Phosphorylation of H2A at serine 95, a plant-specific modification in the histone core domain, is required for flowering time regulation and deposition of the histone variant H2A.Z in Arabidopsis.
Abstract
Phosphorylation of histone H3 affects transcription, chromatin condensation, and chromosome segregation. However, the role of phosphorylation of histone H2A remains unclear. Here, we found that Arabidopsis thaliana MUT9P-LIKE-KINASE (MLK4) phosphorylates histone H2A on serine 95, a plant-specific modification in the histone core domain. Mutations in MLK4 caused late flowering under long-day conditions but no notable phenotype under short days. MLK4 interacts with CIRCADIAN CLOCK ASSOCIATED1 (CCA1), which allows MLK4 to bind to the GIGANTEA (GI) promoter. CCA1 interacts with YAF9a, a co-subunit of the Swi2/Snf2-related ATPase (SWR1) and NuA4 complexes, which are responsible for incorporating the histone variant H2A.Z into chromatin and histone H4 acetylase activity, respectively. Importantly, loss of MLK4 function led to delayed flowering by decreasing phosphorylation of H2A serine 95, along with attenuated accumulation of H2A.Z and the acetylation of H4 at GI, thus reducing GI expression. Together, our results provide insight into how phosphorylation of H2A serine 95 promotes flowering time and suggest that phosphorylation of H2A serine 95 modulated by MLK4 is required for the regulation of flowering time and is involved in deposition of the histone variant H2A.Z and H4 acetylation in Arabidopsis.
INTRODUCTION
Histone phosphorylation occurs on serine, threonine, and/or tyrosine residues at H3 and is generally associated with the regulation of transcription or entry into mitosis/meiosis. Phosphorylation of H3 at serine 10, serine 28, threonine 3, and threonine 11 is evolutionarily conserved in yeast, mammalian cells, and plants (Cerutti and Casas-Mollano, 2009; Banerjee and Chakravarti, 2011; Rossetto et al., 2012). The phosphorylation of H3 at serine 10 (H3S10ph) by MAP kinase signaling stimulates the acetylation of lysine 14 in H3, which in turn recruits Transcription Factor II D to activate gene expression (Cheung et al., 2000; Lo et al., 2000; Hirota et al., 2005). H3 serine28 phosphorylation (H3S28ph), generated by MITOGEN- AND STRESS-ACTIVATED PROTEIN KINASE1, not only prevents recruitment of the gene-silencing Polycomb-repressive complexes and their methylation of H3K27, but it also induces a switch from methylation to acetylation at H3K27 (Lau and Cheung, 2011). In mammalian cells and plants, phosphorylation of H3 at threonine 3 (H3T3ph) generated by the mitosis-activated Haspin kinase functions in chromosome alignment and centromere cohesion (Dai et al., 2005; Cerutti and Casas-Mollano, 2009). However, the phosphorylation of H3 at threonine 3 by the kinase MUT9p is associated with transcriptional silencing in Chlamydomonas reinhardtii (Ivaldi et al., 2007).
In contrast to the phosphorylation of histone H3, the phosphorylation sites of histone H2A have not been well defined, and these modifications are usually associated with DNA damage. In yeast, phosphorylation of histone H2A at serine 129 by kinases Mec1 (Mitosis entry checkpoint protein 1) and Tel1 (Telomere length regulation protein 1) is required for efficient DNA double-strand break repair by nonhomologous end joining (Rogakou et al., 1998; Downs et al., 2000). Phosphorylation of histone H2A at serine 129 enhances the recruitment of the INO80 complex to facilitate the repair of double-strand DNA breaks (van Attikum et al., 2004). In mammalian cells, the Williams Syndrome Transcription Factor-SNF2H (Sucrose Non-Fermentable 2H) chromatin remodeling complex plays a vital role in the DNA damage response and phosphorylates tyrosine 142 of H2A.X (Xiao et al., 2009). In addition to DNA damage, phosphorylation of H2A at serine 121 by Bub1 (Budding Uninhibited By Benzimidazoles 1) regulates the localization of shugoshin to prevent chromosomal instability (Kawashima et al., 2010), and phosphorylation of histone H2A at tyrosine 56 by Casein Kinase II (CK2) regulates transcriptional elongation (Basnet et al., 2014). In maize (Zea mays), the phosphorylation of H2A at threonine 133 is associated with centromere function and maintenance during meiosis (Dong and Han, 2012).
Flowering in Arabidopsis thaliana is promoted by long summer days and is repressed by short winter days. The photoperiod pathway in the leaves controls this response via a signaling cascade involving GIGANTEA (GI) and the transcriptional regulator CONSTANS (CO) (Fornara et al., 2010). CO is required to promote flowering under long-day (LD) but not short-day (SD) conditions (Putterill et al., 1995). CO encodes a B-box-type zinc-finger transcription factor that directly activates the expression of FLOWERING LOCUS T (FT) (Yanovsky and Kay, 2002). The expression of CO is repressed by CYCLING DOF FACTOR1 and activated by FLAVIN BINDING, KELCH REPEAT, F-BOX1 and GI (Imaizumi et al., 2005). The expression of CO and GI mRNAs is controlled by the circadian clock (Fowler et al., 1999; Park et al., 1999; Suárez-López et al., 2001; Mizoguchi et al., 2002).
The Swi2/Snf2-related ATPase (SWR1) complex is an ATP-dependent complex required for deposition of the histone variant H2A.Z, whereas NuA4 is a histone acetylation complex that modifies H4, H2A, and H2A.Z in Saccharomyces cerevisiae (Krogan et al., 2004). These two complexes share four subunits, Yaf9, Swc4, Arp4, and Act1, which function together to regulate chromatin structure and gene transcription (Krogan et al., 2004; Zhang et al., 2004). Yaf9 is a YEAST domain protein, which is evolutionarily conserved in yeast, mammals, and plants. In yeast, the Yaf9 components of the SWR1 and NuA4 complexes are required for gene expression, H4 acetylation, and Htz1 replacement at specific genes (Zhang et al., 2004; Wang et al., 2009; Altaf et al., 2010). There are two genes encoding Yaf9 in Arabidopsis: YAF9a (TAF14b) and YAF9b (TAF14). The loss of YAF9a function results in early flowering in LD via reduced H4 acetylation at FLOWERING LOCUS C (FLC) and the promotion of acetylation at FT, whereas the phenotype is not obvious in SD (Zacharaki et al., 2012).
Casein kinase I, a serine/threonine protein kinase, is a multifunctional protein kinase found in most eukaryotic cells. In mammalian cells, casein kinase I is involved in vesicular trafficking, DNA repair, the circadian rhythm, and morphogenesis (McKay et al., 2001; Milne et al., 2001). In the alga Chlamydomonas, MUT9p, which is related to casein kinase I, is capable of phosphorylating H3 at threonine 3 (Casas-Mollano et al., 2008). The Arabidopsis genome encodes four proteins (MLK1, MLK2, MLK3, and MLK4) related to MUT9p (Cerutti and Casas-Mollano, 2009; Wang et al., 2015; Huang et al., 2016). These four proteins were copurified with the evening complex, which is composed of EARLY FLOWERING4 (ELF4), ELF3, and LUX ARRHYTHMO (Huang et al., 2016). MLK1 and MLK2 also function as kinases to phosphorylate H3 at threonine 3, and the loss of MLK1 and MLK2 function results in hypersensitivity to osmotic stress (Wang et al., 2015). In rice (Oryza sativa), EARLIER FLOWERING1 encodes a casein kinase I protein that phosphorylates the DELLA protein SLR1 (Dai and Xue, 2010).
Histone modifications play critical roles in development, but the patterns of development differ dramatically between plants and animals. One fundamental outstanding question is how plant dialects of the histone modification code function in developmental regulation. In particular, H2A is more divergent than H3 and H4, and its modifications (especially phosphorylation) have been less well studied. Here, we report that MLK4 phosphorylates H2A at serine 95 (H2AS95ph), a plant-specific site, both in vitro and in vivo in Arabidopsis. MLK4 interacts with CIRCADIAN CLOCK ASSOCIATED1 (CCA1) and targets GI directly. In addition, CCA1 binds to YAF9a, a co-subunit of the SWR1 and NuA4 complexes, which are responsible for incorporating the histone variant H2A.Z into chromatin and for histone H4 acetylase activity. The loss of MLK4 function results in late flowering, as well as a reduction in H2AS95ph, H2A.Z accumulation, H4 acetylation, and the expression of GI in LD conditions in Arabidopsis.
RESULTS
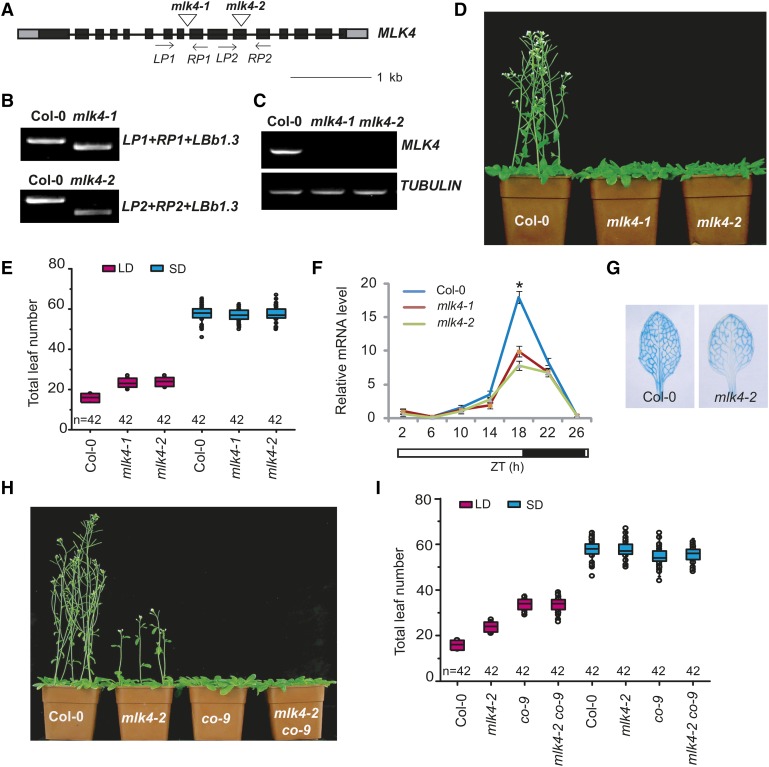
MLK4 Is Involved in the Photoperiod Response Pathway
To characterize the function of MLK4, we identified two Arabidopsis mlk4 mutants containing T-DNA insertions. Genotypic analyses revealed a T-DNA insertion in intron 8 and exon 11 of mlk4-1 and mlk4-2, respectively (Figures 1A and 1B). No full-length MLK4 mRNA was detected in the mlk4-1 or mlk4-2 mutants (Figure 1C), indicating that both are null alleles. Defects in MLK4 resulted in late flowering in LD conditions, whereas the phenotype was not notable under SD, indicating that MLK4 is necessary for the LD response and participates in the photoperiod pathway (Figures 1D and 1E, Table 1). These results are consistent with a previous report (Huang et al., 2016). The late-flowering phenotype was completely complemented by transforming the mutants with a full-length MLK4 cDNA fused to the HA tag driven by the MLK4 promoter (ProMLK4:HA-MLK4) (Supplemental Figure 1). The overexpression of MLK4 driven by the 35S promoter led to early flowering under LD, but no obvious phenotype under SD photoperiods (Supplemental Figures 2A and 2B; Table 1).
