The inability of plants physically to move necessitates that they have mechanisms in place to rapidly respond to adverse changes in the environment. The lipid-derived hormone jasmonic acid (JA) is one such component of a robust plant immune system that regulates gene expression in response to pathogens and insects. JA is a key player in defense against necrotrophic pathogens, such as the gray mold-causing fungus Botrytis cinerea, and it is also involved in accumulation of feeding deterrent secondary metabolites in response to attack by insect herbivores (Pieterse et al., 2012). In addition, JA and its methylated ester, methyl jasmonate (MeJA), have been shown to control aspects of flowering, root development, and responses to various abiotic stresses (Huang et al., 2017). Understanding hormone action requires a detailed insight into the underlying gene network that effects rapid signaling to bring about changes in plant metabolism, including but not limited to, biosynthesis of defense-related proteins and secondary metabolites. However, most studies on JA to date have been restricted to only a few time points and therefore have failed to provide a comprehensive view of the dynamic transcriptomic response controlled by this phytohormone.
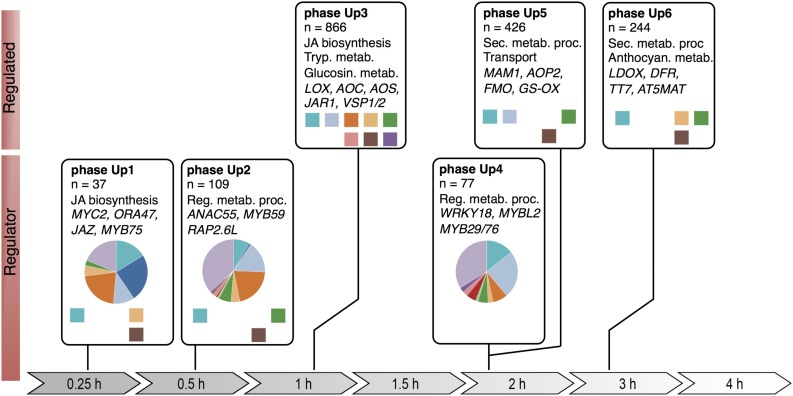
In recent work, Hickman et al. (2017) describe a dense time-series experiment where they conduct thorough computational analyses on RNA-seq data collected from 14 consecutive time points on MeJA-treated leaf tissue of Arabidopsis thaliana, specifically the sixth true leaf. Interestingly, they observed that sets of coexpressed genes are expressed in distinctive temporal patterns; upregulation of genes occurs six times (see figure) and downregulation follows in four phases. Differentially expressed genes (DEGs) that control transcriptional processes are induced rapidly in the first 0.5 h after treatment, while their downstream target genes follow by 1 h after treatment. In addition, the majority of the upregulated genes were enriched for Gene Ontology terms such as “response to wounding” and “JA defense responses” in accordance with the observed ∼50% transcriptional overlap with insect-infested Arabidopsis leaves.
Major upregulated transcriptional phases in the JA gene regulatory network. Transcriptional phases are indicated by boxes aligned on the timeline. DEGs were assigned to the phases according to the time point they first became differentially expressed; indicated are overrepresented functional categories and representative genes. Colored squares indicate known TF DNA binding motifs overrepresented in gene promoters; pie charts indicate the proportion of TF gene families. (Reprinted from Hickman et al. [2017], Figure 4.)
Cluster analyses and enrichment for transcription factors identified a predominance of bHLH, ERF, and MYB TFs in MeJA upregulated genes, many of which were previously uncharacterized or missing from Arabidopsis microarrays. Furthermore, characterization of T-DNA mutants in these putative regulators identified four novel genes (bHLH27, ERF16, MYB59, and ANAC056) with altered sensitivity to either B. cinerea or the generalist insect Mamestra brassicae, supporting their role in JA-mediated plant defense. However, the double mutant myb48 myb59 showed greater resistance to M. brassicae and more severe disease symptoms with B. cinerea. Subsequent RNA-seq on myb48 myb59 revealed 399 DEGs, with key JA-responsive biosynthesis and marker genes, such as VSP2, showing significantly higher expression in the mutant. This confirmed the notion that the coordinated activity of MYB48 and MYB59 is required for JA-mediated positive regulation of resistance to necrotrophic pathogens and negative regulation of insect resistance.
This comprehensive look at the JA-controlled network in plants constitutes a valuable resource for future researchers aiming to identify control points and key players in JA-mediated immune responses and plant growth and development.
Footnotes
Articles can be viewed without a subscription.
References
- Hickman R., et al. (2017). Architecture and dynamics of the jasmonic acid gene regulatory network. Plant Cell 29: 2086–2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Liu B., Liu L., Song S. (2017). Jasmonate action in plant growth and development. J. Exp. Bot. 68: 1349–1359. [DOI] [PubMed] [Google Scholar]
- Pieterse C.M., Van der Does D., Zamioudis C., Leon-Reyes A., Van Wees S.C. (2012). Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 28: 489–521. [DOI] [PubMed] [Google Scholar]