Abstract
Data commons collocate data, storage, and computing infrastructure with core services and commonly used tools and applications for managing, analyzing, and sharing data to create an interoperable resource for the research community. An architecture for data commons is described, as well as some lessons learned from operating several large-scale data commons.
With the amount of available scientific data being far larger than the ability of the research community to analyze it, there’s a critical need for new algorithms, software applications, software services, and cyberinfrastructure to support data throughout its life cycle in data science. In this article, we make a case for the role of data commons in meeting this need. We describe the design and architecture of several data commons that we’ve developed and operated for the research community in conjunction with the Open Science Data Cloud (OSDC), a multipetabyte science cloud that the nonprofit Open Commons Consortium (OCC) has managed and operated since 2009.1 One of the distinguishing characteristics of the OSDC is that it interoperates with a data commons containing over 1 Pbyte of public research data through a service-based architecture. This is an example of what is sometimes called “data as a service,” which plays an important role in some science-as-a-service frameworks.
There are at least two definitions for science as a service. The first is analogous to the software-as-a-service2 model, in which instead of managing data and software locally using your own storage and computing resources, you use the storage, computing, and software services offered by a service provider, such as a cloud service provider (CSP). With this approach, instead of setting up his or her own storage and computing infrastructure and installing the required software, a scientist uploads data to a CSP and uses preinstalled software for data analysis. Note that a trained scientist is still required to run the software and analyze the data. Science as a service can also refer more generally to a service model that relaxes the requirement of needing a trained scientist to process and analyze data. With this service model, specific software and analysis tools are available for specific types of scientific data, which is uploaded to the science-as-a-service provider, processed using the appropriate pipelines, and then made available to the researcher for further analysis if required. Obviously these two definitions are closely connected in that a scientist can set up the required science-as-a-service framework, as in the first definition, so that less-trained technicians can use the service to process their research data, as in the second definition. By and large, we focus on the first definition in this article.
There are various science-as-a-service frameworks, including variants of the types of clouds formalized by the US National Institute of Standards and Technology (infrastructure as a service, platform as a service, and software as a service),2 as well as some more specialized services that are relevant for data science (data science support services and data commons):
data science infrastructure and platform services, in which virtual machines (VMs), containers, or platform environments containing commonly used applications, tools, services, and datasets are made available to researchers (the OSDC is an example);
data science software as a service, in which data is uploaded and processed by one or more applications or pipelines and results are stored in the cloud or downloaded (general-purpose platforms offering data science as a service include Agave,3 as well as more specialized services, such as those designed to process genomics data);
data science support services, including data storage services, data-sharing services, data transfer services, and data collaboration services (one example is Globus4); and
data commons, in which data, data science computing infrastructure, data science support services, and data science applications are collocated and available to researchers.
Data Commons
When we write of a “data commons,” we mean cyberinfrastructure that collocates data, storage, and computing infrastructure with commonly used tools for analyzing and sharing data to create an interoperable resource for the research community.
In the discussion below, we distinguish among several stakeholders involved in data commons: the data commons service provider (DCSP), which is the entity operating the data commons; the data contributor (DC), which is the organization or individual providing the data to the DCSP; and the data user (DU), which is the organization or individual accessing the data. (Note that there’s often a fourth stakeholder: the DCSP associated with the researcher accessing the data.) In general, there will be an agreement, often called the data contributors agreement (DCA), governing the terms by which the data is managed by the DCSP and the researchers accessing the data, as well as a second agreement, often called the data access agreement (DAA), governing the terms of any researcher who accesses the data.
As we describe in more detail later, we’ve built several data commons since 2009. Based on this experience, we’ve identified six main requirements that, if followed, would enable data commons to interoperate with each other, science clouds,1 and other cyberinfrastructure supporting science as a service:
Requirement 1, permanent digital IDs. The data commons must have a digital ID service, and datasets in the data commons must have permanent, persistent digital IDs. Associated with digital IDs are access controls specifying who can access the data and metadata specifying additional information about the data. Part of this requirement is that data can be accessed from the data commons through an API by specifying its digital ID.
Requirement 2, permanent metadata. There must be a metadata service that returns the associated metadata for each digital ID. Because the metadata can be indexed, this provides a basic mechanism for the data to be discoverable.
Requirement 3, API-based access. Data must be accessed by an API, not just by browsing through a portal. Part of this requirement is that a metadata service can be queried to return a list of digital IDs that can then be retrieved via the API. For those data commons that contain controlled access data, another component of the requirement is that there’s an authentication and authorization service so that users can first be authenticated and the data commons can check whether they are authorized to have access to the data.
Requirement 4, data portability. The data must be portable in the sense that a dataset in a data commons can be transported to another data commons and be hosted there. In general, if data access is through digital IDs (versus referencing the data’s physical location), then software that references data shouldn’t have to be changed when data is rehosted by a second data commons.
