A gibberellin 20-oxidase plays a pivotal role in transitioning the axillary meristem to a stolon or floral shoot and hence the trade-off between vegetative propagation and fruit yield in strawberry.
Abstract
Asexual and sexual reproduction occur jointly in many angiosperms. Stolons (elongated stems) are used for asexual reproduction in the crop species potato (Solanum tuberosum) and strawberry (Fragaria spp), where they produce tubers and clonal plants, respectively. In strawberry, stolon production is essential for vegetative propagation at the expense of fruit yield, but the underlying molecular mechanisms are unknown. Here, we show that the stolon deficiency trait of the runnerless (r) natural mutant in woodland diploid strawberry (Fragaria vesca) is due to a deletion in the active site of a gibberellin 20-oxidase (GA20ox) gene, which is expressed primarily in the axillary meristem dome and primordia and in developing stolons. This mutation, which is found in all r mutants, goes back more than three centuries. When FveGA20ox4 is mutated, axillary meristems remain dormant or produce secondary shoots terminated by inflorescences, thus increasing the number of inflorescences in the plant. The application of bioactive gibberellin (GA) restored the runnering phenotype in the r mutant, indicating that GA biosynthesis in the axillary meristem is essential for inducing stolon differentiation. The possibility of regulating the runnering-flowering decision in strawberry via FveGA20ox4 provides a path for improving productivity in strawberry by controlling the trade-off between sexual reproduction and vegetative propagation.
INTRODUCTION
Asexual and sexual reproduction are inseparable in the life history of plants and take place jointly in a large number of angiosperms (Abrahamson, 1980). Asexual reproduction produces offspring that are genetically identical to the parent. A high diversity of mechanisms is involved in this process, including the production of tubers, rhizomes, corms, bulbs, and stolons (Klimeš et al., 1997). In natural populations, clonal reproduction likely provides ecological and evolutionary benefits to flowering plants (Vallejo-Marín et al., 2010). Natural vegetative propagation has also been harnessed for food production in several crop species. A major representative of these crop species is potato (Solanum tuberosum), in which tubers are storage organs derived from an underground stolon (elongated stem). In strawberry (Fragaria spp), a major fleshy fruit-bearing crop that undergoes inbreeding depression (Darrow, 1929; Kaczmarska et al., 2016), daughter plants or ramets (clonal plants) produced from an aerial stolon (runner) are essential for the clonal propagation of commercial varieties. Notably, vegetative reproduction can occur at the expense of fruit yield (Barrett, 2015), which depends on the number of inflorescences in the plant and on the duration of the flowering period (Costes et al., 2014). Thus, new insights into the mechanisms underlying the plant decision to produce either stolons or shoots with inflorescences are crucial for improving strawberry productivity.
Strawberry is an herbaceous perennial crop species from the Rosaceae family. Natural perpetual flowering (PF) mutants in which flowering occurs all along the vegetative cycle instead of once a year in spring (seasonal flowering [SF]) have been identified in cultivated (F. x ananassa) and woodland (F. vesca) strawberries (Iwata et al., 2012; Gaston et al., 2013). This trait allows the flowering period to be extended and increases fruit yield. In F. vesca, this phenotype is caused by the recessive tfl1 mutation (Brown and Wareing, 1965) in TERMINAL FLOWER1 (FveTFL1), a floral repressor gene (Iwata et al., 2012). In this species, a runnerless recessive mutant (r) has been discovered (Brown and Wareing, 1965) and the locus was mapped onto linkage group II (Sargent et al., 2004). In F. x ananassa, we recently showed that the genetic control of these two major characters is different from that in F. vesca and that the dominant PFRU mutation has an opposite effect on flowering and runnering (Gaston et al., 2013; Perrotte et al., 2016a). The basis for this antagonism is poorly understood. In the two species, stolons are produced from axillary meristems (AXMs) and this process is regulated by various endogenous and environmental factors including age, daylength, temperature, and hormones (Hytönen and Elomaa, 2011). To date, the molecular mechanisms underlying the differentiation of an AXM to a stolon remain elusive.
To search for genetic factors controlling stolon induction from AXM, we investigated the F. vesca recessive r mutant. Here, we provide evidence that AXM fate is responsible for the trade-off between flowering and runnering using a population segregating for the r and tfl1 mutations. We further show, using classical and mapping-by-sequencing strategies, that a deletion in a gibberellin 20-oxidase (GA20ox) gene expressed in AXM and in the developing stolon underlies the runnerless [r] phenotype in F. vesca. In addition, we show that stolon formation can be rescued by the exogenous application of bioactive gibberellin (GA) onto the r mutant. This work highlights the pivotal role of a specific AXM-expressed GA20ox enzyme, which catalyzes a rate-limiting step in the synthesis of bioactive GAs, in the induction of stolon differentiation in AXM and therefore in the trade-off between vegetative propagation and flowering in strawberry.
RESULTS
The Regulation of Axillary Meristem Fate Governs the Trade-Off between Flowering and Runnering
Strawberry is a perennial rosette-forming herbaceous plant. The primary shoot or primary crown (PC) is composed of leaves and AXMs at the axils of leaves and is terminated by an inflorescence (Figure 1A). The fate of the AXM depends on its location in the plant. When the shoot apical meristem (SAM) becomes floral, the AXM at the axil of the uppermost leaf below the terminal inflorescence develops into a shorter secondary shoot or branch crown (BC), leading to sympodial branching. AXMs along the PC can develop into either a BC (Sugiyama et al., 2004) or a stolon, which is a specialized and highly elongated stem (Savini et al., 2008), or stay dormant. This fate is controlled by genotypic and environmental factors (Hytönen et al., 2008, 2009).
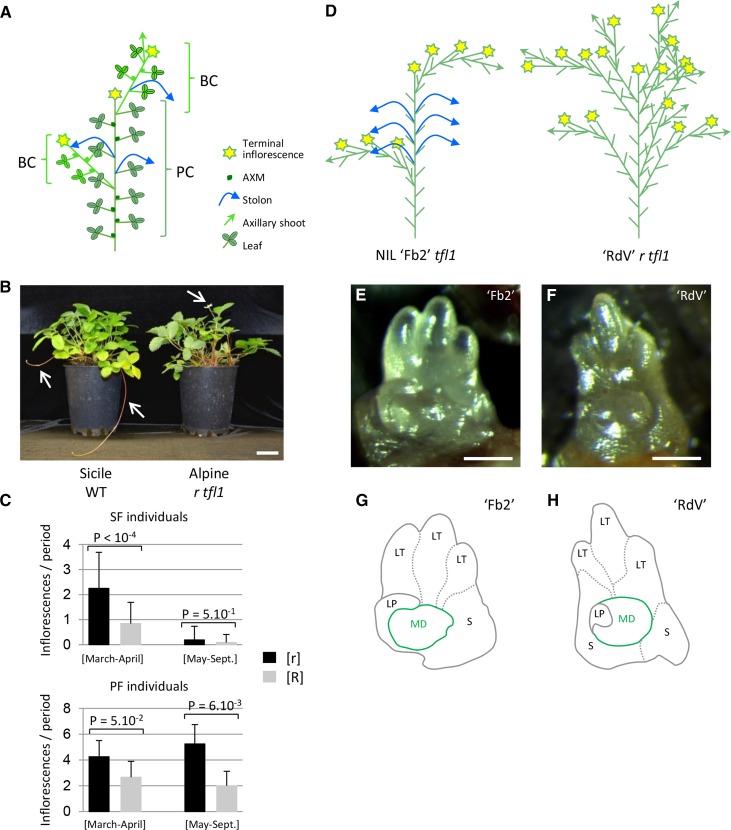
Figure 1.
r Mutant Phenotype and Flowering/Runnering Trade-Off in F. vesca.
(A) Architecture of strawberry plant. The primary shoot or PC is composed of leaves, with AXMs at the axils of leaves and is terminated by an inflorescence. Along the PC, AXMs can develop in either BCs or stolons or stay dormant. Green arrow, axillary shoot; blue arrow, stolon; yellow star, inflorescence.
(B) The ‘Sicile’ wild type (WT) develops stolons typical of a wild-type plant ([R] phenotype). ‘Alpine’ is a natural runnerless r mutant ([r] phenotype) carrying the tfl1 perpetual flowering mutation. Arrows, stolons and flower.
(C) Flowering/runnering trade-off in F. vesca. Runnering reduces the number of inflorescences in both seasonal [SF] and perpetual flowering [PF] individuals. Data are shown for the second year of phenotyping. n = 18, 80, 5, and 9 for [SF r], [SF R], [PF r], and [PF R], respectively, ±sd. Mann-Whitney test is considered statistically significant at P < 0.05.
(D) Architecture of ‘Reine des Vallées’ (‘RdV’) r tfl1 and NIL ‘Fb2’ tfl1. In ‘Fb2’, AXMs produce either stolons (blue arrows) or BCs (green arrows) terminated by an inflorescence (star). In ‘RdV’, almost all AXMs produce BCs terminated by inflorescences, leading to a bushy plant. Short green line represents developed leaf. Three 5.5-month-old plants were analyzed per genotype.
(E) to (H) AXMs of ‘Fb2’ tfl1 (E) and ‘RdV’ r tfl1 (F) and their schematic drawing ([G] and [H]). LT, leaflet; LP, leaf primordium; S, stipule; MD, meristematic dome.
Bars = 5 cm in (B) and 100 µm in (E) and (F).
