red; boxed areas in b,g) and underlying ppl leading edge cells (green) in transplanted MZoep (c; n=3 embryos) and transplanted MZoep embryos overexpressing CA-Mypt within the YSL (h; n=3 embryos); 6-8hpf; vertical dashed line in (h) indicates start of vegetal-directed movements of ppl cells; error bars, s.e.m.
(d,i) 3D directional correlation between leading edge ppl and ecto cells in a transplanted MZoep (d; 6.7hpf) and transplanted MZoep mutant embryo overexpressing CA-Mypt within the YSL (i; 6.7hpf); color-coded correlation ranging from 1 (red, highest) to -1 (white, lowest); red arrows indicate local averaged ecto velocities; boxed areas were used for measurements in (e,j).
(e, j) 3D average directional correlation between leading edge ppl and adjacent ecto cells (boxed areas in d, i) in transplanted MZoep (e; n=3 embryos) and transplanted MZoep embryos overexpressing CA-Mypt within the YSL (j; n=3 embryos); 6-8hpf; vertical dashed line (j) as in (h); error bars, s.e.m.
All scale bars, 100µm.
To test if slowing down neurectoderm epiboly movements would restore ppl-induced redirection of neurectoderm cells, we reduced the speed of vegetal-directed neurectoderm cell movements in transplanted MZoep mutant embryos by overexpressing CA-Mypt specifically within the YSL of those embryos (Fig. 4f and Supplementary Fig. 5h-k) and monitored the interaction between transplanted ppl cells and adjacent neurectoderm cells. We found that ppl cells showed increased animal-directed movements when vegetal-directed neurectoderm cell movements were slowed down in MZoep embryos (Fig. 4g, h and Supplementary Video 8). Moreover, neurectoderm cells adjacent to the transplanted ppl cells transiently reoriented their movement towards the animal pole (Fig. 4h), resulting in temporary high animal-directed movement alignment between transplanted ppl and adjacent neurectoderm cells (CL (t80) = 0.53 ± 0.04 (s.e.m., n=3 embryos); Fig. 4i, j). These observations strongly support the notion that the difference in movement speed along the AV axis between ppl and neurectoderm cells determines whether ppl influences neurectoderm cell movements or vice versa.
Friction forces between neurectoderm and ppl mediate the effect of ppl on neurectoderm morphogenesis
We hypothesized that the observed large-scale cellular rearrangements within the neurectoderm might be generated by friction forces arising at the tissue interface between ppl and neurectoderm. To test this hypothesis, we formulated a theoretical model based on the physical principles of viscous fluid motion at low Reynolds numbers (Supplementary Note). In this model, we considered the neurectoderm as a thin layer of viscous compressible fluid exposed to external friction arising at its interface with EVL and/or yolk cell and being in contact with ppl cells, which were modelled as a rectangular element exerting a friction force on the neurectoderm (Supplementary Note).
We first aimed at comparing neurectoderm velocity profiles along the animal-vegetal axis with a simplified, effectively 1D theory (Fig. 5a). To assess the effect of ppl cells on neurectoderm movement, we measured unperturbed neurectoderm epiboly movements in MZoep mutants devoid of ppl cells and subtracted those epiboly movements from the overall neurectoderm flow field in wt embryos (Supplementary Fig. 6a). Consistent with our experimental observations (Fig. 5b), we assumed that the velocity of the calculated neurectoderm flow vanishes at the ventral and dorsal margins of the neurectoderm in both the presence and absence of the ppl. In our model, the external friction force acting outside of the ppl domain causes the velocity profile to decay exponentially away from ppl cells, on a length scale that decreases when the friction coefficient increases (Supplementary Note), while in the absence of such external friction, this decay is linear (Supplementary Fig. 6b). When performing a fit to experimentally obtained neurectoderm flow profiles in wt, we obtained a very close agreement between the calculated and experimentally observed flow profiles for a very low external friction force (Fig. 5b and Supplementary Fig. 6c).
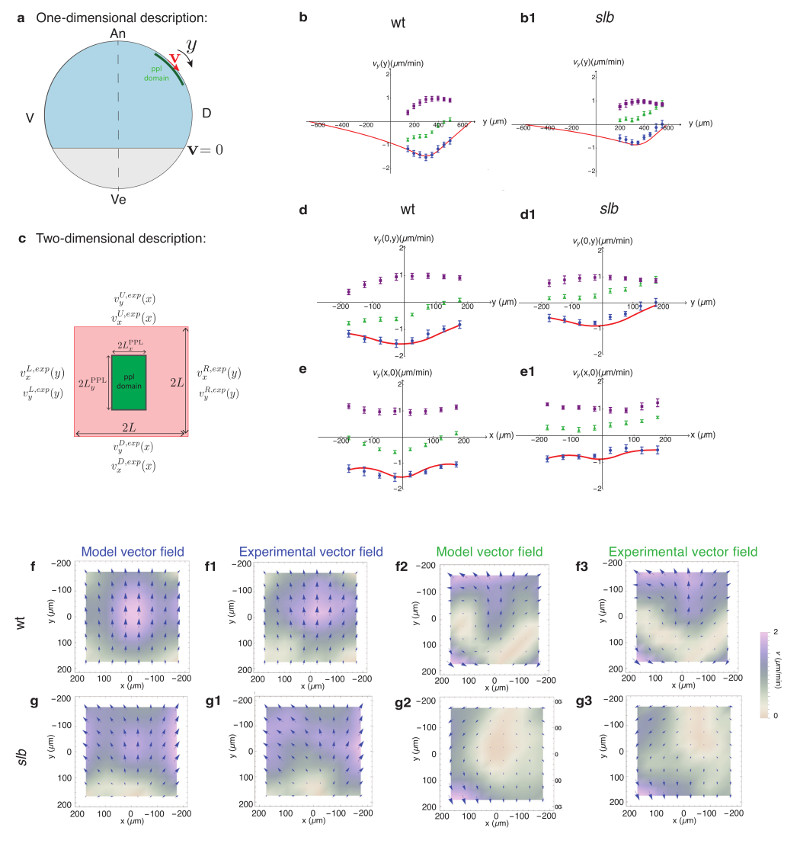
Figure 5. Hydrodynamic model description of the influence of prechordal plate (ppl) on neurectoderm (ecto) cell flows through friction forces at the tissue interface.
(a) Illustration of 1D ecto flow description along the tissue midline axis; ppl domain exerts an animal-directed force on the ecto; the differential velocity v of ecto tissue equals 0 at tissue boundaries.
(b,b1) 1D analysis of ecto flow velocity (vy) along the tissue midline axis in wt (b) and slb morphant (b1) embryos; predicted flow profile (red), experimentally obtained flow velocities in wt and slb morphant embryos subtracted by the flows in MZoep mutants (blue), non-subtracted flow profiles in wt and slb morphant embryos (green), and flows in MZoep mutants (purple) are shown; values of the 1D model parameters used for each experimental case are listed in Supplementary Table 1 (Supplementary Note).
(c) Illustration of 2D ecto flow description within the experimental image plane; velocities at the up (U), down (D), right (R) and left (L) boundaries of the image plane are taken from experimental measurements; a uniform force density is exerted on the ecto within the ppl domain.
(d-e1) 2D analysis of ecto flow velocities for wt (d,e) and slb morphant (d1,e1) embryos; upper panels show the vy velocities along the ecto tissue midline axis and lower panels (e, e1) the vy velocities along the mediolateral extent of the ecto; color labeling of curves as in (b,b1); values of the 2D model parameters used for each experimental case are listed in Supplementary Table 1 (Supplementary Note).
(f-g3) 2D vector density plots for the theoretical and experimental ecto flow velocity fields of wt (f-f3) and slb morphant embryos (g-g3); subtracted flow fields for wt (f,f1) and slb morphant (g,g1) embryos; non-subtracted total flow velocity fields for wt (f2,f3) and slb morphant (g2,g3) embryos generated by adding corresponding experimental MZoep velocities to the theoretical flow profiles; direction (arrows) and color-coded velocities from 0 (white, lowest) to 2 (purple, highest).
