Abstract
Acinetobacter baumannii is a bacterial pathogen with increasing impact in healthcare settings, due in part to this organism’s resistance to many antimicrobial agents, with pneumonia and bacteremia as the most common manifestations of disease. A significant proportion of clinically-relevant A. baumannii strains are resistant to killing by normal human serum (NHS), an observation supported here by showing that 12 out of 15 genetically diverse strains of A. baumannii are resistant to NHS killing. To expand our understanding of the genetic basis of A. baumannii serum resistance, a transposon-sequencing (Tn-seq) approach was used to identify genes contributing to this trait. An ordered Tn-library in strain AB5075 with insertions in every non-essential gene was subjected to selection in NHS. We identified 50 genes essential for the survival of A. baumannii in NHS, including already known serum resistance factors, and many novel genes not previously associated with serum resistance. This latter group included the maintenance of lipid asymmetry (mla) genetic pathway as a key determinant in protecting A. baumannii from the bactericidal activity of NHS via the alternative complement pathway. Follow up studies validated the role of 8 additional genes identified by Tn-seq in A. baumannii resistance to killing by NHS but not by normal mouse serum, highlighting the human-species specificity of A. baumannii serum resistance. The identification of a large number of genes essential for serum resistance in A. baumannii indicates the degree of complexity needed for this phenotype, which might reflect a general pattern pathogens rely on to cause serious infections.
INTRODUCTION
Acinetobacter baumannii is a Gram-negative, opportunistic pathogen responsible for approximately 2–10% of all hospital-acquired infections, including pneumonia, bacteremia, urinary tract infections, meningitis, and wound infections (1–4). A. baumannii infections are notoriously difficult to treat due to intrinsic and acquired antimicrobial resistance, often limiting effective therapeutic options (5). Another essential aspect of these infections is the ability of A. baumannii strains to resist the killing action of NHS (6–8).
The highly complex, multifactorial nature of the complement (C) system provides a myriad of targets for potential interference by bacterial factors. It is therefore not surprising that due to the long co-evolution of mammals and microbes, some of the most successful pathogens have developed effective mechanisms for attenuating or escaping C attack (9). As a countermeasure, targeting microbial factors that promote serum resistance, such as the factor H (FH)-binding protein (fHbp) in Neisseria meningitidis, with vaccines can provide protective immunity by a combination of mechanisms including bactericidal activity and increased C susceptibility (10).
To gain additional insights into the mechanisms of C resistance in A. baumannii, we took advantage of the availability of complete microbial genome sequences and transposon (Tn) mutant libraries to investigate the contribution of the products of all non-essential A. baumannii genes to survival in NHS. This screen identified 50 genetic loci needed for C resistance, and prominent among these were genes encoding the Mla proteins needed for the maintenance of outer membrane (OM) lipid asymmetry, which provided an opportunity for an in depth analysis of mechanisms of A. baumannii serum resistance. These studies showed the Mla system provided resistance to the alternative pathway of C and deposition of the corresponding C components, suggesting that this pathway could potentially be targeted for chemotherapeutic or immunotherapeutic interventions.
MATERIALS AND METHODS
Bacterial strains and growth conditions
Ethics statement. All animal experiments performed in this study were carried out in accordance with a protocol approved by the Harvard or Brigham and Women’s Hospital Institutional Biosafety Committees and Institutional Animal Care and Use Committees (IACUC). These guidelines were established by the Institute of Laboratory Animal Resources and approved by the Governing Board of the U.S. National Research Council.
All strains and plasmids as well as oligonucleotide primers used in this study are described in Supplemental Tables II and III respectively. A. baumannii, Escherichia coli and Pseudomonas aeruginosa strains were routinely cultured in lysogeny broth (LB; Lennox; Teknova) and, when required, supplemented with of tetracycline (5 μg/ml), gentamicin (150 μg/ml) or apramycin (50 μg/ml).
Transposon mutant library screening in pooled human serum
The A. baumannii AB5075 mutant library (11) was used in this study. Strain AB5075 is a clinical isolate from a human case of osteomyelitis that exhibits multiple antibiotic resistances and high virulence in a number of animal models of infection (12). The library includes Tn inserts in 3,470 genes inactivated with a Tn conferring resistance to tetracycline (T26) and 447 genes inactivated by a Tn insert encoding hygromycin resistance (T101). Only the 3,470 AB5075 genes inactivated with Tn T26 (tetracycline resistant) were investigated in this study.
To prepare the A. baumannii AB5075 Tn library for selection in human sera, we first generated a “total Tn mutant library pool” by replica plating the entire Tn mutant library onto rectangular LB agar plates using a 96-pin replicator (Thermo Scientific). After overnight growth, colonies from individual plates were harvested with 10 ml of LB+20% glycerol and 1 ml samples from each plate mixed together, aliquoted and stored at −80°C for further use.
Four individual vials of the total Tn library were thawed at room temperature and used to inoculate quadruplicate 50 ml LB cultures that were grown with aeration at 37°C to an optical density at 650 nm (OD650 nm) of 0.4. Bacterial cultures were then chilled on ice for 15 min and 1 ml aliquots containing approximately 1.5×108 CFUs centrifuged and washed with 1 ml of gelatin veronal buffer with added Ca2+ and Mg2+ (GVB++). Washed cells were pelleted, resuspended in 100 μl of GVB++ and used to inoculate 4 × 900 μl aliquots of intact- or heat-inactivated (HI)-NHS (56°C × 20 min) and incubated at 37°C for 2 h with shaking (250 rpm). After this time 50 μl aliquots from each of these 8 library samples (4 incubated in intact- and 4 in HI-NHS) were removed for viable count determinations by serial dilution and plating and the remaining 950 μl of the sample spread onto 245 × 245 mm square LB agar plates (Corning) supplemented with 5 μg/ml of tetracycline. Plates were incubated at 37°C for 16 h after which time bacterial colonies were harvested with 20 ml of LB-20% glycerol broth, pelleted and stored at −80°C until further use.
Preparation of DNA for high-throughput sequencing
Transposon-chromosome junctions were prepared and amplified for high-throughput sequencing following protocols described by Fu et al. (13).
Genomic DNA was extracted from 0.5 ml aliquots of each Tn library (8 in total: 4 intact- and 4 HI-NHS samples), sheared into ~500-bp fragments by ultrasonication using a Covaris M220 system, end-repaired (Quick Blunting Kit; NEB) and A-tailed by Taq polymerase (NEB). Adaptor DNA sequences (Adapter M1.0 and Adapter M2.0; Supplemental Table III) were then ligated to the fragmented DNA and PCR reactions carried out to amplify DNA fragments containing the Tn-chromosomal junction sequences along with the appropriate sequences required for Illumina sequencing (such as P5 and P7 hybridization sequences and barcodes) using primers Tn5-PCR F and Multiplex index 1–12, depending on the number of samples pooled for sequencing (Supplemental Table III). The final PCR products were purified on a 2% agarose gel and DNA fragments of 200–400-bp in length isolated. Equimolar DNA fragments from each library were combined and sequenced using the MiSeq Reagent Kit v2 50 cycles (Illumina) along with the custom sequencing primer Tn5-Seq (Supplemental Table III).
Sequencing analysis
Reads from the Illumina sequencing run were first error-corrected by Musket (14) and then mapped to the A. baumannii AB5075-UW genome using the Burrows-Wheeler Alignment tool (15), allowing for 0 mismatches. Genes that had less than 10 mapped reads in samples incubated with HI-NHS were excluded from further analysis. The combined read counts per gene were further normalized per million mapped reads to account for the differences in sizes of mapped reads among sequenced libraries. Then a genomic location bias correction was performed in each library as described by (16). During bacterial growth and replication, the amount of DNA close to the origin of replication (Ori) increases, which results in more DNA around the Ori becoming available for sequencing. This generally translates into a significant Tn positional bias that becomes apparent as a V-shape in the read counts per Tn relative to their genomic location, with an overall higher average reads close to the Ori. Therefore an initial positional bias correction was performed on each sequenced Tn pool to generate normalized sets of data that no longer possessed a skew towards the origin of the chromosome.
To normalize for chromosomal replication biases in read counts in all 8 sequenced libraries, the entire AB5075 genome was first divided into 10 kb nonoverlapping windows starting with windows centered around the Ori. Then the number of reads within each of the windows was normalized based on a factor calculated from the average read depth within the window divided by the average depth for the chromosome.
After the genomic location bias correction was performed in each library the average number of normalized, position-corrected reads in all four intact and HI-NHS replicate studies was calculated for each gene and these values used to calculate a HI-NHS/intact-NHS ratio, which corresponds to the fold change. Genes from intact NHS samples with < 1 mapped read/gene were assigned an arbitrary value of 1 in order to calculate fold change values. Finally, t tests were used to assess the statistical significance of changes in the frequency of reads in HI- and intact-NHS libraries among all 4 replicate studies. Significance was defined by a p value of ≤0.05.
Protein sequences were mapped on the COG (cluster of orthologous genes) database (prokaryotic proteins) via RPSBLAST to identify COG families and classes using WebMGA server (17).
