Abstract
GTP is a major regulator of multiple cellular processes, but tools for quantitative evaluation of GTP levels in live cells have not been available. Here we report characterization of genetically encoded GTP sensors, constructed by inserting cpYFP into a region of the bacterial FeoB G-protein that undergoes a GTP-driven conformational change. GTP binding to these sensors results in a ratiometric change in their fluorescence, thereby providing an internally normalized response to changes in GTP levels while minimally perturbing those levels. Mutations introduced into FeoB to alter its affinity for GTP allowed generation of sensors with a wide dynamic range. Critically, in mammalian cells the sensors show consistent changes in fluorescence intensity ratios upon depletion or restoration of GTP pools. These sensors are suitable for detecting spatio-temporal changes in GTP levels in living cells, and for the development of high throughput screenings of molecules modulating intracellular GTP levels.
Keywords: Guanosine tri-phosphate (GTP), G-Proteins, Ratiometric Sensors, circularly permutated Yellow Fluorescent Protein (cpYFP), Melanoma
Introduction
GTP, regulates a large array of cellular processes ranging from protein synthesis1 to cell signaling2,3, including activation of G-proteins, dysfunction of which underlies a variety of human diseases4-6, most notably cancer7. While the need for a method measuring GTP levels in live cells is clear, no such method currently exists.
We report here the construction of genetically encoded GTP sensors based on fluorescent protein (FP) technology. FP-based methods are widely use, which should facilitate utilization of these novel sensors by researchers interested in GTP regulated processes. These sensors exhibit a rapid, internally normalized ratiometric change in fluorescence upon changes in GTP concentration in vitro and GTP levels in living cells. They also reveal heterogeneity in the intracellular distribution of GTP, raising the possibility that such variation could regulate G-protein activity in different cellular compartments.
Results
Construction of a FeoB-cpYFP fusion that changes fluorescence upon binding GTP
The G-protein domain of E. coli iron transport protein FeoB exhibits a loop (aa 35-40) that undergoes a conformational change upon GTP binding8 (Fig. 1a, b). As a bacterial protein, FeoB is unlikely to interact specifically with eukaryotic proteins that could confound its function as a GTP sensor. FeoB GTP on and off rates are ∼106 M-1sec-1 and 12 sec-1, respectively9, so it can respond quickly to changes in GTP concentration, and its GTP hydrolysis rate is only 0.0015 sec-1 so its hydrolysis activity should not reduce local GTP pools.
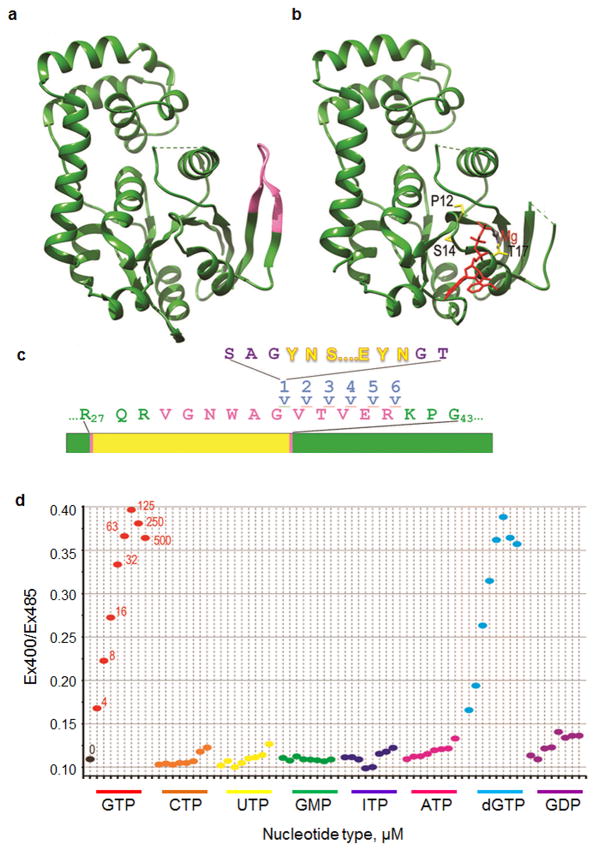
Figure 1. Construction and nucleotide selectivity of a GTP sensor.
(a) The G-protein domain of E.coli FeoB without ligand (pdb 3HYR)8. The V29-R29 switch I region that undergoes a conformational change upon ligand binding is highlighted in pink. (b) The G-protein domain of E.coli FeoB protein with a bound GTP analogue (in red with the magnesium ion in grey; pdb3HYT). Amino acid side chains mutated to alter GTP affinity (P12, S14, T17) are shown in stick representation and colored yellow. The switch I region is not visible in the crystal structure because it becomes disordered upon binding GTP. (c) Sensor construction. Twenty-four distinct fusions were made by inserting the cpYFP (yellow) at 6 different positions (after residues 35-40) within the switch I region (pink) of the FeoB G-protein domain (green), either with or without SAG or GT linkers (purple) at the N- or C-terminal fusion points, respectively. The lines indicate insertion of the cpYFP after residue 35. (d) Ratio of emission intensity when excited at 405nm vs. 485nm (Ex405/Ex485) is plotted for FY5a+5a in the presence of no nucleotide (“0”; black); 4, 8, 16, 32, 65, 125, 250, 500μM GTP (in red), 4-500μM CTP (orange), ), 4-500μM UTP (yellow), ), 4-500μM GMP (green), ), 4-500μM ITP (blue), ), 4-500μM ATP (magenta), ), 4-500μM dGTP (cyan) and ), 4-500μM GDP (purple).
DNA encoding a circularly permuted yellow fluorescent protein (cpYFP) was inserted into DNA encoding residues 35-40 of the FeoB G-protein domain (Fig. 1c). Fusion nomenclature followed the form “FY1a+1a”, where the number designates the insertion site, with “1” corresponding to insertion after residue 35 and “6” to insertion after residue 40, and where an “a” indicates the presence of a ser-ala-gly or gly-thr linker at, respectively, the cpYFP N- or C-terminus. Most of the 24 fusions generated changed fluorescence upon binding GTP, with three (FY1+1a, FY5a+5a and FY6+6a) showing a >2-fold decrease in emission when excited at 485nm (Supplementary Fig. S1). However, only FY5a+5a showed a ratiometric change in which GTP caused a decrease in fluorescence when excited at <450nm, and an increase when excited at >450nm (Supplementary Fig. S2a), resulting in a large change in the ratio of emission intensities when excited at 405nm vs. 485nm (Ex405/Ex485; Fig. 1d). Such ratiometric changes are critical because, by measuring the ratio of fluorescence at two wavelengths, the signal is normalized and corrected for variations in sensor levels. The ratiometric signal of FY5a+5a is large, ranging from 1.7 to 5.3 across the relevant pH range (Supplementary Fig. S3). By comparison, the Perceval sensor, which operates by similar principles to sense ATP/ADP ratios exhibits a maximal ratiometric signal of ∼2.010, while the ATeam sensors, which utilize a FRET-mechanism to report on ATP, exhibit maximal ratiometric signals of 1.7 to 2.7 across a similar pH range11.
