Abstract
Changes in behavior are necessary to apply genomic discoveries to practice. We prospectively studied medication changes made by providers representing eight different medicine specialty clinics whose patients had submitted to preemptive pharmacogenomic genotyping. An institutional clinical decision support (CDS) system provided pharmacogenomic results using traffic light alerts: green/genomically favorable, yellow/genomic caution, red/high risk. The influence of pharmacogenomic alerts on prescribing behaviors was the primary endpoint. 2279 outpatient encounters were analyzed. Independent of other potential prescribing mediators, medications with high pharmacogenomic risk were changed significantly more often than prescription drugs lacking pharmacogenomic information (odds ratio [OR]=26.2 [9.0–75.3], p<0.0001). Medications with cautionary pharmacogenomic information were also changed more frequently (OR=2.4 [1.7–3.5], p<0.0001). No pharmacogenomically high-risk medications were prescribed during the entire study when physicians consulted the CDS tool. Pharmacogenomic information improved prescribing in patterns aimed at reducing patient risk, demonstrating that enhanced prescription decision-making is achievable through clinical integration of genomic medicine.
Keywords: pharmacogenomics, implementation, clinical decision support, prescribing behavior, precision medicine
INTRODUCTION
Approximately two million patients per year experience adverse drug reactions, and 100,000 deaths occur as a direct result(1). Moreover, common drug efficacy rates are only 50–60%(2). Genetic variation has been shown to contribute to inter-individual differences in adverse reactions and efficacy for hundreds of drugs(3–5). Leveraging this information may be a potential path to safer and more effective prescribing. Because of this, pharmacogenomics has been a logical arena in which to explore and solve larger barriers to genomic implementation in the era of precision medicine(6). Obstacles include lack of routine availability of genetic tests, limited provider education about genomics, and the need for facile clinical decision support (CDS) tools that integrate into workflows(7, 8).
Many groups are currently working to address these barriers, leading to various, often customized, implementation solutions(9–20). Inclusion of genetic information within Food and Drug Administration (FDA) labels and the development of pharmacogenomic guidelines (pharmgkb.org/page/cpic) are important forward strides(21). As with any clinical innovation, the true utility of an implementation intervention is measured by physician adoption and user response(22). For example, in a study examining physician responses to interruptive drug-drug interaction alerts, 93% were overridden or ignored(23). Underlying causes of physician reluctance to follow clinical guidance include limitations in awareness, lack of agreement, and clinical inertia(24). Understanding clinician prescribing behaviors in response to genomic information is an essential next step.
Our institutional genomic implementation program, The 1200 Patients Project, was initiated five years ago to overcome obstacles to clinical translation of pharmacogenomics by preemptively genotyping patients and making available to treating physicians patient-specific results at clinical visits, accompanied by CDS(25, 26). Utilizing this approach, we aimed to examine prospectively the impact of available pharmacogenomic information on physician prescribing behaviors. We hypothesized that pharmacogenomic results delivered via a CDS tool would significantly alter prescribing decisions, especially for high-risk medications. We also sought to identify limitations to physician use of genomic information to guide future iterations of implementation efforts.
RESULTS
Participants, Clinic Visits, and Medication Changes
Seventeen providers representing a diverse cohort of eight different medicine primary care and subspecialty clinics from two metropolitan outpatient locations were approached for participation, and every provider who was invited to participate agreed. These providers, along with study staff, then recruited patients to the study over a four-year period. The percentage of patients who agreed to participate when approached was 87%. Of these 1,108 consenting patients, 930 gave a blood sample for genotyping (the remaining 178 patients who consented but never provided a blood sample consisted almost entirely of patients who signed consent at a ‘new patient’ (first visit) with a study physician at our institution, yet never returned to our medical center for any further medical care thus never submitting a blood sample) and 868 patients had results available to providers at the time of data cut-off. The final patients included for analysis were those who returned for at least one clinic visit with a study provider after their pharmacogenomic results became available (n=547). All such clinic visits for these patients were analyzed over a three-year period (October 1, 2012–September 11, 2015). This yielded 2279 clinic visits by these patients with their enrolling providers during the study period, with an average of 3.8 (range 1–26) visits per patient-provider pair. This corresponded to 415 of the 547 patients having multiple visits with their treating provider during the study. Additionally, 47 patients had visits with more than one study provider. Detailed demographic and clinical information about the participating patients and providers is shown in Table 1. Measured characteristics of the provider group suggest that these physicians had modest baseline knowledge of and minimal to no experience with using pharmacogenomics prior to this study, characteristics which mirror those described for a recently-surveyed 10,000 U.S. physician cohort(7).
Table 1.
Patient and Provider Demographics
| Patient Baseline Demographics | ||
|---|---|---|
| Characteristic | Number of Patients (n=547) (%) | |
| Age in years, mean | 60 (range 19–95) | |
| Female/male | 290/257 (53.0/47.0) | |
| Racea | ||
| White | 324 (59.0) | |
| Black or African American | 164 (30.0) | |
| Asian | 25 (4.5) | |
| Unknown | 25 (4.5) | |
| More than one race/ other | 13 (2.3) | |
| American Indian/ Alaskan Native/ Native Hawaiian | 3 (0.005) | |
| Ethnicitya | ||
| Not Hispanic or Latino | 319 (58.0) | |
| Hispanic or Latino | 20 (3.6) | |
| Not Answered/ Unknown | 201 (36.7) | |
| Most Prevalent Diseases | ||
| Hypertension | 277 (50.6) | |
| Hyperlipidemia | 275 (50.0) | |
| Mechanical Joint Disorders | 111 (20.0) | |
| GERD | 105 (19.0) | |
| Obesity | 105 (19.0) | |
| Coronary Artery Disorder (Arteriosclerosis) | 87 (15.9) | |
| Diabetes Mellitus | 69 (12.6) | |
| Hypothyroidism | 67 (12.2) | |
| Sleep Apnea | 65 (11.8) | |
| Most Prevalent Medications (at Study Enrollment) | ||
| Aspirin | 237 (43.3) | |
| Atorvastatin | 134 (24.5) | |
| Hydrochlorothiazide | 104 (19.0) | |
| Lisinopril | 89 (16.3) | |
| Amlodipine | 86 (15.7) | |
| Levothyroxine | 78 (14.3) | |
| Metoprolol | 68 (12.4) | |
| Fluticasone Propionate | 59 (10.8) | |
| Acetaminophen | 55 (10.3) | |
| Visits by Specialty (number of providers) | Number Clinic Visits | Number Patients Seen |
| Primary Care (7) | 1359 | 267 |
| Cardiology (2) | 449 | 138 |
| Oncology (3) | 208 | 60 |
| Gastroenterology (1) | 107 | 53 |
| Executive Health (1) | 49 | 24 |
| Nephrology (1) | 52 | 20 |
| Hepatology (1) | 25 | 22 |
| Pulmonology (1) | 30 | 10 |
| Total (17) | 2279 | 594b |
| Provider Baseline Demographics | ||
| Characteristic | Number of Providers (n=17) (%) | |
| Female/male | 7/10 (41.2/58.8) | |
| Years in practice, mean | 21.4 (range 3–46) | |
| Patients seen per clinic, mean | 9.1 (range 6–16) | |
| Ordered a PGx test in the preceding six monthsc | 5 (29.4) | |
| PGx impacted prescribing in the preceding six monthsc | ||
| Never | 11 (64.7) | |
| Almost never | 2 (11.8) | |
| Sometimes | 3 (17.6) | |
| Frequently | 1 (5.9) | |
| Almost always | 0 (0.0) | |
| “How informed do you feel about PGx?”d | ||
| Very well informed | 2 (11.8) | |
| Somewhat informed | 8 (47.0) | |
| Somewhat under-informed | 5 (29.4) | |
| Very under-informed | 2 (11.8) | |
| “I believe there is insufficient PGx information for most drugs”d | ||
| Agree strongly | 6 (35.3) | |
| Agree somewhat | 7 (41.2) | |
| Not sure | 1 (5.9) | |
| Disagree somewhat | 3 (17.6) | |
| Disagree strongly | 0 (0.0) | |
| “PGx evidence is relevant to prescribing decisions for most of my patients”d | ||
| Agree strongly | 2 (11.8) | |
| Agree somewhat | 4 (23.5) | |
| Not sure | 6 (35.3) | |
| Disagree somewhat | 3 (17.6) | |
| Disagree strongly | 2 (11.8) | |
Race and ethnicity were self-reported by patients.