Figure 1.
Mutations in MLK4 Result in Late Flowering under a Long-Day Photoperiod.
(A) Gene structure of MLK4, indicating exons (boxes), introns (lines), and T-DNA insertions (triangles). The primers used for genotype analysis are marked with arrows.
(B) Genotypic analysis of the mlk4 mutants. The genotype was analyzed with the left genomic primer (LP), right genomic primer (RP), and the vector primer (LBb1.3). The positions of LP1, RP1, LP2, and RP2 are indicated in (A).
(C) The presence of full-length MLK4 transcript in the mlk4 mutants was examined by RT-PCR.
(D) The 32-d-old mlk4 mutants exhibit a delayed-flowering phenotype under a LD photoperiod.
(E) The total leaf number of Col-0 and mlk4 mutants under a LD photoperiod and SD photoperiod. Flowering time was assessed by counting the number of rosette leaves and cauline leaves at bolting under LD and SD. Values shown are mean ± sd of total leaves; 42 plants were scored for each line.
(F) The relative level of CO mRNA was examined in Col-0 and mlk4 mutants. The black bars indicate the dark period, and the white bars indicate the light period. Experiments were repeated at least three times, and each data point indicates the mean ±se, n = 3 replicates. ZT, Zeitgeber time. Asterisks indicate significant difference from Col-0 using Student’s t test (P < 0.05).
(G) The stable expression of ProCO:GUS was examined by staining in the leaves of Col-0 and mlk4-2 plants for GUS activity.
(H) Representative image of 40-d-old Col-0, mlk4-2, co9, and mlk4-2 co9 double mutant plants under a LD photoperiod.
(I) The total leaf number of Col-0, mlk4-2, co-9, and mlk4-2 co-9 plants under LD and SD. Flowering time was assessed by counting the number of rosette leaves and cauline leaves at bolting under LD and SD. Values shown are mean ± sd of total leaves; 42 plants were scored for each line.
Table 1. Primary Leaf Number at Bolting of mlk4, co, gi, and cca1 Mutants in LD and SD.
|
LD |
SD |
|||
|---|---|---|---|---|
| Mutant | Rosette Leaves | Cauline Leaves | Rosette Leaves | Cauline Leaves |
| Col-0 | 12.2 ± 1.0 | 3.6 ± 0.5 | 49.4 ± 4.0 | 8.5 ± 1.1 |
| mlk4-1 | 17.0 ± 1.1 | 6.4 ± 0.8 | 48.3 ± 3.1 | 8.9 ± 1.1 |
| mlk4-2 | 17.5 ± 1.2 | 6.3 ± 0.7 | 49.1 ± 3.7 | 8.7 ± 1.0 |
| OX5 | 10.4 ± 0.8 | 4.3 ± 1.0 | 45.2 ± 6.0 | 11.0 ± 1.3 |
| OX4 | 10.2 ± 0.8 | 4.3 ± 0.7 | 47.9 ± 5.4 | 9.2 ± 1.2 |
| co-9 | 25.5 ± 2.0 | 8.1 ± 0.9 | 46.7 ± 4.1 | 8.1 ± 1.1 |
| gi-1 | 27.4 ± 1.7 | 8.3 ± 0.8 | 48.3 ± 4.1 | 8.9 ± 1.2 |
| mlk4-2/co-9 | 25.4 ± 2.8 | 8.2 ± 0.8 | 46.9 ± 3.1 | 8.7 ± 1.1 |
| mlk4-2/gi-1 | 26.9 ± 2.8 | 7.9 ± 1.1 | 48.5 ± 3.0 | 8.6 ± 1.1 |
| OX4/co-9 | 25.9 ± 1.8 | 8.4 ± 0.8 | 46.8 ± 3.5 | 8.7 ± 1.1 |
| OX4/gi-1 | 27.5 ± 1.9 | 8.6 ± 0.8 | 48.7 ± 5.1 | 8.9 ± 1.1 |
| cca1-21 | 10.9 ± 0.8 | 3.7 ± 0.6 | ||
| cca1-22 | 10.8 ± 0.9 | 3.7 ± 0.8 | ||
| mlk4-2/cca1-22 | 17.1 ± 1.6 | 6.1 ± 0.7 | ||
| soc1-6 | 19.5 ± 1.3 | 5.9 ± 0.8 | ||
| mlk4-2/soc1-6 | 25.1 ± 2.3 | 8.7 ± 0.9 | ||
| flc-3 | 10.9 ± 0.7 | 3.0 ± 0.9 | ||
| mlk4-2/flc-3 | 14.7 ± 0.8 | 5.9 ± 1.0 | ||
| Complemented line 8 | 12.5 ± 0.7 | 3.1 ± 0.5 | ||
| Complemented line 36 | 12.5 ± 0.7 | 3.2 ± 0.5 | ||
Values shown are mean number ± sd of rosette and cauline leaves; 40 plants were scored for each line. MLK4-overexpressing lines 4 and 5 are indicated as OX4 and OX5, respectively.
CO is the central regulator promoting flowering in the LD pathway. To investigate the relationship between MLK4 and CO, we measured the transcript levels of CO and FT by qRT-PCR, revealing that CO and FT expression was reduced in the mlk4 mutants (Figure 1F; Supplemental Figure 2C). This result was further validated by monitoring the expression of stable transgenic GUS driven by the CO promoter; ProCO:GUS was highly expressed in the vascular tissues of the wild-type plants but had limited expression in mlk4-2 vascular tissue (Figure 1G). We then investigated whether MLK4 exhibits a circadian rhythm-dependent expression pattern. The transcript levels of MLK4 displayed a peak at the end of the 16-h light period, which is similar to the expression patterns of CO and FT (Supplemental Figure 2D).
To investigate the genetic relationship between CO and MLK4, we generated the co mlk4 double mutant by crossing co-9 with mlk4-2. The double mutant had a similar flowering time to co-9 under LD and SD photoperiods (Figures 1H and 1I, Table 1). Next, the MLK4-overexpressing line was crossed to co-9, which resulted in a late-flowering phenotype and similar flowering time to co-9 under LD and SD photoperiods (Supplemental Figure 2E; Table 1). These results indicate that MLK4 and CO are involved in the same pathway and that flowering time modulated by MLK4 depends on CO. The involvement of MLK4 in the photoperiod pathway was further validated by examining the mlk4-2 flc-3 and mlk4-2 soc1-6 double mutants. The mlk4 flc3 double mutant displayed late flowering compared with flc-3, but early flowering compared with mlk-2. However, the flowering time of the mlk4 soc1 double mutant was later than that of the mlk4-2 and soc1-6 single mutants under LD (Table 1). We therefore conclude that MLK4 is involved in the photoperiod pathway.
MLK4 Phosphorylates Histone H2A at Serine 95
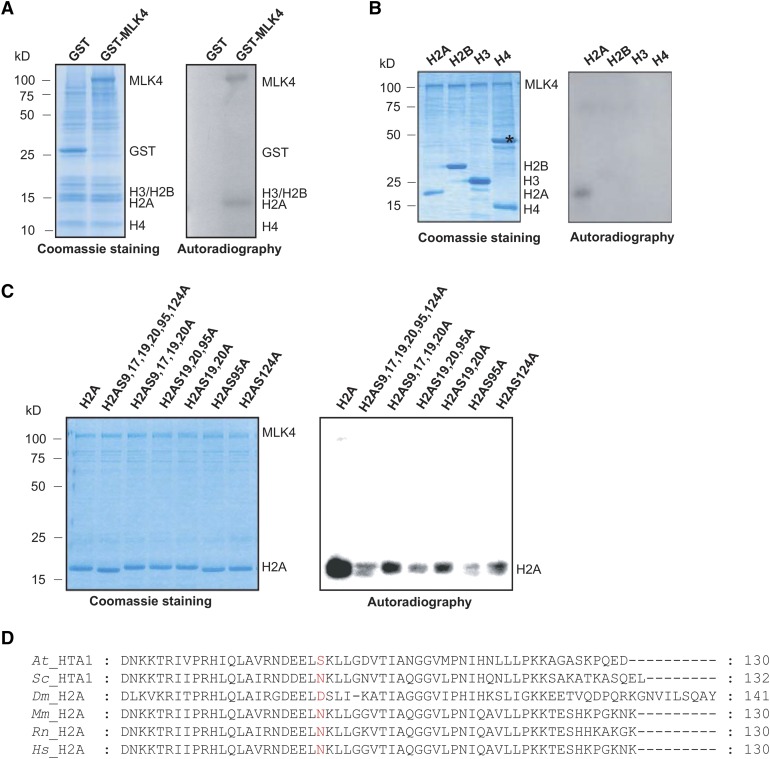
We investigated the MLK4 kinase activity on histones by performing a phosphorylation assay using glutathione S-transferase (GST)-fused MLK4 and GST alone. GST alone did not exhibit any histone kinase activity, but MLK4 specifically phosphorylated cauliflower histone H2A, but not histones H2B, H3, or H4, indicating that MLK4 is a kinase specific to H2A (Figure 2A). To confirm these results, we investigated the activity of MLK4 on purified Arabidopsis histones H2A, H2B, H3, and H4 from Escherichia coli. H2A, but not the other histones, was phosphorylated by MLK4 (Figure 2B).
Figure 2.
Phosphorylation of H2A at Serine 95 by MLK4.
(A) The kinase activity of MLK4 on cauliflower core histones was assessed. The GST-MLK4 purified from E. coli incubated with cauliflower core histone to test in vitro phosphorylation activity. The positions of H2A, H2B, H3, H4, MLK4, and GST alone are indicated.
(B) MLK4 phosphorylates H2A. The ability of MLK4 to phosphorylate H2A, H2B, H3, and H4 was assessed. Asterisk indicates a nonspecific band.
(C) The activity and specificity of the MLK4 kinase were assessed using different substrates. The substrates were H2A fused with His and were either wild-type H2A or had serine-to-alanine substitutions at various residues.
For (A) to (C), the left panel shows the Coomassie blue-stained gel, and the positions of the different histones and MLK4 are indicated on the right. Autoradiography (right panel) shows kinase activity and specificity. Molecular mass markers in kilodaltons are indicated on the left.
(D) Alignment of histone H2A proteins from Arabidopsis, yeast (S. cerevisiae), fruit fly (Drosophila), mouse (Mus musculus), Rat (Rattus norvegicus), and human (Homo sapiens). The serine 95 of Arabidopsis H2A and corresponding sites from different species are marked in red. The sequences of H2As were aligned using ClustalW and examined with MEGA5.
