Abstract
T-cell acute lymphoblastic leukemia (T-ALL) is an aggressive hematopoietic neoplasm resulting from the malignant transformation of T-cell progenitors, and comprises approximately 15% and 25% of pediatric and adult ALL cases respectively. It is well-established that activating NOTCH1 mutations are the major genetic lesions driving T-ALL in most patients, but efforts to develop targeted therapies against this pathway have produced limited success in decreasing leukemic burden and come with significant clinical side effects. A finer detailed understanding of the genetic and molecular mechanisms underlying T-ALL is required identify patients at increased risk for treatment failure and the development of precision medicine strategies. Generation of genetic models that more accurately reflect the normal developmental history of T-ALL are necessary to identify new avenues for treatment. The DNA methyltransferase enzyme DNMT3A is also recurrently mutated in T-ALL patients, and we show here that inactivation of Dnmt3a combined with Notch1 gain-of-function leads to an aggressive T-ALL in mouse models. Moreover, conditional inactivation of Dnmt3a in mouse hematopoietic cells leads to an accumulation of immature progenitors in the thymus which are less apoptotic. These data demonstrate that Dnmt3a is required for normal T-cell development, and acts as a T-ALL tumor suppressor.
INTRODUCTION
T-cell acute lymphoblastic leukemia (T-ALL) arises from the accumulation of genomic abnormalities that induce aberrant proliferation, increased cell survival, and impaired differentiation of immature T-cell progenitors. Like in many cancers, the mechanisms of T-ALL transformation are similar to those regulating normal developmental processes. Thymocyte development progresses through defined stages of differentiation, beginning with the earliest thymic progenitors (ETPs; Lineage- c-Kit+ CD25−) and progressing through double-negative 2 (DN2; Lineage- c-Kit+ CD25+) and 3 (DN3; Lineage- c-Kit− CD25+) stages before generating mature T-cells (1). The Notch signaling pathway is fundamental for T-lymphopoiesis, and an absolute requirement for T-cell commitment from lymphoid precursors (2). Notch1 is a transmembrane receptor that functions as a ligand-activated transcription factor. Ligand binding to Notch1 receptors leads to cleavage catalyzed by the γ-secretase complex, resulting in release of Notch Intracellular Domain (NICD) which translocates to the nucleus (3) to activate transcription of downstream target genes (4–6). Demonstrating the close link between T-cell development and oncogenesis, the most prevalent genetic lesions in T-ALL patients are activating mutations of NOTCH1, which occur in over 50% of cases (7). The oncogenic capacity of activated NOTCH1 is readily demonstrated in murine systems by the rapid development of T-ALL in retroviral-driven NICD bone marrow transplant models (8).
Epigenetic dysfunction plays a central role in the pathology of many human cancers. Genome sequencing studies have revealed that the de novo DNA methyltransferase enzyme DNMT3A is one of the most recurrently mutated genes across almost all hematopoietic cancers (9, 10), including T-lineage neoplasms such as T-ALL (11, 12) and T-cell lymphoma (13) where it is mutated in 10–18% of patients. The mutation spectrum of DNMT3A in T-ALL is different from that seen in myeloid neoplasms, suggesting distinct mechanisms of transformation (9–11, 14, 15). In T-ALL, DNMT3A mutations frequently associate with NOTCH1 mutations and predict poor clinical outcomes (14). Understanding the synergistic activity between dysregulated epigenetics and signaling pathways could highlight windows of therapeutic vulnerability for precision medicine. In this study, we elucidated the role of Dnmt3a in T-cell development and transformation using genetic mouse models.
METHODS
Mice
Animal procedures were approved by the Institutional Animal Care and Use Committee (IACUC) and conducted in accordance with Washington University School of Medicine institutional guidelines. Mice were C57Bl/6 background, Mx1-Cre:Dnmt3afl/fl and Mx1-Cre:Dnmt3afl/fl;Dnmt3bfl/fl mice have been described (16). Recombination of floxed alleles was mediated by six intraperitoneal injections (300μg/mouse) of polyinosinic-polycytidylic acid (pIpC; Sigma, St. Louis, MO, USA) in PBS every other day. The following strains were obtained from the Jackson Laboratory (Bar Harbor, ME, USA) - Lck-CRE (003802), CD8a-CRE (008766), Rosa26NICD (008159), Rosa26EYFP (006148) and Nr4a1KO (006187).
Viral Transduction and Transplantation
Retroviral transduction of c-Kit-enriched bone marrow (anti-CD117 microbeads; Miltenyi Biotec, Auburn, CA, USA) with NICD in the MSCV-IRES-GFP (MIG) retrovirus (8) was performed as previously described (17). Recipient mice were transplanted by retro-orbital injection following a split dose of 11.0 Gy of irradiation. For T-ALL rescue, Nr4a1 and Dnmt3a were cloned into MSCV-IRES-mCherry (MIC) to transduce primary GFP+ T-ALL cells. 4×104 GFP+mCherry+ cells were transplanted into secondary recipients. For secondary transplantations, leukemic cells were transplanted into sublethally irradiated (6.0 Gy) mice. The TtRMPVIR retroviral backbone (Addgene, Cambridge, MA, USA) was modified to place Nr4a1-2A-mCherry under the control of a tetracycline-responsive promoter. Sample size was calculated based on published studies for NICD transduction and transplantation (8, 18) to provide at least 80% power to compare a median survival difference of 25% based on two-sided two-sample test for proportions (p<0.05). NICD transduction and transplantation was performed independently for primary mice five times.
Cell Purification and Flow Cytometry
Single cell suspensions were stained with antibodies at 4°C and analyzed on FACSAria, LSRFortessa, or LSR II platforms (BD, Franklin Lakes, NJ, USA). Lineage marker cocktail consisted of Gr-1, Mac-1, B220, Ter119, CD4 and CD8a. The following antibody (clones) were used (eBioscience, San Diego, CA, USA or Biolegend, San Diego, CA, USA) - Gr-1 (RB6-8C5), Mac-1 (M1/70), B220 (RA3-6B2), Ter119 (TER119), CD4 (GK1.5), CD8 (53-6.7), CD3 (145-2C11), Sca-1 (D7), c-Kit (2B8), CD150 (TC15-12F12.2), CD48 (HM48-1), CD45.1 (A20), CD45.2 (104), Il7rα (A7R34). Apoptosis analysis was performed with the AnnexinV Apoptosis Detection Kit (eBioscience). To detect TCR rearrangement, genomic DNA was isolated using the PureLink Genomic DNA kit (Invitrogen, Carlsbad, CA, USA) and amplified with the following primers; TCR Vb5 Fwd – CCCAGCAGATTCTCAGTCCAACAG, TCR Jb2 Rev – TGAGAGCTGTCTCCTACTATCGATT. Real-time PCR was performed with Taqman probes (Applied Biosystems, Foster City, CA, USA).
Cell Culture and Western Blot
P12-Ichikawa cells (DSMZ, Braunschweig, Germany) were cultured in RPMI-1640 (Gibco, Carlsbad, CA, USA) supplemented with 10% heat inactivated fetal bovine serum (FBS; EMD Millipore, Billerica, MA, USA) and penicillin/streptomycin. All shRNAs expressed in pLKO.1-GFP lentivirus with the following target sequences – DNMT3A#1 (CCCAAGGTCAAGGAGATTATT), DNMT3A#2 (CCGGCTCTTCTTTGAGTTCTA), DNMT3A#3 (GCCTCAGAGCTATTACCCAAT), DNMT3A#4 (CCAGATGTTCTTCGCTAATAA), DNMT3A#5 (CCACCAGAAGAAGAGAAGAAT), Dnmt3b#1 (CCAGGAGTATTTGAAGATGAT), Dnmt3b#2 (GCTCTGATATTCTAATGCCAA), Dnmt3b#3 (CCCAAGTTGTACCCAGCAATT), Dnmt3b#4 (GCACTTTAATCTGGCTACCTT), Dnmt3b#5 (CAAGAAAGACATCTCAAGATT). For Dnmt3b shRNAs, plasmids were pooled in equimolar ratios, and lentivirus produced using the pooled DNA. OP9-DL1 stromal cells were cultured in α-MEM (Gibco) with 20% FBS and penicillin/streptomycin. For T-cell assay, HSCs were isolated, transduced and sorted onto OP9-DL1 cells along with recombinant mFlt3L (5 ng/mL; Miltenyi Biotec) and mIL-7 (1 ng/mL; Miltenyi Biotec). Doxycycline (Sigma) and Cytosporone B (Sigma) was prepared as per manufacturer’s instructions. Protein samples were prepared using RIPA Lysis Buffer (Santa Cruz, Dallas, TX, USA). Primary antibodies were used to detect NICD (TA5000078; OriGene, Rockville, MD, USA), DNMT3A (SC-20703; Santa Cruz), NR4A1 (TA314017; OriGene), β-actin (SC-47778; Santa Cruz). Blots were developed using ECL substrate WBKLS010 (EMD Millipore). Chromatin immunoprecipitation with DNMT3A or control IgG antibodies was performed as previously described (19). ChIP-qPCR reactions were performed using Power SYBR Green Mastermix (Applied Biosystems) on the StepOnePlus Real-Time PCR System (Applied Biosystems) with the following primers: NR4A1 Fwd - AACGTTTGGAGTAGCTGGGA, NR4A1 Rev – AGGAGTTTGAGACCAGCCTG. ACTB ChIP-grade primers were ordered from Diagenode (C17011048; Denville, NJ, USA).