The total number of patients seen across the various specialties includes 47 patients with visits to multiple providers. For example, a patient with a visit to a primary care physician as well as a cardiologist was counted separately in each specialty.
Prior to participating in this study.
Opinions of pharmacogenomics from each provider were assessed before joining the study.
PGx = pharmacogenomics
At 25% of visits, at least one medication change occurred, representing 812 total medication changes (221 discontinuations/395 medications started/196 dose changes). The most frequent changes are in Table 2.
Table 2.
Applicability of Pharmacogenomics to Most Frequently Changed Medications
| Medication | Prescriptions, n (at baseline) | Medication Changes,a n (%)b | Influencedc by PGx, n (%) |
|---|---|---|---|
| Atorvastatin | 301 | 35 (4.3) | 14 (40.0) |
| Lisinoprild | 240 | 35 (4.3) | n/ad |
| Amlodipine | 232 | 34 (4.2) | 10 (29.4) |
| Hydrochlorothiazide | 258 | 34 (4.2) | 9 (26.5) |
| Omeprazole | 144 | 26 (3.2) | 12 (46.2) |
| Chlorthalidoned | 42 | 23 (2.8) | n/ad |
| Aspirin | 524 | 22 (2.7) | 4 (18.2) |
| Esomeprazole | 73 | 22 (2.7) | 9 (40.9) |
| Fluticasone Propionate | 136 | 20 (2.5) | 2 (10.0) |
| Metoprolol | 196 | 20 (2.5) | 6 (30.0) |
includes new medications, dose changes, and discontinuations;
reflects each drug’s changes as a percentage of all total medication changes in the study;
see Methods for determination of prescriptions influenced by PGx;
no clinically actionable pharmacogenomic information existed for this drug, so attribution of influence of pharmacogenomics for this drug was not applicable (n/a);
PGx = pharmacogenomics
CDS Tool Accessions to Consider Pharmacogenomic Information
The first component of adoption that we sought to establish was physician accession of pharmacogenomic information. Of evaluable visits, 69% were associated with a dedicated login into our institutional pharmacogenomic results CDS system, also called the Genomic Prescribing System (GPS). Composite login rates were 83%, 74%, and 59% in years 1, 2, and 3 of the study, respectively. Each enrolled provider accessed the system on multiple occasions. Interestingly, the likelihood of a physician accessing the GPS was significantly higher at visits where those providers were making a medication change (Odds Ratio [OR]=1.61 [1.24–2.09], p<0.0001), although causality cannot be concluded from this association and it is equally possible that the cause and effect were actually the opposite.
Analysis of physician and patient factors influencing GPS access identified that physicians on busier clinical days were significantly less likely to log-in (mean 10.6 patients/clinic for no login visits versus 8.4 patients/clinic for login visits, p<0.0001). Other analyzed factors were not compelling (Supplementary Table 1).
Physician Responses to Pharmacogenomic Alerts
Table 3 summarizes the pharmacogenomic information viewed by providers during the study. In total, 34.2% of all medications on patients’ active drug lists had associated alerts (i.e., either a favorable green light [20.9%], cautionary yellow light [12.8%], or high-risk red light [0.5%]). These alerts reflect the prevalence of clinical pharmacogenomic information that was available about the medications being taken by patients throughout the study. The remaining medications (65.8% of drugs on patients’ active drug lists) had no known actionable pharmacogenomic information.
Table 3.
Provider Responses to Delivered Pharmacogenomic Alerts
| Alert Color | Total Alerts | Distinct Alerts | |||||
|---|---|---|---|---|---|---|---|
| Delivered | Viewed (%) | Changed (%) | Delivered | Viewed (%) | Changed (%) | Odds Ratio (95% CI, p value)a | |
| Red (High Risk) | 45 | 40 (89%) | 11 (24%) | 20 | 20 (100%) | 10 (50%) | 26.15 (9.0–75.3, p<0.0001) |
| Yellow (Cautionary) | 1072 | 706 (66%) | 53 (5%) | 385 | 287 (75%) | 50 (13%) | 2.4 (1.7–3.5, p<0.0001) |
| Green (Favorable) | 1752 | 410 (23%) | 52 (3%) | 563 | 228 (40%) | 45 (8%) | 1.12 (0.77–1.63, p=0.55) |
| Total PGx Alerts | 2869 | 1156 (40%) | 116 (4%) | 968 | 535 (55%) | 105 (11%) | -- |
| No PGx Infob | 5510 | n/a | 181 (3%) | 1970 | n/a | 149 (8%) | reference |
Notes: In total, 2869 pharmacogenomic alerts were viewed by physicians about drugs on patients’ current medication lists (34% of all medications on drug lists had associated pharmacogenomic alerts). Correspondingly, 5510 medications (66% of all medications on these lists) were denoted as lacking actionable pharmacogenomic information (“no PGx info”). Because providers who had multiple visits with a given patient were provided with duplicate alerts, removal of these repeat deliveries yielded 968 distinct delivered pharmacogenomic alerts (for visits at which physicians logged into the Genomic Prescribing System [GPS]). The frequency of distinct delivered alerts viewed (i.e., clicked to receive detailed clinical decision support [CDS]) reflected the alert color, with 100% red, 75% yellow, and 40% green alerts clicked to receive CDS (p<0.0001, chi-square). Analysis of medication changes as a percentage of truly distinct alert deliveries provided the most descriptive illustration of the effect of pharmacogenomic information on prescribing since many alerts (especially green and yellow alerts) for drugs that the prescriber made a decision not to change were delivered numerous times to the same provider for the same patient during the study.
odds ratios and 95% confidence intervals (CI) describing the likelihood of a medication change were calculated for each pharmacogenomic result type (red, yellow, and green respectively), with the reference (comparator) group consisting of all other medications for which no actionable pharmacogenomic information existed.
indicates the number of drugs for which no actionable pharmacogenomic information was available within the results delivery system. Medication changes involving these drugs represented the basal rate of medication changes occurring irrespective of pharmacogenomic influences, thus serving as the reference group for the analyses;
PGx = pharmacogenomics
To evaluate the primary study endpoint, we sought to measure the composite prescribing influence of available, potentially actionable pharmacogenomic information compared against the prescribing of drugs for which there was no pharmacogenomic guidance. To evaluate this, we examined how frequently providers changed medications when pharmacogenomic information was viewed, and we stratified the analysis by degree of pharmacogenomic risk. Importantly, we found that the OR of a red light medication being changed was 26.2 [9.0–75.3] (p<0.0001) compared to a drug without any pharmacogenomic information (Table 3). Similarly, when compared to green light medications, the OR of a red light medication being changed was 23.3 [7.8–69.5] (p<0.0001). Yellow risk medications had an OR of change=2.4 [1.7–3.5] (p<0.0001) compared to drugs without pharmacogenomic information (Table 3). When compared to green light medications, the OR of a yellow light medication being changed was 2.1 [1.4–3.4] (p=0.001). The only variable that differed between these medication changes was the presence (or absence) of potentially actionable pharmacogenomic information, meaning that the observed differences in rates of prescribing changes were likely directly due to the pharmacogenomic information provided. There was no difference in the rate of change for green light drugs compared to drugs without pharmacogenomic information, suggesting that physicians interpreted green lights (genetically favorable) as not warranting modification.
Interestingly, medication changes in response to pharmacogenomic risk alerts did not always occur the first time the provider viewed the alert, instead requiring repeat delivery before inducing a prescribing change. For example, 51% (27/53) of yellow alert changes occurred at the first delivery, while 49% (26/53) happened at a subsequent delivery.
Among the most frequently changed drugs, a significant proportion of the changes were influenced by available pharmacogenomic information (Table 2, far-right column). Nearly half of all changes in omeprazole and atorvastatin—two frequently prescribed drugs in our cohort—were influenced by pharmacogenomic results. Simvastatin (69%) and rabeprazole (50%), though not among the most frequently changed drugs in our study, had the highest overall percentages of changes influenced by pharmacogenomic CDS.
