Abstract
Background
The findings from studies on the relationship between aldehyde dehydrogenases(ALDH) gene Glu504Lys polymorphism and colorectal cancer(CRC) were inconsistent.
Objectives
The aim of this meta-analysis was to assess ALDH gene Glu504Lys polymorphism and CRC risk.
Methods
All of the relevant studies were identified from PubMed and Embase database. Statistical analyses were conducted with STATA 12.0 software. Odds ratio (OR) with 95% confidence interval (CI) values were applied to evaluate the strength of the association. Nine studies with 2779 cases and 4533 controls were included.
Results
No significant variation in CRC risk was detected in any of the genetic models overall. To explore the sources of heterogeneity,we performed further sub-group analyses by ethnicity and quality assessment of these studies. In the sub-group analysis by race, significant associations between ALDH gene Glu504Lys polymorphism and CRC risk were found in China(Glu/Lys vs Glu/Glu: OR = 0.70, 95%CI = 0.57–0.85; the dominant model: OR =0.69, 95%CI =0.48–0.98) and Japan(Lys/Lys vs Glu/Glu:OR =0.72, 95%CI =0.55–0.95).
Conclusion
This meta-analysis suggests that the ALDH2 Glu504Lys polymorphism may be associated with susceptibility to CRC. Furthermore, large and well-designed studies are needed to confirm these conclusions.
Keywords: Aldehyde dehydrogenases, colorectal cancer, polymorphism, susceptibility
Introduction
Colorectal cancer (CRC) is one of the most prevailing cancers occurring in the digestive system,and it is ranked as the third worldwide primary cancer-causing death worldwide1. The highest morbidity of CRC is discovered in Australia, Europe, and North America. In addition, the morbidity of CRC is rapidly increasing in many countries within Eastern Asia2. CRC is a multi-step process and multiple factors are involved. Although etiology of CRC is still not fully understood, cumulative evidence demonstrates that the environment, life style, diet habit, and other environmental factors contribute to the pathogenesis of CRC in different degrees3. In recent years, molecular epidemiological studies have manifested that single nucleotide polymorphisms (SNPs) in genes play a key role in CRC development and progression4. Genome-wide association studies (GWASs) have showed that genetic factors accounted for 33% of CRC cases in the world5,6.
The primary enzymes involved in alcohol metabolism are alcohol dehydrogenases (ADH) and aldehyde dehydrogenases(ALDH)7. ALDH2 is the key enzyme responsible for acetaldehyde elimination, and its polymorphic variants determine blood acetaldehyde concentrations after drinking. Therefore, genetic variants that result in functional differences in enzyme activity lead to differences in acetaldehyde exposure between drinkers.Human ALDH2 gene is located on chromosome 12q24.2 and composed of 13 exons, spanning 46,031 bp8. A common SNP is at exon 12 results from the substitution of glutamate(-Glu) to lysine (Lys) at residue 487 appropriately named Glu504Lys(NCBI ID: rs671), which encodes a catalytically inactive subunit of ALDH2. The variant 504lys allele decreases the activity of ALDH29, and ALDH2 Glu/Lys genotype has only 6.25% of the normal ALDH2 Glu proteins10. Previous meta-analysis suggested that ALDH2 Glu504Lys polymorphism may play crucial roles in the pathogenesis of gastric cancer11.
To date, many studies have investigated the interrelation between ALDH2 Glu504Lys polymorphism and CRC risk. However, the results have been inconsistent. One of the most potential reasons is that the statistical power is limited for the individual studies. The present comprehensive meta-analysis of currently published studies aimed to identify the relationships between the ALDH2 Glu504Lys polymorphism and the risk of CRC.
Materials and methods
Search strategy
Extensive literature was retrieved using the PubMed and EMBASE databases for relevant studies published (the last search update was June 15, 2016) with the following key words: (colorectal cancer odds ratio [OR] colorectal carcinoma) AND (ALDH or aldehyde dehydrogenases) AND (polymorphisms OR variant). The search was not limited to language.We contacted the corresponding author for specific raw data if the data presented in the article were not sufficient.When overlapping data existed, only the latest study with the largest sample was searched for this meta-analysis.
