HDT1 and HDT2 fine-tune GA metabolism in Arabidopsis root tips to control the switch from cell division to expansion, and in this way determine meristem cell number and regulate root growth rate.
Abstract
Root growth is modulated by environmental factors and depends on cell production in the root meristem (RM). New cells in the meristem are generated by stem cells and transit-amplifying cells, which together determine RM cell number. Transcription factors and chromatin-remodeling factors have been implicated in regulating the switch from stem cells to transit-amplifying cells. Here, we show that two Arabidopsis thaliana paralogs encoding plant-specific histone deacetylases, HDT1 and HDT2, regulate a second switch from transit-amplifying cells to expanding cells. Knockdown of HDT1/2 (hdt1,2i) results in an earlier switch and causes a reduced RM cell number. Our data show that HDT1/2 negatively regulate the acetylation level of the C19-GIBBERELLIN 2-OXIDASE2 (GA2ox2) locus and repress the expression of GA2ox2 in the RM and elongation zone. Overexpression of GA2ox2 in the RM phenocopies the hdt1,2i phenotype. Conversely, knockout of GA2ox2 partially rescues the root growth defect of hdt1,2i. These results suggest that by repressing the expression of GA2ox2, HDT1/2 likely fine-tune gibberellin metabolism and they are crucial for regulating the switch from cell division to expansion to determine RM cell number. We propose that HDT1/2 function as part of a mechanism that modulates root growth in response to environmental factors.
INTRODUCTION
Root architecture and growth is regulated through the activity of root meristems. The root growth rate correlates with the cell number in the root meristem (RM), as this determines the number of cells that can differentiate in a given time. The RM includes the stem cell niche (SCN), which is composed of the quiescent center (QC) and its surrounding stem cells (van den Berg et al., 1995; Scheres, 2007). The daughter cells of the stem cells divide a few times before they switch to expansion. These dividing cells are the transit-amplifying cells and together with the SCN they form the RM, whereas the expanding cells form the elongation zone (Moubayidin et al., 2009; Perilli et al., 2010). The RM cell number is determined by the activity of stem cells, as well as by the number of times transit-amplifying cells divide before switching to expansion.
The genes and mechanisms involved in regulating the stem cell activity or the switch from cell division to expansion are becoming clear (Helariutta et al., 2000; Sabatini et al., 2003; Aida et al., 2004; Dello Ioio et al., 2007, 2008; Galinha et al., 2007; Bennett and Scheres, 2010; Moubayidin et al., 2010). In Arabidopsis thaliana roots, the stem cell activity depends on two independent pathways controlled by transcription factors; the SHORT-ROOT (SHR)/SCARECROW (SCR) pathway and the PLETHORA (PLT) pathway (Helariutta et al., 2000; Sabatini et al., 2003; Aida et al., 2004; Galinha et al., 2007). The expression domains of SHR and SCR overlap with the distal PLT transcript accumulation area to position the SCN (Aida et al., 2004). Loss-of-function mutations in SHR, SCR, or PLT1/PLT2 cause a loss of stem cell maintenance, and this results in a reduced RM cell number (Helariutta et al., 2000; Sabatini et al., 2003; Aida et al., 2004; Galinha et al., 2007). The switch from cell division to expansion is controlled by transcription factors ARR12, ARR1, and SHY2 (Dello Ioio et al., 2007; Bennett and Scheres, 2010; Moubayidin et al., 2010). ARR1 and ARR12 activate the expression of SHY2, a transcriptional repressor that triggers the switch from cell division to expansion (Dello Ioio et al., 2008). Gain-of-function mutations in ARR1 or SHY2 cause a reduced RM cell number due to an earlier switch to expansion (Dello Ioio et al., 2008; Moubayidin et al., 2010).
Beside transcription factors, chromatin-remodeling factors also contribute to transcriptional reprogramming by creating an active or silent chromatin configuration (Strahl and Allis, 2000; Jenuwein and Allis, 2001; Kouzarides, 2007; Shahbazian and Grunstein, 2007). Some of them are essential for maintaining stem cell activity in Arabidopsis. For example, the chromatin-remodeling ATPase BRAHMA is indispensable to maintain the SCN by controlling PIN gene expression (Yang et al., 2015). GCN5, a histone acetyltransferase, positively regulates PLT-mediated SCN maintenance (Kornet and Scheres, 2009). In loss-of-function mutants of brm or gcn5, RM cell number is reduced due to gradual loss of stem cells.
Recently, it has been demonstrated that the switch from cell division to expansion in Arabidopsis roots is accompanied by changes in level of histone acetylation (Rosa et al., 2014). Histone acetylation level is regulated by histone acetyltransferases and histone deacetylases (Shahbazian and Grunstein, 2007). In the region where the switch from cell division to expansion is initiated, among the highest expressed histone acetylation genes in Arabidopsis roots are the four plant-specific histone deacetylase genes, named HDT1-HDT4 (Supplemental Data Set 1; Birnbaum et al., 2003). This suggests that these HDTs may play a role in controlling the switch from cell division to expansion. However, the function of HDTs in root development has not been investigated. It has been shown that HDTs are involved in responses to biotic and abiotic stress (Sridha and Wu, 2006; Bourque et al., 2011; Ding et al., 2012; Luo et al., 2012), and they can repress the expression of defense-related genes by altering their chromatin acetylation status (Bourque et al., 2011; Ding et al., 2012). We studied whether HDTs regulate the switch from cell division to expansion and in this way contribute to the mechanisms controlling RM cell number and subsequently root growth.
Here, we investigated the RM phenotype of hdt mutants. We show that two members of the HDT family, HDT1/2, determine the RM cell number by affecting the switch from cell division to expansion. Downregulation of their expression (hdt1,2i) reduces RM cell number and results in a markedly changed transcriptome. Genetic analyses indicate that the strongly increased expression of C19-GIBBERELLIN 2-OXIDASE2 (GA2ox2) in hdt1,2i is a cause of this reduced RM cell number. HDT1/2 negatively regulate the level of histone H3 acetylation of GA2ox2 and possibly in this way repress the transcription of this gene. These data indicate that HDT1/2 repression of GA2ox2 expression contributes to regulation of the switch from cell division to expansion in Arabidopsis roots.
RESULTS
HDT1 and HDT2 Control Root Growth
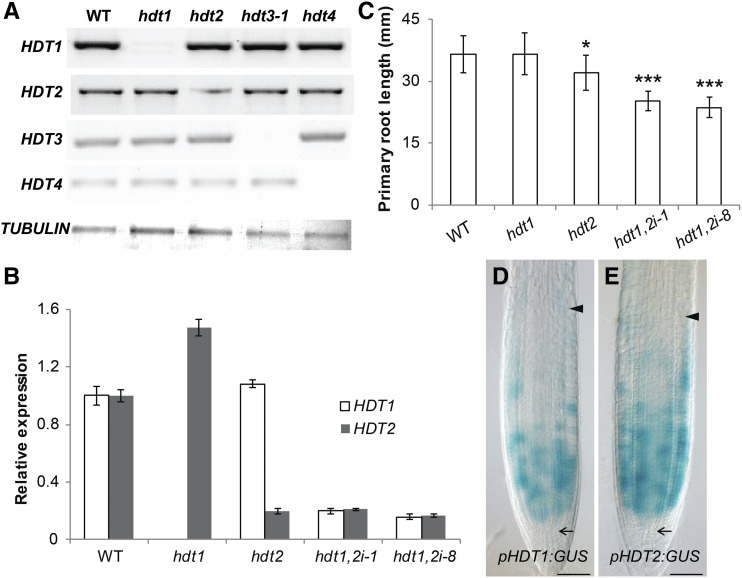
To test whether HDTs control Arabidopsis root growth, we analyzed T-DNA insertion mutants of all four HDT genes (designated hdt1, 2, 3-1, 3-2 [Luo et al., 2012], and 4; Supplemental Figure 1). RT-PCR analyses showed that hdt1, 3-1, and 4 were null mutants, as transcript of the mutated genes was not detectable (Figure 1A). hdt2, however, was a knockdown mutant that retained 20% of transcript (Figures 1A and 1B). hdt1, hdt3-1, hdt3-2, and hdt4 displayed no root phenotype. However, hdt2 showed a 12% reduced root length, compared with the wild type (Figure 1C; Supplemental Figure 2), at 7 d after germination (DAG).
Figure 1.
Silencing of HDT1 and HDT2 Leads to Reduced Root Growth.
(A) Expression of HDT genes in the wild type and hdt mutants. RT-PCR analysis was performed with cDNA prepared from seedlings at 6 DAG. The TUBULIN gene was used as a loading control.
(B) RT-qPCR analyses of HDT1 and HDT2 expression in wild-type, hdt1, hdt2, hdt1,2i-1, and hdt1,2i-8 root tips at 6 DAG. All panels show mean ± se values determined from three independent experiments.
(C) Primary root length of wild-type, hdt1, hdt2, hdt1,2i-1, and hdt1,2i-8 seedlings at 7 DAG. Data shown are average ± sd (n > 20). Asterisks indicate significant differences compared with the wild type (*P < 0.05, ***P < 0.001; Student’s t test).
(D) and (E) Expression patterns of pHDT1:GUS ([D], n = 22) and pHDT2:GUS ([E], n = 25) in root tips of seedlings at 6 DAG. Arrows indicate the SCN and arrowheads indicate the boundary between the RM and elongation zone. Bars = 50μm.
To determine if the other three HDTs contribute to root growth in the hdt2 mutant background, we crossed hdt2 with hdt1, 3-1, and 4 to generate double mutants. hdt2 hdt3-1 and hdt2 hdt4 homozygous plants were morphologically indistinguishable from hdt2 (Supplemental Figure 2), suggesting that neither HDT3 nor HDT4 contributes to root growth. In contrast, selfing of HDT1/hdt1 HDT2/hdt2 did not result in any homozygous hdt1 hdt2 double mutants among more than 200 daughter plants tested. This indicates that loss of function of both HDT1 and HDT2 is lethal, by which the role of HDT1 in root growth could not be studied. In order to study this role, we first determined the HDT1/2 expression patterns in roots, by creating HDT1/2 promoter/GUS fusions (pHDT1:GUS and pHDT2:GUS) as well as translational GFP fusions (pHDT1:HDT1-GFP and pHDT2:HDT2-GFP). The latter two constructs were functional, as they complemented the hdt1 hdt2 lethal phenotype and fully restored root growth in the corresponding background (Supplemental Figure 2). This indicates that both promoter regions are sufficient to drive genes expression at the right place. Transgenic pHDT1:GUS and pHDT2:GUS plants showed that both promoters were especially active in the RM. They displayed patchy expression patterns, suggesting a cell cycle phase dependent expression (Figures 1D and 1E). Their high activity in the RM is consistent with transcriptome data (Birnbaum et al., 2003) and supports the idea that both HDT genes are involved in controlling RM activity. To study this, we made use of the root-specific ROOT CLAVATA HOMOLOG1 (RCH1) promoter to knockdown both HDT1/2. The RCH1 promoter is specifically active in the RM and it is first activated during the torpedo stage of embryo development (Casamitjana-Martínez et al., 2003). So if HDT1/2 are essential for developmental processes preceding this embryonic stage, these will not be affected. Five independent lines (designated hdt1,2i-1, 4, 6, 8, and 9) were generated. The levels of HDT1/2 transcript, in these lines, were between 15% and 40% of that of the wild type (Figure 1B; Supplemental Figure 3). In comparison with hdt2, all hdt1,2i lines showed more severely reduced root growth (Figure 1C; Supplemental Figure 2), indicating that both HDT1 and HDT2 control root growth. The level of root length reduction in these hdt1,2i lines correlates with decreasing levels of HDT1/2 transcript (Figures 1B and 1C; Supplemental Figures 2 and 3), indicating that HDT1/2 positively regulate root growth.