Requirement 5, data peering. By “data peering,” we mean an agreement between two data commons service providers to transfer data at no cost so that a researcher at data commons 1 can access data commons 2. In other words, the two data commons agree to transport research data between them with no access charges, no egress charges, and no ingress charges.
Requirement 6, pay for compute. Because, in practice, researchers’ demand for computing resources is larger than available computing resources, computing resources must be rationed, either through allocations or by charging for their use. Notice the asymmetry in how a data commons treats storage and computing infrastructure. When data is accepted into a data commons, there’s a commitment to store and make it available for a certain period of time, often indefinitely. In contrast, computing over data in a data commons is rationed in an ongoing fashion, as is the working storage and the storage required for derived data products, either by providing computing and storage allocations for this purpose or by charging for them. For simplicity, we refer to this requirement as “pay for computing,” even though the model is more complicated than that.
Although very important for many applications, we view other services, such as those for providing data provenance,5 data replication,6 and data collaboration,7 as optional and not core services.
OSDC and OCC Data Commons
The OSDC is a multipetabyte science cloud that serves the research community by collocating a multidisciplinary data commons containing approximately 1 Pbyte of scientific data with cloud-based computing, high-performance data transport services, and VM images and shareable snapshots containing common data analysis pipelines and tools.
The OSDC is designed to provide a long-term persistent home for scientific data, as well as a platform for data-intensive science, allowing new types of data-intensive algorithms to be developed, tested, and used over large sets of heterogeneous scientific data. Recently, OSDC researchers have logged about two million core hours each month, which translates to more than US$800,000 worth of cloud computing services (if purchased through Amazon Web Services’ public cloud). This equates to more than 12,000 core hours per user, or a 16-core machine continuously used by each researcher on average.
OSDC researchers used a total of more than 18 million core hours in 2015. We currently target operating OSDC computing resources at approximately 85 percent of capacity, and storage resources at 80 percent of capacity. Given these constraints, we can determine how many researchers to support and what size allocations to provide them. Because the OSDC specializes in supporting data-intensive research projects, we’ve chosen to target researchers who need larger-scale resources (relative to our total capacity) for data-intensive science. In other words, rather than support more researchers with smaller allocations, we support fewer researchers with larger allocations. Table 1 shows the number of times researchers exceeded the indicated number of core hours in a single month during 2015.
Table 1.
Data-intensive users supported by the Open Science Data Cloud.
| No. core hours per month | No. users |
|---|---|
| 20,000 | 120 |
| 50,000 | 34 |
| 100,000 | 23 |
| 200,000 | 5 |
The OSDC Community
The OSDC is developed and operated by the Open Commons Consortium, a nonprofit that supports the scientific community by operating data commons and cloud computing infrastructure to support scientific, environmental, medical, and healthcare-related research. OCC members and partners include universities (University of Chicago, Northwestern University, University of Michigan), companies (Yahoo, Cisco, Infoblox), US government agencies and national laboratories (NASA, NOAA), and international partners (Edinburgh University, University of Amsterdam, Japan’s National Institute of Advanced Industrial Science and Technology). The OSDC is a joint project with the University of Chicago, which provides the OSDC’s datacenter. Much of the support for the OSDC came from the Moore Foundation and from corporate donations.
The OSDC has a wide-reaching, multicampus, multi-institutional, interdisciplinary user base and has supported more than 760 research projects since its inception. In 2015, 470 research groups from 54 universities in 14 countries received OSDC allocations. In a typical month (November 2015), 186 of these research groups were active. The most computational-intensive group projects in 2015 included projects around biological sciences and genomics research, analysis of Earth science satellite imagery data, analysis of text data in historical and scientific literature, and a computationally intensive project in sociology.
OCC Data Commons
The OCC operates several data commons for the research community.
OSDC data commons
We introduced our first data commons in 2009. It currently holds approximately 800 Tbytes of public open access research data, including Earth science data, biological data, social science data, and digital humanities data.
Matsu data commons
The OCC has collaborated with NASA since 2009 on Project Matsu, a data commons that contains six years of Earth Observing-1 (EO-1) data, with new data added daily, as well as selected datasets from other NASA satellites, including NA-SA’s Moderate Resolution Imaging Spectrometer (MODIS) and the Landsat Global Land Surveys.
The OCC NOAA data commons
In April 2015, NOAA announced five data alliance partnerships (with Amazon, Google, IBM, Microsoft, and the OCC) that would have broad access to its data and help make it more accessible to the public. Currently, only a small fraction of the more than 20 of data that NOAA has available in its archives is available to the public, but NOAA data alliance partners have broader access to it. The focus of the OCC data alliance is to work with the environmental research community to build an environmental data commons. Currently, the OCC NOAA data commons contains Nexrad data, with additional datasets expected in 2016.