Inclusion criteria
The included studies met all the following inclusion criteria:( a) evaluated the associations of ALDH2 Glu504Lys polymorphism and CRC risk; (b) used case-control or cohort design; and (c) provide sufficient data for calculation of odds ratio (OR) with 95% confidence interval (CI). In addition,the following exclusion criteria were also used: (a) no usable data reported; (b) the study only involved a case population; (c) not concerned with CRC risk; and (d) duplicate publications.
Data extraction
The following information was extracted from each study: name of the first author; year of publication; country of origin; ethnicity; number of subjects in each category, and the counts of persons with different genotypes in cases and controls. Information on the Hardy-Weinberg equilibrium test (HWE) was also calculated.
Quality assessment
The quality of these articles was also evaluated independently by the two investigators according to the predefined quality assessment rules in Table 112. The criteria cover the representativeness of patients, source of controls, ascertainment of cancer, total sample size, quality control of genotyping methods, and Hardy-Weinberg equilibrium(HWE) in the control population. Disagreements were resolved by consensus. The total score ranged from 0 (worst) to 15 (best). Papers scoring <10 were classified as “low quality” and those scoring ≥10 as “high quality.”
Table 1.
Scale for quality assessment
| Criteria | Score |
| Source of cases | |
| Selected from population or cancer registry Selected from hospital Selected from pathological archives, but without a description Not described |
3 2 1 0 |
| Source of controls | |
| Population-based Blood donors or volunteers Hospital-based (cancer-free patients) Not described |
3 2 1 0 |
| Specimens obtained from patients to determine genotypes | |
| White blood cells or normal tissues Tumor tissues or exfoliated cells of tissue |
3 0 |
| Hardy-Weinberg equilibrium in controls | |
| Hardy-Weinberg equilibrium Hardy-Weinberg disequilibrium |
3 0 |
| Total sample size | |
| ≥ 1000 ≥ 500 but < 1000 ≥ 200 but < 500 > 0 but < 200 |
3 2 1 0 |
Statistical analysis
All the data was analyzed using Stata 12.0 software (Stata Corp., LP, http://www.stata.com; Stata Corporation, College Station, TX). The association strength between ALDH2 Glu504Lys polymorphism and CRC risks was measured by OR with 95%CI).We examined ALDH2 Glu504Lys polymorphism using the homozygote comparison (Lys/Lys vs Glu/Glu), heterozygote comparison (Glu/Lys vs Glu/Glu),dominant (Lys/Lys+Glu/Lys vs Glu/Glu) and recessive (Lys/Lys vs Glu/Lys+Glu/Glu) models. We also quantified the effects of heterogeneity using the I2 test. When a significant I2>50% indicated heterogeneity across studies,either the random effects model or the fixed effects model was performed13. Subgroup analyses were performed by ethnicity and quality assessment. A sensitivity analysis was used to evaluate the stability of the results.Publication bias was investigated using Begg's funnel plot and Egger's regression test (p<0.05 was considered to be statistically significant).
Results
Study characteristics
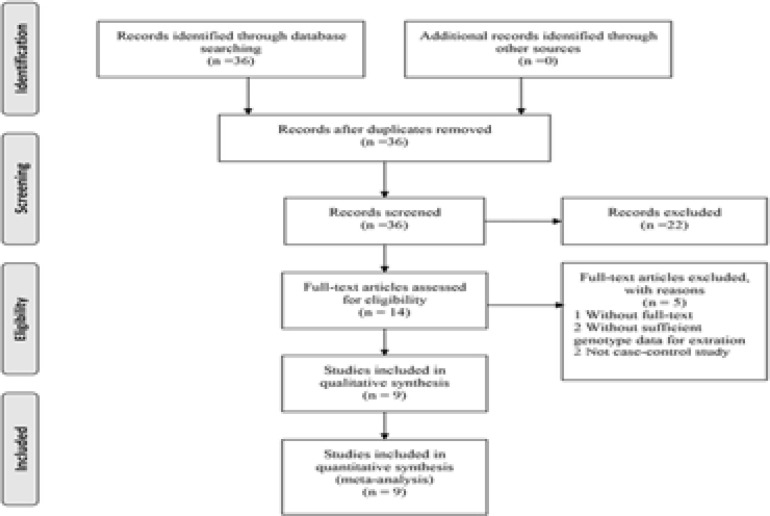
The search strategy searched 36 potentially relevant articles. According to the inclusion criteria and exclusion criteria, nine studies with full text were included in the meta-analysis and 27 studies were excluded14–22. These 9 case-control studies selected included a total of 2779 cases and 4533 healthy controls. A flow chart of the study selection process is summarized in Figure 1.