Reduced Root Growth of hdt1,2i Involves a Lower RM Cell Number
To investigate how HDT1/2 affect root growth, the kinematic growth of roots was analyzed in wild-type and two hdt1,2i lines, hdt1,2i-1 and hdt1,2i-8, which had the strongest reduction of HDT1/2 mRNA levels. Root growth rate from 6 to 7 DAG was determined (see Methods). In comparison with the wild type, hdt1,2i-1 or hdt1,2i-8 roots grew 56% and 60% slower, respectively (Table 1; Supplemental Table 1). Root growth rate is determined by number of cells that are added from the RM to elongation zone within a defined time period and to what extent these cells subsequently expand (De Veylder et al., 2001). Therefore, the reduced root growth rate of hdt1,2i could be due to fewer cells being added to the elongation zone or reduced expansion of newly added cells. The length of cortical cells in the differentiated zone of both hdt1,2i-1 and hdt1,2i-8 roots was measured and shown to be the same as in the wild type. However, the number of cells added to the elongation zone was reduced in hdt1.2i-1 and hdt1,2i-8 (Table 1; Supplemental Table 1). This reduced cell number could be caused either by the presence of fewer cells in hdt1,2i RM or by a prolonged cell cycle duration. We inferred cell cycle duration from kinematic growth analyses and this showed that it was only 7% or 6% longer in hdt1,2i-1 and hdt1,2i-8 compared with the wild type. However, RM cell number was reduced by 54% in hdt1,2i-1 and by 59% in hdt1,2i-8. This reduction equals the difference in root growth rate (Table 1; Supplemental Table 1).
Table 1. Kinematic Analyses of Root Growth in Wild-Type and hdt1,2i-1 Seedlings.
| Parameter | Wild Type | hdt1,2i-1 | Difference (%) | P Value |
|---|---|---|---|---|
| Root growth rate (μm/h, M) | 320.6 ± 35.0 | 141.8 ± 24.7 | −56 | <0.001 |
| Fully expanded cell length (μm, N) | 191.0 ± 9.2 | 195.2 ± 9.0 | – | NS |
| Cell production per day (X) | 40.3 ± 4.3 | 17.4 ± 2.9 | −57 | <0.001 |
| Cell number in the RM (Y) | 38.0 ± 3.9 | 17.3 ± 1.7 | −54 | 0.001 |
| Cell cycle duration (h, Z) | 22.8 ± 2.4 | 24.4 ± 4.7 | 7 | 0.107 |
The analyses were performed based on wild-type and hdt1,2i-1 root cortical cell file. Data shown are average ± sd. P values were determined by Student’s t test (n > 20). NS, no statistical significance. X = 24*M/N and Z = 24*Y/X.
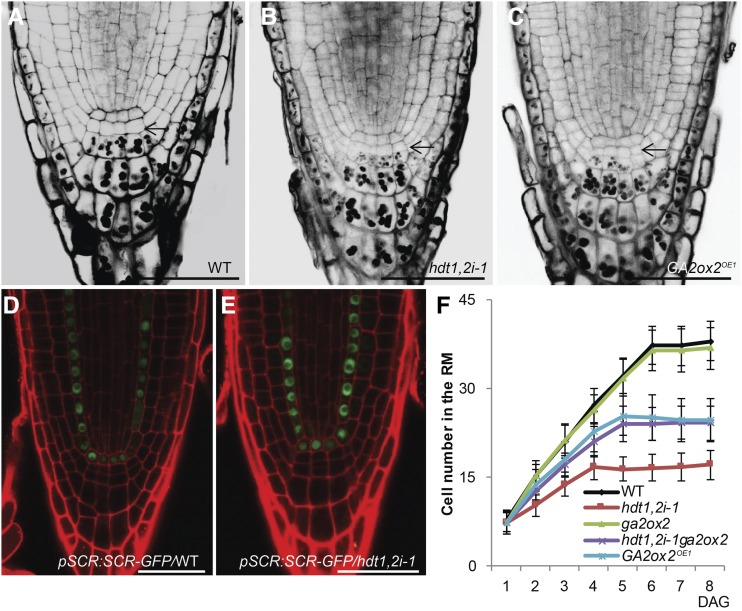
The reduced cell number in RM can be caused either by the loss of the SCN or by a reduced number of divisions of the transit-amplifying cells. To distinguish between these two possibilities, we first investigated whether the SCN was lost in hdt1,2i. The columella stem cells in hdt1,2i-1 and hdt1,2i-8 were present; these are the cells adjacent to the QC and not containing starch granules, which occur in differentiated columella cells (Figures 2A and 2B; Supplemental Figure 4A). Also, SCR, which is required for SCN maintenance (Sabatini et al., 2003), had a wild-type-like expression pattern in the QC and endodermis of hdt1,2i-1 roots (Figures 2D and 2E). QC divisions were more frequently observed in hdt1,2i-8, but not in hdt1,2i-1 (Supplemental Figures 4A and 4I). However, expression of WOX5, another QC marker (Sarkar et al., 2007), was maintained in hdt1,2i-8, like in the wild type and hdt1,2i-1 (Supplemental Figures 4D to 4F). This suggests that QC cells are maintained in hdt1,2i-8. In addition, the RMs of hdt1,2i-1 and hdt1,2i-8 were still present at 21 DAG (Supplemental Figures 4J to 4L). Together, these data indicate that the reduced cell number in hdt1,2i RM is not caused by loss of the SCN.
Figure 2.
Reduced Cell Number in hdt1,2i-1 and GA2ox2OE1 RM Is Caused by an Earlier Switch from Cell Division to Expansion.
(A) to (C) mPS-PI-stained root tips of the wild-type (A), hdt1,2i-1 (B), and GA2ox2OE1 (C) seedlings at 6 DAG. Arrows indicate columella stem cells which did not display any starch granule.
(D) and (E) Expression pattern of the pSCR:SCR-GFP in the wild-type (D) and hdt1,2i-1 (E) root tips at 7 DAG (n = 10; representative images are shown). Identical confocal microscope settings were used in (D) and (E).
(F) Cell number of the RM in wild-type, ga2ox2, hdt1,2i-1, ga2ox2 hdt1,2i-1, and GA2ox2OE1 seedlings measured daily after germination during 8 d. Data shown are average ± sd (n > 20).
Bars = 50 μm in (A) to (E).
RM cell number increases after germination, and it reaches a maximum when the number of cells added by cell division is equal to the number of cells that switch to expansion (Dello Ioio et al., 2008). By monitoring at what time the maximum RM cell number is reached, we determined when cell division and cell expansion have reached a balance. The maximum RM cell number was established at 4 DAG in hdt1,2i-1, whereas this was first reached at 6 DAG in the wild type (Figure 2F). Similar results were also obtained for the hdt1,2i-8 line (Supplemental Figure 4C). This demonstrates that HDT1/2 are involved in regulating the switch from cell division to expansion. The reduced RM cell number indicates that an earlier switch occurs after fewer divisions of the transit-amplifying cells in hdt1,2i.
Possible Targets of HDT1/2 Revealed by Transcriptome Analyses
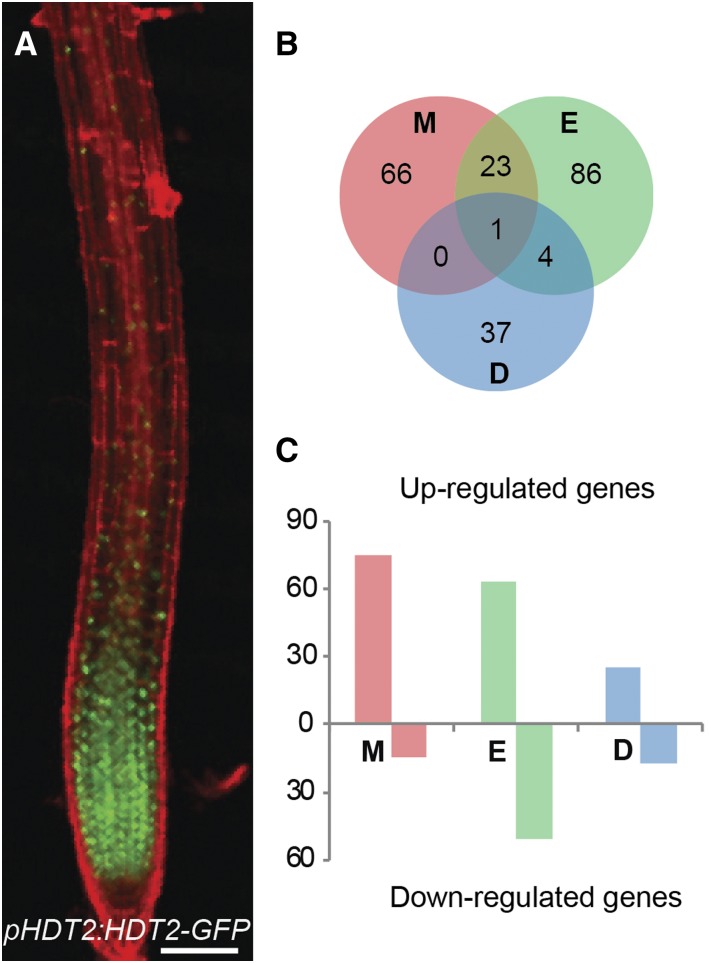
In the RM and elongation zone, HDT1/2 were located in both nucleolus and nucleoplasm (Supplemental Figures 5B and 5C), where they could alter the expression level of genes by changing chromatin acetylation levels (Wu et al., 2003; Zhou et al., 2004; Ding et al., 2012). We performed Illumina RNA sequencing to determine the transcriptome differences between the wild type and hdt1,2i-1 to obtain insight in how RM cell number might be reduced in hdt1,2i-1. HDT1/2 proteins accumulated highest in the meristem, markedly lower in the elongation zone and were hardly detectable in the differentiated zone (Figure 3A; Supplemental Figure 5A). We compared transcriptomes from the wild type and hdt1,2i-1 in these three zones (see Methods). This resulted in 90, 114, and 42 differentially expressed genes (DEGs) in the meristem, elongation, and differentiated zones, respectively (Figure 3B; Supplemental Data Set 2).