All error bars s.d.
Next, we extended our analysis to slb morphant embryos, where the effect of changing the relative velocity between neuroectoderm and ppl cells was clearly detectable throughout the timeframe of our analysis (6-8 hpf). When analyzing slb morphant embryos, we made the assumption that if the ppl cells were to exert a dynamic friction force that is linearly dependent on the relative velocities between the two tissues, we would expect the force to be decreased by the same amount as the relative velocity between neurectoderm and ppl (20% reduction). Indeed, we found that calculated neurectoderm flow profiles with a ppl friction force reduced by 20% (Supplementary Note, Table 1) closely resembled the experimentally obtained flow profiles in slb morphants (Fig. 5b1). Together, our 1D analysis of neurectoderm flows in wt and slb morphant embryos revealed a remarkable quantitative similarity between the model predictions and experimental observations, supporting the notion that friction forces mediate the effect of ppl on neurectoderm motion.
We further asked whether our 1D description could also account for 2D neurectoderm flow patterns within the domain of observation. Since our 1D analysis revealed that external friction outside of the ppl domain is very low, we neglected its contribution to neurectoderm flows in our 2D analysis. We also assumed for simplicity that the bulk and shear viscosity of the fluid are equal. We then solved the equations in the 2D rectangular domain of our experimental observations, imposing the experimentally measured velocities on the boundaries of the domain (Fig. 5c; Supplementary Note) and compared predicted to experimentally observed neurectoderm velocities. We first analyzed alterations in neurectoderm movement in wt embryos displaying normal ppl cell movements. By adjusting the ratio between the force density and neurectoderm bulk viscosity (f /ηb) (Supplementary Note, Table 1), we found that the predicted neurectoderm velocity profile in wt embryos matched well the magnitude and shape of the experimentally determined velocity profile (Fig. 5d,e, f-f3). Assuming that the force exerted by ppl cells originates from dynamic friction between these two moving tissues with a friction coefficient ξ, we further obtained the ratio of internal viscosity to friction against ppl cells ηb/ξ (Supplementary Note, Table 1) and, given previous measurements of tissue viscosity, 21 a value of the friction coefficient in the order of ξ~1 pN.s/μm3 (Supplementary Note). Applying the same logic to slb morphant embryos produced 2D neurectoderm flow patterns very closely matching the experimentally observed ones (Fig. 5d1,e1,g-g3), suggesting that the friction force density at the neurectoderm-ppl interface is critical for the effect ppl cells have on neurectoderm cell movements.
Friction forces between neurectoderm and ppl depend on transient E-cadherin mediated cell-cell contacts at the tissue interface
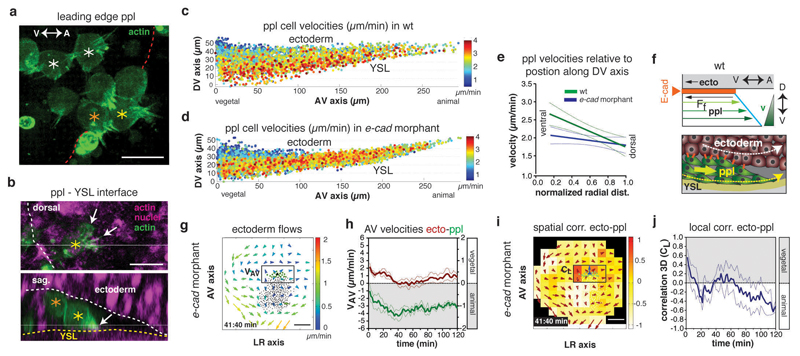
To understand how friction can arise at this interface, we first analyzed ppl cell migration and protrusive activity by visualizing the actin cytoskeleton of migrating ppl cells (Fig. 6a, b). We found that ppl cells were organized in a shingled array along the dorsal-ventral (DV) axis of the ppl (Fig. 6a, f and Supplementary Video 9) with cells at the leading edge displaying protrusions typically adhering to the YSL surface, which served as a substrate for their migration (Fig. 6b and Supplementary Video 10). Moreover, ppl progenitors close to the interface with the neurectoderm usually trailed behind cells positioned further away from this interface along the DV extent of the ppl (Fig. 6a, b and Supplementary Video 9), consistent with the possibility that friction at this interface slows down their animal-directed migration. To further test this assumption, we plotted the velocities of ppl progenitors in wt embryos along the DV axis of the ppl (Fig. 6c). We found a linear velocity gradient along this axis with its minimum at the interface to the overlying neurectoderm (Fig. 6e), as expected for friction forces at the neurectoderm-ppl interface most strongly slowing down the movement of ppl cells directly adjacent to this interface (Fig. 6f).
Figure 6. E-cadherin-mediated friction forces between prechordal plate (ppl) and neurectoderm (ecto) determine ecto morphogenesis.
(a,b) Confocal images of leading edge (red dots) ppl donor cells expressing lifeact-GFP (actin, green) transplanted in host embryo labeled with Utrophin-Cherry (actin, purple) and H2A-mCherry (nuclei, purple); asterisks, ppl cells at YSL interface (yellow), between YSL and ecto (orange), and at ecto interface (white); dorsal view as maximal z-stack projection (a); dorsal (top) and sagittal (bottom) confocal sections with ppl protrusions (arrows) and interfaces to YSL (yellow dots) and ecto (white dots) indicated (b).
(c,d) Average instantaneous velocities of migrating ppl cells in wt (c) and e-cadherin morphant embryo (d) along the AV and DV axis color-coded from 0 (blue) to 4 (red) µm/min.
(e) Linear regression lines of binned mean velocities of ppl cells along the normalized radial distance of the DV axis from ventral (0) to dorsal (1) for wt (green; P=0.0006, n=6 embryos) and e-cadherin morphant embryos (blue; P=0.15; n= 4 embryos); P values from F-test with null hypothesis; P > 0.05, slope equals zero; error bars s.e.m.
(f) Schematic illustrating ppl (yellow arrow) dragging ecto cells (white arrow) and friction forces slowing down ppl cells at the ppl-ecto interface (bottom), leading to a linear velocity gradient within ppl (top); Ff, friction force; E-cadherin, orange line/dots.
(g) 2D tissue flow map indicating velocities of ecto cell movements along the AV (VAP) and left-right (LR) (VLR) axis at the dorsal side of a e-cadherin morphant embryo at 6.7hpf; local average ecto velocities indicated and color-coded from 0 (blue) to 2 (red) µm/min; positions of all/leading edge ppl cells, black/green dots; boxed area used for measurements in (h).
(h) Mean velocities along the AV axis (VAV) of ecto (red; boxed area in g; right y-axis) and underlying ppl leading edge cells (green, left y-axis) in e-cadherin morphant embryos (n=4 embryos); 6-8hpf; error bars, s.e.m.
(i) 3D directional correlation between leading edge ppl and adjacent ecto cells in a e-cadherin morphant embryo at 6.7hpf; correlation color-coded from 1 (red, highest) to -1 (white, lowest); red arrows, local averaged ecto velocities; boxed area used for measurements in (j).
(j) 3D directional correlation values between leading edge ppl and adjacent ecto cells (boxed area in i) in e-cadherin morphant (n=4 embryos); 6-8hpf; error bars, s.e.m.