Microarray and RNA-seq
RNA was isolated using the NucleoSpin RNA XS kit (Macherey-Nagel, Bethlehem, PA, USA). For microarray analysis, RNA was processed using the Pico Ovation Kit (NuGen, San Carlos, CA, USA) and hybridized to Mouse Gene 2.0 arrays (Affymetrix, Santa Clara, CA, USA). Affymetrix chip data was normalized and analyzed using Transcriptome Analysis Console 2.0 (Affymetrix). For RNA-seq, library preparation was performed with the SMARTer Ultra Low RNA kit (Takara, Mountain View, CA, USA) and sequenced on HiSeq2500 (Illumina, San Diego, CA, USA). RNA-seq reads were aligned to the mouse genome (mm10) with STAR version 2.4.2a. Gene-level transcript counts were then imported into the R/Bioconductor package EdgeR. Genes not expressed in any sample were excluded from further analysis. Differentially expressed genes were filtered for fold-change >2 with false-discovery rate (FDR) <0.05.
Enhanced Representation Bisulfite Sequencing and Analysis
eRRBS was performed as previously described (20). Libraries were prepared from 25 ng of genomic DNA and sequenced on a HiSeq2500 sequencer (Illumina). Low quality sequence were trimmed by using Picard tools, and filtered reads were aligned to mouse genome (mm9) using Bismark version 0.14.5 running Bowtie2 version 2.1.0. CpG methylation was calculated using bismark_methylation_extractor. For identification of DMRs, the genome was split into 100 bp windows with 50bp sliding step. High quality windows were defined as containing at least 3 CpG sites (>10X coverage) in each biological replicate. The significance of DNA methylation difference in each comparison was tested using non-paramedic Wilcoxon singed-rank test followed by Benjamini & Hochberg correction. DMRs were defined as windows with FDR q-value <0.05 with averaged DNA methylation difference >25%. Genome annotations were downloaded from the UCSC Genome Browser. Promoters were defined as the 2.5 kb surrounding the TSS (−500bp/+2000bp) of RefSeq genes. Mouse bone marrow superenhancer data was downloaded from dbSUPER data base. Mouse bone marrow H3K27ac ChIP-seq peaks (ENCFF061JZQ) was downloaded from ENCODE, the active enhancers were defined as H3K27ac peaks that locating out of gene promoters. For hypergeometric distribution calculation, the probability mass function of hypergeometric distribution is calculated as:
Where N is the number of non-overlapping 150bp windows (average length of DMRs) in mouse genome, K is the number of non-overlapping 150bp windows of each genomic features in mouse genome, n is the number of DMRs, and k is the number of DMRs that overlapping to each genomic features. Data have been uploaded to the NCBI Gene Expression Omnibus under accession numbers GSE83769, GSE83755, GSE83759.
Statistics
Student t-test and ANOVA’s were used for statistical comparisons where appropriate. Survival curves were analyzed by Mantel-Cox logrank test. Significance is indicated on the figures using the following convention: *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001. Error bars on graphs represent the S.E.M.
RESULTS
Dnmt3a loss-of-function co-operates with Notch1 gain-of-function to accelerate T-ALL
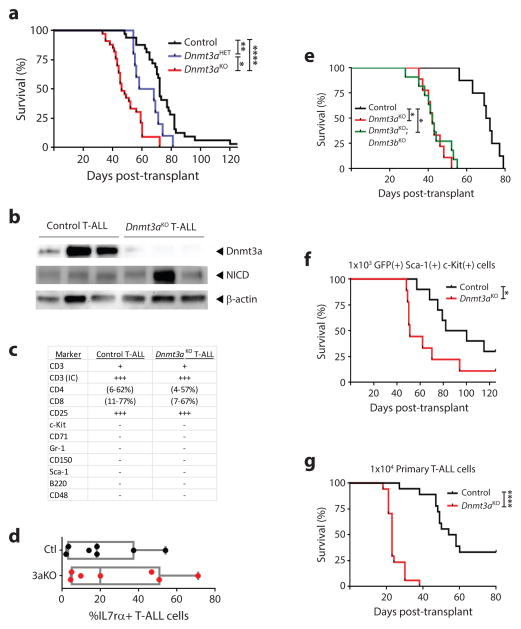
We used mouse models to examine genetic co-operation between Dnmt3a and Notch1 in T-ALL. Mx1-Cre:Dnmt3a+/+ (control) and Mx1-CRE:Dnmt3afl/fl (Dnmt3aKO) mice were treated with pIpC and six-weeks after the last injection, bone marrow progenitor cells were transduced with a retrovirus expressing NICD. Transplantation of 1×105 bulk c-Kit+ cells with comparable transduction levels revealed a much shorter latency to T-ALL in mice receiving Dnmt3aKO NICD+ cells (median survival control = 72-days, Dnmt3aKO = 46-days; Figure 1a). Dnmt3a was effectively eliminated from the Dnmt3aKO T-ALLs, and NICD was expressed at comparable levels (Figure 1b). The rate of T-ALL development was dose dependent as transplantation of Mx1-Cre:Dnmt3afl/+ (Dnmt3aHET) NICD+ cells produced an intermediate latency (median survival = 63-days, Figure 1a). Loss of Dnmt3a did not influence the morphological phenotype of the final disease (Figure 1c), including markers of more immature T-ALL subtypes such as IL7rα (Figure 1d). The enhanced pathogenesis of Dnmt3aKO T-ALL was not due to compensatory Dnmt3b function as transplantation of Mx1-CRE:Dnmt3afl/fl;Dnmt3bfl/fl (Dnmt3aKO;Dnmt3bKO) NICD+ cells produced the same survival as Dnmt3aKO alone (Figure 1e). To exclude the possibility that accelerated leukemogenesis was due to a higher content of T-ALL initiating cells in the Dnmt3aKO bolus, 1×103 NICD+ c-Kit+ Sca-1+ cells were isolated two-days post-transduction and transplanted. Latency to T-ALL was again significantly accelerated in a Dnmt3aKO background (Figure 1f). To confirm the phenotype was cell intrinsic, secondary transplantation was performed. Limiting dilution determined that secondary transfer of 1×104 primary Dnmt3aKO NICD+ cells was sufficient to transmit T-ALL to 100% of recipients (data not shown). 1×104 control or Dnmt3aKO NICD+ cells from four primary T-ALLs were transplanted into 4–5 secondary recipients per tumor (Figure 1g). Primary Dnmt3aKO NICD+ cells transferred T-ALL rapidly (median survival = 23-days), with a marked reduction in latency and increase in penetrance compared to control NICD+ T-ALL cells (33% of secondary recipients still alive 80-days post-transplant, median survival = 56-days).
Figure 1. Dnmt3a loss-of-function co-operates with Notch1 gain-of-function to accelerate T-ALL.