Wild-type woodland strawberries such as ‘Sicile’ produce stolons and are seasonal flowering [SF] (Figure 1B). The ‘Alpine’ r mutant fails to produce stolons. In addition, ‘Alpine’ carries the natural tfl1 allele conferring a PF phenotype (Iwata et al., 2012; Koskela et al., 2012) and therefore flowers continuously from spring to late fall. In [SF] genotypes, floral induction at the SAM is triggered in fall by short days and by low temperature (Heide et al., 2013) and, consequently, the inflorescences emerge in the following spring (Perrotte et al., 2016b). We crossed ‘Alpine’ r tfl1 with ‘Sicile’ wild type to produce the ‘Ilaria_F2’ population, which segregated for both flowering and runnering. Segregation for runnerless [r] (≤3 stolons per plant per year) and runnering [R] (>3 stolons per plant per year) phenotypes was consistent with the 3:1 ([R]/[r]) ratio (Supplemental Figure 1) expected for the recessive r mutation (Brown and Wareing, 1965). Segregation for [PF] was consistent with the 3:1 ([SF]/[PF]) ratio expected for the recessive tfl1 mutation (Iwata et al., 2012). In the second year after planting of the ‘Ilaria_F3’ population, [SF] individuals flowered in March-April, while [PF] individuals flowered continuously from March to September. In both [SF] and [PF] subpopulations, we clearly observed an increase in the number of inflorescences produced in the r background (Figure 1C) that is reminiscent of the trade-off controlled by the single PFRU locus in cultivated strawberry F. x ananassa (Gaston et al., 2013). These results further suggest that the genetic regulation of AXM fate as either stolons or BCs bearing inflorescences is central for controlling strawberry productivity.
We therefore examined the fate of AXMs in the r and wild-type genotypes. For this purpose, we introgressed the runnering wild-type allele from F. bucharica into F. vesca ‘Reine des Vallées’ (‘RdV’) r tfl1 to create the near-isogenic line (NIL) ‘Fb2:39-63’ (‘Fb2’) tfl1 (Urrutia et al., 2015). In ‘Fb2’, AXMs at the 6th leaf and above produced stolons, while the uppermost AXM produced a BC. In ‘RdV’, all AXMs produced BCs or remained dormant (Figure 1D). The same effect of the r mutation on plant architecture was observed in ‘Alpine’ r tfl1 when compared with ‘Sicile’ wild type (Supplemental Figure 2). We further examined AXMs in more detail in ‘RdV’ and ‘Fb2’ (Figures 1E to 1H). Regardless of their fate, AXM morphology and shape were similar between ‘Fb2’ and ‘RdV’. Thus, in the r mutant, the fate of an AXM that does not produce stolon is to remain dormant or to generate a BC terminated by an inflorescence. This is likely the cause of the trade-off between runnering and flowering and, thus, of the differences in productivity observed in the ‘Ilaria_F3’ population segregating for the r mutation (Figure 1C).
Homozygous Deletion in FveGA20ox4 Is Strictly Linked to the [r] Phenotype
The R (RUNNERING) locus carrying the r mutation was previously mapped onto LGII within a 989-kb range (Sargent et al., 2004) (Figure 2A). Using microsatellites in the ‘Ilaria_F2’ population enlarged with 1350 additional individuals, we mapped the r mutation and reduced the region to 87.5 kb (Figure 2A). Whole-genome sequencing (WGS) (Supplemental Table 1) of pools of [r] recombinants (homozygous for runnerless) allowed the identification of 23 SNPs/InDels linked to the r mutation in a 63.28-kb region. We further checked the status of these SNPs/InDels in two genotypes, i.e., F. vesca displaying either [R] (‘Pawtuckaway’) or [r] (‘Baron Solemacher’) phenotypes, using genome sequence data available in a public database (Jung et al., 2014), hypothesizing a single origin for the r mutation in the available F. vesca germplasm. We made this assumption because [r] has been used for decades in F. vesca breeding, like [PF] (Iwata et al., 2012). Only one deletion (DEL) and 10 SNPs are linked to the r mutation, encompassing a 59.11-kb region containing eight genes (Figures 2B and 2C; Supplemental Table 2). To further reduce the R locus, we used [R] heterozygous recombinants. We investigated the heterozygosity of the R locus through phenotyping of seedlings issued from selfing. Among the [R] recombinants, one named alpha5-58 was found to be informative because it was heterozygous for runnering, according to the phenotype of its progeny, and it recombined just after the DEL in FveGA20ox4 (Figure 2D). By combining phenotyping and genotyping data from the 26 F3 lines issued from selfing, we unambiguously identified the DEL at position 25,536,553 on chromosome 2 as the r mutation; it is located in a GA20ox gene (cited as FveGA20ox4; Mouhu et al., 2013) (gene09034) (Figures 2D and 2E). DEL in FveGA20ox4 is an in-frame 9-bp deletion located in the second exon of the gene (Figure 2E) leading to the production of a shorter protein (ΔFvega20ox4) missing the Cys268Val269Lys270 amino acids (Figure 2F).
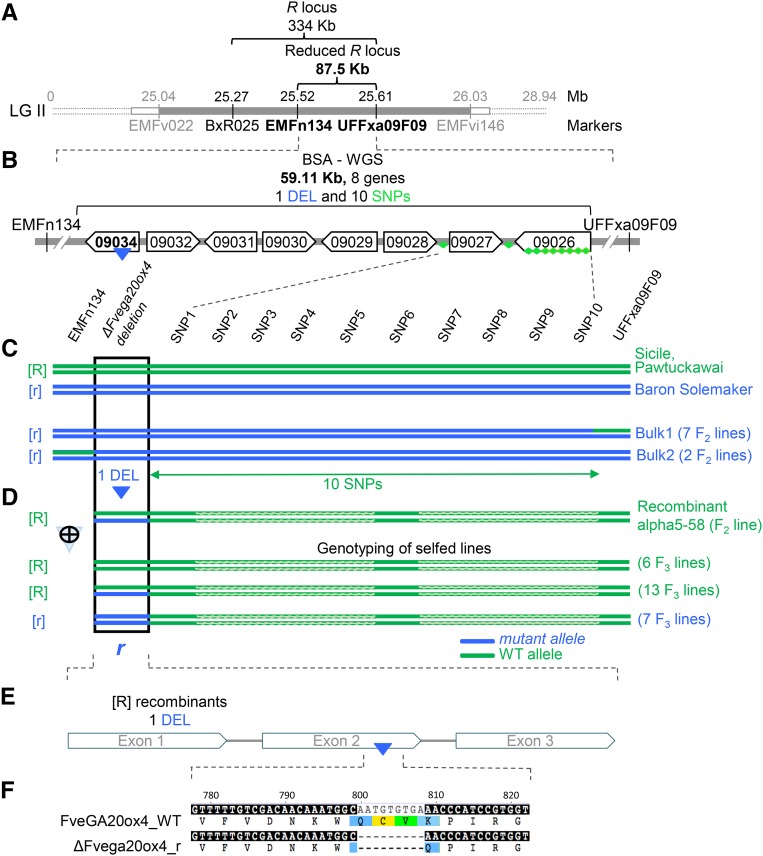
Figure 2.
Mutation in FveGA20ox4 Underlies the R Locus.
(A) Map-based cloning of the R locus (in gray) in linkage group 2 (LGII) (Sargent et al., 2004). The R locus was reduced to 334 kb using microsatellite markers (in black) and to 87.5 kb by recombinant analysis of 1350 individuals.
(B) R locus reduced to 59.11 kb by bulk segregant analysis-WGS. This region includes eight genes. Of the 11 SNPs/InDels linked to the runnerless phenotype identified, one deletion (DEL, blue triangle) is in a predicted GA20ox (gene09034) and 10 SNPs (green diamonds) are either in intergenic regions or in predicted genes (Supplemental Table 2).
(C) Identification of the r mutation responsible for the runnerless trait by screening a large number of recombinant lines (1350 individuals) from the Ilaria F2 population to identify runnerless ([r] phenotype) lines recombining between the two markers (EMFn134 and UFFxa09F09) flanking the r mutation. The nine recombinant runnerless lines identified were grouped into two bulks according to the position of the breaking point: left to marker UFFxa09F09 (Bulk1 with seven F2 lines) and right to marker EMFn134 (Bulk2 with two F2 lines). WGS of these bulks and of Sicile (runnering [R] phenotype) (25× to 35× genome coverage) allowed the identification of all polymorphisms (23 SNPs/InDels) in the chromosomal region carrying r. Comparison with genome sequences of ‘Pawtuckawai’ ([R] phenotype) and ‘Baron Solemacher’ ([r] phenotype) available at https://www.rosaceae.org/ further allowed the reduction of the candidate polymorphisms to 11 SNPs/InDels. The deletion (DEL) in gene FveGA20ox4 was the most likely candidate due to the function of the protein.
(D) Unequivocal identification of DEL in FveGA20ox4 as the causal mutation. Recombinant lines were screened to identify one runnering line ([R] phenotype) named alpha5-58, which was heterozygous at the FveGA20ox4 locus and recombined just after the DEL in FveGA20ox4. After selfing, 26 F3 lines were obtained, seven of which displayed a runnerless [r] phenotype. All seven lines were homozygous for the DEL in FveGA20ox4, recombined after this mutation, and were homozygous wild type thereafter. The 10 SNPs carried by the chromosomal segment after the DEL in FveGA20ox4 were therefore excluded as candidate polymorphisms. The recessive DEL in FveGA20ox4 (ΔFvega20ox4) was confirmed as the causal mutation for the runnerless [r] phenotype.
(E) DEL of nine nucleotides occurs in exon 2 of GA20ox (gene09034).
(F) A shorter protein with a deletion of three amino acids (Cys268Val269Lys270) is produced in the r mutant (ΔFvega20ox4_r) in comparison with FveGA20ox4_WT.
The unequivocal genetic evidence that FveGA20ox4 controls the decision of the AXM to generate a stolon correlates well with existing data showing that exogenously applied bioactive gibberellins (GA3) affect stolon production in strawberry (Thompson and Guttridge, 1959; Hytönen et al., 2009). We further checked its genetic status in the Ilaria_F2 population (Figure 3A) and confirmed that all [r] individuals were homozygous for the deleted allele, ΔFvega20ox4. Plants homozygous or heterozygous for the wild-type allele were [R] (Figure 3A). Additionally, we analyzed 25 [r] and [R] accessions of F. vesca from different geographical origins to determine if runnering depends on the allelic status of FveGA20ox4 (Figure 3B) and if the genetic structure of the population is linked to the runnering trait (Figure 3C). In all 25 accessions, the runnering behavior was consistent with the FveGA20ox4 genotype: A runnering [R] phenotype was associated with a homozygous wild-type allele and runnerless [r] was associated with a homozygous mutant allele (Figure 3B). We further analyzed genome-wide population structure using Structure (Pritchard et al., 2000). The ΔK statistic (Evanno et al., 2005), designed to identify the most relevant number of clusters (K) in a population, was the highest for K = 2. By using a membership probability threshold (Q-value) of 0.7, all accessions were clearly assigned to one group that corresponds to the FveGA20ox4 allelic status R or r (Figure 3C). This result suggests that the r mutation contributed to the genetic structure of these accessions. Because all runnerless mutants are arranged in a single cluster, these results are in agreement with the single origin of the mutation.