Arabidopsis H2A contains six serines, four threonines, and three tyrosines, which are all potential phosphorylation sites. These amino acids in H2A were individually replaced with alanines and the mutated histones were used as substrates to test the kinase activity of MLK4. The H2A substrate in which the serines were replaced by alanines (H2ASA) was nearly devoid of MLK4-modulated phosphorylation, while the substrates with threonine-to-alanine (H2ATA) or tyrosine-to-alanine (H2AYA) substitutions were still phosphorylated normally by MLK4, indicating that serines are the predominant phosphorylation sites by MLK4 in Arabidopsis H2A (Supplemental Figure 3).
Next, the individual serine sites were investigated for phosphorylation by MLK4. The six serines of H2A are located at amino acid positions 9, 17, 19, 20, 95, and 124. The H2A histones containing serine-to-alanine substitutions at 9, 17, 19, and 20 were strongly phosphorylated, but this phosphorylation activity was impaired when serine 19, serine 20, and serine 95 were substituted by alanine (Figure 2C). Further substitution analyses revealed that the phosphorylation activity of MLK4 was reduced when serine 95, but not serine 19 or serine 20, was replaced by alanine (Figure 2C); therefore, MLK4 has phosphorylation activity at H2A serine 95.
Serine 95 of H2A Is Unique to Plants
The Arabidopsis genome has five genes encoding H2A, three encoding H2A.Z, three encoding H2A.W, and two encoding H2A.X. Phylogenetic alignments of the H2A variants showed that serine 95 exists in most of the H2As and the H2A.Xs, but not in H2A.Z, H2A.W, or in the H2A protein HTA4 (Supplemental Figure 4A). Moreover, serine 95 is conserved in land plants from moss to angiosperms, and the Charophyte green alga Klebsormidium, but not in the Chlorophyte green alga Chlamydomonas (Supplemental Figure 4B), suggesting that serine 95 is likely specific to streptophytes. The serine 95 residue was also not observed in yeast, Drosophila melanogaster, mouse, or humans, indicating that serine 95 is unique to plants (Figure 2D).
H2AS95ph Is Essential for Promoting Flowering Time
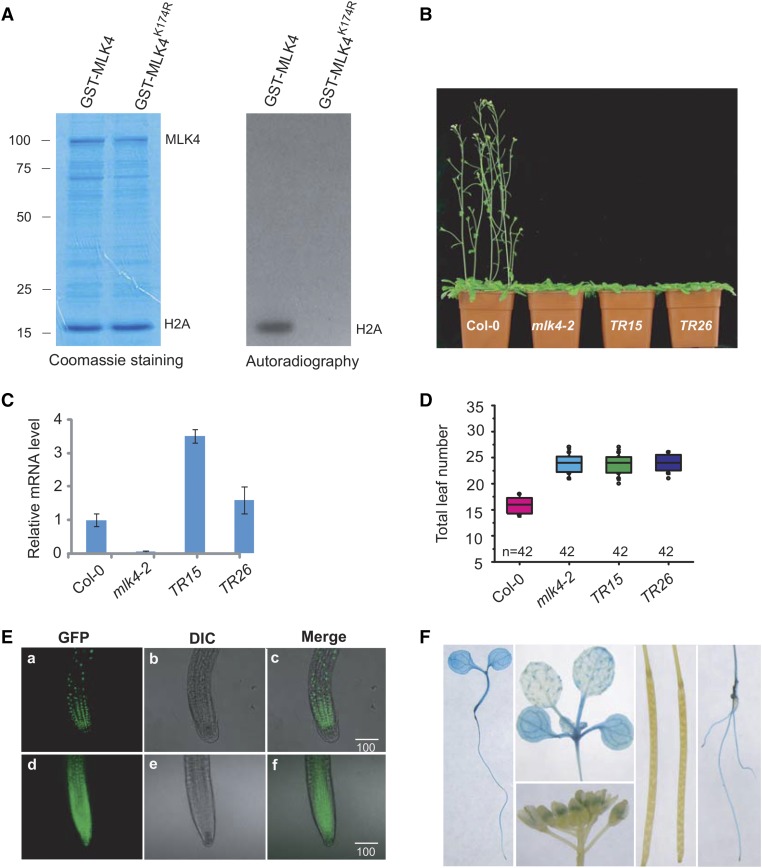
MLK4 has a characteristic conserved glycine-rich ATP binding loop with the consensus sequence GXGXXG (a structural hallmark of protein kinases and nucleotide binding proteins) and an invariant lysine, which gives the enzyme the correct structure for phosphoryl-transfer (Steinberg, 2008). The invariant lysine 174 of MLK4 was mutated to arginine (MLK4K174R) to investigate whether it is required for MLK4 function. We expressed and purified MLK4 K174R from E. coli and tested its Arabidopsis H2A phosphorylation activity. MLK4, but not MLK4 K174R, phosphorylated H2A, indicating that lysine 174 is critical for the H2A phosphorylation activity of MLK4 (Figure 3A).
Figure 3.
Complementation of mlk4-2 with Mutated MLK4K174R and Subcellular Localization of MLK4.
(A) The conserved lysine 174 of MLK4 was converted to arginine (MLK4K174R) to investigate its necessity for phosphorylation of Arabidopsis H2A in vitro. MLK4K174R and H2A were expressed and purified in E. coli. MLK4 was used as a positive control. The Coomassie-stained gel (left) shows protein loading; the autoradiograph (right) shows phosphorylation. Molecular mass markers in kilodaltons are indicated on the left.
(B) Phenotypic analysis of flowering time in Col-0, mlk4-2, and two transgenic lines harboring ProMLK4:MLK4K174R (TR15 and TR26) under LD. Two complemented lines with ProMLK4:MLK4K174R in mlk4-2 are indicated as TR15 and TR26.
(C) Relative transcript levels of MLK4 in the wild-type (Col-0), mlk4-2, TR15, and TR26 plants. Experiments were repeated at least three times, and the data from the representative experiments shown are presented as means ± se, n = 3 replicates.
(D) The total leaf number of Col-0, mlk4-2, TR15, and TR26 under LD. Flowering time was assessed by counting the number of rosette leaves and cauline leaves at bolting under LD and SD. Values shown are mean ± sd of total leaves; 42 plants were scored for each line.
(E) MLK4 localizes to the nucleus. Subcellular localization of GFP fusion proteins was observed in 7-d-old Arabidopsis seedlings stably expressing GFP or GFP fusion proteins. Root tip cells overexpressing GFP-MLK4 (panels a to c) and GFP (panels d to f) observed by confocal laser scanning microscopy and bright-field microscopy (differential interference contrast [DIC]) and merged images are shown. Bars = 100 µm.
(F) The expression pattern of MLK4. GUS staining from stably expressed ProMLK4:GUS was observed in cotyledons, hypocotyls, roots, and pollen. Staining was not observed in siliques.
We then used a complementation test to determine whether MLK4 K174R had MLK4 function in vivo. To that end, a vector containing MLK4 K174R fused with the HA tag driven by the MLK4 promoter (ProMLK4:HA-MLK4K174R) was transformed into the mlk4-2 mutant. MLK4 K174R did not complement the late-flowering phenotype of mlk4-2 despite having a similar level of MLK4 transcript to the wild type (Figures 3B to 3D), suggesting that H2AS95ph modulated by MLK4 might play a critical role in the regulation of flowering time.
Subcellular Location of MLK4 and Expression Pattern of MLK4
Since MLK4 has phosphorylation activity for histone H2A, we reasoned that it is a nuclear protein. We generated transgenic plants expressing MLK4 fused to green fluorescent protein (GFP) or GFP alone, finding that MLK4 indeed localized to the nucleus, whereas GFP alone was observed in the nucleus and the cytoplasm (Figure 3E; Supplemental Figure 5A). This result was confirmed in transformed Arabidopsis protoplasts. MLK4 fused to GFP colocalized with 4′,6-diamidino-2-phenylindole (Supplemental Figure 5B), suggesting that MLK4 is a nuclear protein.
Next, we investigated the expression pattern of MLK4 by transforming plants with a construct containing GUS driven by the MLK4 promoter (ProMLK4:GUS). ProMLK4:GUS activity was mainly observed in the vascular tissues of the cotyledons, hypocotyls, and roots, as well as pollen (Figure 3F). No GUS activity was detected in the siliques.
MLK4 Interacts with CCA1 in Vitro and in Vivo
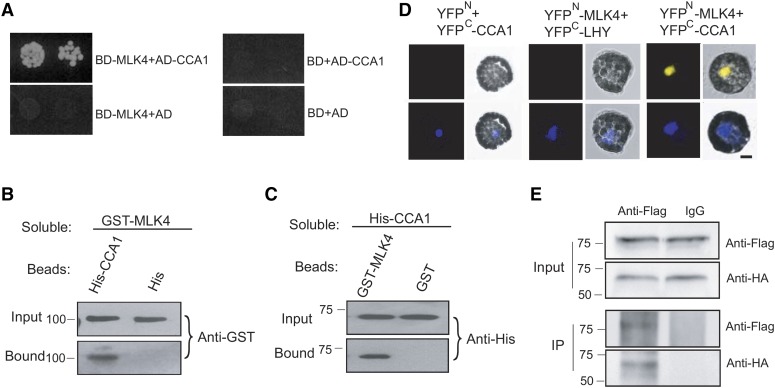
To elucidate its effects on the genetic regulation of flowering, we next investigated the interaction of MLK4 with flowering network proteins using the yeast two-hybrid system. Yeast two-hybrid analysis revealed that CCA1, but not the other proteins in the photoperiod pathway, interact with MLK4 (Figure 4A). These results were confirmed using a protein pull-down assay; MLK4 fused to GST was observed to bind beads containing His fused to CCA1, but not His beads alone (Figure 4B). In a complementary experiment, bead-bound GST-MLK4, but not the GST control, pulled down the soluble His-tagged CCA1 (Figure 4C).
Figure 4.
MLK4 Interacts with CCA1.
(A) A yeast two-hybrid assay revealed an interaction between MLK4 and CCA1. The growth of two dilutions (2 × 10−2 and 2 × 10−3) of the yeast culture on SD medium lacking Trp, Leu, His, and adenine is shown.
(B) Beads containing a His tag (His) or His-fused CCA1 were assayed for their ability to bind to soluble GST-fused MLK4. The input and bound proteins were detected with an antibody to GST (anti-GST).
(C) Beads containing a GST tag or GST-fused MLK4 were assessed for their ability to bind to soluble His-fused CCA1 and detected with antibody to His (anti-His).
(D) Either MLK4 fused to the N terminus of YFP or the N terminus of YFP alone were tested for their ability to bind to the C terminus of YFP fused to LATE ELONGATED HYPOCOTYL (LHY) or the C terminus of YFP fused to CCA1. Yellow fluorescence and a bright-field image were recorded and the resulting images were merged. Twenty-five cells were examined for each transformation. Bar = 10 µm.
(E) Coimmunoprecipitation of MLK4 and CCA1. FLAG-CCA1 and HA-MLK4 were cotransformed into Arabidopsis protoplasts, and the expressed proteins were immunoprecipitated using an anti-FLAG antibody and detected with anti-Flag and anti-HA.
In (A) to (E), experiments were repeated at least three times, and representative experiments are shown. In (B) to (E), molecular mass markers in kilodaltons are indicated on the left; the sizes of the bands are as expected.