(a) Kaplan-Meier plot showing time to morbidity for mice transplanted with control (n = 31), Dnmt3aHET (n = 10), or Dnmt3aKO (n = 33) hematopoietic progenitor cells transduced with the NICD-expressing retrovirus. (b) Protein expression in control and Dnmt3aKO NICD+ T-ALL blasts. (c) Immunophenotypes of control and Dnmt3aKO T-ALLs. For expression level, − = <1% positive cells, + = 1–5% positive cells, ++ = 5–50% positive cells, +++ = >50% positive cells. (d) Level of IL7rα on cell surface of T-ALL blasts. (e) Kaplan-Meier plot for mice transplanted with control (n = 8), Dnmt3aKO (n = 9), or Dnmt3aKO;Dnmt3bKO (n = 11) hematopoietic progenitor cells transduced with the NICD-expressing retrovirus. (f) Kaplan-Meier plot for mice transplanted with 1×103 Sca-1+ c-Kit+ GFP+ control (n = 10) or Dnmt3aKO (n = 9) NICD-transduced hematopoietic progenitor cells. (g) Survival of mice transplanted with 1×104 control (n = 18) or Dnmt3aKO (n = 17) primary T-ALL cells. Four individual primary T-ALLs for each genotype were transplanted into 4–5 sublethally irradiated mice. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
Dnmt3aKO T-ALL is characterized by DNA hypomethylation at enhancers
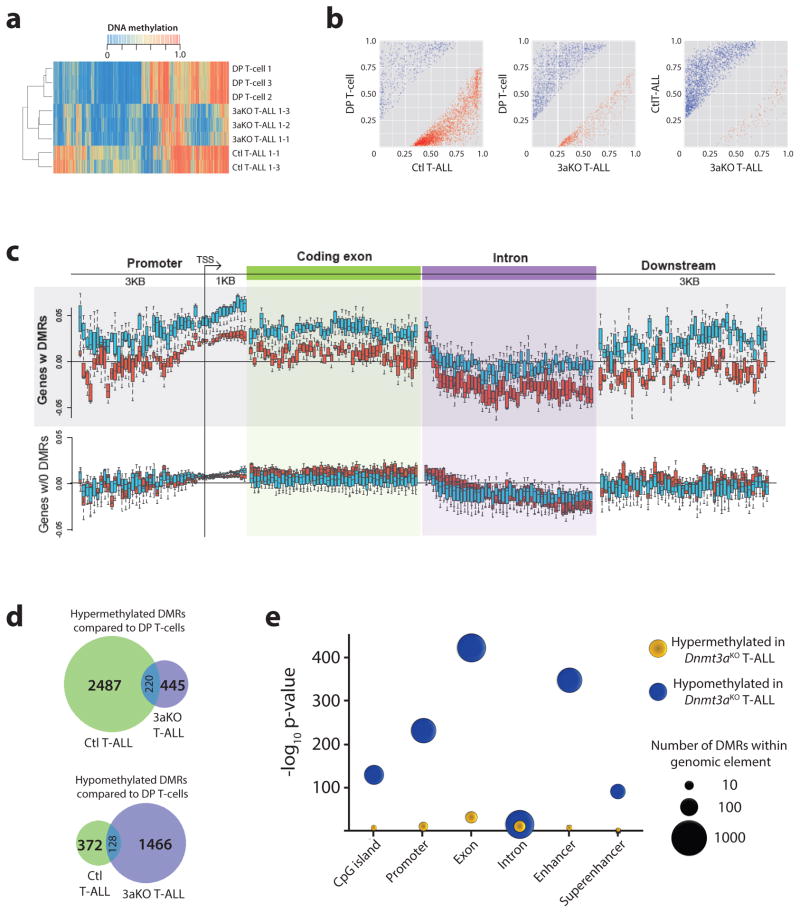
To understand how altered DNA methylation might contribute to the pathogenesis, DNA methylation profiles were generated for control and Dnmt3aKO NICD+ T-ALL blasts from moribund mice using enhanced reduced representation bisulfite sequencing (eRRBS). DNA methylation changes were grouped into differentially methylated regions (DMRs), defined as 100 bp regions containing at least three CpGs with a DNA methylation difference of >25% in the same direction (Supplementary Table 1). Hierarchical clustering by DNA methylation showed that T-ALLs clustered based on genotype, and independently of normal CD4+CD8a+ double-positive (DP) T-cells (Figure 2a). Plotting pair-wise differences in DMR methylation level between samples showed there were substantially more hypermethylated DMRs in control T-ALL cells compared to normal DP T-cells (Figure 2b). Conversely, Dnmt3aKO T-ALLs contained more hypomethylated DMRs in comparison to both normal DP T-cells and control T-ALL (Figure 2b). For genes with DMRs in T-ALL samples compared to DP T-cells, Dnmt3aKO samples were consistently hypomethylated compared to control T-ALL in all genomic contexts (Figure 2c). Surprisingly, some elements such as promoters showed some gains in DNA methylation in Dnmt3aKO T-ALLs compared to normal DP T-cells (Figure 2c), although not to the degree of control T-ALL samples. Loss of Dnmt3a influenced the loci impacted by DNA methylation changes as there was only partial overlap between the hypo- and hypermethylated DMRs in both T-ALL genotypes compared to normal DP T-cells (Figure 2d). The most significant hypomethylation in Dnmt3aKO T-ALLs occurred in exons and enhancers (Figure 2e), a consistent feature of tumors driven by loss of Dnmt3a (21, 22).
Figure 2. Dnmt3aKO T-ALL is characterized by DNA hypomethylation at enhancers.
(a) Hierarchical clustering of specimens based on DNA methylation level of differentially methylated regions (DMRs). (b) DNA methylation levels of DMRs between pairwise sample comparisons. (c) Average DNA methylation level change in genomic context for control (blue bars) and Dnmt3aKO (red bars) T-ALL compared to normal DP T-cells for genes with and without DMRs. (d) Overlap of hypo- and hyper-methylated DMRs for control (Ctl) and Dnmt3aKO (3aKO) T-ALLs in comparison to normal DP T-cells. (e) Hypergeometric test showing enrichment for hypomethylated DMRs in Dnmt3aKO T-ALL in exons and enhancers.
Dnmt3aKO NICD+ T-ALL exhibits a gene expression profile resembling ETP-ALL
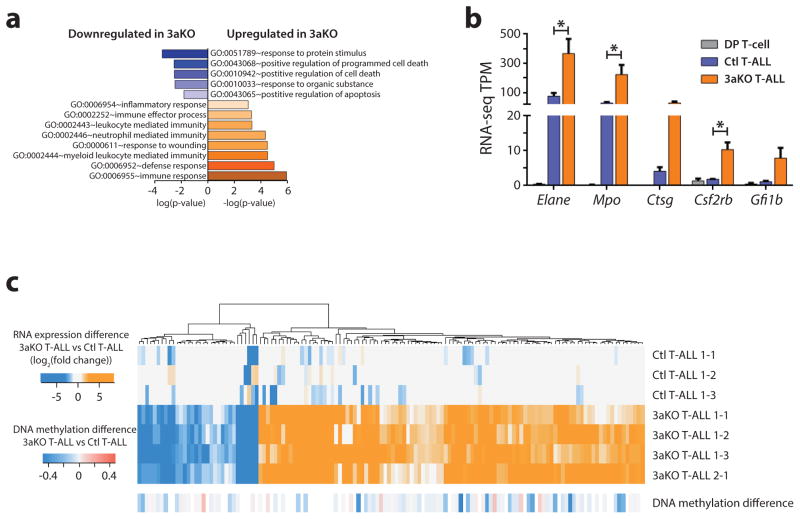
RNA-seq gene expression signatures were generated for the same samples used for DNA methylation profiling. There were actually relatively few statistically significant differences between control and Dnmt3aKO blasts (FDR <0.05, fold-change >2.0-fold), with 283 and 97 genes upregulated and downregulated respectively in Dnmt3aKO T-ALL (Supplementary Table 2). Gene ontology analysis showed that the genesets downregulated in Dnmt3aKO T-ALLs were predominantly associated with positive regulation of apoptosis and programmed cell death (Figure 3a). Conversely, the genesets upregulated in Dnmt3aKO T-ALLs were associated with myeloid cell function and immune/inflammatory response, including genes characteristic of myeloid cells such as Elane, Mpo, Ctsg, Csf2rb and Gfi1b (Figure 3b). Expression of myeloid genes in T-ALL is characteristic of the ETP-ALL subtype. ETP-ALL comprises up to 15% of adult T-ALL cases, and is associated with a high risk of treatment failure (23, 24). Genetically, ETP-ALL is distinguished by a high prevalence of DNMT3A mutations (25). To determine the impact of DNA methylation changes on gene expression, promoter DNA methylation levels of the differentially expressed genes were examined. Similar to our previous studies (16, 19), gene expression changes between control and Dnmt3aKO T-ALL were not associated with consistent DNA methylation differences (Figure 3c).
Figure 3. Dnmt3aKO NICD+ T-ALL exhibits a gene expression profile resembling ETP-ALL.
(a) Gene ontology analysis showing functional gene categories enriched in genes differentially expressed between control and Dnmt3aKO (3aKO) T-ALL. (b) Expression level of myeloid lineage genes in normal DP T-cells, control (Ctl) T-ALL, and Dnmt3aKO (3aKO) T-ALL. (c) Promoter DNA methylation levels of genes differentially expressed between control (Ctl) and Dnmt3aKO (3aKO) T-ALLs.