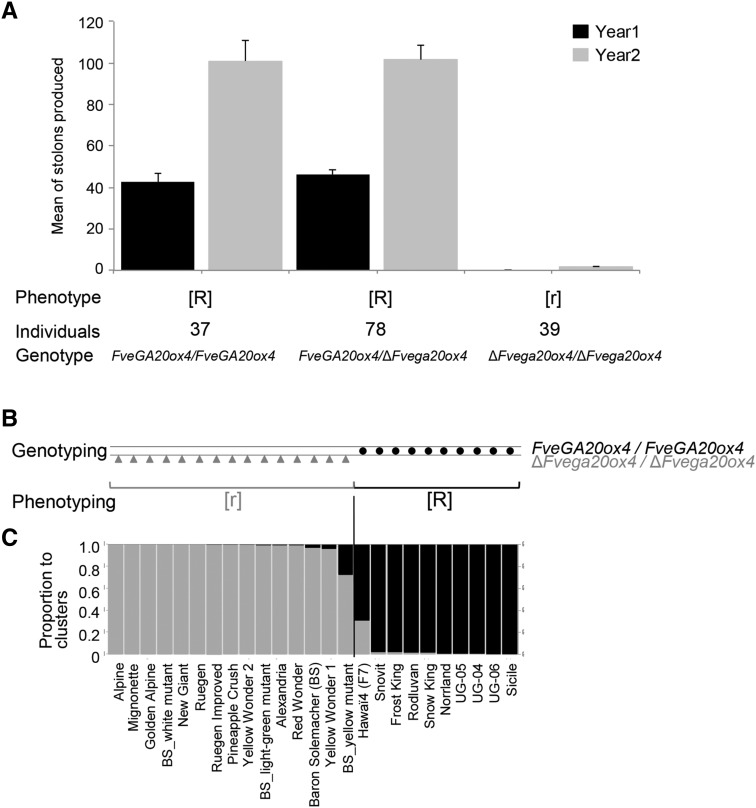
Figure 3.
Distribution of r Mutation in Ilaria_F2 Segregating Population and in F. vesca Germplasm.
(A) Allelic status of FveGA20ox4 in the Ilaria_F2 segregating population. The 154 plants were scored as runnering [R] or runnerless [r] when the number of stolons was >3 or ≤3, respectively. The FveGA20ox4 wild-type allele confers [R] phenotype when present in simple or double dose. The ΔFvega20ox4 mutant allele confers [r] phenotype when presents in double dose. Error bars show se.
(B) Genotyping of FveGA20ox4 and phenotyping of runnering. FveGA20ox4 is the wild-type gene, and ΔFvega20ox4 is the deleted mutant version. Circle, homozygous FveGA20ox4/FveGA20ox4; triangle, homozygous ΔFvega20ox4/ΔFvega20ox4. [R], runnering phenotype; [r], runnerless phenotype.
(C) Genetic structure of 25 F. vesca accessions. Genetic structure was determined by examining 37 microsatellite loci distributed throughout the genome. Each accession is represented by a single vertical line, which is partitioned into gray and black segments in proportion to the estimated membership in the two clusters (K = 2).
FveGA20ox4 Is Strongly Expressed in the AXM
Strawberry GA20ox predicted on the basis of their homology with known GA20ox include FveGA20ox1, FveGA20ox2, FveGA20ox3, and FveGA20ox4, as well as the more distantly related FveGA20ox5 (Kang et al., 2013; Mouhu et al., 2013). Phylogenetic analysis indicated clustering of FveGA20ox4 with AtGA20ox from Arabidopsis thaliana (Supplemental Figure 3A). In the ‘Fb2’ wild-type and ‘RdV’ r genotypes, FveGA20ox2 and FveGA20ox5 were preferentially expressed in the PC, FveGA20ox3 was expressed in all vegetative aerial organs (Supplemental Figure 4), and FveGA20ox1 was not detected. In contrast, FveGA20ox4 was strongly and preferentially expressed in the stolon (Figures 4A and 4B). In situ hybridization of ‘Fb2’ wild type detected transcript accumulation in the AXM dome as well as the leaf primordia and leaflet (Figures 4E to 4H), but not in the SAM (Figures 4C and 4D).
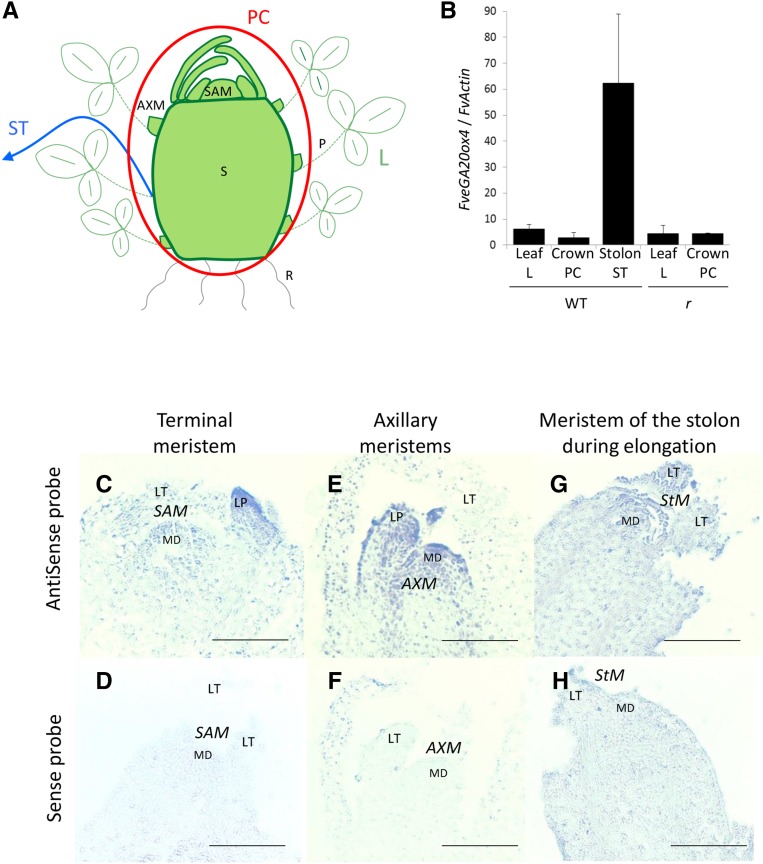
Figure 4.
FveGA20ox4 Transcripts Are Localized in the Axillary Meristem and Stolon.
(A) Tissue sampling of PC (in red circle), leaf (L), and stolon (ST; blue arrow). PC includes stem (S), SAM, and AXMs. R, roots.
(B) Relative expression of FveGA20ox4 in leaf, PC, and stolon of ‘Fb2’ wild type and ‘RdV’ r mutant. Normalization with FveActin. n = 3 ± se.
(C) to (H) In situ hybridization of FveGA20ox4 on the ‘Fb2’ tfl1 SAM ([C] and [D]), AXM ([E] and [F]), and meristem of the stolon during elongation ([G] and [H]). Upper panel of (C), (E), and (G), antisense probe; lower panel of (D), (F), and (H), sense probe. Samples were processed side-by-side with sense and antisense probes. MD, meristematic dome; LT, leaflet; LP, leaf primordium; ST, stolon; StM, stolon meristem. Bar = 0.1 mm.
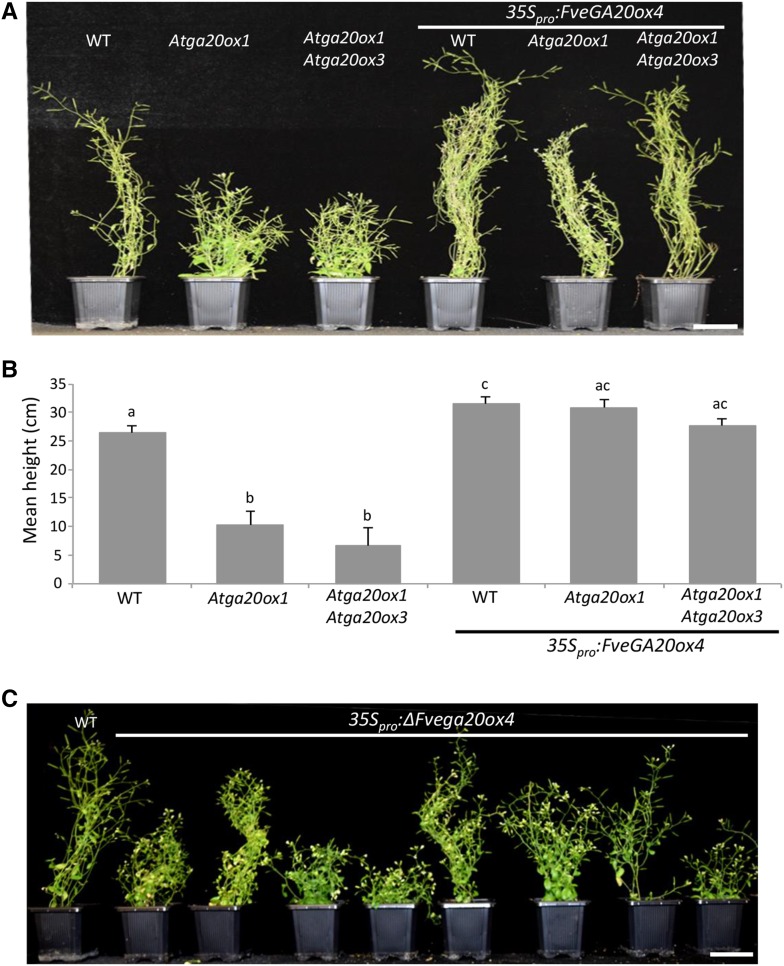
We overexpressed FveGA20ox4 in the Arabidopsis Atga20ox1 single mutant and in the Atga20ox1 Atga20ox3 double mutant, which display a dwarf phenotype (Phillips et al., 1995; Rieu et al., 2008; Plackett et al., 2012), to further explore its functional role. The expression of FveGA20ox4 rescued the dwarf phenotype of both the single and double mutants and increased plant height in the wild type (Figures 5A and 5B), thus validating in planta the predicted gibberellin biosynthesis function of FveGA20ox4. The overexpression of ∆Fvega20ox4 mostly produced dwarf phenotypes in the wild-type background and was unsuccessful in the single and double mutant background, i.e., no seed germination (Figure 5C), suggesting that the mutated protein has a dominant-negative effect.