Figure 3.
HDT1/2 Orchestrate Transcriptional Reprogramming.
(A) Expression pattern of pHDT2:HDT2-GFP that complements hdt1 hdt2 lethal phenotype in root of 7 DAG seedling. Bar = 100 μm.
(B) DEGs overlapped between the meristem zone (M), elongation zone (E), and differentiated zone (D) of wild-type and hdt1,2i-1 roots.
(C) The number of upregulated and downregulated DEGs in meristem, elongation, and differentiated zone of hdt1,2i-1 roots compared with the wild type.
In the meristem zone, 75 genes were upregulated in hdt1,2i-1, representing ∼83% of DEGs in this zone. This is consistent with a function of HDTs in repressing gene expression (Wu et al., 2003; Zhou et al., 2004). In the elongation and differentiated zones, the number of upregulated genes was 63 and 25, representing ∼55% and 60% of DEGs, respectively (Figure 3C). Twenty-four DEGs were shared between the meristem and elongation zones, five DEGs were shared between the elongation and differentiated zones, and only one DEG was shared between the meristem and differentiated zones. In total, 217 DEGs were identified (Figure 3B; Supplemental Data Set 2).
Gene Ontology (GO) enrichment analyses for the 217 DEGs showed that the upregulated genes in hdt1,2i-1 were enriched in genes involved in “response to chemical/stimulus.” The downregulated genes were enriched in genes involved in regulating root development (Supplemental Figure 6). As HDT1/2 most likely repress the expression of genes (Wu et al., 2003; Zhou et al., 2004; Ding et al., 2012), we expected that the earlier switch to expansion of transit-amplifying cells in hdt1,2i-1 was at least in part caused by genes with increased expression. The most upregulated gene from the hdt1,2i-1 meristem zone encodes a MATE protein that belongs to a proton-dependent efflux transporter family, which contains at least 54 members in Arabidopsis (Eckardt, 2001). However, the expression level of several paralogs was very high in the wild type and was not changed in hdt1,2i-1 (Supplemental Data Set 2). Assuming functional redundancy of these genes, the upregulation of one MATE gene probably did not have a major effect on root growth. GA2ox2 was the second most upregulated gene (Supplemental Data Set 2). This gene, the highest expressed member from its family in root tips, was a good candidate to contribute to the reduced RM cell number in hdt1,2i-1. GA2ox2 encodes an oxidase that inactivates bioactive C19 gibberellins via 2-oxidation. This is a major gibberellin (GA) inactivating pathway in Arabidopsis, and GA delays the switch from cell division to expansion in Arabidopsis roots (Rieu et al., 2008; Ubeda-Tomás et al., 2008, 2009; Moubayidin et al., 2010).
hdt1,2i-1 Root Tips Have a Gibberellin-Deficient Phenotype
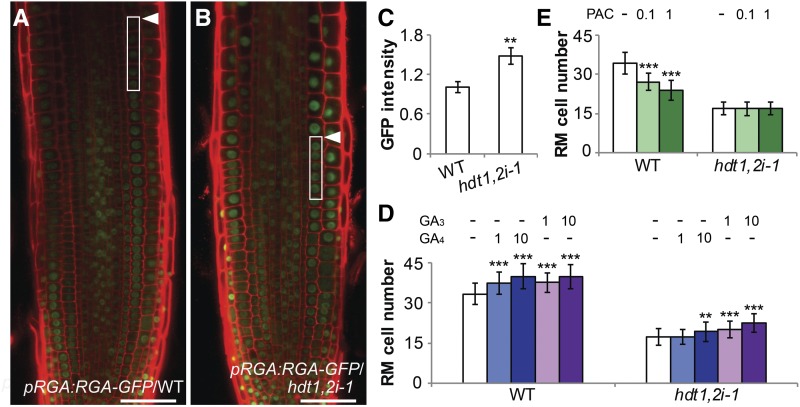
To determine whether GA level is indeed reduced in hdt1,2i-1 root tips, we made use of a DELLA reporter construct (pRGA:GFP-RGA). GA destabilizes DELLA proteins such as RGA (REPRESSOR OF ga1-3) (Peng et al., 1997; Silverstone et al., 2001). hdt1,2i-1 plants were crossed with pRGA:GFP-RGA to generate homozygous plants. RGA protein level was quantified by measuring GFP intensity in wild-type (n = 11) and hdt1,2i-1 (n = 12) roots. In each root, GFP intensity in nuclei of five cortical cells, at the transition from division to elongation, were measured and the average values were determined. This showed that in hdt1,2i-1, RGA protein level was around 50% increased (Figures 4A to 4C), indicating that hdt1,2i-1 root tips have a reduced GA level. Consistent with this, PIN1 and PIN2 levels, which are positively regulated by GA (Willige et al., 2011), were considerably reduced in hdt1,2i-1 compared with the wild type (Supplemental Figures 7A to 7D).
Figure 4.
The hdt1,2i-1 Root Tip Has a Gibberellin-Deficient Phenotype.
(A) and (B) Expression of pRGA:GFP-RGA in wild-type (A) and hdt1,2i-1 (B) root tips at 6 DAG. Arrowheads indicate the boundary between the RM and elongation zone. Close-up of boxed areas show five cortical cells that were used for quantification in (C). Identical confocal microscope settings were used to image pRGA:GFP-RGA proteins in (A) and (B); representative images are shown. Bars = 50 μm.
(C) Quantification of fluorescence intensity of pRGA:GFP-RGA in the wild type (n = 11) and hdt1,2i-1 (n = 12). Arbitrary unit was used for GFP intensity. Root cells used for quantification are indicated in (A) and (B). Data shown are mean ± se. Asterisk indicates significant difference between the wild type and hdt1,2i-1 (**P < 0.01, Student’s t test).
(D) and (E) Cell number in the wild type and hdt1,2i-1 RM after treatment with 1 μM and 10 μM GA4/GA3 (D) or with 0.1 μM and 1 μM PAC (E) at 6 DAG. Data shown are average ± sd (n > 40). Asterisks indicate significant differences compared with the mock (−; **P < 0.01, ***P < 0.001; Student’s t test).
To investigate whether the reduced GA level in hdt1,2i-1 root tips is caused by the upregulation of GA2ox2, wild-type and hdt1,2i-1 seedlings were treated with exogenous GA4 or GA3. GA4 is a substrate of GA2ox2, whereas GA3 is not (Thomas et al., 1999; Hedden and Phillips, 2000; Yamauchi et al., 2007). RM cell number in both the wild type and hdt1,2i-1 was increased significantly after GA3 (1 and 10 μM) application (Figure 4D). A similar increase in RM cell number was also observed in the wild type after GA4 (1 and 10 μM) application in the wild type. However, in hdt1,2i-1, RM cell number was not affected by applying 1 μM GA4, and it was only increased slightly by application of 10 μM GA4. This suggests that the upregulated GA2ox2 in hdt1,2i-1 root tips rapidly degrades exogenously applied GA4 but not GA3.
In line with the reduced GA level in hdt1,2i-1 root tips, the exogenously applied paclobutrazol (PAC; 0.1 and 1 μM), a GA biosynthesis inhibitor (Wang et al., 1986), significantly reduced RM cell number in the wild type. Whereas RM cell number in hdt1,2i-1 was not affected (Figure 4E). This suggests that the GA level in hdt1,2i-1 is so low that PAC has no additional inhibitory effect on RM cell number. Collectively, these data indicate that the upregulation of GA2ox2 reduces GA level in hdt1,2i-1 root tips and this contributes to the decreased RM cell number.
HDT2 Regulates the Expression of GA2ox2 Directly
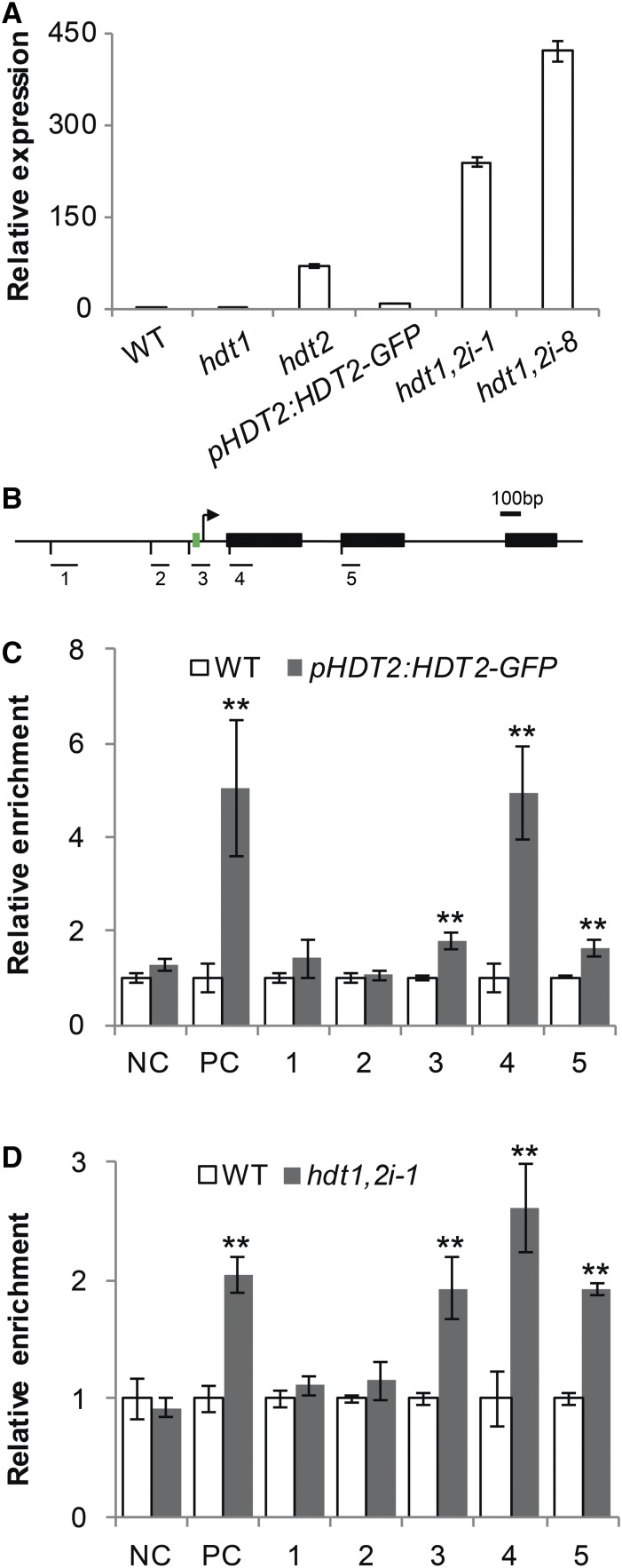
To examine whether there is a correlation between the expression levels of HDT1/2 and GA2ox2, we compared the GA2ox2 mRNA level in root tips of several hdt mutants/lines. This included the hdt1 and hdt2 mutants, the hdt1 hdt2 double mutant complemented by pHDT2:HDT2-GFP, hdt1,2i-1, and hdt1,2i-8. The hdt1 and the complemented line had a similar GA2ox2 mRNA level as the wild type, whereas in hdt2 and two hdt1,2i lines, the expression level of GA2ox2 was increased (Figure 5A). Lower level of HDT2 mRNA (Figure 1B) correlates with higher expression level of GA2ox2. This inverse correlation suggests that the expression of GA2ox2 is negatively regulated by HDT2.