This protein pull-down interaction was confirmed in vivo by a bimolecular fluorescence complementation (BiFC) analysis. Functional YFP in the nucleus was observed when MLK4-YFPN (MLK4 fused with the yellow fluorescent protein N terminus) was coexpressed with CCA1-YFPC (CCA1 fused with the YFP C terminus) in Arabidopsis protoplasts, but not with the respective controls, thereby providing further evidence that MLK4 binds directly to CCA1 (Figure 4D). This interaction was further validated by coimmunoprecipitation (co-IP). FLAG-MLK4 and HA-CCA1 were cotransformed into Arabidopsis protoplasts, followed by immunoprecipitation using the anti-FLAG antibody. MLK4, but not the control, was observed to coprecipitate with CCA1 (Figure 4E). These results suggest that MLK4 interacts with CCA1 in vitro and in vivo.
CCA1 Binds the GI Promoter in Vitro and in Vivo
CCA1 is phosphorylated by CK2 (Sugano et al., 1998); therefore, we investigated whether MLK4 could phosphorylate CCA1. However, MLK4 was not observed to phosphorylate CCA1, whereas CK1A, one of the α-subunits of CK2, was able to phosphorylate CCA1, indicating that MLK4 is not involved in the posttranslational modification of CCA1 (Supplemental Figure 5C).
To evaluate the genetic relationship between CCA1 and MLK4, we isolated two cca1 null alleles (Supplemental Figures 6A to 6C). The loss-of-function cca1 mutants flowered earlier than the wild type (Figures 5A and 5B, Table 1). We generated the mlk4-2 cca1-22 double mutant and found that it flowered later than the wild type under LD conditions, similar to the mlk4-2 mutant (Figures 5A and 5B, Table 1). These results suggest that MLK4 acts in the same pathway as CCA1.
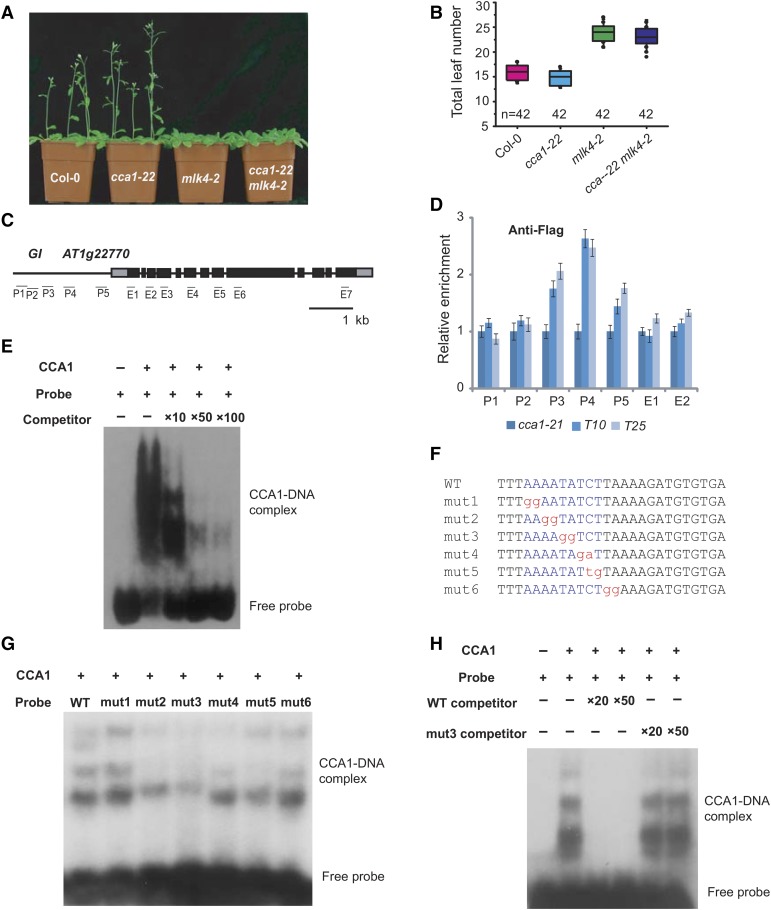
Figure 5.
CCA1 Binds the Promoter of GI in Vitro and in Vivo.
(A) Representative images of 30-d-old Col-0, cca1-22, mlk4-2, and the cca1-22 mLk4-2 double mutant under a LD photoperiod.
(B) The total leaf number of Col-0, cca1-22, mlk4-2, and cca1-22 mLk4-2 plants under LD. Flowering time was assessed by counting the number of rosette leaves and cauline leaves at bolting under LD. Values shown are mean ± sd of total leaves; 42 plants were scored for each line.
(C) Gene structure of GI, indicating exons (boxes) and introns (lines). The locations of the gene regions analyzed by ChIP-PCR are marked. P1 to P5 indicate regions in the GI promoter, and E1 to E7 indicate the coding region.
(D) The amounts of CCA1 at different regions of GI were determined using ChIP-PCR. T10 and T25 are shown in Supplemental Figure 6. The y axis denotes enrichment relative to UBIQUITIN. Experiments were repeated at least three times, and the data from the representative experiments shown are presented as means ± se, n = 3 replicates.
(E) Gel shift assay with CCA1 and fragments of the GI promoter region. The binding ability of CCA1 to fragments of the GI promoter (indicated in Supplemental Figure 7) labeled with 32P was assessed, and this binding specificity was tested by adding unlabeled competitor probe.
(F) The sequences of probes used for EMSA. The conserved motif, AAAATATCT, is marked in blue, and the mutated nucleotides are marked in red, lowercase letters.
(G) and (H) Gel shift assay with CCA1 and different probes. The binding ability of CCA1 to different mutated probes labeled with 32P was assessed, and this binding specificity was tested by adding unlabeled wild-type competitor probe or mutated probe 3.
CCA1 encodes a MYB-related transcription factor that binds a conserved DNA motif, AAAATATCT (Alabadí et al., 2001). The GI promoter region contains three of these motifs in the P3 and P4 regions (Supplemental Figure 7; Figure 5C). To examine the binding of CCA1 to GI, we produced a construct with full-length CCA1 cDNA fused to a FLAG tag driven by the CCA1 promoter (ProCCA1:FLAG-CCA1). When this construct was transformed into the cca1-22 mutant, it rescued the short hypocotyl phenotype, indicating that FLAG-CCA1 retains CCA1 function (Supplemental Figures 6D and 6E). We then assessed the distribution of CCA1 using a chromatin immunoprecipitation (ChIP) analysis with a specific FLAG antibody, followed by a quantitative PCR analysis of the DNA enrichment at multiple points along the GI promoter. CCA1 was strongly enriched at the GI promoter in two complemented lines, suggesting that CCA1 binds to the GI promoter in vivo (Figures 5C and 5D). This interaction was confirmed by an electrophoretic mobility shift assay (EMSA). A retarded band was observed when CCA1 was present, but was abolished when the reaction included specific competitor probes (Figures 5E and 5F; Supplemental Figure 7). We used the mutated probes to test the binding specificity of CCA1, finding that the bands were reduced in assays with the mutated probes, except for mutated probe 1 (Figures 5F and 5G). The mutated probes failed to reduce the binding between CCA1 and wild-type probe (Figure 5H). These results indicate that CCA1 specifically binds the GI promoter in vitro and in vivo.
MLK4 Is Required for the Deposition of H2AS95ph at GI
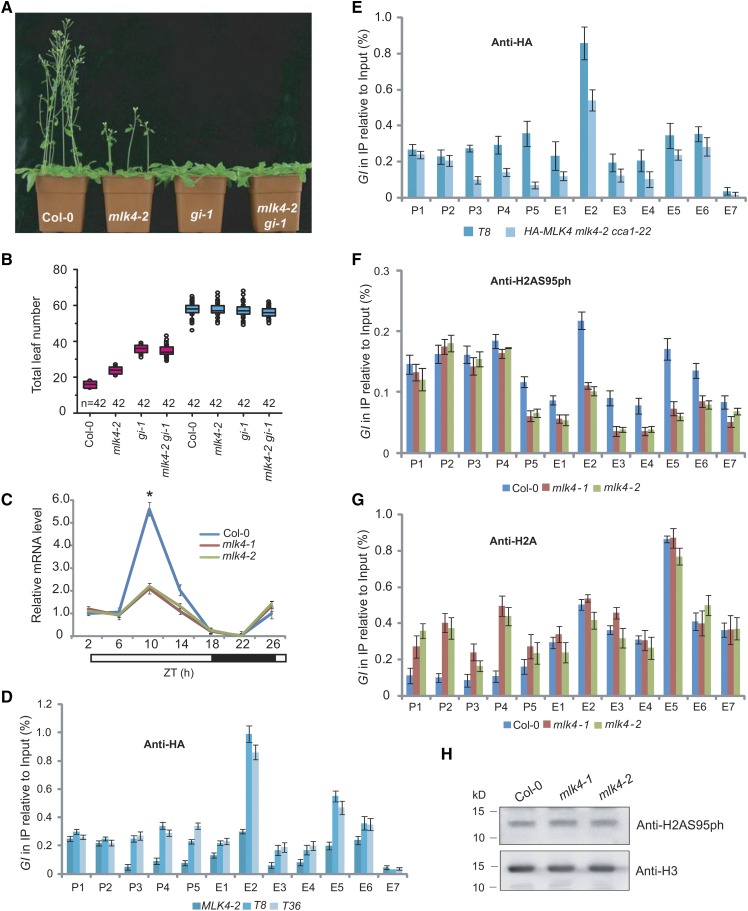
To elucidate the genetic interaction between MLK4 and GI, we generated the mlk4-2 gi-1 double mutant, which had a similar phenotype to the gi-1 mutant under LD and SD conditions (Figures 6A and 6B). Next, we crossed the MLK4-overexpressing line to gi-1, which resulted in a late-flowering phenotype, like that of gi-1 (Supplemental Figures 8A and 8B; Table 1). The level of GI expression was reduced in the mlk4 mutants (Figure 6C). These observations indicate that MLK4 acts in the same pathway as GI.
Figure 6.
MLK4 Is Required for H2A Serine 95 Phosphorylation at GI.
(A) Representative images of 30-d-old Col-0, mlk4-2, gi-1, and the mlk4-2 gi-1 double mutant under a LD photoperiod.
(B) The total leaf number of Col-0, mlk4-2, gi-1, and mlk4-2 gi-1 mutants under LD and SD. Flowering time was assessed by counting the number of rosette leaves and cauline leaves at bolting under LD and SD. Values shown are mean ± sd of total leaves; 42 plants were scored for each line.
(C) The relative mRNA levels of GI were determined in Col-0 and the mlk4 mutants. The black bar indicates the dark period, and the white bars indicate the light periods. ZT, Zeitgeber time. Experiments were repeated at least three times, and the representative experiments shown indicate the mean ± se, n = 3 replicates. Asterisks indicate significant difference from Col-0 using Student’s t test (P < 0.05).
(D) The amounts of MLK4 at different regions of GI were determined in mlk4-2 complemented lines T8 and T36 (see Supplemental Figure 1).
