Abstract
The extracellular matrix (ECM) constituting up to 20% of the organ volume is a significant component of the brain due to its instructive role in the compartmentalization of functional microdomains in every brain structure. The composition, quantity and structure of ECM changes dramatically during the development of an organism greatly contributing to the remarkably sophisticated architecture and function of the brain. Since fetal brain is highly plastic, we hypothesize that the fetal brain ECM may contain cues promoting neural growth and differentiation, highly desired in regenerative medicine. Thus, we studied the effect of brain-derived fetal and adult ECM complemented with matricellular proteins on cortical neurons using in vitro 3D bioengineered model of cortical brain tissue. The tested parameters included neuronal network density, cell viability, calcium signaling and electrophysiology. Both, adult and fetal brain ECM as well as matricellular proteins significantly improved neural network formation as compared to single component, collagen I matrix. Additionally, the brain ECM improved cell viability and lowered glutamate release. The fetal brain ECM induced superior neural network formation, calcium signaling and spontaneous spiking activity over adult brain ECM. This study highlights the difference in the neuroinductive properties of fetal and adult brain ECM and suggests that delineating the basis for this divergence may have implications for regenerative medicine.
Keywords: fetal, brain, extracellular matrix, matricellular, 3D cell culture, neurons, decellularized
Graphical abstract
INTRODUCTION
Adult human brain extracellular matrix (ECM), which constitutes about 20% of the organ volume, is highly organized and unique in composition amongst the ECM of other tissues1,2. Unlike other tissues in the body, brain ECM contains low levels of fibrous proteins like collagen, fibronectin and vitronectin, resulting in a ratio of glycosaminoglycans (GAGs) to collagen around 10:13,4. The brain ECM is mainly composed of a family of aggregating proteoglycans called lecticans, which consists of four members: neurocan, brevican, versican and aggrecan3,5. The lectican core proteins interact with secreted ECM glycoproteins like tenascin-R/C through their C-terminal domains, while their N-terminal domains bind to a major non-sulfated GAG component of brain ECM, hyaluronic acid (HA) through link proteins2–3. Other GAG chains including heparin sulfate and chondroitin sulfate connect to the attachment sites in the middle portion of the lectican core proteins3. The high level of specialization of brain ECM provides specific biochemical cues and maintains homeostatic functions by clustering of soluble or membrane-associated signaling molecules in microdomains surrounding neurons and glia1. As an example, the perineuronal net (PNN), an ECM structure composed of chondroitin sulfate proteoglycans (CSPGs), HA, tenascin-C and tenascin-R, is exclusively found in the brain. The link proteins and their interactions with proteoglycans vary depending upon brain region and the maturity of the brain ECM5. The heterogeneity in the molecular composition of the brain ECM enables its functional role in the integration and differentiation of stem cells in embryonic brain or the neurogenic niche in the adult brain1. Furthermore, the ECM helps in maintaining soluble growth factors, in the organization of ion channels, and axonal and pre/post-synaptic proteins6,1. In addition, there exist astrocyte-secreted non-structural brain ECM-associated proteins called matricellular proteins, which are known to help reshape cell-ECM/ECM-ECM interactions, and aid in synaptogenesis and in circuit rewiring following injury7. Therefore, the heterogeneity present in the complex ECM of brain tissue is important to consider while designing substrates for neuronal culture, by maximizing the inclusion of ECM components from the specific tissue type8.
Tissue-specific biologic scaffolds have been prepared from the ECM of several tissues including urinary bladder, myocardium, muscle, tendon and esophagus9,10,11. These have all been shown to promote functional tissue remodeling due to the presence of innate 3D ultrastructure of the source tissue, unique distribution of ligands, bioactive molecular cues produced due to scaffold degradation, recruitment of endogenous progenitor cells and innervation10. Recently, methods to decellularize and solubilize rodent and porcine brain-derived ECM have been reported12,13,14. The hydrogel formulations of these matrix scaffolds had neurotrophic potential via retention of growth factors and proteins, site appropriate differentiation effects on mouse neuroblastoma cells, neural stem cells (NSCs) and induced pluripotent stem cell (iPSC)-derived neurons, and enhanced maturation effects in comparison to conventional substrates such as collagen type I and laminin15,16,17. The proliferation and differentiation effects, gelation kinetics and mechanical properties of brain-derived ECM have been studied in isolation, but not in composite systems that combine mechanical stability with bioactivity14. Most importantly, the differential effects of fetal versus adult brain ECM in in vitro systems for neurobiological studies are needed. Fetal ECM is likely to be more conducive to the growth of primary neurons in vitro. Indeed, a recent study comparing ECM from different developmental stages of heart showed the beneficial effects of decellularized fetal heart ECM in comparison to neonatal or adult heart ECM18. A similar investigation into the brain ECM would be informative, considering the immense complexity of these tissues.
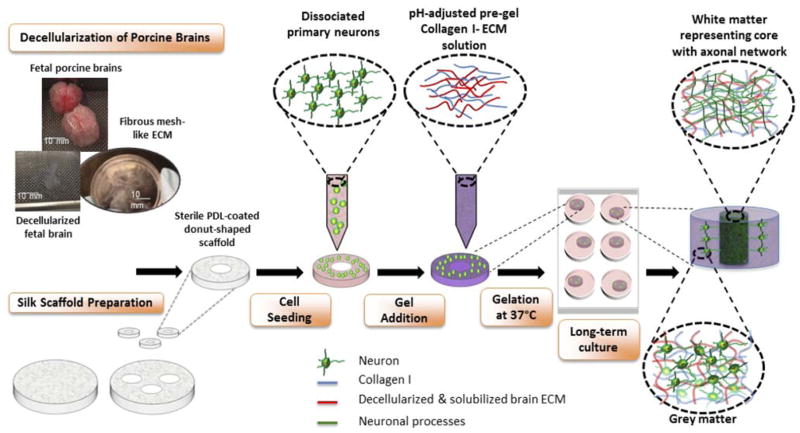
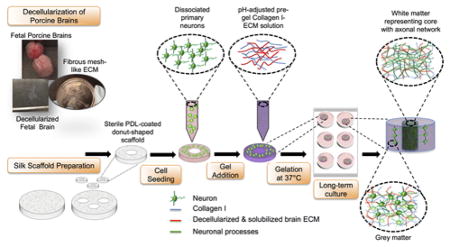
The goal of this study was to investigate how changes of the brain ECM occurring during development impact the growth and functionality of neurons. Building upon our recently developed tissue model of in vitro 3D brain cortex, our aim was to generate 3D physiological cultures that mimic the structural, functional and biochemical properties of native brain tissue19,20. The design of the donut-shaped constructs provides an excellent framework for studying axonal in-growth in vitro because of the presence of an axon-rich middle region resembling white matter, surrounded by neuronal soma-containing silk porous protein scaffold representative of grey matter19,20 (Schematic 1). The model was enriched with fetal and adult brain ECM, and additionally supplemented with astrocyte-secreted matricellular proteins to study their effect on neuronal network formation. To our knowledge, this is the first study to compare the proneuronal activity of adult and fetal mammalian brain ECM using the 3D composite hydrogel systems.
Schematic 1.