Figure 5.
HDT2 Directly Regulates the Expression of GA2ox2.
(A) RT-qPCR analyses of GA2ox2 expression in root tips of the wild type, hdt1, hdt2, and pHDT2:HDT2-GFP complemented hdt1 hdt2 mutant, hdt1,2i-1, and hdt1,2i-8 lines at 6 DAG.
(B) Schematic representation of the GA2ox2 genomic region. Black boxes represent exons, and green box indicates TATA box. The bent arrow indicates the transcription start site. Bars below with numbers indicate regions used in ChIP-qPCR experiments. Bar = 100 bp.
(C) and (D) ChIP-qPCR analyses of pHDT2:HDT2-GFP complemented hdt1 hdt2 and wild-type seedlings using anti-GFP antibody (C), and hdt1,2i-1 and wild type roots using anti-H3ac antibody (D). PC or NC represents positive or negative control.
Data shown in (A), (C), and (D) are mean ± se values determined from three independent experiments. Asterisks in (C) and (D) indicate significant differences compared with the wild type (**P < 0.01, Student’s t test).
HDTs can mediate transcriptional repression by deacetylating genes (Ding et al., 2012). To investigate whether HDT2 binds to the GA2ox2 locus, we performed chromatin immunoprecipitation-quantitative PCR (ChIP-qPCR) on wild-type and pHDT2:HDT2-GFP complemented seedlings using an anti-GFP antibody. Five different DNA fragments were amplified spanning a region from 850 bp upstream to 618 bp downstream of the GA2ox2 start codon. These included three fragments from the promoter region and a part of the first and the second exon, respectively (Figure 5B). The MATE gene (AT2G04050) was used as positive control. It is also upregulated in hdt1,2i-1 (Supplemental Data Set 2) and HDT2 binds to its first exon (David Latrasse, personal communication). A randomly selected Arabidopsis intergenic region of chromosome 5 was used as negative control. A marked enrichment of HDT2 on the positive control, as well as on the first exon of GA2ox2 was found in pHDT2:HDT2-GFP-complemented seedlings, compared with wild-type seedlings. In addition, a statistically significant enrichment was also found for one promoter fragment (TATA box) and the second exon of GA2ox2. No enrichment was observed for the negative control and the other two fragments of GA2ox2 (Figure 5C). This indicates that HDT2 binds to GA2ox2.
Subsequently, we determined whether knockdown of HDT1/2 affects the acetylation level of the HDT2-bound GA2ox2 regions. Since histone H3ac generally marks actively transcribed chromatin (Kouzarides, 2007), we determined H3ac levels of GA2ox2 in hdt1,2i. We performed ChIP-qPCR on hdt1,2i-1 and wild-type roots (see Methods) using an anti-H3ac antibody. An increase in H3ac level was found for the positive control in hdt1,2i-1 roots. Furthermore, at the HDT2 binding sites, a 2.6-fold increase was found for the first exon of GA2ox2 and a statistically significant increase in H3ac level was also found for the TATA box region (1.9-fold) and the second exon (1.9-fold) (Figure 5D). Furthermore, there was no increase in H3ac level at the negative control locus and the HDT2 nonbinding sites in hdt1,2i-1. This shows that knockdown of HDT1/2 results in an increased H3ac level at regions to which HDT2 binds. Taken together, these results indicate that HDT1/2 repress the expression of GA2ox2 likely by deacetylating histone H3 at specific regions within the GA2ox2 locus.
GA2ox2 Is a Determinant in HDT1/2-Regulated Switch from Cell Division to Expansion
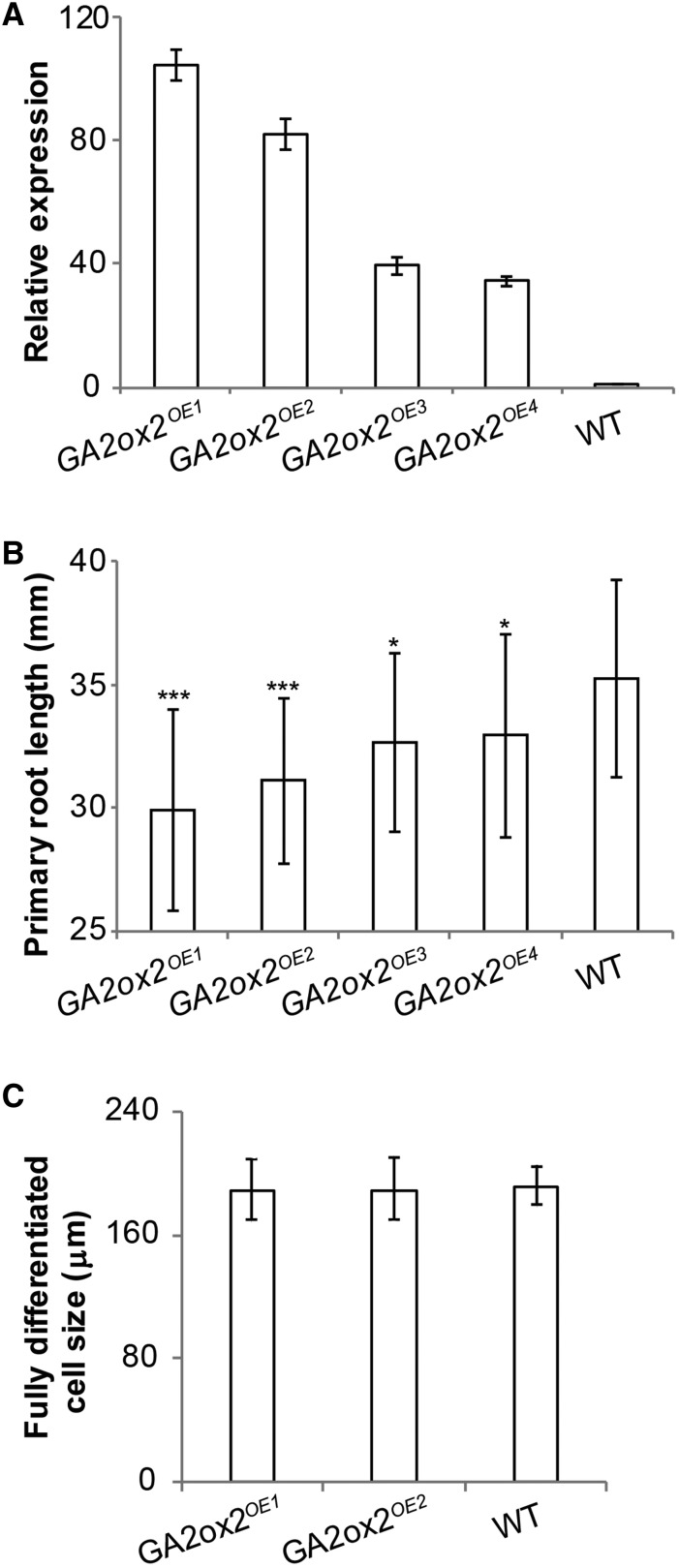
To further study whether the reduced root growth of hdt1,2i plants is caused by the upregulation of GA2ox2, we generated four GA2ox2 overexpression lines driven by the root-specific RCH1 promoter (GA2ox2OE1-OE4) (Casamitjana-Martínez et al., 2003). In these lines, the level of GA2ox2 transcripts was 30 to 100 times higher than that of the wild type (Figure 6A). All four GA2ox2OE lines formed shorter roots than the wild type. The level of reduction was correlated with the level of upregulation of GA2ox2 (Figure 6B). To test whether overexpression of GA2ox2 causes the short root phenotype in a similar way as hdt1,2i, the SCN and the length of fully differentiated cells were analyzed, the RM cell number was determined daily from 1 to 8 DAG in GA2ox2OE1 and GA2ox2OE2. In both lines, the SCN was shown not to be disturbed at 6 DAG and expression of WOX5 was wild type like (Figures 2A and 2C; Supplemental Figures 4B, 4D, and 4G to 4I). This was also supported by the maintenance of the RMs till 14 DAG (Supplemental Figures 4J, 4M, and 4N). The length of fully differentiated cortical cells in both GA2ox2OE1 and GA2ox2OE2 roots was measured and shown to be identical to the wild type (Figure 6C). The maximum RM cell number was established at 5 DAG in both GA2ox2OE1 and GA2ox2OE2 (Figure 2F; Supplemental Figure 4C), whereas in the wild type, this was first reached at 6 DAG. RM cell number of GA2ox2OE1 and GA2ox2OE2 was reduced significantly at 6 DAG, compared with that of the wild type (Figures 2F, 7A, and 7C; Supplemental Figure 4C). These results suggest that the switch from cell division to expansion occurs earlier in GA2ox2OE lines, like in hdt1,2i lines. The similarity of the GA2ox2OE RM to that of hdt1,2i further supports that lower GA levels in hdt1,2i-1 root tips contribute to RM size regulation.
Figure 6.
GA2ox2 Controls Root Growth.
(A) RT-qPCR analyses of GA2ox2 expression in root tips of the wild type and four independent GA2ox2 overexpression lines at 6 DAG. All panels show mean ± se values determined from three independent experiments.
(B) Primary root length of the four GA2ox2 overexpression lines and the wild type at 7 DAG. Data shown are average ± sd (n > 20). Asterisks indicate significant differences between mutants and the wild type (*P < 0.05, ***P < 0.001; Student’s t test).
(C) The length of fully differentiated cortical cells in the wild type, GA2ox2OE1, and GA2ox2OE2 at 7 DAG. Data shown are average ± sd (n > 20) with no significant difference between the wild type and mutants (Student’s t test).
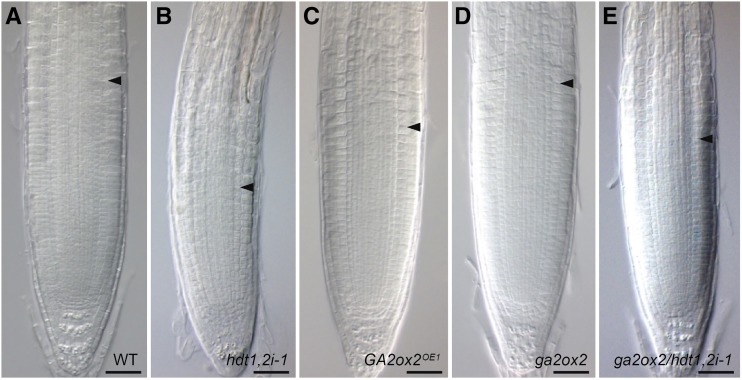
Figure 7.