(E) The amounts of MLK4 in different regions of GI were determined in T8 and HA-MLK4 mLk4-2 cca1-22.
(F) The amounts of H2AS95ph at different regions of GI were tested in the wild type and mlk4 mutants.
(G) The amounts of H2A in different regions of GI were tested in the wild type and mlk4 mutants.
From (D) to (G), the y axis denotes enrichment relative to input. Experiments were repeated at least three times, and the data from representative experiments shown are presented as means ± se, n = 3 replicates. P1 to P5 indicate regions in the GI promoter, and E1 to E7 indicate the coding region (see Figure 5C).
(H) The global H2AS95ph levels were measured in the wild type and mlk4 mutants. H3 is shown as an internal control. The molecular weight is indicated on the left; the sizes of the bands are as expected. Experiments were repeated at least three times, and representative experiments are shown.
Given that CCA1 interacts with MLK4 and binds to the GI promoter, we reasoned that MLK4 might regulate GI directly, as does CCA1. We investigated the distribution of MLK4 at GI using two lines carrying the ProMLK4:HA-MLK4 transgene in the mlk4-2 background, which complemented the mutant phenotype (Supplemental Figure 1). We measured the binding of MLK4 using a ChIP assay with the specific HA antibody, followed by quantitative PCR analysis of the amount of DNA enrichment. MLK4 was strongly enriched at the GI promoter and along the gene body in the two complemented lines, suggesting that MLK4 directly targets GI in vivo (Figure 6D). Of note, the broader enrichment of MLK4 at GI in comparison to the binding of CCA1 mainly to the promoter region might partly be due to MLK4 spreading from the promoter region. To this end, we conclude that MLK4 promotes CO expression via directly targeting GI.
We asked whether MLK4 could bind the GI promoter in vitro. We performed an EMSA and observed a retarded band when CCA1, but not MLK4, was present (Supplemental Figure 8C). To investigate whether the targeting of GI by MLK4 is dependent on CCA1 in vivo, we crossed ProMLK4:HA-MLK4/mlk4-2 into cca1-22 to generate the HA-MLK4/mlk4-2 cca1-22 mutant. We measured the profile of MLK4 at GI using ChIP-PCR. The enrichment of MLK4 was reduced in the HA-MLK4/mlk4-2 cca1-22 mutant (Figure 6E), suggesting that the targeting of GI by MLK4 is dependent on CCA1 in vivo.
We generated an antibody specific to H2AS95ph and found that it was able to recognize H2A phosphorylated at serine 95, but not unphosphorylated H2A (Supplemental Figure 9). We measured the distribution of H2AS95ph at GI using ChIP with the H2AS95ph-specific antibody, followed by quantitative PCR analysis. The enrichment of H2AS95ph was observed at the GI promoter and along the coding region of the gene, whereas the enrichment of H2AS95ph was reduced in the mlk4 mutants (Figure 6F), indicating that the deposition of H2AS95ph depends on MLK4. To confirm that reduced H2AS95ph is not caused by a reduction of H2A in the mlk4 mutants, we measured the deposition of H2A via ChIP-PCR. High H2A deposition at GI was observed in the mlk mutants, and the H2A deposition at the GI coding region in the mlk4 mutants was as high as that of the wild type (Figure 6G), suggesting the reduced H2AS95ph is caused by phosphorylation of H2A, but not by H2A density. We next investigated whether H2AS95ph was attenuated in mlk4 mutants using protein gel blot analysis. No notable changes of H2AS95ph were observed in the wild type and mlk4 mutants (Figure 6H), suggesting MLK4 is likely involved in specific gene regulation. We thereby conclude that MLK4 is required for the deposition of H2A95ph and that H2AS95ph is associated with transcriptional activity.
CCA1 Interacts with YAF9a, a Co-Subunit of the SWR1 and NuA4 Complexes
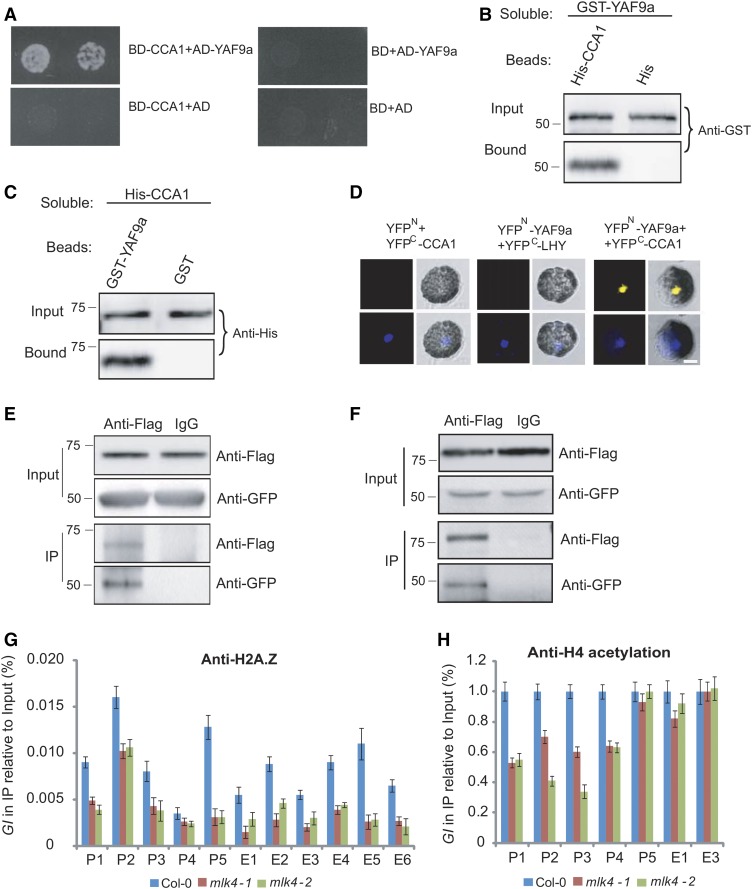
The positive relationship between the H2AS95ph modification and the transcript level of GI suggests that the histone acetylation or chromatin remodeling complexes may interact with MLK4. In a yeast two-hybrid screening, CCA1 and MLK4 were fused with the BD and used to investigate associations with histone acetyltransferases, histone deacetylases, and components of the SWR1 and NuA4 complexes. CCA1, but not MLK4, was observed to bind to YAF9a, a component of the NuA4 and SWR1 complex (Figure 7A).
Figure 7.
MLK4 Is Required for H2A.Z Deposition and H4 Acetylation.
(A) A yeast two-hybrid assay revealed interaction between CCA1 and YAF9a. The growth of two dilutions (2 × 10−2 and 2 × 10−3) of the yeast culture on SD medium lacking Trp, Leu, His, and adenine is shown.
(B) Beads containing a His tag (His) or His-fused CCA1 were assayed for their ability to bind a soluble GST-fused YAF9a. The input and bound protein was detected with an antibody to GST (anti-GST).
(C) Beads containing a GST tag or a GST-fused YAF9a were assessed for their ability to bind a soluble His-fused CCA1 and detected with an antibody to His (anti-His).
(D) YAF9a fused to the N terminus of YFP or the N terminus of YFP alone were tested for the ability to bind to the C terminus of YFP fused to LHY or the C terminus of YFP fused to CCA1. Twenty-five cells were examined for each transformation. Bar = 10 µm.
(E) Coimmunoprecipitation of CCA1 and YAF9a. FLAG-CCA1 and GFP-YAF9a were cotransformed into Arabidopsis protoplasts, immunoprecipitated using an anti-FLAG antibody, and detected with anti-FLAG and anti-GFP.
(F) Coimmunoprecipitation of MLK4 and YAF9a, FLAG-MLK4 and GFP-YAF9a, were cotransformed into Arabidopsis protoplasts, immunoprecipitated using an anti-FLAG antibody, and detected with anti-FLAG and anti-GFP.
In (A) to (F), experiments were repeated at least three times, and representative experiments are shown. Molecular mass markers in kilodaltons are indicated on the left, and the bands agree with expectation.
(G) and (H) The amounts of H2A.Z and H4 acetylation at different regions of GI were determined using ChIP-PCR. The y axis denotes enrichment relative to input or UBIQUITIN. Experiments were repeated at least three times, and the data from the representative experiments shown are presented as means ± se, n = 3 replicates.
We confirmed these interactions via a protein pull-down assay; GST-bound YAF9a was observed to bind beads containing His-fused CCA1, but not to bind His beads alone (Figure 7B). In a complementary experiment, bead-attached GST-YAF9a, but not the GST control, was observed to bind the soluble His-tagged CCA1 (Figure 7C). We further confirmed this interaction using BiFC. A functional YFP was observed in the nucleus following the coexpression of YAF9a-YFPN with CCA1-YFPC, but not with the controls, thereby providing further evidence that YAF9a binds directly to CCA1 (Figure 7D). We validated these interactions by Co-IP. GFP-YAF9a and FLAG-CCA1 were cotransformed into Arabidopsis protoplasts, then immunoprecipitated using an anti-FLAG antibody, further demonstrating that YAF9a, but not the control, binds to CCA1 (Figure 7E). These results indicate that YAF9a interacts with CCA1 in vitro and in vivo.
MLK4 Is Required for Deposition of H2A.Z and Acetylation of H4 at GI
We next investigated whether YAF9a forms a protein complex with MLK4 in vivo. GFP-YAF9a and FLAG-MLK4 were cotransformed into protoplasts, then immunoprecipitated using an anti-FLAG antibody. YAF9a was observed bind to MLK4, suggesting that MLK4 and YAF9a are involved in a protein complex (Figure 7F).
Given that YAF9 is conserved between yeast, mammalian, and plant cells, we next considered the possibility that there may be changes in the level of H2A.Z accumulation and H4 acetylation in mlk4 plants. We investigated the profile of H2A.Z using ChIP with an H2A.Z-specific antibody, followed by quantitative PCR analysis of DNA enrichment. In addition to peak 5 at the transcriptional start site of GI, H2A.Z was highly enriched at the GI promoter and along the GI coding sequence in wild-type plants, but not in the mlk4 mutants (Figure 7G). Using an antibody specific to acetylated H4, we found that H4 acetylation was reduced at the promoter of GI, but not in the gene body of GI in the mlk4 mutants (Figure 7H). Thus, we conclude that MLK4 is required for the deposition of H2A.Z and the acetylation of H4 at GI.
The yaf9a mutant was first identified based on its early flowering phonotype in LD, whereas its phenotype is not obvious in SD (Zacharaki et al., 2012), suggesting that YAF9a is at least partially involved in the photoperiod pathway. To investigate the function of YAF9a at GI, we examined the yaf9a-1 mutant and found that the expression of GI was reduced in this mutant (Supplemental Figures 10A to 10C). Examination of H2A.Z and H4 acetylation revealed that H2A.Z and H4 acetylation were reduced in the yaf9a mutants compared with the wild type (Supplemental Figures 10C and 10D), indicating that YAF9a is required for the expression, H2A.Z deposition, and H4 acetylation of GI. Collectively, these results indicate that MLK4 modulates the expression of GI via reducing H2A.Z deposition and H4 acetylation of GI.