A stem cell origin for Dnmt3a loss-of-function Notch1 gain-of-function T-ALL
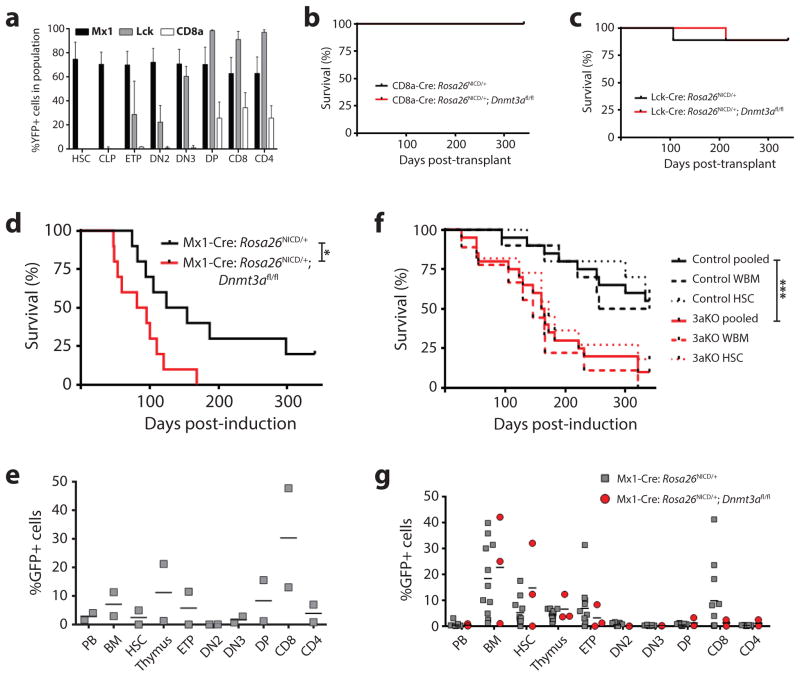
Given the gene expression similarities between Dnmt3aKO T-ALL and human ETP-ALL, perhaps the shorter latency to T-ALL in a Dnmt3aKO background is predicated more by the developmental context of the leukemia-initiating cells rather than DNA methylation changes. How Dnmt3a inactivation influences the T-ALL cell-of-origin was examined using genetic mouse models permitting induction of oncogenic mutations at different stages of T-cell development. Dnmt3afl/fl mice were crossed to Rosa26NICD mice, in which mouse NICD (amino acids 1749–2293) followed by IRES-GFP is knocked into the Rosa26 locus (26), preceded by a floxed STOP cassette. Inactivation of Dnmt3a and activation of NICD occurs simultaneously in Rosa26NICD/+;Dnmt3afl/fl mice upon Cre activation. We crossed these mice to Cre drivers expressed in different hematopoietic compartments – Mx1-Cre (HSCs and all hematopoietic cells after pIpC), Lck-Cre (ETPs and thymic progenitors) and CD8a-Cre (mature T-cells). Expression specificity was confirmed by crossing Cre drivers to Rosa26EYFP mice which express EYFP after Cre-excision of a floxed STOP cassette (Figure 4a).
Figure 4. A stem cell origin for Dnmt3a loss-of-function Notch1 gain-of-function T-ALL.
(a) Percentage of YFP+ cells in hematopoietic populations in lox-stop-lox Rosa26EYFP mice crossed to the indicated Cre driver genotypes (n = 4–5 per genotype). Survival of mice transplant with P0 liver cells from Rosa26NICD/+ or Rosa26NICD/+;Dnmt3afl/fl pups crossed to (b) CD8a-Cre (n = 6 per genotype), (c) Lck-Cre (n = 9 per genotype), or (d) Mx1-CRE (n = 10 per genotype with recipients induced with pIpC four-weeks post-transplant). (e) Chimerism of GFP+ cells in hematopoietic compartments of surviving control mice from (d) one-year post-transplant. (PB = peripheral blood, BM = bone marrow, CD8 = CD8a+CD4− T-cells, CD4 = CD4+CD8a− T-cells). (f) Survival of mice transplanted with either whole bone marrow (WBM) or purified HSCs from pIpC-treated Mx1-Cre:Rosa26NICD/+ (control, n = 10 HSC, 10 WBM) or Mx1-Cre:Rosa26NICD/+;Dnmt3afl/fl (3aKO, n = 11 HSC, 9 WBM) mice. Pooled = combined survival of WBM plus HSC transplanted mice for each genotype. (g) Chimerism of GFP+ cells in hematopoietic compartments of surviving mice from (f) one-year post-transplant.
Treatment of Mx1-Cre:Rosa26NICD/+;Dnmt3afl/fl and Mx1-CRE:Rosa26NICD/+ (control) mice with pIpC produced to an off-target hyperinflammatory response with infiltration of mixed immune cells into the skin. To avoid this, we transplanted 1×106 liver cells from individual P0 mice into 2–4 recipients per donor. The Cre’s were not expressed in the P0 donor cells which ensured that donor-derived T-cell development occurred only in the recipient mice (Mx1-Cre expression was induced four-weeks post-transplant by pIpC). No mice transplanted with CD8a-Cre:Rosa26NICD/+ cells with or without Dnmt3a generated T-ALL (Figure 4b). Only 1/9 mice transplanted with Lck-Cre:Rosa26NICD/+;Dnmt3afl/fl or Lck-Cre:Rosa26NICD/+ cells developed T-ALL (Figure 4c). Transplantation of P0 Mx1-Cre:Rosa26NICD/+ cells generated robust T-ALL, which again was accelerated with loss of Dnmt3a (Figure 4d). This suggests that T-ALL mutations must occur in a cell more primitive than ETPs for development of this leukemia. 2/10 mice transplanted with Mx1-Cre:Rosa26NICD/+ cells were still alive after one-year. Analysis of survivors showed that GFP+ cells could be detected in most hematopoietic compartments, but were conspicuously absent in DN2 thymic progenitors (Figure 4e). The stem cell origin of this T-ALL was further highlighted by transplantation of purified HSCs (Lineage- Sca-1+ c-Kit+ CD48− CD150+) from adult Mx1-Cre:Rosa26NICD/+ and Mx1-Cre:Rosa26NICD/+;Dnmt3afl/fl mice. 1×106 WBM cells or 150 purified HSCs from donor mice were transplanted and recipients were treated with pIpC four-weeks post-transplant. Acceleration of T-ALL in a Dnmt3aKO background was produced by transplantation of either WBM or purified HSCs (Figure 4f). Analysis of surviving mice after one-year showed that surprisingly, robust chimerism of GFP+ cells could be detected in many hematopoietic cell compartments without T-ALL transformation. However, we noted a striking trend in that there was complete absence of GFP+ cells from both genotypes in the DN2 cell compartment (Figure 4g).
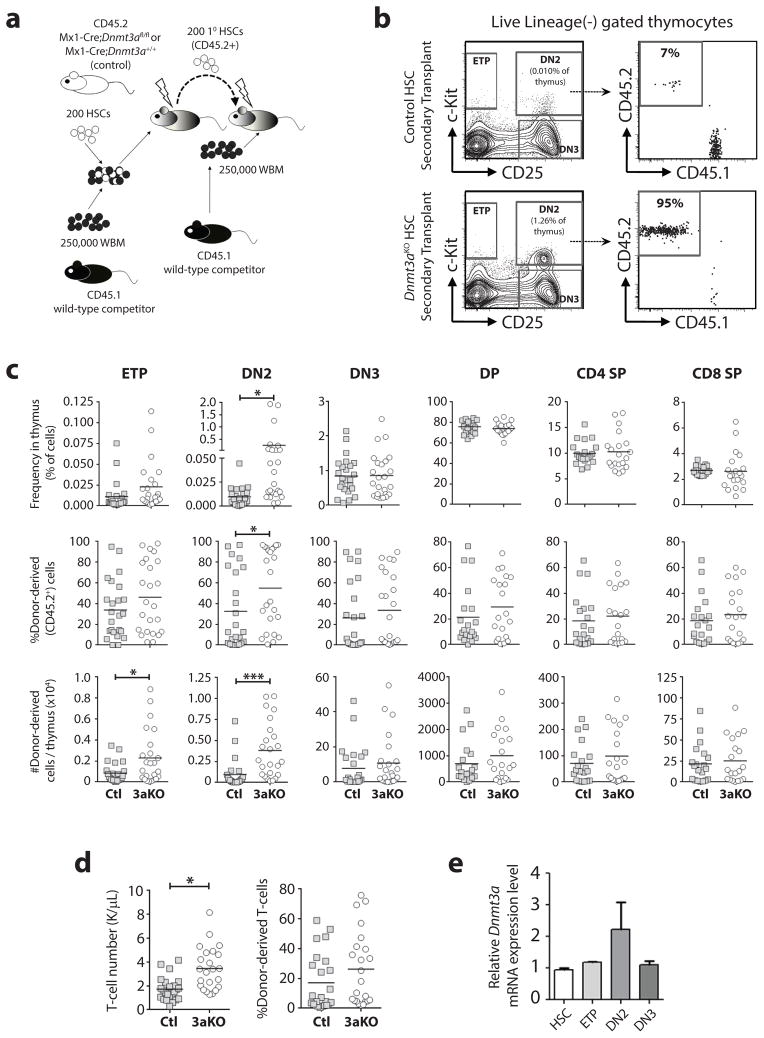
Loss of Dnmt3a causes developmental arrest of thymic progenitors
The T-ALL results suggested that Dnmt3a loss-of-function might have context-specific effects for T-cell progenitors that influence the leukemic cell-of-origin. To determine the consequences of loss of Dnmt3a for T-cell progenitor development, we performed serial HSC transplantation of 200 pIpC-treated control (Mx1-Cre:Dnmt3a+/+) or Dnmt3aKO HSCs (Figure 5a), and analyzed T-cell development in secondary recipients 18-weeks post-transplant. As in previous studies (16), floxing efficiency of Dnmt3a alleles in the HSCs used for secondary transplant was almost 100% (data not shown). Thymus analysis (Figure 5b) showed that a number of mice transplanted with Dnmt3aKO HSCs showed a significant accumulation of Dnmt3aKO DN2 thymic progenitors (Figure 5c). However, this phenotype was not fully penetrant with a >5-fold expansion of donor-derived DN2 cells present in only 38% (9/24) of secondary recipient mice transplanted with Dnmt3aKO HSCs (and was not observed in any of the primary recipients), suggesting some type of selection event or epigenetic stochasticity in those particular mice. This suggests Dnmt3a may be required for normal thymocyte progenitor maturation, even if peripheral blood output is maintained by the cells that do progress through DN2 (Figure 5d). Dnmt3a is most highly expressed in DN2 thymic progenitors (Figure 5e), implicating a unique requirement for Dnmt3a activity at this developmental stage.