National Cancer Institute’s (NCI’s) genomic data commons (GDC)
Through a contract between the NCI and the University of Chicago and in collaboration with the OCC, we’ve developed a data commons for cancer data; the GDC contains genomic data and associated clinical data from NCI-funded projects. Currently, the GDC contains about 2 Pbytes of data, but this is expected to grow rapidly over the next few years.
Bionimbus protected data cloud
We also operate two private cloud computing platforms that are designed to hold human genomic and other sensitive biomedical data. These two clouds contain a variety of sensitive controlled-access biomedical data that we make available to the research community following the requirements of the relevant data access committees.
Common software stack
The core software stack for the various data commons and clouds described here is open source. Many of the components are developed by third parties, but some key services are developed and maintained by the OCC and other working groups. Although there are some differences between them, we try to minimize the differences between the software stacks used by the various data commons that we operate. In practice, as we develop new versions of the basic software stack, it usually takes a year or so until the changes can percolate throughout our entire infrastructure.
OSDC Design and Architecture
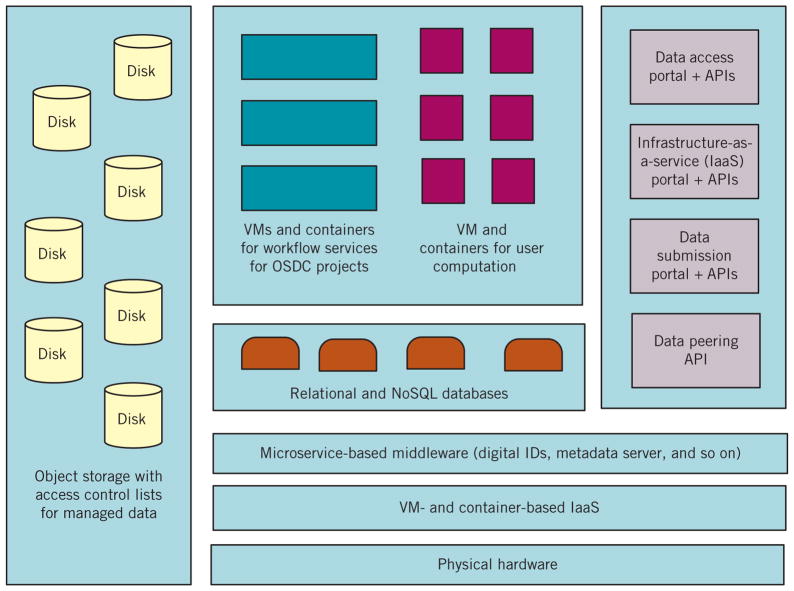
Figure 1 shows the OSDC’s architecture. We are currently transitioning from version 2 of the OSDC software stack1 to version 3. Both are based on OpenStack8 for infrastructure as a service. The primary change made between version 2 and version 3 is that version 2 uses GlusterFS9 for storage, while version 3 uses Ceph10 for object storage in addition to OpenStack’s ephemeral storage. This is a significant user-facing change that comes with some tradeoffs. Version 2 utilized a POSIX-compliant file system for user home directory (scratch and persistent) data storage, which provides command-line utilities familiar for most OSDC users. Version 3’s object storage, however, provides the advantage of an increased level of interoperability, as Ceph’s object storage has an interface compatible with a large subset of Amazon’s S3 RESTful API in addition to OpenStack’s API.
Figure 1.
The Open Science Data Cloud (OSDC) architecture. The various data commons that we have developed and operate share an architecture, consisting of object-based storage, virtual machines (VMs), and containers for on-demand computing, and core services for digital IDs, metadata, data access, and access to computing resources, all of which are available through RESTful APIs. The data access and data submission portals are applications built using these APIs.
In version 3, there’s thus a clearer distinction between the way users interface with scratch data and intermediate working results on ephemeral storage, which is simple to use and persists only until VMs are terminated. This results in longer-term data on object storage, which requires the small extra effort of curating through the API interface. Although there’s a learning curve required in adopting object storage, we’ve noticed that it’s small and easily overcome with examples in documentation. It also tempers increased storage usage that could stem from unnecessary data that isn’t actively removed.
The OSDC has a portal called the Tukey portal, which provides a front-end Web portal interface for users to access, launch, and manage VMs and storage. The Tukey portal interfaces with the Tukey middleware, which provides a secure authentication layer and interface between various software stacks. The OSDC uses federated login for authentication so that academic institutions with InCommon, CANARIE, or the UK Federation can use those credentials. We’ve worked with 145 academic universities and research institutions to release the appropriate attributes for authentication. We also support Gmail and Yahoo logins, but only for approved projects when other authentication options aren’t available.