DISCUSSION
In this study, we identified a modification site in the histone core domain, the serine 95 of H2A, which is phosphorylated. Our data provide insights into the mechanism by which the histone-phosphorylating enzyme MLK4 affects photoperiod-dependent flowering time in Arabidopsis. MLK4 has H2A serine 95 phosphorylation activity in vitro and in vivo. The loss of MLK4 function results in late-flowering plants with reduced transcript and H2AS95ph levels of GI. The substitution of MLK4 lysine 174 with arginine led to a loss of H2A phosphorylation activity in vitro, and MLK4K174R failed to rescue the late-flowering phenotype of mlk4 mutants, demonstrating that H2AS95ph is vital for flowering time. Taken together, these results suggest that MLK4 might be critical for H2AS95ph and the regulation of the downstream developmental processes in Arabidopsis. We observed MLK4 expression in roots and anthers, suggesting that MLK4 might also be involved in root and pollen development.
Our results indicate that MLK4 phosphorylates multiple sites on H2A, but prefers serine 95. The MLK4 activity on H2A containing serine-to-alanine substitutions at serine 95 was similar to its activity on H2A containing serine-to-alanine substitutions at 9, 17, 19, 20, 95, and 124, suggesting that serine 95 is the dominant site for MLK4 activity. However, MLK4 still had partial phosphorylation acidity when all serines in H2A were replaced with alanine, suggesting that MLK4 might phosphorylate other sites in H2A. We observed no notable changes of H2AS95ph in the wild type or the mlk4 mutants, suggesting that other kinases might phosphorylate serine 95 of H2A. MLK4 also has autophosphorylation activity, but whether this autophosphorylation activity affects its functions remains unclear. A recent study showed that MLK4 copurified with components of the evening complex of the circadian clock in a phytochrome B-dependent manner (Huang et al., 2016), suggesting that MLK4 might phosphorylate the components of this complex, which may, in turn, regulate GI expression. CCA1 binds to GI and represses its expression (Lu et al., 2012). Although mutations in CCA1 resulted in slightly early flowering, the cca1 mLk4 double mutant exhibited late flowering, suggesting that the appropriate flowering time requires both CCA1 and MLK4.
Our results also provide insight into the specificity of MLK4 for its target gene. Most histone modification enzymes have not been observed to bind DNA, except for REF6 and MLL1, which contain a zinc finger domain and CXXC domain, respectively (Cierpicki et al., 2010; Cui et al., 2016). The fundamental question of how these histone modification proteins target specific genes to carry out their functions remains to be explored. Our study showed that CCA1 physically interacts with MLK4 and allows MLK4 to target GI.
Although Arabidopsis MLK4 has a high amino acid sequence similarity to Chlamydomonas MUT9p, MLK4 targets H2AS95ph, whereas MUT9p has H3T3ph activity, although MUT9p has also been shown to have phosphorylation activity on H2A in vitro (Casas-Mollano et al., 2008). Arabidopsis has four MLK proteins that are closely related to MUT9p. Phosphorylation activity on H3 threonine 3 was not observed for MLK1 or MLK2 in vitro; however, defects in mlk1 mLk2 result in the reduction of H3T3ph, indicating that MLK1 or MLK2 has H3T3ph activity in vivo (Wang et al., 2015). The kinase functions of MUT9p may have diverged to produce new functions during the evolution of the land plants, leading to the separation of phosphorylation activity for H3T3 and H2AS95. The sequences of H3 and H4 are conserved among species, whereas the histone H2A and H2B sequences are diverse. Correspondingly, the phosphorylation sites at H2A are much more divergent than those of H3 between yeast, mammals, and plants. The phosphorylation of H2A at serine 129 in yeast and humans is correlated with DNA damage (Rogakou et al., 1998); however, these conserved phosphorylation sites were not observed in Arabidopsis. The alignment of H2A sequences showed that serine 95 exists in plants, but not in yeast, fly, mouse, or mammalian cells, suggesting that the phosphorylation of H2A at serine 95 is specific to plants. This serine 95 of H2A was not observed in Chlamydomonas, suggesting this amino acid residue and MLK4 activity likely evolved de novo in the green algal and plant lineage. Unlike Chlamydomonas, the life cycle of land plants contains a vegetative stage and reproductive stage. Defective H2AS95 phosphorylation modulated by MLK4 resulted in late flowering, indicating that this histone phosphorylation site might promote the transition from the vegetative to the reproductive stage.
A loss of MLK4 function results in the reduced expression and H2AS95ph levels of GI, indicating that the level of H2AS95ph is correlated with the level of GI transcription. H3S10ph facilitates the release of RNA polymerase II from promoter-proximal pausing in Drosophila, while H3S28ph modulates RNA polymerase III-dependent transcription (Ivaldi et al., 2007; Zhang et al., 2011). In our study, the reduced H2AS95ph caused by a loss of MLK4 function resulted in the downregulation of H2A.Z accumulation and H4 acetylation, which are involved in transcriptional activity. In yeast, histone variant H2A.Z is a component of euchromatin that functions to antagonize the opposite chromatin state and is selectively present at the vast majority of gene promoter regions (Raisner et al., 2005). In contrast to yeast, H2A.Z binding levels are correlated with gene activity in humans (Barski et al., 2007), but in plants, the presence of H2A.Z within genes is correlated with lower transcription levels and a higher variability in expression patterns across tissue types and environmental conditions (Coleman-Derr and Zilberman, 2012). In yeast, the Swr1 complex is required for the deposition of histone H2A.Z at specific chromosome locations (Mizuguchi et al., 2004). Arabidopsis contains the corresponding subunits of SWC2 and PHOTOPERIOD-INDEPENDENT EARLY FLOWERING1 (PIE1), which are the homologs of yeast Swc2 and Swr1, respectively. These subunits interact with three Arabidopsis homologs of H2A.Z proteins, HTA8, HTA9, and HTA11 (Choi et al., 2007), indicating that the SWR1 complex is involved in H2A.Z deposition. The loss-of-function pie1 mutant flowers early because PIE1 is required for the deposition of H2A.Z at FLC, MAF4, and MAF5 (Deal et al., 2007). NuA4, the only essential histone acetyltransferase complex in S. cerevisiae, acetylates the N-terminal tails of histones H4 and H2A, which directly stimulates the incorporation of H2A.Z by the SWR1 complex (Altaf et al., 2010). Yaf9 is a component of the NuA4 and SWR1 complexes, can bind histones H3 and H4 in vitro, and is required for histone variant H2A.Z deposition and for H2A.Z and H4 acetylation (Zhang et al., 2004; Altaf et al., 2010). Our results show that Arabidopsis YAF9a is required for H4 acetylation and the deposition of H2A.Z at GI, indicating that Arabidopsis YAF9a might be the counterpart of yeast Yaf9 and that YAF9a is involved in chromatin and transcriptional regulation at GI. MLK4 and YAF9a form a protein complex. Defects in MLK4 might destroy this complex and thus result in the reduction of H2A.Z deposition and H4 acetylation, which indicates that H2AS95ph modulated by MLK4 might be involved in H4 acetylation and the deposition of H2A.Z. Mutations in MLK4 also resulted in enhanced H2A occupancy at the GI promoter; this might be antagonistic to H4 acetylation and H2A.Z deposition. Together, our results provide important insights into the mechanism by which MLK4-modulated H2AS95ph regulates flowering time and promotes gene transcription by enhancing H2A.Z accumulation and H4 acetylation in Arabidopsis.
METHODS
Plant Materials
The Arabidopsis thaliana ecotype Col was grown at 22°C under a LD photoperiod in a 16-h-light/8-h-dark cycle and light intensity of 170 µmol m−2 s−1 or a SD photoperiod with 8 h light/16 h dark. The mutant strains obtained from the SALK collection were as follows: mlk4-1, CS472560; mlk4-2, SALK_201615c; cca1-21, SALKseq_120169; cca1-22, SALKseq_123282; co-9, CS870084; and yaf9a-1, SALK_106430.
Plasmid Constructs
The plasmids were constructed with the DNA primers and protocols described in Supplemental Data Set 1. All cloned DNAs were confirmed by DNA sequencing.
Histone Extraction from Cauliflower
The histone extraction was performed using an extraction kit following the manufacturer’s instructions (Epigenteck; OP-0006-100). Briefly, 1 g surface tissue of cauliflower was chopped and homogenized with pre-lysis buffer. The solution was transferred into a conical tube and centrifuged at 3000 rpm for 5 min at 4°C. The pellet was resuspended in lysis buffer and incubated on ice for 30 min, followed by centrifugation at 12,000 rpm for 5 min at 4°C. The supernatant fraction was balanced with balance buffer and stored for future use.
Yeast Two-Hybrid Assay
The yeast two-hybrid assay was performed according to the manufacturer’s protocol (Clontech; user manual 630489). Briefly, the Saccharomyces cerevisiae strain Y190 was transformed with the bait constructs pGBKT-MLK4 and pGBKT-CCA1, then transformed with pGADT7-CCA1 or pGADT7-YAF9a. Vectors lacking coding region insertions were used as negative controls. The yeast was scored for protein interaction based on their ability to grow on synthetic defined medium lacking Trp, Leu, His, and adenine. The primers used to generate the constructs are shown in Supplemental Data Set 1.
Transient Expression in Arabidopsis Protoplasts and BiFC
For BiFC, MLK4, YAF9a, and CCA1 were cloned into the pUC-SPYCE (amino acids 156–239) or pUC-SPYNE (amino acids 1–155) vectors. Arabidopsis mesophyll protoplast isolation and transformation were performed as described previously (Yoo et al., 2007). Briefly, the leaves of 3-week-old Arabidopsis plants were detached onto double-sided tape, digested with enzymes, and washed with MMG buffer (0.4 M mannitol, 15 mM MgCl2, 4 mM MES, pH 5.7). The protoplasts were cotransformed with the corresponding constructs and then detected under a confocal laser scanning microscope (Zeiss LSM700) or immunoprecipitated with specific antibodies at 1:100 to 200 dilution.
Protein Pull-Down Assays, Co-IP, and Immunoblot Assays
For the pull-down assay, beads were incubated with 3 µg of the fusion protein, washed, and incubated with 3 µg of soluble protein overnight at 4°C. Mock controls included extracts prepared from either the His-Tag or GST vectors. The beads were washed five times with a solution containing 20 mM Tris (pH 7.4), 150 mM NaCl, and 0.05% Tween 20, separated on an SDS-PAGE gel, and analyzed by immunoblot using an anti-GST antibody (GenScript; A00866-100, lot 13D000626) or an anti-His antibody (Abmart; M30111M, lot 273884).