Construction of A. baumannii mlaD∷Tn-pmla and mlaA∷Tn-pmlaA complemented strains
To construct the A. baumannii mlaD∷Tn-pmla complemented strain, we first amplified the entire mlaFEDCB operon including 15 bp upstream of its putative start codon using primer pair XmaI-mlaFEDCB-F/PstI-mlaFEDCB-R (Supplemental Table III). The 3,257 bp-long amplified PCR product was then column purified (Zymo research) and digested with XmaI/PstI restriction enzymes (NEB) overnight at 37°C. The digested PCR product was then ligated into XmaI/PstI-restricted pMJG120 plasmid using T4 DNA ligase (NEB) to yield the complementation plasmid pmla. Plasmid pmla was transformed into electro-competent A. baumannii AB5075 mlaD∷Tn, to yield the A. baumannii mlaD∷Tn-pmla complementation strain.
Similar protocols were used for the construction of A. baumannii mlaA∷Tn-pmlaA complementation strain. In brief the primer pair XmaI-mlaA-F/PstI-mlaA-R (Supplemental Table III) was used to amplify a 915 bp amplicon corresponding to the mlaA gene plus 15 bp upstream of its putative start codon. The PCR product was then digested with XmaI/PstI restriction enzymes, ligated into pMJG120 (18) as outlined above to yield plasmid pmlaA and transformed into A. baumannii mlaA∷Tn mutant strain to yield the complemented strain A. baumannii mlaA∷Tn-pmlaA.
The correct sequence of the mlaFEDCB operon and mlaA gene inserts from plasmids pmla and pmlaA respectively was verified by sequencing using primer pair AN180/AN181 (Supplemental Table III).
Serum killing assays with NHS, C1q- and factor B-depleted sera and normal mouse serum
Overnight cultures of wild type (WT) A. baumannii AB5075 as well as mlaA∷Tn and mlaD∷Tn mutant strains were diluted 1:100 into fresh LB and grown to an OD650 nm of 0.4. In the case of mlaA∷Tn-pmlaA and mlaD∷Tn-pmla, these strains were first grown from overnight cultures to an OD650 nm of 0.4 then induced for 2 h with 10 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) at which point the OD650 nm of the cultures was measured then diluted back to OD650 nm of 0.4 with fresh LB in order to match the cell densities of WT AB5075 and mlaA∷Tn and mlaD∷Tn mutant strains. One ml aliquots of these cultures were pelleted by centrifugation (16,000 g × 5 min), washed once with 1 ml of GVB++ and cells suspended in 100 μl of GVB++. The serum-killing assay was performed by combining 45 μl of intact- or HI-NHS with 5 μl of bacteria prepared as before in a V-bottom, 96-well plate (Corning Incorporated). Samples were incubated for 2 h at 37°C with shaking, then the number of CFU/ml estimated by serial dilution and plating.
The susceptibility of A. baumannii AB5075, mlaD∷Tn and mlaD∷Tn-pmla complemented strains was also evaluated against C1q- and factor B (FB)-depleted sera (Complement Technology) following exactly the protocol outlined above for NHS. C1q- and FB-depleted sera were also reconstituted by adding back purified C1q- or FB at the physiological concentrations of 70 and 200 μg/ml respectively and tested against the mlaD∷Tn as outlined above.
In some studies, serum susceptibility of A. baumannii Tn mutant strains was tested in normal mouse serum (NMS) following the protocols previously described. Due to the unstable nature of the mouse classical pathway components (19) all serum assays involving mouse C, were carried out with unfrozen NMS prepared from blood freshly collected from male C57BL/6J mice (The Jackson Laboratory, Bar Harbor, ME) following recommendations described by Lachmann (20).
Serum killing with cobra venom factor-depleted human sera
NHS was treated with 25 U/ml of cobra venom factor (CVF) (21) (Quidel Corporation) following the manufacture’s recommendations or with an equal volume of PBS as control for 1 h at 37°C with shaking (200 rpm). PBS-NHS samples were then split in two aliquots, one HI, the other used directly as active sera and 45 μl from each of these three serum samples tested in the standard serum killing assay as outlined above against WT A. baumannii AB5075, mlaD∷Tn and mlaD∷Tn-pmla complementation strains.
C4 deposition ELISA
The A. baumannii mlaD∷Tn-pmla complemented strain was diluted 1:100 from an overnight culture, grown to an OD650 nm of 0.4 and induced for 2 h with IPTG (10 mM) at which point the culture reached OD650 nm of approximately 1. Similarly, overnight cultures of WT A. baumannii AB5075 and mlaD∷Tn mutant strains were diluted 1:100 into fresh LB and grown to OD650 nm of 1 to match the final OD650 nm reached by the mlaD∷Tn-pmla complemented strain. Then AB5075, mlaD∷Tn and mlaD∷Tn-pmla cultures were diluted back OD650 nm of 0.4 with fresh LB. Equal volumes of these bacterial suspensions were centrifuged (16,000 g × 5min), pellets resuspended in the same volume of coating buffer (40 mM sodium phosphate buffer; pH 7) and used to coat high-binding ELISA plates (Santa Cruz Biotechnology) with 50 μl of bacterial suspension/well. As a positive control for activation of the lectin pathway, additional wells were coated with 50 μl of a 10 μg/ml solution of mannan (Sigma-Aldrich) in coating buffer. After overnight incubation at 4°C, ELISA plates were blocked with a 0.5% BSA solution in TBS (10 mM Tris-HCl, 140 mM NaCl, pH 7.4) at 37°C for 1 h then washed three times with 0.05% Tween 20 in PBS and incubated with two-fold serial dilutions of C1q-depleted human sera in GVB++ for 1 h at 37°C. Plates were washed as above, and wells incubated with polyclonal goat anti-human C4 Abs (Complement Technology, Inc) diluted 1:1,000 in dilution buffer (0.05% Tween 20, 5 mM CaCl2, 0.5% BSA in TBS) for 1 h at 37°C. Plates were then washed as above and incubated with secondary rat-anti-goat alkaline phosphatase-conjugated Abs (Sigma-Aldrich) diluted 1:1,000 in dilution buffer for 1 h at 37°C. Bound Ab was then quantified using the colorimetric substrate p-nitrophenyl phosphate (Santa Cruz Biotechnology) measuring optical density at OD405 nm.
Quantification of C C5b-9 binding to A. baumannii by flow cytometry
One ml cultures of WT A. baumannii AB5075, mlaD∷Tn mutant and mlaD∷Tn-pmla complemented strain, prepared as above for serum killing assays, were centrifuged at 16,000 g × 5 min, pelleted cells washed once with 1 ml GVB++ buffer and concentrated 10-fold in GVB++. Fifty microliter aliquots of each of the three washed strains were then incubated with 0.45 ml of a 30% solution of either intact- or HI-NHS in GVB++ at 37°C for 1h 15 min in a rotating shaker. After this time, the C reaction was stopped by pelleting cells as above and washing cell suspensions once with 1 ml of ice-cold PBS.
To detect deposition of C5b-9, bacterial suspensions were pelleted by centrifugation and cells stained with a 50 μl solution containing the fluorescent nucleic acid stain, SYTO-62, and a mouse mAb to a C9 neoantigen conjugated to FITC (clone aE11) at a final concentration of 30 μM and 10 μg/ml, respectively, in PBS for 1 h at 37°C with shaking (200 rpm). Control samples included unstained cells and cells stained with either SYTO-62 or FITC-conjugated mAb alone. The stained cells were then washed once with 0.5 ml of PBS, fixed with 10 % neutral buffered formalin and analyzed in a MACSQuant (Miltenyi Biotec) flow cytometer. Results were analyzed using FlowJo software (Tree Star, Inc).
Virulence of A. baumannii mlaD∷Tn strain in a mouse model of bloodstream infection in immunocompetent and C-depleted mice
The animal model of bloodstream infection used was previously described (22). In brief, C57BL/6J mice (n=13–16/group, male, 10 weeks of age; The Jackson Laboratory, Bar Harbor, ME) were injected i.p. with 15 μg of CVF (Complement Technology) or 100 μl of PBS as control 36, 24, and 12 h prior and i.v. challenge with 7.6×105 CFU of A. baumannii mlaD∷Tn strain. Two hours after infection mice were sacrificed, blood collected via heart stick and numbers of surviving bacteria determined by serial diluting and plating of blood samples.
C3 quantification in pooled sera collected from PBS- and CVF-treated mice was performed using a C3 sandwich ELISA (Genway Biotech) following manufacture’s instructions.
Phospholipase C treatment
Phospholipase C treatment and TLC were performed using protocols previously described by Nakamura et al 2011 (23). Five hundred ml of WT A. baumannii AB5075, mlaD∷Tn mutant and mlaD∷Tn-pmla complemented strains were grown in LB as described above. Cultures (OD650 nm of 0.4) were then centrifuged at 6,000 × g for 30 min, pellets, washed once with 20 ml of a solution of 1 × PBS, 0.4 M sucrose, 15 mM MgCl2, pH 7.5 and resuspended in 1 ml of the same buffer. Cell suspensions were then treated with 25 U of phospholipase C from Bacillus cereus (Sigma-Aldrich) for 1 h at 37°C while mixing on a rotating shaker. After this time bacterial cells were pelleted at 16,000 g × for 5 min and the lipids were extracted by the method of Bligh and Dyer (24). Lipid samples were then separated by TLC on glass backed silica gel 60 plates (Merck) with chloroform-acetone 94:4 (v/v) solvent system, stained by iodine vapor for 30 min and the intensity of spots quantified using ImageJ software. The standard was 100 μg of 1,2-dipalmitoyl-sn-glycerol (Sigma-Aldrich).