Figure 5.
Overexpression of FveGA20ox4 in Dwarf Atga20ox Arabidopsis Mutants Restores Plant Growth, Whereas Overexpression of ΔFvega20ox4 Causes a Dwarf Phenotype in the Wild Type.
(A) and (B) Arabidopsis Colombia-0 (WT), Atga20ox1 single mutant, and Atga20ox1 Atga20ox3 double mutant overexpressing FveGA20ox4.
(A) Representative image of controls and transgenic plants transformed with 35Spro:FveGA20ox4. Bar = 5 cm.
(B) Height of the transgenic Arabidopsis lines overexpressing FveGA20ox4. n = 6 for controls and 10 for transgenic plants ±se. Values with different letters differ significantly (Mann-Whitney test) (P < 0.05).
(C) Arabidopsis Colombia-0 (WT) overexpressing ΔFvega20ox4. Representative image of controls and transgenic plants transformed with 35Spro:ΔFvega20ox4. Bar = 5 cm.
Because FveGA20ox4 transcript abundance was not affected in the r mutant (Figure 4B), we further investigated whether the three amino acid deletion in FveGA20ox4 protein was responsible for the loss of gibberellin biosynthetic activity. Comparison of FveGA20ox4 with other GA20ox from Arabidopsis indicated that the deletion was within the predicted catalytic domain of the enzyme (Lange et al., 1997; Huang et al., 2015), thus possibly affecting its activity (Supplemental Figure 3B). GA20ox enzymes catalyze several reactions in the GA biosynthetic pathway to produce precursors that are further converted into bioactive GAs by subsequent enzymes in the pathway (Figure 6A). We produced recombinant FveGA20ox4 and ΔFvega20ox4 proteins and showed that, while wild-type FveGA20ox4 converted [14C] GA12 to [14C] GA9 (Figure 6B), as do most other GA20ox enzymes analyzed to date (Pimenta Lange et al., 2013), the mutated ΔFvega20ox4 enzyme was not able to convert the GA12 substrate, indicating that the deleted version of the protein was not functional (Figure 6C).
Figure 6.
Deletion Leads to a Loss of FveGA20ox4 Catalytic Activity.
(A) Simplified GA biosynthetic pathway. Intermediates (blue), bioactive (green), and inactive (red) GAs are represented for the last steps of the pathway. Enzymes are framed and colored according to the product of the reaction.
(B) and (C) HPLC-radiochromatograms of products from incubations of recombinant FveGA20ox4 wild type (B) and ΔFvega20ox4 mutant (C) proteins with 14C-labeled GA12. Identities of substrate and product were confirmed as methyl esters trimethylsilyl ethers by gas chromatography-mass spectrometry by comparison of their mass spectra with published spectra.
Gibberellin Regulates AXM Fate
Because of the preferential transcript accumulation of FveGA20ox4 in AXM and in the meristem of the stolon during elongation (Figures 4E and 4G) and the effect of its loss of function on AXM fate, we investigated the endogenous GA levels in stolon, leaf, and PC tissue from two runnering genotypes (‘Sicile’ and ‘Fb2’) and, for comparison, in leaf and PC tissue from two runnerless genotypes (‘Alpine’ and ‘RdV’). The 13-hydroxylated pathway, which leads to the production of the biologically active form GA1 and of the inactive catabolic forms GA29 and GA8 (Hedden and Thomas, 2012) (Figure 6A), is the predominant GA biosynthetic pathway in strawberry (Taylor et al., 1994). GA1 levels were higher in the leaves than in BCs and stolons (Supplemental Table 3). Strikingly, GA8 accumulated strongly in the stolon, supporting the hypothesis of high GA20ox activity driven by the strong transcript accumulation of FveGA20ox4 in this organ and further suggesting that the stolon should also have high GA 3-oxidase activity to produce GA1 from GA20 and high GA 2-oxidase activity to fine-tune GA1 levels via GA1 to GA8 catabolism (Hedden and Thomas, 2012).
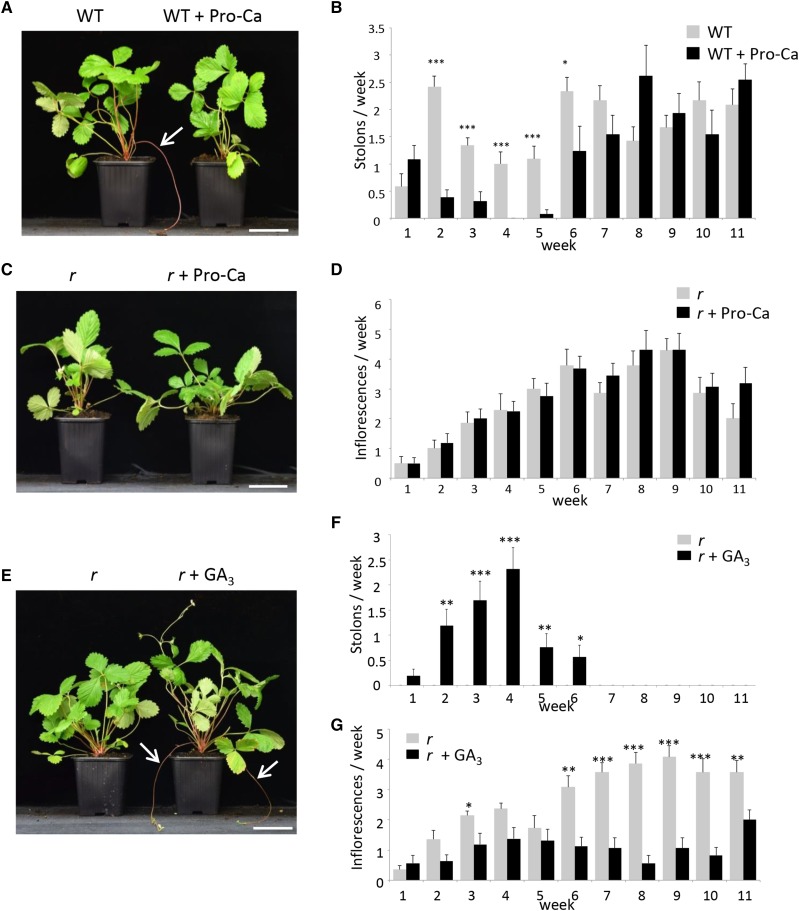
As expected from these results and from previous reports (Hytönen et al., 2009), treatment of a wild-type genotype (‘Rodluvan’) in the spring with prohexadione-calcium (Pro-Ca), an inhibitor of GA oxidases (Rademacher, 2000), led to the severe reduction and eventually (4 weeks after treatment) the arrest of stolon emergence (Figures 7A and 7B). Stolon production resumed thereafter to its normal level 5 weeks after the last treatment. The number of inflorescences produced was unaffected in the Pro-Ca-treated ‘Alpine’ r tfl1 mutant (Figures 7C and 7D). Conversely, treatment of the ‘Alpine’ r tfl1 mutant with the bioactive gibberellin GA3 restored stolon production (Guttridge and Thompson, 1964) to levels similar to those of the wild type at 4 weeks after treatment, after which the effect of GA3 vanished (Figures 7E and 7F). Conversely, GA3 treatment had a significant negative effect on the production of inflorescences (Figure 7G).
Figure 7.
Treatment of the Wild Type and the r Mutant with GA Inhibitor (Pro-Ca) and Bioactive GA.
(A) and (B) Treatment of the wild type genotype with Pro-Ca.
(A) Representative image showing stolon production (white arrow) on week 3. Bar = 6.5 cm.
(B) Pro-Ca treatment significantly reduces the number of stolons produced between weeks 2 and 6. n = 13 or 12 ± se.
(C) and (D) Treatment of the r mutant with Pro-Ca.
(C) Representative image on week 3. Bar = 6.5 cm.
(D) Pro-Ca treatment has no effect on the number of inflorescences. n = 16 or 14 ± se.
(E) to (G) Treatment with bioactive GA restores runnering in the r mutant.
(E) Representative image showing stolon production (white arrows) in the r mutant after GA3 treatment on week 3. Bar = 6.5 cm.
(F) Quantification of stolon production after GA3 treatment. Mock-treated plants never produce stolon.
(G) Quantification of inflorescence production after GA3 treatment. n = 16 and 14 for GA-treated and mock-treated plants ± se. Treatments were performed twice a week during the first two weeks (last treatment on week 1). Asterisks indicate significant differences (Mann-Whitney test): *P < 0.05, **P < 0.01, and ***P < 0.001.
DISCUSSION
In plant species displaying both sexual reproduction and vegetative propagation through stolons, such as strawberry, the AXM is indeterminate and can produce a stolon or a secondary shoot and inflorescence. In this work, we show that, in the diploid strawberry F. vesca, a member of the GA20ox family (which catalyzes the rate-limiting steps in bioactive GA biosynthesis) is specifically expressed in the AXM where it plays a pivotal role in the decision of the meristem to produce either a stolon or an inflorescence-bearing shoot. Depending on the allelic state of FvGA20ox4, the AXM either produces a stolon (active allele), remains dormant, or produces a secondary branch crown terminated by an inflorescence (inactive allele). The resulting modification in strawberry architecture translates into a change in inflorescence number in both seasonal and perpetual flowering strawberry genotypes. We therefore identified a way to control the balance between flowering and runnering, both of which are essential for strawberry fruit production, by modifying a key enzyme that can switch propagation type from sexual to asexual reproduction.