For the Co-IP, MLK4, YAF9a, and CCA1 were fused with FLAG, GFP, or HA and cloned into the pUC19 vector. The co-IP was performed as described previously (Zhang et al., 2015). Briefly, 1 × 106 Arabidopsis protoplasts were lysed with PEN-140 buffer (140 mM NaCl, 2.7 mM KCl, 25 mM Na2HPO4, 1.5 mM KH2PO4, 0.01 mM EDTA, and 0.05% CA-630). The supernatant was precleared with Protein G and precipitated with anti-FLAG (Sigma-Aldrich; H6908, lot SLBQ7119V) or anti-HA (Sigma-Aldrich; H9658, lot 095M4778V) antibodies. The protein complexes were isolated by binding to protein G beads (Santa Cruz; Sc-2002) and washed five times with PEN-400 buffer (400 mM NaCl, 2.7 mM KCl, 25 mM Na2HPO4, 1.5 mM KH2PO4, 0.01 mM EDTA, and 0.05% CA-630). The samples were analyzed by protein gel blot analysis using an anti-GFP (Clontech Laboratories; JL-8, lot A5033481), anti-HA, or anti-FLAG antibody.
Phosphorylation Reaction in Vitro
The phosphorylation reaction assay was performed as described (Demidov et al., 2005; Lu et al., 2017). Briefly, 10 µg core histone or 2 µg of purified protein was incubated in reaction buffer (50 mM Tris-HCl, pH 7.4, 10 mM MgCl2, 50 mM NaCl, 1 mM DTT, 2 mM EDTA, and 50 µM ATP) with 2.5 µCi γ-32P ATP at 30°C for 1 h. The reaction products were separated by SDS-PAGE and autoradiographed with x-ray film.
Complementation Assay
For mlk4 complementation, the ProMLK4:HA-MLK4 construct was generated by fusing the full-length cDNA of MLK4 to HA and then inserting the fusion into a pCambia1300 vector harboring the 2500 bp MLK4 promoter. The construct was then transformed into the mlk4-2 mutant by the floral dip method (Clough and Bent, 1998).
For cca1 complementation, the ProCCA1:FLAG-CCA1 construct was generated by fusing the full-length cDNA of CCA1 to FLAG, then inserting the fusion into a pCambia1300 vector harboring the 2500 bp CCA1 promoter. The construct was then transformed into the cca1 mutant.
Subcellular Location of MLK4 and Promoter-GUS Assay
For the Pro35S:GFP-MLK4 construct, the full-length cDNA of MLK4 was fused with GFP and transformed into pAVA321, then the GFP-MLK4 cassette was subcloned into pCambia 1300. After transformation and selection, the roots of stable transgenic plants were analyzed for GFP subcellular localization under a confocal microscope (Zeiss LSM700).
For the ProMLK4:GUS and ProCO:GUS constructs, over 2500 bp of MLK4 promoter or CO promoter, respectively, were inserted into pCambia 1391z. After transformation and selection, the stable transgenic plants were crossed into different backgrounds and stained with staining buffer [100 mM K4Fe(CN)6 3H2O, 100 mM K3Fe(CN)6, 100 mM X-Gluc, 10% Triton X-100, and 500 mM NaPO4 buffer, pH 7.2] at 37°C overnight, then washed with an ethanol series (30%, 50%, 70%, and 100%) and recovered with water. Over 40 independent transgenic lines were obtained, and the representative plants were used for further GFP or GUS staining assays.
EMSA
The EMSA was performed as described (Hellman and Fried, 2007). Briefly, 1 to 2 µg purified protein was mixed with 4 pmol γ-32P ATP labeled probe with or without various dosages of unlabeled probe. After separation in a 4.5% native nondenaturing acrylamide gel, the gel was exposed with X-film overnight. The sequence of the probe is shown in Supplemental Data Set 1.
Reverse Transcription and RT-PCR
Total RNA was isolated from the leaves of 3-week-old seedlings and reverse transcribed with oligo(dT) primers (Promega), and the amounts of individual gene transcripts were measured with gene-specific primers. RT-PCR analysis was performed with the CFX real-time PCR instrument (Bio-Rad) and SYBR Green mixture (Roche). The relative expression of the genes was quantitated with the 2−ΔΔCT Ct calculation, using UBIQUITIN as the reference housekeeping gene for the expression analyses. The enrichment of DNA at specific genes was quantified with the 2−ΔΔCT Ct calculation, using UBIQUITIN as the reference housekeeping gene or relative to the input DNA (10% of starting chromatin is used for input) for chromatin immunoprecipitation assays. See Supplemental Data Set 1 for primers.
ChIP Assay
The ChIP was performed as described (Lu et al., 2017). Briefly, 3 g of 2-week-old seedlings was fixed with 1% formaldehyde for 10 min and quenched in 0.125 M glycine. The leaves were ground in a mortar and pestle in buffer I (0.4 M sucrose, 10 mM Tris, pH 8.0, 5 mM β-mercaptoethanol, 0.1 mM PMSF, and protease inhibitor cocktail) and filtered through Miracloth. After centrifugation, the pellet was extracted with buffer II (0.25 M sucrose, 10 mM Tris, pH 8.0, 10 mM MgCl2, 1% Triton X-100, 5 mM β-mercaptoethanol, 0.1 mM PMSF, and protease inhibitor cocktail) and then with buffer III (1.7 M sucrose, 10 mM Tris, pH 8.0, 10 mM MgCl2, 1% Triton X-100, 5 mM β-mercaptoethanol, 0.1 mM PMSF, and protease inhibitor cocktail). The nuclei were then lysed in lysis buffer (50 mM Tris pH 8.0, 10 mM EDTA, 1% SDS, 5 mM β-mercaptoethanol, 0.1 mM PMSF, and protease inhibitor cocktail), and the extract was sonicated to fragment the DNA to a size range of 300 to 500 bp. After centrifugation, the supernatant was diluted using dilution buffer (1.1% Triton X-100, 1.2 mM EDTA, 16.7 mM Tris, pH 8.0, 167 mM NaCl, 0.1 mM PMSF, and protease inhibitor cocktail) and precleared with protein A or protein G magnetic beads. Specific antibodies anti-FLAG (Sigma-Aldrich; F3165, lot SLBQ7119V), anti-HA (Sigma-Aldrich; H9658, lot 095M4778V), anti-H2A.Z (Abcam; ab4174, lot GR292900-1), anti-H2A (Abcam; ab18255, lot GR208654-1), and anti-H4 acetylation (Millipore; 06-866, lot 2768452) or control IgG serum were added to the precleared supernatants for an overnight incubation at 4°C. The antibody protein complexes were isolated by binding to protein A or protein G beads. The washed beads were heated at 65°C for 8 h with proteinase K to reverse the formaldehyde cross-linking and digest proteins. The sample was then extracted with phenol/chloroform and the DNA was precipitated in ethanol and resuspended in water. The purified DNA was analyzed by PCR with the gene-specific primers shown in Supplemental Data Set 1.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under the following accession numbers: MLK1 (At5g18190), MLK2 (At3g03940), MLK3 (At2g25760), MLK4 (At3g13670), Arabidopsis thaliana H2A (AED96521.1), Arabidopsis lyrata H2A (EFH40608.1), Populus H2A (EEE97577.2), Sorghum bicolor H2A (EER97345.1), Oryza sativa H2A (BAD01189.1), Selaginella moellendorffii H2A (EFJ34776.1), Physcomitrella patens H2A (EDQ63096.1), Klebsormidium flaccidum H2A (GAQ82823.1), Chlamydomonas reinhardtii H2A (EDO96247.1), Saccharomyces cerevisiae HTA1 (KZV12463.1), Drosophila melanogaster H2A (AGB96377.1), Mus musculus H2AX (CAA84585.1), Rattus norvegicus H2A (NP_001102761.1), Homo sapiens H2A (NP_778235.1), Arabidopsis thaliana HTA1 (AED96521.1), Arabidopsis thaliana HTA2 (AEE85314.1), Arabidopsis thaliana HTA3 (AEE33135.1), Arabidopsis thaliana HTA4 (AEE83295.1), Arabidopsis thaliana HTA5 (AEE28361.1), Arabidopsis thaliana HTA6 (AAO39897.1), Arabidopsis thaliana HTA7 (AED93713.1), Arabidopsis thaliana HTA8 (ABH04530.1), Arabidopsis thaliana HTA9 (AEE32847.1), Arabidopsis thaliana HTA10 (AEE32617.1), Arabidopsis thaliana HTA11 (AAO63269.1), Arabidopsis thaliana HTA12 (ABF59030.1), and Arabidopsis thaliana HTA13 (AEE76410.1).
Supplemental Data
Supplemental Figure 1. Complementation of mlk4-2 with ProMLK4:HA-MLK4.
Supplemental Figure 2. Overexpression of MLK4 results in early flowering under long-day conditions.
Supplemental Figure 3. The specificity of phosphorylation by MLK4 for serines of H2A.
Supplemental Figure 4. Alignment of H2A from different species.
Supplemental Figure 5. Subcellular localization of MLK4 and phosphorylation of CCA1 by MLK4.
Supplemental Figure 6. Complementation of cca1-22 with ProCCA1:FLAG-CCA1.
Supplemental Figure 7. Identification of CCA1 binding sites in the GI promoter.
Supplemental Figure 8. Phenotypic analysis of flowering time in 5-week-old Col-0, OX4, gi-1, and the OX4 gi-1 double mutant.
Supplemental Figure 9. The specificity of the antibody for H2AS95 phosphorylation.
Supplemental Figure 10. YAF9a is required for deposition of H2A.Z and H4 acetylation.
Supplemental Data Set 1. Plasmids and primers used in this study.
Supplemental File 1. Alignments used in Figure 2D and Supplemental Figure 4.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (grants 31371306, 31571315, and 91435101 to Y.D.) and the Strategic Priority Research Program “Molecular Mechanism of Plant Growth and Development” of CAS (Grant XDPB04).
AUTHOR CONTRIBUTIONS
Y.S. and Y.D. conceived the study and designed the experiments. Y.S. performed most of experiments and all authors took part in the interpretation of results and preparation of the manuscript. Y.D. wrote the manuscript.
Glossary
- LD
long-day
- SD
short-day
- BiFC
bimolecular fluorescence complementation
- co-IP
coimmunoprecipitation
- ChIP
chromatin immunoprecipitation
- EMSA
electrophoretic mobility shift assay
Footnotes
Articles can be viewed without a subscription.