We instrument all the resources that we operate so that we can meter and collect the data required for accounting and billing each user. We use Salesforce.com, one of the components of the OSDC that isn’t open source, to send out invoices. Even when computing resources are allocated and no payment is required, we’ve found that receipt of these invoices promotes responsible usage of OSDC community resources. We also operate an interactive support ticketing system that tracks user support requests and system team responses for technical questions. Collecting this data lets us track usage statistics and build a comprehensive assessment of how researchers use our services.
While adding to our resources, we’ve developed an infrastructure automation tool called Yates to simplify bringing up new computing, storage, and networking infrastructure. We also try to automate as much of the security required to operate the OSDC as is practical.
The core OSDC software stack is open source, enabling interested parties to set up their own science cloud or data commons. The core software stack consists of third-party, open source software, such as OpenStack and Ceph, as well as open source software developed by the OSDC community. The latter is licensed under the open source Apache license. The OSDC does use some proprietary software, such as Salesforce.com to do the accounting and billing, as mentioned earlier.
OCC Digital ID and Metadata Services
The digital ID (DID) service is accessible via an API that generates digital IDs, assigns key-value attributes to digital IDs, and returns key-value attributes associated with digital IDs. We also developed a metadata service that’s accessible via an API and can assign and retrieve metadata associated with a digital ID. Users can also edit metadata associated with digital IDs if they have write access to it. Due to different release schedules, there are some differences in the digital ID and metadata services between several of the data commons that we operate, but over time, we plan to converge these services.
Persistent Identifier Strategies
Although the necessity of assigning digital IDs to data is well recognized,11,12 there isn’t yet a widely accepted service for this purpose, especially for large datasets.13 This is in contrast to the generally accepted use of digital object identifiers (DOIs) or handles for referencing digital publications. An alternative to a DOI is an archival resource key (ARK), a Uniform Resource Locator (URL) that’s also a multipurpose identifier for information objects of any type.14,15 In practice, DOIs and ARKs are generally used to assign IDs to datasets, with individual communities sometimes developing their own IDs. DataCite is an international consortium that manages DOIs for datasets and supports services for finding, accessing, and reusing data.16 There are also services such as EZID that support both DOIs and ARKs.17
Given the challenges the community is facing in coming to a consensus about which digital IDs to use, our approach has been to build an open source digital ID service that can support multiple digital IDs, support “suffix pass-through,”13 and that can scale to large datasets.
Digital IDs
From the researcher viewpoint, the need for digital IDs associated with datasets is well appreciated.18,19 Here, we discuss some of the reasons that digital IDs are important for a data commons from an operational viewpoint.
First, with digital IDs, data can be moved from one physical location or storage system within a data commons to another without the need to change any code that references the data. As the amount of data grows, moving data between zones within a data commons or between storage systems becomes more and more common, and digital IDs allow this to take place without impeding researchers.
Second, digital IDs are an important component of the data portability requirement. More specifically, datasets can be moved between data commons, and, again, researchers don’t need to change their code. In practice, datasets can be migrated over time, with the digital IDs’ references updated as the migration proceeds.
Signpost is the digital ID service for the OSDC. Instead of using a hard-coded URL, the primary way to access managed data via the OSDC is through a digital ID. Signpost is an implementation of this concept via JavaScript Object Notation (JSON) documents.
The Signpost digital ID service integrates a mutable ID that’s assigned to the data with an immutable hash-based ID that’s computed from the data. Both IDs are accessible through a REST API interface. With this approach, data contributors can make updates to the data and retain the same ID, while the data commons service provider can use the hash-based ID to facilitate data management. To prevent unauthorized editing of digital IDs, an access control list (ACL) is kept by each digital ID specifying the read/write permissions for different users and groups.
User-defined identities are flexible, can be of any format (including ARKs and DOIs), and provide a layer of human readability. They map to hashes of the identified data objects, with the bottom layer utilizing hash-based identifiers, which guarantee data immutability, allow for identification of duplicated data via hash collisions, and allow for verification upon retrieval. These map to known locations of the identified data.
Metadata Service
The OSDC metadata service, Sightseer, lets users create, modify, and access searchable JSON documents containing metadata about digital IDs. The primary data can be accessed using Signpost and the digital ID. At its core, Sightseer provides no restrictions on the JSON documents it can store. However, it has the ability to specify metadata types and associate them with JSON schemas. This helps prevent unexpected errors in metadata with defined schemas. Sightseer has similar abilities as Signpost to provide ACLs to specify users that have write/read access to the specific JSON document.
Case Studies
Two case studies illustrate some of the projects that can be supported with data commons.
Matsu
Project Matsu is a collaboration between NASA and the OCC that’s hosted by the University of Chicago, processes the data produced each day by NASA’s EO-1 satellite, and makes a variety of data products available to the research community, including flood maps. The raw data, processed data, and data products are all available through the OSDC. Project Matsu uses a framework called the OSDC Wheel to ingest raw data, process and analyze it, and deliver reports with actionable information to the community in near real time.20 Project Matsu uses the data commons architecture illustrated in Figure 1.