Scale bars 20µm (a,b) and 100µm (g,i); arrows; AV and DV axes
We then asked which molecular players might be involved in building up friction at the neurectoderm-ppl interface. Our estimation of the friction coefficient between neurectoderm and ppl cells being in the order of ξ~1 pN.s/ μ m3 (Supplementary Note) argues against the possibility of interstitial fluid alone generating this friction, given that the friction coefficient for a fluid with the viscosity of water and a layer of thickness h = 100 nm (corresponding to the estimated distance between ppl and neurectoderm) would be in the order of ξ ~0.01 pN.s/μm3. In contrast, our initial analysis indicated that the cell-cell adhesion receptor E-cadherin, unlike ECM components, (Fig. 1f, Supplementary Fig. 1a), might be a likely candidate generating friction between these tissues. We therefore tested whether E-cadherin is needed to sustain coupling of ppl with neurectoderm cell movements by lowering the amount of E-cadherin expressed within the gastrulating embryo using e-cadherin (cdh1) MOs22. Consistent with previous reports22–24, we found that in e-cadherin morphant embryos neurectoderm cell movements towards the vegetal pole were strongly reduced whereas the EVL/YSL advanced normally in direction of the vegetal pole (Supplementary Fig. 7a, b). Additionally we observed that under reduced E-cadherin levels, ppl progenitor cells display less correlated movements towards the animal pole with slightly diminished velocity (Supplementary Fig. 7c-f). Notably, the influence of animal-directed ppl cell migration on the overlying adjacent neurectoderm cells was drastically diminished showing nearly exclusively vegetal directed movements of neurectoderm cells (Fig. 6g, h; Supplementary Video 11). Consequently, the directional correlation between ppl and neurectoderm cell movements was nearly completely abolished (CL(t120) = -0.14 ± 0.03 (s.e.m., n=4 embryos); Fig. 6i, j; Supplementary Video 12), and the DV velocity gradient of cells within the ppl was much less pronounced (Fig. 6d, e, and Supplementary Fig. 7g). This suggests that E-cadherin is critical for building up friction at the neurectoderm-ppl interface. Notably, the loss of correlation of directional movement between ppl and neurectoderm cells in e-cadherin morphant embryos was accompanied by a more vegetal positioning of the anterior neural anlage (Supplementary Fig. 7l, m), underlining the importance of coupling neurectoderm to ppl cell movement for positioning of the neural anlage.
E-cadherin has previously been suggested to be required for radial cell intercalations within the ectoderm and ectoderm epiboly movements23,24. To exclude that the observed loss of ppl and neurectoderm tissue coupling in e-cadherin morphant embryos is merely due to a failure of E-cadherin function within the ectoderm, we transplanted e-cadherin morphant ppl cells into MZoep mutants which were beforehand injected with CA-Mypt mRNA into the YSL to increase the effect of ppl cells on adjacent neurectoderm cells as observed earlier (Fig. 4f-j). We found that animal-directed movements of e-cadherin morphant ppl cells were nearly unaltered compared to transplanted wt ppl cells (compare Supplementary Fig. 7i with Fig. 4h). However, the effect of morphant ppl cells on rearrangement of cell movements within the neurectoderm was significantly diminished and shorter lived compared to their wt counterparts (Supplementary Fig. 7h, i), leading to an overall reduced alignment of movements between neurectoderm and ppl cells (CL(t80) = 0.22 ± 0.05 (s.e.m., n=4 embryos) ; Supplementary Fig. 7j, k). Together, our analysis of e-cadherin morphant embryos and cells support a critical function for E-cadherin in generating friction between ppl and neurectoderm, which is essential for force transduction between those tissues and, consequently, positioning of the neural anlage.
E-cadherin mediated friction between neurectoderm and ppl is sufficient to reorient neurectoderm cell movements
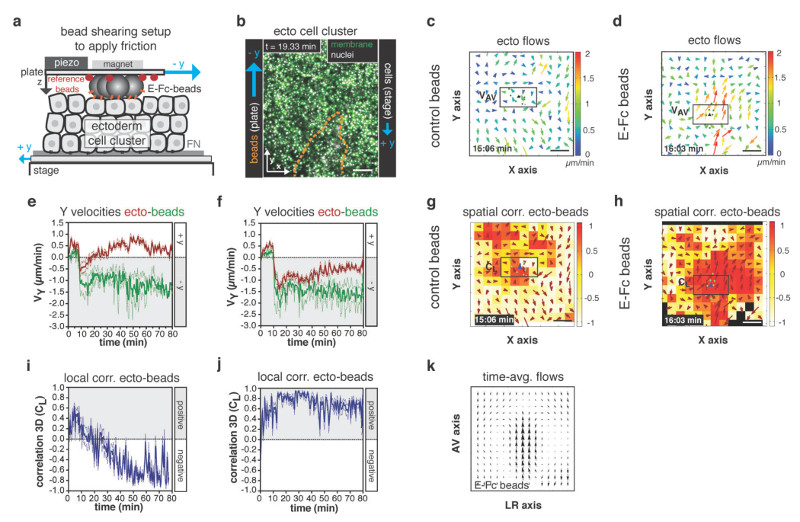
Vertical signaling from the axial mesendoderm to the overlying neurectoderm has been shown to be an important determinant of axial patterning within the forming neural keel4,25 and thus could in principle contribute to the observed influence of ppl on neurectoderm movement. To address this possibility, we thought to test if E-cadherin mediated friction alone might be sufficient to explain the observed effect of ppl on neurectoderm movement in the embryo. Hence, we designed an in vitro experiment where we substituted ppl cells with a layer of E-cadherin-coated beads and sheared them over a cluster of ex vivo cultured ectoderm progenitor cells to create friction between them (Fig. 7a). To mimic similar conditions as in the embryo, we moved the labeled ectoderm cell cluster (GPI-GFP membrane and H2A-mCherry nuclei) in one direction (stage movement ~ 0.5 µm/min) and the E-cadherin-coated beads, mimicking the ppl, with higher velocities (top plate ~ 1.5 µm/min) in the opposite direction of the ectoderm cell cluster (Fig. 7a, b; Supplementary Fig. 8a-c). Remarkably, we observed that this movement of E-cadherin-coated beads was sufficient to reorient cell movements within the ectoderm cell cluster (Fig. 7d, f), leading to high local correlation between bead and ectoderm cell movements (Fig. 7h, i) and to generate double vortex flows within the ectoderm cluster reminiscent of the situation in vivo (Fig. 7k). In contrast, no such effects were obtained when using uncoated control beads (Fig. 7c, e, g, i), suggesting that E-cadherin mediated contact between beads and ectoderm cells is critical for the beads to reorient ectoderm cell movements. Collectively, these findings support the view that E-cadherin mediated friction between ppl and neurectoderm cells is sufficient to explain the observed effect of ppl on neurectoderm movement in the embryo.
Figure 7. E-cadherin-mediated friction is sufficient to reorient neurectoderm (ecto) cell movements in vitro.
(a) Illustration of parallel plate setup for application of friction on ecto cells in vitro; uncoated control or coated with E-cadherin/Fc (E-Fc) polystyrene beads were sheared uniaxial (- y) over a cluster of opposing moving ecto cells, (+ y) to create friction; fluorescent reference beads (red) absorbed to top plate were used to track position and movement of adjacent polystyrene beads; E-cadherin receptors (orange) mediating friction indicated.
(b) Maximum projection confocal image of ecto cell cluster expressing GPI-GFP (membrane, green) and H2A-mCherry (nuclei, white) plated onto a fibronectin-coated dish; directions of cell/stage movement (+y; velocity ~0.5µm/min) and E-Fc-coated beads/top plate movement (-y; velocity ~1.5µm/min) indicated; position of cluster of beads above ecto cells outlined (orange dashed line).
(c,d) 2D tissue flow map indicating average velocities of ecto cell movements along the Y (VY) and X (VX) axis after application of friction using control (c) or E-Fc-coated (d) beads at a representative time point; local average ecto velocities indicated and color-coded ranging from 0 (blue) to 2 (red) µm/min; positions of leading edge polystyrene beads are marked by green dots; boxed area was used for measurements in (e,f).
(e,f) Mean velocities along the Y axis (Vy) of leading edge control (e; n=3 experiments) or E-Fc-coated (f; n=3 experiments) beads (green) and adjacent ecto cells (boxed area in c,d; red curve) plotted before (t = 0-10min) and after (t = 10-80min) application of friction; error bars, s.e.m.