Additionally, by studying the allelic status of the FvGA20ox4 gene in runnering and runnerless F. vesca varieties, we provide answers to old questions on the origin of the runnerless trait in cultivated woodland strawberry. Before the breeding of the cultivated hybrid octoploid strawberry Fragaria x ananassa in the 1750s via a cross between two American species, strawberry species of wild origin were widely cultivated throughout Europe. Among these was F. vesca. According to Duchesne (1766), the runnerless trait in woodland strawberry was first described by Furetiere in his Dictionnaire printed in 1690. Furetiere highlighted the rare occurrence of runnerless strawberry plants at that time. Remarkably, Duchesne (1766) already confirmed the genetic origin of the runnerless trait by analyzing more than 30 plants grown from seeds and further observed that runnerless plants had more branch crowns than standard plants. He recognized the F. vesca origin of the runnerless mutant, named it F. eflagellis, and indicated that it spread from a garden from Burgundy province in France to several gardens in the Paris area. The runnerless trait has been retained from early varieties until today (Darrow, 1966). More recent varieties such as ‘Reine des Vallées’ and ‘Baron Solemacher’ combining runnerless and perpetual flowering traits are still successfully cultivated. We show here that the runnerless trait found in these varieties and in the other runnerless F. vesca varieties studied has a single origin that goes back more than three centuries.
GAs are central regulators of many developmental processes (Depuydt and Hardtke, 2011; Davière and Achard, 2013). To date, the role of GA in shoot meristems has primarily been studied in the SAM. Bioactive GAs are produced in the primordia by the key GA biosynthetic enzymes GA20ox to enable organ differentiation and growth. Because GAs promote cell differentiation and SAM indeterminacy must be maintained, bioactive GAs are excluded from the meristematic dome through several mechanisms (Galinha et al., 2009; Veit, 2009). In the cells surrounding the meristematic dome, bioactive GAs can be converted to inactive GAs by GA 2-oxidase. Depending on the species, the photoperiod, and the meristem fate, both GA20ox and GA 2-oxidase genes are further regulated by several transcription factors including KNOX (Sakamoto et al., 2001; Hay et al., 2002; Rosin et al., 2003; Jasinski et al., 2005; Bolduc and Hake, 2009), SHORT VEGETATIVE PHASE, and SOC1 (Li et al., 2008; Tao et al., 2012; Andrés et al., 2014). By contrast, the regulation of the induction of stolon differentiation in the AXM by GAs remains poorly understood (Hytönen et al., 2009). In potato, exogenous application of bioactive GAs promotes the production of stolons from AXMs, while the inactivation of bioactive GAs reduces stolon emergence (Kumar and Wareing, 1974; Kloosterman et al., 2007). In strawberry, treatment with bioactive GAs in long days (LDs) inhibits flowering but enhances stolon production (Thompson and Guttridge, 1959; Hytönen et al., 2009); inhibition of GA biosynthesis has the opposite effect (Hytönen et al., 2009). One hypothesis is that GA precursors or bioactive GAs synthesized in the leaf under LD move to the AXM to promote stolon induction (Eriksson et al., 2006; Regnault et al., 2015). This idea is further supported by the finding that overexpressing FveSOC1 in F. vesca induces both stolon production and FveGA20ox4 transcript accumulation in the leaf (Mouhu et al., 2013).
Actually, our data demonstrate that FveGA20ox4 is strongly expressed in the AXM, whereas we detected weak transcript accumulation in other vegetative tissues. Moreover, transcriptome analysis of reproductive tissues at various developmental stages failed to detect FveGA20ox4 transcripts in expanding fruit (Kang et al., 2013). Stolon differentiation is not induced when FveGA20ox4 is inactivated, strongly suggesting that the bioactive GAs controlling stolon induction are specifically synthesized in the AXM and are not transported from the leaf to the AXM. Taking as model the photoperiodic activation of GA biosynthesis in the SAM during flowering in Arabidopsis (Andrés et al., 2014), which is a LD species unlike the short-day (SD) strawberry (Heide et al., 2013), it is tempting to speculate that, under inductive LD conditions, FLOWERING LOCUS T (Mouhu et al., 2013) upregulates SOC1 expression specifically in the strawberry AXM, where in turn it induces the high FveGA20ox4 expression required for stolon differentiation. Because FveGA20ox4 transcript accumulation is not restricted to the flanks of the AXM but also occurs in the meristematic dome, an additional level of control would be necessary to maintain a pool of undifferentiated cells in the meristem of the growing stolon. Such control could be fulfilled by GA 2-oxidase, which converts bioactive GA1 to the inactive GA8 that accumulates to high levels in the stolon.
Our data further demonstrate that the production of BCs is the default developmental program of the AXMs present in the PC. When FveGA20ox4 is inactivated, the AXM eventually shifts from stolon to BC production, which is consistent with previous observations on the fate of stolon tips under LDs (i.e., stolon production) and under SDs (i.e., BC production) (Hytönen et al., 2009). As shown here, this has considerable influence on strawberry productivity in both seasonal flowering and perpetual flowering strawberry genotypes. Due to strong inbreeding depression (Kaczmarska et al., 2016), it is necessary to produce daughter plants in order to vegetatively propagate strawberry varieties, hence excluding the commercial use of strict runnerless mutants. It is technically feasible to use bioactive GAs and GA inhibitors to modulate runnering/flowering (Hytönen et al., 2008), but such products are currently not registered for commercial use for vegetative propagation in many countries. The identification of FveGA20ox4 as a breeding target thus provides the opportunity to modulate daughter plant production/fruit yield in octoploid cultivated strawberry by screening strawberry genetic resources for weak alleles of FveGA20ox4 and introgressing them into commercial varieties via marker-assisted selection. In addition, the newly developed CRISPR/Cas9 system for gene editing, which is well adapted for use in polyploid species (Wang et al., 2014) and has already proved its utility for improving crop yields via altering plant architecture (Krieger et al., 2010; Park et al., 2014), can be used to target one or several FveGA20ox4 homoeoalleles of F. x ananassa and thus modulate the runnering/flowering balance in cultivated strawberry.
METHODS
Plant Material and Growth Conditions
Diploid strawberry (Fragaria vesca) genotypes were used in this study. Seeds were sown in a mixture of two-thirds loam and one-third grit and grown at 16-h/8-h day/night at 22°C/18°C using supplementary light at 150 µmol m−2 s−1 (high pressure sodium lamp; Philips 400W). Plants were transplanted to 1-liter pots containing the same mixture and were maintained in a greenhouse under natural conditions.
The wild-type ‘Sicile’ and ‘Rodluvan’ (wild type) (seasonal flowering [SF] and runnering [R] phenotypes) varieties and the ‘Alpine’ and ‘Reine des Vallées’ (‘RdV’) (perpetual flowering [PF] and runnerless [r] phenotypes) varieties, which carry the r mutation in the tfl1 background (r tfl1 double mutant), were used for plant architecture analysis, r mutation mapping, and/or hormonal treatments. For r mutation mapping, ‘Alpine’ was crossed with ‘Sicile’ to generate an F2 population of 154 individuals named Ilaria_F2. For fine mapping, 1350 additional Ilaria_F2 individuals were produced. The trade-off between flowering and runnering was analyzed in an F3 population of 112 individuals named Ilaria_F3 issued from selfing of one Ilaria_F2 individual. The segregation ratios of r and tfl1 in Ilaria_F2 and F3 were as expected for recessive mutations.
The runnerless ‘RdV’ r tfl1 genotype and the runnering NIL ‘Fb2:39-63’ (‘Fb2’) tfl1 genotype were used for plant architecture analysis, meristem imaging, in situ hybridization, and qRT-PCR. ‘Fb2’ tfl1 was obtained by introgression of the wild-type F. bucharica runnering locus into ‘RdV’. The 3.7-Mb introgression is located between positions 18.950.307 and 22.663.853 of F. vesca genome v1.1 (Urrutia et al., 2015).
Seeds of the Arabidopsis thaliana Atga20ox1 single mutant (SALK_016701C) and the Atga20ox1-3/Atga20ox3-1 double mutant (Rieu et al., 2008; Plackett et al., 2012) were provided by The Nottingham Arabidopsis Stock Centre. Columbia-0 (Col-0) ecotype was used as a control. Plants were grown under 16-h/8-h day/night at 22°C/18°C with 70/62% humidity in a growth chamber. Light level was 100 µmol m−2 s−1 (Philips 400W).
Plant Phenotyping
Phenotyping of the Ilaria_F2 and F3 populations was performed during two successive years after sowing. Seeds from Ilaria_F2 and F3 were respectively sown in October and March and plants were grown in LD conditions under 16-h/8-h day/night and placed in a greenhouse under natural conditions in April. To quantify stolons in the Ilaria_F2 and Ilaria_F3 populations, the stolons in each individual were counted weekly during two successive years and were removed after counting. In addition, to analyze the trade-off between inflorescence and stolon production in the Ilaria_F3 population, inflorescences in each individual were counted weekly from March to September during two successive years. Stolons and inflorescences were counted when they visually emerged. The seasonal flowering period occurred during March and April and was a consequence of floral initiation in the previous autumn (Perrotte et al., 2016b), whereas the perpetual flowering period occurred from May to September (end of phenotyping). The F. vesca genetic resources were phenotyped by the National Clonal Germplasm Repository (USDA) for the presence or absence of stolons during a 2-year period. To evaluate the impact of GA3 and Pro-Ca on inflorescence emergence and stolon production, the number of inflorescences and stolons was counted weekly until 11 weeks after treatment in ‘Rodluvan’ wild type and ‘Alpine’ r tfl1 at 3 months old.
Plant Architecture, Meristem Observation, and in Situ Hybridization
Plant architecture of ‘Sicile’ wild type, ‘Alpine’ and ‘RdV’ r tfl1, and ‘Fb2’ tfl1 was analyzed using 5.5-month-old plants grown in the greenhouse. The plants were dissected and the different organs, including leaves, inflorescences, and stolons, were organized according to their position on the PC and its BCs.