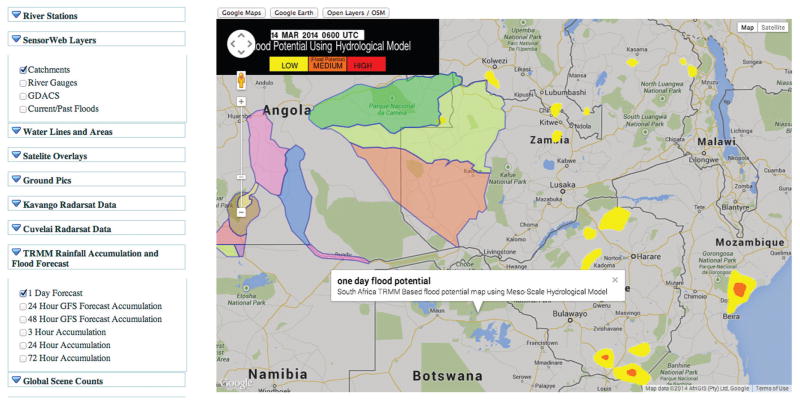
As part of Project Matsu, we host several focused analytic products with value-added data. Figure 2 shows a screenshot from one of these focused analytic products, the Project Matsu Namibia Flood Dashboard,20 which was developed as a tool for aggregating and rapidly presenting data and sources of information about ground conditions, rainfall, and other hydrological information to citizens and decision makers in the flood-prone areas of water basins in Namibia and the surrounding areas. The tool features a bulletin system that produces a short daily written report, a geospatial data visualization display using Google Maps/Earth and OpenStreetMap, methods for retrieving NASA images for a region of interest, and analytics for projecting flood potential using hydrological models. The Namibia Flood Dashboard is an important tool for developing better situational awareness and enabling fast decision making and is a model for the types of focused analytics products made possible by collocating related datasets with each other and with computational and analytic capabilities.
Figure 2.
A screenshot of part of the Namibia Flood Dashboard from 14 March 2014. This image shows water catchments (outlined and colored regions) and a one-day flood potential forecast of the area from hydrological models using data from the Tropical Rainfall Measuring Mission (TRMM), a joint space mission between NASA and the Japan Aerospace Exploration Agency.
Bionimbus
The Bionimbus Protected Data Cloud21 is a petabyte-scale private cloud and data commons that has been operational since 13 March 2013. Since going online in 2013, it has supported more than 152 allocation recipients from over 35 different projects at 29 different institutions. Each month, Bionimbus provides more than 2.5 million core hours to researchers, which at standard Amazon AWS pricing would cost over $500,000. One of the largest users of Bionimbus is the Cancer Genome Atlas (TCGA)/International Cancer Genome Consortium (ICGC) PanCancer Analysis of Whole Genomes working group (PCAWG). PCAWG is currently undertaking a large-scale analysis of most of the world’s whole genome cancer data available to the cancer community through the TCGA and ICGA consortia using several clouds, including Bionimbus.
Bionimbus also uses the data commons architecture illustrated in Figure 1. More specifically, the current architecture uses OpenStack to provide virtualized infrastructure, containers to provide a platform-as-a-service capability, and object-based storage with an AWS compatible interface. Bionimbus is a National Institutes of Health (NIH) Trusted Partner22 that interoperates with both the NIH Electronic Research Administration Commons to authenticate researchers and with the NIH Database of Genotypes and Phenotypes system to authorize users access to specific controlled access datasets, such as the TCGA dataset.
Discussion
Three projects that are supporting infrastructures similar to the OCC data commons are described in the sidebar. With the appropriate services, data commons support three different but related functions. First, data commons can serve as a data repository or digital library for data associated with published research. Second, data commons can store data along with computational environments in VMs or containers so that computations supporting scientific discoveries can be reproducible. Third, data commons can serve as a platform, enabling future discoveries as more data, algorithms, and software applications are added to the commons.
Data commons fit well with the science-as-a-service model: although data commons allow researchers to download data, host it themselves, and analyze it locally, they also allow current data to be reanalyzed with new methods, tools, and applications using collocated computing infrastructure. New data can be uploaded for an integrated analysis, and hosted data can be made available to other resources and applications using a data-as-a-service model, in which data in a data commons is accessed through an API. A data-as-a-service model is enhanced when multiple data commons and science clouds peer so that data can be moved between them at no cost.
Challenges
Perhaps the biggest challenge for data commons, especially large-scale data commons, is developing long-term sustainability models that support operations year after year.
Over the past several years, funding agencies have required data management plans for the dissemination and sharing of research results, but, by and large, they haven’t provided funding to support this requirement. What this means is that a lot of data is searching for data commons and similar infrastructure, but very little funding is available to support this type of infrastructure.