(g,h) 3D directional correlation between ecto cells and adjacent control (g) or E-Fc-coated beads (h) at a representative time point; correlation color-coded ranging from 1 (red, highest) to -1 (white, lowest); red arrows indicate local averaged ecto velocities; position of all/leading edge ppl cells marked by white/green dots; blue arrowhead indicates average velocity of ppl leading edge cells; boxed area was used for measurements in (i,j).
(i,j) 3D average directional correlation between ecto cells (boxed area in g,h) and leading edge control (i; n=3 experiments) or E-Fc-coated beads (j; n=3 experiments) before (t = 0-10min) and after (t = 10-80min) application of friction.
(k) Time-averaged tissue flow map (over 70 (10-80) min from 3 experiments) of ecto cell movements along the y (Vy) and x (Vx) axis after application of friction using E-Fc-coated beads; error bars, s.e.m. Scale bars, 100µm (b,c,d,g,h).
Friction forces lead to distinct tissue deformations within the neurectoderm
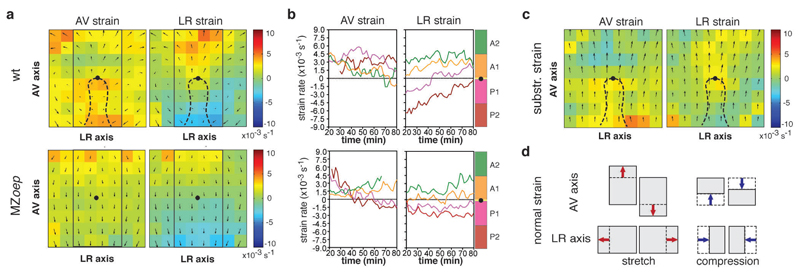
To further corroborate on the role of frictional forces on neurectoderm morphogenesis, we asked whether neurectoderm cells become compressed in front of the ppl cells and stretched behind, as one would expect if the ppl cells would be pushing the overlying neurectoderm towards the animal pole. To this end, we compared neurectoderm tissue deformation in wt versus MZoep mutant embryos by calculating normal tissue domain strain rates along the AV and left-right (LR) axes (Fig. 8d; normal strain) and shear strain rates (Supplementary Fig. 8h) from measured neurectoderm cell velocities. We observed that both normal and shear strain rates appeared considerably different between wt and MZoep mutant embryos during the course of gastrulation (Fig. 8a, b; Supplementary Fig. 8e, f). In particular, when subtracting the time-averaged strain rates of MZoep from wt embryos (Fig. 8c), we found a pronounced elongation of the neurectoderm along the AV axis behind the ppl leading edge, while ahead of the ppl the neurectoderm was compressed along the AV axis and elongated along the LR axis. Moreover, we found inverse shear deformations of the neurectoderm in areas left and right of the ppl in wt, but not in MZoep mutant embryos (Supplementary Fig. 8e-g). Together, these distinct tissue deformations in wt compared to MZoep mutant embryos are consistent with the notion that the ppl pushes the overlying neurectoderm towards the animal pole.
Figure 8. Friction forces trigger tissue deformations within the neurectoderm (ecto).
(a) Ecto tissue deformations along the AV and LR axes of wt (upper panels; n=3) and MZoep (lower panels; n=3) embryos plotted as time-averaged strain values for each domain (50x50µm); average normal strain rate is color coded according to amount of stretch [minimum green (0) to maximum red (10 x10-3s-1)] or compression [minimum green (0) to maximum blue (-10 x10-3s-1)]; tissue flows of ecto are indicated as time-averaged velocities; dashed line indicates ppl position and black dot marks ppl leading edge as reference point in wt and MZoep; rectangle outlines area used for defining sectors along the AV axis in (b).
(b) Mean normal strain rates of ecto tissue along the AV (left panels) and LR (right panels) axes of wt (upper panels; n=3 embryos) and MZoep (lower panels; n=3 embryos) embryos in defined sectors (100x200µm) of the ecto (A1 and A2 anterior and P1 and P2 posterior of ppl leading edge; for detailed description refer to Supplementary Fig. 1e) as a function of time during gastrulation (plotted from 6.3-7.3 in 10min intervals); amount of stretch/compression within each sector is plotted along the y-axis;
(c) Ecto tissue strain rate maps derived by subtraction of AV (left panel) and LR (right panel) time-averaged strain values of wt from MZoep mutant embryos (n=3 embryos); color-code as in (a); tissue flows of ecto are indicated as time-averaged velocities; black dot marks ppl leading edge as reference point.
(d) Illustration of kind and direction of tissue deformation in the ecto derived from normal strain; arrows indicate direction of stretch or compression of a tissue domain along the AV and LR axes dependent on the direction and magnitude of ecto movements.
Discussion
Biochemical signals from the axial mesendoderm have long been thought to trigger cell fate specification and patterning of the developing neural anlage4,25. Our finding that the generation of friction forces at the neurectoderm-mesendoderm interface is critical for proper positioning of the neural anlage during gastrulation shows that alongside biochemical, also mechanical signals play an important role for mesendoderm influencing neural plate development. Interestingly, the extent by which mesendoderm affects neurectoderm morphogenesis depends on the magnitude of friction force generated at the tissue interface, which again is determined by the speed difference between those tissues. This suggests that the regulation of differential speed between neurectoderm and mesendoderm during gastrulation constitutes an important factor determining neurectoderm morphogenesis.
Our data also suggest that friction forces at the neurectoderm-mesendoderm interface are generated by transient E-cadherin-mediated heterotypic contacts between ppl and neurectoderm cells. While the friction coefficient (~1 pN.s/μm3) at the neurectoderm-ppl interface estimated from our simulations is in principle compatible with this notion, directly confirming this by comparing an estimated with an expected friction coefficient due to E-cadherin mediated transient interactions between ppl and neurectoderm cells remains difficult as long as number, lifetime and elasticity of E-cadherin bonds at the tissue interface are unknown. Still, our genetic and biophysical experiments, and in particular the E-cadherin loss-of-function approaches, strongly argue in favor of a decisive function of E-cadherin in friction generation at the neurectoderm-ppl interface.
Mechanical coupling between mesendoderm and neurectoderm, potentially mediated by ECM accumulation at the interface between these tissues, has previously been speculated to be required for coordination of their convergence movements during neurulation7,8. Yet, whether and how such potential mechanical coupling between these tissues might lead to the generation of forces at their interface, and how such forces are transmitted between the tissues is yet unclear. Furthermore, large vortex cell flows have previously been observed within the chicken epiblast during primitive streak formation26,27. Yet, given that these large-scale tissue flows occur before ingression of mesoderm and endoderm progenitors and are thought to be driven by cell shape changes and cell intercalations within the epiblast itself28,29, the role of friction forces in this process remains uncertain.
Our observations that ppl mesoderm directly affects the movements of the neurectoderm through friction forces at their interface unravel an important yet unrecognized mechanical function of the mesendoderm in neurectoderm morphogenesis. Friction forces emerging at the interface between fluids and the plasma membrane have previously been implicated in vascular development30 and tissue deformation during Drosophila gastrulation31. Our findings demonstrate that the generation of friction forces between forming tissues sliding against each other constitutes a key regulatory mechanism of embryo morphogenesis in development.
Methods
Embryo staging and fish line maintenance
Zebrafish (danio rerio) were maintained as described32. Embryos were raised at 28-31°C in E3 buffer and staged according to morphological criteria33. Zebrafish lines (female and male between the age of 0.5-2.5 years) used for obtaining embryos: wild type (WT) strains TL and AB and following transgenic and mutant lines were used: MZoeptz257/tz2579 9, Tg(dharma:eGFP)34, MZoep;Tg(dharma:eGFP), Tg(gsc:GFP)35, Tg(gsc:GFP-CAAX), Tg(actb2:Cherry-UtrCH) and Tg(actb1:lifeact-GFP)18. No cell lines were used in this study. All animal experiments were carried out along the guidelines of the Ethics and Animal Welfare Committee (ETK) in Austria.