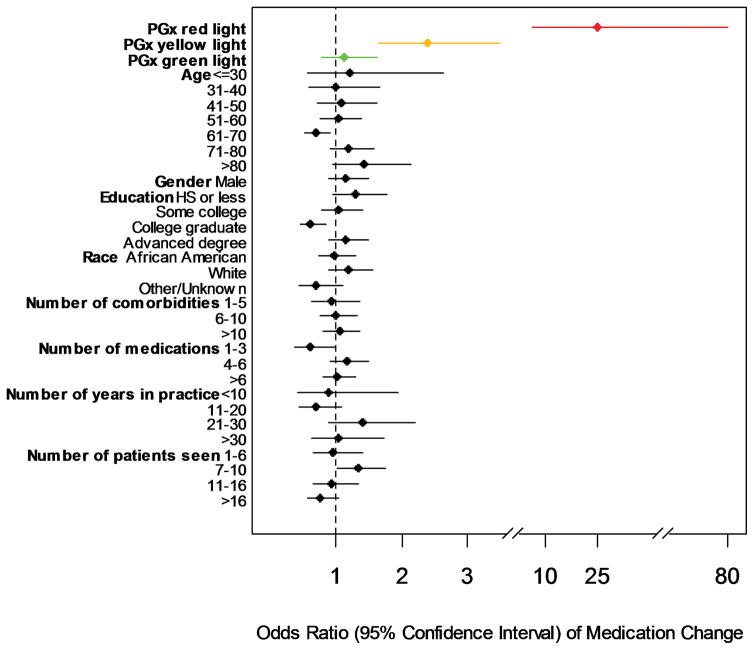
To contextualize the above findings, we assessed for contributions of specific patient and physician factors on associations with medication changes. In contrast to pharmacogenomic information, almost none of these factors showed a reliable association with whether a patient was likely to have a medication change occur (Figure 1). The only exception was that patients on the fewest number of medications (1–3) were less likely than other patients to have their provider make a prescription change. This latter finding likely logically reflects the fact that patients on fewer medications simply had less opportunities for their physicians to change one of those medications compared to patients taking a larger number of prescriptions.
Figure 1. Analysis of Patient, Physician Practice, and Pharmacogenomic Factors on Likelihood of a Medication Change.
Odds ratios (OR) and 95% confidence intervals are displayed for each patient, physician practice, and pharmacogenomic risk variable analyzed. In contrast to the large impact of pharmacogenomic information, almost none of the other analyzed clinical factors showed a reliable association with medication changes occurring at visits. The only exception was that patients on the fewest number of total medications (1–3 medications) were less likely than other patients to have a prescription change. Note: it is noted that single sub-group variables (like “patients aged 61–70 years”, or “college graduates”) within some of the broader evaluated clinical categories had individual, statistically significant findings, but we did not consider these as significant results at the level of the overall clinical variable because the relationship was not retained across the entire category (e.g., age was not associated with medication change likelihood across the full range of analyzed age-decade sub-groups; and educational level as a variable was not associated with medication change likelihood overall, in fact, those with advanced degrees beyond college trended in the opposite direction as college graduates). PGx=pharmacogenomic; HS=high school.
Pharmacogenomic Information Appropriately Impacts Major Prescription Decisions
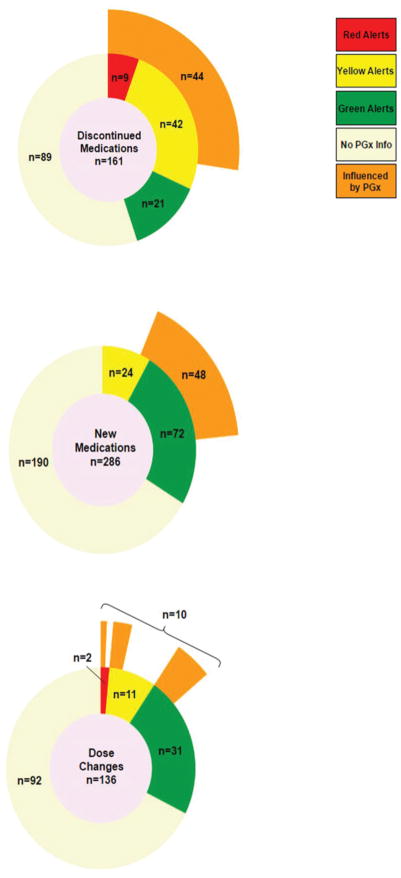
To further explore the extent to which pharmacogenomic results were driving medication change behaviors, we analyzed specific drug changes and their corresponding influence (‘attribution’) scores at all visits where GPS was accessed. Drug discontinuations were informative as a first category. As shown in Figure 2A, when pharmacogenomic information was available, a large majority of provider decisions to stop drugs were influenced by pharmacogenomic recommendations (61%). All of the pharmacogenomically-impacted drug discontinuations occurred for drugs with red and yellow risk alerts. Providers frequently (at least 8 unique times) discontinued statins in patients with increased genetic myopathy risk(27) (SLCO1B1 rs4149056 C-allele carriers, yellow light in GPS) and in patients with drug-specific genomically-predicted suboptimal lipid lowering(28, 29), in exchange for alternative statins. For proton pump inhibitors (PPIs), a very commonly observed maneuver (at least 15 different occurrences) was for providers to discontinue first-generation PPIs in CYP2C19 ultrarapid metabolizers (red lights) in favor of newer-generation PPIs, which have less susceptibility to genomic variability at the CYP2C19 locus(30, 31). For hypertension treatment, physicians frequently (at least 12 different times) noted difficulty achieving blood pressure optimization with one or more antihypertensive agents, and would use pharmacogenomic information to discontinue one antihypertensive in favor of a genomically favorable alternative. To do so, clinicians used a disease-based search function(32)—receiving all pharmacogenomic information about antihypertensives for a given patient—or the “pharmacogenomic alternatives” CDS information to choose a genomically favorable replacement. The latter behavior—selection of a genetically favorable pharmacogenomic alternative medication to replace an existing higher risk drug—in many instances was facilitated by the provision of a list of all pharmacogenomically annotated (red/yellow/green) medications in the tool, provided on the same screen alongside the ‘current medications’ list (see Supplementary Figure 1). This feature, enabling this observed behavior among providers, illustrated an important advantage of preemptive genotyping in our implementation model. The use of such comparative information also importantly illuminated the observation that study providers found pharmacogenomic information to be of high utility in situations where there was apparent clinical indifference among several therapy choices that could be prevailed upon by genomic information.
Figure 2. Available Pharmacogenomic Information Impacts Major Prescription Decisions.
Medication changes at all study visits where providers accessed pharmacogenomic results via GPS are depicted. In the center of each diagram, the total number of medication changes of each type are shown (2A=visits with drug discontinuations; 2B=visits with new medications prescribed; 2C=visits with dose adjustments). The first concentric circle then divides the total number of medication changes into categories based on whether pharmacogenomic information was available within the GPS (beige represents drugs without pharmacogenomic information; green/yellow/red represent drugs with each of these respective pharmacogenomic alert types). The outermost concentric layer (orange) indicates the proportion of those medication changes, stratified by green/yellow/red alert level, that were attributed through a formal evaluation process as being influenced by the pharmacogenomic information. 2A - Drug Discontinuations. There were 161 drug cessations during the study period at visits where providers accessed the GPS. While about half (n=89, 55%) of these discontinued medications did not have pharmacogenomic information, when pharmacogenomic signals were available (9 + 42 + 21=72), a large majority (44/72=61%) of provider decisions to stop drugs were influenced by the provided pharmacogenomic recommendations. 2B – New Medications Prescribed. There were 286 new drugs prescribed during study visits where providers accessed the GPS. While the majority of these new prescriptions did not have pharmacogenomic information (n=190), for those that did (24+72=96), physicians reported that the decision to prescribe the ultimately chosen drug was affirmatively influenced by pharmacogenomic information in 50% (48/96) of cases. 2C - Dose Adjustments. There were 136 dose changes at visits when providers accessed the GPS during the study period. The majority of dose-adjustments that occurred in drugs with pharmacogenomic information were for green light drugs (n=31), although yellow (n=11) and red light medication (n=2) dose-adjustments were observed. Altogether, 23% (10/44) provider decisions to make dose changes in drugs with viewed pharmacogenomic information were influenced by GPS recommendations. GPS=Genomic Prescribing System.