RESULTS
A. baumannii is highly resistant to killing by NHS
We evaluated the susceptibility to C-mediated killing in NHS of 15 A. baumannii strains, including five multi-drug resistant strains from the Singapore General Hospital, (strains S1–S5), five isolates from American soldiers wounded in the Iraqi war conflict (strains I25–I29) and five sequenced strains from multiple geographical locations with various degrees of virulence in animal models of infection (12, 25, 26). Strains were grown to mid-logarithmic phase then incubated for 2 h in either undiluted, intact- or HI-NHS as a control. As depicted in Fig. 1, 12 out of the 15 strains were highly resistant to killing by NHS, indicating serum resistance is a common feature among virulent A. baumannii strains.
Figure 1.
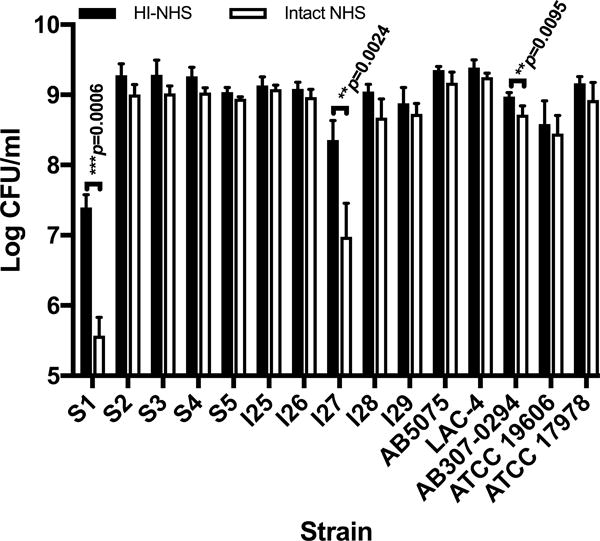
Serum sensitivity of A. baumannii strains in NHS. Fifteen A. baumannii strains were incubated for 2 h at 37°C in the presence of intact- (open columns) or HI-NHS (solid columns). Data represent means ± SD of 3 independent experiments. P values were determined using a two-tailed, unpaired t test on log10-transformed data after normality was confirmed using the Shapiro-Wilk test.
Genetic basis of A. baumannii serum resistance
We used a Tn-library in A. baumannii strain AB5075, created and kindly provided to us by Dr. Colin Manoil (11), to investigate the genetic basis of A. baumannii resistance to killing by human C. AB5075 is a multidrug-resistant global clone 1 strain isolated in 2008 from a patient with an osteomyelitis infection (12). It displays high virulence in various animal models of infection and is amenable to genetic manipulation (12). These characteristics make AB5075 an attractive model strain for studying A. baumannii pathogenicity including serum resistance. The library encompasses a total of 10,762 Tn mutants and includes two to three unique, sequence-verified Tn inserts in virtually all non-essential genes.
A Tn library pool of A. baumannii was prepared then subjected to selection in intact and HI-NHS to generate 4 independent biological replicates for each gene, followed by Illumina sequencing of Tn-chromosome junctions. Reproducibility was determined by comparing the number of mapped reads per Tn mutant among all 4 intact- or HI-NHS independent replicates. Data analysis revealed a Pearson correlation coefficient (R) for the sequenced reads that ranged from 0.88–0.98 for samples in intact NHS and from 0.97–0.98 for samples in HI-NHS (Supplemental Table I), indicating that the Tn-seq results were highly comparable among all four independent experiments. In total 3,241 out of the 3,470 Tn-disrupted genes (93%) were scored in this analysis. The remaining 229 genes could not be analyzed as they either completely failed to be recovered (109 genes) or had < 10 reads per gene in the HI-NHS sample libraries (120 genes), which was established as the cut-off read count needed for further analysis. These 229 genes most likely represent Tn mutants that displayed a competitive growth disadvantage within the total Tn mutant library pool during the inoculum recovery period and therefore were represented at levels below the lower limit of gene detection by Tn-seq analysis. Alternatively these genes may encode bacterial factors needed for the survival of A. baumannii to killing by some yet uncharacterized, complement-independent, heat-stable serum factors.
The average number of normalized reads in each intact- or HI-NHS data set was determined for every gene and these values used to calculate a ratio of reads between HI- and intact-NHS. Genes needed for serum resistance were defined as those that had ≥ 3-fold decrease in mapped reads after dividing HI- by intact-NHS values and p values ≤ 0.05 (n=4; 2-tails; unpaired t test). A total of 50 A. baumannii AB5075 genes were identified as needed for serum resistance (Table I and Fig. 2, top-left quadrant). Of these, there were 3 genes with >100-fold decrease (6% of total scored; Fig. 2, green symbols) and 26 with a 10-100-fold decrease (52% of total scored; Fig. 2, red symbols) in Tn insert read abundance in HI- versus intact-NHS. We also found an additional 21 genes with a 3–10 fold decrease in Tn insert read abundance (42% of total scored; Fig. 2, blue symbols). The remaining Tn insertions in 3,191 genes did not meet the criteria for conferring serum resistance because of either limited decrease in read abundance (< 3-fold decrease in HI- versus intact-NHS) and/or lack of statistical differences shared among all four replicate studies (p values ≥ 0.05).
Table I.
A. baumannii AB5075 Tn mutants with decreased survival in intact- versus HI-NHS.
| AB5075 Locus Tag | Reads in HI-NHS | Reads in Intact NHS | Fold Change: Reads in HI-NHS/Intact-NHS | p-value a | Gene Name | Annotation |
|---|---|---|---|---|---|---|
| Amino acid transport and metabolism | ||||||
| ABUW_2418 | 246.53 | 46.70 | 5.28 | 4.97E-06 | Lysine exporter protein | |
| ABUW_2456 | 41.56 | 10.58 | 3.93 | 4.67E-03 | Hydroxymethylglutaryl-CoA lyase | |
| Carbohydrate transport and metabolism | ||||||
| ABUW_1134 | 115.01 | 16.66 | 6.90 | 7.45E-07 | Major facilitator superfamily MFS_1 | |
| ABUW_3041 | 194.12 | 9.64 | 20.15 | 4.77E-05 | algC | Phosphomannomutase |
| Cell wall/membrane/envelope biogenesis | ||||||
| ABUW_0090 | 14.50 | 2.02 | 7.17 | 8.20E-05 | glmU | Bifunctional protein GlmU |
| ABUW_0383 | 170.98 | 4.48 | 38.13 | 2.31E-04 | mlaF | Uncharacterized ABC transporter, ATP-binding protein YrbF |
| ABUW_0384 | 103.29 | 13.90 | 7.43 | 2.92E-04 | mlaE | Uncharacterized ABC transporter, permease component YrbE |
| ABUW_0385 | 189.42 | 3.18 | 59.52 | 7.44E-05 | mlaD | Putative phospholipid ABC transporter-binding protein MlaD |
| ABUW_0386* | 304.05 | 5.83 | 52.14 | 8.47E-05 | mlaC | Putative phospholipid-binding protein MlaC |
| ABUW_1868 | 89.41 | 9.87 | 9.06 | 1.25E-03 | Hypothetical protein | |
| ABUW_3259 | 156.77 | 1.43 | 109.68 | 9.43E-05 | mlaA | VacJ family lipoprotein |
| ABUW_3448 | 31.65 | 3.07 | 10.31 | 1.75E-04 | Glycosyl transferase, group 1 | |
| ABUW_3638 | 26.16 | 1 | 26.16 | 2.60E-03 | pbpG | D-alanyl-D-alanine carboxypeptidase family protein |
| ABUW_3820 | 21.82 | 1 | 21.82 | 1.18E-03 | Nucleotide sugar epimerase/dehydratase | |
| ABUW_3824 | 64.65 | 1 | 64.65 | 2.31E-05 | Family 1 glycosyl transferase | |
| ABUW_3828 | 15.97 | 1 | 15.97 | 3.79E-04 | Hypothetical protein | |
| ABUW_3829 | 47.91 | 1 | 47.91 | 1.84E-06 | Hypothetical protein | |
| ABUW_3830 | 27.28 | 1.39 | 19.56 | 4.43E-05 | UDP-glucose/GDP-mannose dehydrogenase | |