Microinjections of mRNA morpholino antisense oligonucleotides and dextran
Capped mRNA for injection was synthetized using the SP6 mMessage mMachineKit (Ambion). For ubiquitous mRNA overexpression, 100 pg h2afva-mCherry, 75 pg h2afva-tagBFP36 and 100 pg membrane RFP (mRFP) mRNA was injected into 1-cell stage embryos. To generate mesoderm progenitors, one-cell-stage Tg(gsc:GFP), Tg(gsc:GFP-CAAX) or Tg(actb2:lifeact-GFP) embryos were injected with 100 pg cyclops (cyc) mRNA37 and 2ng casanova (cas) morpholino (MO; GeneTools) oligonucleotides19. To down-regulate Myosin-II activity specifically within the YSL, 50-75 pg of mRNA encoding for a constitutively active Myosin-II phosphatase 1 (CA-Mypt) consisting of the N-terminus (1-300aa) of the human myosin phosphatase targeting subunit 1 lacking the inhibitory domain38, together with 100 pg of h2afva-mCherry mRNA, was directly injected into the YSL of embryos between 512K and high stage (2.75 – 3.3 hpf)39. To interfere with prechordal plate (ppl) progenitor cell number, 0.5-1ng of cyc MO (GeneTools) targeted against the ATG start codon of the cyc cDNA was injected into one-cell-stage embryos40. To reduce E-cadherin and Wnt11 expression levels, 3-4 ng e-cadherin MO (GeneTools)22 or 6ng of wnt11 MO (Gene Tools)41, both targeted against the ATG start codon of the respective cDNAs was injected into one-cell-stage embryos. Interstitial fluid was labeled by injection of dextran Alexa Fluor 647 (10000 MW; ThermoFisher Scientific) at high stage (3.3 hpf) into the extracellular space at the animal pole of the developing embryo.
Sample preparation for live cell imaging
Embryos were mounted in 0.5% low-melting-point (LMP) agarose (Invitrogen) into agarose molds inside a petri dish and covered with E3 medium with the dorsal side of the embryo facing upwards. For imaging of cell-division-inhibited embryos, aphidicolin and hydroxyurea were added into the 0.5% LMP agarose solution.
High-resolution multiphoton imaging
For in vivo fluorescence imaging, embryos where mounted at 60% epiboly (6 hpf) and subsequently imaged on an upright multiphoton microscope (TrimScope II, LaVision) equipped with a W Plan-Apochromat 20 x 1.0 NA dipping lens (Zeiss). GFP, mCherry/RFP, Dextra-647 and BFP fluorescence were imaged at 900nm, 1100nm and 810nm excitation wavelength, respectively, using a Ti-Sapphire femtosecond laser system (Coherent Chameleon Ultra) combined with optical parametric oscillator (Coherent Chameleon Compact OPO) technology. Excitation intensity profiles were adjusted to tissue penetration depth and Z-sectioning for imaging was set between 2-4.8 µm. For long-term imaging, movies were acquired for 100-140min with a frame rate between 95-166 seconds. All embryos were imaged with a temperature control unit set to 28.5°C, and embryos were checked for normal development after imaging.
Confocal imaging
For whole embryo confocal imaging, embryos were imaged using a Leica SP5 confocal microscope equipped with a Leica 25x 0.95 NA water dipping lens. The temperature during imaging was kept constant at 28.5 °C using a temperature chamber. To analyze YSL ring advancement, consecutive z-stacks (up to 150 µm depth) of Tg(actb2:Cherry-UtrCH) embryos throughout the course of epiboly were recorded. YSL ring advancement was determined by PIV analysis on maximum z-projections of acquired time-lapse z-stacks using a custom-designed Matlab script18.
For whole-mount imaging, embryos probed with anti-Fibronectin antibody were imaged with a Zeiss LSM510 Meta confocal microscope, using a 40x/NA1.0 water-dipping lens. Live cell imaging of in vitro experiments were performed on a Leica SP5 confocal microscope equipped with a 20x/NA0.7 air objective. Embedded sections of E-cadherin stained embryos were imaged with a Leica SP5 TCS microscope, using a 63x/NA1.4 oil immersion objective. Live cell imaging of in vitro experiments were performed on a Leica SP5 TCS microscope equipped with a 20x/NA0.7 air objective.
Transplantation assays
For cell transplantation experiments, donor and host embryos were kept in Danieaus’s solution [58 mM NaCl, 0.7 mM KCl, 0.4 mM MgSO4, 0.6 mM Ca(NO3)2 and 5mM HEPES (pH 7.6)] after dechorionation. Tg(gsc:GFP) or Tg(gsc:GFP-CAAX) donor embryos injected with cyc mRNA and cas MO were checked at 30% epiboly (3 hpf) for GFP expression, indicative of mesoderm induction19. Single or groups of cells (100-200 cells) were then removed from the animal pole of those embryos using a glass transplantation needle (20 µm diameter) and transplanted below the neurectoderm cells at the dorsal side close to the margin of a MZoep host embryo at 60% epiboly (6 hpf). For detecting the dorsal side of the host embryo, MZoep;Tg(dharma:eGFP) embryos were used expressing Dharma:EGFP at their dorsal side. Transplanted embryos were mounted for imaging as described above.
In-situ hybridization assays
Whole mount in-situ hybridizations were performed as described10. For notail (ntl) and otx2 in-situs, antisense RNA probes were synthesized from partial sequences of the respective cDNAs. Images were taken with a dissecting stereo-microscope (Olympus SZX 12) equipped with QImaging Micropublisher 5.0 camera.
Bead shearing on ectoderm aggregates
Beads were prepared as following: 10 µl of a 0.5% w/v solution of magnetic polysterene particles (10 and 20 µm diameter; Spherotech, Inc.) was incubated for 2 hours at 4 °C in either 100µl of 1 x PBS (Hank’s Balanced Salt Solution) for control beads, or 100 µl of recombinant mouse E-cadherin/Fc Chimera (E-Fc) in 1 x PBS (50 µg/µl) to prepare E-cadherin-coated beads. Beads were centrifuged at 3000 x g for 15 min, washed with 1 x HBSS (Hank’s Balanced Salt Solution) containing 1.3 mM CaCl2) buffer, again centrifuged and then re-suspended in 1 x HBSS. Beads with passively absorbed E-Fc were stored up to two weeks at 4 °C. To test E-Fc coupling efficiency, control and E-Fc-coupled beads were boiled in 4x NuPAGE LDS sample buffer (Thermo Scientific), supernatants were loaded on a 4 - 15% protein gel and E-Fc was detected by Western Blot using a rat monoclonal E-cadherin antibody (DECMA-1, Santa Cruz). For shearing E-Fc-coated beads over the surface of ectoderm aggregates, we developed a parallel plate device, consisting of a glass plate controlled by two piezo elements for nanometer-precision movements in y and z direction. The glass plate was assembled on top of a stage containing an inlet for a cell culture dish, which was mounted on a Leica SP5 TCS confocal microscope. Magnetic polysterene beads were assembled into elongated clusters (~100-200 beads mimicking the size and shape of the ppl in vivo) and held in position at the bottom of the glass plate by a fixed magnet on top of the glass plate. As reference points, small fluorescent beads (4µm Tetraspeck Microshperes; Invitrogen) were absorbed to the glass plate to track the location and movement of the magnetic polysterene particles. For preparing ectoderm cell aggregates, MZoep mutant embryos, injected with H2A-mCherry (nuclei) and GPI-GFP (membrane) mRNA at 1-cell stage, were dissociated at 4-5 hpf in DMEM/F12 media mixture, and ectoderm cells were harvested and seeded in clusters on Fibronectin-coated glass dishes. Polysterene particles were positioned on top of the ectoderm cell cluster with slight indentation. To mimic the movements of ppl and ectoderm cells in vivo, ectoderm cells were moved continuously in one direction (~0.5 µm/min) and polysterene particles were sheared over the ectoderm cells (~1.5 µm/min) in the opposite direction, resulting in a comparable velocity ratio (1:3) as in wild type embryos. Velocity profiles and correlations were obtained from nuclei and bead positions and calculated similar to measured flow profiles in the embryo.