Moreover, datacenters are sometimes divided into several “pods” to facilitate their management and build out—for lack of better name, we sometimes use the term cyberpod to refer to the scale of a pod at a datacenter. Cyberinfrastructure at this scale is also sometimes called midscale computing,23 to distinguish it from the large-scale infrastructure available to Internet companies such as Google and Amazon and the HPC clusters generally available to campus research groups. A pod might contain 50 to several hundred racks of computing infrastructure. Large-scale Internet companies have developed specialized software for mid- to large-scale (datacenter-scale) computing,24 such as MapReduce (Google)25 and Dynamo (Amazon),26 but this proprietary software isn’t available to the research community. Although some software applications, such as Hadoop,23 are available to the research community and scale across multiple racks, there isn’t a complete open source software stack containing all the services required to build a large-scale data commons, including the infrastructure automation and management services, security services, and so on24 required to operate a data commons at midscale.
We single out three research challenges related to building data commons at the scale of cyberpods:
Software stacks for midscale computing. The first research challenge is to develop a scalable open source software stack that provides the infrastructure automation and monitoring, computing, storage, security, and related services required to operate at the scale of a cyberpod.
Datapods. The second research challenge is to develop data management services that scale out to cyberpods. We sometimes use the term datapods for data management infrastructure at this scale—that is, data management infrastructure that scales to midscale and larger computing infrastructure.
AnalyticOps. The third challenge is to develop an integrated development and operations methodology to support large-scale analysis and reanalysis of data. You might think of this as the analogy of DevOps for large-scale data analysis.
An additional category of challenges is the lack of consensus within the research community for a core set of standards that would support data commons. There aren’t yet widely accepted standards for indexing data, APIs for accessing data, and authentication and authorization protocols for accessing controlled-access data.
Lessons Learned
Data reanalysis is an important capability. For many research projects, large datasets are periodically reanalyzed using new algorithms or software applications, and data commons are a convenient and cost-effective way to provide this service, especially as the data grows in size and becomes more expensive to transfer.
In addition, important discoveries are made at all computing resource levels. As mentioned, computing resources are rationed in a data commons (either directly through allocations or indirectly through charge backs). Typically, there’s a range of requests for computing allocations in a data commons spanning six to seven or more orders of magnitude, ranging from hundreds of core hours to tens of millions of core hours. The challenge is that important discoveries are usually made across the entire range of resource allocations, from the smallest to the largest. This is because when large datasets, especially multiple large datasets, are collocated, it’s possible to make interesting discoveries even with relatively small amounts of compute.
The tragedy of the commons can be alleviated with smart defaults in implementation. In the early stages of the OSDC, the number of users was smaller, and depletion of shared computational resources wasn’t an urgent concern. As the popularity of the system grew and attracted more users, we noted some user issues (for example, increase in support tickets that noted that larger VM instances wouldn’t launch) as compute core utilization surpassed 85 percent. Accounting and invoicing promotes responsible usage of community resources. We also implemented a quarterly resource allocation system with a short survey to users requiring optin for continued resource usage extending into the next quarter. This provides a more formal reminder every three months to users who are finishing research projects to relinquish their quotas and has been successful for tempering unnecessary core usage. Similarly, as we moved to object storage functionality, we noted more responsible usage of storage, as scratch space is in ephemeral storage and removed by default when the computing environment is terminated. The small extra effort in moving data via an API to the object storage requires more thoughtful curation and usage of resources.
Over the past several years, much of the research focus has been on designing and operating data commons and science clouds that are scalable, contain interesting datasets, and offer computing infrastructure as a service. We expect that as these types of science-as-a-service offerings become more common, there will be a variety of more interesting higher-order services, including discovery, correlation, and other analysis services that are offered within a commons or cloud and across two or more commons and clouds that interoperate.
Today, Web mashups are quite common, but analysis mashups, in which data is left in place but continuously analyzed as a distributed service, are relatively rare. As data commons and science clouds become more common, these types of services can be more easily built.
Finally, hybrid clouds will become the norm. At the scale of a several dozen racks (a cyberpod), a highly utilized data commons in a well-run data-center is less expensive than using today’s public clouds.22 For this reason, hybrid clouds consisting of privately run cyberpods hosting data commons that interoperate with public clouds seem to have certain advantages.
Properly designed data commons can serve several roles in science as a service: first, they can serve as an active, accessible, citable repository for research data in general and research data associated with published research papers in particular. Second, by collocating computing resources, they can serve as a platform for reproducing research results. Third, they can support future discoveries as more data is added to the commons, as new algorithms are developed and implemented in the commons, and as new software applications and tools are integrated into the commons. Fourth, they can serve as a core component in an interoperable “web of data” as the number of data commons begins to grow, as standards for data commons and their interoperability begin to mature, and as data commons begin to peer.
Related Work.