| ABUW_3831 | 10.88 | 1 | 10.88 | 1.12E-04 | wza | Polysaccharide export protein |
| Coenzyme transport and metabolism | ||||||
| ABUW_2736 | 12.96 | 1 | 12.96 | 2.26E-03 | cinA | Competence/damage-inducible protein CinA |
| Energy production and conversion | ||||||
| ABUW_2489 | 13.74 | 1 | 13.74 | 1.13E-04 | azoR | Acyl carrier protein phosphodiesterase |
| Function unknown | ||||||
| ABUW_0135 | 188.72 | 15.50 | 12.17 | 3.32E-05 | Signal peptide protein | |
| ABUW_0986 | 13.49 | 1.60 | 8.42 | 2.83E-03 | Restriction endonuclease | |
| ABUW_1191 | 80.33 | 6.73 | 11.93 | 1.28E-05 | Hypothetical protein | |
| ABUW_1192 | 128.51 | 15.82 | 8.12 | 3.32E-05 | Hypothetical protein | |
| ABUW_2168 | 18.86 | 4.64 | 4.06 | 1.98E-05 | Hypothetical protein | |
| ABUW_2637 | 42.68 | 7.05 | 6.06 | 3.22E-06 | Peptidase M15 | |
| ABUW_2735 | 83.53 | 1 | 83.53 | 1.29E-03 | Hypothetical protein | |
| ABUW_3758 | 69.64 | 1 | 69.64 | 3.23E-04 | dedA | Protein DedA |
| ABUW_3825 | 14.71 | 1 | 14.71 | 5.50E-04 | Hypothetical protein | |
| ABUW_3881 | 11.85 | 1 | 11.85 | 1.74E-05 | Matrixin superfamily | |
| General function prediction only | ||||||
| ABUW_1060 | 102.07 | 22.96 | 4.45 | 1.65E-05 | Hypothetical protein | |
| ABUW_2363 | 66.02 | 1 | 66.02 | 3.95E-06 | Tetratricopeptide repeat family protein | |
| ABUW_3822 | 10.96 | 1 | 10.96 | 3.16E-04 | Bacterial transferase hexapeptide (three repeats) family | |
| Inorganic ion transport and metabolism | ||||||
| ABUW_2898 | 94.75 | 1 | 94.75 | 3.65E-03 | Hypothetical protein | |
| ABUW_3369 | 152.10 | 1 | 152.10 | 2.40E-06 | Rhodanese domain protein | |
| Intracellular trafficking, secretion, and vesicular transport | ||||||
| ABUW_0532 | 44.31 | 1 | 44.31 | 9.32E-05 | yajC | Preprotein translocase, YajC subunit |
| Lipid transport and metabolism | ||||||
| ABUW_0312 | 49.27 | 14.06 | 3.51 | 9.64E-06 | ispH | Hydroxymethylbutenyl pyrophosphate reductase |
| ABUW_0729 | 217.50 | 1 | 217.50 | 1.85E-06 | uppP | Undecaprenyl-diphosphatase UppP |
| ABUW_1737 | 26.29 | 5.25 | 5.01 | 9.58E-05 | uppS | Undecaprenyl diphosphate synthase |
| ABUW_2957 | 181.73 | 59.25 | 3.07 | 4.98E-04 | Enoyl-CoA hydratase/isomerase | |
| Mobilome: prophages, transposons | ||||||
| ABUW_2662 | 57.04 | 7.55 | 7.56 | 5.25E-07 | Hypothetical protein | |
| Posttranslational modification, protein turnover, chaperones | ||||||
| ABUW_3385 | 54.74 | 13.50 | 4.05 | 1.86E-03 | prc | Carboxy-protease |
| ABUW_3642 | 36.05 | 3.83 | 9.42 | 2.85E-04 | Putative periplasmic carboxyl-terminal protease | |
| Signal transduction mechanisms | ||||||
| ABUW_3302 | 35.38 | 7.14 | 4.95 | 7.79E-04 | relA | GTP pyrophosphokinase (ppGpp synthetase I) |
| ABUW_3408 | 67.30 | 20.19 | 3.33 | 1.86E-04 | Sel1 domain protein | |
| ABUW_3639 | 11.93 | 2.76 | 4.31 | 6.46E-03 | gacA | Response regulator |
| ABUW_3832 | 13.56 | 1 | 13.56 | 4.00E-03 | ptp | Protein-tyrosine-phosphatase ptp |
| Translation, ribosomal structure and biogenesis | ||||||
| ABUW_1608 | 13.74 | 1 | 13.74 | 3.75E-03 | miaA | tRNA delta(2)-isopentenylpyrophosphate transferase |
| ABUW_1736 | 43.68 | 5.96 | 7.33 | 9.91E-06 | frr | Ribosome recycling factor |
t test was used to compare normalized mapped reads between HI- and intact NHS samples among four independent experimentsand derive corresponding p values.
Gene ABUW_0386 (mlaC) belongs to the lipid transport and metabolism category but as part of the mlaFEDCB operon was placed here for clarity.
Figure 2.
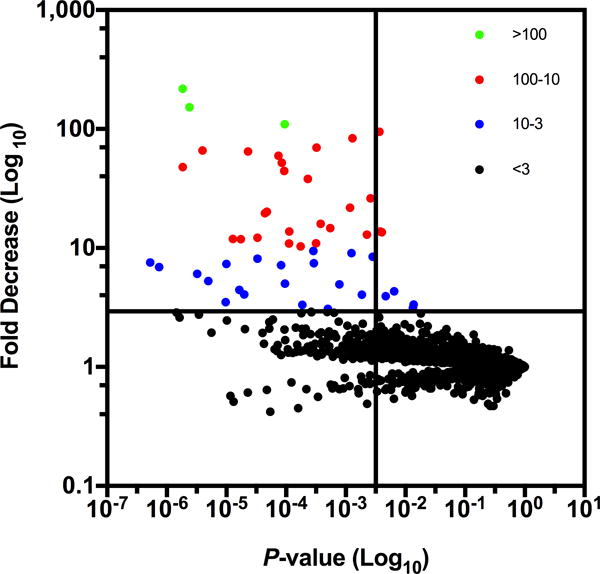
Fold decrease and p value for survival of A. baumannii Tn mutants in human serum based on ratio of normalized mapped reads in HI- versus intact-NHS from 4 independent experiments. Each point represents a different Tn mutant and fold change. Mutants with > 100, 100-10, 10-3 and <3 -fold decreases in survival are shown in green, red, blue and black color respectively. The vertical and horizontal lines in the graph are drawn at p=0.05 and 3-fold change respectively.
Predicted functions of gene products required for A. baumannii serum resistance
All genes identified as important for A. baumannii survival in human serum were subjected to bioinformatic analysis and classified into functional categories. The complete list of the functional categories of A. baumannii AB5075 serum resistance genes is presented in Table I. Genes belonging to the “cell wall/membrane/envelope biogenesis” category had the most impact on serum resistance accounting for 28% of all genes identified (Fig. 3A and Table I). Among these we found 4 genes, ABUW_0383 (mlaF) ABUW_0384 (mlaE), ABUW_0385 (mlaD), and ABUW_3259 (mlaA), annotated as encoding proteins for the “maintenance of lipid asymmetry” (mla) pathway (27). We also identified the gene ABUW_0386 from the lipid transport and metabolism category, which encodes a putative phospholipid (PL)-binding protein MlaC that is part of the Mla pathway, as needed for serum resistance. In E. coli the products of the mlaFEDCB operon and mlaA gene function together to remove PLs from the outer leaflet of the OM which can accumulate under certain stress conditions (27, 28). The Mla pathway therefore restores the asymmetric lipid distribution of the OM with LPS and PLs in the outer and inner leaflets respectively (27).
Figure 3.
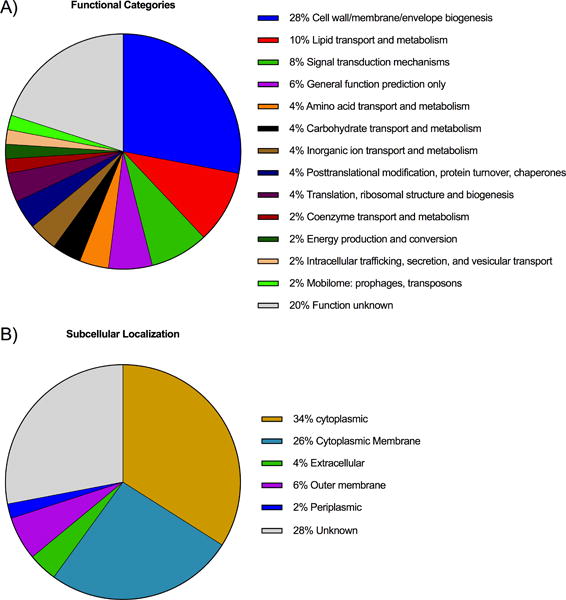
A) Distribution into functional categories of 50 serum resistance genes in A. baumannii AB5075 identified by Tn-seq. Functional annotation was performed using WebMGA tool that uses the RPSBLAST algorithm with the COG database for prokaryotic proteins. B) Predicted subcellular localization of protein products encoded by Tn mutants identified by Tn-seq. Subcellular localization prediction was performed with the PSORTb v.3.0 server.
As expected, genes in the “cell wall/membrane/envelope biogenesis” category-encoding proteins needed for capsular polysaccharide biosynthesis, as well as their surface transport and retention, were identified as factors strongly contributing to serum resistance, as it is known the capsule plays an important role in this phenotype (29). Validation studies carried out with two mutants in genes ABUW_3831 (wza) and ABUW_2898 (wzi homolog), which encode predicted OM proteins required for the surface transport and retention of capsule, respectively, demonstrated the critical role of capsule for A. baumannii survival in intact NHS. Tn-inserts in these genes led to 7.6- and 3.8-log10 killing of ABUW_3831 and ABUW_2898 mutants, respectively when comparing intact- versus HI-NHS (both ****p<0.0001; Fig. 4A).
Figure 4.