Whole-mount immunohistochemistry and antibodies
For whole-mount immunohistochemistry, embryos were fixed for 2 hours with 2% paraformaldehyde in 1x PBS, washed twice after fixation with 50 mM glycine in 1x PBS, 0.05% Triton-X, 0.05% Tween (PBSTT), and pre-blocked with 5% purified Bovine Serum Albumin (BSA, Sigma Aldrich, A9418) in PBSTT. For Fibronectin immunohistochemistry, embryos were fixed at 60%, 80% and 90% epiboly and Fibronectin was detected using a primary rabbit anti-fibronectin antibody (Sigma-Aldrich, F3648; 1/100 dilution). For E-cadherin immunohistochemistry, embryos were fixed at 75-80% epiboly and E-cadherin was detected using a primary rabbit antibody against zebrafish E-cadherin (antibody facility MPI-CBG Dresden; 1/200 dilution). Incubation with primary antibodies was performed overnight in PBSTT with 2% purified BSA at 4°C. Embryos were consequently washed with PBSTT 4 x for 30 min and incubated overnight with secondary antibody (Alexa 488-conjugated goat anti-rabbit, ThermoFisher Scientific, A-11008; 1/5000 dilution) and rhodamine-phalloidin for F-actin staining (ThermoFisher Scientific, R415; 1/200 dilution). Embryos were washed 4 x for 30 min with PBSTT and nuclei were stained with DAPI nuclei acid stain (ThermoFisher Scientific, D1306).
Preparation of histological sections
For tissue sectioning, pre-stained embryos were re-fixed in 2% PFA, embedded in increasing concentrations of OCT medium (Tissue-Tek® O.C.T. Compound, Sakura® Finetek; 0%-10%-30%-50%-70%-90% diluted in 30% sucrose/PBS), shock-frozen in 90% OCT solution on dry ice, and cryo-sectioned at thickness of 20 µm before embedding in ProLong Gold antifade mountant (ThermoFisher Scientific, P36930).
Calculations of tissue strain rates
Strain rates were calculated within the neurectoderm close to the animal-vegetal axis in the wild type (wt) and MZoep mutant embryos. To calculate tissue strain rates, the neurectoderm tissue was subdivided into small domains of 50 µm2 boxes in xy (~ 50 cells/domain/time point) and the velocity of cells within each box was averaged to calculate its instantaneous average ensemble. The strain rates were then calculated similar to a previous approach41, by using spatial derivatives of the velocities within the neighboring boxes along the animal-vegetal (AV) and lateral (LR) axes of the embryo, considering the adjacent domains. To distinguish direction and kind of strain, we calculated normal strain rates along the AV, LR axes and shear strain rates. Normal strain rates determine the stretch (positive value) or compression (negative value) across the tissue domain along a specific direction (AV or LR) and shear strain rates capture the change of angle when the tissue deforms, whereby the angle of the domain (unit of the tissue) can shrink (positive value) or enlarge (negative value) during deformation. Strain rates were calculated as follows:
where x and y are mathematical representations of the LR and AV axes, u and v are the velocities in these directions respectively and ε resembles strain rate.
Image and data processing for flow and correlation analysis
Images acquired from multiphoton live cell imaging were initially processed with ImSpector software (LaVison Bio Tec) to compile channels from imaging data, and the exported files were further processed using Imaris software (Bitplane) to visualize the recorded channels in 3D. Full data sets containing all the channels from live cell imaging were used for identifying embryo landmarks needed to align all embryos in the same way for comparison of different experiments. Each imaged embryo was rotated and aligned along the AV axis at the dorsal side of the embryo using the gsc:GFP, or in the case of MZoep mutant embryos, the dharma:EGFP signal as dorsal marker for correct embryo orientation. Ppl progenitor nuclei were extracted by surface masking of the gsc:GFP signal within prechordal plate progenitors. Neurectoderm cell nuclei were calculated from non-surface masked areas, and nuclei of remaining deep cells including paraxial mesoderm, endoderm and YSN were identified by their characteristic positions/movements and then manually subtracted. Nuclei positions of ppl progenitors and neurectoderm cells in xyz-dimensions were extracted for each time point and used for further quantitative analysis.
Cell tracking data containing nuclei positions over time were analyzed with custom made Perl scripts. From the 3D cell positions instantaneous velocity of a cell i at time t was calculated as follows:
where 𝒗i(t) is the velocity vector of cell i at time t and ri(t) is the position of cell i at time t and ∆t is the elapsed time between two consecutive 3D image set in the time lapse movie.
Analysis and visualization of ppl progenitor cell movements
In this section we delineate different analysis types that are based on methods generally used to quantify collective motion behavior43,44 to characterize collective cell migration.
To visualize how individual cell movements correlate with the average movement of the ppl cell collective, we calculated correlation values between the direction of movement of each individual cell and the average movement direction of the collective as follows:
where 𝒗̂i(t) is the normalized 3D velocity vector of cell i and
is the average normalized 3D velocity vector of the group at time t and N(t) is the number of ppl cells at time t. We calculated Ci(t) for every time point for each cell. Ci(t) can take values between 1 (if a cell movement is perfectly aligned with the average movement of the ppl collective) and -1 (if a cell is moving in the opposite direction of the collective). Correlation values of individual cells Ci(t) are indicated in images for a representative time point by the color of the velocity vectors (Supplementary Fig. 2b).
To quantify the alignment of cell movements, we defined the “order parameter” as the absolute value of the averaged normalized velocity as follows45:
where 𝒗̂i(t) is the normalized velocity of a cell and N(t) is the total number of cells at time t. 𝜑 (t) can take values between 0 (if cells move randomly, disordered movement) and 1 (if cells move uniformly in the same direction, highly ordered movement; Supplementary Fig. 2c).
Calculations and visualization of neurectoderm flows
To quantify and visualize neurectoderm cell motion without ppl progenitor cells, we calculated velocity flow maps in spatially defined areas of the embryo. Three dimensional cell velocity vectors were averaged in 50 by 50 µm2 sectors in xy planes and over the full z direction for every time point to create a grid covering the whole area of the visualized embryo. For visualization we plotted xy projections of the 3D velocity vectors for every time point and reconstructed a velocity flow map. Magnitude of the average velocity, namely
where N is the number of cells in a sector at time t, was calculated and is indicated by the color of the arrow in each sector. The center point of the grid was determined as the middle point of the imaged area.
To quantify and visualize neurectoderm cell motion with underlying ppl progenitor cells, we determined the center 0,0 point of the above-mentioned grid as the average position of the first 20 leading cells. We used this 0,0 grid position as reference point to compare the cell movements from different embryos. For each time point of image acquisition, the 0,0 grid position was superimposed to keep the reference point fixed for every time frame and the 50x50 µm2 sectors, containing the average instantaneous cell velocities, were back projected around this reference point (Fig. 2c, Supplementary Fig. 2e, f). This method allowed us to quantify cell velocities in the adjacent neurectoderm in a relative position to the leading ppl progenitor cells and to directly compare different data sets with each other.
Averaged neurectoderm velocity fields were calculated by making both time and ensemble averages for each experimental case. Time averages were taken over the whole duration of image acquisition (typically 120min) for wt, cyc morphant, slb morphant, CA-Mypt and MZoep mutant embryos and visualized as time-averaged neurectoderm flow velocity fields (Fig 2g).
Directional correlation analysis of neurectoderm and prechordal plate progenitor cell movements
In order to quantify the correlation between the movement of neurectoderm and underlying ppl progenitor cells, we calculated their directional correlation in every sector of the grid (grids were positioned the same way as described above) for each time point as follows:
where and are the normalized averaged 3D velocity vectors in a grid at time t of neurectoderm and ppl progenitor cells, respectively (Supplementary Fig. 2g). Cecto−pp (t) can take values between 1 (ectoderm and ppl progenitor cells move in the same direction) and -1 (they move in opposite directions). Cecto−pp (t) value of each grid sector is indicated in images and movies by the color of the sector (Fig. 2e, Supplementary Figure 2g, h).