The sensor is selective for GTP and dGTP
While 4μM GTP was sufficient to induce a large change in FY5a+5a fluorescence (Fig. 1d, Supplementary Fig. S2a), even 500μM UTP, CTP, and GMP had little effect and these nucleotides did not significantly interfere with the GTP-induced change when in 5-fold excess of GTP (Fig. 1d and Supplementary Fig. S2b-g). High concentrations of ITP and ATP had small effects on fluorescence but, even at 500μM, induced less than 10% of the change in the Ex400/Ex485 ratio induced by 100 μM GTP and did not markedly affect the changes induced by GTP even when in 5-fold excess of the latter (Fig. 1d and Supplementary Fig. S2h-k).
In contrast, dGTP had effects indistinguishable from GTP (Fig. 1d, Supplementary Fig. S2l). However, even in proliferating cells, dGTP levels are 100-fold lower than GTP12, so the potential for dGTP to interfere with GTP sensing is negligible.
Fluorescence was also affected by GDP, though the change in Ex400/Ex485 ratio induced by GDP was ∼5-fold smaller, and required ∼5-fold higher concentrations, than that induced by GTP (Fig. 1d, Supplementary Fig. S2m). Because GDP has the greatest potential to interfere with GTP sensing, we carried out more extensive interference experiments with GDP:GTP ratios varying from 1:10 to 10:1. Under conditions where GTP addition induced a 3-fold change in Ex400/Ex485, addition of 4-fold excess GDP reduced this change by less than 10%, and addition of even higher amounts of GDP reduced this change by a maximum of only 25% (Supplementary Fig. S4). Since, even under extreme starvation conditions, cellular GTP levels are several-fold in excess of GDP12,13, these data indicate that GDP will not interfere with intracellular sensing of GTP.
The sensor responds rapidly to changes in GTP levels
The kinetics of the response to GTP binding were measured by reacting GTP-free FY5a+5a with GTP in a stop-flow fluorometer. Both the GTP-induced decrease in fluorescence at Ex485 and the increase at Ex400 were fast, with half-lives of 6 and 10msec at, respectively, 500 and 100μM GTP (Fig. 2a). To measure the rate of the GTP-dissociation dependent response, FY5a+5a pre-mixed with 100 μM GTP was reacted with 2.5mM GDP. Because GDP was in 25-fold excess, sensors that release GTP usually rebind GDP, so GTP release is monitored by following the reversal of the fluorescence changes induced by the triphosphate. The GTP-dissociation dependent changes were also fast, with half-lives of ∼120msec (Fig. 2b). To determine bleaching kinetics, FY5a+5a, with and without GTP, was continuously exposed to either 400nm or 485nm light. After more than one hour, emission had decreased by less than 0.5% (Ex485) or 0.2% (Ex400) in the absence of GTP and even less (<0.1%) in the presence of GTP (Fig. 2c).
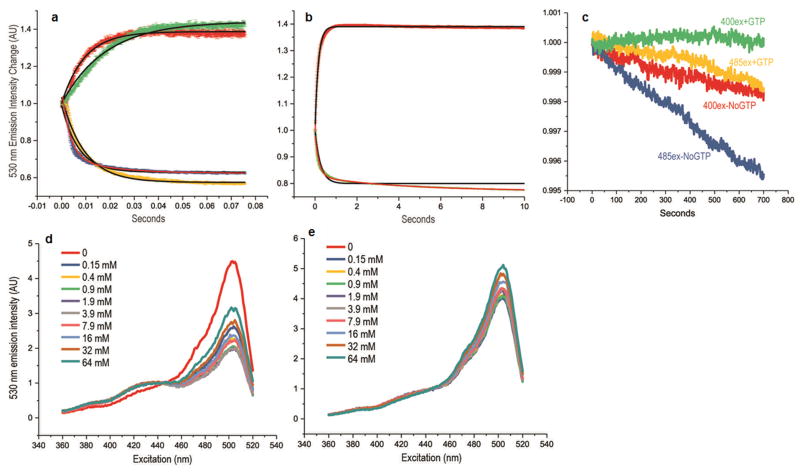
Figure 2. Kinetics and Mg++ dependence of GTP induced fluorescence changes.
(a) Emission intensity (530 nm) upon excitation at 400nm (red and green traces) or 485nm (blue and yellow traces) vs. time for reactions in which GTP-free FY5a+5a fusion was reacted with 100 (green and yellow) or 500 (red and blue) μM GTP. Initial fluorescent intensities normalized to a value of 1.0. Trace thickness shows +/-SEM for n=4. Black lines show fits to single-phase exponential association (red and green traces) or exponential decay (blue and yellow traces) functions. The high GTP concentration 485ex data (blue trace) was not well-fit by a single-phase exponential so a fit with a double-phase exponential decay function (red line) is also shown. (b) Fluorescence emission intensity (530nm) upon excitation at 400nm (green trace) or 485nm (red trace) vs. time for reactions in which FY5a+5a with 100μM GTP was reacted with 2.5mM GDP. As in panel (a), the black lines show single-phase exponential fits while the red line shows a double-phase fit. While it's unclear why the data do not always obey single-phase exponential functions, they do show that the fluorescent responses are fast, occurring with half-lives of ∼10 and ∼100msec in response to, respectively, increases or decreases in GTP concentrations. (c) Fluorescent emission intensity (530nm) vs. excitation for FY5a+5a continuously illuminated at either 400nm (green and red traces) or 485nm (yellow and blue traces) either with (green and yellow traces) or without (red and blue traces) GTP. Initial intensity is normalized to 1.0 for all traces. Emission intensity decreases (bleaching) are all <0.5% over more than hour of constant excitation. (d) Fluorescent emission intensity (530nm) vs. excitation wavelength for FY5A+5A with 100μM GTP. Reactions contained 0. mM EDTA and either 0 (red trace), 0.25mM (dark blue), 0.5mM (yellow), 0.9mM (green), 1.9mM (purple), 3.9mM (grey), 7.9mM (pink), 16mM (light blue), 32mM (brown), or 64mM (dark green) Mg++ (Mg++ concentrations specified in the figure are expressed to 2 significant figures and correspond to free Mg++ concentrations due to chelation by EDTA). (e) As in (d) but with GTP-free FY5a+5a fusion.
The sensor signal is effectively constant over physiological magnesium concentration ranges
Since GTP binds FeoB in complex with Mg++, variations in Mg++ could affect sensor fluorescence. We measured the effect of varying Mg++ concentrations with and without GTP: with 0.1mM EDTA and no Mg++, GTP failed to induce a change in sensor fluorescence (Fig. 2d). Addition of 0.25mM Mg++ partially restored the GTP-induced change and 0.5 mM restored 85% of the change. The Ex400/Ex485 ratios varied less than 5% when free Mg++ ranged from 0.9mM to 3.9mM, but higher Mg++ (>7.9mM) reduced this ratio due to increases in fluorescence at higher excitation wavelengths (Fig. 2d). Without GTP, excitation spectra didn't vary with Mg++ concentrations in the 0 to 3.9mM range, but showed emission increases at higher excitation wavelengths with Mg++>7.9mM (Fig. 2e). These data indicate that Mg++ is required for the GTP-induced change in fluorescence and that very high Mg++ concentrations affect fluorescence independently of GTP. However, both with and without GTP, fluorescence is effectively constant over the 0.5-5mM Mg++ concentration ranges prevailing in vivo14.