Figure 5. Loss of Dnmt3a causes developmental arrest of DN2 thymic progenitors.
(a) Competitive HSC transplantation scheme. (b) Representative flow cytometry plots of thymus samples from secondary transplant recipients showing gating for live, lineage-negative thymocytes. (c) Complete thymus analysis from secondary transplant recipients showing frequency of cell populations, the proportion of donor-derived cells (CD45.2+) in each population, and the absolute number of donor-derived cells for each population per thymus (n = 20–28, DP = CD4+CD8a+, CD4+ SP = CD4+CD8a−, CD8+ SP = CD8a+CD4−). (d) Absolute number and donor-derived chimerism of peripheral blood T-cells in secondary recipient mice (e) Relative mRNA expression of Dnmt3a in wild-type HSCs and T-cell progenitors (n = 3 pools of 4 mice, mean ± S.E.M). *p<0.05, ***p<0.001.
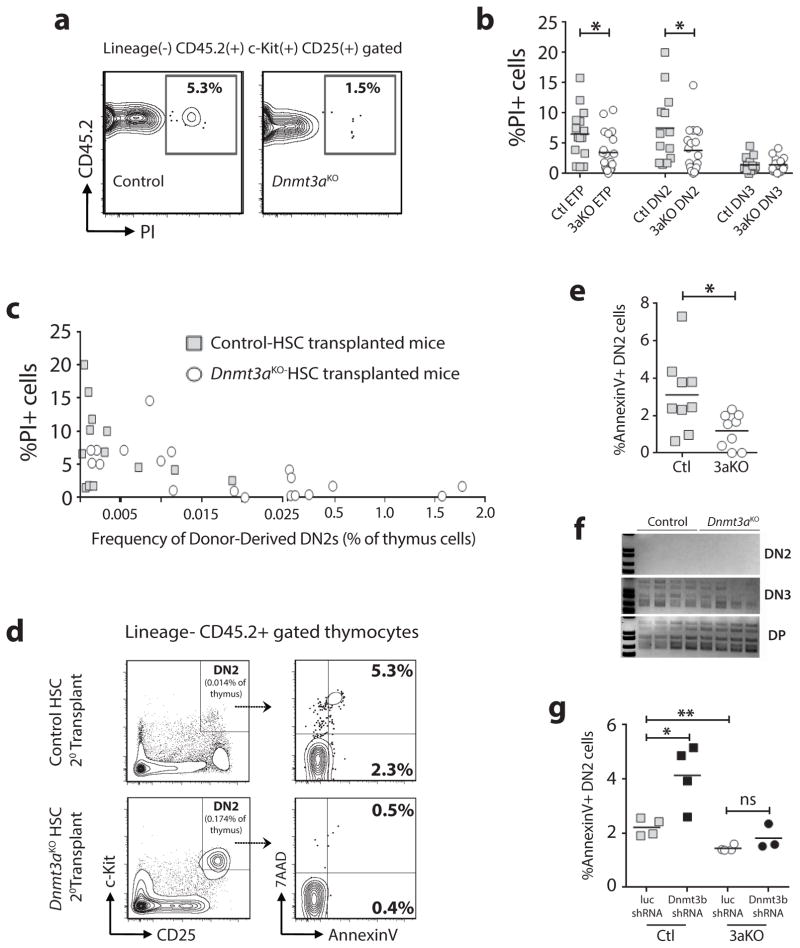
Dnmt3aKO DN2 thymic progenitors are less apoptotic
Apoptosis is a critical and highly regulated process in T-cell development necessary for elimination of autoreactive thymus progenitors during negative selection (27, 28). Propidium iodide (PI) staining (Figure 6a) showed a significant reduction of PI+ (non-viable) Dnmt3aKO ETP and DN2 cells in the thymus of secondary transplant recipients (Figure 6b). There was an inverse correlation between the degree of expansion of the Dnmt3aKO DN2 population and the proportion of PI+ cells (Figure 6c). Independent AnnexinV staining (Figure 6d) confirmed a lower proportion of apoptotic cells in the Dnmt3aKO DN2 population (Figure 6e). Reduced apoptosis of Dnmt3aKO thymic progenitors was not due to defective T-cell receptor (TCR) rearrangement (Figure 6f), or due to aberrant function of Dnmt3b in the absence of Dnmt3a. While Dnmt3b knockdown in control in vitro-derived DN2 cells increased apoptosis, this effect was abrogated without Dnmt3a (Figure 6g). Cumulatively, these data suggest that Dnmt3aKO DN2 progenitors accumulate due to reduced apoptosis.
Figure 6. Dnmt3aKO DN2 thymic progenitors are less apoptotic.
(a) Representative flow cytometry plots showing proportion of non-viable cells in donor-derived DN2 population of secondary transplant recipients. (b) Quantification of non-viable (PI+) cells in thymocyte progenitor populations from secondary transplant recipients (n = 13–20). (c) Frequency of donor-derived DN2 cells versus the percent non-viable cells in individual secondary recipient mice with greater than 10% chimerism. (d) Representative flow cytometry plots showing thymus AnnexinV staining of secondary transplant recipients. (e) Quantification of apoptotic (AnnexinV+) DN2 cells from control- (Ctl, n = 9) and Dnmt3aKO- (3aKO, n = 9) HSC transplanted mice. (f) PCR for TCRβ5 rearrangement in indicated cell populations showing normal TCR recombination in Dnmt3aKO populations. (g) Quantification of AnnexinV in in vitro-derived DN2 cells following transduction of control or Dnmt3aKO HSCs with luciferase (luc) or Dnmt3b shRNAs. *p<0.05, **p<0.01.
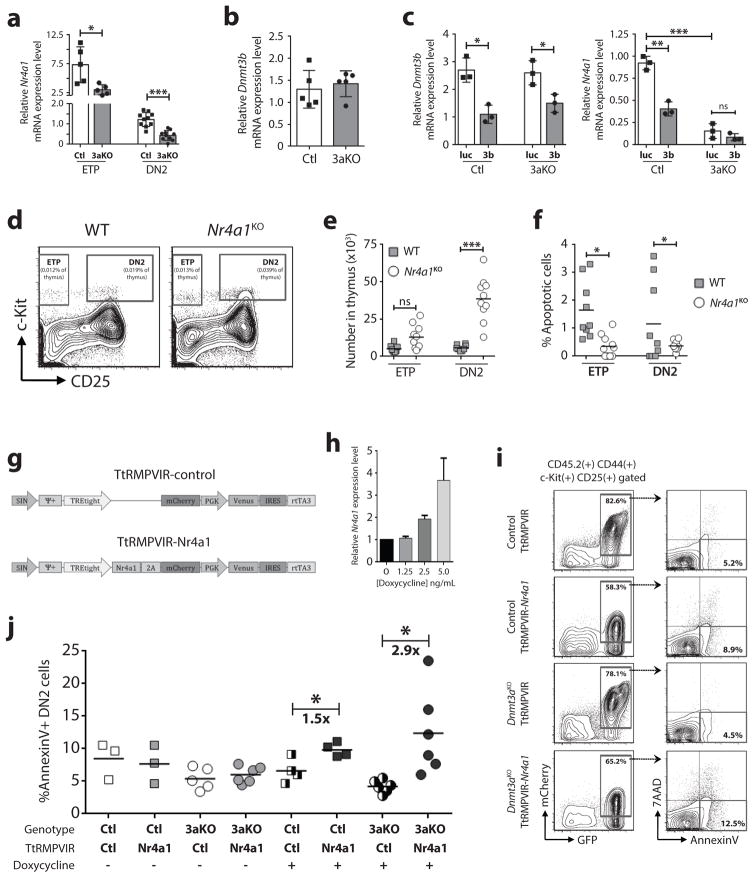
Nr4a1 is downregulated in Dnmt3aKO DN2 thymic progenitors
To investigate molecular mechanisms underlying the developmental arrest of Dnmt3aKO DN2 thymocytes, microarray gene expression profiling was performed. Despite some robust phenotypic differences, there was a high transcriptional similarity between control and Dnmt3aKO DN2 cells (Supplementary Table 3), with only 180 transcripts differentially expressed (ANOVA p-value <0.05, fold-change >1.50-fold). We curated the lists to identify potential gene expression differences that might explain the lower apoptotic index of Dnmt3aKO DN2 cells. In microarray results, the orphan nuclear receptor Nr4a1 (also called Nur77) was 2.3-fold downregulated in Dnmt3aKO DN2 cells. This observation was more significant in an extended targeted analysis, which also showed Nr4a1 downregulation in Dnmt3aKO ETPs (Figure 7a). Dnmt3b was not aberrantly expressed in Dnmt3aKO DN2 cells (Figure 7b), and inhibition of Dnmt3b in a Dnmt3aKO background did not alter Nr4a1 levels in in vitro-derived DN2 cells (Figure 7c).