Preparation of 3D in vitro bioengineered brain constructs infused with decellularized brain ECM-collagen I gel. Process starts with decellularization of porcine brains and silk scaffold preparation. Aqueous silk scaffold is punched into 6mm diameter constructs with 2mm diameter central hole to create donut-shaped scaffolds. The poly-D-lysine coated scaffolds are seeded with dissociated embryonic day 18 primary neurons. Decellularized ECM is mixed with Collagen I solution and added to the donut-shaped scaffolds seeded with cells. The cell-seeded silk-scaffolds are flooded with media after complete gelation of ECM-collagen I. After long-term culture, the center of the construct shows a dense axonal network representing white matter, surrounded by the neuronal cell body-containing grey matter
MATERIALS AND METHODS
Fetal and adult porcine brain procurement
Young adult (4–6 months old) porcine brains were ordered from Lampire Biological Laboratories (Pipersville, PA). The porcine brains were collected from normal porcine at a U.S.D.A inspected abattoir and supplied frozen. For fetal porcine brain extraction, freshly frozen fetal pigs (Nebraska Scientific, Omaha, NE), embryonic-days 80–90 (E80–90), were used.
Optimization of brain decellularization: Age dependant ECM
Adult porcine brain was decellularized using a previously established method16. The dura mater was stripped from the thawed adult porcine brain, which was then chopped into smaller pieces (~2 cm) and incubated in distilled water overnight at 4°C (Schematic 1b). Briefly, the decellularization process consisted of incubation in the following solutions: 0.05% trypsin-EDTA (Life Technologies Corporation, Carlsbad, CA, USA) in phosphate-buffered saline (PBS) at 37°C for 1h, 3% triton X-100 (Sigma-Aldrich, Milwaukee, WI, USA) in PBS for 1h, 1M sucrose (Sigma-Aldrich, Milwaukee, WI, USA) solution for 15 min, distilled water for 15 min, 4% sodium deoxycholate (Sigma) in distilled water for 1h, 0.1% peracetic acid (Sigma-Aldrich, Milwaukee, WI, USA) in 4% ethanol for 2h and finally, in distilled water overnight at 4°C. All the incubation solutions were supplemented with 1% antibiotic-antimycotic (Life Technologies Corporation, Carlsbad, CA, USA) and the steps were performed at room temperature unless otherwise mentioned. The protocol was modified in terms of the incubation times and concentrations of solutions for effective decellularization of fetal porcine brains. The incubation steps that worked best for fetal brains included: distilled water overnight at 4°C, 0.05% trypsin-EDTA with 0.2% DNase I (Roche, Indianapolis, IN, USA) in PBS at 37°C for 30mins, 3% Triton X-100 with 0.2% DNase I in PBS for 30mins, 1M sucrose solution for 15mins, distilled water for 15mins, 1% sodium deoxycholate in PBS for 15min, 0.1% peracetic acid in 4% ethanol for 2hrs and finally, in distilled water overnight at 4°C. The decellularized fetal and adult brain tissues were lyophilized and stored at −20°C for use in experiments.
Characterization of fetal and adult brain ECM
DAPI staining
Before lyophilization, the decellularized brain samples were mounted on a slide using fluoroshield mounting medium with DAPI (Abcam, Cambridge, MA, USA) to test for the presence of nuclei and compared against a sample that was partially decellularized.
Pico green
The DNA content of the brain ECM was quantified using Quant-iT™ PicoGreen® dsDNA Assay Kit (Life Technologies Corporation, Carlsbad, CA, USA), and compared against the DNA content of adult and fetal porcine whole brains, and a commercially available matrix, Matrigel (Corning, Bedford, MA, USA). The whole brain samples were lyophilized to be able to measure their dry weight. DNA was extracted from all the lyophilized samples using PureLink® Genomic DNA extraction kit (Thermo Fischer Scientific, Waltham, MA USA). Briefly, the lyophilized samples were digested in Proteinase K, followed by RNA digestion and DNA spin columns were used to bind and elute the genomic DNA. Following DNA extraction, each sample was tested for DNA content in Pico Green assay, using three different dilutions (1:20, 1:50, 1:100).
Collagen and sGAG assays
Collagen and sulfated GAG (sGAG) content of the brain ECM was measured using Sircol soluble collagen and Blyscan sGAG assays (Accurate Chemical & Scientific Corporation, Westbury, NY, USA). The ECM samples were solubilized in 1 mg/ml pepsin from porcine gastric mucosa (Sigma-Aldrich, Milwaukee, WI, USA) in 0.1N hydrochloric acid (Sigma-Aldrich, Milwaukee, WI, USA) at room temperature for 24–48h and diluted 1:20 (v:v) in distilled water for use in collagen assay. For the sGAG assay, the samples were digested in 2mg/ml Proteinase K (Sigma-Aldrich, Milwaukee, WI, USA) at 55°C for 3h and diluted 1:500 (v:v) in distilled water. Briefly, the diluted samples and standards provided with the assay kit were mixed with dye reagents, which bound to either collagen or sGAG. The bound dye was eventually released using dissociation reagents. The absorption was measured at 555 nm and 656 nm for the dye released from collagen and sGAG, respectively. Standard curves were used to calculate the amounts of collagen and sGAG present in the ECM samples.
Protein gel electrophoresis
In order to determine the protein profiles of the extracted ECM, the lyophilized samples were digested with 1 mg/ml pepsin from porcine gastric mucosa (Sigma-Aldrich, Milwaukee, WI, USA) in 0.1N hydrochloric acid (Sigma-Aldrich, Milwaukee, WI, USA). Following digestion, the ECM samples were diluted in distilled water to 2 mg/ml, neutralized and mixed at 1:1 (v:v) ratio with NuPAGE® (Thermo Fischer Scientific, Waltham, MA USA) LDS loading buffer containing reducing agent. An equal quantity (10 μg) of neutralized ECM samples and HiMark™ unstained protein standard (Novex, Thermo Fischer Scientific, Waltham, MA USA) were loaded in separate lanes of a 3–8% NuPAGE® Tris-Acetate gel and the gel was run at a constant voltage of 200V for 2h. The resolved proteins were stained with Coomassie Blue R-250 (Thermo Fischer Scientific, Waltham, MA USA) for 1h, de-stained for another 1h and imaged against the proteins bands (36–400 kDa) of the HiMark™ standard.
Primary rat neuron isolation
Time-pregnant Sprague–Dawley rats were obtained from Charles River Laboratories and housed in the animal facility. Experiments were performed as approved by the Institutional Animal Care and Use Committee at Tufts University, Medford, MA, USA (M2015-28) and conducted in accordance with the National Institutes of Health Guidelines for the Care and Use of Laboratory Animals. Freshly micro-dissected whole rat cortices at embryonic-day 18 (E18) were incubated with 0.25% Trypsin (Life Technologies Corporation, Carlsbad, CA, USA) containing 0.3% DNase (Roche, Indianapolis, IN, USA) for 20 minutes at 37°C, followed by addition of trypsin inhibitor (Sigma-Aldrich, Milwaukee, WI, USA). The tissue was disrupted by pipetting up and down 20 times with a 10 mL serological pipette to generate a single cell suspension. The cell suspension was centrifuged for 5 minutes at 1200 rpm (Centrifuge 5810 R, Eppendorf, Hamburg, Germany) and the pellet was resuspended in Neurobasal medium (Life Technologies Corporation, Carlsbad, CA, USA) supplemented with 2% B27 (Life Technologies Corporation, Carlsbad, CA, USA), 0.5 mM GlutaMax (Life Technologies Corporation, Carlsbad, CA, USA) and 1% penicillin/streptomycin (Sigma-Aldrich, Milwaukee, WI, USA). Total cell yield was determined using a hemocytometer.