Construction of a set of GTP sensors with a wide dynamic range
The GTP Kd of FeoB is ∼4-15μM12,15, but total cellular GTP is typically 300-800μM12. The tight GTP binding of FeoB could render the sensor insensitive to GTP concentration changes across physiologically relevant ranges. We therefore created five FY5a+5a variants by introducing mutations to weaken or eliminate GTP binding. We characterized the effective affinities of these fusions for GTP and the most structurally similar cellular ligands (ATP, GDP; Supplementary Fig. S5 and Table 1). The Keff values (GTP concentrations required to obtain 50% of the maximal ratiometric signal) for the five GTP-binding variants ranged from 30 to 2000μM. The variant mutated to abolish GTP binding showed no response to GTP. We name these fusions GEVALs (GTP EVALuators), with a numbered suffix indicating their GTP Keff value (Table 1), and with the non-binding variant designated GEVALNull.
Table 1. GTP and GDP Keff values for the ligand dependent ratiometric fluorescence changes of mutated sensors.
| FeoB Mutation | GTP Keff μM | GDP Keff μM |
|---|---|---|
| WT (GEVAL30) |
33.2±0.3 | 100±6 |
| P12G (GEVAL260) |
260±20 | 1000±200 |
| P12G/S14A (GEVAL530) |
528±20 | 1280±200 |
| P12G/T17S (GEVAL1150) |
1150±100 | 2770±300 |
| P12G/S14A/T17S (GEVAL2300) |
2300±100 | 3700±300 |
| P12A/S14A/T17A (GEVALNull) |
N/A | N/A |
GEVAL fluorescence is affected by pH (Supplementary Figs. S3, S6, S7, S8a), a feature common to FP-based ligand sensors16. FP pH sensors or pH-sensitive dyes are often deployed in parallel with ligand sensors to determine if a change in fluorescence of the latter is due to a change in pH or ligand concentration10. For GEVALs, the most effective control in this respect is GEVALNull, since it doesn't respond to GTP but should respond exactly like the other GEVALs to pH or any other change in cell environment. If GEVAL fluorescence changes are observed under conditions where GEVALNull fluorescence is constant, then they can be attributed to changes in GTP levels rather than other effects. To confirm that GEVALNull would respond to pH identically to the GTP-sensing fusions we measured all their Ex400/Ex485 ratios as a function of pH and GTP concentration (Supplementary Fig. S7). Without GTP, all the GEVALs exhibited equal Ex400/Ex485 ratios that varied identically as pH was varied from 5.5-8.5. Upon GTP addition the ratios increased for all the GEVALs, with the largest increases seen for the sensors with the highest GTP affinity (Supplementary Fig. S7a-e). However, the Ex400/Ex485 ratio for GEVALNull remained invariant upon GTP addition (Supplementary Fig. S7). The GEVAL sensors were expressed well in E. coli with no indication of aggregation, but to evaluate this further we measured their apparent MW and polydispersity by dynamic light scattering and found them to be monomeric and monodisperse in solution (Supplementary Fig. S8b).
GEVAL sensors' response to pharmacological depletion of GTP pools
DNAs encoding GEVAL30, GEVAL530 and GEVALNull, representing, respectively the high and medium GTP affinity sensors, and the unresponsive negative control, were cloned into lentiviral expression vectors and transduced into SK-Mel-103 human melanoma cells. Equivalent expression levels of all sensors were verified by immunofluorescence and immunoblotting (Fig. 3a). Cells were then transiently transfected with the pHRed17 vector (with over 90% efficiency) to control for pH variations (Supplementary Fig. S9a).
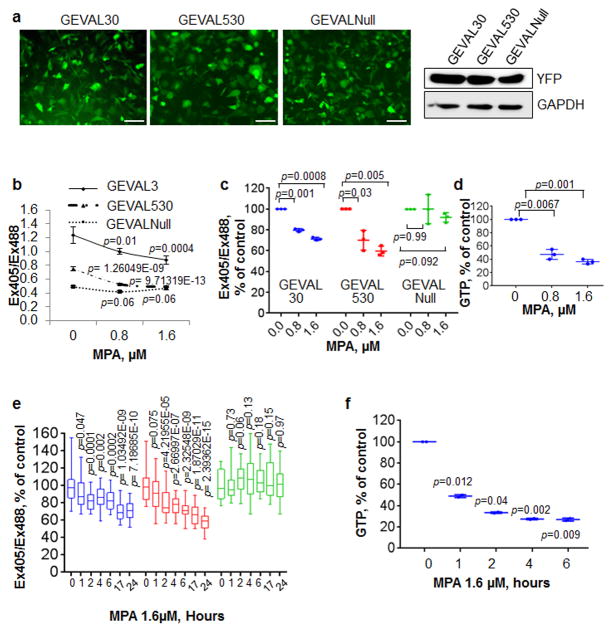
Figure 3. Validation of the GEVAL sensors GTP-sensing activity in mammalian cells.
(a) Human metastatic melanoma SK-Mel-103 cells were transduced with lentiviral supernatant to express GEVAL30, GEVAL530, and GEVALNull. The sensor expression was evaluated by both immunofluorescence and immunoblot to YFP (GAPDH was used to verify equal loading). Scale bar, 100μm. (b) Cells as in (a) were plated on coverslip chambers, treated for 48hrs with the indicated concentration of MPA, and imaged as described in Material and Methods. 30 cells/population were analyzed with ImageJ to calculate the Ex405/Ex488 ratio. The average -/+ SEM of the ratio for each condition within one experiment is reported. Statistics performed by two-tailed Student-t-Test. (c) Dot-plot of three independent experiments as in (b) normalized over the untreated control in each group; horizontal bars represent the average, vertical lines are standard deviation. Statistics performed by two-tailed Student-t-Test. (d) GTP quantification by HPLC measurement in SK-Mel-103 treated with the same concentrations of MPA as in (b and c). The data is express as the dot-plot of 3 independent experiments; horizontal bars represent the average, vertical lines are standard deviation. Statistics performed by two-tailed Student-t-Test. (e) Populations of SK-Mel-103 cells as in (b,c) were imaged (0h) and then treated with 1.6μM MPA and imaged at the indicated time points. Bar and whisker plots of 30 cells from two independent experiments normalized over the untreated control (0h) in each group; horizontal bars represent the average, whiskers represent minimum and maximum values, boxes represent the 1st and 3rd quartiles. Statistics performed by two-tailed Student-t-Test. (f) GTP quantification by HPLC measurement in SK-Mel-103 treated with 1.6μM MPA for the indicated time-points. The data is expressed as the dot-plot of 2 independent experiments; horizontal bars represent the average, vertical lines are standard deviation. Statistics performed by two-tailed Student-t-Test.
To evaluate potential autofluorescence and bleed-through, untransduced cells and cells transduced with only pHRed or one of the GEVAL sensors were imaged as described in Materials and Methods, using sequential excitation with 405 nm and 488nm for the GEVAL sensors and 458nm and 594nm for the pHRed sensor. Untransduced cells could not be visualized with any of the fluorescent lights; GEVAL-infected cells gave a robust signal when excited at 405nm and 488nm, and showed a minimal bleedthrough in the 458nm channel of pHRed. Cells transfected with pHRed displayed robust signals at pHRed-specific excitation wavelengths and demonstrated a <9% bleed-through in the 405nm channel for the GEVAL sensors (Supplementary Fig. S9a). The GEVAL sensors were also photostable (Supplementary Fig. S9b).