References
- Alabadí D., Oyama T., Yanovsky M.J., Harmon F.G., Más P., Kay S.A. (2001). Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293: 880–883. [DOI] [PubMed] [Google Scholar]
- Altaf M., Auger A., Monnet-Saksouk J., Brodeur J., Piquet S., Cramet M., Bouchard N., Lacoste N., Utley R.T., Gaudreau L., Côté J. (2010). NuA4-dependent acetylation of nucleosomal histones H4 and H2A directly stimulates incorporation of H2A.Z by the SWR1 complex. J. Biol. Chem. 285: 15966–15977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee T., Chakravarti D. (2011). A peek into the complex realm of histone phosphorylation. Mol. Cell. Biol. 31: 4858–4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A., Cuddapah S., Cui K., Roh T.-Y., Schones D.E., Wang Z., Wei G., Chepelev I., Zhao K. (2007). High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837. [DOI] [PubMed] [Google Scholar]
- Basnet H., Su X.B., Tan Y., Meisenhelder J., Merkurjev D., Ohgi K.A., Hunter T., Pillus L., Rosenfeld M.G. (2014). Tyrosine phosphorylation of histone H2A by CK2 regulates transcriptional elongation. Nature 516: 267–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casas-Mollano J.A., Jeong B.R., Xu J., Moriyama H., Cerutti H. (2008). The MUT9p kinase phosphorylates histone H3 threonine 3 and is necessary for heritable epigenetic silencing in Chlamydomonas. Proc. Natl. Acad. Sci. USA 105: 6486–6491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerutti H., Casas-Mollano J.A. (2009). Histone H3 phosphorylation: universal code or lineage specific dialects? Epigenetics 4: 71–75. [DOI] [PubMed] [Google Scholar]
- Cheung P., Tanner K.G., Cheung W.L., Sassone-Corsi P., Denu J.M., Allis C.D. (2000). Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol. Cell 5: 905–915. [DOI] [PubMed] [Google Scholar]
- Choi K., Park C., Lee J., Oh M., Noh B., Lee I. (2007). Arabidopsis homologs of components of the SWR1 complex regulate flowering and plant development. Development 134: 1931–1941. [DOI] [PubMed] [Google Scholar]
- Cierpicki T., Risner L.E., Grembecka J., Lukasik S.M., Popovic R., Omonkowska M., Shultis D.D., Zeleznik-Le N.J., Bushweller J.H. (2010). Structure of the MLL CXXC domain-DNA complex and its functional role in MLL-AF9 leukemia. Nat. Struct. Mol. Biol. 17: 62–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Coleman-Derr D., Zilberman D. (2012). Deposition of histone variant H2A.Z within gene bodies regulates responsive genes. PLoS Genet. 8: e1002988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui X., et al. (2016). REF6 recognizes a specific DNA sequence to demethylate H3K27me3 and regulate organ boundary formation in Arabidopsis. Nat. Genet. 48: 694–699. [DOI] [PubMed] [Google Scholar]
- Dai C., Xue H.W. (2010). Rice early flowering1, a CKI, phosphorylates DELLA protein SLR1 to negatively regulate gibberellin signalling. EMBO J. 29: 1916–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J., Sultan S., Taylor S.S., Higgins J.M. (2005). The kinase haspin is required for mitotic histone H3 Thr 3 phosphorylation and normal metaphase chromosome alignment. Genes Dev. 19: 472–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deal R.B., Topp C.N., McKinney E.C., Meagher R.B. (2007). Repression of flowering in Arabidopsis requires activation of FLOWERING LOCUS C expression by the histone variant H2A.Z. Plant Cell 19: 74–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demidov D., Van Damme D., Geelen D., Blattner F.R., Houben A. (2005). Identification and dynamics of two classes of aurora-like kinases in Arabidopsis and other plants. Plant Cell 17: 836–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Q., Han F. (2012). Phosphorylation of histone H2A is associated with centromere function and maintenance in meiosis. Plant J. 71: 800–809. [DOI] [PubMed] [Google Scholar]
- Downs J.A., Lowndes N.F., Jackson S.P. (2000). A role for Saccharomyces cerevisiae histone H2A in DNA repair. Nature 408: 1001–1004. [DOI] [PubMed] [Google Scholar]
- Fornara F., de Montaigu A., Coupland G. (2010). SnapShot: control of flowering in Arabidopsis. Cell 141: 550–550. [DOI] [PubMed] [Google Scholar]
- Fowler S., Lee K., Onouchi H., Samach A., Richardson K., Morris B., Coupland G., Putterill J. (1999). GIGANTEA: a circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J. 18: 4679–4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellman L.M., Fried M.G. (2007). Electrophoretic mobility shift assay (EMSA) for detecting protein-nucleic acid interactions. Nat. Protoc. 2: 1849–1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota T., Lipp J.J., Toh B.-H., Peters J.-M. (2005). Histone H3 serine 10 phosphorylation by Aurora B causes HP1 dissociation from heterochromatin. Nature 438: 1176–1180. [DOI] [PubMed] [Google Scholar]
- Huang H., Alvarez S., Bindbeutel R., Shen Z., Naldrett M.J., Evans B.S., Briggs S.P., Hicks L.M., Kay S.A., Nusinow D.A. (2016). Identification of evening complex associated proteins in Arabidopsis by affinity purification and mass spectrometry. Mol. Cell. Proteomics 15: 201–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi T., Schultz T.F., Harmon F.G., Ho L.A., Kay S.A. (2005). FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science 309: 293–297. [DOI] [PubMed] [Google Scholar]
- Ivaldi M.S., Karam C.S., Corces V.G. (2007). Phosphorylation of histone H3 at Ser10 facilitates RNA polymerase II release from promoter-proximal pausing in Drosophila. Genes Dev. 21: 2818–2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima S.A., Yamagishi Y., Honda T., Ishiguro K., Watanabe Y. (2010). Phosphorylation of H2A by Bub1 prevents chromosomal instability through localizing shugoshin. Science 327: 172–177. [DOI] [PubMed] [Google Scholar]
- Krogan N.J., et al. (2004). Regulation of chromosome stability by the histone H2A variant Htz1, the Swr1 chromatin remodeling complex, and the histone acetyltransferase NuA4. Proc. Natl. Acad. Sci. USA 101: 13513–13518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau P.N.I., Cheung P. (2011). Histone code pathway involving H3 S28 phosphorylation and K27 acetylation activates transcription and antagonizes polycomb silencing. Proc. Natl. Acad. Sci. USA 108: 2801–2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo W.-S., Trievel R.C., Rojas J.R., Duggan L., Hsu J.-Y., Allis C.D., Marmorstein R., Berger S.L. (2000). Phosphorylation of serine 10 in histone H3 is functionally linked in vitro and in vivo to Gcn5-mediated acetylation at lysine 14. Mol. Cell 5: 917–926. [DOI] [PubMed] [Google Scholar]
- Lu C., Tian Y., Wang S., Su Y., Mao T., Huang T., Xu Z. (2017). Phosphorylation of SPT5 by CDKD;2 is required for VIP5 recruitment and normal flowering in Arabidopsis thaliana Plant Cell 29: 277–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S.X., Webb C.J., Knowles S.M., Kim S.H., Wang Z., Tobin E.M. (2012). CCA1 and ELF3 Interact in the control of hypocotyl length and flowering time in Arabidopsis. Plant Physiol. 158: 1079–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay R.M., Peters J.M., Graff J.M. (2001). The casein kinase I family: roles in morphogenesis. Dev. Biol. 235: 378–387. [DOI] [PubMed] [Google Scholar]
- Milne D.M., Looby P., Meek D.W. (2001). Catalytic activity of protein kinase CK1 δ (casein kinase 1δ) is essential for its normal subcellular localization. Exp. Cell Res. 263: 43–54. [DOI] [PubMed] [Google Scholar]
- Mizoguchi T., Wheatley K., Hanzawa Y., Wright L., Mizoguchi M., Song H.-R., Carré I.A., Coupland G. (2002). LHY and CCA1 are partially redundant genes required to maintain circadian rhythms in Arabidopsis. Dev. Cell 2: 629–641. [DOI] [PubMed] [Google Scholar]
- Mizuguchi G., Shen X., Landry J., Wu W.-H., Sen S., Wu C. (2004). ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303: 343–348. [DOI] [PubMed] [Google Scholar]
- Park D.H., Somers D.E., Kim Y.S., Choy Y.H., Lim H.K., Soh M.S., Kim H.J., Kay S.A., Nam H.G. (1999). Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science 285: 1579–1582. [DOI] [PubMed] [Google Scholar]
- Putterill J., Robson F., Lee K., Simon R., Coupland G. (1995). The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80: 847–857. [DOI] [PubMed] [Google Scholar]
- Raisner R.M., Hartley P.D., Meneghini M.D., Bao M.Z., Liu C.L., Schreiber S.L., Rando O.J., Madhani H.D. (2005). Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell 123: 233–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou E.P., Pilch D.R., Orr A.H., Ivanova V.S., Bonner W.M. (1998). DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273: 5858–5868. [DOI] [PubMed] [Google Scholar]
- Rossetto D., Avvakumov N., Côté J. (2012). Histone phosphorylation: a chromatin modification involved in diverse nuclear events. Epigenetics 7: 1098–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinberg S.F. (2008). Structural basis of protein kinase C isoform function. Physiol. Rev. 88: 1341–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suárez-López P., Wheatley K., Robson F., Onouchi H., Valverde F., Coupland G. (2001). CONSTANS mediates between the circadian clock and the control of flowering in Arabidopsis. Nature 410: 1116–1120. [DOI] [PubMed] [Google Scholar]
- Sugano S., Andronis C., Green R.M., Wang Z.-Y., Tobin E.M. (1998). Protein kinase CK2 interacts with and phosphorylates the Arabidopsis circadian clock-associated 1 protein. Proc. Natl. Acad. Sci. USA 95: 11020–11025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Attikum H., Fritsch O., Hohn B., Gasser S.M. (2004). Recruitment of the INO80 complex by H2A phosphorylation links ATP-dependent chromatin remodeling with DNA double-strand break repair. Cell 119: 777–788. [DOI] [PubMed] [Google Scholar]
- Wang A.Y., Schulze J.M., Skordalakes E., Gin J.W., Berger J.M., Rine J., Kobor M.S. (2009). Asf1-like structure of the conserved Yaf9 YEATS domain and role in H2A.Z deposition and acetylation. Proc. Natl. Acad. Sci. USA 106: 21573–21578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Casas-Mollano J.A., Xu J., Riethoven J.-J.M., Zhang C., Cerutti H. (2015). Osmotic stress induces phosphorylation of histone H3 at threonine 3 in pericentromeric regions of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 112: 8487–8492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao A., et al. (2009). WSTF regulates the H2A.X DNA damage response via a novel tyrosine kinase activity. Nature 457: 57–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanovsky M.J., Kay S.A. (2002). Molecular basis of seasonal time measurement in Arabidopsis. Nature 419: 308–312. [DOI] [PubMed] [Google Scholar]
- Yoo S.-D., Cho Y.-H., Sheen J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2: 1565–1572. [DOI] [PubMed] [Google Scholar]
- Zacharaki V., Benhamed M., Poulios S., Latrasse D., Papoutsoglou P., Delarue M., Vlachonasios K.E. (2012). The Arabidopsis ortholog of the YEATS domain containing protein YAF9a regulates flowering by controlling H4 acetylation levels at the FLC locus. Plant Sci. 196: 44–52. [DOI] [PubMed] [Google Scholar]
- Zhang H., Richardson D.O., Roberts D.N., Utley R., Erdjument-Bromage H., Tempst P., Côté J., Cairns B.R. (2004). The Yaf9 component of the SWR1 and NuA4 complexes is required for proper gene expression, histone H4 acetylation, and Htz1 replacement near telomeres. Mol. Cell. Biol. 24: 9424–9436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Zhong Q., Evans A.G., Levy D., Zhong S. (2011). Phosphorylation of histone H3 serine 28 modulates RNA polymerase III-dependent transcription. Oncogene 30: 3943–3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., et al. (2015). C-terminal domains of a histone demethylase interact with a pair of transcription factors and mediate specific chromatin association. Cell Discov. pii: 15003. [DOI] [PMC free article] [PubMed] [Google Scholar]