Generation of Hydrogels
Hydromatrix (Sigma-Aldrich, Milwaukee, WI, USA) hydrogel was formed using the manufacturer instructions. Briefly, Hydromatrix was diluted to 0.3% (w/v) with isosmotic sucrose (20%) and kept on ice till applying on the silk scaffold. Cell culture medium was applied immediately to initiate gel formation. Puramatrix (Corning, Bedford, MA, USA) hydrogel was formed using the manufacturer instructions. Briefly, Hydromatrix was diluted to 0.3% (w/v) with isosmotic sucrose (20%) and kept on ice till applying on the silk scaffold. Cell culture medium was applied immediately to initiate gel formation. Fibrin gel was prepared by mixing 20 mg/ml fibrinogen (Sigma-Aldrich, Milwaukee, WI, USA) with 10U/ml thrombin (Sigma-Aldrich, Milwaukee, WI, USA) at a ratio of 2:5. Matrigel (Corning, Bedford, MA, USA) was added to the silk scaffold seeded with cells, and its gelation was induced with the increase in temperature. HyStem C (Sigma-Aldrich, Milwaukee, WI, USA) hydrogel was formed using the manufacturer instructions. Briefly, the lyophilized components, thiol-modified gelatin, thiol-modified hyaluronan and the crosslinker PEGDA were reconstituted in distilled water provided in the kit. The reconstituted gelatin and hyaluronan were mixed at 1:1 (v:v) ratio, followed by the addition of the crosslinker. Collagen I hydrogel was prepared at 3 mg/ml using rat tail collagen I (Corning, Bed-ford, MA, USA). Depending on the required final volume, the supplied collagen I solution was diluted with 10x PBS and distilled water. Finally, the pH was increased using 1N NaOH to start the gelation process. For combination hydrogels with collagen I, fibronectin (Sigma-Aldrich, Milwaukee, WI, USA) and laminin (Roche, Indianapolis, IN, USA) were mixed at concentration of 200 μg/ml with the diluted collagen solution before increasing the pH.
For the generation of ECM-collagen I hydrogels, the lyophilized ECM was solubilized with 1 mg/ml pepsin from porcine gastric mucosa (Sigma-Aldrich, Milwaukee, WI, USA) in 0.1N hydrochloric acid (Sigma-Aldrich, Milwaukee, WI, USA). The solubilization time for fetal and adult ECM at room temperature was approximately 24hrs and 48hrs, respectively. Once solubilized, the ECM was mixed with Neurobasal media (Life Technologies Corporation, Carlsbad, CA, USA) at a 1:1 ratio and neutralized using 1M NaOH (Sigma-Aldrich, Milwaukee, WI, USA). The neutralized ECM solution was mixed with 3 mg/ml rat tail collagen I (Corning, Bedford, MA, USA) at an ECM concentration range of 50–1000 μg/ml and the gelation process started using NaOH. The ECM-collagen solution was kept on ice until complete gelation was required. The solution was used within 2h of preparation.
Assembly of 3D bioengineered cortical brain tissue model
The assembly of bioengineered cortical tissue was performed as previously described20. Briefly, the silk porous 3D scaffolds were coated with 0.1 mg/mL poly-D-lysine (PDL) (Sigma-Aldrich, Milwaukee, WI, USA) either overnight at 4°C or for 2hrs at room temperature. The scaffolds were washed with PBS and incubated in media at 37°C for 30min in order to equilibrate the scaffolds for cell seeding. Subsequently, 1x106 cells were seeded per scaffold. Upon overnight incubation, the unattached cells were washed away with Neurobasal media (Life Technologies Corporation, Grand Island, NY, USA) and the cell-seeded scaffolds were filled with either 3 mg/ml rat tail collagen I (Corning, Bedford, MA, USA) or collagen-fetal/adult ECM composite hydrogel. For the matrix screen experiment, commercially available hydrogels-matrigel (Corning, Bedford, MA, USA), Hydromatrix (Sigma-Aldrich, Milwaukee, WI, USA), Puramatrix (3D Matrix, Waltham, MA, USA), HystemC (Sigma-Aldrich, Milwaukee, WI, USA), fibrin (Sigma-Aldrich, Milwaukee, WI, USA) were added to the cell-seeded scaffolds instead of collagen I. Additionally, laminin (Roche, Indianapolis, IN, USA) and fibronectin (Sigma-Aldrich, Milwaukee, WI, USA) at a concentration of 50 μg/ml, were also tested in combination with collagen. Upon gelation of the hydrogels the constructs were placed in 2 mL of medium/scaffold. Medium change was performed every 3–4 days. For the experiments with matricellular proteins, SPARC (secreted protein acidic and rich in cysteine) (Sigma-Aldrich, Milwaukee, WI, USA), Hevin (R& D Systems, Minneapolis, MN, USA) and Thrombospondin 2 (TSP2) (Abcam, Cambridge, MA) were added to the culture media at concentrations of 50 nM, 15 nM and 4 nM, respectively.
Viability assay
Cell proliferation reagent WST-1 assay was performed at one week time point according to the instructions provided by the manufacturer. Briefly, the samples were incubated for 1hr with WST-1 reagent diluted 1:10 (v:v) in culture medium, followed by a reading of the medium absorbance with plate reader (Molecular Devices, Sunnyvale, CA, USA) at 450 and 600 nm as the reference wavelength. The fresh medium was used as a baseline control and its average absorbance value was subtracted from the value of the samples. Relative viability was calculated by dividing the values of treated samples with values of control samples. A total of N=5–6 samples/condition were used in the assay.
Glutamate assay
Glutamate assay was performed at one week time point according to the instructions provided by the manufacturer. Briefly, the supernatant medium samples were incubated for 30 minutes at 37°C with the assay buffer supplemented with developer and enzyme mix followed by a reading of the medium absorbance with plate reader at 450 nm. The fresh medium was used as a baseline control and its average absorbance value was subtracted from the value of the samples. Relative glutamate amount was calculated by dividing the values of treated samples with values of control samples. A total of N=5–6 samples/condition were used in the assay.
Immunostaining
The samples were fixed at either one week or two week time points with 4% paraformaldehyde (PFA) solution in PBS (Santa Cruz Biotechnology, Santa Cruz, CA, USA). The cells were stained with either both Polyclonal Anti-Glial Acidic Protein (GFAP) antibody produced in rabbit (Sigma-Aldrich, St. Luis, MO, USA) and Monoclonal Anti-β-Tubulin III antibody produced in mouse, clone 2G10 (Sigma-Aldrich, St. Luis, MO, USA) or single-stained with only Anti-β-Tubulin III antibody. The secondary antibodies used included Alexa 488 donkey-anti-mouse and Alexa 647 donkey-anti-rabbit (Life Technologies Corporation, Grand Island, NY, USA).
Imaging and 3D Network Analysis
The dual-stained samples of the 3D cortical brain tissue model consisting of ECM-collagen gels were imaged with a 20x (0.45 NA) objective using an inverted fluorescence microscope (Keyence, BZ-X700 series, Itasca, IL). Z-stacks were taken at 1.2 μm step and projected onto a full focus image using the BZ-analyzer software. The Alexa-488 stained samples were imaged in the axon-rich central region of the donut-shaped scaffold with a multi-photon ready confocal microscope (SP2, Leica, Wetzlar, Germany) equipped with a Ti-Sapphire laser. Images were acquired using a 20x (0.7 NA) objective at 760nm excitation and 525±25nm emission, respectively. Z-image stacks were acquired over 150–200μm with 1 μm step. To visualize the thin neurites, a resolution of 0.73 microns per pixel was used (1024x1024, 750x750μm field of view). Axonal network density analysis was performed on the z-stacks with a custom automated image analysis code. Briefly, each 2D plane within a z-stack was filtered to remove noise and cell bodies. The filtered images were then binarized to produce a neurite mask. Regions of low signal where neurite extensions were not observed were identified and excluded from the analysis by dilating the neurite mask with increasing radii until a minimal (<=5) number of connected components was achieved, giving a total area mask. The percent area covered by the neurites per 2D plane of the corresponding z-stack was determined by dividing the number of positive pixels in the neurite mask with the number of positive pixels in the total area mask. The mean percent area was finally computed over the entire z-stack for each sample resulting in average neurite network density.