Meristems and emerging stolons from the PCs of 4-month-old ‘Fb2’ tfl1 and/or ‘RdV’ r tfl1 plants were dissected under a stereomicroscope for photography and in situ hybridization. For in situ hybridization, PCs including either the SAM or AXM with two young leaves were fixed according to standard protocols (Sicard et al., 2008). In situ hybridization experiments were performed as described previously (Bisbis et al., 2006). Digoxygenin-UTP-labeled antisense RNAs for exon 1 of FveGA20ox4 were transcribed with T7 RNA polymerase and used as probes. FveGA20ox4_ISH primers are used (forward, CATCAAAGCTTCATGCTCG; reverse, AGTATCTTCAAAACCATTCC, 5′-3′).
Map-Based Cloning of the r Mutation Followed by a Combination of Bulk Segregant Analysis and WGS Analyses
The R locus, which was previously mapped on LGII within a 989-kb region (Sargent et al., 2004), was first narrowed down to a 334-kb region between markers BxR025 (forward, TAACCGGAATCGGAGAGATG; reverse, ACAGCTTCATTTGCGCTTTT, 5′-3′) and UFFxa09F09 (Sargent et al., 2006) using new or previously developed (https://www.rosaceae.org) microsatellites in the Ilaria_F2 population of 154 individuals. For fine mapping, the Ilaria_F2 population was enlarged to 1350 additional individuals, which were screened for recombination between BxR025 and UFFxa09F09. Phenotyping and genotyping of the recombinants with new microsatellites allowed the R locus to be reduced to a region between EMFn134 and UFFxa09F09 (Sargent et al., 2006). Genotyping was performed using previously published procedures for DNA extraction (Qiagen) and microsatellite polymorphism identification (Till et al., 2003).
A strategy combining bulk segregant analysis and WGS was developed to reduce the number of SNPs/InDels linked to the r mutation. Nine homozygous [r] individuals recombining between markers EMFn134 and UFFxa09F09 were selected and pooled into two bulks with genotypes recombining either in the left part of the locus (seven genotypes; bulk1) or the right part of the locus (two genotypes; bulk2). Illumina paired-end shotgun indexed libraries were obtained for bulks 1 and 2 r mutants and ‘Sicile’ wild type and sequenced to a depth of 35× and 25×, respectively, on an Illumina HiSeq 2500 at GeT-PlaGe (INRA) per the manufacturer’s instructions (Illumina). Raw fastq files were mapped to the strawberry reference genome sequence F. vesca v2.0.a1 (Shulaev et al., 2011; Tennessen et al., 2014) using BWA version 0.7.12 (Li and Durbin, 2009) (http://bio-bwa.sourceforge.net/). Variant calling (SNPs and InDels) was performed using SAMtools version 1.2 (Li et al., 2009) (http://htslib.org/). Variant analysis and comparison between r bulks and ‘Sicile’ wild type were performed as previously described (Garcia et al., 2016; Petit et al., 2016). The SNPs identified were further analyzed using WGS of one [r] genotype, ‘Baron Solemacher’, and one [R] genotype, ‘Pawtuckaway’, available in GDR (https://www.rosaceae.org/).
Genetic Structures of 25 F. vesca Accessions
The relatedness of 25 F. vesca accessions was studied with 37 neutral microsatellite loci (Supplemental Table 4) distributed throughout the genome using the Bayesian assignment approach from Structure v2.3.4 (Pritchard et al., 2000). The most relevant number of subpopulations (clusters, K) in the population was determined according to the criterion of Evanno, using the ΔK method (Evanno et al., 2005). Leaves used for DNA extraction were obtained from the National Clonal Germplasm Repository (USDA). Genotyping was performed using the KASP method (Smith and Maughan, 2015).
Phylogenetic Analysis, RNA Isolation, and qRT-PCR
Phylogenetic analysis was performed with GA20ox proteins, GA 2-oxidase proteins, and GA 3-oxidase proteins from F. vesca strawberry and Arabidopsis (Supplemental Table 5). The suffix used for F. vesca is Fve according to Jung et al. (2015). GA20ox proteins included the five predicted FveGA20ox proteins already identified (http://bioinformatics.towson.edu/Strawberry/default.aspx). Multiple sequence alignments were generated via ClustalW (Supplemental File 1; Thompson et al., 1997) using BLOSUM matrix with default parameter setting (gap cost between 0.1 and 10). A phylogenetic tree was produced with the Geneious Tree Builder (http://www.geneious.com/) from 1000 bootstrap replicates by applying the neighbor-joining method with Jukes-Cantor like genetic distance model. Parameter settings were the following: no gap penalty, no outgroup, random seed of 1000, support threshold of 25%.
All tissue samples were collected at the end of April from 4-month-old ‘Fb2’ tfl1 and ‘RdV’ r tfl1 mutant plants. Leaves, PCs, and stolons were dissected and frozen in liquid N2 before RNA isolation with a Spectrum Plant Total RNA kit (Sigma-Aldrich). cDNA synthesis was performed with 1 µg of total RNA with an iScript cDNA synthesis kit (Bio-Rad) following the manufacturer’s instructions. RT-PCR was performed using 5 μL of the resulting cDNA product (1/10 dilution) and 10 mM of each primer in a final volume of 20 μL with GoTaq qPCR Master Mix (Promega). For each tissue sample (leaves, PCs, or stolons), three biological replicates, each resulting from the pooling of tissues from two plants, and three technical replicates per biological replicate were analyzed using a CFX96 real-time system (Bio-Rad). FveACTIN (gene26612) was used as a reference gene. The primers used are described in Supplemental Table 4.
Overexpression in Arabidopsis and Enzymatic Activity Assays
A 1696- and a 1687-bp DNA fragment corresponding to the open reading frame of FveGA20ox4 and ∆Fvega20ox4 were PCR-amplified from ‘Sicile’ wild type and ‘Alpine’ r mutant, respectively, using BxFveGA20ox4_C primers (forward, ATGCTTCCTATTCTTCTTTC; reverse, TCAATTGACTGATTTGGATTC, 5′-3′) and cloned through LR reaction (Gateway) into the pK7WG2D plasmid. The resulting vectors were introduced into Agrobacterium tumefaciens (strain GV3101) by electroporation. Five Arabidopsis plants of each genotype, Col-0, Atga20ox1, and Atga20ox1-3/Atga20ox3-1, were transformed by the floral dip method (Clough and Bent, 1998). For plant height measurements, six plants were phenotyped for controls and 10 for transgenic lines when possible.
The cDNAs (FveGA20ox4 gene09034-v1.0-hybrid) corresponding to the FveGA20ox4 wild-type protein (FveGA20ox4) and the mutated form (ΔFvega20ox4) were synthesized (Integrated DNA Technologies), PCR-amplified, and cloned behind a 6xHis tag into Champion pET300/NT-DEST vector through the LR reaction (Gateway). Recombinant plasmids were transformed into Escherichia coli strain Rosetta 2 (DE3; Novagen). Protein production was induced with 1 mM isopropyl β-d-1-thiogalactopyranoside for 2 h at 30°C in 2× YT medium.
Recombinant protein production, enzyme assays, and enzyme product purification and analysis were performed essentially as described previously (Lange, 1997; Pimenta Lange et al., 2013). The identities of substrate and products were confirmed as their methyl esters trimethylsilyl ethers by gas chromatography-mass spectrometry by comparing their mass spectra with published spectra (Gaskin and MacMillan, 1992): [1-,7-,12-,18-14C]GA12 mass/charge (% relative abundance), M+352(5), 344(3), 320(11), 312(6), 306(8), 298(4), 290(34), 284(20), 245(100), 239(52), 201(47), 195(22); [1-,7-,12-,18-14C]GA9, M+338(6), 330(3), 306(75), 298(45), 276(100), 270(54), 251(60), 243(42), 233(66), 232(78), 227(38), 226(48), 189(48), 183(36).
GA Analysis and Treatments
GA content of leaves, PCs, and stolons (when present) was characterized in four genotypes: wild-type ‘Sicile’ and ‘Fb2’ and ‘Alpine’ and ‘RdV’ r mutants. Tissues were dissected from twelve 4-month-old plants. For ‘RdV’ and ‘Fb2’, two biological replicates (each resulting from the pooling of tissues from three plants) were analyzed for each tissue. For ‘Sicile’ and ‘Alpine’, one sample was analyzed for each tissue. GAs were analyzed using 100 mg dry weight of freeze-dried sample that was spiked with 17,17-d2-GA standards (1 ng each, from L. Mander, Canberra, Australia) as previously described (Lange et al., 2005).
For GA3 and Pro-Ca treatments, 3-month-old ‘Rodluvan’ wild type and ‘Alpine’ r mutant plants were sprayed twice a week during the first 2 weeks, weeks 0 and 1, with either GA3 (50 mg/L) or Pro-Ca (100 mg/L), as previously described (Mouhu et al., 2013). On the first day of the experiment, stolons of ‘Rodluvan’ wild type were removed. Plants were grown under natural conditions in the greenhouse under long days (June). Stolons were systematically removed after counting.
Statistical Analysis
When appropriate, a two-sided Mann-Whitney test was performed.
Accession Numbers
Arabidopsis Genome Initiative locus identifiers (https://www.arabidopsis.org/) used in this study are as follows: At4G25420 (AtGA20ox1), At5G51810 (AtGA20ox2), At5G07200 (AtGA20ox3), At1G60980 (AtGA20ox4), At1G44090 (AtGA20ox5), At1G78440 (AtGA2ox1), At1G30040 (AtGA2ox2), At2G34555 (AtGA2ox3), At1G47990 (AtGA2ox4), At1G02400 (AtGA2ox6), At1G50960 (AtGA2ox7), At4G21200 (AtGA2ox8), At1G15550 (AtGA3ox1), At1G80340 (AtGA3ox2), At4G21690 (AtGA3ox3), and At1G80330 (AtGA3ox4). F. vesca locus identifiers can be found in the Strawberry Genomic Resources database (http://bioinformatics.towson.edu/strawberry/) under the following accession numbers: gene13360 (FveGA20ox1), gene19438 (FveGA20ox2), gene19437 (FveGA20ox3), gene09034 (FveGA20ox4), gene10825 (FveGA20ox5), gene05020 (FveGA2ox1), gene00852 (FveGA2ox2), gene03182 (FveGA2ox3), gene07935 (FveGA2ox4), gene19549 (FveGA2ox5), gene06004 (FveGA3ox1), gene01056 (FveGA3ox2), gene01058 (FveGA3ox3), gene01059 (FveGA3ox4), gene01060 (FveGA3ox5), and gene11192 (FveGA3ox6).