Next, cells were transduced with 1× and 5× viral titers for each sensor followed by evaluation of YFP levels by immunoblot (Supplementary Fig. S9c). NTP levels were assessed by HPLC and equal amounts of GTP were detected in all cell populations, similarly to untransduced control cells (Supplementary Fig. S9d), indicating that GEVAL levels do not affect intracellular GTP pools. Proliferation and viability of 1× and 5× cells were assessed via trypan blue assay 48hrs post-treatment with 0, 0.2μM or 2μM of mycophenolic acid (MPA), an inhibitor of inosine monophosphate dehydrogenases (IMPDH), that causes depletion of intracellular GTP levels18,19. Uninfected and GEVAL-expressing cells responded similarly to the tested MPA concentrations, (Supplementary Fig. S9e,f), indicating that the GEVAL sensors do not affect cells with depleted GTP levels.
To assess sensors' sensitivity, cells expressing GEVAL and pHRed sensors were treated with increasing concentrations of MPA for 48hrs, and the Ex405/Ex488 ratio for the GTP sensors and Ex594/Ex458 ratio for pHRed were calculated. To transform the Ex594/Ex458 ratios into pH values, we interpolated data provided by Dr. Gary Yellen (see Material and Methods). The analysis showed no difference in overall cell pH among all populations (Supplementary Fig. S10a,b). The functionality of the pHRed sensor was verified by monitoring its response to changes in the cell media pH via addition of NH4Cl (20mM final), followed by acetic acid (10mM final) as previously published17 (Supplementary Fig. S10c,d).
Importantly, fluorometric ratios of GEVAL30 and GEVAL530 decreased in response to MPA, while GEVALNull did not show any significant difference (Figs. 3b,c). Similar results were obtained in 1× and 5× cells (Supplementary Fig. S9c,g). Depletion of GTP pools by MPA was verified in parallel by HPLC (Fig.3d).
The Ex405/Ex488 ratio of GEVAL30 in the “no MPA” treatment was the highest (1.24), suggesting that a sensor is effectively GTP-replete in vivo. This ratio decreased in parallel with GTP depletion to 1.0 (77% of the original value). The GEVAL530 ratio in untreated cells was lower than GEVAL30 (0.75 vs 1.24) suggesting that this weaker sensor being only partially replete with GTP. The GEVAL530 sensor became apparently emptied of GTP as the concentration of MPA was increased, reaching the same ratio value as the GEVALNull sensor (0.5), which exhibited the lowest Ex405/Ex488 ratio, irrespective of MPA treatment (Fig. 3b). Similar results were obtained with another IMPDH1/2 inhibitor, mizoribine20 (Supplementary Fig. S11a,b)
Spatio-temporal changes in GEVAL sensors activity in response to GTP depletion
GEVAL-expressing cell populations were imaged at different times after treatment with 1.6μM MPA (Fig. 3e). GEVAL30 and GEVAL530 quickly responded to MPA, reaching values similar to the long-term treatment (48hrs) within 4-6hrs (compare to Fig. 3c). GEVALNull activity was unaffected (Fig. 3e). HPLC measurements were performed in parallel up to 6hrs (Fig. 3f), at which point GTP depletion was comparable to the one achieved at longer time points (Fig. 3d).
Next, individual GEVAL-expressing cells were imaged at 30min intervals post-MPA treatment. The Ex405/Ex488 ratios for GEVAL30 and GEVAL530 decreased rapidly over time (Fig. 4a, and Supplementary Fig. S11d). Similar results were obtained using mizoribine (Fig 4b and Supplementary Fig. S11c,e).
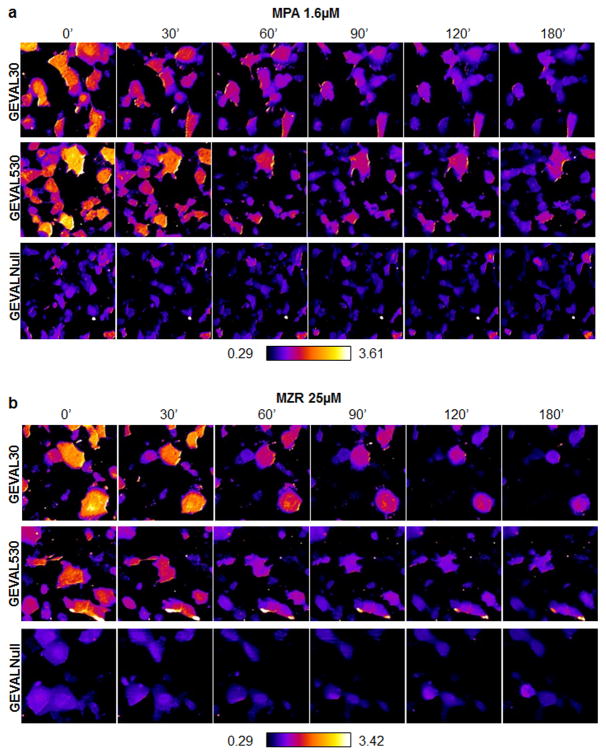
Figure 4. Detection of spatio-temporal variations of GTP in cells.
SK-Mel-103 cells were transduced with lentiviral supernatant to express the GEVAL30, GEVAL530, or GEVALNull. Cells were plated on coverslip chambers and imaged with a Leica AOBS SP5 confocal microscope as described above (0h). Cells were then treated with 1.6μM MPA and imaged at the indicated time points. Pixel-by-pixel ratiometric images were generated within ImageJ with the RatioPlus Plugin for each time point. All the ratios were then combined in a stack to ensure a uniform application of contrast enhancement and false-coloring (with the FIRE_LUT). (a) Representative ratiometric images of cell populations for each sensor over time. (b) Cells as in (a) but treated with 25μM MZR. Quantification of Ex405/Ex488 in 10 cells/population over time for cells treated as above is presented in Supplementary Fig. S11d,e.
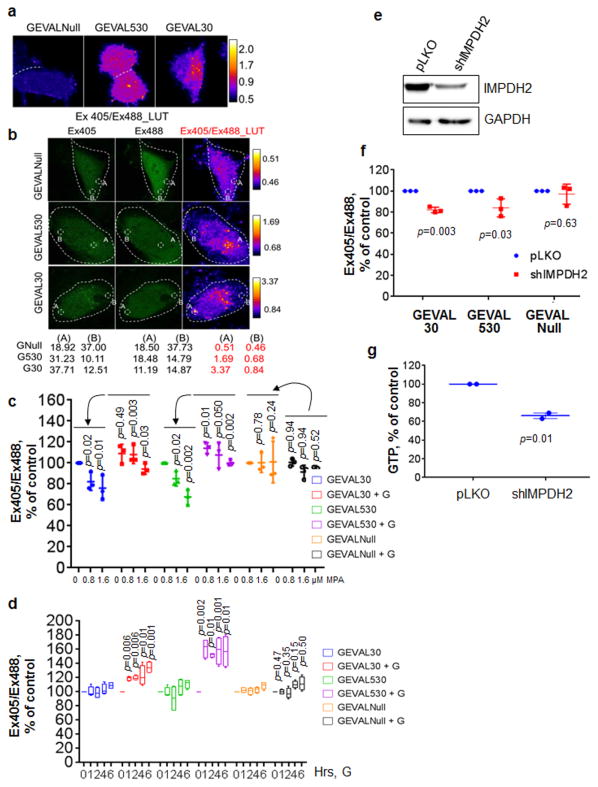
To assess intracellullar variations in GEVAL sensor activity, cells were imaged as described, followed by pixel-by-pixel calculation of the Ex405/Ex488 ratio and false-coloring. GEVALNull shows a uniformly low ratio throughout the cell, while both GEVAL530 and GEVAL30 reveal the existence of pockets with higher and lower ratios (Fig. 5a). Individual sensor analysis was also performed to reveal the dynamic range of each sensor, allowing us to calculate the exact ratio for different subcellular regions displaying high (A) or low (B) Ex405/Ex488 ratio (Fig. 5b).