Figure 1.
PRISMA Flow Diagram of the meta-analysis.
The publication years ranged from 1998 to 2012. All the studies were published in English. The genotype distributions in the controls of all studies were in agreement with HWE. General characteristics and the allele and genotype distributions in the published articles included in this meta-analysis are shown in Table 2.
Table 2.
Characteristics of the included studies for meta-analysis
| Study | Area | Cases/Controls | Genotypes for Genotypes for controls cases |
HWE test |
Quality scores |
|||||
| Glu/Glu Glu/Lys Lys/Lys |
Glu/Glu Glu/Lys Lys/Lys | |||||||||
| Chiang 2012 |
China | 545/103 | 304 | 218 | 23 | 43 | 53 | 7 | 0.08 | 11 |
| Yang 2009 |
China | 426/785 | 274 | 119 | 33 | 489 | 261 | 35 | 0.98 | 12 |
| Gao 2008 | China | 190/222 | 131 | 54 | 5 | 123 | 90 | 9 | 0.13 | 12 |
| Yin 2007 | Japan | 685/778 | 400 | 257 | 28 | 416 | 309 | 53 | 0.67 | 13 |
| Matsuo 2006 |
Japan | 257/768 | 129 | 104 | 24 | 383 | 314 | 71 | 0.57 | 12 |
| Otani 2005 |
Japan | 106/224 | 61 | 36 | 9 | 137 | 72 | 15 | 0.20 | 10 |
| Kuriki 2005 |
Japan | 72/116 | 45 | 24 | 3 | 64 | 44 | 8 | 0.91 | 9 |
| Hirose 2005 |
Japan | 452/1050 | 299 | 137 | 16 | 605 | 390 | 55 | 0.44 | 12 |
| Yokoyama 1998 |
Japan | 46/487 | 36 | 10 | 0 | 443 | 44 | 0 | 0.30 | 11 |
Meta-analysis
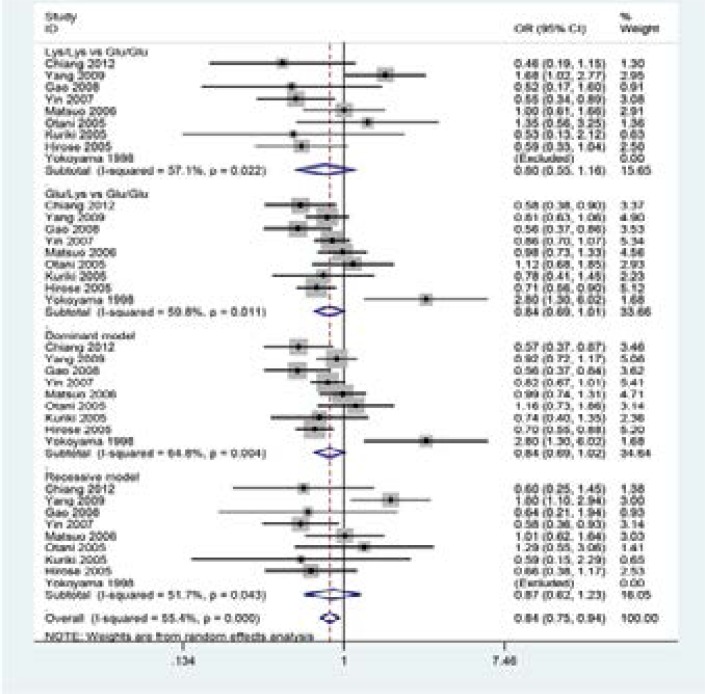
A summary of the meta-analysis findings of the association between ALDH2 Glu504Lys polymorphism and CRC risk is shown in Figure 2–4 and Table 3. When all eligible studies were pooled into one dataset for the meta-analysis,we found no statistical association between the ALDH2 Glu504Lys polymorphism and CRC risk(Lys/Lys vs Glu/Glu: OR =0.80, 95%CI =0.55–1.16; Glu/Lys vs Glu/Glu: OR = 0.84, 95%CI = 0.69–1.01; the dominant model: OR =0.84, 95%CI =0.69–1.02; the recessive model: OR =0.87, 95%CI = 0.62–1.23). Sensitivity analyses were performed by altering the statistic models. No material alteration was detected, indicating that our results were statistically robust.