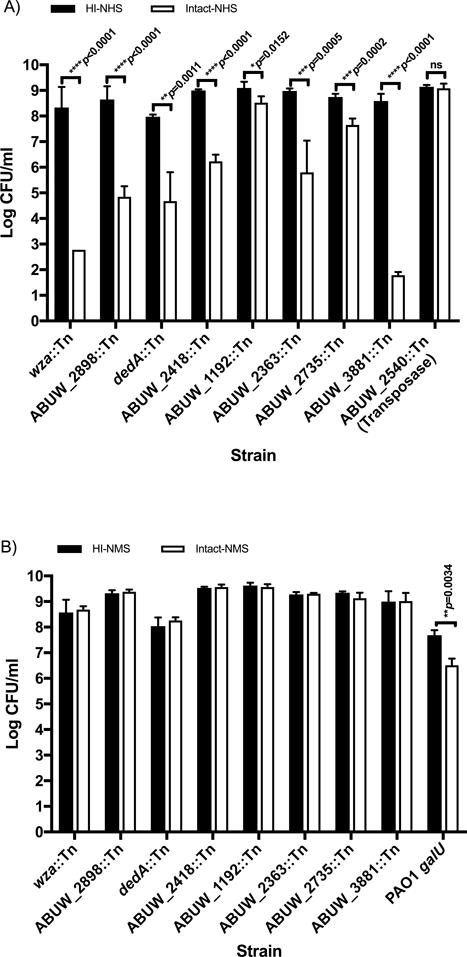
Validation of 8 A. baumannii AB5075 genes identified by Tn-seq as required for human serum resistance. Susceptibility of Tn mutants tested in A) NHS and B) NMS using intact (open columns) or HI-sera (solid columns). Two strains, A. baumannii ABUW_2540∷Tn and P. aeruginosa PA01 galU mutant were included as negative and positive controls in the assays performed with NHS and NMS, respectively. Data represents the means ± SD of 3–5 independent experiments. P values were calculated using a two-tailed, unpaired t test on log10-transformed data after normality was confirmed using the Shapiro–Wilk test.
Genes annotated in the categories of general function prediction, unknown function and signal transduction mechanisms that are needed for capsule production were also identified following serum selection (ABUW_3822, ABUW_3825, homologs of qhbA and wzy respectively and ABUW_3832 (ptp)). In addition, we identified a significant number of genes (20%) needed for serum resistance with unknown functional predictions (Fig. 3A and Table I).
Analysis of the subcellular localization of A. baumannii serum resistance gene products revealed that 34% of all products were predicted to be localized to the bacterial cell envelope (either cytoplasmic membrane, periplasm or OM) and an additional 4% predicted to reside in an extracellular compartment. Thirty four percent of the gene products had a predicted cytoplasmic localization and 28% had unknown bacterial cell localizations (Fig. 3B). The fact that 38% of the genes encoding proteins were predicted to be either a cell wall or secreted factor indicates that the selection and sequencing strategies employed correctly identified many of the essential A. baumannii serum resistance factors, inasmuch as cell envelope components maintaining bacterial OM and cell surface integrity are known to promote resistance to killing by C system components.
Validation of serum resistance genes not previously associated with this phenotype
Two additional Tn mutants encoding the cytoplasmic membrane proteins, DedA, and a L-Lysine permease (ABUW_2418), not previously associated with serum resistance, were selected for further validation in the serum sensitivity assay. DedA belongs to a partially characterized, highly conserved family of proteins that appears to play a role in multiple cellular processes including cell division, temperature sensitivity, membrane lipid composition, envelope-related stress responses, loss of proton motive force and virulence (30). The L-lysine exporter protein is involved in the efflux of excess L-lysine to control for intracellular levels of L-lysine (31, 32). Transposon mutants in the DedA protein and L-lysine permease were decreased 69.6- and 5.2-fold respectively in HI- versus intact-NHS (Table I). As seen in Fig. 4A incubation of DedA protein and L-Lysine permease Tn mutants in intact NHS resulted in approximately 3.3- and 2.8-log10 of killing when compared to the HI-NHS controls.
Transposon mutants in four proteins annotated in the categories of unknown or general function predictions (ABUW_1192, ABUW_2363 and ABUW_2735 and ABUW_3881) were additionally selected for validation in the C killing assay. Transposon insertions in these genes showed a strong decrease in viability in intact- versus HI-NHS by Tn-seq (8.1, 66, 83.5 or 11.8-fold reductions for ABUW_1192∷Tn, ABUW_2363 ∷Tn, ABUW_2735:Tn and ABUW_3881 Tn mutants respectively) and upon validation resulted in 0.6-, 3.2-, 1- and 7.7-log10 reductions in CFU in intact- versus HI-NHS (Table I and Fig. 4A). As a negative control, we tested a Tn mutant strain, ABUW_2540∷Tn, with an unchanged number of mapped reads between HI- and intact-NHS by Tn-seq (fold change=1) and, as expected, it remained completely serum resistant. Taken together these results highlight the accuracy of Tn-seq to predict the requirement of specific A. baumannii genes needed for human serum resistance.
In addition to human sera we also investigated susceptibility of these 8 selected A. baumannii Tn mutants to killing by freshly prepared, unfrozen NMS. As shown in Fig. 4B all 8 A. baumannii Tn mutant strains previously shown to be susceptible to NHS (Fig. 4A), were fully resistant to killing by intact NMS. We used a P. aeruginosa LPS-rough mutant strain, PAO1 galU, as a control to demonstrate functional activity of the NMS, as this strain is significantly killed by mouse C (Fig. 4B).
Detailed analysis of the mla pathway and A. baumannii serum resistance
We next focused our attention on a set of five genes belonging to the Mla pathway. The genetic organization of the mla genes in A. baumannii AB5075 is depicted in Supplemental Fig. 1A. In E. coli this pathway is comprised of 5 proteins (the periplasmic chaperone MlaC and the inner membrane (IM) MlaFEDB ATP-binding cassette family transporter that functions together with OM lipoprotein MlaA, encoded elsewhere on the chromosome, to prevent the accumulation of PLs in the outer leaflet of the OM (27). Mutations in the mla genes are not lethal but result in increased OM permeability (33–35). Specific Tn abundance reductions in HI- versus intact-NHS were as follows: (fold reduction are indicated in brackets); mlaF (38.1); mlaE, (7.4); mlaD, (59.5); mlaC (52.1) mlaB (0.8) and mlaA (109.6).
To investigate the role of mla homolog genes in A. baumannii OM lipid asymmetry we directly compared the content of surface exposed PLs of WT AB5075, mlaD∷Tn mutant and the complemented strain mlaD∷Tn-pmla following whole cell treatment with phospholipase C and detection of diacylglycerol (DAG) released from cells using TLC as described by Nakamura et al (23). As seen in Supplemental Fig. 1B, the overall amount of DAG released from the mlaD∷Tn-mutant strain was higher (spot density increase of 107%) than that extracted from WT AB5075 and mlaD∷Tn-pmla complemented strain (spot density increase of 120%). This increase in surface exposed PLs exhibited by the mlaD∷Tn-mutant strain was accompanied by a significant increased susceptibility of this strain to the detergent SDS when compared to WT AB5075 and mlaD∷Tn-pmla strains (Supplemental Fig. 1C) but not against a sublethal concentration SDS (0.0075%) plus increasing concentration of EDTA, ranging from 1.5–0.00073 mM (data not shown). This indicates that similar to E. coli mutants in the mla pathway, the A. baumannii mla mutants have an increase in the OM permeability, rendering the cells susceptible to detergent by an increase in surface-exposed PLs (27).
To validate the predicted role of the Mla pathway in A. baumannii resistance to human C, we compared the susceptibility in the serum killing assay of WT AB5075 strain and two Tn mutants in the mlaD and mlaA genes. We also tested two strains complemented with either the entire mla operon, mlaD∷Tn-pmla or the mlaA gene, mlaA∷Tn-pmlaA. As shown in Fig. 5A, incubation of A. baumannii mlaA∷Tn in intact NHS resulted in ~1.5 log10 of killing when compared to WT AB5075 (****p<0.0001) and the complemented strain, mlaA∷Tn-pmlaA (****p<0.0001) tested under the same conditions or the same strain incubated in HI-NHS (****p<0.0001). Similar results were seen when mlaD∷Tn mutant and mlaD∷Tn-pmla were tested in the assay. These results demonstrate the critical importance of the mla pathway system in maintaining full resistance of A. baumannii to killing by human sera.
Figure 5.
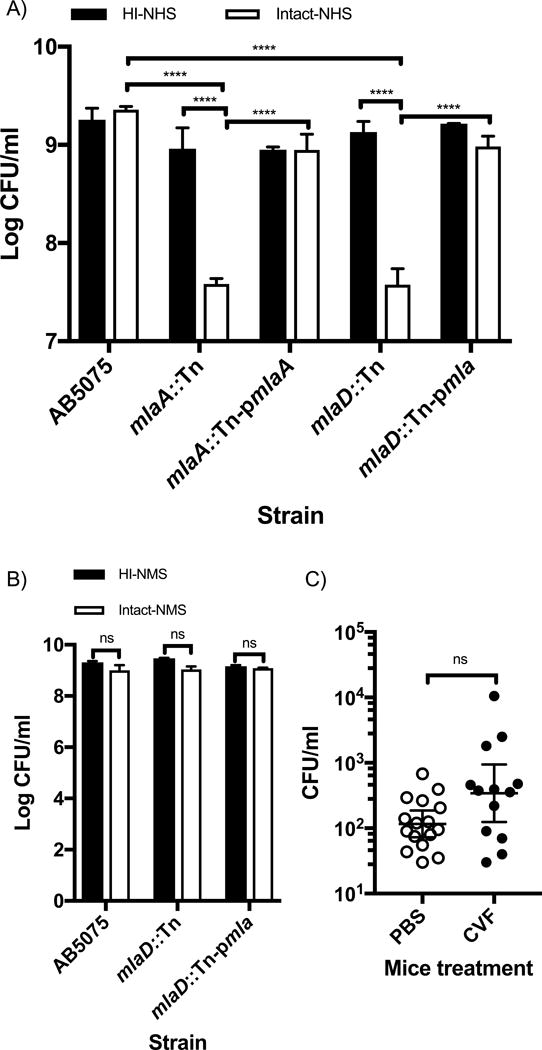
A) Susceptibility of WT AB5075, mlaA∷Tn and mlaD∷Tn mutants as well as mlaA∷Tn-pmlaA and mlaD∷Tn-pmla complemented strains in intact NHS (open columns) or HI-NHS (solid columns). Columns represent the mean ± SD of 3 independent experiments. B) Susceptibility of AB5075, mlaD∷Tn mutant and mlaD∷Tn-pmla complemented strains in intact- (open columns) and HI-NMS (solid columns). Columns represent the mean ± SD of 3 independent experiments. C) Survival of A. baumannii mlaD∷Tn mutant strain in murine model of bloodstream infection in C-depleted mice via treatment with CVF (solid symbols) or control animals given PBS (open symbols); (13–16 mice/group). Normality was confirmed using the Shapiro-Wilk test on log10-transformed data, and statistical significance calculated using one way ANOVA (p<0.0001) followed by a Tukey post-hoc test; ****p<0.0001 (A) or two-tailed, unpaired t test (B). Statistical comparison between PBS and CFV-treated mice was performed using the 2-tailed Mann Whitney test (C).