Figure 5. GEVAL sensor detection of GTP gradients in cells and response to GTP repletion or genetic inhibition.
(a) SK-Mel-103 cells transduced with the three sensors were imaged and false-colored pixel-by-pixel ratiometric images were created as described in the Material and Methods section.(b) Cell images were manipulated as in (a) but in this case the contrast enhancement and false-coloring were applied independently to each sensor population. Indicated areas of high (A) and low (B) ratio within each image were analyzed with ImageJ to obtain numerical ratio values for the scaling within each image. (c) SK-Mel-103 cells transduced with the three sensors where treated for 48hrs with MPA (0.8μM and 1.6μM) in the absence of presence of 100μM guanosine (+G). Cells were then imaged and the Ex405/Ex488 ratio was calculated for 30cells/population. The data represents dot-plot of 3 independent experiments normalized over the untreated control in each group; horizontal bars represent the average, vertical lines are standard deviation. Statistics performed by two-tailed Student-t-Test. (d) Cells as in (c) were pre-treated over-night with 1.6μM MPA and imaged at time 0 (0h), then cells were treated or not with 100μM G and populations of cells were imaged at the indicated time points. The Ex405/Ex488 ratio was calculated for 30cells/population and the data is reported as bar and whisker plots of 4 independent experiments normalized over the untreated control (0h) in each group; horizontal bars represent the average, whiskers represent minimum and maximum values, boxes represent the 1st and 3rd quartiles. Statistics performed by two-tailed Student-t-Test. SK-Mel-103 cells were transduced with empty vector or shRNA to IMPDH2 (e) IMPDH2 suppression was verified by immunoblot and GAPDH was used to verify equal loading. (f) cells as in (e) were imaged as described above and in the material and methods section. The data is presented as dot-plot of 3 independent experiments normalized over the untreated control in each group; horizontal bars represent the average, vertical lines are standard deviation. Statistics performed by two-tailed Student-t-Test. (g) GTP quantification by HPLC measurement in SK-Mel-103 manipulated as in (e). The data is expressed as the dot-plot of 2 independent experiments; horizontal bars represent the average, vertical lines are standard deviation. Statistics performed by two-tailed Student-t-Test.
The distributions of per-pixel signal ratios within individual cells harboring GEVAL30 and GEVAL530 demonstrated statistically significant higher skewness and excess kurtosis values as compared to the cells expressing GEVALNull (Supplementary Fig. S12a,b), further suggesting that the distribution of GTP in the cell is heterogeneous. This heterogeneity was not due to pH variations, since similar pH values were observed between high and low GEVAL activity areas (Supplementary Fig. S13).
GEVAL sensors' response to increased GTP levels
Cells were imaged after MPA treatment +/- 100μM guanosine supplementation (to replenish intracellular GTP levels21-23). MPA treatment caused up-to 34% and 42% decrease in the Ex405/Ex488 ratios for the GEVAL30 and GEVAL530, respectively, which was fully counteracted by guanosine supplementation (Fig. 5c). Thus, the GEVAL sensors detect both depletion and replenishment of GTP levels.
In parallel, intracellular GTP pools were pre-depleted overnight with MPA followed by imaging and treatment with either vehicle or 100μM guanosine. Guanosine increased the activity of GEVAL30 and GEVAL530 but not GEVALNull already within 1 hour (Fig. 5d). Addition of guanosine restored sensors' activity following GTP depletion (Fig. 5c,d) but did not increase the basal activity of the GEVAL30 sensor and only modestly increased that of the GEVAL530 sensor when GTP pools were at their physiological levels (Fig. 5c). These data suggest that GEVAL30 is GTP replete at the normal GTP concentrations found in these cells, and that GEVAL530 is only partially replete.
GEVAL sensors' response to genetic depletion of GTP
To evaluate GEVAL's activity in response to genetic depletion of GTP, cells were transduced with an IMPDH2 shRNA24 (Fig. 5e). IMPDH2 depletion reduced the Ex405/Ex488 ratio of GEVAL30 and GEVAL530 by 16%-18%, while GEVALNull's ratio was unaffected (Fig. 5f). The smaller reduction when compared to that in MPA inhibition was consistent with HPLC data (Fig. 5g, 34% decrease versus Fig. 3d, 53% decrease at 0.8μM MPA) and could be due to incomplete shRNA depletion of IMPDH2. Importantly, activity of GEVAL sensors in cells expressing shRNA-resistant IMPDH2 cDNA was not affected by the above IMPDH2 shRNA (Supplementary Fig. S14a and b).
GEVAL sensors are suitable for high throughput screening
To assess sensors suitability for high throughput (HTP) screening, cells expressing GEVAL30 or GEVAL530 were seeded in 26 wells of a glass-bottom 96-well plate and treated with either vehicle or 1.6μM MPA for 24hrs. The center of each well was imaged and ten cells/well were analyzed and average of their Ex405/Ex488 ratios was used as the average ratiometric intenisity of each well. Z' values of 0.51 and 0.15 were calculated for GEVAL30 and GEVAL530, respectively, (Supplementary Fig. S14c,d) suggesting that GEVAL sensors can be developed for HTP applications.
Discussion
Monitoring cellular GTP currently relies on techniques like HPLC25 or mass spectroscopy (MS)26, which cannot be performed in live cells, result in loss of spatial resolution, and are prone to artifacts because of the lability of NTPs during extraction25,27. A nanosensor for guanine nucelotides has been constructed by coupling of a dye to an aptamer fused to a graphite sheet28. However, this sensor cannot be expressed in cells, and does not differentiate between guanine, GMP, GDP or GTP. Nor does it quantitatively assess GTP levels, as it undergoes only an “off-to-on” conversion upon binding guanine nucleotides.
The GEVAL sensors deployed in conjunction with pHRed sensors are capable of detection not only temporal changes in GTP in living cells but also changes in subcellular GTP distribution.
In addition HPLC- or MS-based measurements cannot distinguish between total and free GTP levels. Several recent publications including ours strongly suggest that free GTP levels may be below total levels21,29,30. This is consistent with our observation that GEVAL30 is sensitive to GTP depletion, even though we had expected that the low KGTP of this sensor might render it insensitive. It's also supported by observations that ∼90% of labeled GTP injected into xenopus oocytes rapidly becomes sequestered into large MW complexes (Dr. Bruce Nicholson, personal communication).
Together, our data reveal that the GEVAL sensors represent a novel and unique tool that allows semiquantitave assessment of changes in GTP levels within cell populations, and in the intracellular distribution of GTP in individual cells.