Figure 2.
Forest plot of CRC risk associated with ALDH2 Glu504Lys polymorphism in the total population.
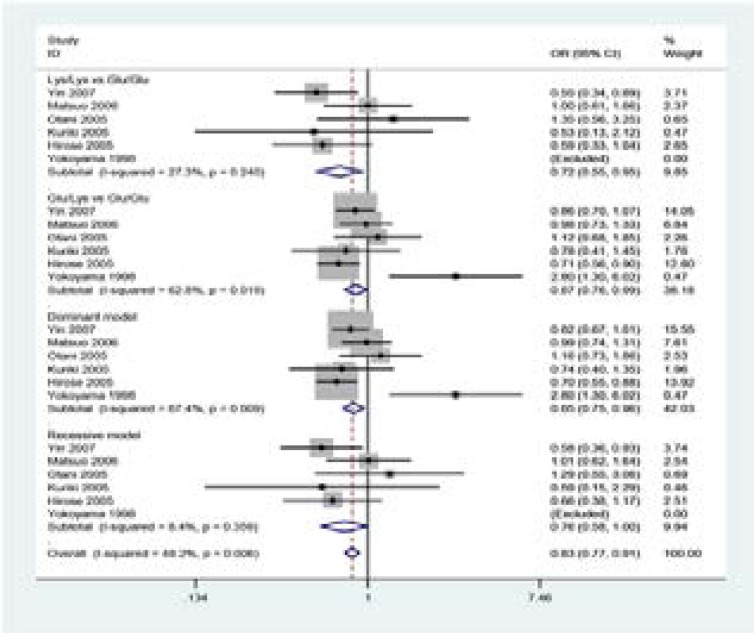
Figure 4.
Forest plot of CRC risk associated with ALDH2 Glu504Lys polymorphism in Japan
Table 3.
Summary ORs and 95%CI of ALDH2 Glu504Lys polymorphism with colorectal cancer risk.
| Variables | N a | Lys/Lys vs Glu/Glu | Glu/Lys vs Glu/Glu | Dominant model | Recessive model |
| OR(95%CI) I2(%) | OR(95%CI) I2(%) | OR(95%CI) I2(%) | OR(95%CI) I2(%) | ||
| Total | 9 | 0.80(0.55–1.16) 57.1 | 0.84(0.69–1.01) 59.8 | 0.84(0.69–1.02) 64.8 | 0.87(0.62–1.23) 51.7 |
| Ethnicity | |||||
| China | 3 | 0.81(0.31–2.08) 75.0 | 0.70(0.57–0.85) 33.0 | 0.69(0.48–0.98) 67.7 | 0.97(0.43–2.20) 67.6 |
| Japan | 6 | 0.72(0.55–0.95) 27.3 | 0.95(0.74–1.21) 62.8 | 0.94(0.73–1.20) 67.4 | 0.76(0.58–1.00) 8.4 |
| Quality | |||||
| High-quality | 8 | 0.81(0.55–1.21) 62.3 | 0.84(0.69–1.03) 64.8 | 0.85(0.69–1.04) 69.1 | 0.89(0.61–1.28) 57.4 |
| Low-quality | 1 | / | / | / | / |
Number of comparisons.
Figure 3.
Forest plot of CRC risk associated with ALDH2 Glu504Lys polymorphism in China.