Calcium signaling
Cells seeded on 3D scaffolds after one week of culture were immersed in extracellular solution: NaCl 140 mM, KCl 2.8 mM, CaCl2 2 mM, MgCl2 2 mM, HEPES 10 mM, glucose 10 mM, pH = 7.4, (all reagents from Sigma-Aldrich, St. Luis, MO, USA). The Fluo-4 (Life Technologies Corporation, Carlsbad, CA, USA) calcium sensitive dye was mixed 1:1 with 20% Pluronic F127 (Life Technologies Corporation, Carlsbad, CA, USA). Next, Fluo-4 was diluted to a final concentration 1 μM in the extracellular buffer pre-warmed to 37°C. The Fluo-4 1 μM solution was applied on the scaffold and incubated at 37°C for 1 hour. Upon incubation the constructs were washed with the extracellular buffer to remove excess dye. The constructs were imaged using an Olympus MVX10 macroscope (Olympus, Shinjuku, Tokyo, Japan) (20x magnification) and Hamamatsu ORCA-Flash4.0 camera (Hamamatsu, Hamamatsu City, Japan). The images were taken with following setup: 15 ms exposure, 50 ms frame frequency, 512x512 pixels, 4x4 binning, 1200 frames/minute at room temperature. The movie was created using ImageJ software (NIH). Maximal amplitude was calculated as fluorescence at time t (Ft) subtracted from initial fluorescence at time 0 (F): (Ft−F0).
Extracellular Field Potential Recording
After a period of incubation (2 weeks), the electrical spiking activities of the neurons in 3D scaffolds were recorded extracellularly at room temperature by sharp glass microelectrodes using an NPI Bridge Amplifier (BA-03X, Tamm, Germany). The 3D scaffolds were immersed in extracellular solution (140 mM NaCl, 2.8 mM KCl, 2 mM CaCl2, 2 mM MgCl2, 10 mM HEPES, and 10 mM D-glucose, with pH adjusted to 7.4 with NaOH). Sharp glass microelectrodes were pulled using Sutter Borosilicate Glass (BF150-86-10) with a Sutter Micropipette Puller (P-87). The microelectrodes were filled with extracellular solution and had a resistance of 60–80 Mohm. The 3D scaffolds were visualized with a Wild M3C Dissecting Microscope (Switzerland). The recording electrodes were positioned close to the cultured tissues. Fast local field potential changes (spikes) were recorded with the NPI amplifier at a bandwidth of 0.3–10 kHz and were further amplified with an A-M Systems Differential AC amplifier (Model 1700) to a combined total gain of 10,000x. The signals were digitized at 10 KHz by a Molecular Devices digitizer (Digidata 1550) using a Dell Optiplex GX620 computer with pClamp 10 software (Molecular Devices). Spikes were defined as sharp changes in local field potential in sub millisecond ranges, and were counted as events when they crossed the detection threshold set at one time of the average peak-to-peak amplitude of baseline noise. The baseline noises of the recordings were between 0.25 to 0.3 mV, and the detection thresholds were set accordingly between 0.25 to 0.3 mV. The amplitudes of the events were in a continuing scale from the detection threshold (0.25–0.3 mV) to the data acquisition’s max amplitude of 1 mV, due to our amplification factor of x10,000. Images showing the measurement setup consisting of scaffolds and electrodes were captured using an AmScope microscope digital camera (MU1000-CK).
Statistics
The graphs were prepared using GraphPad 5 software (GraphPad, CA, USA). Statistical analysis was performed with JMP12 software (SAS, Cary NC, USA). One-way analysis of variance (ANOVA) was the primary analysis method, using Dunnet’s post hoc test with the collagen group as control. The data was log transformed before the variance analysis was conducted. Levels of significance were determined at p<0.05 for the viability assay and at ***p=0.001, **p<0.005, *p<0.007 for the rest of the experiments.
RESULTS AND DISCUSSION
Extracellular matrix screening for primary rat neurons
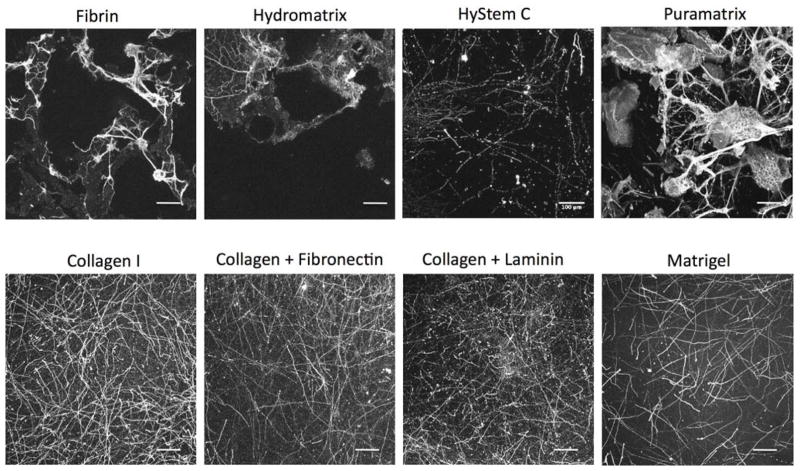
The 3D bioengineered model of cortical brain tissue, recently developed by our group, employed collagen type I as a 3D microenvironment for the growth of primary rat neuronal cells19. In order to achieve superior neuronal survival and growth, we aimed to increase the relevant ECM components related to native brain ECM, where collagen type I is not a major constituent. Thus, initially the effect of different commercially available matrices such as Matrigel, fibrin, Hydromatrix, HyStemC and Puramatrix were evaluated in terms of impact on density of neuronal networks in 3D (Fig. 1). Additionally, collagen type I combinations were tested with laminin and fibronectin, two proteins that are part of the brain ECM5. Puramatrix and Hydromatrix are both synthetic, crosslinked hydrogels consisting of three repeating amino acid sequences that have been shown to promote the proliferation of CNS cells21. Hystem C contains denatured porcine collagen and hyaluronic acid, a major part of brain ECM. However, collagen I consistently resulted in superior growth of primary rat neurons in the 3D tissue system used here (Fig. 1). The neuronal network in the axon-rich compartment of the scaffold was imaged with a confocal microscope, except for the fibrin, Puramatrix and Hydromatrix conditions, which did not allow for axonal network development due to their rapid degradation. Accordingly, the images of the stained neurons were taken within the grey matter region of the porous scaffold for the fibrin, Puramatrix and Hydromatrix conditions (Fig. 1). As evident, some of the features visible within the porous region are due to fluorescence of the silk scaffold.
Figure 1.
Extracellular matrix screening for primary rat neurons in the 3D bioengineered brain model. Different commercial hydrogels (fibrin, hydromatrix, hystemC, puramatrix, matrigel) tested in place of collagen I to fill the pores of the donut-shaped silk scaffold. Combination blends of collagen I-laminin and collagen I-fibronectin tested as well. Scale bars: 100μm
Matrigel has been used for multiple 3D in vitro brain models including the cerebral organoid model and is mainly composed of laminin and collagen IV22. However, Matrigel is not fully defined and moreover is derived from non-brain tissue source, i.e. the basement membrane of murine tumor epithelia23. In addition to the defined ECM system, such as the inclusion of single protein molecules like laminin or fibronectin to collagen type I, other undefined brain-derived ECM components may be needed to better promote neuronal network formation in vivo. As a first step towards identification of brain-specific ECM, we incorporated protein extracts from decellularized brain tissue and matricellular proteins in the silk/collagen donut-shaped model, and examined their effects on neuronal network formation and function.