New prescriptions also revealed important findings. Figure 2B shows that for new prescriptions with pharmacogenomic information (n=96), the decision to prescribe the chosen drug was affirmatively influenced by pharmacogenomic information in 50% of cases. The vast majority of newly-prescribed drugs with pharmacogenomic information were genetically compatible green lights. Most importantly, not one pharmacogenomically high-risk (red alert) medication was prescribed during the entire study when physicians consulted the CDS tool. Even when yellow light drugs were chosen during GPS-utilized visits, those new medications often represented suitable choices when considering clinical context (e.g., yellow light drugs started as replacements for red lights, or started at modified initial doses in accordance with CDS recommendations).
Dose-adjustment to mitigate risk represented a final category of observed prescribing behavior in response to pharmacogenomic information (Figure 2C). Altogether, these composite results show that the ultimate prescribing decision for a considerable proportion of drug changes at visits where pharmacogenomic CDS was utilized were influenced by pharmacogenomics.
Missed Opportunities
Some implementation/adoption aspects tempered more universal impact (Supplementary Figure 2). First, because we chose a passive resulting approach, providers did not always see potentially important alerts (Supplementary Figure 2-legend). Secondly, when new medications were initiated, only 16% were actively searched in GPS prior to being prescribed. Pharmacogenomic information would have been available for 27% of these unsearched instances. Both delivery-model limitations of our implementation and physician behavior inconsistencies represented opportunities for ongoing iteration of our model and potential behavioral intervention to increase adoption.
DISCUSSION
To our knowledge, this is the first comprehensive prospective examination of physician prescribing behaviors in the context of available, broad preemptive pharmacogenomic CDS. We found that physician desire for genomic-based prescribing was robust, and that pharmacogenomic information impacted prescribing in a pattern consistent with greatest reduction of pharmacogenomic risk for patients. Our findings represent a key milestone for encouraging broader integrations of genomic medicine into clinical practice.
The successful clinical translation of genomic information requires not only technological adaptations but also, importantly, behavioral adoption into the clinical decision calculus. Our findings demonstrated that both practical barriers (point-of-care availability, physician time constraints) as well as enactment barriers (inertia of previous practice, lack of familiarity)(24) can be surmounted. In particular, we identified (through physician electronic medical record [EMR] documentation during this study) a consistent theme that outcome expectancy—the expectation that a given behavior will lead to a particular consequence—was the strongest driver of prescribing behavior change among our early adopters. In other words, if utilizing genomic information meant increasing the likelihood that adverse events could be avoided or drug responses could be augmented, then physicians were highly likely to adopt such information during prescribing. The stronger the cause-effect relationship for a pharmacogenomic variant, the greater the likelihood of adoption. Indeed, we found that red (highest risk) alerts were much more likely to alter behavior change than yellow (moderate) alerts, indicating that the stakes associated with non-consideration of genomic information mattered greatly to our early adopters. This implies that implementation models focused around the pharmacogenomic variants with the greatest potential for risk-avoidance might be the most initially successful. Two recent prior studies(33, 34) focused solely on clopidogrel—an ideal high-risk pharmacogenomic example—support this idea. Our findings also suggest that as physicians have increasing opportunities to consider and utilize genomic information—gaining thereby experience in actual practice about whether prescribing precision is improved because of the information—then this will serve to have a large effect on whether adoption will take hold and expand, or not. In this sense, one could strongly argue that future examinations of pharmacogenomic implementation—including perhaps pragmatic clinical trials—might not only need to measure patient outcome benefits for individual drug-gene pairs, but also should consider physician utilization as a potential indicatory benchmark of utility. The role of the patient in potentially influencing physician prescribing behavior must also be considered(35).
This study illuminated potentially important findings about pharmacogenomic CDS software as it pertains to adoption. Rogers(36) previously posited that five primary aspects affect the diffusion and uptake of an innovation—relative advantage, compatibility, complexity, trialability, and observability(37). The nature of a CDS at the point-of-care has the potential to directly affect at least three of these aspects. First, we used a passive resulting approach because it allowed us to directly study questions of compatibility (the degree to which an innovation fits with the needs of potential adopters(37)) and alert thresholds. We found a high GPS accession rate, which was enhanced when physicians made medication changes, indicating that physicians are perhaps used to utilizing ancillary electronic sources to receive medical information(38) and will in fact seek such information if the information is perceived as useful. In fact, our accession rate was higher than anticipated (we had hypothesized an overall login rate of 50%) and this was despite the fact that GPS was not integrated within the EMR during the study. (We have since integrated GPS into our institutional EMR—a move likely critical for sustaining and expanding adoption and almost certainly necessary for implementation success in larger, real-world contexts). Secondly, we designed our pharmacogenomic CDS to minimize complexity, enabling providers to understand implications for prescribing without necessarily understanding genomics. The traffic light alerts did this with a recognizable iconography, but additionally, none of the more detailed CDS contained more than 30 seconds of readable text. Finally, our implementation emphasized trialability, allowing users to decide on relevance on a case-by-case basis in a way that system-wide interruptive alerts do not. Other studies have suggested that infrequent alerts containing embedded clinical recommendations prove more useful and are less likely to be ignored(39). This also diminishes the problems of alert fatigue(40). We do not, however, propose that an entirely passive pharmacogenomic CDS approach is optimal, and in fact, we are currently studying ways to re-engineer our system to combine passive and active(41) alerts to maximize adoption. These concepts deserve close attention as institutions develop pathways and procedures for the delivery of genomic results in increasingly crowded EMRs(42–44).
Our study had limitations. The number of physicians analyzed for prescribing behavior was relatively small. However, the participating physicians represented a diverse group drawn from multiple various medicine disciplines and specialties, and their baseline attitudes and perceptions and minimal to no prior experience with pharmacogenomics suggested that they may not be dissimilar from most contemporary U.S. physicians who share similar general enthusiasm for, but remarkable inexperience with, pharmacogenomics(7). It is nevertheless acknowledged that this study’s providers likely had a bias towards pharmacogenomics because of their willingness to participate, and additionally all were formally included as study co-investigators as well as study subjects with full knowledge at enrollment that their behaviors would be examined, meaning results were subject to the Hawthorne effect. We attempted to minimize the magnitude of this effect by monitoring physician use of pharmacogenomic information over a three-year period—and across >2200 clinic visits—a fact which in our view speaks to the consistency of the behavioral changes and points favorably for next-wave implementations among other provider groups. Indeed, early adopters like the physicians represented in this study are exactly the types who will lead future precision medicine adoption curves. Therefore, studying behavior change in this group was an important step to inform wider dissemination and implementation efforts. The number of red alerts in this study was relatively low, reflecting the rarer incidence of these genomic signals in the human population but also likely reflecting some pre-study trial and error that led to avoidance of these drugs in some patients. Moreover, the total number of drug changes in response to potentially actionable alerts was relatively modest (60 prescription changes out of 405 potentially actionable red and yellow alerts over the entire study), suggesting that there is still considerable room to improve on the adoption curve for pharmacogenomic implementation. Nevertheless, these factors did not limit our statistical power to observe important differences in physician behavior in response to risk alerts. Finally, our study was not powered to detect changes in the rates of specific adverse drug reactions. Clinical outcomes will be important to quantify in future disseminations.
Pharmacogenomic CDS repeatedly and favorably influenced prescribing decisions in a manner that reduced patient risk, with near universal avoidance of genomically-identified high-risk prescriptions when pharmacogenomic information was considered. These findings represent crucial landmarks for the wider implementation of genomic medicine interventions as essential components of the advancement of precision medicine.
METHODS
Participants
Physicians were individually approached for enrollment through a process of direct stakeholder engagement and informed consent. Invited providers from two physically separate outpatient sites were chosen because of busy clinical practices, and to achieve representation across different medicine subspecialties. Providers gave permission for their medication decisions to be analyzed. Physicians were not mandated to view or utilize pharmacogenomic results, nor required to make any pre-specified prescribing choices. Physicians could leave the study at any time. Prior to having access to pharmacogenomic results, enrolled physicians participated in a semi-structured interview about pharmacogenomics with the study principal investigator (P.H.O.), completed a baseline questionnaire, and received a ~15-minute training session on use of the CDS tool, the GPS.