To explore the sources of heterogeneity, we performed further sub-group analyses by ethnicity and quality assessment of included studies. In sub-group analysis by ethnicity, the studies included were divided into China and Japan,and significant association was found between ALDH2 Glu504Lys polymorphism and CRC in China(-Glu/Lys vs Glu/Glu: OR = 0.70, 95%CI = 0.57–0.85; the dominant model: OR =0.69, 95%CI =0.48–0.98) and Japan(Lys/Lys vs Glu/Glu: OR =0.72, 95%CI =0.55–0.95). When stratified according to high-quality studies, ALDH2 Glu504Lys polymorphism was not associated with CRC(Table 3).
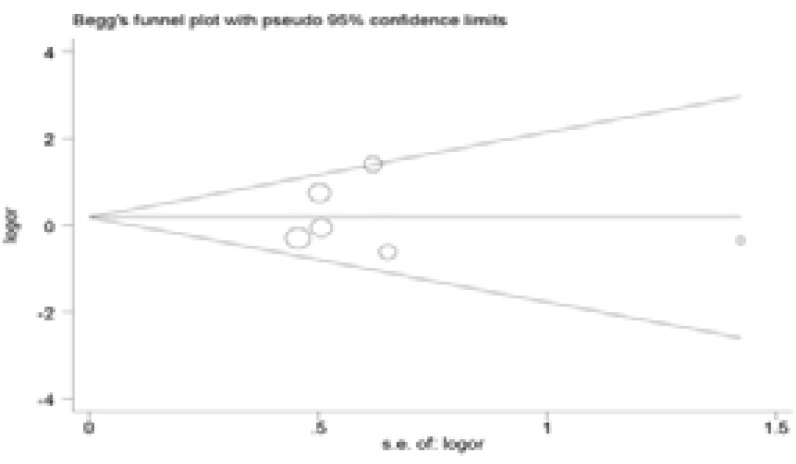
Publication bias
The Begg's funnel plot was performed to assess the publication bias in the reports included for meta-analysis. The shapes of the funnel plots in all genetic models did not reveal any evidence of obvious asymmetry (Figure 5). And Egger's line regression test indicated no significant publication bias (P > 0.05).
Figure 5.
Begg's funnel plot for evaluation of publication bias.
Discussion
CRC is one of the most common malignancies and represents a significant cause of morbidity and mortality worldwide. Previous epidemiological evidence demonstrated that alcohol intake is associated with increased CRC risk23. After alcohol is absorbed, the concentration of alcohol in the colon is as high as that found in the blood24. Most of the acetaldehyde generated during alcohol metabolism is promptly eliminated by ALDH. Previous animal experiments have shown that when ALDH activity was inhibited, tumorigenesis was observed in the colon of rats25. Yokoyama et al. first examined the association between ALDH2 Glu504Lys and colon cancer among Japanese alcoholics and reported a 3-fold increased risk among heterozygotes22. Later, several studies, but not all, showed an association of ALDH2 Glu504Lys polymorphism with increased risk for CRC. Furthermore, these results above were based on a small sample size, it is possible that the observed associations reflect chance observations rather than true associations. To help resolve these conflicting results, we conducted this meta-analysis to get a more authentic result.
The aim of our meta-analysis was to combine the same kind of studies to increase the sample size and statistical power, and thereby get a more authentic result. Finally, 9 case-control studies were included and assessed, involving a total of 2779 cases and 4533 healthy controls. Eventually, the results of the present meta-analysis revealed that ALDH2 Glu504Lys polymorphism is not associated with increased or decreased risk of CRC in the overall population. Nevertheless, considering that other potential confounding factors would influence the result, we further conducted several sub-group analyses. In the stratified analysis by ethnicity, results showed significant association in both China and Japan. This might account for ethnic differences and diverse live environment. The mechanism by which the Glu504Lys polymorphism affects CRC risk is currently unclear. As with other diseases, the development of CRC is due to the joint effect of multiple genes and gene-environment interactions. Matsuo et al. found that heavier drinking, risk genotypes(ADH2 47His and ALDH2 487Lys), and low folate consumption might synergistic increase the risk of CRC18. As the eligible study number was limited in the meta-analysis, these results still need further investigation.