Characterization of fetal and adult brain ECM
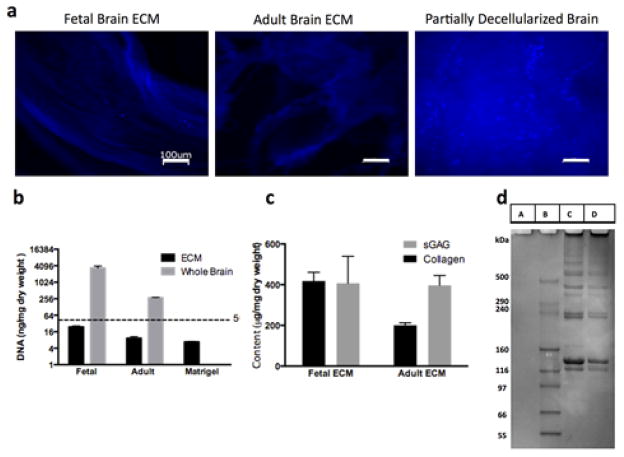
Before using the ECM in the in vitro cell culture experiments, a thorough evaluation of ECM quality in terms of residual DNA content and compositional complexity was performed. DAPI staining showed little to no presence of DNA in the completely decellularized fetal and adult porcine brain ECM in contrast to a partially decellularized brain (Fig. 2a). Further quantification of dsDNA content confirmed the sufficient decellularization of both the fetal and adult brain tissues, with less than 30 ng/mg of DNA in the lyophilized ECM, against the acceptable threshold of <50ng/mg dry weight of tissue24 (Fig. 2b). The dsDNA levels measured in fetal ECM, adult ECM and Matrigel were 25±3.8 ng/mg, 9.3±2.2 ng/mg and 6.8±0.3 ng/mg, respectively. As expected, these levels were significantly lower than those present in the adult and fetal whole brain tissues. Moreover, the DNA content of the decellularized brain tissues was comparable with the DNA content measured in a commercially available matrix, Matrigel. Interestingly, the fetal brain showed higher levels of DNA (Fig. 2b). During prenatal life, there is an overproduction of neurons, glial cells and synaptic connections in the fetal brain25. For example, >50% of neurons are lost early during the postnatal period26. The result of lower levels of DNA content measured in adult brain versus the fetal brain is consistent with the neuronal pruning during brain development.
Figure 2.
Characterization of decellularized fetal and adult brain ECM. (a) DAPI images taken with an inverted fluorescence microscope (Keyence, BZ-X700 series, Itasca, IL) indicating the effective removal of cells from the fetal and adult porcine brains. Little to no presence of cell nuclei seen in the decellularized samples in comparison to a partially decellularized sample. Scale bars: 100μm (b) DNA content in the decellularized brain tissues versus adult and fetal whole brains quantified by Pico green assay. DNA content was found to be less than 30ng/mg dry weight, close to the values found in commercially available matrigel. (c) Collagen and sGAG content of the fetal and adult porcine brain ECM. The overall collagen-sGAG content was measured to be higher for fetal ECM, although the ratio of sGAG to collagen was greater for adult brain ECM. (d) Protein gel showing A, B, C, D corresponding to Pepsin, Marker, Fetal and Adult ECM, respectively. Multiple bands in the high molecular weight range of the marker, indicative of the complexity of brain ECM with many peptides.
The collagen content for fetal and adult ECM was measured at 417.8±42.9 μg/mg and 200.9±11.9 μg/mg, respectively; whereas, the sGAG was 406.6±132.4 μg/mg and 397±48.4 μg/mg, respectively. The ratio of sGAG to collagen was higher for decellularized adult brain ECM, which is characteristic of mature brain ECM3 (Fig. 2c). However, the overall collagen and sGAG content was higher in decellularized fetal ECM (Fig. 2c). The ECM is condensed with the increasing maturity of the brain, rendering it denser than that of the fetal brain2. Also, the data demonstrates that the fetal brain may contain more equal levels of collagen and sGAG. GAG-to-collagen ratio of a tissue plays an important role in determining its swelling capability and mechanical stability27. Enhanced GAG levels render higher water absorption, while the stiff collagen confers mechanical stability27. The difference in GAG-to-collagen ratios as observed with fetal and adult decellularized brain ECM, could be exploited to create in vitro constructs with an appropriate composition for balanced water retention and stability.
The complete protein profiles of fetal and adult porcine brain ECM as tested with protein gel electrophoresis showed several bands in the high molecular weight region, >116 kDa (Fig. 2d). The presence of multiple bands for the decellularized ECM is indicative of the complex composition of the ECM. Indeed it is estimated that brain ECM is composed of many different proteins with varying forms and expression levels depending on age and brain region28. The bands near 129kDa, 139kDa and 250kDa range corresponding to the chains of collagen type I, were observed29. The intensity of these bands is seen to be higher for the fetal ECM than adult ECM, suggesting higher amount of collagen type I in the extracted fetal ECM, validating the result obtained from the Sircol soluble collagen assay. The dark band near the loaded wells and smear observed in the region >500 kDa for the fetal and adult brain ECM is expected to be a contribution of GAG-containing proteoglycans. Such proteoglycans are likely to move shorter distances through the gel due to their highly branched structures. Detailed mass spectroscopy studies can be conducted in the future to characterize the differences of fetal and adult brain ECM proteins. Thus, through the characterization of the DNA and protein content, it was confirmed that the quality of the extracted fetal and adult brain ECM was on par with commercially available matrices like Matrigel and collagen type I, with the additional benefit of compositional similarity to native brain ECM.
Concentration range for fetal and adult brain ECM
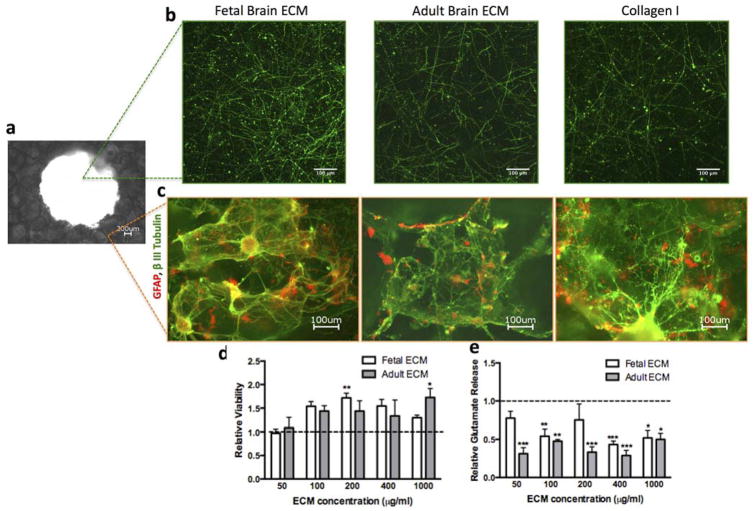
In order to determine an effective concentration of both fetal and adult brain-derived ECM for use in the 3D cultures, a range of 50–1,000 μg/ml was blended with collagen type I (3 mg/ml) hydrogels before addition to the cell-laden silk scaffolds in the 3D culture systems. Throughout this concentration range, the primary rat neurons formed healthy-looking axonal networks within the central region of the donut-shaped scaffolds (Fig. 3a, Fig. 3b). In addition, neurons formed mini-networks within the porous region of the scaffold and were found to co-exist with astrocytes as seen along the walls of the pores by one week (Fig. 3a, Fig. 3c). In our culture system, we used selective media conditions that are designed for enrichment of neurons with minimal support for non-neuronal cells such as astrocytes30. Nevertheless, a small population of astrocytes was found in the direct proximity of neuronal cell bodies. Astrocytes are known to actively participate in controlling neuronal survival and circuit formation31. Thus, this nominal presence of astrocytes in our 3D in vitro model is expected to aid in neuronal connectivity without overtaking the culture.