Adult patients receiving care from an enrolled physician were correspondingly approached for participation by their physician or by study personnel using informed consent. Eligibility criteria were previously described(26). The study was approved by the Institutional Review Board of the University of Chicago, and was registered at clinicaltrials.gov (#NCT01280825).
Genotyping
Enrolled patients were genotyped across a broad custom pharmacogenomic panel comprised of variants selected for their pharmacogenomic importance. Sequenom custom MassARRAYs (Agena Bioscience, San Diego, CA) were used prior to May 5, 2015, while custom OpenArrays from Life Technologies (ThermoFisher, Waltham, MA) were used thereafter. All patients were additionally genotyped across a custom CYP2D6 panel developed in conjunction with Hologic(45) with supplemental CYP2D6 Taqman copy number assessment. All genotyping was conducted in Clinical Laboratory Improvement Amendments (CLIA)-certified and College of American Pathologists (CAP)-accredited laboratories.
Pharmacogenomic Results Delivery
GPS(25, 26, 32) is a web-accessible, password-protected portal delivering preemptive pharmacogenomic results utilizing green/yellow/red “traffic light” alerts(25, 46) to inform physicians of pharmacogenomic drug safety or efficacy (Supplementary Figure 1 shows representative screen shots). Alerts are based on published pharmacogenomic information, including, when available, meta-analyses, clinical guidelines, and FDA label information(25, 47). The list of drugs for which the GPS provided CDS during the study period is shown in Supplementary Table 2. Green lights indicate genetic compatibility for a drug, while yellow and red denote increased risk of adverse reactions or non-response. Detailed clinical information about each pharmacogenomic result is included within result-translation synopses that comprise each drug-based CDS. These CDS include assessment(32, 47) of the quantity and strength of the composite published evidence. New drug/variant-specific CDS were indeed pushed into GPS during the course of the study if published literature became available to support a given genetic variant being migrated to clinically actionable (‘CDS deliverable’) status. Similarly, if new published data changed the clinical recommendation or level of evidence rating for an existing CDS, then that CDS was modified/updated in real-time during the study (this happened with warfarin, for example).
On any given clinic day, a provider would see a number of patients, only some of whom were participating in the study and had GPS information available. In order to remind providers of which patients were study participants with results available in the GPS, study staff emailed providers the day before study patients’ clinic visits. Additionally, reminder stickers were placed next to study patients’ names on providers’ daily paper schedules in clinic workrooms, denoting which patients had pharmacogenomic results available. During the early phases of the project, members of the clinical research team were embedded in study providers’ clinics to provide assistance with GPS. The presence of these staff potentially also served as a reminder to providers to login to the GPS.
GPS was not integrated into the EMR during the period of this study. When providers logged into GPS, each patient’s home screen displayed all current medications for that patient with any associated pharmacogenomic traffic lights. Patients’ current medication lists were manually updated in the GPS by clinical research staff at least 24 hours before each clinic visit to match the current medications listed in the EMR. Drugs without pharmacogenomic information were denoted “no information” (colorless traffic light symbol). A physician’s action of “clicking” on an alert or searching for a drug by name or disease(32) yielded the relevant pharmacogenomic result and CDS.
Evaluating Utilization of Pharmacogenomic CDS
The study was conducted between October 1, 2012–September 11, 2015 (data cut-off). Evaluable clinic visits during this period included all visits at which a preemptively genotyped patient was seeing an enrolled provider, meaning their pharmacogenomic results were available for consideration through GPS. Click log (“use”) data detailing physician navigations into and within GPS were recorded. Data captured included login date/time, names and alert colors of prescribed drugs at each visit, and whether individual alerts were clicked to view more detailed CDS synopses. Logins within (+/−)72 business hours of a clinic visit were considered “associated” with that visit. To correlate GPS use behaviors to prescribing actions, we aligned GPS data by date and provider-patient pair with EMR clinical documentation including pre-visit/post-visit medication lists. All prescription drug changes (new prescriptions/discontinuations/dose changes) made during each evaluable visit were recorded. Medication changes involving topicals/ear or eye drops/vitamins/stool softeners/probiotics and over-the-counter medications were excluded, as were changes involving short-term prescription drugs (defined as use for ≤14 days; e.g., short antibiotic courses) given that the focus of the study was longitudinal long-term prescribing management.
Role of Pharmacogenomic Results in Informing Medication Changes
To rigorously assess the degree, if any, to which the available pharmacogenomic information influenced medication changes, we employed a modified version of the widely used scale developed by the National Cancer Institute’s Cancer Therapy Evaluation Program for attribution of drug toxicity(48) (http://ctep.cancer.gov/protocolDevelopment/electronic_applications/docs/newadverse_2006.pdf). The formal scoring algorithm which was first pretested and then implemented is shown in Supplementary Figure 3. In this scoring system, 1=a medication change “unrelated” to the availability of pharmacogenomic results, 2=“unlikely related”, 3=“possibly related”, 4=“likely related”, and 5=“definitely related”. EMR documentation, GPS click logs, post-visit physician surveys, and post-visit patient surveys were used to assign attributions. An initial subset of attribution scores underwent comparative analysis by at least two independent raters (P.H.O./N.W./B.A.B./J.P.H./C.K./S.H.) until consensus on the algorithm was reached. At least 20% of all final scores were also rated by at least two independent raters to ensure agreement (κ ≥ 0.95), and any disagreeing scores were reviewed by a third reviewer until consensus. We categorized medication changes with scores ≥4 as “influenced by pharmacogenomic information” for the subsequent analyses—a conservative cutoff that was even more stringent than that used in clinical trials for ascribing adverse drug events(49).
Statistical Analyses
For the primary analysis, the influence of available pharmacogenomic information on composite physician prescribing behaviors was assessed by modeling the delivery of a pharmacogenomic alert and a physician’s behavior of enacting (or not) a medication change. Generalized linear mixed-effects models based on the binomial distribution were used including two random effects, one for repeated viewing of alerts for the same patient and the other for patients nested within providers. We included all medications on each patient’s ‘current medications’ list anytime during the study. ORs and 95% confidence intervals for each pharmacogenomic result (red/yellow/green respectively) were estimated referenced to the (comparator) group of all other medications for which no pharmacogenomic information existed.
We also evaluated patient factors (age/gender/education/race/number of comorbidities/number of medications) and provider factors (number of years in practice, number of patients seen that day in clinic) as potential mediators and moderators of medication change behaviors. For these analyses, ORs were calculated in univariate fashion employing a mixed-effects model, using all other possible categories within a given factor as the comparator group (for example, patients taking “1–3 medications” were compared against all other patients taking 4+ medications). Statistical analyses were performed using Stata, version 13.0. P values <0.05 were considered significant.
Supplementary Material
Supplementary Figure 1A shows an example results homepage for an individual patient as viewed by the provider. Detailed clinical decision supports (CDS) (screen shown in Supplementary Figure 1B) were provided in an on-demand fashion when the provider “clicked” on any of the traffic-light result signals from the patient homepage. Alerts were categorized into three stratification levels of evidence, with level 1 recommendations having the strongest supporting evidence (pharmacogenomic information present in the FDA label and/or the presence of a published pharmacogenomic guideline). The quantity of the clinical evidence (total number of patients studied, number of positive studies) and quality (study design, controls, replication) distinguish the remaining level 2 and level 3 alerts. However, all reported, actionable recommendations were based on strong published human clinical studies demonstrating the association of the pharmacogenomic variant with clinical outcomes. Note that, because a comprehensive preemptive genotyping approach was used, physicians had the ability to immediately identify genetically compatible alternative medications in the “pharmacogenomic alternatives” column (see far right of screen shot in Supplementary Figure 1A).
The flow diagram shows the process used to assign attribution (“influence”) scores for each individual medication change throughout the entire study period. In this scoring system, 1 represents a medication change that was “unrelated” to the availability of pharmacogenomic results, 2=“unlikely related”, 3=“possibly related”, 4=“likely related”, and 5=“definitely related”. Physician electronic medical record documentation, Genomic Prescribing System (GPS) click logs, and post-visit physician and patient surveys were used to assign attributions using a standardized, formal process. At least 20% of all final attribution scores were rated by at least two independent raters to ensure agreement (κ ≥ 0.95). Medication changes with scores ≥4 were categorized as being “influenced by pharmacogenomic information” for all subsequent analyses.