GA2ox2 Is a Gene Repressed by HDT1/2 to Determine RM Cell Number.
Root tip phenotypes of the wild type (A), hdt1,2i-1 (B), GA2ox2OE1 (C), ga2ox2 (D), and ga2ox2 hdt1,2i-1 (E) at 6 DAG. Arrowheads indicate the boundary between the RM and elongation zone. Bars = 50 μm.
We next determined whether knockout of GA2ox2 (ga2ox2; Supplemental Figure 1) could rescue the RM defect in hdt1,2i-1. ga2ox2 plants were crossed with hdt1,2i-1 plants to generate ga2ox2 hdt1,2i-1 homozygous plants. At 6 DAG, ga2ox2 plants had a similar RM cell number as the wild type (Figures 2F, 7A, and 7D). In ga2ox2 hdt1,2i-1 plants, the averaged RM cell number was 24.0. This is a substantially increased cell number in comparison to 16.6 in hdt1,2i-1 (Figure 2F, 7B, and 7E). The maximum cell number was established at 5 DAG in ga2ox2 hdt1,2i-1, compared with 4 DAG in hdt1,2i-1 plants (Figure 2F). Therefore, knockout of GA2ox2 rescues (in part) the RM defect of hdt1,2i-1. These findings suggest that HDT1/2 repress GA2ox2 expression to fine-tune gibberellin homeostasis in order to determine RM cell number.
GA2ox2 Expression Is Increased in All Transit-Amplifying Cells in hdt1,2i-1
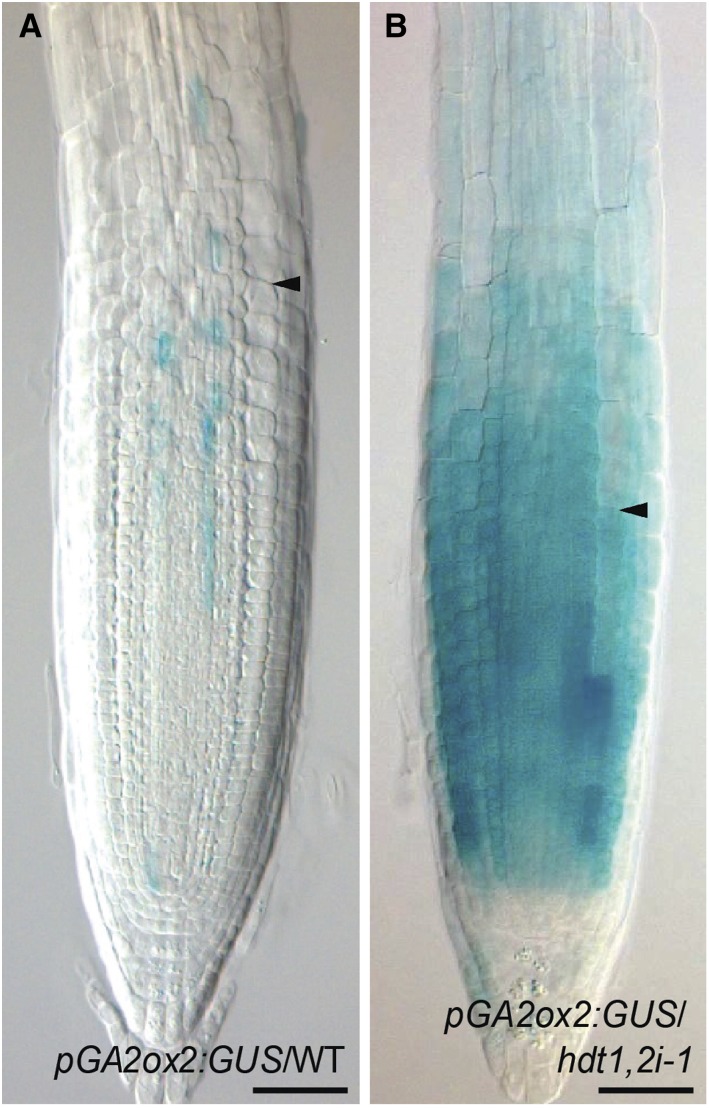
In roots of hdt1,2i-1 plants, the switch from cell division to expansion occurs earlier than in the wild type, as caused by the upregulation of GA2ox2. Therefore, we determined in which cells GA2ox2 expression was increased. We created a GA2ox2 promoter/GUS reporter line (pGA2ox2:GUS) and crossed it with hdt1,2i-1 to generate homozygous pGA2ox2:GUS hdt1,2i-1 plants. In wild-type roots, a low level of expression of pGA2ox2:GUS occurred in vascular tissue and pericycle at the transition from the RM to elongation zone, but it was not detected in endodermis, cortex, and epidermis (Figure 8A). This expression pattern is consistent with the gene expression map of the Arabidopsis roots (Birnbaum et al., 2003). In hdt1,2i-1, GUS activity was markedly increased in both RM and the distal part of elongation zone, and it was not restricted to vascular tissue and pericycle. Instead, we observed high GUS activity in the RM within all cell files (Figure 8B). The level of GUS activity gradually decreased from the distal to the proximal elongation zone where it became undetectable. This is consistent with the expression level of GA2ox2 in these zones in hdt1,2i-1 (Supplemental Data Set 2). These data indicate that HDT1/2 determine GA2ox2 expression level in the RM and elongation zone.
Figure 8.
Enhanced Expression of GA2ox2 in hdt1,2i-1 Root Tip.
Expression pattern of pGA2ox2:GUS in root tips of the wild-type (A) and hdt1,2i-1 (B) seedlings at 6 DAG (n = 15 for each; representative images are shown). Arrowheads indicate the boundary between the RM and elongation zone. Bars = 50 μm.
DISCUSSION
In this study, we demonstrate that HDT1/2 influence the switch from cell division to expansion in the root tip by repressing GA2ox2 expression. Knockdown of HDT1/2 causes an earlier switch to cell expansion and results in a reduced RM cell number. The upregulation of GA2ox2 is a contributing factor to this phenotype. We show that HDT2 binds to several regions of GA2ox2, and at these binding sites the acetylation level is negatively regulated. Upregulation of GA2ox2 in hdt1,2i most likely causes a reduced GA level that results in an earlier switch from cell division to expansion of transit-amplifying cells.
pHDT1:GUS and pHDT2:GUS exhibit a patchy accumulation pattern suggesting cell cycle-dependent expression. However, HDT1/2-GFP fusion proteins are present at an equal level in transit-amplifying cells. It seems unlikely that they have a function in a specific stage of the cell cycle. This is consistent with the observation that cell cycle duration is not affected in hdt1,2i-1. We expect that HDT proteins are especially made when the chromatin is duplicated, similar to histone transcription and translation. The equal levels of HDTs might be caused by the relatively stable nature of these proteins. This is supported by the occurrence of HDT1/2-GFP in expanding cells, whereas GUS is not detectable. An alternative explanation is that HDT1/2 are transported to adjacent cells. This could explain why low levels of HDT1/2-GFP are detected in the SCN, whereas GUS is below the detection level (n > 20).
We show that HDT1 and HDT2 determine the RM cell number. Notably, treatment with trichostatin A (TSA), an inhibitor of certain zinc binding motif containing histone deacetylases (Finnin et al., 1999), increases the RM cell number (Rosa et al., 2014). This seems in disagreement with our finding that decreased HDT activity leads to a reduced cell number. However, although HDTs contain a zinc binding domain, it has not been demonstrated that their activity is targeted by TSA. Furthermore, other studies show that application of TSA inhibits root growth and triggers QC divisions (Nguyen et al., 2013; Pi et al., 2015). To determine whether TSA could target HDT1/2, we compared the effect of TSA on QC cell division in the wild type and hdt1,2i-1. This showed that a TSA treatment and HDT1/2 knockdown increase the frequency of QC divisions in a similar way (Supplemental Figure 4I), indicating that TSA-mediated QC divisions do not involve HDT1/2 but involve other targets.
Overexpression of GA2ox2 causes a similar RM phenotype as HDT1/2 knockdown (Figures 2F, 7B, and 7C). Therefore, in hdt1,2i, the upregulation of GA2ox2 (in part) causes the decreased RM cell number. Upregulation of GA2ox2 in hdt1,2i root tips (Figures 8A and 8B) causes most likely a rapid degradation of GA. In agreement with this, the hdt1,2i-1 RM cell number is partially restored by GA3 and hardly affected by GA4 (Figure 4D). In addition, PAC, which decreases RM cell number in the wild type that can be restored by GA3 (Ubeda-Tomás et al., 2009), has no additional effect on hdt1,2i-1 RM cell number (Figure 4E). This suggests that GA level in hdt1,2i-1 is very low. The most direct way to show that GA level is reduced in hdt1,2i-1 would be by GA quantification, which is technically difficult. However, the increased accumulation of RGA, as well as the decreased PIN1/2 abundance in hdt1,2i-1 root tips (Figures 4A to 4C; Supplemental Figure 7) are consistent with a reduced GA level. Similar phenotypes are also observed in other GA-deficient mutants or PAC-treated seedlings (Ubeda-Tomás et al., 2009; Moubayidin et al., 2010). A transit-amplifying cell exits the mitosis as it enters into the elongation zone (Blilou et al., 2002; Vanstraelen et al., 2009). GA is required to promote mitotic activity (Peng et al., 1997; Silverstone et al., 2001; Achard et al., 2009; Ubeda-Tomás et al., 2009). In line with this, GA biosynthesis mutants ga1 and ga3ox1 ga3ox2, as well as a non-GA-degradable DELLA mutant, have a reduced RM cell number due to an earlier exit from the mitotic cell cycle (Ubeda-Tomás et al., 2009; Moubayidin et al., 2010). We postulate that in hdt1,2i, the upregulation of GA2ox2 causes most likely a reduced GA level that results in an earlier switch from division to expansion and in this manner (in part) decreases RM cell number.
Previously, it has been shown that GA2ox6 transcript level in Arabidopsis roots is higher than that of GA2ox2 (Dugardeyn et al., 2008). However, GA2ox6 is only expressed in the differentiated zone (Supplemental Data Set 2). In the wild type, both genes are hardly expressed in root tips, whereas in hdt1,2i-1, GA2ox2 is remarkably upregulated in RM and its expression starts to decrease in the distal elongation zone and becomes undetectable in the proximal elongation zone and differentiated zone (Figure 8B; Supplemental Data Set 2). This suggests that GA levels will only be disturbed in RM and the distal elongation zone. GA controls the size of fully differentiated root cells (Band et al., 2012; Shani et al., 2013). This is in agreement with the observation that the size of fully differentiated root cells is not affected (Table 1; Supplemental Table 1).