To investigate the effect of ppl movement on neurectoderm cells that are positioned anterior to the leading edge of the ppl, we calculated the directional correlation between the average normalized velocity vector of the first 20 leading ppl cells as follows:
Cecto−leading(t) value of each grid sector is indicated in images and movies by the color of the sector (Fig. 1k).
Instantaneous speed, directionality and polarity analyses of ppl cells
Cell speed 𝑣cell was calculated from single cell trajectories with positions
Data were selected every frame to obtain instantaneous velocities
with
and tlag the time interval between successive frames. Instantaneous velocities values 𝑣 calculated from single trajectories were averaged to obtain the cell speed 𝑣cell (Supplementary Fig. 2d).
To calculate directionality values, single cell trajectories were split into segments of equal length (5 frames). Directionality indices for single trajectories were calculated via a sliding window as the ratio of start-to-end distance versus the summed distance between successive frames in a segment. Obtained directionality values were averaged over all segments in a single trajectory. This analysis yielded values between [0,1], with higher movement directionality closer to 1. (Supplementary Fig. 2d).
For the calculation of speed gradients along the DV axis bright-field images of embryos were obtained to measure the embryo radius RE. The value RE was used as an input parameter to fit (x,y,z) coordinates of pp cells to a sphere with radius RE to obtain the center of the embryo (x0, y0, z0). Ppl cell coordinates were shifted to the origin by linear translation
and transformed to spherical coordinates (x′ ,y′ ,z′) → (R,𝜃,𝜑). In this reference frame the DV axis is represented by the R coordinate (the AP axis and LR axis by 𝜃 and 𝜑 coordinates respectively). 2D velocity maps along the DV and AV axis were generated from instantaneous velocity values 𝑣(R,𝜃) calculated as described above and plotted at interpolated cell positions between consecutive frames (Fig. 6c, d). Speed gradients along the DV axis were obtained from instantaneous velocities 𝑣(R) and were binned and averaged to calculate mean instantaneous velocity values (Fig. 6e).
Direction of protrusion formation of transplanted ppl progenitor cells was analyzed for each movie in steps of 15-20 min with Fiji software using the angle measurement tool. Angle between detected protrusion and the AV axis was measured, where an angle of 0 ° corresponds to the animal and an angle of 180 ° to the vegetal pole, and a 90 and 270 degree angle for the right and left axis respectively. Angles were plotted in a polar plots using IgorPro software (WaveMetrics) (Supplementary Fig. 5d).
Quantification of neural plate positioning
After whole-mount in situ hybridization, the embryos were imaged using a dissecting stereoscope with Leica Imaging Application. Fiji software was utilized to fit a circle around the embryo and to measure the angle between the vegetal pole and the anterior border of the otx2 expression domain by using the built in angle tool. Box-whisker plots were generated to compare angles from different developmental stages (Supplementary Fig. 2m).
Statistical analysis and Reproducibility
Statistical analysis was performed using software Prism 5 (GraphPad) and R (Bell Laboratories). To compare the mean values, unpaired Student’s t-tests were used to calculate two-tailed P values for two groups comparison and Student’s t-tests with Benjamini-Hochberg correction for multiple group testing correction were used to calculate False Discovery Rate for each P value. The data meet the assumption of the tests and the variance is estimated to be similar between groups that are compared. We used a non-parametric unpaired test because we assume that the data follow a Gaussian distribution and that in the compared groups the individual values were not paired or matched with one another. All P values are reported within the figure legends. All n-values report biological replicates (embryos) if not stated otherwise. No statistical method was used to predetermine the sample size. The experiments were not randomized. The investigators were not blinded to allocating during experiments and outcome assessment. For interpreting linear regression slopes we performed an F-test to report the P value (two-tailed) testing the null hypothesis that the overall slope is zero (P>0.05). Where whisker box plots were used, the center of the box shows the median (the additional red dot shows the mean), the whiskers show the minimum and maximum measured value, respectively (capped by 1.5 interquartile range; values measured outside are shown as outliners). Representative stereoscope/confocal/multiphoton images of zebrafish embryos, or ex vivo isolated cells, or Western blot in Fig. 1a, c, f, Fig. 2a, b, i, j, Fig. 3a, g, m, s, Fig. 6a, b, Fig. 7a, b, and Suppl. Fig. 1a-e, Suppl. Fig. 2a, i, k, l, Suppl. Fig. 3a, e, i, j, Suppl. Fig. 4b, d, i, Suppl. Fig. 5b-d, i, j, Suppl Fig. 7a, b, l, and Suppl. Fig. 8a-c have been successfully repeated between 3 and 6 times.
Supplementary Material
Acknowledgements
We would like to thank R. Hauschild for technical assistance and the scientific service units at the IST Austria for continuous support. This work was supported by grants from the OMAA Ernst Mach Predoctoral Scholarship to Z.A., the Cancer Research UK to M.T., the Spanish Ministry of Economy and Competitiveness “Centro de Excelencia Severo Ochoa 2013-2017 and CERCA Programme/Generalitat de Catalunya to V.R., the European Union’s Seventh Framework Program/ERC Grant Agreement (306589) to B.H. and (COLLMOT project 227878) to T.V., the Francis Crick Institute which receives its core funding from Cancer Research UK (FC001317), the UK Medical Research Council (FC001317) and the Wellcome Trust (FC001317) to S.G. and G.S., and the Austrian Science Foundation (FWF): I930-B20 to C.-P.H.
Footnotes
Code availability. Relevant computational codes used for data processing are available from the authors on request.
Data availability. All data that support the conclusions in the study are available from the authors on reasonable request.