Patterns of suboptimal utilization are shown—representing both delivery-model limitations of our implementation and physician behavior inconsistencies—suggesting opportunities for ongoing iteration and intervention to increase future adoption. First, because we chose a passive resulting approach, providers sometimes did not see potentially important alerts, primarily because of lack of log-in. Because of this, across the 711 visits at which physicians did not log into GPS (31% of all study visits), 1298 total potential alerts were not delivered, with 39% of those representing yellow or red alerts (only n=7 were distinct red lights, with six being non-response PPI signals). Most prominently among the undelivered alerts were eight unique yellow lights for simvastatin, and one yellow light for clopidogrel. In order to mitigate this problem, during the course of this study a small number (4/17) of our early adopters themselves instituted programs within their clinics wherein they had clinic assistants always pre-print pharmacogenomic results from the CDS system for their manual review while seeing patients. Interestingly, when these physicians employed this type of assistance from ancillary clinic personnel for pharmacogenomic integration, the number of ‘missed alerts’ was almost zero. As one primary example of subsequent implementation iteration, and in part because of these results, the previously stand-alone GPS has now been integrated with our institutional EMR. Secondly, providers only rarely proactively utilized the GPS during new medication prescribing. Pharmacogenomic information would have been available for 27% of these unsearched instances. Fortunately, none of the unsearched new medications were prescribed to patients who would have had a red alert for the chosen drug. Lastly, several use constraints were identified that caused behavioral inconsistencies for some providers. Notes: aOut of 286 new medications that were prescribed when physicians logged in, the search functionality/alternative medication column was used 62 times. That meant 224 new medications were prescribed during a login visit without utilizing search functionalities. bFour physicians routinely accessed pharmacogenomic information printed out from the GPS by ancillary staff. cOut of the 1568 login visits during the study period, a disease search was conducted 26 times. dOf the visits where a provider logged in, there were 98 distinct risk alerts that were viewed but not clicked to view the more detailed clinical decision supports. eOver the course of the study period, 103 new medications were prescribed without a login. Pharmacogenomic information was available for 32 of these drugs. fThere were 253 distinct red and yellow alerts that were never delivered due to lack of login.
Supplementary Table 1. Factors Potentially Impacting Physician Accession of Genomic Prescribing System.
Supplementary Table 2. Drugs Available in GPS during Study Period.
STUDY HIGHLIGHTS.
What is the current knowledge on the topic?
Many groups have worked to address barriers to genomic implementation. As with any clinical innovation, the true utility of an intervention is measured by adoption and user response. Understanding clinician prescribing behaviors in response to genomic information is an essential step.
What question did this study address?
We hypothesized that preemptively-obtained pharmacogenomic results delivered to physicians via a point-of-care clinical decision support (CDS) tool would significantly improve prescribing.
What this study adds to our knowledge
Pharmacogenomic information impacted physicians to change prescribing in patterns aimed at reducing patient risk. Medication changes occurred among drugs with known pharmacogenomic risk significantly more often than for drugs with no pharmacogenomic information. When genomic information was available and viewed, the majority of decisions around medication discontinuations and new prescriptions were influenced by pharmacogenomic results.
How this might change clinical pharmacology or translational science
This is the first comprehensive prospective examination of physician prescribing behaviors in the context of available, broad preemptive pharmacogenomic CDS. The findings represent key milestones for encouraging broader integrations of genomic medicine into clinical practice.
Acknowledgments
This research was supported by NIH K23 GM 100288-01A1 (P.H.O.), NIH/National Heart, Lung, and Blood Institute grant 5 U01 HL105198-09 (M.J.R. and P.H.O.), The University of Chicago Pritzker Summer Research Program (NIH 2T35D062719-26), The Conquer Cancer Foundation of the American Society for Clinical Oncology (M.J.R.), The William F. O’Connor Foundation, The University of Chicago Comprehensive Cancer Center support grant, The University of Chicago Bucksbaum Institute for Clinical Excellence Pilot Award (P.H.O.), and the Central Society for Clinical and Translational Research - Early Career Development Award (P.H.O.).
Footnotes
CONFLICT OF INTEREST/DISCLOSURE
P.H.O., K.D., and M.J.R. are named as co-inventors on a pending patent application for the Genomic Prescribing System. M.J.R. is a co-inventor holding patents related to pharmacogenetic diagnostics and receives royalties related to UGT1A1 genotyping, although no royalties were received from the genotyping performed in this study. P.H.O., K.D., and M.J.R. are co-founders and stockholders of PrescriptIQ, Inc. R.B.A. is a founder of, stockholder of, and consultant to Personalis, Inc.
AUTHOR CONTRIBUTIONS
P.H.O., N.W., and B.A.B. wrote the manuscript; P.H.O., L.P.M., R.B.A., O.I.O., W.M.S., D.O.M., and M.J.R. designed the research; P.H.O., N.W., K.D., B.A.B., J.P.H., C.K., S.H., M.S., M.J.S., A.M.D., Y.A.S., R.N., T.S.P., J.L.K., D.L.B., K.L., D.T.R., C.M., M.E.S., W.H., A.S.C., B.P., K-T.J.Y., E.K.Y.L., W.M.S., and M.J.R. performed the research; P.H.O., N.W., K.D., B.A.B., S.M.L., J.P.H., C.K., S.H., and M.J.R. analyzed the data; P.H.O., K.D., L.P.M., K-T.J.Y., E.K.Y.L., S.L.V., D.O.M., and M.J.R. contributed new reagents/analytical tools.