SHY2 is an AUX/IAA-type transcriptional repressor that triggers the switch from cell division to expansion (Dello Ioio et al., 2008; Moubayidin et al., 2010). SHY2 expression is induced in this transition zone by ARR1 and ARR12, two cytokinin-responsive transcription factors, which are expressed in the transition zone as well. GA represses the expression of ARR1 and in this manner delays the accumulation of SHY2 (Mason et al., 2004, 2005; Dello Ioio et al., 2008). However, we did not observe any effect on ARR1/SHY2 transcript levels in hdt1,2i-1 RM and elongation zone. Possibly the reduced GA level in hdt1,2i-1 transit-amplifying cells causes ARR1/SHY2 transcript to accumulate so the proteins will reach the threshold level required to induce the switch from cell division to expansion earlier than in the wild type. This most likely does not affect their transcript levels in total root extracts (Dello Ioio et al., 2007, 2008), as the size of the region where they are expressed is most likely not affected.
The fact that hdt1,2i lines have a stronger phenotype than the hdt2 mutant points to an additive effect of HDT1 and HDT2 in controlling root growth (Figures 1B and 1C; Supplemental Figure 2). They both presumably repress GA2ox2 expression, as in hdt1,2i-1/8 lines this gene is induced much higher than in hdt2 (Figure 5A), whereas HDT2 mRNA levels in these mutants are very similar (Figure 1B). This could mean that HDT1 and HDT2 repress the expression of same genes, but together have to be present at a sufficient level. This is supported by the observation that the hdt1 mutant has no root phenotype, but has an increased HDT2 expression level (Figures 1B and 1C).
GA2ox2 is only one gene out of 217 DEGs in hdt1,2i-1. However, knockout of GA2ox2 rescues hdt1,2i-1 root growth significantly and GA2ox2 overexpression phenocopies hdt1,2i-1 (Figures 2F, 7B, and 7E). This suggests that GA2ox2 upregulation is a cause of the short-root phenotype in hdt1,2i. HDT2 binds to two exons of GA2ox2, as well as to the TATA box (Figures 5B and 5C). The identified HDT2 binding sites from GA2ox2 are also potential HDT1 binding sites, as HDT1 and HDT2 are most likely functionally redundant. Binding to the TATA box appears to be important as pGA2ox2:GUS is markedly enhanced in hdt1,2i-1 background. This reporter construct does contain the TATA box but not the two exons (see Methods). The TATA box is considered to be the binding site of basal transcriptional machinery (Patikoglou et al., 1999; Bernard et al., 2010; Liu et al., 2015a). Knockdown of HDT1/2 results in increased histone acetylation levels of the GA2ox2 locus. We postulate that this decrease in histone acetylation increases the accessibility of the GA2ox2 promoter to basal transcription factors.
Remarkably, the DEGs in hdt1,2i-1 are enriched in genes involved in response to various stimuli and root development (Supplemental Figure 6). This suggests that HDT1/2 play a role in adaptation of root development to environmental stimuli. It has been shown that mild abiotic stress conditions in general lead to transiently reduced growth that is reversible. The hormone abscisic acid (ABA) plays a key role in regulating stress responses and exogenously applied ABA and salt reduce RM cell number in Arabidopsis seedlings (Yang et al., 2014; Liu et al., 2015b). Expression of the four HDTs is repressed by ABA and salt (Luo et al., 2012). We postulate that the HDTs may be part of a mechanism that modulates root growth upon response to abiotic stress. Repression of HDT1/2 results in retarded root growth by reducing the number of transit-amplifying cells. However, the SCN is well maintained (Figures 2A and 2B; Supplemental Figure 4A); thus, the root can reestablish growth upon release of the stress.
METHODS
Plant Materials and Growth Conditions
The Arabidopsis thaliana accession Columbia-0 was used as the wild type. The T-DNA insertion lines used are the following: GK355_H03 for HDT1 (At3g44750), SAIL_1247_A02 for HDT2 (At5g22650), SAIL_240_C08 and SALK_129799 (Luo et al., 2012) for HDT3 (At5g03740), GK_279_D04 for HDT4 (At2g27840), and SALK_051749 for GA2ox2 (At1g30040) (Alonso et al., 2003; Rieu et al., 2008). All seeds were obtained from the Nottingham Arabidopsis Stock Centre. The T-DNA insertions were verified by genotyping with primers listed in Supplemental Table 2.1. The Arabidopsis GFP reporter lines used are pSCR:SCR-GFP (Helariutta et al., 2000), pRGA:GFP-RGA (Silverstone et al., 2001), pPIN1:PIN1-GFP (Benková et al., 2003), and pPIN2:PIN2-GFP (Blilou et al., 2005).
Plants were grown vertically on 0.5× Murashige and Skoog medium including vitamins (Duchefa) and 1% sucrose under long-day conditions (16 h light/8 h dark) at 22°C. For the GA and PAC treatment, 2-d-old seedlings were transferred to the same medium containing GA3, GA4, or PAC (Duchefa).
Root Growth Phenotype Analyses
Root length of Arabidopsis plants was measured at 7 DAG. The cell number in root meristem was determined by counting cortical cells from the cortical-endodermal initial cell to the first expanding cortical cell daily from 1 to 8 DAG. For the kinematic growth analyses, root tips of ∼30 seedlings were marked at 6 and then at 7 DAG, and the length increase within this 24-h interval was determined by measuring the distance between these two marks. The size of fully differentiated cells was determined by measuring the length of cortical cells in the differentiated zone. All measurements were done by using Image Lab 2.0 software (Bio-Rad).
Histology and Microscopy
Arabidopsis roots were mounted on slides in chloral hydrate solution and analyzed under Axio Imager A1 microscope (Zeiss) with Nomarski optics. GUS activity was visualized after incubation of transgenic plants for 1 h at 37°C in 0.1 M NaH2PO4-Na2HPO4 (pH 7) buffer including 3% sucrose, 0.05 mM EDTA, 0.5 mg/mL X-gluc, 2.5 mM potassium ferrocyanide, and potassium ferricyanide (Jefferson et al., 1987). All confocal images were acquired using Leica SP2 confocal laser scanning microscope. Fresh transgenic roots were mounted on slides with 10 μM propidium iodide for cell wall staining. GFP was detected with an excitation wavelength of 488 nm and propidium iodide was detected with an excitation wavelength of 543 nm. GFP intensity was measured by using Image Lab 2.0 software (Bio-Rad). To visualize starch in columella cells, mPS-PI staining was performed according to Truernit et al. (2008).
Constructs and Plant Transformation
To generate pHDT1:HDT1-GFP and pHDT2:HDT2-GFP constructs, the genomic sequences of HDT1 and HDT2 including their putative promoters (1.5 kb for HDT1 and 0.5 kb for HDT2) were PCR amplified from genomic DNA using Phusion High-Fidelity DNA Polymerase (Finnzymes) and directionally cloned into pENTR-D-TOPO (Invitrogen). Subsequently, these pENTR-D-TOPO constructs, the pENTR 4-1 vector (Invitrogen), and the pENTR 2-3 vector (Invitrogen) containing a GFP open reading frame and a CaMV 35S terminator were recombined into the binary destination pBnRGW vector by Multisite Gateway reaction. The pBnRGW vector is a modified vector based on PKGW (Karimi et al., 2002) including the pNAP:RFP expression cassette from pFluar101 (Stuitje et al., 2003) for easy selection of red fluorescent transgenic seeds. For pHDT1:GUS, pHDT2:GUS, and pGA2ox2:GUS constructs, the promoters (1.8 kb for GA2ox2) were cloned into the pENTR-D-TOPO vector. The pENTR-D-TOPO constructs, the pENTR4-1 vector (Invitrogen), and the pENTR 2-3 vector containing a GUS were recombined into the binary destination pBnRGW vector by Multisite Gateway reaction. The primers used for making these constructs are listed in Supplemental Table 2.2.
To build the pRCH1:HDT1HDT2 RNAi construct (hdt1,2i), the CaMV 35S promoter in pK7GWIWG2 vector (Limpens et al., 2005) was replaced with RCH1 promoter [pK7GWIWG2(II)]. The RCH1 promoter was first amplified on genomic DNA and cloned into pENTR-D-TOPO. Then it was cut out from the vector by HindIII (partial digestion) and XbaI and recombined with two fragments of the pK7GWIWG2(II) vector in a three-point ligation. The two fragments of pK7GWIWG2(II) vector were obtained by digestion either with HindII and NcoI, or with SpeI (compatible with XbaI) and HindIII. The whole RNAi cassette including the RCH1 promoter was cut out from the vector using ApaI and HindIII and ligated into the pBnRGW binary vector digested with the same enzymes and in such way creating pBnRRGWIWG vector. The RNAi target sequences of HDT1 and HDT2 coding sequences (0.6 kb for each) were combined in one amplicon using a two-step PCR, following the procedure described by Franssen et al. (2015). In brief, the first step PCRs introduced short overlaps (15 bp) in PCR fragments of HDT1 and HDT2 coding sequences with HDT1rnai-F and HDT1rnai-R primers, or with HDT2rnai-F and HDT2rnai-R primers. These two fragments were used as templates in the second-step PCR with HDT1rnai-F and HDT2rnai-R primers. The final PCR fragment was introduced into pENTR-D-TOPO vector and recombined in inverse-repeat orientation into the pBnRRGWIWG binary vector by a LR Gateway reaction (Invitrogen). The primers used for RNAi are presented in Supplemental Table 2.3.
To construct pRCH1:GA2ox2 (GA2ox2OE), the RCH1 promoter was cut out of the pENTR-D-TOPO vector using the NotI and AscI restriction sites, and then ligated with a BsaI-digested pENTR4-1 vector. GA2ox2 cDNA was cloned into pENTR-D-TOPO vector. These two vectors and the CaMV 35S terminator containing pENTR 2-3 vector were recombined in a Multisite Gateway reaction into the binary destination pBnRGW vector. The primers used are shown in Supplemental Table 2.4.
All constructed binary vectors were introduced into Arabidopsis through Agrobacterium tumefaciens (strain C58)-mediated transformation by floral dipping method (Clough and Bent, 1998). The Arabidopsis (Columbia-0) plants were transformed with pHDT1:GUS, pHDT2:GUS, pGA2ox2:GUS, pRCH1:HDT1HDT2 RNAi, and pRCH1:GA2ox2 constructs. To test whether pHDT1:HDT1-GFP and pHDT2:HDT2-GFP could complement the hdt1 hdt2 mutant, homozygous line for hdt1 and heterozygous for HDT2 plants were transformed with pHDT1:HDT1-GFP and pHDT2:HDT2-GFP. The primers used for confirming the complementation are listed in Supplemental Table 2.5.
Gene Expression Analyses
For RT-PCR, ∼10 wild-type and hdt seedlings were collected at 6 DAG. For study of gene expression by RT-qPCR, ∼100 root tips (including meristem and elongation zone) were collected at 6 DAG, and this was done in three independent experiments (three biological replicates). The collected plant material was used for total RNA extraction using the E.Z.N.A. Plant RNA kit (Omega). cDNA was synthesized from 1 μg of RNA by reverse transcription with random hexamer primers using the iScript Select cDNA synthesis kit (Bio-Rad) according to the manufacturer’s instructions. RT-qPCR was performed in a 10-μL reaction with MyiQ SYBR Green Super-mix (Bio-Rad). Each sample was quantified in triplicates using CFX3.0 software (Bio-Rad) and normalized using TUBULIN as a reference. The normalization using Actin as a reference shows similar results. The primers used for RT-PCR or RT-qPCR are listed in Supplemental Table 2.6 or 2.7.