Limitations
Some limitations of our meta-analysis should be addressed. First of all, studies included in the present meta-analysis mainly provided data towards Asians, other ethnicities including Caucasian, and others should be investigated in future studies. Second, most of the men had a high frequency of drinking, while the majority of women had a low frequency of drinking, which may affect the final results. Further stratification analysis should aim at gender. Third, the existence of gene-gene and gene-environment interactions may affect the accuracy of our results. Lack of the relevant data limited our further analysis of potential interactions among gene-gene and gene-environment.
Conclusion
This meta-analysis suggested that the ALDH2 Glu504Lys polymorphism may be associated with CRC risk. Large- scale case-control and population-based association studies are warranted to validate the risk identified in the current meta-analysis and investigate the potential gene-gene and gene-environment interactions on CRC risk.
Conflict of interest
None to declare.
References
- 1.Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin. 2014;64:104–117. doi: 10.3322/caac.21220. PubMed. [DOI] [PubMed] [Google Scholar]
- 2.Potter JD. Colorectal cancer: molecules and populations. J Natl Cancer Inst. 1999;91:916–932. doi: 10.1093/jnci/91.11.916. PubMed. [DOI] [PubMed] [Google Scholar]
- 3.Jemal A1, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. PubMed. [DOI] [PubMed] [Google Scholar]
- 4.Gao X, Duan H, Zhu Z. Association between PTEN IVS4 polymorphism and cancer risk: a meta-analysis. Cancer Biomark. 2013;13:465–470. doi: 10.3233/CBM-130386. PubMed. [DOI] [PubMed] [Google Scholar]
- 5.Lichtenstein P, Holm NV, Verkasalo PK, et al. Environmental and heritable factors in the causation of cancer-analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000;343:78–85. doi: 10.1056/NEJM200007133430201. PubMed. [DOI] [PubMed] [Google Scholar]
- 6.Whiffin N, Hosking FJ, Farrington SM, et al. Identification of susceptibility loci for colorectal cancer in a genome-wide meta-analysis. Hum Mol Genet. 2014;23:4729–4737. doi: 10.1093/hmg/ddu177. PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edenberg HJ. The genetics of alcohol metabolism: role of alcohol dehydrogenase and aldehyde dehydrogenase variants. Alcohol Res Health. 2007;30:5e13. [PMC free article] [PubMed] [Google Scholar]
- 8.Yoshida A, Rzhetsky A, Hsu LC, Chang C. Human aldehyde dehydrogenase gene family. Eur J Biochem. 1998;251:549–557. doi: 10.1046/j.1432-1327.1998.2510549.x. PubMed. [DOI] [PubMed] [Google Scholar]
- 9.Wang RS, Nakajima T, Kawamoto T, Honma T. Effects of aldehyde dehydrogenase-2 genetic polymorphisms on metabolism of structurally different aldehydes in human liver. Drug Metab Dispos. 2002;30:69e73. doi: 10.1124/dmd.30.1.69. [DOI] [PubMed] [Google Scholar]
- 10.Crabb DW, Edenberg HJ, Bosron WF, Li TK. Genotypes for aldehyde dehydrogenase deficiency and alcohol sensitivity. The inactive ALDH2(2) allele is dominant. J Clin Invest. 1989;83:314–316. doi: 10.1172/JCI113875. PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang HL, Zhou PY, Liu P, Zhang Y. ALDH2 and ADH1 genetic polymorphisms may contribute to the risk of gastric cancer: a meta-analysis. PLoS One. 2014;9:e88779. doi: 10.1371/journal.pone.0088779. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 12.Jiang DK, Ren WH, Yao L, et al. Meta-analysis of association between TP53 Arg72Pro polymorphism and bladder cancer risk. Urology. 2012;76:765, e1–e7. doi: 10.1016/j.urology.2010.04.044. PubMed. [DOI] [PubMed] [Google Scholar]