Material and Methods
GTP sensor construction
DNA coding for FeoB8 was synthesized with codons optimized for mammalian expression and cloned into pET15b (Genscript, NJ). DNA encoding FeoB switch I region was engineered to contain NgoMIV and SmaI sites at codons 34/35 and 41/42. FeoB DNA was PCR amplified with forward and reverse primers containing, respectively, XbaI and HindIII restriction sites, and ligated into XbaI/HindIII digested pRSET A (Invitrogen) from which a single NgoMIV site had been removed by site-directed mutagenesis. DNA coding for cpYFP was PCR amplified from the ATP/ADP ratio sensor Perceval31 (Addgene). For insertion into each position of the FeoB switch I region, a total of 4 PCR products were created from 2 forward and 2 reverse primers that differed in encoding either an additional 3 (SAG) or 2 (GT) linker residues at, respectively, the N- or C-terminal points of insertion of the cpYFP. NgoMIV digested PCR products were ligated into NgoMIV+SmaI digested pRSET (-Ngo) FeoB plasmid. Since the pRSET vectors did not give strong protein expression, FeoB-cpYFP DNAs were PCR amplified with primers containing NdeI and XhoI restriction sites, digested with NdeI and XhoI and ligated into NdeI/XhoI digested pET32 (EMDMillipore). To generate point mutations in the FeoB component of the sensor to alter its affinity for GTP we used PCR-mediated mutagenesis with mutagenic primers as described in the Quick-Change Mutagenesis Kit (Stratagene).
Protein Expression and Purification
pET32 expression vectors were transformed into BL21-DE3 cells and single colonies were grown overnight in Luria Broth with 50μg/ml ampicillin and used to inoculate larger volumes of 2× YT media at 1:100 dilutions. Cells were grown at 37°C to an OD600 of 0.6-0.8 and induced with 1mM IPTG, transferred to 30°C and grown for 12-18hrs. Cells were harvested by centrifugation at 4°C and the bright yellow-green cell pellets were re-suspended in 50mM Tris-HCl, pH 8.3, 200mM NaCl, 0.1% Tween-20, 10mM imidazole (lysis buffer). Prior to lysis by sonication (Misonix, CT), PMSF was added to 1mM. Lysates were clarified by 30min centrifugation at 15,000g and the protein was purified from the supernatants on Ni-NTA resin (Qiagen, CA).
Fluorometry
Fluorescence spectra for purified proteins were measured on Horiba JobinYvon Fluoromax-3 or Shimadzu RF-5301PC spectrofluorometers. GEVAL sensors at 1μM concentration in 100mM KCl, 50mM PIPES pH 7.0, 5mM MgCl2, and 10% glycerol (buffer A), and in the presence of varying concentrations of different nucleotides (as specified in individual figure legends; mixed and incubated for 5-10 minutes at room temperature), were excited at 320nm to 520nm at room temperature while emission at 530nm was measured. For experiments in which the effects of Mg++ concentration were measured, 0.1mM EDTA was added to buffer A and 1M MgCl2 was added to give total Mg++ concentrations of 0, 0.25, 0.5, 1, 2, 4, 18, 16, 32 and 64mM. Stop-flow fluorometry data were collected at room temperature on an Applied Photosystems stopped-flow fluorometer equipped with an excitation monochromator and 515nm log-pass cutoff filter, with excitation at either 400 or 485nm, and emission measured at 530nm. Syringes feeding the stop-flow reactant chambers were filled with either 2μM GEVAL30 alone or 2μM GEVAL30+100 μM GTP in buffer A, and then reacted with an equal volume of either 200μM GTP, 1mM GTP or 2.5mM GDP, as indicated in figure legends.
Ligand binding assays
Binding assays were done on a Biotek Synergy 2 plate reader at 28 °C. 10μl aliquots of ligand (GTP, GDP, ATP) in binding buffer at 10× final concentration were pipetted into a 96-well fluorescence plate and 90μl of the GTP sensor protein solution at 1.12μM was added to give a protein concentration of 1μM. The plate was read with excitation/emission at 485/528 and 400/528nm. The ratio of the fluorescence from excitation at the 2 wavelengths was fit to:
where FR=fluorescence ratio (F360/F485) at a given ligand concentration, F0=fluorescence ratio (F360/F485) at zero ligand, FN=fluorescence ratio normalized to 1.0 at zero ligand concentration, Kd=ligand dissociation constant, R=(F0/ F∞), F∞= fluorescence ratio (F360/F485) at infinite ligand concentration. Kinetic assays were carried out similarly, but with the plate reader set to dispense the sensor and immediately start reading fluorescence at 2 second intervals.
pH dependence of the GTP sensors
The pH dependence of the GTP sensors was measured in 100mM KCl, 5mM MgCl2, 10% glycerol with the following buffering agents in the specified pH ranges: MES (pH 5.5-6.5), PIPES (pH 7), Tris-HCl (pH 7.5-8.5), and Glycine-NaOH (pH 9-10); 2μl of 50μM protein solutions in water were added to 98μl of the various pH buffers to give a protein concentration of 1μM (+/- 1mM GTP), and fluorescence was measured as described for the ligand binding assays.
Dynamic Light Scattering
Dynamic light-scattering measurements were done on a DynaPro Protein Solutions instrument using 50μl of a 20μM protein solution and 104 seconds of acquisition time. Hydrodynamic radii (Rh) and MW were calculated using the DynaPro software assuming an isotropic sphere model and buffer viscosity set to phosphate-buffered saline (PBS) at 25° C.
Cell Lines
SK-Mel-103 human melanoma cell lines were obtained from Sloan Kettering Memorial Cancer Center; HEK293FT were purchased from Clontech (Palo Alto, CA, USA). Cells were cultured in DMEM (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal calf serum, 2mM glutamine and penicillin-streptomycin antibiotics. Cells were kept at 37°C under an atmosphere of 5% carbon dioxide. For cell imaging, 1hr prior to the imaging cells were rinsed once with PBS and supplemented with an immunofluorescence-compatible media (IFM): FluoroBrite DMEM (Invitrogen) supplemented with 4mM Glutamine, 0.5% FBS, and 20mM Hepes pH7.4. Cell lines have been recently authenticated and verified for being mycoplasma-free using MycoAlert mycoplasma detection Kit purchased from Lonza (Allendale, NJ, USA, Cat # LT07-318).
Antibodies and other reagents
Mycophenolic acid, mizoribine, and guanosine were purchased from Sigma-Aldrich (St. Louis, MO, USA). The following antibodies were used: mouse monoclonal to GAPDH (Cat. # AM4300, Ambion, Austin,TX, USA); rabbit polyclonal to IMPDH2 (Cat # HPA001400, Sigma-Aldrich); and mouse monoclonal to GFP/YFP (Cat # sc-9996, Santa Cruz Biotechnology, Santa Cruz, CA, USA). Nunc Lab-Tek chambered coverglass 1.5mm were purchased from Thermo Fisher Scientific (Fair Lawn, NJ, USA); 1.5mm glass bottom 96well culture plates were purchased from Mattek (Ashland, MA).
Immunoblotting
Whole cell extracts were prepared and analyzed as previously described21.
Plasmids and Infection
The GW1-pHRed, pCMVdeltaR8.2 and pCMV-VSV-G vectors were purchased from Addgene (Cambridge, MA, USA). The pLV-SV4-puro lentiviral vector was obtained from Dr. Peter Chumakov, Cleveland Clinic (Cleveland, OH, USA). Lentiviral vectors encoding short-harpin RNA (shRNA) to IMPDH2 along with a non-silencing control vector were purchased from Sigma and described previously21. The GEVAL_sensors were PCR-amplified from the bacterial expression vectors with the following primers and cloned into the pLVp lentiviral expression system.