Figure 7. Nr4a1 is downregulated in Dnmt3aKO DN2 thymic progenitors.
(a) Relative expression of Nr4a1 in control (Ctl) and Dnmt3aKO (3aKO) ETP and DN2 cells by real-time PCR (n = 5–10). (b) Relative expression of Dnmt3b in control and Dnmt3aKO DN2 cells (n = 5). (c) Relative expression of Nr4a1 in in vitro-derived DN2 cells following transduction of control or Dnmt3aKO HSCs with control luciferase (luc) or Dnmt3b (3b) shRNAs (n = 3). (d) Representative flow cytometry plots of live, lineage-negative thymocytes from eight-week old wild-type (WT) and Nr4a1KO mice. (e) Quantification of ETP and DN2 thymocytes from adult WT (n = 9) and Nr4a1KO (n = 10) mice. (f) Quantification of apoptotic (AnnexinV+) DN2 cells from WT (n = 9) and Nr4a1KO (n = 10) mice. (g) Schematic representation of doxycycline-inducible retroviruses. (h) Expression level of Nr4a1 in T-cell progenitors transduced with TtRMPVIR-Nr4a1 with increasing concentrations of doxycycline. (i) Representative flow cytometry plots showing AnnexinV staining of in vitro-derived DN2 cells following transduction of control and Dnmt3aKO HSCs with inducible retroviruses and induction with doxycycline. (j) Quantification of apoptotic (AnnexinV+) in vitro-derived DN2 cells following transduction of HSCs with a control or Nr4a1-expressing inducible retrovirus. Transduced cells were cultured for 13-days to reach DN2 stage of development, and then induced with doxycycline for two-days prior to AnnexinV analysis. *p<0.05, **p<0.01, ***p<0.001.
Nr4a1 acts in the apoptotic process during negative selection of T-cell progenitors (27, 28). In autoreactive T-cells, Nr4a1 undergoes phosphorylation-dependent export from the nucleus to the mitochondria, where it conformationally alters Bcl-2 to promote apoptosis (29–31). Thus, decreased Nr4a1 expression in Dnmt3aKO DN2 cells could be responsible for the reduced apoptosis. To investigate if Nr4a1 ablation phenocopies Dnmt3a inactivation in T-cell progenitors, thymus analysis of Nr4a1 knockout (Nr4a1KO) mice (32) was performed (Figure 7d). Adult Nr4a1KO mice showed an expansion of DN2s (Figure 7e), which were less apoptotic (Figure 7f), reminiscent of Dnmt3aKO DN2 cells.
We used a retroviral rescue model to determine if increasing Nr4a1 in Dnmt3aKO thymic progenitors could restore apoptotic competence. To specifically investigate the function of Nr4a1 at the DN2 stage, we generated an inducible retrovirus in which the tetracycline trans-activator protein (rtTA3 = Tet-ON) is constitutively expressed with bicistronic expression of Venus fluorescent protein, and on the same plasmid Nr4a1 expression is under the regulation of a tetracycline-responsive promoter element linked to mCherry expression via a 2A peptide (TtRMPVIR-Nr4a1, control empty vector = TtRMPVIR; Figure 7g). We transduced HSCs and allowed them to develop on OP9-DL1 stroma (33) for 13-days, then added 2.5 ng/mL doxycycline (a dose titrated to induce a two-fold increase in Nr4a1 expression; Figure 7h) and evaluated apoptosis two-days later (Figure 7i). In the absence of doxycycline, there was no difference in AnnexinV+ staining in DN2 cells carrying the control or Nr4a1-expressing vector between the control and Dnmt3aKO genotypes. While induction of Nr4a1 did induce a significant increase in the proportion of AnnexinV+ control DN2 cells, the effect was much more robust in a Dnmt3aKO background (Figure 7j).
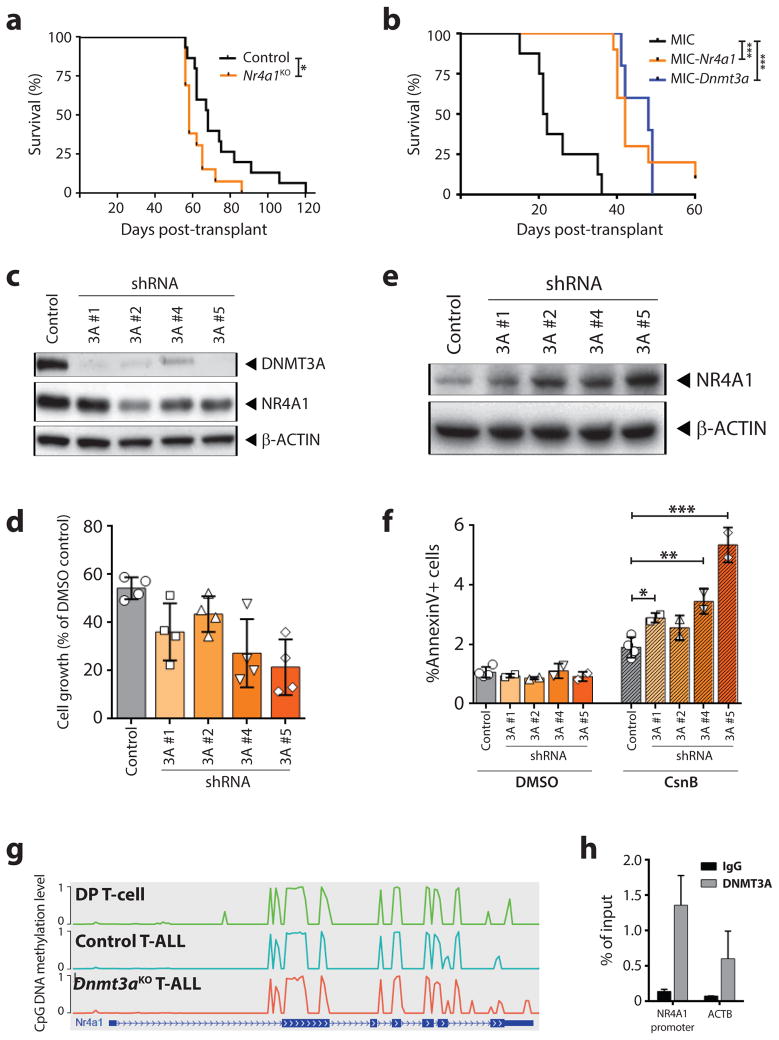
Nr4a1 downregulation is a downstream effector of Dnmt3a loss-of-function in T-ALL
We examined the function of Nr4a1 in Dnmt3aKO T-ALL leukemogenesis. Nr4a1 expression levels were reduced in T-ALLs lacking Dnmt3a (Supplementary Table 2). Retroviral expression of NICD in Nr4a1KO bone marrow progenitors phenocopied the Dnmt3aKO shorter latency to T-ALL (Figure 8a). Furthermore, retroviral rescue of Dnmt3aKO primary T-ALL with ectopic Nr4a1 extended survival over control empty vector transduced cells in secondary recipients to a similar extent as rescue with Dnmt3a (Figure 8b). To determine if NR4A1 may be a therapeutic target in DNMT3A-mutant T-ALL, we created stable P12-Ichikawa T-ALL cell lines (NOTCH1 mutation) with reduced DNMT3A function by shRNA knockdown. 4/5 shRNAs tested substantially decreased DNMT3A protein expression, and NR4A1 was also reduced in these cells (Figure 8c). These cells were then treated with Cytosporone B (CsnB), a natural agonist of NR4A1 (34). We determined the IC50 of CsnB for control P12 to be 0.04 mM. At this dose, P12 cells carrying DNMT3A shRNAs showed retarded growth (Figure 8d). The same dose of CsnB induced a stronger upregulation of NR4A1 in DNMT3A-shRNA cell lines (Figure 8e) and increased apoptosis (Figure 8f), suggesting that DNMT3A loss-of-function makes the cells more sensitive to increased NR4A1 levels. As a mechanism leading to downregulation of Nr4a1 in Dnmt3a loss-of-function cells, we could not identify DNA methylation differences at the Nr4a1 locus in Dnmt3aKO T-ALL cells (Figure 8g). But DNMT3A binding was enriched over genome background levels at the NR4A1 promoter in control P12 cells (Figure 8h), suggesting DNMT3A may recruiting transcriptional activating factors to this locus in T-cells.