Several projects share many of the goals of data commons in general, and the Open Commons Consortium (OCC) data commons in particular. Here, we discuss three of the most important: the National Institutes of Health (NIH) Big Data to Knowledge (BD2K) program, the Research Data Alliance (RDA), and the National Data Service (NDS).
The work described in the main text is most closely connected with the vision for a commons outlined by the BD2K program at the US National Institutes for Health.1 The commons described in this article can be viewed partly as an implementation of a commons that supports the principles of findability, accessibility, interoperability, and reusability2 which are key requirements of the data-sharing component of the BD2K program.
Of the three projects mentioned, the largest and most mature is the RDA,3 the goals of which are to create concrete pieces of infrastructure that accelerate data sharing and exchange for a specific but substantive target community; adopt the infrastructure within the target community; and use the infrastructure to accelerate data-driven innovation.3
The goals of the NDS are to implement core services for discovering data; storing persistent copies of curated data and associated metadata; accessing data; linking data with other data, publications, and credit for reuse; and computing and analyzing data (www.nationaldataservice.org). Broadly speaking, the goals of the commons described here are similar to the NDS and, for this reason, they can be considered as some of the possible ways to implement the services proposed for the NDS.
The OCC and Open Science Data Cloud (OSDC) started in 2008, several years before BD2K, RDA, and NDS, and have been developing cloud-based computing and data commons services for scientific research projects ever since. Roughly speaking, the goals of these projects are similar, but the OSDC is strictly a science service provider and data commons provider, whereas the RDA is a much more general initiative. The BD2K program is focused on biomedical research, especially for NIH-funded researchers, while the NDS is a newer effort that involves the National Science Foundation supercomputing centers, their partners, and their users.
References
- 1.Bonazzi V. NIH Commons Overview, Framework & Pilots. 2015 https://datascience.nih.gov/commons.
- 2.Wilkinson MD, et al. The FAIR Guiding Principles for Scientific Data Management and Stewardship. Scientific Data. 2016;3:160018. doi: 10.1038/sdata.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berman F, Wilkinson R, Wood J. Building Global Infrastructure for Data Sharing and Exchange through the Research Data Alliance. D-Lib Magazine. 2014;20 www.dlib.org/dlib/january14/01guest_editorial.html. [Google Scholar]
Acknowledgments
This material is based in part on work supported by the US National Science Foundation under grant numbers OISE 1129076, CISE 1127316, and CISE 1251201 and by National Institutes of Health/Leidos Biomedical Research through contracts 14X050 and 13XS021/HHSN261200800001E.
Biographies
Robert L. Grossman is director of the University of Chicago’s Center for Data Intensive Science, a professor in the Division of Biological Sciences at the University of Chicago, founder and chief data scientist of Open Data Group, and director of the nonprofit Open Commons Consortium. Grossman has a PhD from Princeton University from the Program in Applied and Computational Mathematics. He’s a Core Faculty and Senior Fellow at the University of Chicago’s Computation Institute. Contact him at robert.grossman@uchicago.edu.
Allison Heath is director of research for the University of Chicago’s Center for Data Intensive Science. Her research interests include scalable systems and algorithms tailored for data-intensive science, specifically with applications to genomics. Heath has a PhD in computer science from Rice University. Contact her at aheath@uchicago.edu.
Mark Murphy is a software engineer at the University of Chicago’s Center for Data Intensive Science. His research interests include the development of software to support scientific pursuits. Murphy has a BS in computer science engineering and a BS in physics from the Ohio State University. Contact him at murphymarkw@uchicago.edu.
Maria Patterson is a research scientist at the University of Chicago’s Center for Data Intensive Science. She also serves as scientific lead for the Open Science Data Cloud and works with the Open Commons Consortium on its Earth science collaborations with NASA and NOAA. Her research interests include cross-disciplinary scientific data analysis and techniques and tools for ensuring research reproducibility. Patterson has a PhD in astronomy from New Mexico State University. Contact her at mtpatter@uchicago.edu.
Walt Wells is director of operations at the Open Commons Consortium. His professional interests include using open data and data commons ecosystems to accelerate the pace of innovation and discovery. Wells received a BA in ethnomusicology/folklore from Indiana University and is pursuing an MS in data science at CUNY. Contact him at walt@occ-data.org.
Contributor Information
Robert L. Grossman, University of Chicago
Allison Heath, University of Chicago.
Mark Murphy, University of Chicago.
Maria Patterson, University of Chicago.
Walt Wells, Center for Computational Science Research.