Supplemental Data
Supplemental Figure 1. Frequency distribution of the number of stolons produced in the Ilaria_F2 segregating population.
Supplemental Figure 2. Architecture of the wild type and r mutants.
Supplemental Figure 3. Comparison between FveGA20ox4 and GA oxidases of Arabidopsis thaliana.
Supplemental Figure 4. Expression of three additional FveGA20ox found in F. vesca.
Supplemental Table 1. Illumina paired-end shotgun indexed libraries obtained for bulks 1 and 2 r mutants and ‘Sicile’ wild type.
Supplemental Table 2. List of the 11 SNPs/InDel in linkage disequilibrium with r allele.
Supplemental Table 3. Endogenous GA contents of ‘Sicile’ and ‘Fb2’ wild type, ‘Alpine’, and ‘RdV’ r.
Supplemental Table 4. List of primers used in the manuscript.
Supplemental Table 5. Gene identifiers for phylogenetic analysis.
Supplemental File 1. Alignment used to produce the phylogenetic tree in Supplemental Figure 3.
Acknowledgments
This research was funded by the French Ministry of Research. We thank Nahla Bassil (USDA-ARS, NCGR, Corvallis, OR) for diversity resources and Peter Hedden (Rothamsted Research, UK) for Arabidopsis mutant seeds. We thank Johann Petit for photos, Frédéric Delmas for plant transformation, Norbert Bollier for recombinant protein production, Jean-Philippe Mauxion for genotyping, and Alain Bonnet for help with phenotyping. CNRGV Toulouse confirmed the sequence of Fragaria diploid in the R locus by BAC sequencing. WGS data were produced by GeT-PlaGe Toulouse, France. This work was supported by Région Aquitaine (REGAL Project) and by the French Academy of Agriculture (Dufrenoy Grant to T.T., 2015) and was carried out under the auspices of the EU-FP7-KBBE-2010-4 Project (Grant 265942).
AUTHOR CONTRIBUTIONS
T.T., C.R., and B.D. designed the experiments. C.B. performed next-generation sequencing bioinformatics analysis. M.J.P.L. and T.L. performed enzymatic activity and GA endogenous assays. A.M. produced the NIL genotype. M.L. performed architecture experiments. M.H. conducted in situ hybridization. T.T. performed all the other experiments. T.T., C.R., and B.D. wrote the manuscript. All authors discussed the results.
Glossary
- PF
perpetual flowering
- SF
seasonal flowering
- AXM
axillary meristem
- GA
gibberellin
- PC
primary crown
- SAM
shoot apical meristem
- BC
branch crown
- NIL
near-isogenic line
- WGS
whole-genome sequencing
- LD
long day
- SD
short day
- Pro-Ca
prohexadione-calcium
Footnotes
Articles can be viewed without a subscription.
References
- Abrahamson W.G. (1980). Demography and vegetative reproduction. In Demography and Evolution in Plant Populations, O.T Solbrig, ed (Berkeley, CA: University of California Press), pp. 89–106. [Google Scholar]
- Andrés F., Porri A., Torti S., Mateos J., Romera-Branchat M., García-Martínez J.L., Fornara F., Gregis V., Kater M.M., Coupland G. (2014). SHORT VEGETATIVE PHASE reduces gibberellin biosynthesis at the Arabidopsis shoot apex to regulate the floral transition. Proc. Natl. Acad. Sci. USA 111: E2760–E2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett S.C.H. (2015). Influences of clonality on plant sexual reproduction. Proc. Natl. Acad. Sci. USA 112: 8859–8866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisbis B., Delmas F., Joubès J., Sicard A., Hernould M., Inzé D., Mouras A., Chevalier C. (2006). Cyclin-dependent kinase (CDK) inhibitors regulate the CDK-cyclin complex activities in endoreduplicating cells of developing tomato fruit. J. Biol. Chem. 281: 7374–7383. [DOI] [PubMed] [Google Scholar]
- Bolduc N., Hake S. (2009). The maize transcription factor KNOTTED1 directly regulates the gibberellin catabolism gene ga2ox1. Plant Cell 21: 1647–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown T., Wareing P.F. (1965). The genetical control of the everbearing habit and three other characters. Euphytica 14: 97–112. [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Costes E., Crespel L., Denoyes B., Morel P., Demene M.-N., Lauri P.-E., Wenden B. (2014). Bud structure, position and fate generate various branching patterns along shoots of closely related Rosaceae species: a review. Front. Plant Sci. 5: 666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darrow G.M. (1929). Development of runners and runner plants in the strawberry. No. 157352, Technical Bulletins, USDA, Economic Research Service, http://EconPapers.repec.org/RePEc:ags:uerstb:157352. [Google Scholar]
- Darrow G.M. (1966). The Strawberry: History, Breeding and Physiology. (New York: Holt Rinehart Winston; ). [Google Scholar]
- Davière J.-M., Achard P. (2013). Gibberellin signaling in plants. Development 140: 1147–1151. [DOI] [PubMed] [Google Scholar]
- Depuydt S., Hardtke C.S. (2011). Hormone signalling crosstalk in plant growth regulation. Curr. Biol. 21: R365–R373. [DOI] [PubMed] [Google Scholar]
- Duchesne N. (1766) Histoire Naturelle des Fraisiers. (Paris: Didot Panckoucke).
- Eriksson S., Böhlenius H., Moritz T., Nilsson O. (2006). GA4 is the active gibberellin in the regulation of LEAFY transcription and Arabidopsis floral initiation. Plant Cell 18: 2172–2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evanno G., Regnaut S., Goudet J. (2005). Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol. Ecol. 14: 2611–2620. [DOI] [PubMed] [Google Scholar]
- Galinha C., Bilsborough G., Tsiantis M. (2009). Hormonal input in plant meristems: A balancing act. Semin. Cell Dev. Biol. 20: 1149–1156. [DOI] [PubMed] [Google Scholar]
- Garcia V., et al. (2016). Rapid identification of causal mutations in tomato EMS populations via mapping-by-sequencing. Nat. Protoc. 11: 2401–2418. [DOI] [PubMed] [Google Scholar]
- Gaskin P., MacMillan J. (1992). GC-MS of the Gibberellins and Related Compounds: Methodology and a Library of Spectra. (Bristol, UK: Cantock’s Enterprises; ). [Google Scholar]
- Gaston A., Perrotte J., Lerceteau-Köhler E., Rousseau-Gueutin M., Petit A., Hernould M., Rothan C., Denoyes B. (2013). PFRU, a single dominant locus regulates the balance between sexual and asexual plant reproduction in cultivated strawberry. J. Exp. Bot. 64: 1837–1848. [DOI] [PubMed] [Google Scholar]
- Guttridge C.G., Thompson P.A. (1964). The effect of gibberellins on growth and flowering of Fragaria and Duchesnea. J. Exp. Bot. 15: 631–646. [Google Scholar]
- Hay A., Kaur H., Phillips A., Hedden P., Hake S., Tsiantis M. (2002). The gibberellin pathway mediates KNOTTED1-type homeobox function in plants with different body plans. Curr. Biol. 12: 1557–1565. [DOI] [PubMed] [Google Scholar]
- Hedden P., Thomas S.G. (2012). Gibberellin biosynthesis and its regulation. Biochem. J. 444: 11–25. [DOI] [PubMed] [Google Scholar]
- Heide O.M., Stavang J.A., Sønsteby A. (2013). Physiology and genetics of flowering in cultivated and wild strawberries - A review. J. Hortic. Sci. Biotechnol. 88: 1–18. [Google Scholar]
- Huang Y., Wang X., Ge S., Rao G.Y. (2015). Divergence and adaptive evolution of the gibberellin oxidase genes in plants. BMC Evol. Biol. 15: 207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hytönen T., Elomaa P. (2011). Genetic and environmental regulation of flowering and runnering in strawberry. Genes Genomics 5: 56–64. [Google Scholar]
- Hytönen T., Elomaa P., Moritz T., Junttila O. (2009). Gibberellin mediates daylength-controlled differentiation of vegetative meristems in strawberry (Fragaria x ananassa Duch). BMC Plant Biol. 9: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hytönen T., Mouhu K., Koivu I., Juntilla O. (2008). Prohexadione-calcium enhances the cropping potential and yield of strawberry. J. Hortic. Sci. 73: 210–215. [Google Scholar]
- Iwata H., Gaston A., Remay A., Thouroude T., Jeauffre J., Kawamura K., Oyant L.H., Araki T., Denoyes B., Foucher F. (2012). The TFL1 homologue KSN is a regulator of continuous flowering in rose and strawberry. Plant J. 69: 116–125. [DOI] [PubMed] [Google Scholar]
- Jasinski S., Piazza P., Craft J., Hay A., Woolley L., Rieu I., Phillips A., Hedden P., Tsiantis M. (2005). KNOX action in Arabidopsis is mediated by coordinate regulation of cytokinin and gibberellin activities. Curr. Biol. 15: 1560–1565. [DOI] [PubMed] [Google Scholar]
- Jung S., et al. (2014). The Genome Database for Rosaceae (GDR): Year 10 update. Nucleic Acids Res. 42: 1237–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung S., Bassett C., Bielenberg D.G., Cheng C.H., Dardick C., Main D., Meisel L., Slovin J., Troggio M., Schaffer R.J. (2015). A standard nomenclature for gene designation in the Rosaceae. Tree Genet. Genomes 11: 108. [Google Scholar]
- Kaczmarska E., Garonski J., Jablonska-Rys E., Zalewska-Korona M., Radzki W., Slawinska A. (2016). Hybrid performance and heterosis in strawberry (Fragaria × ananassa Duchesne), regarding acidity, soluble solids and dry matter content in fruits. Plant Breed. 135: 232–238. [Google Scholar]
- Kang C., Darwish O., Geretz A., Shahan R., Alkharouf N., Liu Z. (2013). Genome-scale transcriptomic insights into early-stage fruit development in woodland strawberry Fragaria vesca. Plant Cell 25: 1960–1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klimeš L., Klimesova J., Hendricks R., van Groenendael J. (1997). Clonal plant architecture: a comparative analysis of form and function. In The Ecology and Evolution of Clonal Plants, de Kroon H., van Groenendael J., eds (Leiden, The Netherlands: Backhuys Publishers; ), pp. 1–29. [Google Scholar]