Figure 3.
Concentration range test for fetal and adult brain ECM. (a) Macroscopic view of the donut-shaped scaffold taken using 4x objective (b) Representative images showing the growth of primary cortical rat neurons (embryonic day 18) in 3D silk scaffold infused with collagen I gel or collagen I supplemented with fetal or adult brain ECM within the axon-rich central region of the scaffold, stained by β-III Tubulin at 1wk time point. (c) Representative images showing the growth of primary cortical rat neurons (embryonic day 18) in 3D silk scaffold infused with collagen I gel or collagen I supplemented with fetal or adult brain ECM within the donut-shaped scaffold stained by β-III Tubulin and GFAP, respectively at 1wk time point. (d) WST1 viability assay at 1wk over a concentration range of 50–1000 μg/ml ECM showed increased viability relative to collagen (set at 1); **p=0.015, *p=0.034; 4≤n≤6 (e) Glutamate release tested at 1 wk over a concentration range of 50–1000 μg/ml ECM relative to collagen showed a decreased level of glutamate released from the cells cultured in the presence of ECM; ***p<0.001, **P≤0.005, *p<0.007; 4≤n≤6
Cell viability and glutamate release relative to collagen I-based model was determined for the following concentrations of ECM: 50, 100, 200, 400 and 1,000 μg/ml. An overall increase in cell viability was observed for all concentrations of ECM, with the exception of 50 μg/ml fetal and adult ECM, which yielded similar level as collagen I alone (Fig. 3d). The ECM additive consistently lowered the level of glutamate release along the entire concentration range as compared to collagen I alone (Fig. 3e). This can be explained as a result of additional glutamate uptake by potentially more glial cells in the ECM conditions. The extracellular glutamate levels were within the tonic levels in the nM range32. Such ambient levels of extracellular glutamate are known to be controlled by mechanisms from glia32.
For further experiments, a single concentration of 1,000 μg/ml ECM was chosen for the additions of both fetal and adult ECM, since the neuronal cell viability and glutamate release outcome matched best at this concentration. These results suggest that both fetal and adult ECM significantly improve neuronal survival in a concentration-dependent manner, further supporting the benefit of improved substrates for neuronal cultures.
Growth of primary rat neurons in the presence of fetal/adult brain ECM and matricellular proteins
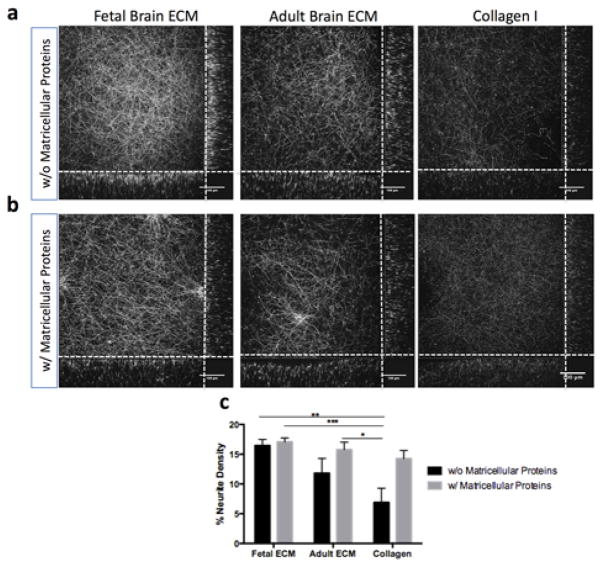
The primary rat neurons grown in the 3D bioengineered silk constructs with brain ECM-collagen type I gels showed increased viability and network formation. Therefore, further evaluation of axonal ingrowth over longer duration via analysis of axonal network density was pursued. Over two weeks, the primary neurons formed dense axonal connections in the center of the construct filled with brain ECM-collagen type I combination gels (Fig. 4a). The quantification of network density confirmed a significant improvement in network formation in the presence of fetal ECM and an adult ECM when compared to collagen I alone (Fig. 4c).
Figure 4.
Primary rat neurons in silk-collagen 3D bioengineered brain model in the presence of ECM and matricellular proteins. (a) Growth of primary cortical rat neurons (embryonic day 18) in 3D silk scaffold infused with collagen I gel or collagen I supplemented with fetal or adult brain ECM at a concentration of 1000μg/ml. Neurites stained by β-III Tubulin at 2wk time point. (b) Growth of primary cortical rat neurons in 3D silk scaffold infused with collagen I gel or collagen I supplemented with fetal or adult brain ECM at 1000μg/ml of gel. The culture media was additionally supplemented with astrocyte-released matricellular proteins (SPARC, Hevin, TSP2). Neurites stained by β-III Tubulin at 2wk time point. (c) Average neurite density per 2D plane of corresponding 3D Z-stack as quantified by using a custom image analysis code; ***p=0.001, **p<0.005, *p<0.007; n=5; collagen I as control
It is known that astrocytes secrete non-structural proteins that play important roles in regulating neuron-ECM interactions. In order to examine the astrocyte-associated microenvironment for neuronal network development, we further incorporated astrocyte-released matricellular proteins, SPARC, Hevin and TSP2 to the brain ECM containing model. The matricellular proteins further intensified the effect of brain ECM on network formation in comparison to collagen I alone (Fig. 4b,c).
The differential effects of fetal versus adult brain ECM on primary neuronal growth can be explained by the changes in composition of brain ECM during development. In rodents, a loose embryonic matrix is first deposited in the central nervous system6. This matrix is composed of mainly hyaluronan, neurocan, versican V1, versican V0, tenascin-C and link proteins HAPLN1/Crtl16,25. These protein levels peak during embryonic development and eventually are down-regulated or replaced by different homologous proteins like versican V2, aggrecan, brevican, phosphacan, tenascin-R and link proteins HAPLN2/Bral1 and HAPLN4/Bral2, which form a significantly firmer mature ECM soon after the postnatal period6,25. Some components of the juvenile matrix are maintained at similar levels during adulthood, mostly in the neurogenetically active areas of the brain6, and these are the constituents of greatest interest from the perspective of culturing embryonic neuronal cells in vitro. For instance, tenascin-C is an ECM molecule highly expressed during early developmental stages and later only localized in brain regions with neurogenesis capability, where it plays an important role in modulating cell migration and proliferation33. Interestingly, tenascin-C falls into the category of matricellular proteins34. This presumably explains why the addition of similar matricellular proteins: SPARC, Hevin, and TSP2 helped to maintain healthy cultures of embryonic primary neurons in vitro and substantiated the effect of decellularized brain ECM over longer durations35,36. However, the matricellular proteins had a more prominent effect on the adult ECM and collagen I conditions. It was evident that the exclusive addition of fetal ECM provided the most suitable microenvironment for maintaining healthy 3D cultures of embryonic neuronal cells. These observations suggest that the fetal ECM likely retained proteins that modulate neuron-ECM interactions.