GEVAL_FWD (XhoI) CATTGctcgagATGAAAAAACTGACAATCGGACTGA
GEVAL_REV (NheI) CGGTAgctagcTTAGGACACGACATCGC
The IMPDH2-shRNA resistant vector was created with the Q5 site-directed mutagenesis kit (New England BioLabs, NEB, Ipswich, MA, USA) from our previously generated IMPDH2 lentiviral expression vector24, using the following primers and following the manufacturer's instructions.
FWD primer gtaccgaggaATGGGTTCTCTCGATGCC
REV primer tttttgagtcgGATCCCATCGGAAAAGAAG
The lentiviral infection protocol was described previously32. All infected cells were briefly selected for resistance to puromycin and used in the described assays.
Nucleotide Quantification
Nucleotide quantification was performed as previously described21,33. Briefly, cells were harvested by trypsinization, extracted with 0.4N perchloric acid and neutralized. NTPs were separated and quantified using a strong anion exchange column (Sigma Aldrich, St. Louis, MO) assembled on the Waters 2796 Bioseparation module controlled by Empower 3 software at the Bioanalytics, Metabolomics, and Pharmacokinetics Core Facility at Roswell Park Cancer Institute. Nucleotides were eluted with a linear gradient of 80% 0.005M ammonium phosphate, pH 2.8, and 20% 0.75M ammonium phosphate, pH 3.7, to 20% 0.005M ammonium phosphate, pH 2.8 and 80% 0.75 M, pH 3.7 over 30 minutes which was then maintained for an additional 5 minutes. The mobile phase was then returned to starting conditions over 10 minutes, and equilibrated for an additional 30 minutes before the next injection. Nucleotides were identified based on their UV absorbance spectrum and quantified at either 254 or 281 nm by comparison to the absorbance of a known amount of authentic standard.
Cell imaging acquisition and analysis
Cells were imaged with a Leica AOBS SP5 confocal equipped with a multi-line Argon laser, a 405 diode laser, and a 594 helium-neon laser, with a 40× immersion lens (except for the HTP screening test, were a 20× dry lens was used). The microscope is regularly checked by Leica representatives during visits covered by the service contract. The GTP sensors were excited sequentially with the 405nm wavelength of the diode laser and the 488nm line of the argon laser and fluorescent emission was collected from 502nm to 544nm. The pHRed sensor was excited sequentially with the 458nm line of the argon laser and the 594nm helium-neon laser and fluorescent emission was collected from 598nm to 647nm. Thus, each acquired image contained 4 channels for the 4 excitation/emission combinations: Ex405/Em530 and Ex488/Em530 for the GTP sensors, and Ex458/Em630 and Ex594/Em630 for the pHRed. One hour prior to the imaging, cells were rinsed with PBS and replenished with the immunofluorescence-compatible media (IFM) and when necessary, corresponding amounts of drugs or vehicle control, to equilibrate the cells with the IFM. A minimum of 3 images/population/treatment was captured and the ratiometric analysis was performed on 30 cells/ population/treatment with ImageJ (NIH) by outlining the individual cells and measuring the mean fluorescence for each channel for each cell.. We then calculated the Ex405/Ex488 Ex/Em ratio for the GTP sensors and Ex594/Ex458 Ex/Em ratio for pHRed. The channels were imaged sequentially to minimize the amount of bleed-through. When expressed in conjunction with pHRed, 9% of the Ex458 channel values were subtracted from the Ex405 prior to analysis; however, when normalized on control, corrected and uncorrected images gave nearly identical results. To extrapolate the approximate pH from the Ex594/Ex458 Ex/Em ratio we interpolated data provided by Dr. Gary Yellen to create a regression curve where the “Y” value is the Ex594/Ex458 ratio and the “X” value is the pH based on the wavelengths in use in our system. The calculated ratios were then used to determine the approximate pH for each cell, an approach that has been proven to be quite reliable17. For the gradient imaging, we cropped regions containing single cells and performed a pixel by pixel ratio between the two channels with the RatioPlus plugin, with a background subtraction of 10 in the numerator image and a multiplication factor of 2.0. The ratio images were then subjected to a 0.5% contrast enhancement and false-colored with the FIRE LUT. When comparing ratios across the three sensors or during time course, the images where blended in one stack in ImageJ so that the enhancement manipulation and LUT could be applied uniformly to all images at once. For the short term treatment with MPA, vehicle- and MPA-treated cells were pre-treated over-night with 0.5mM N-acetyl-cysteine (NAC) to prevent potential short-term induction of ROS by MPA34,35. For the bleaching experiment, the same group of cells was sequentially imaged every 30sec. The Ex405/Ex488 ratio for each time point was calculated and normalized on the ratio of t0. For the continuous time-lapse analysis, the values of GEVAL30 and GEVAL530 were normalized by those of GEVALNull in the same conditions. For HTP analysis, the Z' score was calculated using the formula Z'=(1-(3*(σs+σc)/|μs-μc|)36. A Z' factor values within [0.5 and 1], [0 and 0.5] or below 0 indicate an optimal, a marginal or a non-useful assay, respectively.
Statistical Analysis
Each experiment was performed at least two independent times (normally between 2 and 4 independent experiments, the exact “n” is specified in each figure legend). The modality of data representation (i.e. dot-plots of all experiments, bar-and-whiskers graphs, etc) is described in each figure legend. Statistical analysis was performed using two-tailed Student t-Test (or, where indicated, two-tailed Mann-Whitney test). A two-tailed p value<0.05 was considered statistically significant for all analyses and the exact p value for each comparison is reported within each figure.
Data availability
The data that support the findings of this study are available from the corresponding authors upon reasonable request.
Additional information can be found in the associated Life Sciences Reporting Summary.
Supplementary Material
Acknowledgments
This work has been supported by NIH grants CA151128 and GM118933 (to R.S.), CA197996 (to D.J.S.), CA120244, CA193981, and CA190533 (to M.A.N.), Ruth L. Kirschstein National Research Service Award F32CA189622 (to A.B-S.), National Institute of Health 1F99CA21245501 (to H.C.A.); Empire State Development Corporation Krabbe Disease Research Working Capital X561 and Krabbe Disease Research Capital Equipment U446 (M.L.F.), and by the Jennifer Linscott Tietgen Foundation (M.A.N.).
Footnotes
Author contributions: A.B-S., M.R., R.S., and M.A.N. designed the experiments and wrote the manuscript; A.B-S. and M.R. performed most of the experiments and analyzed the data; C.E.F., L.M.R., S.M, K.M., E.E.F, and A.B, performed some of the experiments; H.C.A. performed HPLC analysis, E.H. assisted with the microscopy acquisition and analysis; D.J.S., A.V.B., E.S.K, and M.L.F. supervised part of the study; R.S. and M.A.N conceived the initial hypothesis and supervised the study. A.B-S. and M.R. contributed equally to this study. All authors discussed the results and commented on the manuscript.
Competing financial interests: The authors declare no competing financial interests.