RNA-Seq
Root tips of ∼1000 wild-type and ∼2000 hdt1,2i-1 seedlings at 6 DAG were cut and collected in three parts: the meristem zone (from columella till the disappeared lateral root cap, ∼500 or 300 μm in the wild type or hdt1,2i-1), elongation zone (until the appeared root hairs, ∼900 or 800 μm in the wild type or hdt1,2i-1), and differentiated zone (∼1 mm for both, to avoid lateral root primordia). The experiment was performed three times (three biological replicates). Total RNA from each zone was extracted using RNeasy Micro Kit (Qiagen). RNA was sequenced at BGI Tech Solutions (Hong Kong) using low-input HiSeq2000 instrument. This generated ∼12 million 50-bp clean single-end reads for each sample. Sequencing data were analyzed by mapping to the Arabidopsis genome (TAIR10) using a CLC Genomics Workbench (Denmark). Gene expression levels were determined by calculating the reads per kilobase per million mapped reads (Shi et al., 2006). DEGs are defined based on relatively stringent statistics and filtering (fold change > 2, false discovery rate, P value < 0.05) within the CLC. GO enrichment analyses were performed using agriGO v2.0 (Tian et al., 2017).
ChIP Assay
Approximately 1 gram of wild-type and pHDT2:HDT2-GFP (in the hdt1 hdt2 background) complemented seedlings at 6 DAG were used for each anti-GFP ChIP assay. Approximately 0.5 g of wild-type and hdt1,2i-1 roots (∼5 mm from the tip) at 6 DAG were used for each anti-H3ac ChIP assay. For both assays, three independent ChIP experiments were performed as described (Kaufmann et al., 2010), with some modifications. For anti-GFP assays, sonicated chromatin was incubated with GFP-Trap agarose beads (Chromotek) for 90 min at 4°C. For anti-H3ac assays, sonicated chromatin was immunoprecipitated with an anti-H3ac antibody (06-599; Millipore) for 1 h at 4°C and then incubated with Protein A/G PLUS-Agarose beads (Santa Cruz) for 50 min at 4°C. Immunoprecipitated DNA was recovered using the IPure kit v2 (Diagenode) and quantified using the Qubit dsDNA HS Assay Kit (Life Technologies). An aliquot of nontreated sonicated chromatin was processed in parallel and used as total input DNA control. ChIP-qPCR data analyses were performed according to Frank et al. (2001). All ChIP enrichments were calculated as percentage of DNA immunoprecipitated at the locus of interest relative to the corresponding input samples and normalized to the percentage of DNA immunoprecipitated at that locus in wild-type plants. The primers used for ChIP-qPCR are listed in Supplemental Table 2.8.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: HDT1 (At3g44750), HDT2 (At5g22650), HDT3 (At5g03740), HDT4 (At2g27840), MATE (At2g04050), and GA2ox2 (At1g30040).
Supplemental Data
Supplemental Figure 1. Genome Structure of HDT1-4 and GA2ox2 Genes.
Supplemental Figure 2. HDT1/2 Control Root Growth.
Supplemental Figure 3. Expression of HDT1 and HDT2 Genes in Wild-Type and hdt1,2i Lines.
Supplemental Figure 4. Reduced Cell Number in hdt1,2i and GA2ox2OE RM Is Caused by an Earlier Switch from Cell Division to Expansion.
Supplemental Figure 5. Expression Pattern of pHDT1:HDT1-GFP and pHDT2:HDT2-GFP in Roots.
Supplemental Figure 6. Gene Ontology Enrichment Analyses of DEGs in hdt1,2i-1 Roots.
Supplemental Figure 7. PIN1 and PIN2 Proteins Are Reduced in hdt1,2i-1.
Supplemental Table 1. Kinematic Analyses of Root Growth in Wild-Type and hdt1,2i-8 Seedlings.
Supplemental Table 2. Primers Used in This Study.
Supplemental Data Set 1. Expression of Histone Acetylation Genes from Eight Subzones of the Root.
Supplemental Data Set 2. Gene Expression Map in the Wild-Type and hdt1,2i-1 Roots.
Acknowledgments
This research was supported by the European Commission (ERC-2011-AdG-294790) and by the China Scholarship Council. We thank Ben Scheres (Plant Developmental Biology, Wageningen University and Research Center) for critical reading of this article and for the provision of Arabidopsis transgenic GFP reporter lines.
AUTHOR CONTRIBUTIONS
H.L. conceived this project, designed the research, analyzed the data, and wrote the article. H.L., J.T.-G., S.S., and W.Z. conducted the experiments. D.L., M.B., and H.H. contributed to ChIP studies. O.K. and T.B. modified the article.
Footnotes
Articles can be viewed without a subscription.
References
- Achard P., Gusti A., Cheminant S., Alioua M., Dhondt S., Coppens F., Beemster G.T., Genschik P. (2009). Gibberellin signaling controls cell proliferation rate in Arabidopsis. Curr. Biol. 19: 1188–1193. [DOI] [PubMed] [Google Scholar]
- Aida M., Beis D., Heidstra R., Willemsen V., Blilou I., Galinha C., Nussaume L., Noh Y.S., Amasino R., Scheres B. (2004). The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119: 109–120. [DOI] [PubMed] [Google Scholar]
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657. [DOI] [PubMed] [Google Scholar]
- Band L.R., Úbeda-Tomás S., Dyson R.J., Middleton A.M., Hodgman T.C., Owen M.R., Jensen O.E., Bennett M.J., King J.R. (2012). Growth-induced hormone dilution can explain the dynamics of plant root cell elongation. Proc. Natl. Acad. Sci. USA 109: 7577–7582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benková E., Michniewicz M., Sauer M., Teichmann T., Seifertová D., Jürgens G., Friml J. (2003). Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115: 591–602. [DOI] [PubMed] [Google Scholar]
- Bennett T., Scheres B. (2010). Root development-two meristems for the price of one? Curr. Top. Dev. Biol. 91: 67–102. [DOI] [PubMed] [Google Scholar]
- Bernard V., Brunaud V., Lecharny A. (2010). TC-motifs at the TATA-box expected position in plant genes: a novel class of motifs involved in the transcription regulation. BMC Genomics 11: 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnbaum K., Shasha D.E., Wang J.Y., Jung J.W., Lambert G.M., Galbraith D.W., Benfey P.N. (2003). A gene expression map of the Arabidopsis root. Science 302: 1956–1960. [DOI] [PubMed] [Google Scholar]
- Blilou I., Frugier F., Folmer S., Serralbo O., Willemsen V., Wolkenfelt H., Eloy N.B., Ferreira P.C., Weisbeek P., Scheres B. (2002). The Arabidopsis HOBBIT gene encodes a CDC27 homolog that links the plant cell cycle to progression of cell differentiation. Genes Dev. 16: 2566–2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blilou I., Xu J., Wildwater M., Willemsen V., Paponov I., Friml J., Heidstra R., Aida M., Palme K., Scheres B. (2005). The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433: 39–44. [DOI] [PubMed] [Google Scholar]
- Bourque S., Dutartre A., Hammoudi V., Blanc S., Dahan J., Jeandroz S., Pichereaux C., Rossignol M., Wendehenne D. (2011). Type-2 histone deacetylases as new regulators of elicitor-induced cell death in plants. New Phytol. 192: 127–139. [DOI] [PubMed] [Google Scholar]
- Casamitjana-Martínez E., Hofhuis H.F., Xu J., Liu C.M., Heidstra R., Scheres B. (2003). Root-specific CLE19 overexpression and the sol1/2 suppressors implicate a CLV-like pathway in the control of Arabidopsis root meristem maintenance. Curr. Biol. 13: 1435–1441. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Dello Ioio R., Linhares F.S., Scacchi E., Casamitjana-Martinez E., Heidstra R., Costantino P., Sabatini S. (2007). Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Curr. Biol. 17: 678–682. [DOI] [PubMed] [Google Scholar]
- Dello Ioio R., Nakamura K., Moubayidin L., Perilli S., Taniguchi M., Morita M.T., Aoyama T., Costantino P., Sabatini S. (2008). A genetic framework for the control of cell division and differentiation in the root meristem. Science 322: 1380–1384. [DOI] [PubMed] [Google Scholar]
- De Veylder L., Beemster G.T., Beeckman T., Inze D. (2001). CKS1At overexpression in Arabidopsis thaliana inhibits growth by reducing meristem size and inhibiting cell-cycle progression. Plant J. 25: 617–626. [DOI] [PubMed] [Google Scholar]
- Ding B., Bellizzi Mdel.R., Ning Y., Meyers B.C., Wang G.L. (2012). HDT701, a histone H4 deacetylase, negatively regulates plant innate immunity by modulating histone H4 acetylation of defense-related genes in rice. Plant Cell 24: 3783–3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugardeyn J., Vandenbussche F., Van Der Straeten D. (2008). To grow or not to grow: what can we learn on ethylene-gibberellin cross-talk by in silico gene expression analysis? J. Exp. Bot. 59: 1–16. [DOI] [PubMed] [Google Scholar]
- Eckardt N.A. (2001). Move it on out with MATEs. Plant Cell 13: 1477–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnin M.S., Donigian J.R., Cohen A., Richon V.M., Rifkind R.A., Marks P.A., Breslow R., Pavletich N.P. (1999). Structures of a histone deacetylase homologue bound to the TSA and SAHA inhibitors. Nature 401: 188–193. [DOI] [PubMed] [Google Scholar]
- Frank S.R., Schroeder M., Fernandez P., Taubert S., Amati B. (2001). Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes Dev. 15: 2069–2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franssen H.J., Xiao T.T., Kulikova O., Wan X., Bisseling T., Scheres B., Heidstra R. (2015). Root developmental programs shape the Medicago truncatula nodule meristem. Development 142: 2941–2950. [DOI] [PubMed] [Google Scholar]
- Galinha C., Hofhuis H., Luijten M., Willemsen V., Blilou I., Heidstra R., Scheres B. (2007). PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 449: 1053–1057. [DOI] [PubMed] [Google Scholar]
- Hedden P., Phillips A.L. (2000). Gibberellin metabolism: new insights revealed by the genes. Trends Plant Sci. 5: 523–530. [DOI] [PubMed] [Google Scholar]
- Helariutta Y., Fukaki H., Wysocka-Diller J., Nakajima K., Jung J., Sena G., Hauser M.T., Benfey P.N. (2000). The SHORT-ROOT gene controls radial patterning of the Arabidopsis root through radial signaling. Cell 101: 555–567. [DOI] [PubMed] [Google Scholar]
- Jefferson R.A., Kavanagh T.A., Bevan M.W. (1987). GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6: 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T., Allis C.D. (2001). Translating the histone code. Science 293: 1074–1080. [DOI] [PubMed] [Google Scholar]