- 13.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. PubMed. [DOI] [PubMed] [Google Scholar]
- 14.Chiang CP, Jao SW, Lee SP, et al. Expression pattern, ethanol-metabolizing activities, and cellular localization of alcohol and aldehyde dehydrogenases in human large bowel: association of the functional polymorphisms of ADH and ALDH genes with hemorrhoids and colorectal cancer. Alcohol. 2012;46:37–49. doi: 10.1016/j.alcohol.2011.08.004. PubMed. [DOI] [PubMed] [Google Scholar]
- 15.Yang H, Zhou Y, Zhou Z, et al. A novel polymorphism rs1329149 of CYP2E1 and a known polymorphism rs671 of ALDH2 of alcohol metabolizing enzymes are associated with colorectal cancer in a southwestern Chinese population. Cancer Epidemiol Biomarkers Prev. 2009;18:2522–2527. doi: 10.1158/1055-9965.EPI-09-0398. [DOI] [PubMed] [Google Scholar]
- 16.Gao CM, Takezaki T, Wu JZ, et al. Polymorphisms of alcohol dehydrogenase 2 and aldehyde dehydrogenase 2 and colorectal cancer risk in Chinese males. World J Gastroenterol. 2008;14:5078–5083. doi: 10.3748/wjg.14.5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yin G, Kono S, Toyomura K, et al. Alcohol dehydrogenase and aldehyde dehydrogenase polymorphisms and colorectal cancer: the Fukuoka Colorectal Cancer Study. Cancer Sci. 2007;98:1248–1253. doi: 10.1111/j.1349-7006.2007.00519.x. PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsuo K, Wakai K, Hirose K, et al. A gene-gene interaction between ALDH2 Glu487Lys and ADH2 His47Arg polymorphisms regarding the risk of colorectal cancer in Japan. Carcinogenesis. 2006;27:1018–1023. doi: 10.1093/carcin/bgi282. PubMed. [DOI] [PubMed] [Google Scholar]
- 19.Otani T, Iwasaki M, Hanaoka T, et al. Folate, vitamin B6, vitamin B12, and vitamin B2 intake, genetic polymorphisms of related enzymes, and risk of colorectal cancer in a hospital-based case-control study in Japan. Nutr Cancer. 2005;53:42–50. doi: 10.1207/s15327914nc5301_5. PubMed. [DOI] [PubMed] [Google Scholar]
- 20.Kuriki K, Hamajima N, Chiba H, et al. Relation of the CD36 gene A52C polymorphism to the risk of colorectal cancer among Japanese, with reference to with the aldehyde dehydrogenase 2 gene Glu487Lys polymorphism and drinking habit. Asian Pac J Cancer Prev. 2005;6:62–68. [PubMed] [Google Scholar]
- 21.Hirose M, Kono S, Tabata S, et al. Genetic polymorphisms of methylenetetrahydrofolate reductase and aldehyde dehydrogenase 2, alcohol use and risk of colorectal adenomas: Self-Defense Forces Health Study. Cancer Sci. 2005;96:513–518. doi: 10.1111/j.1349-7006.2005.00077.x. PubMed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yokoyama A, Muramatsu T, Ohmori T, et al. Alcohol-related cancers and aldehyde dehydrogenase-2 in Japanese alcoholics. Carcinogenesis. 1998;19:1383–1387. doi: 10.1093/carcin/19.8.1383. PubMed. [DOI] [PubMed] [Google Scholar]
- 23.Seitz HK, Simanowski UA, Garzon FT, et al. Possible role of acetaldehyde in ethanol-related rectal cocarcinogenesis in the rat. Gastroenterology. 1990;98:406–413. doi: 10.1016/0016-5085(90)90832-l. PubMed. [DOI] [PubMed] [Google Scholar]
- 24.Erikson CJ. The role of acetaldehyde in the actions of alcohol (update 2000) Alcohol Clin Exp Res. 2001;25:S15–S32. doi: 10.1097/00000374-200105051-00005. [DOI] [PubMed] [Google Scholar]
- 25.Luo HR, Israel Y, Tu GC, et al. Genetic polymorphism of aldehyde dehydrogenase 2 (ALDH2) in a Chinese population: gender, age, culture, and genotypes of ALDH2. Biochem Genet. 2005;43:223–227. doi: 10.1007/s10528-005-5213-8. PubMed. [DOI] [PubMed] [Google Scholar]