References
- 1.Shoji S, Walker SE, Fredrick K. Ribosomal translocation: one step closer to the molecular mechanism. ACS Chem Biol. 2009;4:93–107. doi: 10.1021/cb8002946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simon MI, Strathmann MP, Gautam N. Diversity of G Proteins in Signal Transduction. Science. 1991;252:802–808. doi: 10.1126/science.1902986. [DOI] [PubMed] [Google Scholar]
- 3.Neves SR, Ram PT, Iyengar R. G protein pathways. Science. 2002;296:1636–1639. doi: 10.1126/science.1071550. [DOI] [PubMed] [Google Scholar]
- 4.MacMicking JD. IFN-inducible GTPases and immunity to intracellular pathogens. Trends Immunol. 2004;25:601–609. doi: 10.1016/j.it.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 5.Kresse A, et al. Analyses of murine GBP homology clusters based on in silico, in vitro and in vivo studies. BMC Genomics. 2008;9:158. doi: 10.1186/1471-2164-9-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vestal DJ, Jeyaratnam JA. The guanylate-binding proteins: emerging insights into the biochemical properties and functions of this family of large interferon-induced guanosine triphosphatase. J Interferon Cytokine Res. 2011;31:89–97. doi: 10.1089/jir.2010.0102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Downward J. Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer. 2003;3:11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- 8.Guilfoyle A, et al. Structural basis of GDP release and gating in G protein coupled Fe2+ transport. Embo J. 2009;28:2677–2685. doi: 10.1038/emboj.2009.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marlovits TC, Haase W, Herrmann C, Aller SG, Unger VM. The membrane protein FeoB contains an intramolecular G protein essential for Fe(II) uptake in bacteria. Proc Natl Acad Sci U S A. 2002;99:16243–16248. doi: 10.1073/pnas.242338299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berg J, Hung YP, Yellen G. A genetically encoded fluorescent reporter of ATP:ADP ratio. Nature methods. 2009;6:161–166. doi: 10.1038/nmeth.1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Imamura H, et al. Visualization of ATP levels inside single living cells with fluorescence resonance energy transfer-based genetically encoded indicators. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:15651–15656. doi: 10.1073/pnas.0904764106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Traut TW. Physiological concentrations of purines and pyrimidines. Molecular and Cellular Biochemistry. 1994;140:1–22. doi: 10.1007/BF00928361. [DOI] [PubMed] [Google Scholar]
- 13.Matsushika A, Nagashima A, Goshima T, Hoshino T. Fermentation of xylose causes inefficient metabolic state due to carbon/energy starvation and reduced glycolytic flux in recombinant industrial Saccharomyces cerevisiae. PLoS ONE. 2013;8:e69005. doi: 10.1371/journal.pone.0069005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Romani AM. Cellular magnesium homeostasis. Archives of biochemistry and biophysics. 2011;512:1–23. doi: 10.1016/j.abb.2011.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Marlovits TC, Haase W, Herrmann C, Aller SG, Unger VM. The membrane protein FeoB contains an intramolecular G protein essential for Fe(II) uptake in bacteria. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:16243–16248. doi: 10.1073/pnas.242338299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Palmer AE, Qin Y, Park JG, McCombs JE. Design and application of genetically encoded biosensors. Trends in Biotechnology. 2011;29:144–152. doi: 10.1016/j.tibtech.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tantama M, Hung YP, Yellen G. Imaging intracellular pH in live cells with a genetically encoded red fluorescent protein sensor. J Am Chem Soc. 2011;133:10034–10037. doi: 10.1021/ja202902d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ransom JT. Mechanism of action of mycophenolate mofetil. Therapeutic drug monitoring. 1995;17:681–684. doi: 10.1097/00007691-199512000-00023. [DOI] [PubMed] [Google Scholar]
- 19.Sievers TM, et al. Mycophenolate mofetil. Pharmacotherapy. 1997;17:1178–1197. [PubMed] [Google Scholar]
- 20.Turka LA, Dayton J, Sinclair G, Thompson CB, Mitchell BS. Guanine ribonucleotide depletion inhibits T cell activation. Mechanism of action of the immunosuppressive drug mizoribine. J Clin Invest. 1991;87:940–948. doi: 10.1172/JCI115101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wawrzyniak JA, et al. A purine nucloetide biosynthesis enzyme guanosine monophosphate reductase is a suppressor of melanoma invasion. Cell Reports. 2013;5:493–507. doi: 10.1016/j.celrep.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gu JJ, et al. Induction of apoptosis in IL-3-dependent hematopoietic cell lines by guanine nucleotide depletion. Blood. 2003;101:4958–4965. doi: 10.1182/blood-2002-08-2547. [DOI] [PubMed] [Google Scholar]
- 23.Laliberte J, Yee A, Xiong Y, Mitchell BS. Effects of guanine nucleotide depletion on cell cycle progression in human T lymphocytes. Blood. 1998;91:2896–2904. [PubMed] [Google Scholar]
- 24.Mannava S, et al. Direct role of nucleotide metabolism in C-MYC-dependent proliferation of melanoma cells. Cell Cycle. 2008;7:2392–2400. doi: 10.4161/cc.6390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buckstein MH, He J, Rubin H. Characterization of nucleotide pools as a function of physiological state in Escherichia coli. J Bacteriol. 2008;190:718–726. doi: 10.1128/JB.01020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen P, et al. A LC-MS/MS method for the analysis of intracellular nucleoside triphosphate levels. Pharmaceutical research. 2009;26:1504–1515. doi: 10.1007/s11095-009-9863-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qiu Y, et al. Mycophenolic acid-induced GTP depletion also affects ATP and pyrimidine synthesis in mitogen-stimulated primary human T-lymphocytes. Transplantation. 2000;69:890–897. doi: 10.1097/00007890-200003150-00038. [DOI] [PubMed] [Google Scholar]
- 28.Wang Y, Tang L, Li Z, Lin Y, Li J. In situ simultaneous monitoring of ATP and GTP using a graphene oxide nanosheet-based sensing platform in living cells. Nat Protoc. 2014;9:1944–1955. doi: 10.1038/nprot.2014.126. [DOI] [PubMed] [Google Scholar]
- 29.Sumita K, et al. The Lipid Kinase PI5P4Kbeta Is an Intracellular GTP Sensor for Metabolism and Tumorigenesis. Molecular cell. 2016;61:187–198. doi: 10.1016/j.molcel.2015.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Takeuchi K, et al. Structural reverse genetics study of the PI5P4Kbeta-nucleotide complexes reveals the presence of the GTP bioenergetic system in mammalian cells. FEBS J. 2016;283:3556–3562. doi: 10.1111/febs.13739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berg J, Hung YP, Yellen G. A genetically encoded fluorescent reporter of ATP:ADP ratio. Nat Methods. 2009;6:161–166. doi: 10.1038/nmeth.1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mannava S, et al. Depletion of deoxyribonucleotide pools is an endogenous source of DNA damage in cells undergoing oncogene-induced senescence. Am J Pathol. 2013;182:142–151. doi: 10.1016/j.ajpath.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bianchi-Smiraglia A, et al. Microphthalmia-associated transcription factor suppresses invasion by reducing intracellular GTP pools. Oncogene. 2016 doi: 10.1038/onc.2016.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huh KH, et al. The role of thioredoxin 1 in the mycophenolic acid-induced apoptosis of insulin-producing cells. Cell Death Dis. 2013;4:e721. doi: 10.1038/cddis.2013.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Malekinejad H, Moradi M, Fink-Gremmels J. Cytochrome C and Caspase-3/7 are Involved in Mycophenolic Acid- Induced Apoptosis in Genetically Engineered PC12 Neuronal Cells Expressing the p53 gene. Iran J Pharm Res. 2014;13:191–198. [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang JH, Chung TD, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding authors upon reasonable request.
Additional information can be found in the associated Life Sciences Reporting Summary.