Genome compaction allows animals and plants to fold their genes inside the tiny space of cell nuclei and relies on the conserved histone proteins. Nucleosomes, the core component of chromatin, consist of a histone octamer around which DNA wraps, ∼150 bp per unit, and are stacked in higher-order complexes that eventually shape into chromosomes. However, with great packing comes limited accessibility to some promoters and genes for the transcription machinery. As a countermeasure, histones can be covalently modified on their tails and core regions by several types of enzymes to change their level of compaction locally.
New findings by Su et al. (2017) reveal a connection between photoperiod-mediated induction of flowering time and phosphorylation of histone H2A at serine 95 in Arabidopsis thaliana.
The kinase MUT9p-LIKE KINASE4 (MLK4) is part of a four-protein family with high similarity to the MUT9p kinase from Chlamydomonas reinhardtii (Casas-Mollano et al., 2008). In the single-cell alga, a mut9 mutant reactivates expression of silenced transgenes and derepresses several transposons, pointing to a role in epigenetic gene silencing. Biochemically, MUT9p phosphorylates the tail of histones H2A and H3, which leads to a downregulation of target genes.
Of the four MUT9p-like genes in Arabidopsis, only loss of MLK4 results in a delay in flowering initiation on its own (Su et al., 2017), although partial redundancy may exist among family members (Huang et al., 2016). Su and colleagues define the regulatory cascade linking MLK4 function to photoperiod-mediated induction of flowering time.
Based on double mutant analyses, the authors show that MLK4 acts upstream of the flowering time proteins CONSTANS (CO) and GIGANTEA (GI), but downstream of the clock protein CIRCADIAN CLOCK ASSOCIATED1 (CCA1). Gene expression levels of CO and GI are consistent with these observations and imply that loss of MLK4 leads to lower GI expression, which in turn results in lower CO mRNA levels.
MLK4 is found in the nucleus and interacts physically with CCA1, although it does not phosphorylate the clock protein itself. CCA1 binds to the GI promoter, where it meets up with MLK4, itself responsible for the phosphorylation of local histones H2A on serine 95. This amino acid residue is conserved in all plant H2A proteins, but not in animals or Chlamydomonas, nor is it found in the histone variant H2A.Z. Global phosphorylation of histone H2A on serine 95 is not affected in mlk4 mutants, stressing the very localized effect of MLK4 on histone modification.
CCA1 also recruits the chromatin remodeling complexes SWR1 and NuA4 at the GI promoter via direct interaction with the protein YAF9a. In animals and plants, NuA4 acetylates histones H2A and H4, which stimulates SWR1 to replace histone H2A with H2A.Z. Accordingly, the histone variant H2A.Z is deposited along the GI locus in wild-type plants, but not in mlk4 mutants. Histone H4 acetylation is also decreased at the GI promoter in mlk4 mutants, contributing to the lower expression of the locus.
This work reveals a linear series of events that link phosphorylation on a single residue of histone H2A with flowering time. A few questions remain open still: It is not clear how MLK4 and CCA1 can physically interact when their expression patterns are out of phase with each other. MLK4 accumulates in the evening, when it can then come in contact with components of the evening complex, whereas CCA1 levels reach their peak in the morning (Huang et al., 2016). Perhaps MLK4-mediated phosphorylation of H2A controls the extent of chromatin packing around the GI locus and therefore modulates CCA1 access to its binding sites within the GI promoter. In addition, do MLK family members affect global patterns of histone H2A and H2A.Z occupancy in response to changes in ambient temperature (Kumar and Wigge, 2010)? Research aimed at elucidating the temporal connection between all the players involved promises to come to bloom in the near future.