References
- 1.Nussbaum RL, McInnes RR, Willard HF, Hamosh A, Thompson MW. Thompson & Thompson genetics in medicine. Philadelphia: Saunders/Elsevier; 2007. Available from: http://site.ebrary.com/lib/uchicago/docDetail.action?docID=10567394. [Google Scholar]
- 2.Spear BB, Heath-Chiozzi M, Huff J. Clinical application of pharmacogenetics. Trends in molecular medicine. 2001;7(5):201–4. doi: 10.1016/s1471-4914(01)01986-4. [DOI] [PubMed] [Google Scholar]
- 3.Klein TE, Altman RB. PharmGKB: the pharmacogenetics and pharmacogenomics knowledge base. The pharmacogenomics journal. 2004;4(1):1. doi: 10.1038/sj.tpj.6500230. [DOI] [PubMed] [Google Scholar]
- 4.Relling MV, Evans WE. Pharmacogenomics in the clinic. Nature. 2015;526(7573):343–50. doi: 10.1038/nature15817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Altman RB. Pharmacogenomics: “noninferiority” is sufficient for initial implementation. Clinical pharmacology and therapeutics. 2011;89(3):348–50. doi: 10.1038/clpt.2010.310. [DOI] [PubMed] [Google Scholar]
- 6.Manolio TA, Chisholm RL, Ozenberger B, Roden DM, Williams MS, Wilson R, et al. Implementing genomic medicine in the clinic: the future is here. Genetics in medicine : official journal of the American College of Medical Genetics. 2013;15(4):258–67. doi: 10.1038/gim.2012.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stanek EJ, Sanders CL, Taber KA, Khalid M, Patel A, Verbrugge RR, et al. Adoption of pharmacogenomic testing by US physicians: results of a nationwide survey. Clinical pharmacology and therapeutics. 2012;91(3):450–8. doi: 10.1038/clpt.2011.306. [DOI] [PubMed] [Google Scholar]
- 8.Wang L, McLeod HL, Weinshilboum RM. Genomics and drug response. The New England journal of medicine. 2011;364(12):1144–53. doi: 10.1056/NEJMra1010600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shuldiner AR, Relling MV, Peterson JF, Hicks JK, Freimuth RR, Sadee W, et al. The Pharmacogenomics Research Network Translational Pharmacogenetics Program: overcoming challenges of real-world implementation. Clinical pharmacology and therapeutics. 2013;94(2):207–10. doi: 10.1038/clpt.2013.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dunnenberger HM, Crews KR, Hoffman JM, Caudle KE, Broeckel U, Howard SC, et al. Preemptive clinical pharmacogenetics implementation: current programs in five US medical centers. Annual review of pharmacology and toxicology. 2015;55:89–106. doi: 10.1146/annurev-pharmtox-010814-124835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Relling MV, Klein TE. CPIC: Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network. Clinical pharmacology and therapeutics. 2011;89(3):464–7. doi: 10.1038/clpt.2010.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weitzel KW, Alexander M, Bernhardt BA, Calman N, Carey DJ, Cavallari LH, et al. The IGNITE network: a model for genomic medicine implementation and research. BMC medical genomics. 2016;9:1. doi: 10.1186/s12920-015-0162-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Swen JJ, Wilting I, de Goede AL, Grandia L, Mulder H, Touw DJ, et al. Pharmacogenetics: from bench to byte. Clinical pharmacology and therapeutics. 2008;83(5):781–7. doi: 10.1038/sj.clpt.6100507. [DOI] [PubMed] [Google Scholar]
- 14.Rasmussen-Torvik LJ, Stallings SC, Gordon AS, Almoguera B, Basford MA, Bielinski SJ, et al. Design and anticipated outcomes of the eMERGE-PGx project: a multicenter pilot for preemptive pharmacogenomics in electronic health record systems. Clinical pharmacology and therapeutics. 2014;96(4):482–9. doi: 10.1038/clpt.2014.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gottesman O, Scott SA, Ellis SB, Overby CL, Ludtke A, Hulot JS, et al. The CLIPMERGE PGx Program: clinical implementation of personalized medicine through electronic health records and genomics-pharmacogenomics. Clinical pharmacology and therapeutics. 2013;94(2):214–7. doi: 10.1038/clpt.2013.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pulley JM, Denny JC, Peterson JF, Bernard GR, Vnencak-Jones CL, Ramirez AH, et al. Operational implementation of prospective genotyping for personalized medicine: the design of the Vanderbilt PREDICT project. Clinical pharmacology and therapeutics. 2012;92(1):87–95. doi: 10.1038/clpt.2011.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoffman JM, Haidar CE, Wilkinson MR, Crews KR, Baker DK, Kornegay NM, et al. PG4KDS: a model for the clinical implementation of pre-emptive pharmacogenetics. American journal of medical genetics Part C, Seminars in medical genetics. 2014;166C(1):45–55. doi: 10.1002/ajmg.c.31391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bielinski SJ, Olson JE, Pathak J, Weinshilboum RM, Wang L, Lyke KJ, et al. Preemptive genotyping for personalized medicine: design of the right drug, right dose, right time-using genomic data to individualize treatment protocol. Mayo Clinic proceedings. 2014;89(1):25–33. doi: 10.1016/j.mayocp.2013.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson JA, Elsey AR, Clare-Salzler MJ, Nessl D, Conlon M, Nelson DR. Institutional profile: University of Florida and Shands Hospital Personalized Medicine Program: clinical implementation of pharmacogenetics. Pharmacogenomics. 2013;14(7):723–6. doi: 10.2217/pgs.13.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eadon MT, Desta Z, Levy KD, Decker BS, Pierson RC, Pratt VM, et al. Implementation of a pharmacogenomics consult service to support the INGENIOUS trial. Clinical pharmacology and therapeutics. 2016;100(1):63–6. doi: 10.1002/cpt.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thorn CF, Klein TE, Altman RB. PharmGKB: the Pharmacogenomics Knowledge Base. Methods in molecular biology. 2013;1015:311–20. doi: 10.1007/978-1-62703-435-7_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spallek H, Song M, Polk DE, Bekhuis T, Frantsve-Hawley J, Aravamudhan K. Barriers to implementing evidence-based clinical guidelines: a survey of early adopters. The journal of evidence-based dental practice. 2010;10(4):195–206. doi: 10.1016/j.jebdp.2010.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yeh ML, Chang YJ, Wang PY, Li YC, Hsu CY. Physicians’ responses to computerized drug-drug interaction alerts for outpatients. Computer methods and programs in biomedicine. 2013;111(1):17–25. doi: 10.1016/j.cmpb.2013.02.006. [DOI] [PubMed] [Google Scholar]
- 24.Cabana MD, Rand CS, Powe NR, Wu AW, Wilson MH, Abboud PA, et al. Why don’t physicians follow clinical practice guidelines? A framework for improvement. Jama. 1999;282(15):1458–65. doi: 10.1001/jama.282.15.1458. [DOI] [PubMed] [Google Scholar]
- 25.O’Donnell PH, Bush A, Spitz J, Danahey K, Saner D, Das S, et al. The 1200 patients project: creating a new medical model system for clinical implementation of pharmacogenomics. Clinical pharmacology and therapeutics. 2012;92(4):446–9. doi: 10.1038/clpt.2012.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O’Donnell PH, Danahey K, Jacobs M, Wadhwa NR, Yuen S, Bush A, et al. Adoption of a clinical pharmacogenomics implementation program during outpatient care--initial results of the University of Chicago “1,200 Patients Project”. American journal of medical genetics Part C, Seminars in medical genetics. 2014;166C(1):68–75. doi: 10.1002/ajmg.c.31385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Group SC, Link E, Parish S, Armitage J, Bowman L, Heath S, et al. SLCO1B1 variants and statin-induced myopathy--a genomewide study. The New England journal of medicine. 2008;359(8):789–99. doi: 10.1056/NEJMoa0801936. [DOI] [PubMed] [Google Scholar]
- 28.Chasman DI, Posada D, Subrahmanyan L, Cook NR, Stanton VP, Jr, Ridker PM. Pharmacogenetic study of statin therapy and cholesterol reduction. Jama. 2004;291(23):2821–7. doi: 10.1001/jama.291.23.2821. [DOI] [PubMed] [Google Scholar]
- 29.Iakoubova OA, Sabatine MS, Rowland CM, Tong CH, Catanese JJ, Ranade K, et al. Polymorphism in KIF6 gene and benefit from statins after acute coronary syndromes: results from the PROVE IT-TIMI 22 study. Journal of the American College of Cardiology. 2008;51(4):449–55. doi: 10.1016/j.jacc.2007.10.017. [DOI] [PubMed] [Google Scholar]
- 30.Furuta T, Sugimoto M, Kodaira C, Nishino M, Yamade M, Ikuma M, et al. CYP2C19 genotype is associated with symptomatic recurrence of GERD during maintenance therapy with low-dose lansoprazole. European journal of clinical pharmacology. 2009;65(7):693–8. doi: 10.1007/s00228-009-0628-5. [DOI] [PubMed] [Google Scholar]
- 31.McNicholl AG, Linares PM, Nyssen OP, Calvet X, Gisbert JP. Meta-analysis: esomeprazole or rabeprazole vs. first-generation pump inhibitors in the treatment of Helicobacter pylori infection. Alimentary pharmacology & therapeutics. 2012;36(5):414–25. doi: 10.1111/j.1365-2036.2012.05211.x. [DOI] [PubMed] [Google Scholar]
- 32.Hussain S, Kenigsberg BB, Danahey K, Lee YM, Galecki PM, Ratain MJ, et al. Disease-Drug Database for Pharmacogenomic-Based Prescribing. Clinical pharmacology and therapeutics. 2016 doi: 10.1002/cpt.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peterson JF, Field JR, Unertl KM, Schildcrout JS, Johnson DC, Shi Y, et al. Physician response to implementation of genotype-tailored antiplatelet therapy. Clinical pharmacology and therapeutics. 2016;100(1):67–74. doi: 10.1002/cpt.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weitzel KW, Elsey AR, Langaee TY, Burkley B, Nessl DR, Obeng AO, et al. Clinical pharmacogenetics implementation: approaches, successes, and challenges. American journal of medical genetics Part C, Seminars in medical genetics. 2014;166C(1):56–67. doi: 10.1002/ajmg.c.31390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carere DA, VanderWeele TJ, Vassy JL, van der Wouden CH, Roberts JS, Kraft P, et al. Prescription medication changes following direct-to-consumer personal genomic testing: findings from the Impact of Personal Genomics (PGen) Study. Genetics in medicine : official journal of the American College of Medical Genetics. 2016 doi: 10.1038/gim.2016.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rogers E. Diffusion of Innovations. New York: Free Press; 1995. [Google Scholar]