References
- 1.Grossman RL, et al. The Design of a Community Science Cloud: The Open Science Data Cloud Perspective. Proc High Performance Computing, Networking, Storage and Analysis. 2012:1051–1057. [Google Scholar]
- 2.Mell P, Grance T. The NIST Definition of Cloud Computing (Draft): Recommendations of the National Institute of Standards and Technology. Nat’l Inst Standards and Tech. 2011 [Google Scholar]
- 3.Dooley R, et al. Software-as-a-Service: The iPlant Foundation API. Proc. 5th IEEE Workshop Many-Task Computing on Grids and Supercomputers; 2012; https://www.semanticscholar.org/paper/Software-as-a-service-the-Iplant-Foundation-Api-Dooley-Vaughn/ccde19b95773dbb55328f3269fa697a4a7d60e03/pdf. [Google Scholar]
- 4.Foster I. Globus Online: Accelerating and Democratizing Science through Cloud-Based Services. IEEE Internet Computing. 2011;3:70–73. [Google Scholar]
- 5.Simmhan YL, Plale B, Gannon D. A Survey of Data Provenance in E-Science. ACM Sigmod Record. 2005;34(3):31–36. [Google Scholar]
- 6.Chervenak A, et al. Wide Area Data Replication for Scientific Collaborations. Int’l J High Performance Computing and Networking. 2008;5(3):124–134. [Google Scholar]
- 7.Alameda J, et al. The Open Grid Computing Environments Collaboration: Portlets and Services for Science Gateways. Concurrency and Computation: Practice and Experience. 2007;19(6):921–942. [Google Scholar]
- 8.Pepple K. O’Reilly. 2011. Deploying OpenStack. [Google Scholar]
- 9.Davies A, Orsaria A. Scale out with GlusterFS. Linux J. 2013;235:1. [Google Scholar]
- 10.Weil SA, et al. Ceph: A Scalable, High-Performance Distributed File System. Proc. 7th Symp. Operating Systems Design and Implementation; 2006; pp. 307–320. [Google Scholar]
- 11.Mayernik MS. Data Citation Initiatives and Issues. Bulletin Am Soc Information Science and Technology. 2012;38(5):23–28. [Google Scholar]
- 12.Duerr RE, et al. On the Utility of Identification Schemes for Digital Earth Science Data: An Assessment and Recommendations. Earth Science Informatics. 2011;4(3):139–160. [Google Scholar]
- 13.Lagoze C, et al. CED 2 AR: The Comprehensive Extensible Data Documentation and Access Repository. Proc. IEEE/ACM Joint Conf. Digital Libraries; 2014; pp. 267–276. [Google Scholar]
- 14.Kunze J. Towards Electronic Persistence Using ARK Identifiers. Proc. 3rd ECDL Workshop Web Archives; 2003; https://wiki.umiacs.umd.edu/adapt/images/0/0a/Arkcdl.pdf. [Google Scholar]
- 15.Kunze JR. The ARK Identifier Scheme. US Nat’l Library Medicine; 2008. [Google Scholar]
- 16.Pollard T, Wilkinson J. Making Datasets Visible and Accessible: DataCite’s First Summer Meeting. Ariadne. 2010;64 www.ariadne.ac.uk/issue64/datacite-2010-rpt. [Google Scholar]
- 17.Starr J, et al. A Collaborative Framework for Data Management Services: The Experience of the University of California. J eScience Librarianship. 2012;1(2):7. [Google Scholar]
- 18.Ball A, Duke M. How to Cite Datasets and Link to Publications. Digital Curation Centre; 2011. [Google Scholar]
- 19.Green T. We Need Publishing Standards for Datasets and Data Tables. Learned Publishing. 2009;22(4):325–327. [Google Scholar]
- 20.Mandl D, et al. Use of the Earth Observing One (EO-1) Satellite for the Namibia Sensor Web Flood Early Warning Pilot. IEEE J Selected Topics in Applied Earth Observations and Remote Sensing. 2013;6(2):298–308. [Google Scholar]
- 21.Heath AP, et al. Bionimbus: A Cloud for Managing, Analyzing and Sharing Large Genomics Datasets. J Am Medical Informatics Assoc. 2014;21(6):969–975. doi: 10.1136/amiajnl-2013-002155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paltoo DN, et al. Data Use under the NIH GWAS Data Sharing Policy and Future Directions. Nature Genetics. 2014;46(9):934. doi: 10.1038/ng.3062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Future Directions for NSF Advanced Computing Infrastructure to Support US Science and Engineering in 2017–2020. Nat’l Academies Press; 2016. [Google Scholar]
- 24.Barroso LA, Clidaras J, Hölzle U. The Data-center as a Computer: An Introduction to the Design of Warehouse-Scale Machines. Synthesis Lectures on Computer Architecture. 2013;8(3):1–154. [Google Scholar]
- 25.Dean J, Ghemawat S. MapReduce: Simplified Data Processing on Large Clusters. Comm ACM. 2008;51(1):107–113. [Google Scholar]
- 26.DeCandia G, et al. Dynamo: Amazon’s Highly Available Key-Value Store. ACM SIGOPS Operating Systems Rev. 2007;41(6):205–220. [Google Scholar]