- Karimi M., Inzé D., Depicker A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195. [DOI] [PubMed] [Google Scholar]
- Kaufmann K., Muiño J.M., Østerås M., Farinelli L., Krajewski P., Angenent G.C. (2010). Chromatin immunoprecipitation (ChIP) of plant transcription factors followed by sequencing (ChIP-SEQ) or hybridization to whole genome arrays (ChIP-CHIP). Nat. Protoc. 5: 457–472. [DOI] [PubMed] [Google Scholar]
- Kornet N., Scheres B. (2009). Members of the GCN5 histone acetyltransferase complex regulate PLETHORA-mediated root stem cell niche maintenance and transit amplifying cell proliferation in Arabidopsis. Plant Cell 21: 1070–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. (2007). Chromatin modifications and their function. Cell 128: 693–705. [DOI] [PubMed] [Google Scholar]
- Limpens E., Mirabella R., Fedorova E., Franken C., Franssen H., Bisseling T., Geurts R. (2005). Formation of organelle-like N2-fixing symbiosomes in legume root nodules is controlled by DMI2. Proc. Natl. Acad. Sci. USA 102: 10375–10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Jin G., Zhou X. (2015a). Modeling the relationship of epigenetic modifications to transcription factor binding. Nucleic Acids Res. 43: 3873–3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Li R.J., Han T.T., Cai W., Fu Z.W., Lu Y.T. (2015b). Salt stress reduces root meristem size by nitric oxide-mediated modulation of auxin accumulation and signaling in Arabidopsis. Plant Physiol. 168: 343–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M., Wang Y.Y., Liu X., Yang S., Lu Q., Cui Y., Wu K. (2012). HD2C interacts with HDA6 and is involved in ABA and salt stress response in Arabidopsis. J. Exp. Bot. 63: 3297–3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason M.G., Li J., Mathews D.E., Kieber J.J., Schaller G.E. (2004). Type-B response regulators display overlapping expression patterns in Arabidopsis. Plant Physiol. 135: 927–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason M.G., Mathews D.E., Argyros D.A., Maxwell B.B., Kieber J.J., Alonso J.M., Ecker J.R., Schaller G.E. (2005). Multiple type-B response regulators mediate cytokinin signal transduction in Arabidopsis. Plant Cell 17: 3007–3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moubayidin L., Di Mambro R., Sabatini S. (2009). Cytokinin-auxin crosstalk. Trends Plant Sci. 14: 557–562. [DOI] [PubMed] [Google Scholar]
- Moubayidin L., Perilli S., Dello Ioio R., Di Mambro R., Costantino P., Sabatini S. (2010). The rate of cell differentiation controls the Arabidopsis root meristem growth phase. Curr. Biol. 20: 1138–1143. [DOI] [PubMed] [Google Scholar]
- Nguyen H.N., Kim J.H., Jeong C.Y., Hong S.W., Lee H. (2013). Inhibition of histone deacetylation alters Arabidopsis root growth in response to auxin via PIN1 degradation. Plant Cell Rep. 32: 1625–1636. [DOI] [PubMed] [Google Scholar]
- Patikoglou G.A., Kim J.L., Sun L., Yang S.H., Kodadek T., Burley S.K. (1999). TATA element recognition by the TATA box-binding protein has been conserved throughout evolution. Genes Dev. 13: 3217–3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng J., Carol P., Richards D.E., King K.E., Cowling R.J., Murphy G.P., Harberd N.P. (1997). The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev. 11: 3194–3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perilli S., Moubayidin L., Sabatini S. (2010). The molecular basis of cytokinin function. Curr. Opin. Plant Biol. 13: 21–26. [DOI] [PubMed] [Google Scholar]
- Pi L., Aichinger E., van der Graaff E., Llavata-Peris C.I., Weijers D., Hennig L., Groot E., Laux T. (2015). Organizer-derived WOX5 signal maintains root columella stem cells through chromatin-mediated repression of CDF4 expression. Dev. Cell 33: 576–588. [DOI] [PubMed] [Google Scholar]
- Rieu I., Eriksson S., Powers S.J., Gong F., Griffiths J., Woolley L., Benlloch R., Nilsson O., Thomas S.G., Hedden P., Phillips A.L. (2008). Genetic analysis reveals that C19-GA 2-oxidation is a major gibberellin inactivation pathway in Arabidopsis. Plant Cell 20: 2420–2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa S., Ntoukakis V., Ohmido N., Pendle A., Abranches R., Shaw P. (2014). Cell differentiation and development in Arabidopsis are associated with changes in histone dynamics at the single-cell level. Plant Cell 26: 4821–4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatini S., Heidstra R., Wildwater M., Scheres B. (2003). SCARECROW is involved in positioning the stem cell niche in the Arabidopsis root meristem. Genes Dev. 17: 354–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar A.K., Luijten M., Miyashima S., Lenhard M., Hashimoto T., Nakajima K., Scheres B., Heidstra R., Laux T. (2007). Conserved factors regulate signalling in Arabidopsis thaliana shoot and root stem cell organizers. Nature 446: 811–814. [DOI] [PubMed] [Google Scholar]
- Scheres B. (2007). Stem-cell niches: nursery rhymes across kingdoms. Nat. Rev. Mol. Cell Biol. 8: 345–354. [DOI] [PubMed] [Google Scholar]
- Shahbazian M.D., Grunstein M. (2007). Functions of site-specific histone acetylation and deacetylation. Annu. Rev. Biochem. 76: 75–100. [DOI] [PubMed] [Google Scholar]
- Shani E., Weinstain R., Zhang Y., Castillejo C., Kaiserli E., Chory J., Tsien R.Y., Estelle M. (2013). Gibberellins accumulate in the elongating endodermal cells of Arabidopsis root. Proc. Natl. Acad. Sci. USA 110: 4834–4839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L., et al. ; MAQC Consortium (2006). The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat. Biotechnol. 24: 1151–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone A.L., Jung H.S., Dill A., Kawaide H., Kamiya Y., Sun T.P. (2001). Repressing a repressor: gibberellin-induced rapid reduction of the RGA protein in Arabidopsis. Plant Cell 13: 1555–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridha S., Wu K. (2006). Identification of AtHD2C as a novel regulator of abscisic acid responses in Arabidopsis. Plant J. 46: 124–133. [DOI] [PubMed] [Google Scholar]
- Strahl B.D., Allis C.D. (2000). The language of covalent histone modifications. Nature 403: 41–45. [DOI] [PubMed] [Google Scholar]
- Stuitje A.R., Verbree E.C., van der Linden K.H., Mietkiewska E.M., Nap J.P., Kneppers T.J. (2003). Seed-expressed fluorescent proteins as versatile tools for easy (co)transformation and high-throughput functional genomics in Arabidopsis. Plant Biotechnol. J. 1: 301–309. [DOI] [PubMed] [Google Scholar]
- Thomas S.G., Phillips A.L., Hedden P. (1999). Molecular cloning and functional expression of gibberellin 2- oxidases, multifunctional enzymes involved in gibberellin deactivation. Proc. Natl. Acad. Sci. USA 96: 4698–4703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian T., Liu Y., Yan H., You Q., Yi X., Du Z., Xu W., Su Z. (2017). agriGO v2.0: a GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 10.1093/nar/gkx382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truernit E., Bauby H., Dubreucq B., Grandjean O., Runions J., Barthélémy J., Palauqui J.C. (2008). High-resolution whole-mount imaging of three-dimensional tissue organization and gene expression enables the study of phloem development and structure in Arabidopsis. Plant Cell 20: 1494–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubeda-Tomás S., Swarup R., Coates J., Swarup K., Laplaze L., Beemster G.T., Hedden P., Bhalerao R., Bennett M.J. (2008). Root growth in Arabidopsis requires gibberellin/DELLA signalling in the endodermis. Nat. Cell Biol. 10: 625–628. [DOI] [PubMed] [Google Scholar]
- Ubeda-Tomás S., Federici F., Casimiro I., Beemster G.T., Bhalerao R., Swarup R., Doerner P., Haseloff J., Bennett M.J. (2009). Gibberellin signaling in the endodermis controls Arabidopsis root meristem size. Curr. Biol. 19: 1194–1199. [DOI] [PubMed] [Google Scholar]
- van den Berg C., Willemsen V., Hage W., Weisbeek P., Scheres B. (1995). Cell fate in the Arabidopsis root meristem determined by directional signalling. Nature 378: 62–65. [DOI] [PubMed] [Google Scholar]
- Vanstraelen M., Baloban M., Da Ines O., Cultrone A., Lammens T., Boudolf V., Brown S.C., De Veylder L., Mergaert P., Kondorosi E. (2009). APC/C-CCS52A complexes control meristem maintenance in the Arabidopsis root. Proc. Natl. Acad. Sci. USA 106: 11806–11811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S.Y., Sun T., Faust M. (1986). Translocation of paclobutrazol, a gibberellin biosynthesis inhibitor, in apple seedlings. Plant Physiol. 82: 11–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willige B.C., Isono E., Richter R., Zourelidou M., Schwechheimer C. (2011). Gibberellin regulates PIN-FORMED abundance and is required for auxin transport-dependent growth and development in Arabidopsis thaliana. Plant Cell 23: 2184–2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K., Tian L., Zhou C., Brown D., Miki B. (2003). Repression of gene expression by Arabidopsis HD2 histone deacetylases. Plant J. 34: 241–247. [DOI] [PubMed] [Google Scholar]
- Yamauchi Y., Takeda-Kamiya N., Hanada A., Ogawa M., Kuwahara A., Seo M., Kamiya Y., Yamaguchi S. (2007). Contribution of gibberellin deactivation by AtGA2ox2 to the suppression of germination of dark-imbibed Arabidopsis thaliana seeds. Plant Cell Physiol. 48: 555–561. [DOI] [PubMed] [Google Scholar]
- Yang L., Zhang J., He J., Qin Y., Hua D., Duan Y., Chen Z., Gong Z. (2014). ABA-mediated ROS in mitochondria regulate root meristem activity by controlling PLETHORA expression in Arabidopsis. PLoS Genet. 10: e1004791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S., Li C., Zhao L., Gao S., Lu J., Zhao M., Chen C.Y., Liu X., Luo M., Cui Y., Yang C., Wu K. (2015). The Arabidopsis SWI2/SNF2 chromatin remodeling ATPase BRAHMA targets directly to PINs and is required for root stem cell niche maintenance. Plant Cell 27: 1670–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C., Labbe H., Sridha S., Wang L., Tian L., Latoszek-Green M., Yang Z., Brown D., Miki B., Wu K. (2004). Expression and function of HD2-type histone deacetylases in Arabidopsis development. Plant J. 38: 715–724. [DOI] [PubMed] [Google Scholar]