Consistent with our previous observations of the lack of NMS bactericidal activity against A. baumannii Tn mutant strains, the A. baumannii mlaD∷Tn strain was also resistant to killing by NMS (Fig. 5B).
Analysis of the virulence of the A. baumannii mlaD∷Tn strain in a mouse model of bloodstream infection, comparing mice with an intact C system to those depleted of C component C3 following treatment with CVF did not result in a significant increase in A. baumannii blood burdens (Fig. 5C) The effectiveness of mouse C3 depletion by CVF treatment was confirmed by demonstrating a 93.5% reduction in C3 in serum of CFV-treated versus PBS-control mice.
The discrepancy in A. baumannii susceptibility to NHS and NMS highlights a limitation of mouse models to evaluate microbial virulence when differences in mouse and human C systems lead to discrepant interactions with microbial pathogens. Moreover similarly to NMS, serum samples obtained from various other animals species including Wister and Lewis rats, rabbit, and guinea pigs also failed to kill the mlaD∷Tn mutant strain (data not shown) corroborating the human-species specificity of our findings.
Role of specific C components in killing of the A. baumannii mlaD∷Tn mutant strain
We analyzed the contribution of the different C system pathways to the susceptibility of WT A. baumannii AB5075, mlaD∷Tn, and mlaD∷Tn-pmla complemented strains to killing by NHS, using human sera selectively depleted of C1q or FB, needed for activation of the classical or alternative C pathways, respectively. We further evaluated the deposition of the C4 component in C1q-depleted serum by an ELISA, indicative of the lectin pathway-mediated killing. As seen in Fig. 6A, left panel, the A. baumannii mlaD∷Tn mutant remained susceptible to killing by human serum that was selectively depleted of C1q when compared to AB5075 and mlaD∷Tn-pmla complemented strains. Furthermore no significant increase in the killing of A. baumannii mlaD∷Tn was observed when the C1q-depleted serum was reconstituted with C1q (70 μg/ml). In FB-depleted human serum the survival of the A. baumannii mlaD∷Tn strain was similar to that of the WT AB5075 and mlaD∷Tn-pmla complemented strains (Fig. 6A, right panel). Serum killing of the mlaD∷Tn strain was however fully restored when FB-depleted sera was reconstituted with FB at 200 μg/ml (mlaD∷Tn vs. mlaD∷Tn+FB; ****p<0.0001). Measuring the relative deposition of the C4 serum component on A. baumannii strains in C1q-depleted human sera to determine if the lectin pathway mediated killing of the mlaD∷Tn strain (Fig. 6B) showed deposition of C4 was overall low and almost identical among all three A. baumannii strains, suggesting minimal activation of the lectin pathway by the mlaD∷Tn mutant. C4 deposition via the lectin pathway was readily detected on the control antigen, mannan. These findings indicate that an aberrant Mla pathway results in susceptibility of A. baumannii to killing via the alternative pathway of C.
Figure 6.

A) Susceptibility of A. baumannii AB5075, mlaD∷Tn mutant and mlaD∷Tn-pmla complemented strains to killing by C1q-depleted (left panel) or FB-depleted (right panel) human sera. A. baumannii strains were tested against intact (open columns) or HI- (solid columns) C1q- or FB-depleted human sera. The susceptibility A. baumannii mlaD∷Tn was also tested against C1q- or FB-depleted sera reconstituted with 70 or 200 μg/ml of purified C1q or FB, respectively either directly or after heat-inactivation. Columns represent the mean ± SD of 4–6 independent experiments. After normality was confirmed using the Shapiro-Wilk test on log10-transformed data, statistical significance was calculated using one way ANOVA (p<0.0001) followed by a Tukey post-hoc test. ***p<0.0001; B) C4 deposition onto A. baumannii. ELISA plates were coated with either A. baumannii AB5075, the mlaD∷Tn mutant or the mlaD∷Tn-pmla complemented strain or with mannan as a positive control, and incubated with serial dilutions of C1q-depleted sera at 37°C for 1 h. Bound C4b was detected with anti-C4 Abs. Results represent the means ± SD of 4 independent experiments.
Due to the relevance of complement FH, as a key negative regulator of the alternative C pathway in both the fluid phase and on cellular surfaces (36) we investigated the ability of WT AB5075 strain to bind FH and its potential contribution to the serum resistance of this strain. Results from FACS studies as well as C killing assays performed with NHS and FH-depleted human sera revealed little to no binding of purified FH, tested at the physiologic concentration of 500 μg/ml, to WT AB5075. There was a similar killing activity of both intact and FH-depleted human sera against this strain (Data not shown). Taken together, these results demonstrated that FH does not play a role in the resistance of AB5075 to killing by human C.
To positively confirm that killing was mediated by the C3-dependent formation of the terminal C complex (TCC) C5b-9, human serum was depleted of C component C3 using CVF, resulting in the complete abrogation of the killing of the mlaD∷Tn strain (Fig. 7) and a susceptibility comparable to that of the same strain incubated in HI-NHS (Fig. 7). Conversely CVF treatment did not disrupt the resistance of the WT AB5075 or mlaD∷Tn-pmla complemented strains to NHS.
Figure 7.
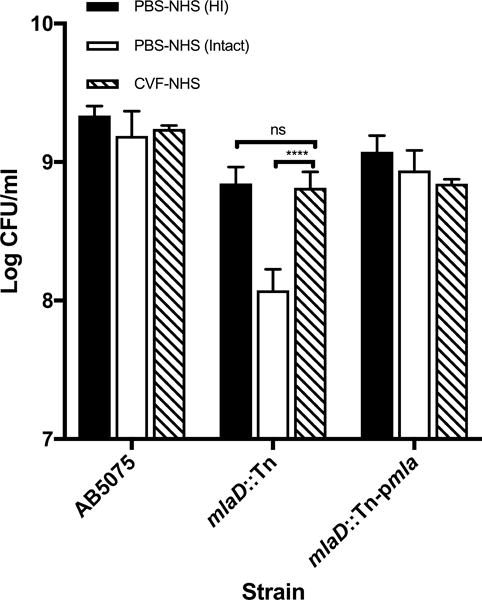
Effect of CVF depletion of C3 on NHS killing activity of WT A. baumannii AB5075, mlaD∷Tn and complemented mlaD∷Tn-pmla strains. Following CVF- (striped columns) or PBS- treatment (open columns), sera were tested against Acinetobacter strains in C killing assays. Also tested in the assay was a PBS-treated NHS sample following HI (solid columns). Data represent means ± SD of 3 independent experiments. Statistical significance was calculated using one way ANOVA (p<0.0001) followed by a Tukey post-hoc test after normality was confirmed using the Shapiro-Wilk test on log10-transformed data. ***p<0.0001.
An evaluation by FACS of the deposition of the TCC components by intact- and HI-NHS on WT AB5075 (Fig. 8; top row), the mlaD∷Tn mutant (Fig. 8; middle row) and the mlaD∷Tn-pmla complemented strain (Fig. 8; bottom row) revealed little deposition of C5b-9 by intact NHS on WT AB5075 cells (17.3% positive cells) or the mlaD∷Tn-pmla complemented strain (8% positive cells) whereas the serum sensitive A. baumannii mlaD∷Tn strain exhibited high levels of surface C5b-9 binding (61.4% of positive cells). There was only very low level of C5b-9 deposition on the cells in HI-NHS samples (Fig. 8; right column). Collectively, these results clearly demonstrate that A. baumannii Mla pathway mutants are killed via the alternative pathway-mediated deposition of the TCC deposition onto bacterial cells.
Figure 8.