Functional evaluation of the constructs with fetal/adult brain ECM
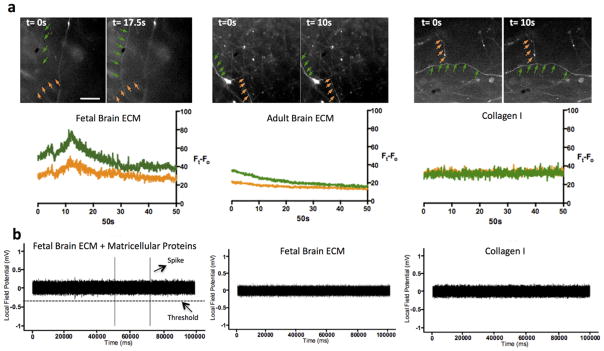
The neuronal function of the 3D tissue models was probed by live calcium imaging and extracellular recordings for single cell-based and network-level activities, respectively. For calcium imaging, 1-week old 3D cultures were stained with calcium indicator, fluo-4 and subjected to time-lapse fluorescence imaging over 1 min at a rate of 50ms/frame. The change in fluorescence intensity over time, indicative of the transient rise in intracellular calcium levels, was recorded as a measure of electrical activity of the neurons37,38. Among the samples tested (2/group, total n=6), only the fetal brain ECM-containing samples showed noticeable spontaneous fluo-4 intensity spikes during all calcium imaging sessions; compared to negligible changes with the adult ECM and collagen-I only samples. Fig. 5a shows representative fluorescence images and the corresponding intensity traces.
Figure 5.
Functional evaluation of the constructs containing brain ECM. (a) Calcium imaging on 1wk long 3D constructs supplemented with fetal and adult ECM in comparison to collagen gel only. Only the fetal ECM condition recorded a spike in fluorescence intensity of Ca dye indicative of neuronal firing. Scale bar is 200 μm and it is the same for all images. (b) Local field potential recording (LFP) of 2wk long 3D cultures containing collagen gel supplemented with fetal and adult brain ECM in comparison to constructs with collagen gel alone. A set of samples additionally supplemented with matricellular proteins (SPARC, Hevin, TSP2) were also tested. Representative trace of the fetal ECM condition along with additional matricellular proteins evoked spontaneous spiking activity as recorded by LFP measurements
To detect neuronal network activities, we used extracellular local field potential (LFP) recording. 2-week old 3D cultures were recorded with a sharp glass microelectrode inserted into the ‘grey matter’ area of the construct. Spontaneous spiking activity, defined as a sub-millisecond voltage change detected at the threshold set at one-fold of baseline noise, was recorded in the samples containing both fetal ECM and matricellular proteins. Interestingly, at present we did not detect spontaneous firing with the fetal ECM-containing samples that were not supplemented with matricellular proteins (Fig. 5b). Also, no spontaneous spiking was observed in any of the adult ECM or control collagen I samples either with or without matricellular proteins (n=6). Thus, a combination of the fetal ECM and matricellular proteins seemed to be necessary to obtain network level activity, whereas inclusion of the fetal ECM was sufficient for generating cell-based calcium signaling.
Considering that in vitro cultured neurons are typically quiescent due to the deprivation of sensory input, the low probability of spontaneous firing, either at single-cell level or network-level, is not surprising. It remains challenging to capture and quantify these rare events in in vitro systems during the homeostatic state, i.e., un-provoked by external stimuli. Nevertheless, even with a small sample size, both calcium imaging and LFP recording have captured spontaneous neuronal spiking in 3D cultures containing fetal brain ECM. These results indicate a potentially more functional and excitable states with this culture system in the presence of fetal brain ECM and the matricellular proteins. The functional evaluation also corroborates the enhanced neuronal network density produced by cortical neurons when cultured in the presence of fetal/adult brain ECM and matricellular proteins.
CONCLUSIONS
Decellularized fetal and adult porcine brain ECM was investigated as a potential matrix for the growth of primary neuronal cells in 3D bioengineered tissue models of cortical brain tissue. The extracted ECM had minimal residual cellular material and retained to a great extent the composition of the native brain ECM, including constituents like sGAG and collagens. A similar extraction protocol in terms of the decellularization agents was followed for both the fetal and adult porcine brains, in order to provide direct comparisons. An overall greater content of sGAG and collagen was observed in fetal ECM in comparison to adult ECM. This is an important finding, since commercially available matrices commonly used for neuronal culture do not contain GAGs, a major constituent of the native brain ECM. The inclusion of brain ECM over a range of concentrations allowed for extensive axonal ingrowth and greater viability of primary neurons in the 3D tissue cultures as opposed to a single component collagen type I matrix. Most importantly, the 3D tissue constructs that incorporated fetal ECM resulted in robust neuronal cultures with dense axonal networks, characterized by spontaneous spiking activity. This study provides a foundation for further investigation of fetal brain-derived ECM and its potential as a material of choice in other 2D and 3D neuronal culture systems.
Acknowledgments
This work was funded by a US National Institutes of Health (NIH) P41 Tissue Engineering Resource Center Grant (EB002520) and the NIH R01 (NS092847). K.C. was supported with Postdoctoral Fellowship from German Research Foundation (DFG) (CH 1400/2-1). The schematic design was brought to completion with the help of Kunal Chawla. We also thank Nina Dinjaski, Olena Tokareva, Benjamin Partlow, Dana Cairns, Corin Williams and Kelly E. Sullivan for helpful discussions and protocols on gel electrophoresis. Thank you also to Karen Christman for early advise on methods.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website.
ACS Publications website at DOI:
Calcium signaling in primary neuron 3D cultures incorporated with fetal brain ECM (.mov). For visualization purposes, the data was 3x time compressed and the video is shown at 50 frames/sec. The field of view corresponds to 875μm by 875μm.