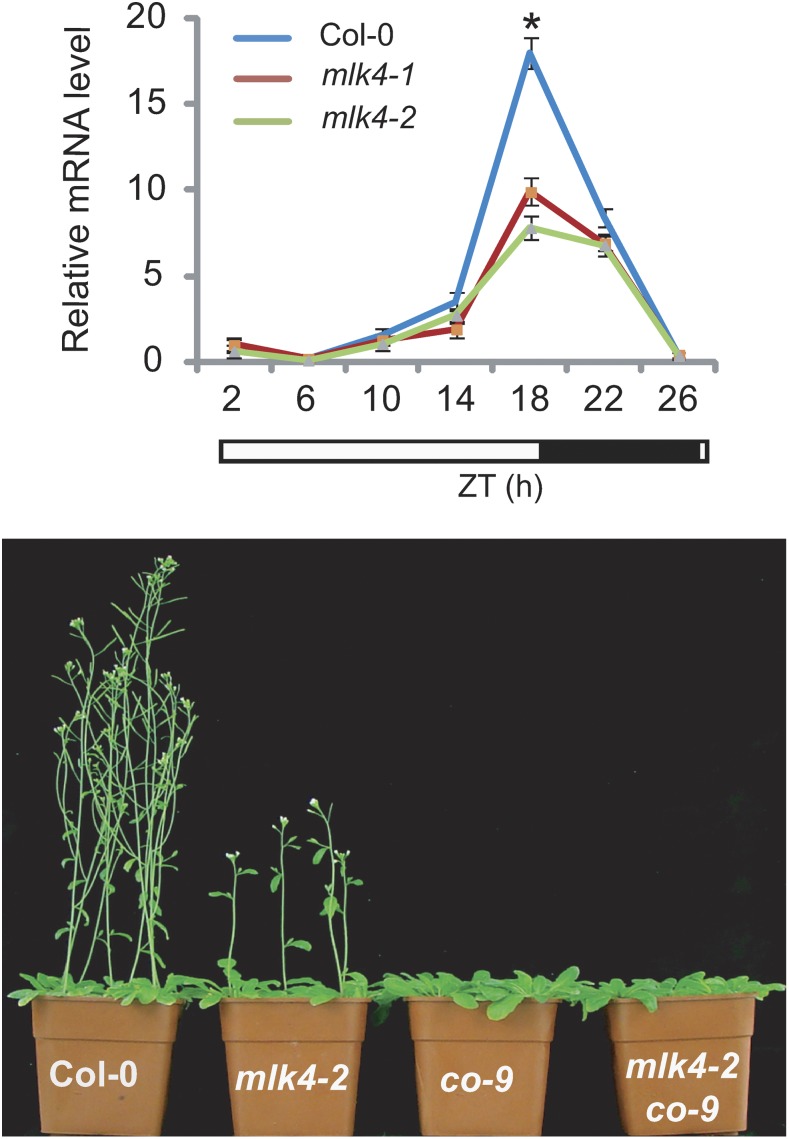
MLK4 contributes to the induction of CONSTANS under long days. Top: Real-time qPCR analysis of CO expression levels in the wild type and mlk4 mutants. Bottom: Epistatic analysis between co and mlk4 mutants demonstrate that CO acts downstream of MLK4. (Reprinted from Su et al. [2017], Figure 1.)
Footnotes
Articles can be viewed without a subscription.
References
- Casas-Mollano J.A., Jeong B.R., Xu J., Moriyama H., Cerutti H. (2008). The MUT9p kinase phosphorylates histone H3 threonine 3 and is necessary for heritable epigenetic silencing in Chlamydomonas. Proc. Natl. Acad. Sci. USA 105: 6486–6491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H., Alvarez S., Bindbeutel R., Shen Z., Naldrett M.J., Evans B.S., Briggs S.P., Hicks L.M., Kay S.A., Nusinow D.A. (2016). Identification of evening complex associated proteins in Arabidopsis by affinity purification and mass spectrometry. Mol. Cell. Proteomics 15: 201–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S.V., Wigge P.A. (2010). H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell 140: 136–147. [DOI] [PubMed] [Google Scholar]
- Su Y., Wang S., Zhang F., Zheng H., Liu Y., Huang T., Ding Y. (2017). Phosphorylation of Arabidopsis histone H2A serine 95: a plant-specific mark involved in flowering time and H2A.Z deposition. Plant Cell 29: 2197–2213. [DOI] [PMC free article] [PubMed] [Google Scholar]