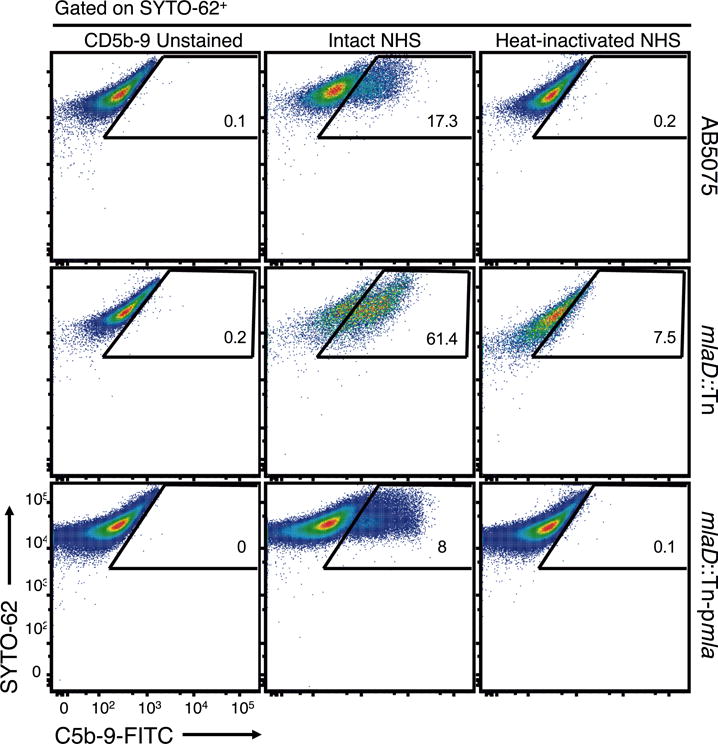
C5b-9 deposition on Acinetobacter strains measured by flow cytometry. Representative flow cytometry dot plots of C5b-9 deposition on WT A. baumannii AB5075 (top row), the mlaD∷Tn mutant (middle row) or the mlaD∷Tn-pmla complemented strain (bottom row). C5b-9 deposition was measured after the incubation of bacterial strains with either 30% intact NHS (middle column) or HI-NHS (right column) for 1 h 15 min followed by labeling with a FITC-conjugated mouse mAb to a C5b-9 neoantigen (X-axis) and the nucleic acid stain SYTO 62 (Y-axis). The percentage of FITC-positive cells in all strain/treatment combinations, which is indicated in every panel, was calculated after gating on a C5b-9-unstained, SYTO 62 positive cell population (left column).
DISCUSSION
The ability to resist C-mediated killing is considered an important determinant of pathogenicity for many Gram-negative bacteria and the results from this study demonstrate that serum resistance is widespread among A. baumannii strains. To expand our current understanding into the molecular mechanisms underlying serum resistance in A. baumannii we conducted a Tn-seq analysis using a mutant library created in the serum resistant strain AB5075. Findings from this screen identified 50 novel C resistance genes of A. baumannii and follow up studies confirmed the role of eight of these genes in resistance to killing by NHS, implicating the remaining genes are also likely to encode proteins providing serum resistance to this organism. Interestingly, parallel studies conducted in NMS revealed that none of these 8 A. baumannii Tn mutant strains were susceptible to killing by mouse C, indicating species specificity of this organism’s interactions with the C components. Additionally, depleting functional C activity in vivo in mouse sera did not enhance the virulence of A. baumannii. This discrepancy in human-mouse serum killing could be explained in terms of concentrations of C proteins, which are significantly lower in mouse than in human sera (37) or differences in C regulatory systems between these species (38, 39). The ability to recruit serum C regulatory proteins (CRP) to microbial surfaces is a prominent mechanism of C evasion used by many pathogens (40) and this binding occurs, for most CRPs, in a species-specific manner (41). Overall, the results here highlight the limitations of murine systems for studying serum resistance and virulence of human pathogens, and also show the utility of working within a human system with the genetic and analytic tools used in our study.
In contrast to genes encoding proteins that synthesize the capsular polysaccharide, Tn mutants in LPS biosynthesis and export genes were not identified as essential for serum resistance in our Tn-seq screen. This absence is consistent with the fact that at least 19 genes belonging to LPS biosynthesis and export pathways in AB5075 were classified as putative essential genes by Gallagher et al (11) in AB5075 and are either absent from the AB5075 Tn library or the Tn inserts are located at the extreme 3′ or 5′ end of coding sequences, suggesting that these inserts do not disrupt the function of the protein encoded by such a gene.
One of the most interesting factors identified in this study contributing to A. baumannii survival in human sera was the mla system. The Mla pathway is an intermembrane PL transport system whose main function is to maintain the asymmetry of the OM of Gram-negative bacteria by preventing the accumulation of PLs in the outer leaflet of the OM by transporting them to the IM (27). In the Mla system the lipoprotein MlaA participates in the extraction of PLs from the outer leaflet of the OM, transferring them to the periplasmic transporter MlaC, which then delivers the substrate to the MlaFEDB transporter complex that finally reinserts them into the IM (27).
Similarly to other bacterial species (23, 27) this works demonstrated the role of the mla pathway of A. bamannii plays in preventing the accumulation of surface exposed PLs and maintaining the barrier function of the OM of this bacteria (Supplemental Figs 1B and C).
Here we validated the contribution of the Mla pathway to A. baumannii resistance to human serum using Tn mutants in their coding sequences, and their corresponding complemented strains, targeting two of the system’s components: the initial OM lipoprotein MlaA and the cytoplasmic membrane protein MlaD which is part of the terminal MlaFEDB transporter complex. Additional C killing assays carried out in NHS pre-treated with CVF, used to selectively deplete C C3 protein, validated the C system as the killing effector of A. baumannii mla mutants. This finding was further supported by FACS studies showing significantly higher levels of the C5b-9 terminal complex deposited by intact NHS on the mlaD∷Tn mutant strain than on WT AB5075 or the mlaD∷Tn-pmla complemented strain or the three strains incubated with HI-NHS. Additional C killing studies conducted with C1q- and FB-depleted human sera, which are defective in the classical and alternative C pathways respectively, as well as ELISA C4-deposition assays on Acinetobacter strains by C1q-depleted sera, used to measure lectin pathway activation, conclusively demonstrated the exclusive role of the alternative pathway in the killing activity of NHS against A. baumannii mlaD∷Tn mutant strain.
Taken together these results highlight the importance of the Mla system of A. baumannii in maintaining full resistance against C killing. Our findings are consistent with a previous study reporting the role of mla homolog genes in maintaining resistance of non-typable Haemophilus influenza (NTHi) to killing by human sera (23). However, while here the expression of the mla genes protects A. baumannii against killing by the alternative C pathway, in NTHi the loss of serum resistance in mla mutants was mediated by the activation of classical pathway via binding of natural IgMs in serum to surface oligosaccharide epitopes (23). The nature of the surface structure(s) of A. baumannii mla mutant strains triggering the activation of the alternative pathway remains to be elucidated. However, since the hallmark of mla mutations in Gram-negative bacteria is the accumulation of surface-exposed PLs it would be possible to speculate that these macromolecules could indeed be playing a key role activating the alternative pathway of the C system in the A. baumannii mlaD∷Tn mutant strain. Future studies however will be needed to further investigate this hypothesis.
The identification of bacterial factors conferring resistance to killing by NHS represents a potentially new approach to vaccine and drug development. The recent licensing of two new vaccines in Europe and the USA against serogroup B N. meningitidis that contain fHbp from N. meningitidis as one of its main active components validates the vaccine development pathway by targeting bacterial factors involved in C-resistance. Alternately, our approach could aid in finding drugs that might have minimal bactericidal activity on their own but would nonetheless be effective in treating infections by enhancing serum sensitivity by binding to and disrupting the function of bacterial factors needed for serum resistance.
In conclusion, the availability of a high-throughput screening technology and data analysis pipeline such as the one used in this study has proven highly successful in identifying novel essential genes and functional factors needed for A. baumannii serum resistance. Future studies will be needed to unravel the exact roles of these uncharacterized genes in serum resistance and investigate their potential as targets for vaccine or antimicrobial development.