References
- 1.Dityatev A, Seidenbecher CI, Schachner M. Compartmentalization from the outside: the extracellular matrix and functional microdomains in the brain. Trends Neurosci. 2010;33(11):503–12. doi: 10.1016/j.tins.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 2.Rauch U. Brain matrix: structure, turnover and necessity. Biochem Soc Trans. 2007;35(Pt 4):656–60. doi: 10.1042/BST0350656. [DOI] [PubMed] [Google Scholar]
- 3.Ruoslahti E. Brain extracellular matrix. Glycobiology. 1996;6(5):489–92. doi: 10.1093/glycob/6.5.489. [DOI] [PubMed] [Google Scholar]
- 4.Yang YL, Sun C, Wilhelm ME, Fox LJ, Zhu J, Kaufman LJ. Influence of chondroitin sulfate and hyaluronic acid on structure, mechanical properties, and glioma invasion of collagen I gels. Biomaterials. 2011;32(31):7932–40. doi: 10.1016/j.biomaterials.2011.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bandtlow CE, Zimmermann DR. Proteoglycans in the developing brain: new conceptual insights for old proteins. Physiol Rev. 2000;80(4):1267–90. doi: 10.1152/physrev.2000.80.4.1267. [DOI] [PubMed] [Google Scholar]
- 6.Zimmermann DR, Dours-Zimmermann MT. Extracellular matrix of the central nervous system: from neglect to challenge. Histochem Cell Biol. 2008;130(4):635–53. doi: 10.1007/s00418-008-0485-9. [DOI] [PubMed] [Google Scholar]
- 7.Kucukdereli H, Allen NJ, Lee AT, Feng A, Ozlu MI, Conatser LM, Chakraborty C, Workman G, Weaver M, Sage EH, Barres BA, Eroglu C. Control of excitatory CNS synaptogenesis by astrocyte-secreted proteins Hevin and SPARC. Proc Natl Acad Sci U S A. 2011;108(32):E440–9. doi: 10.1073/pnas.1104977108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guvendiren M, Burdick JA. Engineering synthetic hydrogel microenvironments to instruct stem cells. Curr Opin Biotechnol. 2013;24(5):841–6. doi: 10.1016/j.copbio.2013.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ott HC, Matthiesen TS, Goh SK, Black LD, Kren SM, Netoff TI, Taylor DA. Perfusion-decellularized matrix: using nature’s platform to engineer a bioartificial heart. Nat Med. 2008;14(2):213–21. doi: 10.1038/nm1684. [DOI] [PubMed] [Google Scholar]
- 10.Brown BN, Badylak SF. Extracellular matrix as an inductive scaffold for functional tissue reconstruction. Transl Res. 2014;163(4):268–85. doi: 10.1016/j.trsl.2013.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crapo PM, Gilbert TW, Badylak SF. An overview of tissue and whole organ decellularization processes. Biomaterials. 2011;32(12):3233–43. doi: 10.1016/j.biomaterials.2011.01.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Waele J, Reekmans K, Daans J, Goossens H, Berneman Z, Ponsaerts P. 3D culture of murine neural stem cells on decellularized mouse brain sections. Biomaterials. 2015;41:122–31. doi: 10.1016/j.biomaterials.2014.11.025. [DOI] [PubMed] [Google Scholar]
- 13.DeQuach JA, Mezzano V, Miglani A, Lange S, Keller GM, Sheikh F, Christman KL. Simple and high yielding method for preparing tissue specific extracellular matrix coatings for cell culture. PLoS One. 2010;5(9):e13039. doi: 10.1371/journal.pone.0013039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Medberry CJ, Crapo PM, Siu BF, Carruthers CA, Wolf MT, Nagarkar SP, Agrawal V, Jones KE, Kelly J, Johnson SA, Velankar SS, Watkins SC, Modo M, Badylak SF. Hydrogels derived from central nervous system extracellular matrix. Biomaterials. 2013;34(4):1033–1040. doi: 10.1016/j.biomaterials.2012.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeQuach JA, Yuan SH, Goldstein LS, Christman KL. Decellularized porcine brain matrix for cell culture and tissue engineering scaffolds. Tissue Eng Part A. 2011;17(21–22):2583–92. doi: 10.1089/ten.tea.2010.0724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crapo PM, Medberry CJ, Reing JE, Tottey S, van der Merwe Y, Jones KE, Badylak SF. Biologic scaffolds composed of central nervous system extracellular matrix. Biomaterials. 2012;33(13):3539–3547. doi: 10.1016/j.biomaterials.2012.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baiguera S, Del Gaudio C, Lucatelli E, Kuevda E, Boieri M, Mazzanti B, Bianco A, Macchiarini P. Electrospun gelatin scaffolds incorporating rat decellularized brain extracellular matrix for neural tissue engineering. Biomaterials. 2014;35(4):1205–14. doi: 10.1016/j.biomaterials.2013.10.060. [DOI] [PubMed] [Google Scholar]
- 18.Williams C, Quinn KP, Georgakoudi I, Black LD. 3rd, Young developmental age cardiac extracellular matrix promotes the expansion of neonatal cardiomyocytes in vitro. Acta Biomater. 2014;10(1):194–204. doi: 10.1016/j.actbio.2013.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang-Schomer MD, White JD, Tien LW, Schmitt LI, Valentin TM, Graziano DJ, Hopkins AM, Omenetto FG, Haydon PG, Kaplan DL. Bioengineered functional brain-like cortical tissue. Proc Natl Acad Sci U S A. 2014;111(38):13811–6. doi: 10.1073/pnas.1324214111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chwalek K, Tang-Schomer MD, Omenetto FG, Kaplan DL. In vitro bioengineered model of cortical brain tissue. Nat Protoc. 2015;10(9):1362–73. doi: 10.1038/nprot.2015.091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thonhoff JR, Lou DI, Jordan PM, Zhao X, Wu P. Compatibility of human fetal neural stem cells with hydrogel biomaterials in vitro. Brain Res. 2008;1187:42–51. doi: 10.1016/j.brainres.2007.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lancaster MA, Renner M, Martin CA, Wenzel D, Bicknell LS, Hurles ME, Homfray T, Penninger JM, Jackson AP, Knoblich JA. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501(7467):373–9. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wong AP, Perez-Castillejos R, Christopher Love J, Whitesides GM. Partitioning microfluidic channels with hydrogel to construct tunable 3-D cellular microenvironments. Biomaterials. 2008;29(12):1853–61. doi: 10.1016/j.biomaterials.2007.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crapo PM, Medberry CJ, Reing JE, Tottey S, van der Merwe Y, Jones KE, Badylak SF. Biologic scaffolds composed of central nervous system extracellular matrix. Biomaterials. 2012;33(13):3539–47. doi: 10.1016/j.biomaterials.2012.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stiles J, Jernigan TL. The basics of brain development. Neuropsychol Rev. 2010;20(4):327–48. doi: 10.1007/s11065-010-9148-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mazarakis ND, Edwards AD, Mehmet H. Apoptosis in neural development and disease. Arch Dis Child Fetal Neonatal Ed. 1997;77(3):F165–70. doi: 10.1136/fn.77.3.f165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Riemer T, Nimptsch A, Nimptsch K, Schiller J. Determination of the glycosaminoglycan and collagen contents in tissue samples by high-resolution 1H NMR spectroscopy after DCl-induced hydrolysis. Biomacromolecules. 2012;13(7):2110–7. doi: 10.1021/bm300485j. [DOI] [PubMed] [Google Scholar]
- 28.Reichardt LF, Tomaselli KJ. Extracellular matrix molecules and their receptors: functions in neural development. Annu Rev Neurosci. 1991;14:531–70. doi: 10.1146/annurev.ne.14.030191.002531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rabotyagova OS, Cebe P, Kaplan DL. Collagen Structural Hierarchy and Susceptibility to Degradation by Ultraviolet Radiation. Mater Sci Eng C Mater Biol Appl. 2008;28(8):1420–1429. doi: 10.1016/j.msec.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brewer GJ. Isolation and culture of adult rat hippocampal neurons. J Neurosci Methods. 1997;71(2):143–55. doi: 10.1016/s0165-0270(96)00136-7. [DOI] [PubMed] [Google Scholar]
- 31.Corty MM, Freeman MR. Cell biology in neuroscience: Architects in neural circuit design: glia control neuron numbers and connectivity. J Cell Biol. 2013;203(3):395–405. doi: 10.1083/jcb.201306099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Herman MA, Jahr CE. Extracellular glutamate concentration in hippocampal slice. J Neurosci. 2007;27(36):9736–41. doi: 10.1523/JNEUROSCI.3009-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Midwood KS, Orend G. The role of tenascin-C in tissue injury and tumorigenesis. J Cell Commun Signal. 2009;3(3–4):287–310. doi: 10.1007/s12079-009-0075-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jones EV, Bouvier DS. Astrocyte-secreted matricellular proteins in CNS remodelling during development and disease. Neural Plast. 2014;2014:321209. doi: 10.1155/2014/321209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bradshaw AD. The role of SPARC in extracellular matrix assembly. J Cell Commun Signal. 2009;3(3–4):239–46. doi: 10.1007/s12079-009-0062-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tan K, Lawler J. The interaction of Thrombospondins with extracellular matrix proteins. J Cell Commun Signal. 2009;3(3–4):177–87. doi: 10.1007/s12079-009-0074-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eiraku M, Watanabe K, Matsuo-Takasaki M, Kawada M, Yonemura S, Matsumura M, Wataya T, Nishiyama A, Muguruma K, Sasai Y. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell. 2008;3(5):519–32. doi: 10.1016/j.stem.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 38.Grienberger C, Konnerth A. Imaging calcium in neurons. Neuron. 2012;73(5):862–85. doi: 10.1016/j.neuron.2012.02.011. [DOI] [PubMed] [Google Scholar]