References and Notes
- 1.Lecuit T, Lenne P-F, Munro E. Force generation, transmission, and integration during cell and tissue morphogenesis. Annu Rev Cell Dev Biol. 2011;27:157–184. doi: 10.1146/annurev-cellbio-100109-104027. [DOI] [PubMed] [Google Scholar]
- 2.Heisenberg C-P, Bellaïche Y. Forces in Tissue Morphogenesis and Patterning. Cell. 2013;153:948–962. doi: 10.1016/j.cell.2013.05.008. [DOI] [PubMed] [Google Scholar]
- 3.Solnica-Krezel L, Sepich DS. Gastrulation: making and shaping germ layers. Annu Rev Cell Dev Biol. 2012;28:687–717. doi: 10.1146/annurev-cellbio-092910-154043. [DOI] [PubMed] [Google Scholar]
- 4.Wilson SW, Houart C. Early steps in the development of the forebrain. Dev Cell. 2004;6:167–181. doi: 10.1016/s1534-5807(04)00027-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Appel B. Zebrafish neural induction and patterning. Dev Dyn. 2000;219:155–168. doi: 10.1002/1097-0177(2000)9999:9999<::aid-dvdy1052>3.3.co;2-b. [DOI] [PubMed] [Google Scholar]
- 6.Woo K, Fraser SE. Order and coherence in the fate map of the zebrafish nervous system. Development. 1995;121:2595–2609. doi: 10.1242/dev.121.8.2595. [DOI] [PubMed] [Google Scholar]
- 7.Araya C, et al. Mesoderm is required for coordinated cell movements within zebrafish neural plate in vivo. Neural Dev. 2014;9:9. doi: 10.1186/1749-8104-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Araya C, Carmona-Fontaine C, Clarke JDW. Extracellular matrix couples the convergence movements of mesoderm and neural plate during the early stages of neurulation. Dev Dyn. 2016;245:580–589. doi: 10.1002/dvdy.24401. [DOI] [PubMed] [Google Scholar]
- 9.Gritsman K, et al. The EGF-CFC protein one-eyed pinhead is essential for nodal signaling. Cell. 1999;97:121–132. doi: 10.1016/s0092-8674(00)80720-5. [DOI] [PubMed] [Google Scholar]
- 10.Montero J-A, et al. Shield formation at the onset of zebrafish gastrulation. Development. 2005;132:1187–1198. doi: 10.1242/dev.01667. [DOI] [PubMed] [Google Scholar]
- 11.Latimer A, Jessen JR. Extracellular matrix assembly and organization during zebrafish gastrulation. Matrix Biology. 2010;29:89–96. doi: 10.1016/j.matbio.2009.10.002. [DOI] [PubMed] [Google Scholar]
- 12.Diz-Muñoz A, et al. Control of directed cell migration in vivo by membrane-to-cortex attachment. PLoS Biol. 2010;8:e1000544. doi: 10.1371/journal.pbio.1000544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dumortier JG, Martin S, Meyer D, Rosa FM, David NB. Collective mesendoderm migration relies on an intrinsic directionality signal transmitted through cell contacts. Proc Natl Acad Sci USA. 2012;109:16945–16950. doi: 10.1073/pnas.1205870109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heisenberg CP, et al. Silberblick/Wnt11 mediates convergent extension movements during zebrafish gastrulation. Nature. 2000;405:76–81. doi: 10.1038/35011068. [DOI] [PubMed] [Google Scholar]
- 15.Ulrich F, et al. Slb/Wnt11 controls hypoblast cell migration and morphogenesis at the onset of zebrafish gastrulation. Development. 2003;130:5375–5384. doi: 10.1242/dev.00758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hatta K, Kimmel CB, Ho RK, Walker C. The cyclops mutation blocks specification of the floor plate of the zebrafish central nervous system. Nature. 1991;350:339–341. doi: 10.1038/350339a0. [DOI] [PubMed] [Google Scholar]
- 17.Thisse C, Thisse B, Halpern ME, Postlethwait JH. Goosecoid expression in neurectoderm and mesendoderm is disrupted in zebrafish cyclops gastrulas. Dev Biology. 1994;164:420–429. doi: 10.1006/dbio.1994.1212. [DOI] [PubMed] [Google Scholar]
- 18.Behrndt M, et al. Forces driving epithelial spreading in zebrafish gastrulation. Science. 2012;338:257–260. doi: 10.1126/science.1224143. [DOI] [PubMed] [Google Scholar]
- 19.Krieg M, et al. Tensile forces govern germ-layer organization in zebrafish. Nat Cell Biol. 2008;10:429–436. doi: 10.1038/ncb1705. [DOI] [PubMed] [Google Scholar]
- 20.Weber GF, Bjerke MA, DeSimone DW. A mechanoresponsive cadherin-keratin complex directs polarized protrusive behavior and collective cell migration. Dev Cell. 2012;22:104–115. doi: 10.1016/j.devcel.2011.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morita H, et al. The physical basis of coordinated tissue spreading in zebrafish gastrulation. Dev Cell. 2017;40:354–366. doi: 10.1016/j.devcel.2017.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Babb SG, Marrs JA. E-cadherin regulates cell movements and tissue formation in early zebrafish embryos. Dev Dyn. 2004;230:263–277. doi: 10.1002/dvdy.20057. [DOI] [PubMed] [Google Scholar]
- 23.Kane DA, McFarland KN, Warga RM. Mutations in half baked/E-cadherin block cell behaviors that are necessary for teleost epiboly. Development. 2005;132:1105–1116. doi: 10.1242/dev.01668. [DOI] [PubMed] [Google Scholar]
- 24.Shimizu T, et al. E-cadherin is required for gastrulation cell movements in zebrafish. Mech Dev. 2005;122:747–763. doi: 10.1016/j.mod.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 25.Wessely O, De Robertis EM. Neural plate patterning by secreted signals. Neuron. 2002;33:489–491. doi: 10.1016/s0896-6273(02)00596-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cui C, Yang X, Chuai M, Glazier JA, Weijer CJ. Analysis of tissue flow patterns during primitive streak formation in the chick embryo. Dev Biology. 2005;284:37–47. doi: 10.1016/j.ydbio.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 27.Fleury V. Clarifying tetrapod embryogenesis by a dorso-ventral analysis of the tissue flows during early stages of chicken development. BioSystems. 2012;109:460–474. doi: 10.1016/j.biosystems.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 28.Rozbicki E, et al. Myosin-II-mediated cell shape changes and cell intercalation contribute to primitive streak formation. Nat Cell Biol. 2015;17:397–408. doi: 10.1038/ncb3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Voiculescu O, Bodenstein L, Lau I-J, Stern CD. Local cell interactions and self-amplifying individual cell ingression drive amniote gastrulation. Elife. 2014;3:e01817. doi: 10.7554/eLife.01817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tzima E, et al. A mechanosensory complex that mediates the endothelial cell response to fluid shear stress. Nature. 2005;437:426–431. doi: 10.1038/nature03952. [DOI] [PubMed] [Google Scholar]
- 31.He B, Doubrovinski K, Polyakov O, Wieschaus E. Apical constriction drives tissue-scale hydrodynamic flow to mediate cell elongation. Nature. 2014;508:392–396. doi: 10.1038/nature13070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Westerfield M. The zebrafish book. A guide for the laboratory use of zebrafish (Danio rerio) 4th edition. 2000. [Google Scholar]
- 33.Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203:253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- 34.Ryu SL, et al. Regulation of dharma/bozozok by the Wnt pathway. Dev Biology. 2001;231:397–409. doi: 10.1006/dbio.2000.0150. [DOI] [PubMed] [Google Scholar]
- 35.Doitsidou M, et al. Guidance of primordial germ cell migration by the chemokine SDF-1. Cell. 2002;111:647–659. doi: 10.1016/s0092-8674(02)01135-2. [DOI] [PubMed] [Google Scholar]
- 36.Compagnon J, et al. The notochord breaks bilateral symmetry by controlling cell shapes in the zebrafish laterality organ. Dev Cell. 2014;31:774–783. doi: 10.1016/j.devcel.2014.11.003. [DOI] [PubMed] [Google Scholar]
- 37.Rebagliati MR, Toyama R, Haffter P, Dawid IB. cyclops encodes a nodal-related factor involved in midline signaling. Proc Natl Acad Sci USA. 1998;95:9932–9937. doi: 10.1073/pnas.95.17.9932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Weiser DC, Row RH, Kimelman D. Rho-regulated myosin phosphatase establishes the level of protrusive activity required for cell movements during zebrafish gastrulation. Development. 2009;136:2375–2384. doi: 10.1242/dev.034892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.D'Amico LA, Cooper MS. Morphogenetic domains in the yolk syncytial layer of axiating zebrafish embryos. Dev Dyn. 2001;222:611–624. doi: 10.1002/dvdy.1216. [DOI] [PubMed] [Google Scholar]
- 40.Karlen S, Rebagliati M. A morpholino phenocopy of the cyclops mutation. Genesis. 2001;30:126–128. doi: 10.1002/gene.1046. [DOI] [PubMed] [Google Scholar]
- 41.Lele Z, Bakkers J, Hammerschmidt M. Morpholino phenocopies of the swirl, snailhouse, somitabun, minifin, silberblick, and pipetail mutations. Genesis. 2001;30:190–194. doi: 10.1002/gene.1063. [DOI] [PubMed] [Google Scholar]
- 42.Blanchard GB, et al. Tissue tectonics: morphogenetic strain rates, cell shape change and intercalation. Nat Methods. 2009;6:458–464. doi: 10.1038/nmeth.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vicsek T, Zafeiris A. Collective motion. Physics Reports. 2012 [Google Scholar]
- 44.Méhes E, Vicsek T. Collective motion of cells: from experiments to models. Integr Biol (Camb) 2014;6:831–854. doi: 10.1039/c4ib00115j. [DOI] [PubMed] [Google Scholar]
- 45.Szabó B, et al. Phase transition in the collective migration of tissue cells: experiment and model. Phys Rev E Stat Nonlin Soft Matter Phys. 2006;74:061908. doi: 10.1103/PhysRevE.74.061908. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.