Supplementary Material
Abbreviations in this article
- CVF
Cobra venom factor
- FB
Factor B
- FH
Factor H
- GVB++
Gelatin veronal buffer with Mg2+ and Ca2+
- HI
Heat-inactivated
- IM
Inner membrane
- IPTG
Isopropyl β-D-1-thiogalactopyranoside
- LB
Lysogeny broth
- mla
Maintenance of lipid asymmetry
- NHS
Normal human serum
- NMS
Normal mouse sera
- OM
Outer membrane
- OD650 nm
Optical density at 650 nm
- Ori
Origin of replication
- PLs
Phospholipids
- TCC
Terminal complement complex
- Tn
Transposon
- Tn-seq
Transposon-sequencing
- WT
Wild type
References
- 1.Lee YT, Kuo SC, Yang SP, Lin YT, Tseng FC, Chen TL, Fung CP. Impact of appropriate antimicrobial therapy on mortality associated with Acinetobacter baumannii bacteremia: relation to severity of infection. Clin Infect Dis. 2012;55:209–215. doi: 10.1093/cid/cis385. [DOI] [PubMed] [Google Scholar]
- 2.Bergogne-Bérézin E, Towner KJ. Acinetobacter spp. as nosocomial pathogens: microbiological, clinical, and epidemiological features. Clin Microbiol Rev. 1996;9:148–165. doi: 10.1128/cmr.9.2.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang M, Hu Z, Hu F. Nosocomial meningitis caused by Acinetobacter baumannii: risk factors and their impact on patient outcomes and treatments. Future Microbiol. 2012;7:787–793. doi: 10.2217/fmb.12.42. [DOI] [PubMed] [Google Scholar]
- 4.De Breij A, Eveillard M, Dijkshoorn L, van den Broek PJ, Nibbering PH, Joly-Guillou ML. Differences in Acinetobacter baumannii strains and host innate immune response determine morbidity and mortality in experimental pneumonia. PLoS ONE. 2012;7:e30673. doi: 10.1371/journal.pone.0030673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Perez F, Endimiani A, Bonomo RA. Why are we afraid of Acinetobacter baumannii? Expert Rev Anti Infect Ther. 2008;6:269–271. doi: 10.1586/14787210.6.3.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.King LB, Swiatlo E, Swiatlo A, McDaniel LS. Serum resistance and biofilm formation in clinical isolates of Acinetobacter baumannii. FEMS Immunol Med Mic. 2009;55:414–421. doi: 10.1111/j.1574-695X.2009.00538.x. [DOI] [PubMed] [Google Scholar]
- 7.King LB, Pangburn MK, McDaniel LS. Serine protease PKF of Acinetobacter baumannii results in serum resistance and suppression of biofilm formation. J Infect Dis. 2013;207:1128–1134. doi: 10.1093/infdis/jis939. [DOI] [PubMed] [Google Scholar]
- 8.Jacobs AC, Hood I, Boyd KL, Olson PD, Morrison JM, Carson S, Sayood K, Iwen PC, Skaar EP, Dunman PM. Inactivation of phospholipase D diminishes Acinetobacter baumannii pathogenesis. Infect Immun. 2010;78:1952–1962. doi: 10.1128/IAI.00889-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rooijakkers S, van Strijp J. Bacterial complement evasion. Mol Immunol. 2007;44:23–32. doi: 10.1016/j.molimm.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 10.Vernikos G, Medini D. Bexsero® chronicle. Pathog Glob Health. 2014;108:305–316. doi: 10.1179/2047773214Y.0000000162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gallagher LA, Ramage E, Weiss EJ, Radey M, Hayden HS, Held KG, Huse HK, Zurawski DV, Brittnacher MJ, Manoil C. Resources for genetic and genomic analysis of emerging pathogen Acinetobacter baumannii. J Bacteriol. 2015;197:2027–2035. doi: 10.1128/JB.00131-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacobs AC, Thompson MG, Black CC, Kessler JL, Clark LP, McQueary CN, Gancz HY, Corey BW, Moon JK, Si Y, Owen MT, Hallock JD, Kwak YI, Summers A, Li CZ, Rasko DA, Penwell WF, Honnold CL, Wise MC, Waterman PE, Lesho EP, Stewart RL, Actis LA, Palys TJ, Craft DW, Zurawski DV. AB5075, a highly virulent isolate of Acinetobacter baumannii, as a model strain for the evaluation of pathogenesis and antimicrobial treatments. mBio. 2014;5:e01076-14–e01076-14. doi: 10.1128/mBio.01076-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fu Y, Waldor MK, Mekalanos JJ. Tn-Seq analysis of Vibrio cholerae intestinal colonization reveals a role for T6SS-mediated antibacterial activity in the host. Cell Host Microbe. 2013;14:652–663. doi: 10.1016/j.chom.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu Y, Schröder J, Schmidt B. Musket: a multistage k-mer spectrum-based error corrector for Illumina sequence data. Bioinformatics. 2013;29:308–315. doi: 10.1093/bioinformatics/bts690. [DOI] [PubMed] [Google Scholar]
- 15.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chao MC, Abel S, Davis BM, Waldor MK. The design and analysis of transposon insertion sequencing experiments. Nat Rev Microbiol. 2016;14:119–128. doi: 10.1038/nrmicro.2015.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu S, Zhu Z, Fu L, Niu B, Li W. WebMGA: a customizable web server for fast metagenomic sequence analysis. BMC Genomics. 2011;12:1. doi: 10.1186/1471-2164-12-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gebhardt MJ, Gallagher LA, Jacobson RK, Usacheva EA, Peterson LR, Zurawski DV, Shuman HA. Joint transcriptional control of virulence and resistance to antibiotic and environmental stress in Acinetobacter baumannii. mBio. 2015;6:e01660–e01615. doi: 10.1128/mBio.01660-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldman MB, Bangalore S, Goldman JN. Functional and biochemical properties of the early classical complement system of mice. J Immunol. 1978;120:216–224. [PubMed] [Google Scholar]
- 20.Lachmann PJ. Preparing serum for functional complement assays. J Immunol Methods. 2010;352:195–197. doi: 10.1016/j.jim.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 21.Van den Berg CW, Aerts PC, Van Dijk H. In vivo anti-complementary activities of the cobra venom factors from Naja naja and Naja haje. J Immunol Methods. 1991;136:287–294. doi: 10.1016/0022-1759(91)90015-8. [DOI] [PubMed] [Google Scholar]
- 22.Maira-Litran T, Kropec A, Goldmann DA, Pier GB. Comparative opsonic and protective activities of Staphylococcus aureus conjugate vaccines containing native or deacetylated Staphylococcal Poly-N-acetyl-beta-(1–6)-glucosamine. Infect Immun. 2005;73:6752–6762. doi: 10.1128/IAI.73.10.6752-6762.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakamura S, Shchepetov M, Dalia AB, Clark SE, Murphy TF, Sethi S, Gilsdorf JR, Smith AL, Weiser JN. Molecular basis of increased serum resistance among pulmonary isolates of non-typeable Haemophilus influenzae. PLoS Pathog. 2011;7:e1001247. doi: 10.1371/journal.ppat.1001247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bligh EGE, Dyer WJW. A rapid method of total lipid extraction and purification. Can J Biochem Phys. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 25.Harris G, Lee RK, Lam CK, Kanzaki G, Patel GB, Xu HH, Chen W. A mouse model of Acinetobacter baumannii–associated pneumonia using a clinically isolated hypervirulent strain. Antimicrob Agents Ch. 2013;57:3601–3013. doi: 10.1128/AAC.00944-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Russo TA, Luke NR, Beanan JM, Olson R, Sauberan SL, MacDonald U, Schultz LW, Umland TC, Campagnari AA. The K1 Capsular polysaccharide of Acinetobacter baumannii strain 307-0294 is a major virulence factor. Infect Immun. 2010;78:3993–4000. doi: 10.1128/IAI.00366-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malinverni JC, Silhavy TJ. An ABC transport system that maintains lipid asymmetry in the gram-negative outer membrane. Proc Natl Acad Sci USA. 2009;106:8009–8014. doi: 10.1073/pnas.0903229106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nikaido H. Restoring permeability barrier function to outer membrane. Chem Biol. 2005;12:507–509. doi: 10.1016/j.chembiol.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 29.Senchenkova SN, Shashkov AS, Popova AV, Shneider MM, Arbatsky NP, Miroshnikov KA, Volozhantsev NV, Knirel YA. Structure elucidation of the capsular polysaccharide of Acinetobacter baumannii AB5075 having the KL25 capsule biosynthesis locus. Carbohydr Res. 2015;408:8–11. doi: 10.1016/j.carres.2015.02.011. [DOI] [PubMed] [Google Scholar]
- 30.Doerrler WT, Sikdar R, Kumar S, Boughner LA. New Functions for the Ancient DedA Membrane Protein Family. J Bacteriol. 2012;195:3–11. doi: 10.1128/JB.01006-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vrljic M, Sahm H, Eggeling L. A new type of transporter with a new type of cellular function: L-lysine export from Corynebacterium glutamicum. Mol Microbiol. 1996;22:815–826. doi: 10.1046/j.1365-2958.1996.01527.x. [DOI] [PubMed] [Google Scholar]
- 32.Aleshin VV, Zakataeva NP, Livshits VA. A new family of amino-acid-efflux proteins. Trends Biochem Sci. 1999;24:133–135. doi: 10.1016/s0968-0004(99)01367-5. [DOI] [PubMed] [Google Scholar]
- 33.Hong M, Gleason Y, Wyckoff EE, Payne SM. Identification of two Shigella flexneri chromosomal loci involved in intercellular spreading. Infect Immun. 1998;66:4700–4710. doi: 10.1128/iai.66.10.4700-4710.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim K, Lee S, Lee K, Lim D. Isolation and characterization of toluene-sensitive mutants from the toluene-resistant bacterium Pseudomonas putida GM73. J Bacteriol. 1998;180:3692–3696. doi: 10.1128/jb.180.14.3692-3696.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Endo R, Ohtsubo Y, Tsuda M, Nagata Y. Identification and characterization of genes encoding a putative ABC-type transporter essential for utilization of gamma-hexachlorocyclohexane in Sphingobium japonicum UT26. J Bacteriol. 2007;189:3712–3720. doi: 10.1128/JB.01883-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Córdoba SR, de Jorge EG. Translational mini-review series on complement factor H: genetics and disease associations of human complement factor H. Clin Exp Immunol. 2008;151:1–13. doi: 10.1111/j.1365-2249.2007.03552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ong GL, Mattes MJ. Mouse strains with typical mammalian levels of complement activity. J Immunol Methods. 1989;125:147–158. doi: 10.1016/0022-1759(89)90088-4. [DOI] [PubMed] [Google Scholar]
- 38.Jacobson AC, Weis JH. Comparative functional evolution of human and mouse CR1 and CR2. J Immunol. 2008;81:2953–2959. doi: 10.4049/jimmunol.181.5.2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pouw RB, Vredevoogd DW, Kuijpers TW, Wouters D. Of mice and men: The factor H protein family and complement regulation. Mol Immunol. 2015;67:12–20. doi: 10.1016/j.molimm.2015.03.011. [DOI] [PubMed] [Google Scholar]
- 40.Hovingh ES, van den Broek B, Jongerius I. Hijacking complement regulatory proteins for bacterial immune evasion. Front Microbiol. 2016;7:2004. doi: 10.3389/fmicb.2016.02004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pan X, Yang Y, Zhang JR. Molecular basis of host specificity in human pathogenic bacteria. Emerg Microbes Infect. 2014;3:e23. doi: 10.1038/emi.2014.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