- Kloosterman B., Navarro C., Bijsterbosch G., Lange T., Prat S., Visser R.G.F., Bachem C.W.B. (2007). StGA2ox1 is induced prior to stolon swelling and controls GA levels during potato tuber development. Plant J. 52: 362–373. [DOI] [PubMed] [Google Scholar]
- Koskela E.A., Mouhu K., Albani M.C., Kurokura T., Rantanen M., Sargent D.J., Battey N.H., Coupland G., Elomaa P., Hytönen T. (2012). Mutation in TERMINAL FLOWER1 reverses the photoperiodic requirement for flowering in the wild strawberry Fragaria vesca. Plant Physiol. 159: 1043–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger U., Lippman Z.B., Zamir D. (2010). The flowering gene SINGLE FLOWER TRUSS drives heterosis for yield in tomato. Nat. Genet. 42: 459–463. [DOI] [PubMed] [Google Scholar]
- Kumar D., Wareing P.F. (1974). Studies on tuberization of Solanum andigena. New Phytol. 73: 833–840. [Google Scholar]
- Lange T. (1997). Cloning gibberellin dioxygenase genes from pumpkin endosperm by heterologous expression of enzyme activities in Escherichia coli. Proc. Natl. Acad. Sci. USA 94: 6553–6558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange T., Kegler C., Hedden P., Phillips A.L., Graebe J.E. (1997). Molecular characterization of gibberellin 20-oxidases: Structure-function studies on recombinant enzymes and chimaeric proteins. Physiol. Plant. 100: 543–549. [Google Scholar]
- Lange T., Kappler J., Fischer A., Frisse A., Padeffke T., Schmidtke S., Pimenta Lange M.J. (2005). Gibberellin biosynthesis in developing pumpkin seedlings. Plant Physiol. 139: 213–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D., Liu C., Shen L., Wu Y., Chen H., Robertson M., Helliwell C.A., Ito T., Meyerowitz E., Yu H. (2008). A repressor complex governs the integration of flowering signals in Arabidopsis. Dev. Cell 15: 110–120. [DOI] [PubMed] [Google Scholar]
- Li H., Durbin R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R.; 1000 Genome Project Data Processing Subgroup (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouhu K., Kurokura T., Koskela E.A., Albert V.A., Elomaa P., Hytönen T. (2013). The Fragaria vesca homolog of suppressor of overexpression of constans1 represses flowering and promotes vegetative growth. Plant Cell 25: 3296–3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.J., Jiang K., Tal L., Yichie Y., Gar O., Zamir D., Eshed Y., Lippman Z.B. (2014). Optimization of crop productivity in tomato using induced mutations in the florigen pathway. Nat. Genet. 46: 1337–1342. [DOI] [PubMed] [Google Scholar]
- Perrotte J., Gaston A., Potier A., Petit A., Rothan C., Denoyes B. (2016a). Narrowing down the single homoeologous FaPFRU locus controlling flowering in cultivated octoploid strawberry using a selective mapping strategy. Plant Biotechnol. J. 14: 2176–2189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrotte J., Guédon Y., Gaston A., Denoyes B. (2016b). Identification of successive flowering phases highlights a new genetic control of the flowering pattern in strawberry. J. Exp. Bot. 67: 5643–5655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit J., Bres C., Mauxion J.-P., Tai F.W., Martin L.B.B., Fich E.A., Joubès J., Rose J.K.C., Domergue F., Rothan C. (2016). The glycerol-3-phosphate acyltransferase GPAT6 from tomato plays a central role in fruit cutin biosynthesis. Plant Physiol. 171: 894–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips A.L., Ward D.A., Uknes S., Appleford N.E.J., Lange T., Huttly A.K., Gaskin P., Graebe J.E., Hedden P. (1995). Isolation and expression of three gibberellin 20-oxidase cDNA clones from Arabidopsis. Plant Physiol. 108: 1049–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimenta Lange M.J., Liebrandt A., Arnold L., Chmielewska S.-M., Felsberger A., Freier E., Heuer M., Zur D., Lange T. (2013). Functional characterization of gibberellin oxidases from cucumber, Cucumis sativus L. Phytochemistry 90: 62–69. [DOI] [PubMed] [Google Scholar]
- Plackett A.R., et al. (2012). Analysis of the developmental roles of the Arabidopsis gibberellin 20-oxidases demonstrates that GA20ox1, -2, and -3 are the dominant paralogs. Plant Cell 24: 941–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard J.K., Stephens M., Donnelly P. (2000). Inference of population structure using multilocus genotype data. Genetics 155: 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rademacher W. (2000). Growth retardants: Effects on gibberellin biosynthesis and other metabolic pathways. Annu. Rev. Plant Physiol. Plant Mol. Biol. 51: 501–531. [DOI] [PubMed] [Google Scholar]
- Regnault T., Davière J.-M., Wild M., Sakvarelidze-Achard L., Heintz D., Carrera Bergua E., Lopez Diaz I., Gong F., Hedden P., Achard P. (2015). The gibberellin precursor GA12 acts as a long-distance growth signal in Arabidopsis. Nat. Plants 1: 15073. [DOI] [PubMed] [Google Scholar]
- Rieu I., Ruiz-Rivero O., Fernandez-Garcia N., Griffiths J., Powers S.J., Gong F., Linhartova T., Eriksson S., Nilsson O., Thomas S.G., Phillips A.L., Hedden P. (2008). The gibberellin biosynthetic genes AtGA20ox1 and AtGA20ox2 act, partially redundantly, to promote growth and development throughout the Arabidopsis life cycle. Plant J. 53: 488–504. [DOI] [PubMed] [Google Scholar]
- Rosin F.M., Hart J.K., Van Onckelen H., Hannapel D.J. (2003). Suppression of a vegetative MADS box gene of potato activates axillary meristem development. Plant Physiol. 131: 1613–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto T., Kamiya N., Ueguchi-Tanaka M., Iwahori S., Matsuoka M. (2001). KNOX homeodomain protein directly suppresses the expression of a gibberellin biosynthetic gene in the tobacco shoot apical meristem. Genes Dev. 15: 581–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargent D.J., Davis T.M., Tobutt K.R., Wilkinson M.J., Battey N.H., Simpson D.W. (2004). A genetic linkage map of microsatellite, gene-specific and morphological markers in diploid Fragaria. Theor. Appl. Genet. 109: 1385–1391. [DOI] [PubMed] [Google Scholar]
- Sargent D.J., Clarke J., Simpson D.W., Tobutt K.R., Arús P., Monfort A., Vilanova S., Denoyes-Rothan B., Rousseau M., Folta K.M., Bassil N.V., Battey N.H. (2006). An enhanced microsatellite map of diploid Fragaria. Theor. Appl. Genet. 112: 1349–1359. [DOI] [PubMed] [Google Scholar]
- Savini G., Giorgi V., Scarano E., Neri D. (2008). Strawberry plant relationship through the stolon. Physiol. Plant. 134: 421–429. [DOI] [PubMed] [Google Scholar]
- Shulaev V., et al. (2011). The genome of woodland strawberry (Fragaria vesca). Nat. Genet. 43: 109–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sicard A., Petit J., Mouras A., Chevalier C., Hernould M. (2008). Meristem activity during flower and ovule development in tomato is controlled by the mini zinc finger gene INHIBITOR OF MERISTEM ACTIVITY. Plant J. 55: 415–427. [DOI] [PubMed] [Google Scholar]
- Smith S.M., Maughan P.J. (2015). SNP genotyping using KASPar assays. Methods Mol. Biol. 1245: 243–256. [DOI] [PubMed] [Google Scholar]
- Sugiyama N., Iwama T., Inaba Y., Kurokura T., Neri D. (2004). Varietal differences in the formation branch crows in strawberry plants. HortScience 73: 216–220. [Google Scholar]
- Tao Z., Shen L., Liu C., Liu L., Yan Y., Yu H. (2012). Genome-wide identification of SOC1 and SVP targets during the floral transition in Arabidopsis. Plant J. 70: 549–561. [DOI] [PubMed] [Google Scholar]
- Taylor D.R., Blake P.S., Browning G. (1994). Identification of gibberellins in leaf tissues of strawberry (Fragaria x ananassa Duch.) grown under different photoperiods. Plant Growth Regul. 15: 235–240. [Google Scholar]
- Tennessen J.A., Govindarajulu R., Ashman T.L., Liston A. (2014). Evolutionary origins and dynamics of octoploid strawberry subgenomes revealed by dense targeted capture linkage maps. Genome Biol. Evol. 6: 3295–3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.D., Gibson T.J., Plewniak F., Jeanmougin F., Higgins D.G. (1997). The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25: 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson P.A., Guttridge C.G. (1959). Effects of gibberellic acid on the initiation of flowers and runners in the strawberry. Nature 184: 72–73. [Google Scholar]
- Till B.J., et al. (2003). Large-scale discovery of induced point mutations with high-throughput TILLING. Genome Res. 13: 524–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urrutia M., Bonet J., Arús P., Monfort A. (2015). A near-isogenic line (NIL) collection in diploid strawberry and its use in the genetic analysis of morphologic, phenotypic and nutritional characters. Theor. Appl. Genet. 128: 1261–1275. [DOI] [PubMed] [Google Scholar]
- Vallejo-Marín M., Dorken M.E., Barrett S.C.H. (2010). The ecological and evolutionary consequences of clonality for plant mating. Annu. Rev. Ecol. Evol. Syst. 41: 193–213. [Google Scholar]
- Veit B. (2009). Hormone mediated regulation of the shoot apical meristem. Plant Mol. Biol. 69: 397–408. [DOI] [PubMed] [Google Scholar]
- Wang Y., Cheng X., Shan Q., Zhang Y., Liu J., Gao C., Qiu J.-L. (2014). Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 32: 947–951. [DOI] [PubMed] [Google Scholar]