- 37.Scott SD, Plotnikoff RC, Karunamuni N, Bize R, Rodgers W. Factors influencing the adoption of an innovation: an examination of the uptake of the Canadian Heart Health Kit (HHK) Implementation science : IS. 2008;3:41. doi: 10.1186/1748-5908-3-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thomairy NA, Mummaneni M, Alsalamah S, Moussa N, Coustasse A. Use of Smartphones in Hospitals. The health care manager. 2015;34(4):297–307. doi: 10.1097/HCM.0000000000000080. [DOI] [PubMed] [Google Scholar]
- 39.Devine EB, Lee CJ, Overby CL, Abernethy N, McCune J, Smith JW, et al. Usability evaluation of pharmacogenomics clinical decision support aids and clinical knowledge resources in a computerized provider order entry system: a mixed methods approach. International journal of medical informatics. 2014;83(7):473–83. doi: 10.1016/j.ijmedinf.2014.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Beeler PE, Eschmann E, Rosen C, Blaser J. Use of an on-demand drug-drug interaction checker by prescribers and consultants: a retrospective analysis in a Swiss teaching hospital. Drug safety. 2013;36(6):427–34. doi: 10.1007/s40264-013-0022-1. [DOI] [PubMed] [Google Scholar]
- 41.Bell GC, Crews KR, Wilkinson MR, Haidar CE, Hicks JK, Baker DK, et al. Development and use of active clinical decision support for preemptive pharmacogenomics. Journal of the American Medical Informatics Association : JAMIA. 2014;21(e1):e93–9. doi: 10.1136/amiajnl-2013-001993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Herr TM, Bielinski SJ, Bottinger E, Brautbar A, Brilliant M, Chute CG, et al. Practical considerations in genomic decision support: The eMERGE experience. Journal of pathology informatics. 2015;6:50. doi: 10.4103/2153-3539.165999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Overby CL, Rasmussen LV, Hartzler A, Connolly JJ, Peterson JF, Hedberg RE, et al. A Template for Authoring and Adapting Genomic Medicine Content in the eMERGE Infobutton Project. AMIA Annual Symposium proceedings / AMIA Symposium AMIA Symposium. 2014;2014:944–53. [PMC free article] [PubMed] [Google Scholar]
- 44.Caraballo PJ, Hodge LS, Bielinski SJ, Stewart AK, Farrugia G, Schultz CG, et al. Multidisciplinary model to implement pharmacogenomics at the point of care. Genetics in medicine : official journal of the American College of Medical Genetics. 2016 doi: 10.1038/gim.2016.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fang H, Liu X, Ramirez J, Choudhury N, Kubo M, Im HK, et al. Establishment of CYP2D6 reference samples by multiple validated genotyping platforms. The pharmacogenomics journal. 2014;14(6):564–72. doi: 10.1038/tpj.2014.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O’Donnell PH, Danahey K, Ratain MJ. The Outlier in All of Us: Why Implementing Pharmacogenomics Could Matter for Everyone. Clinical pharmacology and therapeutics. 2016;99(4):401–4. doi: 10.1002/cpt.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kaufman AL, Spitz J, Jacobs M, Sorrentino M, Yuen S, Danahey K, et al. Evidence for Clinical Implementation of Pharmacogenomics in Cardiac Drugs. Mayo Clinic proceedings. 2015;90(6):716–29. doi: 10.1016/j.mayocp.2015.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trotti A, Byhardt R, Stetz J, Gwede C, Corn B, Fu K, et al. Common toxicity criteria: version 2.0. an improved reference for grading the acute effects of cancer treatment: impact on radiotherapy. International journal of radiation oncology, biology, physics. 2000;47(1):13–47. doi: 10.1016/s0360-3016(99)00559-3. [DOI] [PubMed] [Google Scholar]
- 49.Eaton A, Iasonos A, Gounder MM, Pamer EG, Drilon A, Vulih D, et al. Toxicity Attribution in Phase I Trials: Evaluating the Effect of Dose on the Frequency of Related and Unrelated Toxicities. Clinical cancer research : an official journal of the American Association for Cancer Research. 2016;22(3):553–9. doi: 10.1158/1078-0432.CCR-15-0339. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1A shows an example results homepage for an individual patient as viewed by the provider. Detailed clinical decision supports (CDS) (screen shown in Supplementary Figure 1B) were provided in an on-demand fashion when the provider “clicked” on any of the traffic-light result signals from the patient homepage. Alerts were categorized into three stratification levels of evidence, with level 1 recommendations having the strongest supporting evidence (pharmacogenomic information present in the FDA label and/or the presence of a published pharmacogenomic guideline). The quantity of the clinical evidence (total number of patients studied, number of positive studies) and quality (study design, controls, replication) distinguish the remaining level 2 and level 3 alerts. However, all reported, actionable recommendations were based on strong published human clinical studies demonstrating the association of the pharmacogenomic variant with clinical outcomes. Note that, because a comprehensive preemptive genotyping approach was used, physicians had the ability to immediately identify genetically compatible alternative medications in the “pharmacogenomic alternatives” column (see far right of screen shot in Supplementary Figure 1A).
The flow diagram shows the process used to assign attribution (“influence”) scores for each individual medication change throughout the entire study period. In this scoring system, 1 represents a medication change that was “unrelated” to the availability of pharmacogenomic results, 2=“unlikely related”, 3=“possibly related”, 4=“likely related”, and 5=“definitely related”. Physician electronic medical record documentation, Genomic Prescribing System (GPS) click logs, and post-visit physician and patient surveys were used to assign attributions using a standardized, formal process. At least 20% of all final attribution scores were rated by at least two independent raters to ensure agreement (κ ≥ 0.95). Medication changes with scores ≥4 were categorized as being “influenced by pharmacogenomic information” for all subsequent analyses.
Patterns of suboptimal utilization are shown—representing both delivery-model limitations of our implementation and physician behavior inconsistencies—suggesting opportunities for ongoing iteration and intervention to increase future adoption. First, because we chose a passive resulting approach, providers sometimes did not see potentially important alerts, primarily because of lack of log-in. Because of this, across the 711 visits at which physicians did not log into GPS (31% of all study visits), 1298 total potential alerts were not delivered, with 39% of those representing yellow or red alerts (only n=7 were distinct red lights, with six being non-response PPI signals). Most prominently among the undelivered alerts were eight unique yellow lights for simvastatin, and one yellow light for clopidogrel. In order to mitigate this problem, during the course of this study a small number (4/17) of our early adopters themselves instituted programs within their clinics wherein they had clinic assistants always pre-print pharmacogenomic results from the CDS system for their manual review while seeing patients. Interestingly, when these physicians employed this type of assistance from ancillary clinic personnel for pharmacogenomic integration, the number of ‘missed alerts’ was almost zero. As one primary example of subsequent implementation iteration, and in part because of these results, the previously stand-alone GPS has now been integrated with our institutional EMR. Secondly, providers only rarely proactively utilized the GPS during new medication prescribing. Pharmacogenomic information would have been available for 27% of these unsearched instances. Fortunately, none of the unsearched new medications were prescribed to patients who would have had a red alert for the chosen drug. Lastly, several use constraints were identified that caused behavioral inconsistencies for some providers. Notes: aOut of 286 new medications that were prescribed when physicians logged in, the search functionality/alternative medication column was used 62 times. That meant 224 new medications were prescribed during a login visit without utilizing search functionalities. bFour physicians routinely accessed pharmacogenomic information printed out from the GPS by ancillary staff. cOut of the 1568 login visits during the study period, a disease search was conducted 26 times. dOf the visits where a provider logged in, there were 98 distinct risk alerts that were viewed but not clicked to view the more detailed clinical decision supports. eOver the course of the study period, 103 new medications were prescribed without a login. Pharmacogenomic information was available for 32 of these drugs. fThere were 253 distinct red and yellow alerts that were never delivered due to lack of login.
Supplementary Table 1. Factors Potentially Impacting Physician Accession of Genomic Prescribing System.
Supplementary Table 2. Drugs Available in GPS during Study Period.