Figure 8. Nr4a1 downregulation is a downstream effector of Dnmt3a loss-of-function in T-ALL.
(a) Kaplan-Meier plot showing time to morbidity for mice transplanted with WT (n = 15) or Nr4a1KO (n = 13) NICD+ hematopoietic progenitor cells. (b) Kaplan-Meier plot showing time to morbidity for mice transplanted with primary NICD+ Dnmt3aKO T-ALL cells transduced with control empty vector (MIC; n=8), Nr4a1-expressing (MIC-Nr4a1; n=10), or Dnmt3a-expressing (MIC-Dnmt3a; n=5) retrovirus. (c) Protein expression in P12 cells transduced with shRNAs targeting DNMT3A. (d) Growth rate (relative to DMSO-treated cells) of P12 cells transduced with control or DNMT3A-shRNAs after 72-hour treatment with 0.04 mM Cytosporone B (CsnB). (e) Western blot of control and DNMT3A-shRNA P12 cells after 72-hour treatment with 0.04 mM CsnB. (f) Proportion of apoptotic (AnnexinV+) cells after 72-hour treatment with 0.04 mM CsnB. (g) DNA methylation level of Nr4a1 locus in normal DP T-cells, control T-ALL and Dnmt3aKO T-ALL cells. Height of line indicates level of DNA methylation along the gene. (h) ChIP for DNMT3A in control P12 cells shows enrichment for binding NR4A1 promoter over genome background (ACTB) and IgG control.
DISCUSSION
In addition crucial roles for Dnmt3a in HSC differentiation (16, 19) and myeloid tumor suppression (21, 35), the studies presented here establish important functions for Dnmt3a in T-cell development and evolving T-ALL. While activating NOTCH1 mutations are the major genetic lesion in T-ALL (7), little progress has been made into developing targeted therapies for these patients, likely due to the context of how co-operating mutations shape the leukemia biology. We show in mouse models that Dnmt3a loss-of-function co-operates with Notch1 gain-of-function in driving an aggressive T-ALL. Our results suggest for Dnmt3aKO NICD+ T-ALL, the mutations must be acquired in a cell more primitive than ETPs. However, the observation that leukemia-free mice completely lacked GFP+ cells in the DN2 cell compartment suggests there is something unique about this developmental stage that is required for T-cell transformation. These data support a model whereby oncogenic mutations are acquired in a HSC, which allows dissemination throughout hematopoietic progeny. The HSCs may not necessarily be pathogenic themselves, but merely a vessel to acquire, maintain and propagate the mutations. The mutations may only be transformative when they are passed to a more committed progenitor cell with the right combination of developmental context, epigenetic environment and proliferative capacity.
The mutational spectrum of DNMT3A is markedly different in T-ALL than in myeloid malignancies such as acute myeloid leukemia (AML). While virtually all DNMT3A mutations in AML are heterozygous (9, 10), T-ALL patients typically harbor homozygous or compound heterozygous DNMT3A mutations (11, 14, 15), and there is less prevalence of the “hotspot” DNMT3AR882H dominant negative variant (36, 37). Similarly, Dnmt3a-null mice are predisposed to T-ALL when combined with a Flt3ITD transgene, whereas Dnmt3a-heterozygous mice predominantly develop AML with the same Flt3ITD allele (22). This clearly suggests some dose dependence of DNMT3A levels for the lineage of leukemia generated, with 0% DNMT3A activity preferential for T-lineage disease, and 50–70% DNMT3A activity predisposing for myeloid neoplasia. In AML, heterozygous non-R882 DNMT3A mutant AML generally leads to loss-of-function alleles (one wild-type allele plus one loss-of-function allele = 50% wild-type DNMT3A activity), whereas the DNMT3AR882 dominant negative retains approximately 20% DNA methylation activity of the wild-type protein (one wild-type allele plus one loss-of-function allele = 70% wild-type DNMT3A activity). The functional consequences of the different DNA methylation activity may make lineage-committed more progenitors more susceptible to transformation depending on the nature of the upstream DNMT3A mutation acquired in a HSC (38–40).
We demonstrate that Dnmt3aKO-derived DN2 progenitors accumulate in the thymus and have reduced apoptosis, partially due to downregulation of Nr4a1. This mirrors what happens to HSCs lacking Dnmt3a in the bone marrow - expansion of a functionally-defective progenitor cell population. We show mouse and human T-cells lacking Dnmt3a are sensitive to increases in Nr4a1 levels. Investigation into more specific NR4A1 agonists as a therapy for DNMT3A-mutant T-ALL warrants further study as there is no established standard of care for patients with relapsed or primary refractory T-ALL, and prognosis remain dismal with a median survival of five months (41). The search for molecular drug targets based on a patient’s genetic profile is necessary to improve outcomes. Results from the studies presented here highlight novel therapeutic vulnerabilities of T-ALL driven by distinct combinations of mutations, and are necessary for the development of targeted therapies for this high-risk patient population.
Supplementary Material
Acknowledgments
We thank the Alvin J. Siteman Cancer Center at Washington University School of Medicine for the use of the Siteman Flow Cytometry Core, which provided cell sorting and analysis. The Siteman Cancer Center is supported in part by NCI Cancer Center Support Grant CA91842. We thank the Genome Technology Access Center Washington University School of Medicine for genomic analysis. The Center is partially supported by NCI Cancer Center Support Grant CA91842 and by ICTS/CTSA Grant UL1TR000448 NIH, and NIH Roadmap for Medical Research. Research reported in this publication was supported by the Washington University Institute of Clinical and Translational Sciences grant UL1 TR000448 NIH. The content is solely the responsibility of the authors and does not necessarily represent the official view of the NIH.
This work was supported by grants (to GAC) from Alex’s Lemonade Stand Foundation, the Children’s Discovery Institute (MC-II-2013-286), the American Society of Hematology, and the Sidney Kimmel Foundation. WCW was supported by NIH T32HL008088. HC was supported by an American Society of Hematology scholar award. JN was supported in part by an Alex’s Lemonade Stand Foundation POST award. CM was supported in part by NIH T32HL008088, and NIH DK111058-01. ELO was supported by NIH 5T32CA113275-10.
Footnotes
CONFLICT OF INTEREST
The authors declare no conflict of interest.
AUTHORS’ CONTRIBUTIONS
Designed and performed experiments: ACK, AK, WCW, HC, JN, CM, EE, AM, GAC.
Analyzed data: ELO, MCV, ALY, TED, MEF, BZ, GAC.
Supervised the study and wrote the paper: GAC
References
- 1.Aifantis I, Raetz E, Buonamici S. Molecular pathogenesis of T-cell leukaemia and lymphoma. Nat Rev Immunol. 2008;8:380–90. doi: 10.1038/nri2304. [DOI] [PubMed] [Google Scholar]
- 2.Radtke F, Wilson A, Stark G, Bauer M, van Meerwijk J, MacDonald HR, et al. Deficient T cell fate specification in mice with an induced inactivation of Notch1. Immunity. 1999;10:547–58. doi: 10.1016/s1074-7613(00)80054-0. [DOI] [PubMed] [Google Scholar]
- 3.Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–33. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang YH, Li D, Winoto A, Robey EA. Distinct transcriptional programs in thymocytes responding to T cell receptor, Notch, and positive selection signals. Proc Natl Acad Sci USA. 2004;101:4936–41. doi: 10.1073/pnas.0401133101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jarriault S, Brou C, Logeat F, Schroeter EH, Kopan R, Israel A. Signalling downstream of activated mammalian Notch. Nature. 1995;377:355–8. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- 6.Reizis B, Leder P. Direct induction of T lymphocyte-specific gene expression by the mammalian Notch signaling pathway. Genes Dev. 2002;16:295–300. doi: 10.1101/gad.960702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weng AP, Ferrando AA, Lee W, Morris JPt, Silverman LB, Sanchez-Irizarry C, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–71. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 8.Pear WS, Aster JC, Scott ML, Hasserjian RP, Soffer B, Sklar J, et al. Exclusive development of T cell neoplasms in mice transplanted with bone marrow expressing activated Notch alleles. J Exp Med. 1996;183:2283–91. doi: 10.1084/jem.183.5.2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010;363:2424–33. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cancer Genome Atlas Research. Genomic and epigenomic landscapes of adult de novo acute myeloid leukemia. N Engl J Med. 2013;368:2059–74. doi: 10.1056/NEJMoa1301689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simon C, Chagraoui J, Krosl J, Gendron P, Wilhelm B, Lemieux S, et al. A key role for EZH2 and associated genes in mouse and human adult T-cell acute leukemia. Genes Dev. 2012;26:651–6. doi: 10.1101/gad.186411.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aref S, El Menshawy N, El-Ghonemy MS, Zeid TA, El-Baiomy MA. Clinicopathologic Effect of DNMT3A Mutation in Adult T-Cell Acute Lymphoblastic Leukemia. Clinical lymphoma, myeloma & leukemia. 2016;16:43–8. doi: 10.1016/j.clml.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 13.Couronne L, Bastard C, Bernard OA. TET2 and DNMT3A mutations in human T-cell lymphoma. N Engl J Med. 2012;366:95–6. doi: 10.1056/NEJMc1111708. [DOI] [PubMed] [Google Scholar]
- 14.Grossmann V, Haferlach C, Weissmann S, Roller A, Schindela S, Poetzinger F, et al. The molecular profile of adult T-cell acute lymphoblastic leukemia: mutations in RUNX1 and DNMT3A are associated with poor prognosis in T-ALL. Genes Chromosomes Cancer. 2013;52:410–22. doi: 10.1002/gcc.22039. [DOI] [PubMed] [Google Scholar]
- 15.Van Vlierberghe P, Ambesi-Impiombato A, Perez-Garcia A, Haydu JE, Rigo I, Hadler M, et al. ETV6 mutations in early immature human T cell leukemias. J Exp Med. 2011;208:2571–9. doi: 10.1084/jem.20112239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Challen GA, Sun D, Jeong M, Luo M, Jelinek J, Berg JS, et al. Dnmt3a is essential for hematopoietic stem cell differentiation. Nat Genet. 2012;44:23–31. doi: 10.1038/ng.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Challen GA, Goodell MA. Runx1 isoforms show differential expression patterns during hematopoietic development but have similar functional effects in adult hematopoietic stem cells. Exp Hematol. 2010;38:403–16. doi: 10.1016/j.exphem.2010.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chiang MY, Xu L, Shestova O, Histen G, L’Heureux S, Romany C, et al. Leukemia-associated NOTCH1 alleles are weak tumor initiators but accelerate K-ras-initiated leukemia. J Clin Invest. 2008;118:3181–94. doi: 10.1172/JCI35090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Challen GA, Sun D, Mayle A, Jeong M, Luo M, Rodriguez B, et al. Dnmt3a and Dnmt3b Have Overlapping and Distinct Functions in Hematopoietic Stem Cells. Cell Stem Cell. 2014;15:350–364. doi: 10.1016/j.stem.2014.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Akalin A, Garrett-Bakelman FE, Kormaksson M, Busuttil J, Zhang L, Khrebtukova I, et al. Base-pair resolution DNA methylation sequencing reveals profoundly divergent epigenetic landscapes in acute myeloid leukemia. PLoS Genet. 2012;8:e1002781. doi: 10.1371/journal.pgen.1002781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mayle A, Yang L, Rodriguez B, Zhou T, Chang E, Curry CV, et al. Dnmt3a loss predisposes murine hematopoietic stem cells to malignant transformation. Blood. 2015;125:629–38. doi: 10.1182/blood-2014-08-594648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang L, Rodriguez B, Mayle A, Park HJ, Lin X, Luo M, et al. DNMT3A Loss Drives Enhancer Hypomethylation in FLT3-ITD-Associated Leukemias. Cancer Cell. 2016;29:922–34. doi: 10.1016/j.ccell.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang J, Ding L, Holmfeldt L, Wu G, Heatley SL, Payne-Turner D, et al. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 2012;481:157–63. doi: 10.1038/nature10725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Coustan-Smith E, Mullighan CG, Onciu M, Behm FG, Raimondi SC, Pei D, et al. Early T-cell precursor leukaemia: a subtype of very high-risk acute lymphoblastic leukaemia. The lancet oncology. 2009;10:147–56. doi: 10.1016/S1470-2045(08)70314-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neumann M, Heesch S, Schlee C, Schwartz S, Gokbuget N, Hoelzer D, et al. Whole-exome sequencing in adult ETP-ALL reveals a high rate of DNMT3A mutations. Blood. 2013;121:4749–52. doi: 10.1182/blood-2012-11-465138. [DOI] [PubMed] [Google Scholar]
- 26.Murtaugh LC, Stanger BZ, Kwan KM, Melton DA. Notch signaling controls multiple steps of pancreatic differentiation. Proc Natl Acad Sci USA. 2003;100:14920–5. doi: 10.1073/pnas.2436557100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Calnan BJ, Szychowski S, Chan FK, Cado D, Winoto A. A role for the orphan steroid receptor Nur77 in apoptosis accompanying antigen-induced negative selection. Immunity. 1995;3:273–82. doi: 10.1016/1074-7613(95)90113-2. [DOI] [PubMed] [Google Scholar]
- 28.Zhou T, Cheng J, Yang P, Wang Z, Liu C, Su X, et al. Inhibition of Nur77/Nurr1 leads to inefficient clonal deletion of self-reactive T cells. J Exp Med. 1996;183:1879–92. doi: 10.1084/jem.183.4.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin B, Kolluri SK, Lin F, Liu W, Han YH, Cao X, et al. Conversion of Bcl-2 from protector to killer by interaction with nuclear orphan receptor Nur77/TR3. Cell. 2004;116:527–40. doi: 10.1016/s0092-8674(04)00162-x. [DOI] [PubMed] [Google Scholar]
- 30.Thompson J, Winoto A. During negative selection, Nur77 family proteins translocate to mitochondria where they associate with Bcl-2 and expose its proapoptotic BH3 domain. J Exp Med. 2008;205:1029–36. doi: 10.1084/jem.20080101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang A, Rud J, Olson CM, Jr, Anguita J, Osborne BA. Phosphorylation of Nur77 by the MEK-ERK-RSK cascade induces mitochondrial translocation and apoptosis in T cells. J Immunol. 2009;183:3268–77. doi: 10.4049/jimmunol.0900894. [DOI] [PubMed] [Google Scholar]
- 32.Lee SL, Wesselschmidt RL, Linette GP, Kanagawa O, Russell JH, Milbrandt J. Unimpaired thymic and peripheral T cell death in mice lacking the nuclear receptor NGFI-B (Nur77) Science. 1995;269:532–5. doi: 10.1126/science.7624775. [DOI] [PubMed] [Google Scholar]
- 33.Schmitt TM, Zuniga-Pflucker JC. Induction of T cell development from hematopoietic progenitor cells by delta-like-1 in vitro. Immunity. 2002;17:749–56. doi: 10.1016/s1074-7613(02)00474-0. [DOI] [PubMed] [Google Scholar]
- 34.Zhan Y, Du X, Chen H, Liu J, Zhao B, Huang D, et al. Cytosporone B is an agonist for nuclear orphan receptor Nur77. Nature Chemical Biology. 2008;4:548–56. doi: 10.1038/nchembio.106. [DOI] [PubMed] [Google Scholar]
- 35.Celik H, Mallaney C, Kothari A, Ostrander EL, Eultgen E, Martens A, et al. Enforced differentiation of Dnmt3a-null bone marrow leads to failure with c-Kit mutations driving leukemic transformation. Blood. 2015;125:619–28. doi: 10.1182/blood-2014-08-594564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Russler-Germain DA, Spencer DH, Young MA, Lamprecht TL, Miller CA, Fulton R, et al. The R882H DNMT3A Mutation Associated with AML Dominantly Inhibits Wild-Type DNMT3A by Blocking Its Ability to Form Active Tetramers. Cancer Cell. 2014;25:442–54. doi: 10.1016/j.ccr.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim SJ, Zhao H, Hardikar S, Singh AK, Goodell MA, Chen T. A DNMT3A mutation common in AML exhibits dominant-negative effects in murine ES cells. Blood. 2013;122:4086–9. doi: 10.1182/blood-2013-02-483487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spencer DH, Russler-Germain DA, Ketkar S, Helton NM, Lamprecht TL, Fulton RS, et al. CpG Island Hypermethylation Mediated by DNMT3A Is a Consequence of AML Progression. Cell. 2017;168:801–16. doi: 10.1016/j.cell.2017.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scourzic L, Couronne L, Pedersen MT, Della Valle V, Diop M, Mylonas E, et al. DNMT3A(R882H) mutant and Tet2 inactivation cooperate in the deregulation of DNA methylation control to induce lymphoid malignancies in mice. Leukemia. 2016;30:1388–98. doi: 10.1038/leu.2016.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koya J, Kataoka K, Sato T, Bando M, Kato Y, Tsuruta-Kishino T, et al. DNMT3A R882 mutants interact with polycomb proteins to block haematopoietic stem and leukaemic cell differentiation. Nature Commun. 2016;7:10924. doi: 10.1038/ncomms10924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thomas DA, Kantarjian H, Smith TL, Koller C, Cortes J, O’Brien S, et al. Primary refractory and relapsed adult acute lymphoblastic leukemia: characteristics, treatment results, and prognosis with salvage therapy. Cancer. 1999;86:1216–30. doi: 10.1002/(sici)1097-0142(19991001)86:7<1216::aid-cncr17>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.